Abstract
Widespread resistance to antimicrobial and cancer therapeutics is evolving in every country worldwide and has a direct impact on global health, agriculture and the economy. The specificity and selectivity of bioactive peptide natural products present a possible stopgap measure to address the ongoing deficit of new therapeutic compounds. PepSAVI-MS (Statistically-guided bioActive Peptides prioritized Via Mass Spectrometry) is an adaptable method for the analysis of natural product libraries to rapidly identify bioactive peptides. This pipeline was validated via screening of the cyclotide-rich botanical species Viola odorata and identification of the known antimicrobial and anticancer cyclotide cycloviolacin O2. Herein we present and validate novel bioactivities of the anthelmintic V. odorata cyclotide, cycloviolacin O8 (cyO8), including micromolar anticancer activity against PC-3 prostate, MDA-MB-231 breast, and OVCAR-3 ovarian cancer cell lines and antifungal activity against the agricultural pathogen Fusarium graminearum. A reduction/alkylation strategy in tandem with PepSAVI-MS analysis also revealed several previously uncharacterized putatively bioactive cyclotides. Downstream implementation of ultraviolet photodissociation (UVPD) tandem mass spectrometry is demonstrated for cyO8 as a method to address traditionally difficult-to-sequence cyclotide species. This work emphasizes the therapeutic and agricultural potential of natural product bioactive peptides and the necessity of developing robust analytical tools to deconvolute nature’s complexity.
Keywords: Viola odorata, Violaceae, Bioactive peptides, Antimicrobial peptides, Cyclotides, Mass Spectometry
Graphical abstract
1. Introduction
Antimicrobial peptides (AMPs) have guided the coevolution of countless biological species over hundreds of millions of years, the arsenal of naturally-occurring molecular weapons growing in potency and complexity as organisms compete for resources and real-estate. Conserved across all domains of life, AMPs are ancient defense molecules with broad functions (Ajesh and Sreejith, 2009; Pasupuleti et al., 2012; Peschel and Sahl, 2006; Tassanakajon et al., 2015). In multicellular organisms, highly diverse ribosomally-synthesized AMPs are fundamental to the innate immune response, driving off invading bacterial, fungal, and viral pathogens, whereas prokaryotes may wield narrow-spectrum bactericidal AMPs to procure their place in ecological niches (Ganz, 2003; Kommineni et al., 2015; Loo et al., 2017; Mahlapuu et al., 2016). Acting by physical disruption of target membranes via an induced or permanent amphipathic motif (Sato and Feix, 2006; Zhang and Gallo, 2016), the mechanism of action (MOA) employed by most known AMPs has historically prevented the development of widespread AMP resistance in nature, requiring major remodeling of basic membrane structure (Peters et al., 2010). Antimicrobial peptides often exhibit broad spectrum activity across diverse phyla, however, AMPs can be highly specialized, targeting specific and unique membrane constituents (Herbel and Wink, 2016; Lee and Kim, 2015). Unlike neutrally-charged mammalian cells, both cancer and microbial cells rich with anionic molecules can be selectively targeted by AMPs with cell death caused by either membranolytic or intracellular mechanisms (Brogden, 2005; Guzman-Rodriguez et al., 2015; Lee and Kim, 2015; Mahlapuu et al., 2016; Rotem and Mor, 2009). With the recent surge of clinical multidrug resistant pathogens and cancers, AMPs have become increasingly attractive therapeutic candidates boasting novel and diverse MOAs (Bahar and Ren, 2013; Craik et al., 2017; Mahlapuu et al., 2016).
Bioactive peptides exist in all organisms, however the abundance and diversity of these molecules is vastly underexplored as their identification in a sea of macromolecules and small molecule metabolites presents significant analytical challenges. With the implementation of high resolving power mass spectrometry, sensitive bioactivity assays, tailored and streamlined automated data processing and statistical analyses, identification of AMPs from complex natural extracts is viable via the PepSAVI-MS pipeline (Kirkpatrick et al., 2017; Kirkpatrick et al., 2018). Following this pipeline, peptide libraries are screened for growth inhibition in adaptable, high-throughput bioactivity assays and the relative abundances of peptidyl species in each fraction are determined via mass spectrometry. Statistical analysis via an elastic net regression model ranks the most probable peptidyl species responsible for the observed bioactivity. PepSAVI-MS does not require multiple iterations of fractionation as in bioassay-guided fractionation (Britton et al., 2018), nor knowledge of existing gene clusters as in genome mining (Scheffler et al., 2013). PepSAVI-MS uniquely allows for simultaneous contributions from multiple molecular species contributing to synergistic interactions. The components of peptide libraries can be tailored readily to contain constitutive or inducible AMPs by modifying experimental conditions. While a necessary and unique benefit of PepSAVI-MS, the ability to account for synergistic effects as well as contributions from multiple peptidyl species greatly increases the number of putative bioactive targets. As such, strategies to prioritize target species or focus the analysis on a specific protein class of interest (e.g. cyclotides as presented herein) will further streamline the process of rapid bioactive peptide discovery.
Cyclotides are a class of cyclic, disulfide-rich, plant-derived peptides found primarily in the Violaceae and Rubiaceae families that boast a diverse range of innate bioactivities ranging from antibacterial, anticancer, anti-HIV, mulloscicidal and insecticidal (Burman et al., 2010; Craik et al., 1999; Fensterseifer et al., 2015; Gerlach et al., 2010; Plan et al., 2008; Pranting et al., 2010; Wang et al., 2008) indicating a highly stable, evolvable scaffold. Approximately 400 unique cyclotide sequences have been documented; however, tens of thousands up to 150,000 are estimated to exist in nature (Burman et al., 2015; Gruber et al., 2008; Hellinger et al., 2015; Narayani et al., 2017). The extraordinary stability conferred by the highly complex cyclic cysteine knot (CCK) motif found in all known cyclotides makes the conserved three-dimensional structure of this peptide class an ideal scaffold for protein engineering (Camarero, 2017; White and Craik, 2016; Wong et al., 2012). Naturally-occurring and modified cyclotides have already found application in both medicine and commercial agriculture. Grafting therapeutic peptides into cyclotide scaffolds alleviates the issue of peptide instability in vivo, and has the potential to deliver a variety of highly specific and efficacious peptide-based therapeutics via oral administration and to intracellular targets (Henriques et al., 2015; Wong et al., 2012). Meanwhile, Sero-X, a potent bioinsecticide derived from the cyclotide-containing extract of Clitorea ternatea, was approved for use on cotton plants in Australia in early 2017. Highly effective at controlling insect pests, Sero-X does not harm beneficial insects such as bees and ladybugs, and exemplifies the next level of safe and effective biocontrol that can be achieved with peptide-based treatments (APVMA, 2016).
While mass spectrometry has been used to successfully identify many cyclotides, the standard approach includes pre- and post-reduction and alkylation with iodoacetamide to reveal any peaks with a mass shift of 348.16 Da (Gruber et al., 2008) and linearization using endoproteinase Glu-C via a single conserved glutamic acid residue (Poth et al., 2011), or other enzymes (Chan et al., 2013), enabling efficient fragmentation to obtain primary protein sequence, possibly in tandem with transcriptome-mining (Koehbach et al., 2013). While successful, sample loss from the numerous sample preparation steps hinders the sequence elucidation of less abundant peptidyl species. Likely, the challenges associated with purifying structurally similar cyclotides and cyclotide sequencing are why hundreds of putative cyclotide masses have been identified via reduction/alkylation methods, but only a fraction of these masses have been sequenced (Burman et al., 2015; Narayani et al., 2017). A top-down approach in which cyclotides are analyzed undigested would ameliorate sample loss issues (Mohimani et al., 2011), but has not been demonstrated for cyclotides. While the commonly used collision induced dissociation (CID) is a practical method for short, linear peptides or enzymatically-digested proteins, CID often provides insufficient fragmentation of intact cyclic peptides. Ultraviolet photodissociation (UVPD) is an emerging technique that yields highly complex but rich fragmentation data, generating several possible types of fragmentation events (e.g. a, b, c, x, y, and z) (Supplemental Figure 1) (Shaw et al., 2013). UVPD has been used to fragment circular (stapled) peptides (Crittenden et al., 2016), but no cyclotides have been sequenced with UVPD previously.
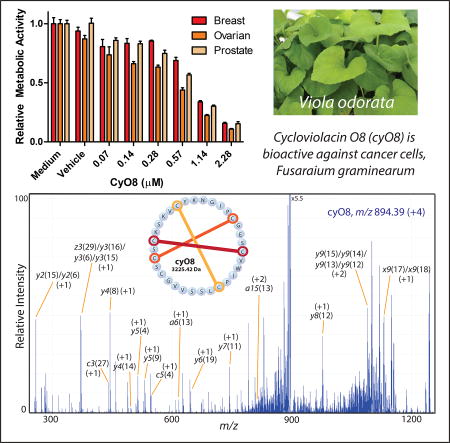
The botanical species Viola odorata L. (Violaceae) abundantly produces >30 known, unique cyclotide sequences with potent and diverse bioactivities (Ireland et al., 2006), and may harbor up to 166 cyclotide species as indicated by mass shift analysis (Narayani et al., 2017). A V. odorata peptide library was screened against breast, prostate, and ovarian cancer cell lines for validation of the PepSAVI-MS pipeline (Kirkpatrick et al., 2017). Constituents of this complex botanical extract demonstrated substantial cancer cell cytotoxicity in all assays, particularly across cyclotide-containing library fractions. While known and novel cycloviolacin O2 (cyO2) activity was verified, our results indicated additional cyclotide species responsible for the activity observed in the cancer cell panel. Further analysis presented herein revealed another cyclotide, cycloviolacin O8 (cyO8), as a putative anticancer peptide (Figure 1). Isolated cyO8 demonstrated micromolar bioactivities against MDA-MB-231 breast, PC-3 prostate, and OVCAR-3 ovarian cancer cell lines. Additionally, the V. odorata library was shown to exhibit robust activity against the filamentous fungus Fusarium graminearum, responsible for the devastating disease Fusarium Head Blight (FHB); however, the bioactive constituents were not identified (Kirkpatrick et al., 2017). Herein, PepSAVI-MS revealed the F. graminearum antifungal activity of cyO8, which was confirmed with isolated peptide. While cy08 has known anthelmintic activity against nematode larvae (Colgrave et al., 2008), this work expands the reported bioactivities to anticancer and antifungal for this cyclotide. Additionally, a simple reduction/alkylation strategy was implemented to further refine molecular targets revealed with PepSAVI-MS, identifying several putative anticancer and antifungal cyclotides that require further molecular characterization. As these masses are low in abundance, traditional MS-based sequencing methods are not sufficient and alternative sequence elucidation techniques are required. Using cyO8 as a representative species, we show the complexity of cyclic-peptide 193 nm UVPD fragmentation and attempt to discern common cyclotide fragmentation patterns to inform application to future de novo sequencing algorithms.
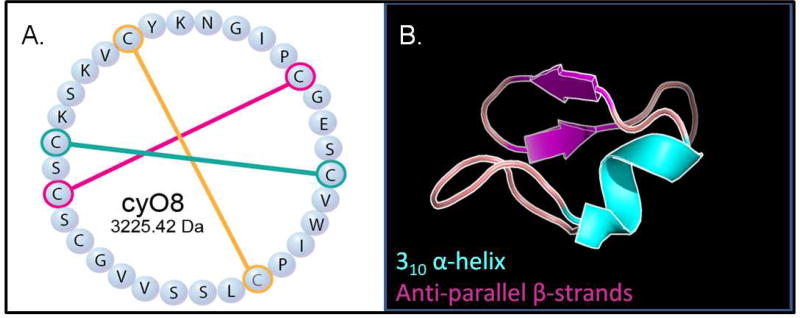
Figure 1.
Cycloviolacin O8 (cyO8), a known anthelmintic cyclotide from Viola odorata. A. CyO8 sequence, MW 3225.42 Da, showing disulfides. B. Predicted three-dimensional cyO8 structure, not showing disulfides. Magenta arrows and cyan domain represent anti-paralled β-sheets and a 310 α-helix, respectively (cybase.org (Mulvenna et al., 2006)); figure generated in Pymol).
2. Results
2.1 Mining V. odorata
Validation of the PepSAVI-MS pipeline produced numerous, previously uncharacterized V. odorata molecular species as highly ranked anticancer and antifungal targets (Kirkpatrick et al., 2017). Further exploration indicated that the original filtering conditions imposed were highly stringent, i.e. filtering out all m/z outside of the bioactivity region excluded peptides that might be detected via mass spectrometry but not yet high enough abundance to be revealed in the bioactivity assessment. As such, we refined the filtering conditions to include one strong cation exchange (SCX) fraction on either side of the bioactivity region and remodeled the cancer panel bioactivity to produce refined lists of ranked putative anticancer compounds with bioactivities against MDA-MB-231 breast, PC-3 prostate, or OVCAR-3 ovarian cancer cell lines (Supplemental Table 1). The V. odorata cyclotide cyO8 (MW 3225.42 Da) was ranked among the top 20 contributors to ovarian and prostate cancer cell cytotoxicity. Additionally, a clear region of bioactivity was seen in the cyclotide-containing fractions 25–31 against F. graminearum (Figure 2). Statistical modeling of the bioactivity region revealed cyO8, in both the +3 and +4 charge states, as a highly ranked antifungal candidate (Supplemental Table 1). Additionally, mass 3152.41 Da was ranked in the top 20 contributors to F. graminearum antifungal activity, and may represent the known cyclotides cycloviolacin O3 (MW 3152.38 Da) or cycloviolacin O7 (MW 3152.41 Da) from V. odorata or Vitri A (MW 3152.38) from Viola tricolor, as some cyclotide sequences are not unique to a single botanical species.
Figure 2.
Viola odorata SCX fraction library versus Fusarium graminearum bioactivity assay. Data are represented as mean ± SD of three replicates. Antifungal bioactivity is seen in V. odorata fractions 25–31.
2.2 Validation of novel bioactivities revealed by PepSAVI-MS
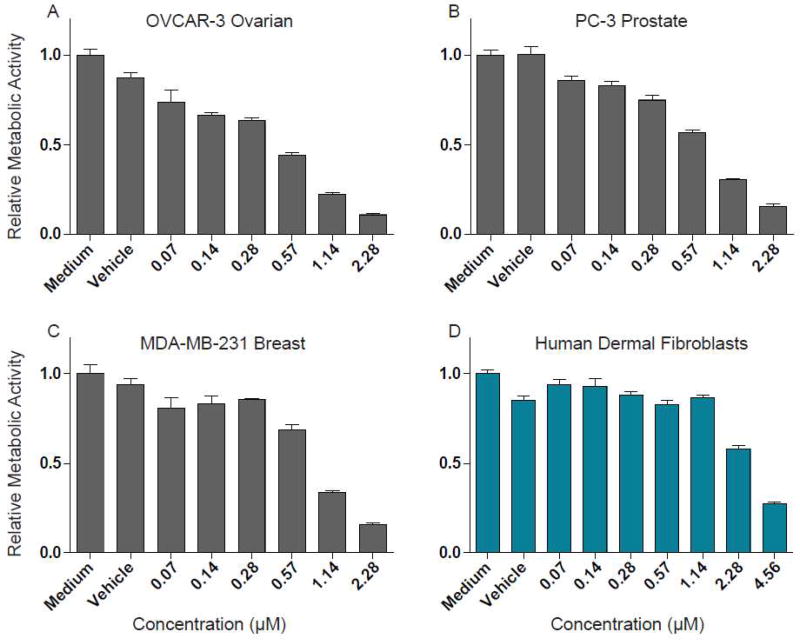
Ranked peptides identified via PepSAVI-MS must be isolated for sequence and structure characterization and verification of putative bioactivities. To confirm the anticipated anticancer and antifungal activities of cyO8, V. odorata SCX fractions were further processed with reversed-phase LC to achieve cyO8 isolation from the complex mixture of plant peptides. Purified cyO8 was shown to be active against the three cancer cell lines tested. Inhibitory concentration (IC50) values were determined for each cell line: MDA-MB-231 breast (1.15 (µM), PC-3 prostate (1.05 µM), and OVCAR-3 ovarian (0.80 µM) (Figure 3). CyO8 cytotoxicity against non-cancerous human dermal fibroblast cells was determined to be ~3X less (3.13 µM) than cancer cell lines (Figure 3). Isolated cyO8 was shown to be active against F. graminearum with an IC50 of 25–30 µM (a two-fold serial dilution was performed and visually assessed for the minimum inhibitory concentration; after 72 h the well containing 24 µM cyO8 showed only the beginnings of fungal growth).
Figure 3.
Inhibitory concentration (IC50) determination of cyO8 against human cancer cell lines. Purified cyO8 tested against human cancer cell lines, OVCAR-3 ovarian (A), PC-3 prostate (B), and MDA-MB-231 breast (C), and against non-cancerous human dermal fibroblasts (D). Vehicle indicates cells are cultured with water in place of cyO8. Data are represented as mean ± SD of three replicates.
2.3 Revealing novel, putatively bioactive V. odorata cyclotides
Taking advantage of the highly conserved three disulfide bonds in cyclotides, reduction and alkylation of V. odorata combined SCX fractions 12–45 produced numerous mass shifts of 348.16 ± 0.05 Da, consistent with the alkylation of three disulfide bonds (Gruber et al., 2008). Interrogation of this data resulted in 25 candidate masses likely belonging to the cyclotide family (Table 1). Eight putative cyclotide species correlated to masses identified in PepSAVI-MS bioactivity screens against cancer cell lines and/or F. graminearum: 3154.38 Da, 3156.36 Da, 3165.49 Da, 3166.48 Da, 3170.38 Da, 3171.38 Da, 3184.39 Da, and 3257.37 Da (Table 1).
Table 1.
Putative cyclotide species identified with a reduction/alkylation mass shift experiment and assays in which mass was ranked as a potential anticancer/antifungal contributor. Masses identified in recent cyclotide screens are indicated. All masses can be matched to known cyclotide masses in other botanical species; however, this is not necessarily an indication of identity.
| Mass (Da) | Assay | Detected in screen |
|---|---|---|
| 3597.55 | Yes | |
| 3568.68 | Yes | |
| 3514.60 | ||
| 3499.54 | ||
| 3497.54 | Yes | |
| 3483.49 | ||
| 3440.51 | ||
| 3364.41 | ||
| 3350.47 | ||
| 3344.68 | Yes | |
| 3332.44 | ||
| 3330.48 | ||
| 3297.37 | Yes | |
| 3269.37 | Yes | |
| 3257.37 | Prostate cancer/Fusarium | |
| 3243.45 | Yes | |
| 3208.41 | ||
| 3186.42 | Yes | |
| 3184.39 | Breast/Prostate/Ovarian/Fusarium | |
| 3171.38 | Ovarian | Yes |
| 3170.38 | Breast | |
| 3166.48 | Breast/Prostate/Fusarium | |
| 3165.49 | Ovarian/Prostate | |
| 3156.36 | Fusarium | Yes |
| 3154.38 | Fusarium |
2.4 Characterization of 3257 Da putative cyclotide
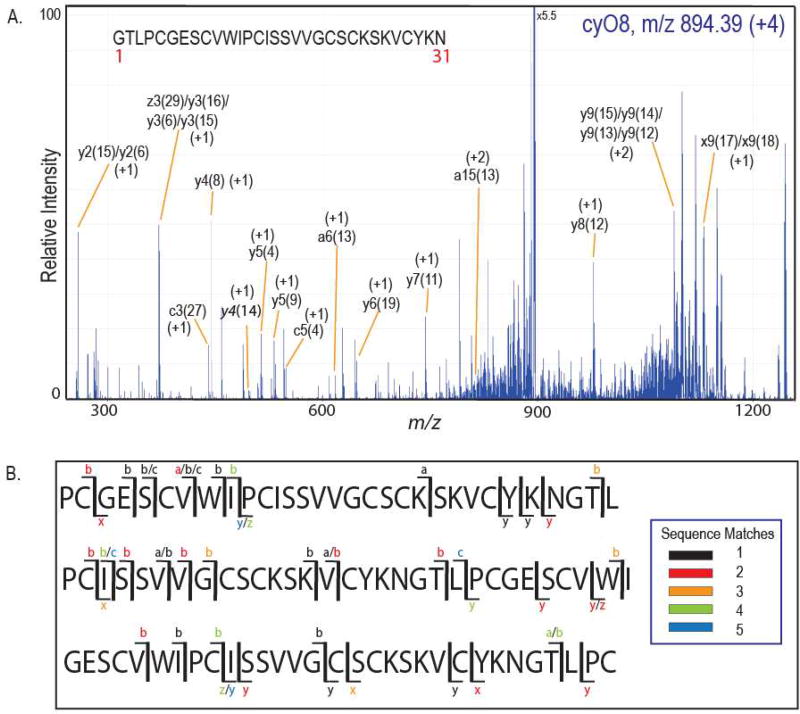
Reduction, alkylation, and linearization with Glu-C of the most abundant 3257.37 Da containing V. odorata SCX fraction resulted in incomplete yet informative CID MS2 spectra (Figure 4). The 32 Da mass difference between the putative cyclotide and the known, highly abundant cyclotide, cyO8, could be localized to a tryptophan residue, showing a mass shift of the peak m/z 533 to m/z 565, and likely represents a double oxidation of the side chain, a known post-translational modification in cyclotides (Hellinger et al., 2015). Of the 24 remaining candidate cyclotide masses, some are likely known cyclotides with singly (oxindolyalanine, oia) or doubly-oxidized tryptophan (N-formylkynurenine, nfk) residues based on mass differences of multiples of 16 Da: 3154.38 Da (cyO2oia), 3184.39 Da (cyO3nfk), and 3170.38 Da (cyO2nfk). However, several putative bioactive cyclotides, 3156.36 Da, 3165.49 Da, 3166.48 Da, and 3171.38 Da remain uncharacterized.
Figure 4.
Collision induced dissociation (CID) fragmentation of cyO8. CID MS2 spectra of reduced, alkylated, gluC digested cyO8 (+4) (top) and reduced, alkylated, gluC digested cyclotide mass 3257.37 Da (+4) (bottom) showing 32 Da double-oxidation mass shift localized to the tryptophan residue of cyO8nfk (cyO8 N-formylkynurenine). Fragment ions are colored the same in both spectra to aid in quick identification.
2.5 UVPD fragmentation of cyO8
Fragmentation of cyclic molecules must occur twice to observe fragment ions, as the first cleavage results only in ring opening and no initial mass change. Incomplete fragmentation of intact cyclotides with traditional tandem MS approaches (e.g. CID, ECD/ETD) and sample loss from numerous reduction, alkylation, and digestion steps has traditionally hindered the sequence determination of low abundance cyclotide species (Burman et al., 2015; Narayani et al., 2017). Generating rich fragmentation data of all ion types (Brodbelt, 2014), high energy UVPD has the potential to circumvent these sequencing challenges, particularly if fragmentation patterns can be established to aid in de novo sequence assignment. Absorption of high energy (193 nm) photons produced during UVPD allows access to excited electronic states that result in extensive fragmentation and production of numerous types of fragment ions. CyO8 fragmentation obtained by 193 nm UVPD using complex V. odorata library fractions yielded the spectrum presented in Figure 5A. Predominant fragmentation events during UVPD occurred at the C-terminus of the two proline residues in the cyO8 sequence; however, fragmentation was seen throughout most sequence iterations (Figure 5B). Peak labels in Figure 5A indicate the residue in position 1 of the identified fragment sequence and the ion type (for example, y4(8) is the sequence that results in an initial fragmentation event between residues 7 and 8 and a second cleavage corresponding to a y4 fragment). The majority of fragment ions were not observed from any one particular sequence iteration, and only about 50% of the cyO8 residues could be confidently assigned. Although the cyO8 precursor mass was abundant, MS2 fragments generated by UVPD were low intensity because the total abundance was distributed among numerous fragment ions. Spectral overlap precluded assignments in the m/z 800–900 and 1000–1200 regions, and isobars or fragment ions within a 30 ppm mass tolerance led to sequence ambiguity, in some cases up to five different sequence possibilities for one observed m/z (Supplemental Tables 2–6).
Figure 5.
Ultraviolet Photodissociation (UVPD) fragmentation of cyO8. A. MS/MS of (+4, m/z 894.39) cyO8 (2 pulses, 2 mJ/pulse) shows abundant, low intensity fragment ions. Fragments are labeled by the fragment ion type and the numeric index (in parentheses) from the sequence map shown indicating the initial cleavage site (e.g. a15(13) indicates an initial cleavage between residues 12 and 13 and a second fragmentation event resulting in an a15 fragment ion). B. Representative cyO8 sequence iterations showing UVPD MS2 coverage. Ion types are color-coded based on the number of ambiguous fragment ion matches.
3. Discussion
Previously, we have shown that cyclotide-containing V. odorata fractions exhibit anti-cancer bioactivities against MDA-MB-231 breast, PC-3 prostate, and OVCAR-3 ovarian cancer cell lines (Kirkpatrick et al., 2017). In addition to the known activity of cyO2, refinement of data filtering criteria in PepSAVI-MS revealed additional known and putative cyclotides with possible antiproliferative activity against cancer cell lines. Specifically, cyO8 which is a known nematocidal cyclotide, was ranked highly in our modeling. Subsequent isolation and targeted bioassays revealed anticancer activities of cyO8 at low micromolar IC50 values. Additionally, cyO8 demonstrated approximately three times less cytotoxicity against human dermal fibroblast cells than to the human cancer cell lines. Commonly used pharmaceuticals targeting breast, ovarian, and prostate cancers exhibit IC50 values from low nanomolar to low micromolar concentrations, pushing cyO8 to the higher end of the treatment range.
While innate antibacterial, anticancer, and antiviral cyclotide bioactivities have been thoroughly explored over the past 35 years, the antifungal properties of cyclotides have seldom been reported, and typically pertain only to common lab strains of Candida sp (Stromstedt et al., 2017; Tam et al., 1999). We are excited to report cyclotide antifungal activity against the agriculturally-relevant, filamentous fungus F. graminearum, with a MIC value of 25–30 µM. Interestingly, several of the V. odorata fractions promoted fungal growth in the F. graminearum bioactivity assays, highlighting a potential issue that may be encountered when screening additional fungal species with the PepSAVI-MS pipeline and the need to filter datasets wider than the observed bioactivity region. Fortunately, cyO8 was a potent fungal inhibitor, as weaker activities may not be observed if growth promotion masks bioactivity. The appearance of the semi-abundant cyO3/cyO7 in the F. graminearum top 20 ranked compounds is consistent with recently published work in which cyO3 was identified as a low micromolar inhibitor of the opportunistic human pathogenic yeast, Candida albicans (Stromstedt et al., 2017). Though exceptionally stable and resistant to harsh conditions, cyclotides make particularly promising topical biofungicides as the eventual, natural degradation of these molecules controls environmental concentrations and prevents unwanted accumulation in soils (Lee and Kim, 2015).
Several putative cyclotide masses identified from the pre- and post- reduction/alkylation analysis were found among the ranked lists for the human cancer cell lines and F. graminearum. Of these masses, we anticipate several may be known cyclotides with oxyindolyalanine or N-formylkynurenine tryptophan modifications. As most of these species are present in very low abundances, we have only been able to confirm the identity of 3257.37 Da as a doubly-oxidized tryptophan cyO8 (cyO8nfk) via CID tandem mass spectrometry. Tryptophan oxidation has been documented previously in cyclotides as the solvent-exposed tryptophan residue is more susceptible to modification, and is thought to play a role in cyclotide degradation pathways in vivo (Burman et al., 2011; Hellinger et al., 2015; Plan et al., 2007; Ravipati et al., 2017). The 24 remaining putative cyclotide species have yet to be characterized. Several of these potential anticancer and antifungal cyclotide species have recently been reported by other groups in large scale reduction/alkylation experiments to identify novel cyclotides; 3154 Da and 3171 Da were reported by Göransson et al. (Burman et al., 2015), whereas similar masses, 3166.23 Da, 3170.36 Da, and 3171.37 Da, were reported by Srivastava et al. (Narayani et al., 2017). Four masses never reported in V. odorata appearing on the human cancer cell line and F. graminearum ranked lists, 3156.36 Da, 3165.49 Da, 3166.48 Da, and 3171.38 Da, are prioritized for sequence elucidation and activity confirmation. Though unknown in V. odorata, all of the proposed cyclotide masses identified herein match known cyclotide sequences in other botanical species and may be identical in sequence. As some cyclotides are shared among numerous plant species, there are also known isobaric cyclotides with unique sequences even within the same plant; further sequence characterization is necessary. By combining a simple reduction/alkylation strategy with the PepSAVI-MS pipeline, we have been able to prioritize ranked m/z species more-likely to be contributing to observed bioactivities.
Traditional mass spectrometry-based cyclotide sequencing approaches utilize a reduction, alkylation, and enzymatic digestion strategy to linearize target peptides as intact cyclotides produce minimal fragmentation upon traditional collision induced dissociation. While linearized cyclotides are easier to fragment, resulting MS/MS spectra are often incomplete and do not allow for confident sequence identification of novel targets, with as little as 20% sequence coverage (Figure 4). Furthermore, increased sample handling steps significantly increase sample loss and result in minimal signal for peptide fragmentation and identification. Herein we demonstrate a top-down UVPD approach, using cyO8 as a model, to reduce sample loss and facilitate the sequence elucidation of less abundant cyclotide species. To estimate the potential complexity of the MS/MS spectra generated for cyclic peptides, the numbers of fragment ions were manually calculated for cyO8 considering a maximal charge of +4. Assuming initial and secondary cut sites at all 31 cyO8 backbone positions and always a b/y initial cleavage, UVPD fragmentation would result in ~22,000 a, b, c, x, y, and z theoretical ions compared to ~11,000 a, b, and y ions with CID (considering 31 residues, each with a 30 fragment ion series, four charge states, and six or three ion types for UVPD or CID, respectively). In comparison, theoretical fragmentation of the 31 residue linear cyO8 peptide results in ~720 a, b, c, x, y, and z fragments for UVPD and ~360 a, b, and y fragments with CID, including fragment ion charge states ranging from +1 to +4 and no internal fragment ions. Even with reduced and alkylated disulfide bridges, both required fragmentation events can occur at any position in the ring and leads to a combinatorial explosion of theoretical fragment ions. As such, the 193 nm UVPD fragmentation of reduced and alkylated cyO8 in V. odorata library fractions resulted in MS2 spectra that were overly data-rich and spectral overlap made peak assignments challenging. Though predominant fragmentation sites were observed at the two highly constrained proline sites, cleavage occurred throughout most cyO8 variations and few other discernible patterns emerged during analysis to inform de novo assignments of uncharacterized cyclotides. Most fragment types, a, b, c, x, y, and z ions, were seen in roughly the same abundances, except possibly a small preferential for b/y ion types. Additionally, ambiguity associated with isobaric fragment ions or fragments similar in m/z introduced uncertainty into the assignment of cyO8 peaks. Ambiguity among similar but non-identical fragment ions may be remedied by producing even higher resolution mass spectra, which may be feasible for targeted experiments where an increase in acquisition time would be insignificant for only one or a few m/z species. UVPD can conceivably be used to verify minor amino acid modifications in known sequences to rapidly aid in the characterization of engineered products. Additionally, UVPD strategies employing lower energy wavelengths (351 nm) in conjunction with chromogenic derivitizing tags may allow for de novo sequencing of unknown cyclotides (Robotham et al., 2016). For future de novo assignments of cyclic molecules fragmented with UVPD, a sophisticated algorithm is necessary in which more common fragmentation patterns are established and used to make decisive fragment assignments based on probability. Multistage mass spectrometry employing MSn coupled with machine learning could lend a tremendous amount of information to aid in de novo sequencing, particularly using UVPD fragmentation; however, this technique requires large datasets for algorithm training, that are as of yet unavailable, and specialized instrumentation (Mohimani et al., 2011).
4. Conclusions
Peptide-based treatments offer new and effective alternatives in drug discovery and sustainable agricultural practices, and cyclotides are promising candidates with enhanced stability and a host of diverse bioactivities. PepSAVI-MS revealed novel anticancer and antifungal activities of the known cyclotide, cyO8, and herein those activities have been validated and characterized. The work described here illuminates the possibility of re-mining known natural product sources via PepSAVI-MS with the likelihood that valuable bioactive constituents were previously overlooked and may be revealed. We also demonstrate the utility of a reduction/alkylation strategy to identify putative cyclotides with results prioritized via PepSAVT-MS to expedite the prioritization and validation of bioactive molecules. UVPD of a cyclotide was demonstrated for the first time using cyO8 as a model sequence and assessed for potential in de novo applications. Several putative cyclotide species with suspected bioactive properties have been identified via PepSAVI-MS; once characterized these novel cyclotides can add to the substantial sequence and functional diversity already seen in this ever-growing peptide family. This work adds to the growing body of knowledge concerning these fascinating knotted molecules whose seemingly boundless bioactivities will undoubtedly be harnessed to combat emerging microbial resistance in agriculture and medicine.
5. Materials and Methods
5.1 Preparation of Plant Material and Library Generation
Viola odorata L. (Violaceae) peptide library was prepared as described previously (Kirkpatrick et al., 2017). Briefly, Viola odorata seeds (Strictly Medicinal Seeds, OR) were grown to mature rosettes in a laboratory greenhouse at 17.5–20.3 °C on a 14/10 h light/dark cycle. Aerial tissue was harvested, manually ground, and proteins extracted in a 10% acetic acid buffer. Centrifugation was used to pellet insoluble material and the remaining extract was filtered (0.45 µm stericup filtration; Millipore), concentrated to remove high-molecular-weight molecules (MWCO 30 kDa; Millipore), and dialyzed into 5 mM ammonium formate (Fluka) pH 2.7 to remove small-molecules (0.1–1 kDa cutoff; SpectrumLabs). The V. odorata peptide library was prepared by fractionating crude extract with a 47 min strong cation exchange (SCX) method (PolySulfethyl A column (100 mm × 4.6 mm, 3 µm particles, PolyLC) using a salt gradient (linear ramp from 5 mM ammonium formate, 20% acetonitrile, pH 2.7 to 500 mM ammonium formate, 20% acetonitrile, pH 3.0). Fractions were collected in one-min intervals, desalted, and stored at 4 °C.
5.2 Bioactivity Assays
V. odorata peptide library was tested against F. graminearum in a 96-well plate format bioactivity assay, as described previously (Kirkpatrick et al., 2017). Each plate well contained 30 µL minimal media, 10 µL F. graminearum spore culture, and 10 µL V. odorata library fraction, antibiotic (Hygromycin B, Corning), or water, and was prepared in triplicate. Minimal media is a modified Czapek-Dox formulation, a standard growth media for laboratory fungus propagation (Correll et al., 1987; Leslie and Summerell, 2006). Plates were incubated at 25 °C, shaking at 300 rpm. After 48 h, BacTiterGlo (Promega) was added to each well to quantify the amount of ATP present, indicative of fungal cell growth, and luminescence was measured with a 500 ms integration time.
5.3 Statistical Modeling
LC-MS/MS data of the V. odorata SCX library fractions (fractions 15–40 for cancer cell lines, 16–33 for Fusarium) was processed using the PepSAVI-MS software and filtered for compounds of interest by applying retention time, mass, and charge-state limits. MS features of the same charge state were binned together within a 0.05 Da window and features with (1) intensities <100 in the activity region, (2) charge states >+10, and (3) intensities >1% of maximum peak intensity in non-activity regions were excluded. Data reduction and modeling were tailored to cancer cell line/Fusarium activity profile. Data filtering of SCX library fractions was as follows: MDA-MB-231 breast (15–24); PC-3 prostate (17–24); OVCAR-3 ovarian (17–23); Fusarium (23–31). Activity regions were defined for each cancer cell line: MDA-MB-231 breast (16–23); PC-3 prostate (18–23); OVCAR-3 ovarian (18–22), Fusarium (25–30). Resulting m/z meeting these criteria were modeled with an elastic net regression with a quadratic penalty parameter specification of 0.001.
5.4 Reversed-Phased Isolation of cyO8
CyO8-containing SCX fractions were subject to reversed-phase isolation on a Waters ACQUITY UPLC H-Class Bio System equipped with an Agilent Zorbax extended C18 column (20k PSI, 300 Å, 5 µm, 4.6 mm × 250 mm). Separation of cyO8 was achieved with a flow rate of 2 mL/min and a linear ramp of 5–25% mobile phase B from 0–5 min and 25–50% mobile phase B from 5–20 min (mobile phase A, water with 0.1% formic acid; mobile phase B, acetonitrile with 0.1% formic acid). Reversed-phase fractions were manually collected, dried down in a vacuum centrifuge, resuspended in water, and fractions containing cyO8 were combined for use in Fusarium conidia germination and cancer/fibroblast cell viability assays. CyO8 purity was assessed via LC-MS, and considering peaks >1% of the abundance of cyO8, purity was deemed to be 95% for the cancer cell line assays and >91% for the Fusarium graminearum assays (separate preparations) with no single impurity exceeding 2% (except doubly oxidized cyO8 at 3.8% in the F. graminearum MIC assay).
5.5 Cancer Cell Line/Fibroblast Viability Assay
MDA-MB-231 breast cancer cells, PC-3 prostate cancer cells, OVCAR-3 ovarian cancer cells cells (all cancer cell lines from ATCC, Manassas, VA), or human dermal fibroblasts (HDF; Lonza Inc., Walkersville, MD, USA) were seeded in triplicate wells of a flat-bottom 96-well plate at a concentration of 5 × 103 cells/well. Cancer cells were in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, 2mM of L-glutamine, and 5 mM of HEPES buffer (all from Invitrogen) while HDFs were in Fibroblast Cell Basal Medium (Lonza) supplemented with 2% fetal bovine serum plus 0.1% insulin, 0.1% recombinant human basic fibroblast growth factor, and 0.1 % gentamicin sulfate/amphotericin, all from a proprietary Fibroblast Cell Medium BulletKit (Lonza). Cells were cultured for 12 h at 37°C in a humidified atmosphere containing 10% CO2 (cancer cells) or 5% CO2 (HDF) in order to form adherent monolayers. The medium was then removed, replaced with fresh complete DMEM with or without cyO8, and the cells were cultured for an additional 24 h. Two hours before the end of culture, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Sigma Aldrich, Oakville, ON, Canada) was added to each well to a final concentration of 0.5 mg/mL to measure mitochondrial succinate dehydrogenase activity. After 2 h, culture supernatants were discarded and formazan crystals were solubilized in 0.1 mL of dimethyl sulfoxide. Absorbance was measured at 570 nm using an ASYS Expert 96 microplate reader (Montreal Biotech Inc., Kirkland, QC, Canada). Average absorbance in each group was normalized relative to the medium control and the percent reduction in viability, as reflected by the change in mitochondrial succinate dehydrogenase activity, was determined.
5.6 Minimum Inhibitory Concentration Assay
MDA-MB-231 breast cancer cells, PC-3 prostate cancer cells, OVCAR-3 ovarian cancer cells, or HDFs were cultured as described above in the absence or presence of different concentrations of cyO8 and inhibitory concentration (IC50) values of cyO8 were calculated from the resulting dose response curves. To determine the cyO8 MIC against Fusarium graminearum, a two-fold serial dilution was performed in a 96-well plate format. CyO8 concentration was determined using a NanoDrop and an extinction coefficient of 7365 M−1 cm−1 In each well, 30 µL minimal media, 10 µL Fusarium spore culture, and 10 µL purified cyO8 was added (for a final concentration of 12, 24, or 48 µM), and the plate was incubated at 22 °C for 72 h, shaking at 250 rpm, and assessed visually for MIC.
5.7 Reduction, Alkylation, and Glu-C digest of V. odorata library
V. odorata peptide library fractions were reduced with 10 mM dithiothreitol (Sigma-Aldrich) at 45 °C, 850 rpm, 30 min, and alkylated with 100 mM iodoacetamide (Sigma –Aldrich) at 25 °C, 850 rpm, 15 min. Pierce C18 spin columns (Thermo Scientific) were used to remove interfering contaminants from the V. odorata samples, which were subsequently dried down in a vacuum concentrator and resuspended in acidified water (0.1% formic acid) for LC-MS/MS analysis. Digested V. odorata samples were reduced and alkylated as described above, then incubated 1:50 enzyme: substrate with endoproteinase Glu-C enzyme (Sigma) at 37 °C for 3 h.
5.8 LC-MS/MS Analysis of V. odorata library
Five microliters of each intact, reduced/alkylated, and reduced/alkylated/glu-C digested V. odorata sample were injected onto a nano-LC-ESI-MS/MS platform: nanoAcquity (Waters, Milford, MA) coupled to a TripleTOF5600 (AB Sciex, Framingham, MA). Samples were initially injected onto a trap column (NanoAcquity UPLC 2G-W/M Trap 5 µm Symmetry C18, 180 µm × 20 mm: Waters) before entering the analytical C18 column (10k PSI, 100 Å, 1.8 µm, 75 µm × 250 mm: Waters). LC separation of peptides was achieved with a flow rate of 0.3 µL/min and a linear ramp of 5%–50% B (mobile phase A, 1% formic acid in water; mobile phase B, 1% formic acid in acetonitrile) over 30 min. The MS was operated in positive-ion, high-sensitivity mode with the MS survey spectrum using a mass range of 350–1600 Da in 250 ms and information-dependent acquisition (IDA) of CID MS/MS data. The first 20 features above 150 counts threshold with a charge state of +2 to +5 were fragmented using rolling collision energy (±5%). Deisotoped peak lists for the intact and reduced/alkylated samples were generated using Progenesis QI for Proteomics software (Nonlinear Dynamics, v.2.0) with a retention time filter of 14–45 min. “Peptide ion data” was exported from Progenesis and analyzed for the characteristic reduction/alkylation mass shift corresponding to three disulfide bonds (348.16 Da) using Python.
5.9 UVPD Analysis of CyO8
Reduced and alkylated cy08 was targeted for analysis following loading of a complex mixture of cyclotides on an in-house packed C-18 (3.5 µm particle size, X-Bridge) trap column (3.5 cm New Objective Integrafrit capillary, 100 µm i.d.) and separation using an in-house packed C-18 (3.5 µm, X-Bridge) analytical column (15 cm New Objective Picofrit capillary, 75 µm i.d.) on a Ultimate 3000 UHPLC system (ThermoFisher Scientific). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid at a net flow rate of 300 nL/min. The applied gradient is as follows: 0–8 min at 2% B; 8–10 min increasing linearly from 2 to 20% B; 10–32 min increasing linearly from 20 to 40% B; rapid increase to 80% B at 32 min and holding until 34 min; 34 min decreasing to 2% B to return the mobile phase to its initial condition. 1 µL of sample at a concentration of approximately 1 mM total protein content was injected on column per experiment. The chromatography system was coupled to Thermo Scientific Orbitrap Fusion Lumos mass spectrometer (San Jose, CA, USA) outfitted with a 193 nm excimer laser (Coherent Excistar 500 Hz, Santa Clara, CA, USA) to allow photodissociation, as implemented and described previously(Klein et al., 2016). UVPD was performed using 2 pulses at 2 mJ per pulse in the low pressure linear ion trap. The Orbitrap mass spectrometer was operated in the positive ion mode with a spray voltage of 1800 V. The 3+ and 4+ charge states of cy08 were targeted during the experiments with a 100 ppm mass tolerance during the MS1 spectrum (60000 FT resolving power, 5 × 105 AGC target). The MS/MS spectra were collected using the following parameters: quadrupole isolation with an isolation width of 4 m/z; 15000 FT resolving power; 2 × 105 AGC target; 2 µscans averaged per spectrum. Theoretical fragment libraries were generated with mMass, an open source mass spectrometry tool to aid in the characterization of cyclic and linear peptides (Niedermeyer and Strohalm, 2012; Strohalm et al., 2008; Strohalm et al., 2010). Theoretical fragments were manually assigned to cyO8 MS2 peaks within 30 ppm error.
Supplementary Material
Highlights.
CyO8 exhibits bioactivity against prostate, ovarian, and breast cancer cell lines
CyO8 is bioactive against the fungal pathogen Fusarium graminearum
193 nm ultraviolet photodissociation mass spectrometry is demonstrated with cyO8
Putative anticancer and antifungal cyclotide masses are identified in Viola odorata
Acknowledgments
L.M.H. acknowledges funding from the National Institutes of Health (1R01GM125814-01). N.C.P. acknowledges support from the NIH Molecular and Cellular Biophysics training grant (T32 GM008570). J.S.B. acknowledges funding from the Welch Foundation (F-1155) and NSF (CHE-1402753).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
N.C.P., C.L.K., C.M.C., D.W.H., and L.M.H. designed the experiments. N.C.P., C.L.K., C.M.C., and J.G.R. performed the experiments. N.C.P., C.L.K., C.M.C., D.W.H., J.S.B., and L.M.H. wrote the paper.
References
- Ajesh K, Sreejith K. Peptide antibiotics: an alternative and effective antimicrobial strategy to circumvent fungal infections. Peptides. 2009;30:999–1006. doi: 10.1016/j.peptides.2009.01.026. [DOI] [PubMed] [Google Scholar]
- APVMA. PUBLIC RELEASE SUMMARY on the evaluation of the new active Clitoria ternatea in the product Sero-X Insecticide. Australian Pesticides and Veterinary Medicines Authority 2016 [Google Scholar]
- Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton ER, Kellogg JJ, Kvalheim OM, Cech NB. Biochemometrics to Identify Synergists and Additives from Botanical Medicines: A Case Study with Hydrastis canadensis (Goldenseal) J Nat Prod. 2018;81:484–493. doi: 10.1021/acs.jnatprod.7b00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbelt JS. Photodissociation mass spectrometry: new tools for characterization of biological molecules. Chemical Society reviews. 2014;43:2757–2783. doi: 10.1039/c3cs60444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Burman R, Herrmann A, Tran R, Kivela JE, Lomize A, Gullbo J, Goransson U. Cytotoxic potency of small macrocyclic knot proteins: structure-activity and mechanistic studies of native and chemically modified cyclotides. Organic & biomolecular chemistry. 2011;9:4306–4314. doi: 10.1039/c0ob00966k. [DOI] [PubMed] [Google Scholar]
- Burman R, Svedlund E, Felth J, Hassan S, Herrmann A, Clark RJ, Craik DJ, Bohlin L, Claeson P, Goransson U, Gullbo J. Evaluation of toxicity and antitumor activity of cycloviolacin O2 in mice. Biopolymers. 2010;94:626–634. doi: 10.1002/bip.21408. [DOI] [PubMed] [Google Scholar]
- Burman R, Yeshak MY, Larsson S, Craik DJ, Rosengren KJ, Goransson U. Distribution of circular proteins in plants: large-scale mapping of cyclotides in the Violaceae. Front Plant Sci. 2015;6:855. doi: 10.3389/fpls.2015.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero JA. Cyclotides, a versatile ultrastable micro-protein scaffold for biotechnological applications. Bioorganic & medicinal chemistry letters. 2017;27:5089–5099. doi: 10.1016/j.bmcl.2017.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, He W, Tan N, Zeng G, Craik DJ, Daly NL. A new family of cystine knot peptides from the seeds of Momordica cochinchinensis. Peptides. 2013;39:29–35. doi: 10.1016/j.peptides.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Colgrave ML, Kotze AC, Ireland DC, Wang CK, Craik DJ. The anthelmintic activity of the cyclotides: natural variants with enhanced activity. Chembiochem. 2008;9:1939–1945. doi: 10.1002/cbic.200800174. [DOI] [PubMed] [Google Scholar]
- Correll JC, Klittich CJR, Leslie JF. Nitrate non-utilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology. 1987;77:1640–1646. [Google Scholar]
- Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- Craik DJ, Lee MH, Rehm FBH, Tombling B, Doffek B, Peacock H. Ribosomally-synthesised cyclic peptides from plants as drug leads and pharmaceutical scaffolds. Bioorg Med Chem. 2017 doi: 10.1016/j.bmc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Crittenden CM, Parker WR, Jenner ZB, Bruns KA, Akin LD, McGee WM, Ciccimaro E, Brodbelt JS. Exploitation of the Ornithine Effect Enhances Characterization of Stapled and Cyclic Peptides. J Am Soc Mass Spectrom. 2016;27:856–863. doi: 10.1007/s13361-016-1355-7. [DOI] [PubMed] [Google Scholar]
- Fensterseifer IC, Silva ON, Malik U, Ravipati AS, Novaes NR, Miranda PR, Rodrigues EA, Moreno SE, Craik DJ, Franco OL. Effects of cyclotides against cutaneous infections caused by Staphylococcus aureus. Peptides. 2015;63:38–42. doi: 10.1016/j.peptides.2014.10.019. [DOI] [PubMed] [Google Scholar]
- Ganz T. The role of antimicrobial peptides in innate immunity. Integrative and comparative biology. 2003;43:300–304. doi: 10.1093/icb/43.2.300. [DOI] [PubMed] [Google Scholar]
- Gerlach SL, Rathinakumar R, Chakravarty G, Goransson U, Wimley WC, Darwin SP, Mondal D. Anticancer and chemosensitizing abilities of cycloviolacin 02 from Viola odorata and psyle cyclotides from Psychotria leptothyrsa. Biopolymers. 2010;94:617–625. doi: 10.1002/bip.21435. [DOI] [PubMed] [Google Scholar]
- Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E, Craik DJ. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Rodriguez JJ, Ochoa-Zarzosa A, Lopez-Gomez R, Lopez-Meza JE. Plant antimicrobial peptides as potential anticancer agents. Biomed Res Int. 2015;2015:735087. doi: 10.1155/2015/735087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinger R, Koehbach J, Soltis DE, Carpenter EJ, Wong GK, Gruber CW. Peptidomics of Circular Cysteine-Rich Plant Peptides: Analysis of the Diversity of Cyclotides from Viola tricolor by Transcriptome and Proteome Mining. J Proteome Res. 2015;14:4851–4862. doi: 10.1021/acs.jproteome.5b00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques ST, Huang YH, Chaousis S, Sani MA, Poth AG, Separovic F, Craik DJ. The Prototypic Cyclotide Kalata B1 Has a Unique Mechanism of Entering Cells. Chem Biol. 2015;22:1087–1097. doi: 10.1016/j.chembiol.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Herbel V, Wink M. Mode of action and membrane specificity of the antimicrobial peptide snakin-2. PeerJ. 2016;4:e1987. doi: 10.7717/peerj.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland DC, Colgrave ML, Craik DJ. A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability. Biochem J. 2006;400:1–12. doi: 10.1042/BJ20060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Broberg CA, McCool EN, Lee WJ, Chao A, McConnell EW, Pritchard DA, Hebert M, Fleeman R, Adams J, Jamil A, Madera L, Stromstedt AA, Goransson U, Liu Y, Hoskin DW, Shaw LN, Hicks LM. The “PepSAVI-MS” Pipeline for Natural Product Bioactive Peptide Discovery. Anal Chem. 2017;89:1194–1201. doi: 10.1021/acs.analchem.6b03625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Parsley NC, Bartges TE, Cooke ME, Evans WS, Heil LR, Smith TJ, Hicks LM. Fungal Secretome Analysis via PepSAVI-MS: Identification of the Bioactive Peptide KP4 from Ustilago maydis. J Am Soc Mass Spectrom. 2018 doi: 10.1007/s13361-017-1880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DR, Holden DD, Brodbelt JS. Shotgun Analysis of Rough-Type Lipopolysaccharides Using Ultraviolet Photodissociation Mass Spectrometry. Anal Chem. 2016;88:1044–1051. doi: 10.1021/acs.analchem.5b04218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehbach J, Attah AF, Berger A, Hellinger R, Kutchan TM, Carpenter EJ, Rolf M, Sonibare MA, Moody JO, Wong GK, Dessein S, Greger H, Gruber CW. Cyclotide discovery in Gentianales revisited--identification and characterization of cyclic cystine-knot peptides and their phylogenetic distribution in Rubiaceae plants. Biopolymers. 2013;100:438–452. doi: 10.1002/bip.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kim BS. Antimicrobial cyclic peptides for plant disease control. The plant pathology journal. 2015;31:1–11. doi: 10.5423/PPJ.RW.08.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium Laboratory Manual. first. Blackwell Publishing; Iowa: 2006. [Google Scholar]
- Loo S, Kam A, Xiao T, Tam JP. Bleogens: Cactus-Derived Anti-Candida Cysteine-Rich Peptides with Three Different Precursor Arrangements. Front Plant Sci. 2017;8:2162. doi: 10.3389/fpls.2017.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front Cell Infect Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Yang YL, Liu WT, Hsieh PW, Dorrestein PC, Pevzner PA. Sequencing cyclic peptides by multistage mass spectrometry. Proteomics. 2011;11:3642–3650. doi: 10.1002/pmic.201000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvenna JP, Wang C, Craik DJ. CyBase: a database of cyclic protein sequence and structure. Nucleic acids research. 2006;34:D192–194. doi: 10.1093/nar/gkj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayani M, Chadha A, Srivastava S. Cyclotides from the Indian Medicinal Plant Viola odorata (Banafsha): Identification and Characterization. J Nat Prod. 2017;80:1972–1980. doi: 10.1021/acs.jnatprod.6b01004. [DOI] [PubMed] [Google Scholar]
- Niedermeyer TH, Strohalm M. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One. 2012;7:e44913. doi: 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- Peters BM, Shirtliff ME, Jabra-Rizk MA. Antimicrobial peptides: primeval molecules or future drugs? PLoS Pathog. 2010;6:e1001067. doi: 10.1371/journal.ppat.1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plan MR, Goransson U, Clark RJ, Daly NL, Colgrave ML, Craik DJ. The cyclotide fingerprint in oldenlandia affinis: elucidation of chemically modified, linear and novel macrocyclic peptides. Chembiochem. 2007;8:1001–1011. doi: 10.1002/cbic.200700097. [DOI] [PubMed] [Google Scholar]
- Plan MR, Saska I, Cagauan AG, Craik DJ. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) Journal of agricultural and food chemistry. 2008;56:5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- Poth AG, Colgrave ML, Philip R, Kerenga B, Daly NL, Anderson MA, Craik DJ. Discovery of cyclotides in the fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem Biol. 2011;6:345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- Pranting M, Loov C, Burman R, Goransson U, Andersson DI. The cyclotide cycloviolacin O2 from Viola odorata has potent bactericidal activity against Gram-negative bacteria. J Antimicrob Chemother. 2010;65:1964–1971. doi: 10.1093/jac/dkq220. [DOI] [PubMed] [Google Scholar]
- Ravipati AS, Poth AG, Troeira Henriques S, Bhandari M, Huang YH, Nino J, Colgrave ML, Craik DJ. Understanding the Diversity and Distribution of Cyclotides from Plants of Varied Genetic Origin. J Nat Prod. 2017;80:1522–1530. doi: 10.1021/acs.jnatprod.7b00061. [DOI] [PubMed] [Google Scholar]
- Robotham SA, Horton AP, Cannon JR, Cotham VC, Marcotte EM, Brodbelt JS. UVnovo: A de Novo Sequencing Algorithm Using Single Series of Fragment Ions via Chromophore Tagging and 351 nm Ultraviolet Photodissociation Mass Spectrometry. Anal Chem. 2016;88:3990–3997. doi: 10.1021/acs.analchem.6b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem S, Mor A. Antimicrobial peptide mimics for improved therapeutic properties. Biochim Biophys Acta. 2009;1788:1582–1592. doi: 10.1016/j.bbamem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Sato H, Feix JB. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic alpha-helical antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1245–1256. doi: 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Scheffler RJ, Colmer S, Tynan H, Demain AL, Gullo VP. Antimicrobials, drug discovery, and genome mining. Applied microbiology and biotechnology. 2013;97:969–978. doi: 10.1007/s00253-012-4609-8. [DOI] [PubMed] [Google Scholar]
- Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J Am Chem Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohalm M, Hassman M, Kosata B, Kodicek M. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun Mass Spectrom. 2008;22:905–908. doi: 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- Strohalm M, Kavan D, Novak P, Volny M, Havlicek V. mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- Stromstedt AA, Park S, Burman R, Goransson U. Bactericidal activity of cyclotides where phosphatidylethanolamine-lipid selectivity determines antimicrobial spectra. Biochim Biophys Acta. 2017;1859:1986–2000. doi: 10.1016/j.bbamem.2017.06.018. [DOI] [PubMed] [Google Scholar]
- Tam JP, Lu YA, Yang JL, Chiu KW. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci U S A. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassanakajon A, Somboonwiwat K, Amparyup P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Developmental and comparative immunology. 2015;48:324–341. doi: 10.1016/j.dci.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Wang CK, Colgrave ML, Gustafson KR, Ireland DC, Goransson U, Craik DJ. Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J Nat Prod. 2008;71:47–52. doi: 10.1021/np070393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AM, Craik DJ. Discovery and optimization of peptide macrocycles. Expert Opin Drug Discov. 2016;11:1151–1163. doi: 10.1080/17460441.2016.1245720. [DOI] [PubMed] [Google Scholar]
- Wong CT, Rowlands DK, Wong CH, Lo TW, Nguyen GK, Li HY, Tam JP. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew Chem Int Ed Engl. 2012;51:5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Gallo RL. Antimicrobial peptides. Current biology : CB. 2016;26:R14–19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.