Abstract
Background
The prognostic value and the best method of testing of topoisomerase II alpha (TOP2A) status have not been established in modern tailored therapy based on breast cancer subtype.
Results
The frequencies of TOP2A overexpression and TOP2A amplified were 55.8% and 9.5%, respectively. TOP2A overexpression correlated strongly with non-luminal A subtype (χ2-test, p < 0.001). TOP2A overexpression was significantly associated with relapse-free survival in luminal B breast cancer (n = 316; log rank test, p < 0.001) but not in other breast cancer subtypes. Cox regression analysis showed that TOP2A overexpression is a significant prognostic factor in luminal B breast cancer (hazard ratio (HR) 4.00, 95% confidence interval (CI) 1.65–9.54, p = 0.002). TOP2A amplified was recognized in HER2 positive breast cancer (p < 0.001). In HER2 positive breast cancer, TOP2A amplified (HR 0.30, 95% CI 0.085–1.07, p = 0.063) appeared to be a better prognostic factor.
Conclusion
In modern tailored therapy, TOP2A overexpression can be a poor prognostic factor in luminal B breast cancer. In contrast, TOP2A amplified could be a better prognostic factor in HER2 positive breast cancer.
Materials and methods
Between May 2005 and April 2015, a total of 643 consecutive non-metastatic invasive breast cancers were evaluated for TOP2A amplified using fluorescence in situ hybridization analysis (FISH) and for TOP2A overexpression using the immunohistochemistry assay. FISH ratios of 2 or higher were designated as TOP2A amplified, and TOP2A staining >10% was defined as TOP2A overexpression. The prognostic values of TOP2A amplified and TOP2A overexpression were retrospectively evaluated.
Keywords: breast cancer, topoisomerase IIA, prognostic value, luminal B
INTRODUCTION
Topoisomerase IIA (TOP2A) is an essential nuclear enzyme that cleaves and recombines double-stranded DNA in processes such as replication, transcription, condensation, and segregation [1]. TOP2A is expressed in proliferating cells in the late S and the G2-M phases, and TOP2A expression is known to be a prognostic factor in various malignancies [2–5]. In breast cancer, TOP2A protein or gene expression is also associated with a poor prognosis [6, 7]; the prognostic value is emphasized in hormone receptor-positive disease [8, 9]. Additionally, TOP2A is a molecular target of anthracycline, and previous reports have shown that TOP2A amplified or deleted are predictive markers of anthracycline-containing adjuvant chemotherapy regimens for early breast cancer [10–13].
Although TOP2A status has several important implications in breast cancer, the standard tools and cutoff values for estimating TOP2A status have not been established [14]. Additionally, previous reports of the predictive and prognostic value of TOP2A status were based on classical adjuvant chemotherapy regimens without taxane and HER2-targeting therapies, which differed from modern clinical practice. The clinical significance and best assessment of TOP2A status need to be re-evaluated under modern tailored therapy based on breast cancer subtype.
At our institution, both TOP2A overexpression and TOP2A amplified had been consecutively evaluated using immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) in patients with breast cancer between May 2005 and April 2015. During this period, adjuvant therapies, including taxane and HER2 targeting therapy, were administered based on breast cancer subtype and the risk for recurrence. We retrospectively evaluated the prognostic value of TOP2A status in these patients with non-metastatic breast cancer.
RESULTS
Patient characteristics and TOP2A status
The median and mean of TOP2A expression were 11.3% and 18.4%, respectively. Histogram of continuous IHC proportion scores of TOP2A is shown in (Supplementary Figure 3). The frequency of TOP2A overexpression was 55.8% (359/643). The numbers for TOP2A amplified, normal, and deleted were 61 (9.5%), 577 (89.7%), and 5 (0.8%), respectively. The associations of the clinicopathological factors with TOP2A overexpression and TOP2A amplified are shown in Tables 1 and 2, respectively. TOP2A overexpression showed a significant correlation with larger T stage (p < 0.001), node positivity (p < 0.001), high nuclear grade (p < 0.001), ER negativity (p < 0.001), PgR negativity (p < 0.001), HER2 positivity (p < 0.001), and high Ki67 index (p < 0.001) using the χ2-test. TOP2A overexpression was recognized in non-luminal A subtype (rate of overexpression, luminal A vs. non-luminal A, 8.3% vs. 65.4%, χ2-test, p < 0.001). Although the associations between TOP2A amplified and several clinicopathological factors, including larger T, high nuclear grade, and high Ki67 index, were significant, almost TOP2A amplified was observed in HER2 positive breast cancers; specifically, the rates of TOP2A amplified were 42.2% (57/135) and 0.8% (4/508) in HER2-positive and -negative cancers, respectively. Thus, the characteristics of TOP2A amplified breast cancer reflected those of HER2 positive breast cancer.
Table 1. Clinicopathological factors and TOP2A overexpression.
| TOP2A overexpression | |||||||
|---|---|---|---|---|---|---|---|
| factor | total | yes | % | no | % | p value | |
| 643 | 359 | 55.8 | 284 | 44.2 | |||
| age | 50≥ | 14 | 87 | 61.7 | 54 | 38.3 | 0.11 |
| 50< | 502 | 272 | 54.2 | 230 | 45.8 | ||
| T | *0 | 2 | 2 | 100 | 0 | 0 | <0.001 |
| 1 | 366 | 163 | 44.5 | 203 | 55.5 | ||
| 2 | 211 | 143 | 32.2 | 68 | 67.8 | ||
| 3 | 36 | 26 | 72.2 | 10 | 27.8 | ||
| 4 | 28 | 25 | 89.3 | 3 | 10.7 | ||
| N | 0 | 422 | 211 | 50 | 211 | 50 | <0.001 |
| 1 | 172 | 111 | 64.5 | 61 | 35.5 | ||
| 2 | 35 | 27 | 77.1 | 8 | 22.9 | ||
| 3 | 14 | 4 | 71.4 | 10 | 28.6 | ||
| Stage | 1 | 302 | 132 | 43.7 | 170 | 56.3 | <0.001 |
| 2 | 262 | 166 | 63.4 | 96 | 33.8 | ||
| 3 | 79 | 61 | 77.2 | 18 | 22.8 | ||
| nuclear grade | 1 | 291 | 83 | 28.5 | 208 | 71.5 | <0.001 |
| 2 | 161 | 103 | 64 | 58 | 36 | ||
| 3 | 263.6 | 173 | 90.6 | 18 | 9.4 | ||
| ER | positive | 498 | 250 | 50.2 | 248 | 49.8 | <0.001 |
| negative | 145 | 109 | 75.2 | 36 | 24.8 | ||
| PgR | positive | 434 | 218 | 50.2 | 216 | 49.8 | <0.001 |
| negative | 209 | 141 | 67.5 | 68 | 32.5 | ||
| HER2 | positive | 134 | 107 | 79.3 | 27 | 20.7 | <0.001 |
| negative | 509 | 252 | 49.6 | 257 | 50.4 | ||
| Ki 67 | 14%> | 211 | 23 | 10.9 | 188 | 89.1 | <0.001 |
| ≧14% | 432 | 336 | 77.8 | 96 | 22.2 | ||
| subtype | luminal A | 108 | 9 | 8.3 | 99 | 91.7 | <0.001 |
| luminal B | 315 | 179 | 56.8 | 136 | 43.2 | ||
| HER2 | 135 | 107 | 79.3 | 28 | 20.7 | ||
| TNBC | 85 | 64 | 75.3 | 21 | 24.7 | ||
| TOP2A amplified | yes | 61 | 48 | 78.7 | 13 | 21.3 | <0.001 |
| no | 582 | 311 | 53.4 | 271 | 46.6 | ||
| Adjuvant chemotherapy | yes | 309 | 235 | 76.1 | 74 | 23.9 | <0.001 |
| no | 334 | 124 | 37.1 | 210 | 62.9 | ||
| Adjuvant radiation therapy | yes | 413 | 232 | 56.2 | 181 | 44.2 | 0.82 |
| no | 230 | 127 | 55.2 | 103 | 44.8 | ||
| recurrence | yes | 73 | 60 | 82.2 | 13 | 17.8 | <0.001 |
| no | 570 | 299 | 52.5 | 271 | 47.5 | ||
p value was evaluated using χ2 test, *T0 means occult breast cancer.
Abbreviations: ER: estrogen receptor; PgR; progesteron receptor; HER2: human epidermal growth factor receptor 2 TNBC: triple negative breast cancer.
Table 2. Clinicopathological factors and TOP2A amplified.
| TOP2A amplified | |||||||
|---|---|---|---|---|---|---|---|
| factor | total | yes | % | no | % | p value | |
| 643 | 61 | 9.5 | 582 | 90.5 | |||
| age | 50≥ | 14 | 17 | 12.1 | 124 | 87.9 | 0.24 |
| 50< | 502 | 44 | 8.8 | 458 | 90.2 | ||
| T | *0 | 2 | 0 | 0 | 2 | 100 | 0.044 |
| 1 | 366 | 28 | 7.7 | 338 | 92.3 | ||
| 2 | 211 | 22 | 10.4 | 189 | 89.6 | ||
| 3 | 36 | 4 | 11.1 | 32 | 88.9 | ||
| 4 | 28 | 7 | 25 | 21 | 75 | ||
| N | 0 | 422 | 37 | 8.8 | 385 | 91.2 | 0.052 |
| 1 | 172 | 15 | 8.7 | 157 | 91.3 | ||
| 2 | 35 | 8 | 22.9 | 27 | 77.1 | ||
| 3 | 14 | 1 | 7.1 | 13 | 92.9 | ||
| Stage | 1 | 302 | 27 | 8.9 | 275 | 91.1 | 0.073 |
| 2 | 262 | 21 | 8 | 241 | 92 | ||
| 3 | 79 | 13 | 16.5 | 66 | 83.5 | ||
| nuclear grade | 1 | 291 | 12 | 4.1 | 279 | 95.9 | <0.001 |
| 2 | 161 | 19 | 11.8 | 142 | 88.2 | ||
| 3 | 263.6 | 30 | 15.7 | 161 | 84.3 | ||
| ER | positive | 498 | 41 | 8.2 | 457 | 91.8 | 0.044 |
| negative | 145 | 20 | 13.8 | 125 | 86.2 | ||
| PgR | positive | 434 | 39 | 9 | 395 | 91 | 0.48 |
| negative | 209 | 22 | 10.5 | 187 | 89.5 | ||
| HER2 | positive | 134 | 57 | 42.2 | 78 | 57.8 | <0.001 |
| negative | 509 | 4 | 0.8 | 504 | 99.2 | ||
| Ki 67 | 14%> | 211 | 13 | 6.2 | 198 | 93.8 | 0.044 |
| ≧14% | 432 | 48 | 10.9 | 384 | 89.1 | ||
| subtype | luminal A | 108 | 0 | 0 | 108 | 100 | <0.001 |
| luminal B | 315 | 1 | 0.3 | 314 | 99.7 | ||
| HER2 | 135 | 57 | 42.2 | 78 | 57.8 | ||
| TNBC | 85 | 3 | 3.5 | 82 | 96.5 | ||
| TOP2A overexpression | yes | 359 | 48 | 13.3 | 311 | 86.7 | <0.001 |
| no | 284 | 13 | 4.6 | 271 | 95.4 | ||
| Adjuvant chemotherapy | yes | 309 | 48 | 15.5 | 261 | 84.5 | <0.001 |
| no | 334 | 13 | 3.9 | 321 | 96.1 | ||
| Adjuvant radiation therapy | yes | 413 | 41 | 9.9 | 372 | 90.1 | 0.61 |
| no | 230 | 20 | 8.7 | 210 | 91.3 | ||
| recurrence | yes | 73 | 4 | 5.5 | 69 | 94.5 | 0.21 |
| no | 570 | 57 | 10 | 513 | 90 | ||
p value was evaluated using χ2 test, *T0 means occult breast cancer.
Abbreviations: ER: estrogen receptor; PgR; progesteron receptor; HER2: human epidermal growth factor receptor 2 TNBC: triple negative breast cancer.
Prognostic value of TOP2A overexpression in each breast cancer subtype
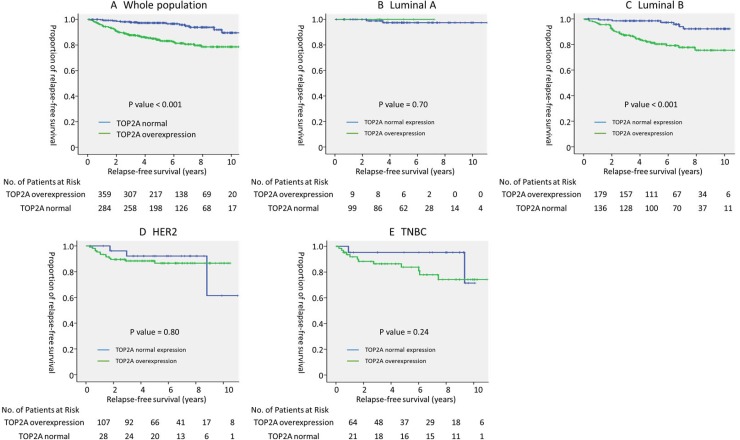
In this study, the median follow-up period was 5.4 years. In the entire population, there was a significant association between TOP2A overexpression and relapse-free survival (5-year RFS; TOP2A overexpression vs. normal expression, 97.3% vs. 84.1%, p < 0.001, log-rank test) (Figure 1A).
Figure 1. Prognostic value of TOP2A overexpression in each breast cancer subtype.
Relapse-free survival of (A) all (n = 643), (B) luminal A (n = 108), (C) luminal B (n = 315), (D) HER2 (n = 135), and (E) TNBC (n = 85) subtypes stratified by TOP2A overexpression. P value was evaluated using the log-rank test. Abbreviations: HER2: human epidermal growth factor receptor 2; TNBC: triple negative breast cancer.
In the subgroup analysis of breast cancer subtypes, TOP2A overexpression was a significant prognostic factor in luminal B breast cancer (5-year RFS; TOP2A overexpression vs. no overexpression, 98.5% vs. 81.3%, p < 0.001, log-rank test); however, no such significant association was observed in other subtypes (Figure 1B–1E). In luminal B breast cancer, Cox regression analysis that included other prognostic factors, such as, age, T factor, N factor, nuclear grade, PgR positivity, and Ki67 index, showed that TOP2A overexpression was the strongest prognostic factor for RFS (hazard ratio [HR] 4.00, 95% confidence interval [CI] 1.65–9.54, p < 0.001) (Table 3). In an exploratory analysis, receiver operating characteristic (ROC) curve of SUVmax was drawn to determine the cutoff value that yielded optimal sensitivity and specificity for prediction of 5-year RFS in luminal B breast cancer based on the Youden index. The ROC curve indicated an optimal TOP2A expression cutoff value of 11.5% for predicting 5-year RFS (area under the curve, 0.74; 95% CI, 0.66–0.82; P < .001; sensitivity, 87.9%; specificity, 59.6%). A continuous value of TOP2A expression was also shown to be the significant prognostic factor in luminal B breast cancer in Cox regression analysis (HR 1.07; 95% CI, 1.02–1.12, P = 0.005).
Table 3. Multivariate Cox regression analysis of clinicopathological factors and TOP2A overexpression, with respect to relapse-free survival among patients with luminal B breast cancer (n = 315).
| factors | n | hazard ratio (95% CI) | p value | |
|---|---|---|---|---|
| age | 50≥ | 69 | 1 | 0.30 |
| 50< | 246 | 0.68 (0.34–1.36) | ||
| T stage | 2 cm≥ | 177 | 1 | 0.036 |
| 2 cm< | 138 | 2.15 (1.05–4.39) | ||
| Nodal status | netative | 202 | 1 | 0.012 |
| positive | 113 | 2.37 (1.21–4.62) | ||
| PgR | positive | 249 | 1 | 0.57 |
| netative | 66 | 1.26 (0.57–2.78) | ||
| nuclear grade | 1, 2 | 236 | 1 | 0.85 |
| 3 | 79 | 1.1 (0.55–2.30) | ||
| Ki67 (continuous value) | 1.02 (1.00–1.04) | 0.11 | ||
| adjuvant chemotherapy | no | 183 | 1 | 0.76 |
| yes | 132 | 0.89 (0.42–1.88) | ||
| adjuvant radiation therapy | no | 105 | 1 | 0.29 |
| yes | 210 | 0.71 (0.31–1.35) | ||
| TOP2A overexpression | no | 136 | 1 | 0.002 |
| yes | 179 | 4.00 (1.65–9.54) |
Abbreviations: PgR; progesteron receptor.
Prognostic value of TOP2A amplified in HER2 positive breast cancer
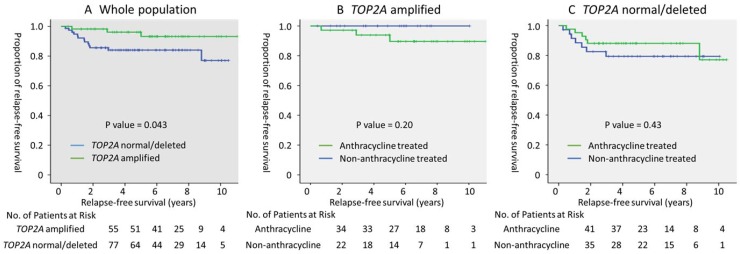
Because TOP2A amplified was strongly associated with HER2 positivity, the prognostic value of TOP2A amplified was examined using the log-rank test in HER2 positive breast cancer. In 135 patients with HER2 positive breast cancer, the rates of administration of adjuvant anthracycline, taxane, and trastuzumab were 56.7%, 64.1%, and 76.1, respectively. TOP2A amplified was associated with better relapse-free survival (5-year RFS; TOP2A amplified vs. normal/deleted, 96.2% vs. 83.8%, p = 0.043, log-rank test, Figure 2A). Cox regression analysis, which included other prognostic factors, confirmed a trend that indicated better prognoses in patients with TOP2A amplified (HR 0.30, 95% CI 0.085–1.07, p = 0.063). As exploratory analysis, the prognostic value of adjuvant anthracycline therapy was examined with respect to the presence of TOP2A amplified; however, adjuvant anthracycline therapy was not associated with survival advantages for patients with or without TOP2A amplified (Figure 2B and 2C).
Figure 2. Prognostic value of TOP2A amplified and adjuvant anthracycline therapy in HER2 positive breast cancer.
(A) Relapse-free survival of the whole population (n = 135), and the (B) TOP2A amplified (n = 57), and (C) TOP2A normal/deleted (n = 78) populations, stratified by adjuvant anthracycline treatment. P value was evaluated using the log-rank test.
DISCUSSION
The treatment strategy for breast cancer has dramatically changed following the introduction of tailored therapy based on breast cancer subtypes [15]. Tailored therapy utilizing endocrine therapy, chemotherapy, and HER2-targeting therapy has significantly improved the prognoses of early and advanced breast cancers [16–20]. The diagnosis of breast cancer subtype was initially developed based on gene-expression signature [21]; however, the hormone receptor expression, HER2 status, and proliferation markers were clinically replaced by multi-gene assay in the analyses [15]. Luminal breast cancer, defined as hormone receptor-positive and HER2 negative breast cancer, is clinically divided into luminal A and luminal B breast cancer using PgR and Ki67 index [22, 23]. Luminal A breast cancer defined as ER and PgR positive with a low Ki67 index has been proven to have an excellent prognosis [24, 25]. In this study, the prognosis of luminal A breast cancer was also excellent, as indicated by the 5-year RFS of 97.6%. Compared with luminal A breast cancer, luminal B breast cancer has significant risks of recurrence, and it has been examined for further risk assessment to refine the adaptation of adjuvant chemotherapy. While multi-gene assays can detect low- or high-risk populations of patients with luminal breast cancers, the value of chemotherapy in the intermediate-risk population is still inconclusive [26, 27]. Additionally, multi-gene assays cannot be performed in individual institutions, and the high cost may prevent these assays from becoming comprehensive. Patient management would benefit from additional and convenient prognostic markers in adjuvant luminal B breast cancer.
TOP2A is a representative proliferation marker, and previous reports have shown that high TOP2A expression was associated with poor prognosis in hormone receptor-positive breast cancer [7, 9, 28]. In our study, we also used IHC analysis to evaluate TOP2A overexpression as a significant prognostic factor in luminal breast cancer; however, the prognostic value was restricted in luminal B breast cancer, defined as tumors with high Ki67 index and/or low PgR expression. This result is different from a previous report in which the prognostic effect of TOP2A gene expression was independent of Ki67 expression in ER positive tumors [8]. A conservative median split of Ki67 gene expression in that study may have led to different prevalence of luminal breast cancer subtype in our study. In this study, luminal breast cancer was subtyped using PgR and Ki67 index of established cut-off values, 20% and 14%, respectively [22, 29]. Our findings supported a previous report of a pooled analysis of four independent gene expression data sets evaluating the prognostic value of TOP2A RNA expression in luminal breast cancer [30]. In that study, the prognostic value of TOP2A expression was evaluated in a population at intermediate-risk for simulated Oncotype DX recurrence sore using proliferation marker components. The authors concluded that TOP2A expression is also useful for identifying those with an intermediate recurrence score who are more likely to relapse. Although TOP2A overexpression was associated with a higher Ki67 index, as shown in previous report [31], TOP2A overexpression remained a significant prognostic factor after adjusting for other clinicopathological factors, including Ki67 index. This result suggests that evaluating the proliferation markers of TOP2A and Ki67 has clinical significance in predicting the risk of relapse in luminal breast cancer. The analysis of TOP2A overexpression by IHC may be superior to the analysis of gene expression with respect to the universality and the convenience. In this study, the cut-off value of TOP2A overexpression was determined as 10%, in accordance with the results of previous studies. In an exploratory analysis, the ROC curve indicated an optimal TOP2A expression cutoff value of 11.5% for predicting 5-year RFS in luminal B breast cancer, which is approximate to 10% of TOP2A expression adopted in this study. Considering the comprehensiveness and the reproducibility of the results of previous studies, 10% of TOP2A expression can be a reasonable cut-off value for predicting recurrence in luminal B breast cancer.
TOP2A is known as a molecular target of anthracycline, and previous meta-analysis has shown that TOP2A amplified was predictive marker for anthracycline-based chemotherapy regimens [11]. A meta-analysis of adjuvant randomized trials comparing anthracycline-based treatment with CMF suggested that TOP2A amplified predicted responsiveness to anthracycline-based chemotherapy in patients with early breast cancer. Patients with TOP2A amplified tumor had higher risk reductions for relapse and survival compared with normal TOP2A tumor by anthracycline-containing chemotherapy. In this study, the prognostic value of TOP2A amplified was evaluated in HER2 positive breast cancer because TOP2A amplified was shown to be strongly associated with HER2 positivity, as shown in previous studies [13, 28]. Unlike above mentioned report, our study failed to show the advantage of anthracycline therapy in patients with TOP2A amplified and HER2 positive breast cancer. Several reasons may account for this discrepancy. First, TOP2A amplified did not affect the prognosis of HER2 positive breast cancer treated with standard adjuvant treatment of trastuzumab-containing chemotherapy [32, 33]. In a single arm study of adjuvant docetaxel and cyclophosphamide plus trastuzumab in patients with HER2-amplified breast cancer, patients with or without TOP2A amplified had excellent prognoses (3-year DFS; TOP2A amplified vs. normal/deleted, 97.2% vs. 96.4%) [33]. In BCIRG-006, HER2 positive breast cancer was treated with anthracycline, taxane, plus trastuzumab showed equivalent prognoses, irrespective of TOP2A status (5-year DFS; TOP2A amplified vs. normal/deleted, 85% vs. 83%) [32]. Second, previous reports showing the predictive value of TOP2A amplified compared anthracycline-based chemotherapy regimens with CMF [11], which is no longer used as a standard treatment regimen for HER2 positive breast cancer. Recent clinical trials have shown excellent prognosis of HER2 positive early breast cancer treated with non-anthracycline regimens consisting of taxane and trastuzumab [33, 34]. Considering the excellent prognosis of HER2-positive breast cancer, to facilitate the decision of adjuvant anthracycline therapy based on the evaluation of TOP2A amplified may lack clinical meaning.
This study has some limitations. First, this retrospective analysis from a single institution could have biases and a multiple-institutional prospective study is needed to confirm our results. Second, PgR expression and Ki67 index were substituted for a multigene expression classifier, such as Oncotype Dx or Mammaprint, to distinguish luminal B from luminal A breast cancer in this study. Some discrepancies between the multi-gene assay and analysis of conventional pathological markers in determining intrinsic subtype may exist. Third, a small number of cases and events in HER2 positive breast cancer is underpowered to make conclusion about the predictive value of TOP2A amplified in anthracycline treatment.
In conclusion, our study indicates that TOP2A overexpression is a marker for poor prognosis, especially in cases of luminal B breast cancer. In contrast, the TOP2A amplified is limited in HER2 positive breast cancer, and the presence of the TOP2A amplified does not influence the survival advantage of adjuvant anthracycline therapy. To tailor the adjuvant therapy based on breast cancer subtype, the assessment of TOP2A overexpression may be considered for risk assessment in luminal B breast cancer.
MATERIALS AND METHODS
Patients and methods
Between May 2005 and Apr 2015, a total of 643 consecutive non-metastatic invasive breast cancers were evaluated at the Kure Medical Center and the Chugoku Cancer Center, Kure, Japan, for their TOP2A status using IHC to analyze TOP2A overexpression and FISH analysis for TOP2A amplified. The prognostic value of TOP2A status was retrospectively evaluated in this study. The Kure Medical Center review board approved this study (29–23). The requirement for informed consents from individual patients was waived because this was a retrospective review of a prospectively maintained patient database.
Clinicopathological factors
The clinicopathological factors in this study, derived from a prospectively maintained database at our institute, included patient age, histological type of cancer, prescribed neoadjuvant therapy, T and N stage according to the TNM classification, nuclear grade, estrogen receptor (ER) status, progesterone receptor (PgR) status, human epidermal growth factor receptor 2 (HER2) status, Ki67 index, breast cancer subtype, prescription of anthracycline and taxane, and recurrence (Table 1). ER and PgR status were determined using IHC assays, and tumors with 1% or more of positively-stained tumor cells were classified as positive for ER and PgR. The HER2 status was determined using IHC and FISH individually or in combination. HER2 positive tumors were defined as those with an IHC score of 3+ or those showing HER2 gene amplification using FISH, in accordance with the ASCO guidelines [35]. Breast cancer subtypes were determined using surrogate markers, including ER, PgR, HER2, and Ki67 index, as described below [15, 22, 23];
Luminal A: ER positive, HER2 negative, Ki67 <14%, and PgR positive ≥20%
Luminal B: ER positive, HER2 negative, and Ki67 ≥14%, or PgR positive <20%
HER2: HER2 positive, and any one of the following: ER, PgR and Ki67
Triple negative breast cancer (TNBC): ER, PgR, and HER2 negative
Adjuvant therapy was essentially determined in accordance with clinical guidelines. Patients with luminal A or B breast cancers were administered adjuvant endocrine therapy for 5 years. The determination of adjuvant chemotherapy for luminal type breast cancer was based on breast cancer recurrence risks and the preference of the patients. One-year adjuvant trastuzumab therapy was administered in cases of HER2 positive breast cancer. Patients with TNBC received adjuvant chemotherapy. Adjuvant chemotherapy essentially consisted of anthracycline- and taxane-based regimens administered individually or in combination. Postoperative follow-up was performed in accordance with the guidelines for medical checks and annual mammography. In cases with signs of recurrence, image tests and biopsies were performed to confirm the status. Recurrence-free survival (RFS) was defined as the elapsed time from the date of surgery until the date of the first event (relapse or death from any cause) or of last follow-up.
Evaluation of TOP2A overexpression and TOP2A amplified in this study
TOP2A overexpression was analyzed by IHC [12]. Briefly, 4-μm-thick paraffin sections were prepared, and the formalin-fixed and paraffin-embedded tumor tissues were stained with monoclonal TOP2A antibodies (Ki-S1, Dako). IHC staining was performed using auto-stainers (Ventana), and the results were stored digitally after examination by virtual microscopy (Hamamatsu Photonics). Only nuclear staining was considered for determining TOP2A positivity. Immunostaining frequency of the tumor cells was automatically evaluated using Genie/Aperio software. Tumors with definitive TOP2A staining in more than 10% of the tumor cells were considered as TOP2A overexpression, in accordance with previous reports [9, 36]. The intensity score of staining was not considered in this analysis. Representative microscopic findings of nuclear staining for TOP2A overexpression and normal breast cancer, and auto-analysis of frequency of TOP2A positive cells are shown in (Supplementary Figure 1A, 1B). TOP2A amplified was evaluated using FISH analysis, as described previously [37]. Briefly, a dual-color probe containing a spectrum orange-labeled TOP2A gene and a spectrum green-labeled centromere control for chromosome 17 were evaluated (DAKO). FISH ratios of 2 or higher, 0.8 to 1.9, and <0.8 were designated as TOP2A amplified, normal, and deleted, respectively. Representative microscopic findings of TOP2A/CEP17 for TOP2A amplified and normal breast cancer are shown in (Supplementary Figure 2A, 2B). The clinical values of TOP2A overexpression and TOP2A amplified were evaluated in accordance with REMARK criteria [38].
Statistical analysis
The association between the clinicopathological factors and TOP2A status was assessed using the χ2 test. Kaplan–Meier survival curves and the log rank test were used to determine the univariate significance of the variables. A Cox regression model was used to examine multiple covariates for survival. Statistical analyses were carried out using SPSS software (version 11 for Windows; 5 SAS Institute, Tokyo, Japan). A p-value of < 0.05 was considered as statistically significant.
SUPPLEMENTARY MATERIALS AND FIGURES
Acknowledgments
The authors would like to thank the Department of Diagnostic Pathology for their technical assistance.
Footnotes
CONFLICTS OF INTEREST
Author have no conflicts of interest.
REFERENCES
- 1.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–37. doi: 10.1038/nrc2608. https://doi.org/10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doussis-Anagnostopoulou IA, Vassilakopoulos TP, Thymara I, Korkolopoulou P, Angelopoulou MK, Siakantaris MP, Kokoris SI, Dimitriadou EM, Kalpadakis C, Matzouranis M, Kaklamanis L, Panayiotidis P, Kyrtsonis MC, et al. Topoisomerase IIalpha expression as an independent prognostic factor in Hodgkin's lymphoma. Clin Cancer Res. 2008;14:1759–66. doi: 10.1158/1078-0432.CCR-07-1395. https://doi.org/10.1158/1078-0432.CCR-07-1395. [DOI] [PubMed] [Google Scholar]
- 3.Shvero J, Koren R, Shvili I, Yaniv E, Sadov R, Hadar T. Expression of human DNA Topoisomerase II-alpha in squamous cell carcinoma of the larynx and its correlation with clinicopathologic variables. Am J Clin Pathol. 2008;130:934–9. doi: 10.1309/AJCPROG61USKCBEI. https://doi.org/10.1309/AJCPROG61USKCBEI. [DOI] [PubMed] [Google Scholar]
- 4.Skotheim RI, Kallioniemi A, Bjerkhagen B, Mertens F, Brekke HR, Monni O, Mousses S, Mandahl N, Soeter G, Nesland JM, Smeland S, Kallioniemi OP, Lothe RA. Topoisomerase-II alpha is upregulated in malignant peripheral nerve sheath tumors and associated with clinical outcome. J Clin Oncol. 2003;21:4586–91. doi: 10.1200/JCO.2003.07.067. https://doi.org/10.1200/JCO.2003.07.067. [DOI] [PubMed] [Google Scholar]
- 5.Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, Koh J, Chan SL, Chan AT, Lai PB, Ching AK, Tong JH, Ng HK, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–52. doi: 10.1002/ijc.23968. https://doi.org/10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen KV, Ejlertsen B, Moller S, Jorgensen JT, Knoop A, Knudsen H, Mouridsen HT. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta Oncol. 2008;47:725–34. doi: 10.1080/02841860801995396. https://doi.org/10.1080/02841860801995396. [DOI] [PubMed] [Google Scholar]
- 7.Fritz P, Cabrera CM, Dippon J, Gerteis A, Simon W, Aulitzky WE, van der Kuip H. c-erbB2 and topoisomerase IIalpha protein expression independently predict poor survival in primary human breast cancer: a retrospective study. Breast Cancer Res. 2005;7:R374–84. doi: 10.1186/bcr1012. https://doi.org/10.1186/bcr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rody A, Karn T, Ruckhaberle E, Muller V, Gehrmann M, Solbach C, Ahr A, Gatje R, Holtrich U, Kaufmann M. Gene expression of topoisomerase II alpha (TOP2A) by microarray analysis is highly prognostic in estrogen receptor (ER) positive breast cancer. Breast Cancer Res Treat. 2009;113:457–66. doi: 10.1007/s10549-008-9964-x. https://doi.org/10.1007/s10549-008-9964-x. [DOI] [PubMed] [Google Scholar]
- 9.Brase JC, Schmidt M, Fischbach T, Sultmann H, Bojar H, Koelbl H, Hellwig B, Rahnenfuhrer J, Hengstler JG, Gehrmann MC. ERBB2 and TOP2A in breast cancer: a comprehensive analysis of gene amplification, RNA levels, and protein expression and their influence on prognosis and prediction. Clin Cancer Res. 2010;16:2391–401. doi: 10.1158/1078-0432.CCR-09-2471. https://doi.org/10.1158/1078-0432.CCR-09-2471. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard KI, Messersmith H, Elavathil L, Trudeau M, O'Malley F, Dhesy-Thind B. HER-2 and topoisomerase II as predictors of response to chemotherapy. J Clin Oncol. 2008;26:736–44. doi: 10.1200/JCO.2007.15.4716. https://doi.org/10.1200/JCO.2007.15.4716. [DOI] [PubMed] [Google Scholar]
- 11.Di Leo A, Desmedt C, Bartlett JM, Piette F, Ejlertsen B, Pritchard KI, Larsimont D, Poole C, Isola J, Earl H, Mouridsen H, O'Malley FP, Cardoso F, et al. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol. 2011;12:1134–42. doi: 10.1016/S1470-2045(11)70231-5. https://doi.org/10.1016/S1470-2045(11)70231-5. [DOI] [PubMed] [Google Scholar]
- 12.Kiyomi T, Nao M, Kazuya K, Akihisa S, Toshinao N, Junichi S, Mieko H, Miho T, Hiroyo T, Kazuaki M, Hironori K. Topoisomerase II-alpha index predicts the efficacy of anthracycline-based chemotherapy for breast cancers: in: HER2 and cancer. Nova Science Publishers, New York. 2011 [Google Scholar]
- 13.Press MF, Sauter G, Buyse M, Bernstein L, Guzman R, Santiago A, Villalobos IE, Eiermann W, Pienkowski T, Martin M, Robert N, Crown J, Bee V, et al. Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol. 2011;29:859–67. doi: 10.1200/JCO.2009.27.5644. https://doi.org/10.1200/JCO.2009.27.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller RE, Parkes RK, Andrulis I, O'Malley FP. Amplification of the TOP2A gene does not predict high levels of topoisomerase II alpha protein in human breast tumor samples. Genes Chromosomes Cancer. 2004;39:288–97. doi: 10.1002/gcc.20008. https://doi.org/10.1002/gcc.20008. [DOI] [PubMed] [Google Scholar]
- 15.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thurlimann B, Senn HJ, Panel M. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–46. doi: 10.1093/annonc/mdv221. https://doi.org/10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, Dowsett M, Ingle J, Peto R. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. doi: 10.1016/S0140-6736(11)60993-8. https://doi.org/10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shigematsu H, Kawaguchi H, Nakamura Y, Tanaka K, Shiotani S, Koga C, Nishimura S, Taguchi K, Nishiyama K, Ohno S. Significant survival improvement of patients with recurrent breast cancer in the periods 2001-2008 vs. 1992-2000. BMC Cancer. 2011;11:118. doi: 10.1186/1471-2407-11-118. https://doi.org/10.1186/1471-2407-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, Cutter D, Darby S, McGale P, Taylor C, Wang YC, Bergh J, Di Leo A, et al. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–44. doi: 10.1016/S0140-6736(11)61625-5. https://doi.org/10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–72. doi: 10.1056/NEJMoa052306. https://doi.org/10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 20.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84. doi: 10.1056/NEJMoa052122. https://doi.org/10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 21.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. https://doi.org/10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 22.Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO, Perou CM. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31:203–9. doi: 10.1200/JCO.2012.43.4134. https://doi.org/10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C, Piccart M. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32:2794–803. doi: 10.1200/JCO.2013.54.1870. https://doi.org/10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 24.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28:1684–91. doi: 10.1200/JCO.2009.24.9284. https://doi.org/10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 25.Ribelles N, Perez-Villa L, Jerez JM, Pajares B, Vicioso L, Jimenez B, de Luque V, Franco L, Gallego E, Marquez A, Alvarez M, Sanchez-Munoz A, Perez-Rivas L, et al. Pattern of recurrence of early breast cancer is different according to intrinsic subtype and proliferation index. Breast Cancer Res. 2013;15:R98. doi: 10.1186/bcr3559. https://doi.org/10.1186/bcr3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi M, Glas AM, Golfinopoulos V, Goulioti T, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med. 2016;375:717–29. doi: 10.1056/NEJMoa1602253. https://doi.org/10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 27.Nitz U, Gluz O, Christgen M, Kates RE, Clemens M, Malter W, Nuding B, Aktas B, Kuemmel S, Reimer T, Stefek A, Lorenz-Salehi F, Krabisch P, et al. Reducing chemotherapy use in clinically high-risk, genomically low-risk pN0 and pN1 early breast cancer patients: five-year data from the prospective, randomised phase 3 West German Study Group (WSG) PlanB trial. Breast Cancer Res Treat. 2017;165:573–583. doi: 10.1007/s10549-017-4358-6. https://doi.org/10.1007/s10549-017-4358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, Smith IE, Dowsett M, Reis-Filho JS. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106:181–9. doi: 10.1007/s10549-006-9492-5. https://doi.org/10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 29.Dowsett M, Nielsen TO, A'Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, McShane L, Paik S, Penault-Llorca F, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–64. doi: 10.1093/jnci/djr393. https://doi.org/10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparano JA, Goldstein LJ, Davidson NE, Sledge GW, Jr, Gray R. TOP2A RNA expression and recurrence in estrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2012;134:751–7. doi: 10.1007/s10549-012-2112-7. https://doi.org/10.1007/s10549-012-2112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch BJ, Guinee DG, Jr, Holden JA. Human DNA topoisomerase II-alpha: a new marker of cell proliferation in invasive breast cancer. Hum Pathol. 1997;28:1180–8. doi: 10.1016/s0046-8177(97)90256-2. [DOI] [PubMed] [Google Scholar]
- 32.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, Pinter T, Valero V, Liu MC, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–83. doi: 10.1056/NEJMoa0910383. https://doi.org/10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones SE, Collea R, Paul D, Sedlacek S, Favret AM, Gore I, Jr, Lindquist DL, Holmes FA, Allison MA, Brooks BD, Portillo RM, Vukelja SJ, Steinberg MS, et al. Adjuvant docetaxel and cyclophosphamide plus trastuzumab in patients with HER2-amplified early stage breast cancer: a single-group, open-label, phase 2 study. Lancet Oncol. 2013;14:1121–8. doi: 10.1016/S1470-2045(13)70384-X. https://doi.org/10.1016/S1470-2045(13)70384-X. [DOI] [PubMed] [Google Scholar]
- 34.Tolaney SM, Barry WT, Dang CT, Yardley DA, Moy B, Marcom PK, Albain KS, Rugo HS, Ellis M, Shapira I, Wolff AC, Carey LA, Overmoyer BA, et al. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer. N Engl J Med. 2015;372:134–41. doi: 10.1056/NEJMoa1406281. https://doi.org/10.1056/NEJMoa1406281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. https://doi.org/10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 36.Erber R, Gluz O, Brunner N, Kreipe HH, Pelz E, Kates R, Bartels A, Huober J, Mohrmann S, Moustafa Z, Liedtke C, Mobus V, Augustin D, et al. Predictive role of HER2/neu, topoisomerase-II-alpha, and tissue inhibitor of metalloproteinases (TIMP-1) for response to adjuvant taxane-based chemotherapy in patients with intermediate-risk breast cancer: results from the WSG-AGO EC-Doc trial. Breast Cancer Res Treat. 2015;150:279–88. doi: 10.1007/s10549-015-3310-x. https://doi.org/10.1007/s10549-015-3310-x. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Hasenclever D, Schaeffer M, Boehm D, Cotarelo C, Steiner E, Lebrecht A, Siggelkow W, Weikel W, Schiffer-Petry I, Gebhard S, Pilch H, Gehrmann M, et al. Prognostic effect of epithelial cell adhesion molecule overexpression in untreated node-negative breast cancer. Clin Cancer Res. 2008;14:5849–55. doi: 10.1158/1078-0432.CCR-08-0669. https://doi.org/10.1158/1078-0432.CCR-08-0669. [DOI] [PubMed] [Google Scholar]
- 38.McShane LM, Hayes DF. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol. 2012;30:4223–32. doi: 10.1200/JCO.2012.42.6858. https://doi.org/10.1200/JCO.2012.42.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.