Abstract
DDB1– and CUL4–associated factor 17 (Dcaf17) is a member of DCAF family genes that encode substrate receptor proteins for Cullin-RING E3 ubiquitin ligases, which play critical roles in many cellular processes. To unravel the function of DCAF17, we performed expression profiling of Dcaf17 in different tissues of wild type mouse by qRT-PCR and generated Dcaf17 knockout mice by gene targeting. Expression profiling of Dcaf17 showed highest expression in testis. Analyses of Dcaf17 transcripts during post-natal development of testis at different ages displayed gradual increase in Dcaf17 mRNA levels with the age. Although Dcaf17 disruption did not have any effect on female fertility, Dcaf17 deletion led to male infertility due to abnormal sperm development. The Dcaf17−/− mice produced low number of sperm with abnormal shape and significantly low motility. Histological examination of the Dcaf17−/− testis revealed impaired spermatogenesis with presence of vacuoles and sloughed cells in the seminiferous tubules. Disruption of Dcaf17 caused asymmetric acrosome capping, impaired nuclear compaction and abnormal round spermatid to elongated spermatid transition. For the first time, these data indicate that DCAF17 is essential for spermiogenesis.
Introduction
Infertility is a global health problem that affects 15% of couples worldwide1,2. Almost half of these infertility cases are attributed to male infertility, where about 75% of all the male factor infertility attributed to defective spermatogenesis3–5. Despite of advancement in the fields of biomedicine and genetics, the underlying causes of male infertility remain largely unknown in about 50% cases and they are categorized as idiopathic infertility cases1.
Mammalian spermatogenesis is a complex and highly regulated process that occurs within the seminiferous tubules, where the pluripotent spermatogonial stem cells proliferate through mitotic cell divisions and differentiate into primary spermatocytes. Primary spermatocytes then undergo two meiotic divisions to produce haploid round spermatids, which subsequently differentiate into highly specialized spermatozoa by a unique metamorphosis process called spermiogenesis6,7. During spermiogenesis, many biochemical and structural changes necessary for the formation of acrosome, nucleus elongation, chromatin condensation, formation of flagellum and removal of cytoplasmic residual body are carried out in an intricate manner8–10. Normal spermatogenesis requires regulated protein homeostasis and disruption of such balance results in male infertility11–13.
The ubiquitin-proteasome system (UPS) plays an important role in numerous developmental processes, including spermatogenesis, by selective and timely protein turnover for proper cellular functions in eukaryotes12,14–16. In UPS, ubiquitin is covalently transferred to one or more lysine residues of the target protein through a series of enzymatic reactions involving ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase enzyme (E3)15,17,18. E3 ubiquitin ligases (E3s) are diverse enzymes with modular, single protein or multi-protein complex structures that provide specificity toward a variety of substrates for the ubiquitination19–21. Cullin (CUL) 4 A and CUL4B, members of cullin-RING finger E3 ligase (CRL) sub-family, have been shown to play distinct and essential roles during spermatogenesis22–25. Both the proteins have displayed complementary expression patterns in adult mouse testis, where CUL4A is mainly present in meiosis-stage spermatocytes, while CUL4B is predominantly expressed in Sertoli cells, spermatogonia and spermatids24. Disruption of Cul4A caused male infertility because of the defective meiotic progression24,25. Whereas, the male infertility phenotype resulted from Cul4B deletion was due to abnormal post-meiotic sperm development22,23.
CUL4 proteins interact with the E2 enzyme via the RING finger protein Hrt1/ROC1/Rbx1 at their C terminus. Whereas at their N terminus, they employ DDB1 (DNA damage-binding protein 1), as an adaptor protein, which in turn recruits different substrate receptor proteins known as DDB1- and CUL4-associated factors (DCAFs)26–28. These DCAFs have been suggested to determine the substrate specificity in different CUL4-DDB1-based E3 ligase complexes27. Mutations in DCAF17 gene caused an extremely rare autosomal recessive disorder known as Woodhouse-Sakati syndrome (WSS) in human29. Patients with WSS display hypogonadism, alopecia, diabetes, mental retardation, deafness and extrapyramidal symptoms29–31. Male patients with DCAF17 mutations showed azoospermia with very less germ cell mass and large number of Sertoli cells in testicular biopsies30,31. However, the role of DCAF17 in the male reproductive system is not clear. To investigate the role of DCAF17 protein in mammalian spermatogenesis, we generated and characterized global Dcaf17 mutant mice. Our data show that Dcaf17 mutant male mice are infertile and the DCAF17 protein is required for normal spermiogenesis process and sperm functionality.
Results
Levels of Dcaf17 mRNA in different tissues of mice
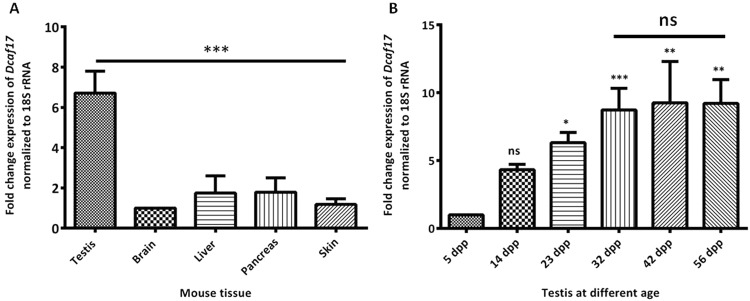
The Dcaf17 mRNA transcript profile was examined by relative qRT-PCR analysis in five different tissues that included brain, liver, pancreas, skin and testis from adult mouse. The level of Dcaf17 transcripts in brain was arbitrarily chosen as a calibrator and standardized to one. The 18S rRNA expression was used as endogenous control. The fold change in the Dcaf17 mRNA levels, normalized to 18S rRNA and relative to brain, is reported (Fig. 1A). Levels of the Dcaf17 mRNA transcript varied among the tissue samples tested. Testis showed highest level of the Dcaf17 mRNA with almost 7 fold higher levels relative to that of the brain (Fig. 1A). Pancreas and liver showed only 1.7 fold higher mRNA levels of Dcaf17 relative to that of the brain. However, the abundance of Dcaf17 mRNA in skin was similar to that in the brain (Fig. 1A).
Figure 1.
Investigating Dcaf17 mRNA levels by real time PCR. (A) Relative expression of Dcaf17 in different tissues of adult mouse. Relative expression values of Dcaf17 mRNA were normalized to 18S rRNA expression and the level of Dcaf17 mRNA in the brain was arbitrarily set at 1. The Dcaf17 is highly expressed in testis, while other tissues show low level of expression. (B) Relative expression of Dcaf17 in testis at different ages. Relative expression values of Dcaf17 mRNA were normalized to 18S rRNA expression and the level of Dcaf17 mRNA in the 5 days postpartum (dpp) testis was arbitrarily set at 1. Expression of Dcaf17 in the testis increased with the age till the age of 32 dpp and then after it remained constant. Asterisks indicate statistical significance (p < 0.0001 for A and p < 0.0036 for B). Error bars represent SEM. Three to five animals were used to investigate the transcript levels of Dcaf17. Experiments were performed in triplicates. Data were analyzed on Graphpad prism 5 software using one-way ANOVA technique followed by post hoc Bonferroni’s (A) or Tukey’s (B) multiple comparison test. ns - not significant.
Further to examine the Dcaf17 mRNA transcript levels in testis at different postnatal developmental stages, relative qRT-PCR was performed on the testes total RNA samples collected at 5, 14, 23, 32, 42 and 56 days postpartum (dpp). The Dcaf17 mRNA level at Day 5 testis was chosen as a calibrator and standardized to one. The 18S rRNA levels were used as endogenous control. The fold change in the Dcaf17 gene expression, normalized to 18S rRNA and relative to 5 dpp testis, is reported (Fig. 1B). The Dcaf17 transcript abundance in testis gradually increased with the age until 32 dpp and plateau thereafter (Fig. 1B).
Generation of Dcaf17 KO mice
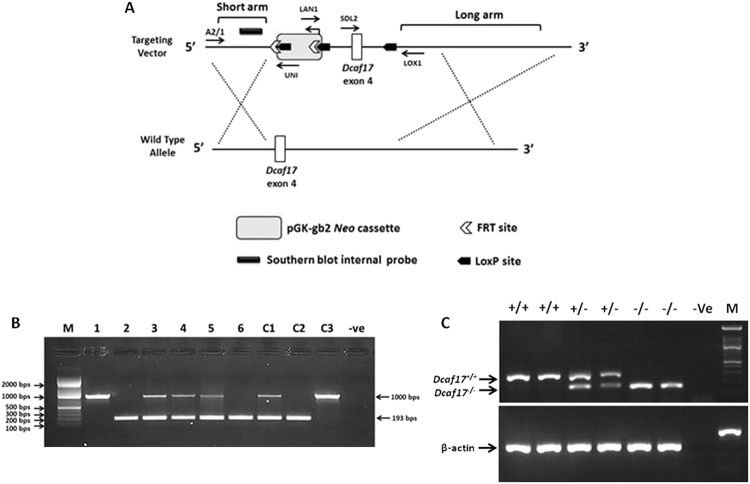
To investigate the role of the Dcaf17 gene in vivo, knockout (KO) mice were created by deleting exon 4 of the Dcaf17 using gene targeting approach (Fig. S1; Fig. S2A,B). The Dcaf17fl/fl mice were confirmed by PCR and Southern blotting (Fig. S2D–F). The Dcaf17−/− mice were generated by crossing Dcaf17fl/fl mice with CMV-Cre transgenic mice to delete exon 4 of Dcaf17 gene. It was found that the mutant allele for Dcaf17 gene could be transmitted from both Dcaf17 heterozygous males and females, and the homozygous mice could be obtained from interbreeding of the heterozygous male and female mice (Fig. 2B,C). Deletion of exon 4 of Dcaf17 gene was confirmed by PCR, RT-PCR (Fig. 2B,C) and cDNA sequencing analyses (Fig. S3). The cDNA sequencing analysis of Dcaf17−/− mice revealed that excision of exon 4 led to a frameshift mutation (NM_001165980.1:c.322_458del:p.G108Vfs*59) with concomitant premature stop codon after 59 residues (Fig. S3). All the conditional, heterozygous and homozygous Dcaf17 KO mice (male and female) were viable and showed no gross abnormalities. Due to male infertility, the colony was maintained through heterozygous mating system and continuous pups’ genotyping.
Figure 2.
Diagrammatic representation (A) of Dcaf17 gene targeting approach in mouse by homologous recombination and genotyping (B,C) of different alleles of Dcaf17 in mice. (A) Homologous recombination strategy in mouse ES cells. The Dcaf17 targeting vector (top) was constructed to replace wild type exon 4 and introduce neomycin drug selection marker, LoxP and FRT sites. (B) Agarose gel image of PCR genotyping of representative Dcaf17 mutant mice. PCR amplification of wild type genotype gives 1 kbps amplicon (1, C3), heterozygous genotype for Dcaf17 mutation gives 1 kbps and 193 bps amplicons (3–5, C1) and homozygous genotype for Dcaf17 mutation gives 193 bps amplicon (2, 6 and C2). 1–6 – genomic DNA samples of different Dcaf17 genotypes; C1-C3 – Different Dcaf17 genotype controls; −Ve – no template control. (C) Agarose gel image of RT-PCR of different Dcaf17 genotypes. PCR products of various Dcaf17 alleles and β-actin were run on the same agarose gel and single image was taken. +/+ - Dcaf17+/+ (WT); +/− - Dcaf17+/− (heterozygous Dcaf17 mutant); −/− - Dcaf17−/− (homozygous Dcaf17 mutant); −Ve – no template control; M – DNA ladder. PCR fragment size for β-actin is 190 bps; for Dcaf17+/+ is 284 bps and for Dcaf17−/− is 148 bps. Gel images were taken using ImageQuant LAS 4000 imaging system.
Male Dcaf17−/− KO mice are infertile
To assess the fertility phenotype of Dcaf17 KO mice, controlled breeding experiments were carried out where adult WT and Dcaf17−/− males were individually mated with WT or Dcaf17−/− adult females. Mating behavior of WT and Dcaf17−/− was monitored by checking the ability to produce vaginal (copulatory) plugs in the female mice. All the females with WT or Dcaf17−/− genotype produced vaginal plugs after mating either with WT or Dcaf17−/− male mice, suggesting normal mating behavior among both the genotypes (Table 1).
Table 1.
Fertility testing of Dcaf17−/− and wild type mice.
| Genotype | Vaginal plug | Total No. of litters | Total No. of pups | Average litter size ± SD | |
|---|---|---|---|---|---|
| Male (n) | Females (n) | ||||
| Dcaf17 +/+ (3) | Dcaf17 +/+ (3) | + | 13 | 46 | 3.54 ± 1.6 |
| Dcaf17 −/− (5) | Dcaf17 +/+ (5) | + | 0 | 0 | 0 |
| Dcaf17 +/+ (3) | Dcaf17 −/− (3) | + | 8 | 25 | 3.13 ± 1.6 |
| Dcaf17 −/− (3) | Dcaf17 −/− (3) | + | 0 | 0 | 0 |
n - Number of animals used; + - Vaginal plug positive; SD - Standard Deviation.
Breeding three WT females and three WT males produced 46 pups in 13 litters with an average litter size of 3.54 pups (Table 1). Whereas, five WT females plugged by five Dcaf17−/− males neither displayed any evidence of pregnancy nor produced any pups within the observation period (Table 1). Similarly, three Dcaf17 homozygous mutant female mated with three individual Dcaf17−/− male did not become pregnant and produce pups. Unlike male Dcaf17−/− mice, the three Dcaf17−/− female mice plugged by three WT males produced 25 pups in 8 litters with an average of 3.13 pups per litter (Table 1). Statistical analysis of the litter size of WT male and WT female mice breeding with that of WT male and Dcaf17 KO female breeding showed no significant difference between the litter size (P value ≤ 0.15), suggesting that there is no effect of Dcaf17 deletion on female fertility in the mice. All the Dcaf17+/− heterozygous KO mice were fertile and produced comparable number of pups with that of WT mice (Data not shown). These data show that Dcaf17 homozygous KO male mice are infertile and the infertility phenotype did not result from abnormal sexual behavior but resulted from defective testicular function.
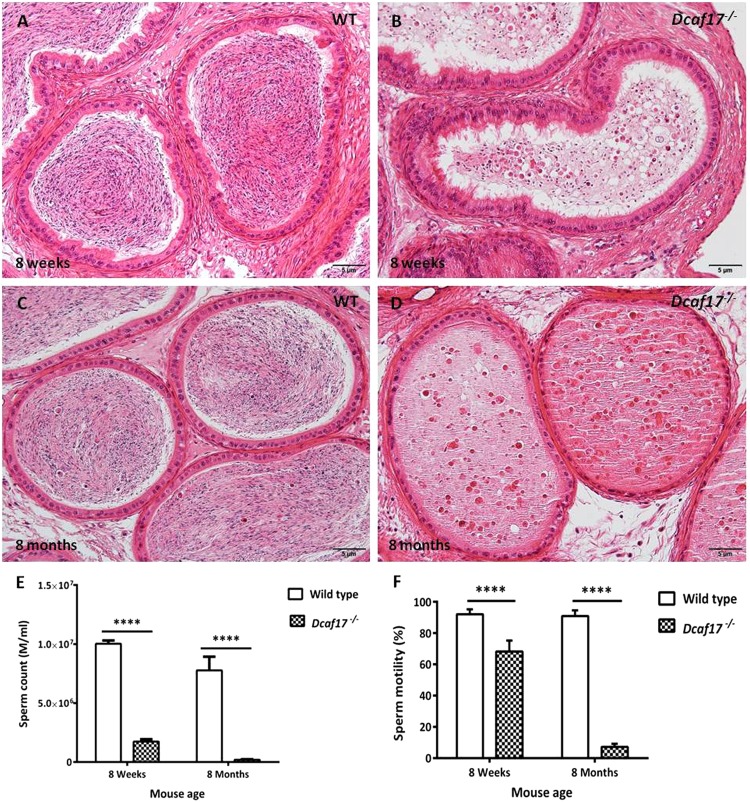
Dcaf17−/− mice have severely reduced sperm production and motility
Breeding experiments of Dcaf17 homozygous KO mice revealed that Dcaf17−/− male mice are infertile. Morphological examination of the wild type and Dcaf17 mutant testes showed no gross differences in the shape, size and weight of the testes from both the genotypes (Fig. S4; Table S1). Further to investigate how Dcaf17 deficiency leads to male infertility, we first examined the cauda epididymal sperm of WT and Dcaf17−/− adult mice. Histological examination of the WT and Dcaf17−/− adult mice cauda epididymides revealed severe reduction in sperm amount within the lumen of Dcaf17−/− cauda epididymis as compared to that of WT adult mice (Fig. 3A–D). Unlike WT cauda epididymis, where the lumen was full of normal mature sperm (Fig. 3A,C), the lumen of Dcaf17−/− cauda epididymis showed very few spermatozoa, most of which were morphologically abnormal, and an unusual occurrence of abnormal cellular debris including numerous deeply stained round structures of various sizes, most likely germ cells that had sloughed off prematurely from the seminiferous tubules (Fig. 3B,D). Moreover, the lumen of cauda epididymis in 8 months old Dcaf17 KO mouse was filled with abnormal cellular debris including numerous deeply stained round structures of various sizes, most likely degrading germ cells (Fig. 3D). No obvious differences in the structure of the epididymal epithelium were apparent between Dcaf17 KO and control mice. Sperm count and motility analyses revealed significant reduction in the number and motility of Dcaf17−/− epididymal sperm in comparison to that of WT mice (Fig. 3E,F). Eight weeks old Dcaf17 KO mice produced almost six fold less sperm (1.7 ± 0.46 × 106) with 68% motility as compared to the WT mice of the same age that produced 10 ± 6.3 × 106 sperm with more than 90% motility (Fig. 3E,F). More reductions in the epididymal sperm count and percentage motility were observed in Dcaf17−/− mice with advancing age. Eight months old Dcaf17 KO mice produced almost 8.5 fold less sperm (0.20 ± 0.1 × 106) compared to that of 8 weeks Dcaf17 KO mice and 35 fold less sperm than their age-matched WT control. Eight months old WT showed about 1.4 fold sperm count reduction (7 ± 2.57 × 106) in comparison to their genotype counterpart at 8 weeks of age (Fig. 3E,F).
Figure 3.
Histology of cauda epididymides and cauda sperm analyses from WT and Dcaf17−/− adult mice. Hematoxylin and eosin (H,E) staining of cauda epididymides sections (5 µm thick) from 8 weeks (A,B) and 8 months (C,D) old WT (A,C) and Dcaf17−/− (B,D) mice. The lumen of WT cauda epididymis (A,C) is full of mature sperm, whereas the lumen of Dcaf17−/− mutant cauda epididymidis (B,D) contains oval shaped sloughed cells of various size. Sperm count (E) and motility (F) analyses of WT and Dcaf17−/− mutant mice at the age of 8 weeks and 8 months old reveal severe and progressive reduction in sperm count (E) and motility (F) in Dcaf17−/− mice compared to WT mice. Around 300 sperm in a total of five fields in each replicate were analyze for sperm motility analysis. Number of animals used for each genotype to analyze sperm count (E) and motility (F) was 5. Data were analyzed on Graphpad prism 5 using two-way ANOVA technique followed by post hoc Bonferroni’s multiple comparison test. For images A to D the magnification is 200X and the scale bars are 5 µm. Error bars in panels E and F represent SEM. Asterisks in E and F indicate statistical significance (p < 0.0001 for E and F).
Dcaf17−/− sperm defects are similar to those found in oligoasthenoteratozoospermia
To examine Dcaf17−/− mutant sperm morphology, sperm smears were prepared and stained with Diff-Quick stain. Sperms from WT male mice showed normal morphology, with characteristic hook-shaped head, midpiece and tail (Fig. 4A). Whereas, all the sperm of Dcaf17−/− mice showed variety of defective morphology with abnormally shaped head containing deformed nucleus (Fig. 4B1–5). Further to analyze Dcaf17−/− sperm defects in more detail, WT and mutant sperm were stained with Mito tracker (red), which labels mitochondria; FITC-labeled lectin PNA (green), which labels acrosome; and DAPI (blue), which labels the nucleus (Fig. 4C and D1–5). As shown in the Fig. 4C, WT sperm displayed hook-shaped head with typical crescent-like acrosome (green) covering the anterior surface of the nucleus (blue), and a mid-piece that is covered by mitochondrial sheath (red). In contrast to WT sperm, the acrosome of Dcaf17−/− sperm failed to acquire the typical crescentic shape and exhibited various defects, including miss-localization and deformation (Figs. 4D1–5). The nucleus (blue) of WT mature sperm was highly condensed occupying most of the head space (Fig. 4C). In the Dcaf17−/− mutant sperm, deformed and less condensed nucleus (blue) was observed (Fig. 4D1–5). The mitochondrial sheath (red), which is responsible for sperm movement, also exhibited various defects in Dcaf17−/− mutant sperm, including mitochondria aggregating near the deformed nucleus, splitting into two separate aggregates and in many cases mid piece coiling around the deformed head (Fig. 4D1–5). We divided the observable sperm head shape defects into three major categories (Fig. 4B1–5, D1–5) and quantified their incidence by observing total number of 150 sperm randomly from Dcaf17 KO mice. Our observation revealed that around 45% sperm had triangular (cupcake) shaped head (Fig. 4B1,D1,D3), 27% of the sperms had oval shaped head (Fig. 4B5,D2,D4,D5) and 28% of the sperms had amorphous head shape (Fig. 4B2,B3). Among all the sperm in the above described head shape defect categories, 80% of the sperm head contained acrosome which was miss-localized and abnormally formed. Acrosome was missing in the remaining 20% of sperm in all three categories analyzed. Tail analysis of the Dcaf17−/− sperm in all three categories showed that in 38% sperm, the midpiece was coiled around sperm head, in 27% sperm had bent head from neck and remaining 35% sperm had normal straight head. Sperm with acrosome, nucleus and midpiece combinatory defects were not observed in WT or heterozygous mice.
Figure 4.
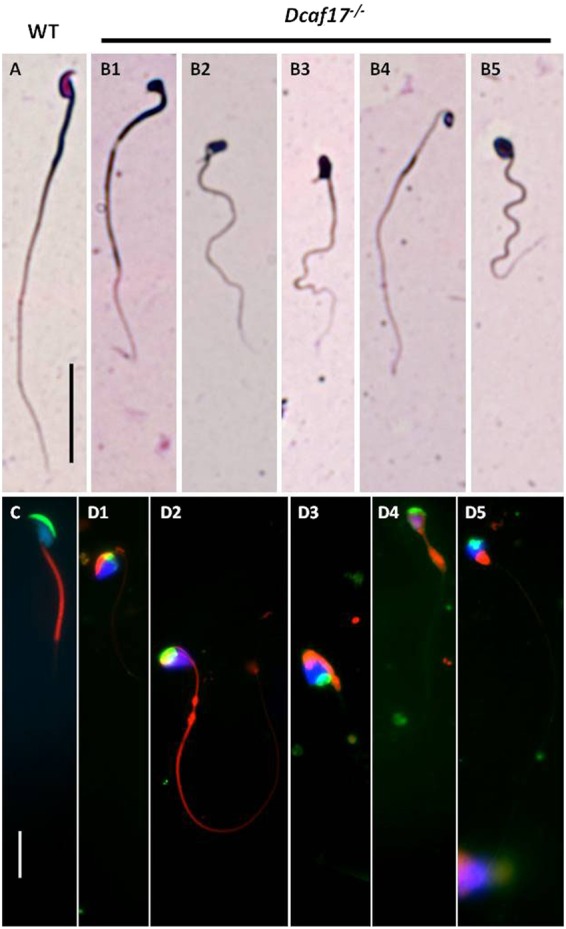
Bright field and fluorescence microscopy of cauda epididymal sperm from WT and Dcaf17−/− adult mice. Representative images of Diff-Quick staining (bright field images, top panels) and MitoTracker staining (fluorescence images, bottom panels) of epididymal sperm spreads from WT (A,C) and Dcaf17−/− (B,D) adult mice. The WT sperm (A,C) show typical hook-shaped head, patent midpiece and tail morphology. Whereas, the Dcaf17−/− sperm (B,D) show variety of morphological defects in head shape, midpiece and tail. To categorize different sperm defects in Dcaf17 KO mice we analyzed total 150 sperms from 3 different mice. Major categories of sperm head defects were triangular (cupcake) (B1,D1,D3), oval (B5,D2,D4,D5) or amorphous (B2,B3) shaped with many times either bent head or coiled midpiece surrounding the head. Fluorescently labeled WT sperm (C) shows normal crescent shaped acrosome (green), uniformly distributed mitochondria (red) along the midpiece and highly condensed nucleus (blue). Dcaf17−/− sperm (D1–5) show spatially dysmorphic sperm structure with abnormal acrosome (green), ectopic localization of mitochondria (red) and diffused chromatin structure (blue). Images C and D1–5 are merged fluorescence images of sperm stained for acrosome (green), midpiece (red) and nucleus (blue). Separate fluorescence images for each sperm acrosome, midpiece and nucleus staining are shown in supplementary figure S5. Magnification of bright images (A,B1–5) is 400X and magnification of fluorescence images (C,D1–5) is 1000X. Scale bar in the image A is 1 mm and in the image C is 10 µm.
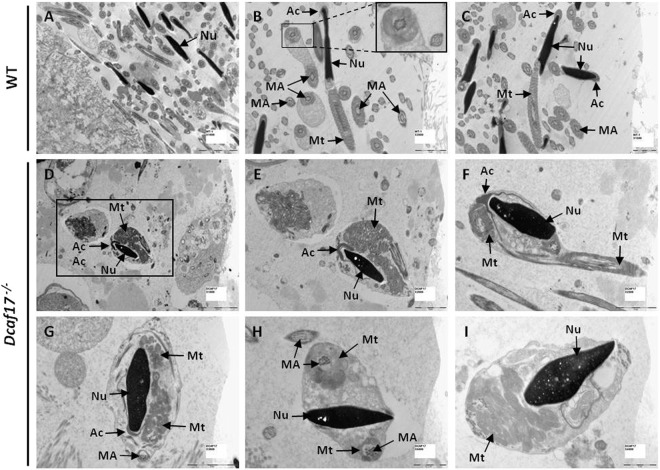
Figure 5.
Transmission electron microscopy (TEM) of cauda epididymides from 8 weeks old WT (A–C) and Dcaf17 −/− (D–I) adult mice. Representative TEM images of WT cauda epididymis (A–C) show spermatozoa with typical elongated nucleus (Nu) with homogeneously condensed chromatin. The acrosome (Ac) in WT sperm is covering the anterior portion of the head and is tightly attached to the nucleus through the acrosome-acroplaxome complex. The mid-piece of WT sperm show spirally arranged mitochondria (Mt) that are enclosed by well-defined mitochondrial sheath. WT sperm tail sections show the typical “9 + 2” pattern of the microtubular axoneme (MA). TEM images of Dcaf17−/− cauda epididymis (D–I) show abnormal sperm with misshaped head containing defective nuclear (Nu) chromatin condensation, malformed and detached acrosome (Ac), disorganized mitochondria (Mt) trapped inside large cytoplasmic droplets and ectopic localization of the microtubular axoneme (MA). Inset image in panel B is enlarged image of squared region of image B. Image E is a higher magnification image of square region shown in the image D. Scale bar are shown at the bottom right corner to each image. Scale bar: A, D 5 µm; B, C, E, F, and G 2 µm; H and I 1 µm.
Further, analyzing the ultrathin section of cauda epididymides from WT and Dcaf17−/− adult mice using TEM showed dramatic differences between the WT and Dcaf17−/− sperm ultrastructure (Fig. 5). Almost all the WT sperm examined displayed normal head, mid-piece and tail structures (Fig. 5A–C). In the WT sperm, the head contained homogeneously condensed chromatin within the nucleus as shown by the density of staining observed in TEM images of WT sperm (Fig. 5A–C). The acrosome cap in the WT sperm was tightly associated with the nucleus through the acrosome-acroplaxome complex and covered the anterior portion of the head (Fig. 5A–C). The mid-piece of WT sperm possessed well-defined mitochondrial sheath enclosing mitochondria arranged in a spiral pattern around the outer dense fibers (ODFs) and the typical “9 + 2” pattern of the axonemal microtubules (Fig. 5B,C). Unlike the WT sperm, the Dcaf17−/− sperm exhibited multiple ultrastructural abnormalities including misshaped head, defective nuclear chromatin condensation, detached acrosome, disorganized mitochondria trapped inside large cytoplasmic droplets and ectopic localization of the flagella (Fig. 5D–I). The head and mid-piece of the mutant sperm were often surrounded by asymmetric mass of residual cytoplasmic droplet in which the mitochondria and the components of flagellar structures were scattered and could not be assembled correctly (Fig. 5D–I). The presence of residual cytoplasm may damage tight associations of the acrosome with the nuclear membrane by expanding sub-acrosomal and perinuclear space that resulted in the detachment of the acrosome (Fig. 5D–G). The nuclei of Dcaf17 KO sperm had chromatin with defective compaction as evident from uneven density and punctate texture of the nuclei (Fig. 5E–I). The mutant spermatozoa often showed partially formed or completely lacking connecting piece (Fig. 5F,G). Sperm from Dcaf17−/− mice displayed disrupted mitochondrial sheath with disorganized ODFs, and axoneme microtubules (Fig. 5E–I). The “9 + 2” microtubular structure was disturbed in the mutant sperm (Fig. 5H). These abnormalities are similar to the defects seen in oligoasthenoteratozoospermia, a human infertility disorder characterized by low number of sperm, poor sperm motility, and abnormal sperm shapes with deformed round heads, distorted nuclei, abnormal acrosomes, and malformed mitochondrial sheaths32,33.
Dcaf17−/− mice have abnormal spermatogenesis
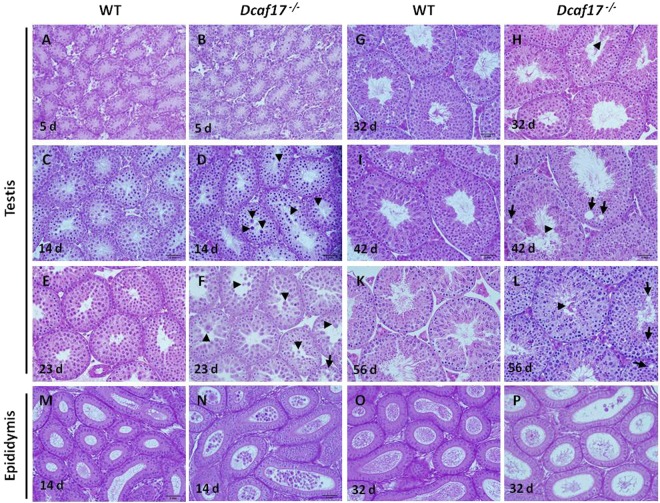
To determine the onset of spermatogenic defects in Dcaf17 KO mice, histological analyses were performed on developing testes from WT and Dcaf17−/− mice at 5, 14, 23, 32, 42 and 56 dpp (Fig. 6). Analysis of testis sections by bright field microscopy at each age showed no differences in the thickness of the seminiferous epithelium between wild-type and Dcaf17 mutant mice testes, indicating that lumen formation by the seminiferous epithelia was unaffected in the absence of DCAF17. At 5 dpp, the seminiferous tubules of WT and Dcaf17−/− testes contained normal Sertoli cells and spermatogonia with no obvious histological differences in seminiferous tubular size and the population of germ cell types (Fig. 6A,B). At 14 dpp, normal spermatocytes were observed in the seminiferous tubules of WT and Dcaf17−/− testes (Fig. 6C,D). However, some of the Dcaf17−/− seminiferous tubules showed abnormal pachytene spermatocytes with less condensed nuclei that were detached from seminiferous epithelium (Fig. 6D). Male mice at 14 days of age were sexually immature; hence, there is no production of mature sperm in the testis and consequently, the cauda epididymal lumen of WT mice were devoid of any spermatogenic cells (Fig. 6M). Interestingly, the cauda epididymal lumen of Dcaf17−/− mice at 14 dpp showed numerous degenerated spermatogenic round cells of various sizes (Fig. 6N). This indicates a defective process of spermatogenesis in Dcaf17 KO mice. Spermatocytes at most advanced stage and round spermatids were observed in the seminiferous tubules of WT testes at 23 dpp (Fig. 6E). Although the seminiferous tubules of Dcaf17−/− testes at 23 dpp showed advanced spermatocytes and round spermatids, many of the tubules contained giant cells, multinucleated cells, vacuoles and sloughed round spermatids (Fig. 6F). At 32 dpp, elongating and elongated spermatids were abundant with normal morphology and chromatin condensation in WT testis (Fig. 6G). In Dcaf17 KO testis at 32 dpp, elongating and elongated spermatids were abnormal in shape and chromatin condensation (Fig. 6H). At 42 and 56 dpp, complete cycle of spermatogenesis within seminiferous tubules depicting various stages of highly organized spermatogenic cells was observed in WT testis sections (Fig. 6I,K). The Dcaf17 KO testes at similar ages showed vacuoles, abnormal spermatogenesis with severe defects in both meiotic spermatocytes and post-meiotic spermatids (Fig. 6J,L). Spermiogenesis defects in the mutant mice were evident from reduced number of elongated spermatids and spermatozoa, abnormal-appearing spermatids, sloughed germ cells and the accumulation of residual bodies in the lumen of Dcaf17−/− testis and epididymides (Fig. 6D,F,H,J,L,N,P).
Figure 6.
Histology of testes and cauda epididymides from WT and Dcaf17 −/− mice at different ages. Sections (5 µm thick) of testes and cauda epididymes from WT and Dcaf17−/− mice at different ages were stained with Hematoxylin and Eosin. Testes of WT (A) and Dcaf17−/− (B) mice at 5 days postpartum (dpp) show normal Sertoli cells with no apparent morphological or histological differences. At 14 dpp, normal spermatocytes are seen in the WT (C) and Dcaf17−/− (D) testes sections. Unlike WT testis at 14 dpp, the lumen of Dcaf17−/− seminiferous tubules (ST) (D) show large shaded cells (arrow heads). Spermatocytes at most advanced stage and round spermatids are observed in the ST of the WT (E) and Dcaf17−/− (F) testes at 23 dpp. The lumen of ST in Dcaf17−/− testis at 23 dpp (F) shows giant, multinucleated and prematurely sloughed cells (arrow heads). Epithelial vacuoles (arrow) are also seen in the Dcaf17−/− testis at 23 dpp. At 32 dpp, the WT testis (G) shows numerous elongating and elongated spermatids with normal morphology and chromatin condensation. Dcaf17−/− testis at 32 dpp (H) shows fewer elongating and elongated spermatids with abnormal shape and chromatin condensation. Testes from 42 dpp (I) and 56 dpp (K) old WT mice show a complete cycle of spermatogenesis with seminiferous tubules depicting various stages of highly organized spermatogenic cells. Testes of Dcaf17−/− mice at 42 dpp (J) and 56 dpp (L) show abnormal spermatogenesis with sever defects in post-meiotic stages of spermatogenesis. Vacuoles (arrows) are observed in the epithelium of some ST of the Dcaf17−/− testis (J,L). The lumen of cauda epididymis (M) from sexually immature (14 dpp) WT mouse is devoid of any degenerated cells. In Dcaf17−/− mouse at 14 dpp, the lumen of cauda epididymis shows numerous prematurely degenerated oval shaped cells of various size. Sexually mature cauda epididymis of WT mouse (32 dpp) has lumen (O) full of normal mature sperm. Prematurely degenerated spermatogenic cells of various size and few abnormal sperm are observed in the lumen of Dcaf17−/− mouse cauda epididymis (P) at 32 dpp. Magnification 400X. Scale bar 2 mm.
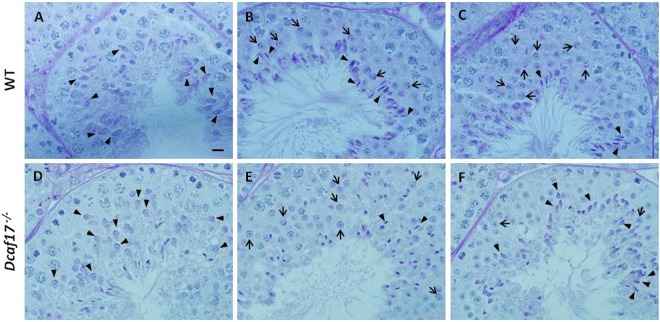
H & E staining of Dcaf17−/− testes at different ages revealed predominantly severe defects in post-meiotic spermatids (Fig. 6). To investigate spermiogenesis defects in detail, Periodic acid–Schiff (PAS) staining of the adult WT and Dcaf17−/− testes sections was performed. WT testis displayed normal acrosomal caps, elongated spermatids and chromatin condensation (Fig. 7A–C). However, Dcaf17−/− testis showed abnormal morphology of elongating and elongated spermatids with defective acrosome and chromatin condensation (Fig. 7D–F).
Figure 7.
Periodic acid-Schiff (PAS) staining of testes sections from 8 weeks old WT and Dcaf17−/− mice. To visualize the glycoproteins/acrosomes (violet) and nuclei (blue), the testis sections from WT (A–C) and Dcaf17−/− (D–F) mice were stained with PAS-stain and hematoxylin counter stain. WT testis sections (A–C) show normal spermatogenesis with well-organized stages of germ cell development, round spermatids with PAS-positive normal acrosomal caps (arrows), elongating and elongated spermatids (arrow heads) and chromatin condensation. Dcaf17−/− testis sections (D–F) show defective spermatogenesis with abnormal acrosomal caps (arrows), distorted elongated spermatids (arrows heads) and chromatin condensation. Magnification – 1000X. Scale bar: 10 µm.
Disruption of Dcaf17 leads to increased germ cells apoptosis
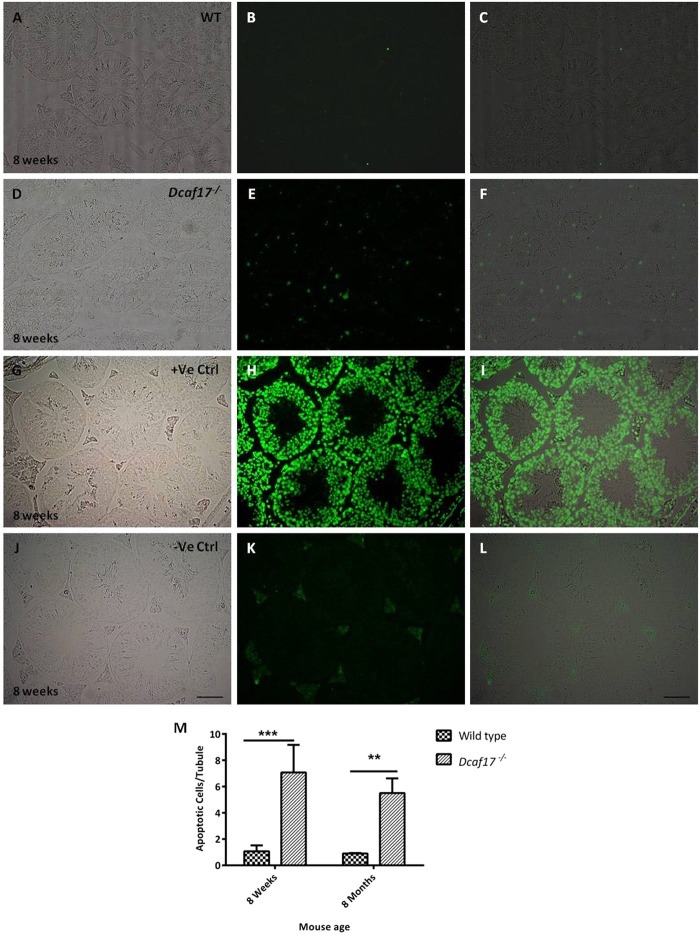
Due to observed reduction in sperm counts and presence of sloughed germs cells in the Dcaf17 KO epididymides, apoptosis was assessed in the seminiferous epithelium of Dcaf17 KO mice and compared with that of WT mice. TUNEL assay on the testes sections of 8 weeks (Fig. 8) and 8 months (Fig. S6) old WT and Dcaf17−/− mice was performed. The results revealed an overall increase in the average number of TUNEL-positive apoptotic cells per seminiferous tubule in Dcaf17−/− testis sections in comparison to that of wild type (P < 0.01value) (Fig. 8D–F,M). Interestingly, there was a variation in the number of TUNEL-positive cells in different Dcaf17−/− tubule sections, suggesting that apoptosis is more pronounced at specific stages of germ cell development (Fig. 8D–F,M). In contrast, WT testes showed fewer number of TUNEL-positive cells per seminiferous tubule within the WT testis sections (Fig. 8A–C,M). Together, these findings suggest that DCAF17 deficiency causing impaired spermatogenesis accompanied by high rate of apoptosis.
Figure 8.
TUNEL staining of testes sections from 8 weeks old WT and Dcaf17−/− mice to assess germ cell apoptosis. Testes sections of 8 weeks old WT (A–C) and Dcaf17−/− (D–F) mice were subjected to TUNEL assay, which detects fragmented DNA in the apoptotic germ cells (green). WT testis section (A–C) shows fewer TUNEL-positive cells (green) (B,C) compare to Dcaf17−/− testis section (E,F). Image H is positive control for TUNEL assay where the testis section was treated with DNaseI enzyme to generate fragmented genomic DNA. Image K is a negative control where testis section was treated only with labelling solution without terminal transferase. Images A,D,G and J are bright field images of respective TUNEL stained fluorescence images B, E, H, and K. Images C,F,I,L are merged images of respective bright field and fluorescence images. Magnification 200X. Scale Bar (J): 50 µm. The number of TUNEL-positive cells (green) per tubule in the testes sections of WT and Dcaf17−/− mice aged 8 weeks and 8 months were plotted in the graph (M). The Histogram (M) represents mean ± SEM of 3 animals analyzed for TUNEL assay from each strain. Asterisks indicate statistical significance (p < 0.001 at 8 weeks and P < 0.01 at 8 month). Data were analyzed on Graphpad prism 5 using two-way ANOVA technique followed by post hoc Bonferroni’s multiple comparison test.
Meiotic progression is not affected in Dcaf17 KO mice
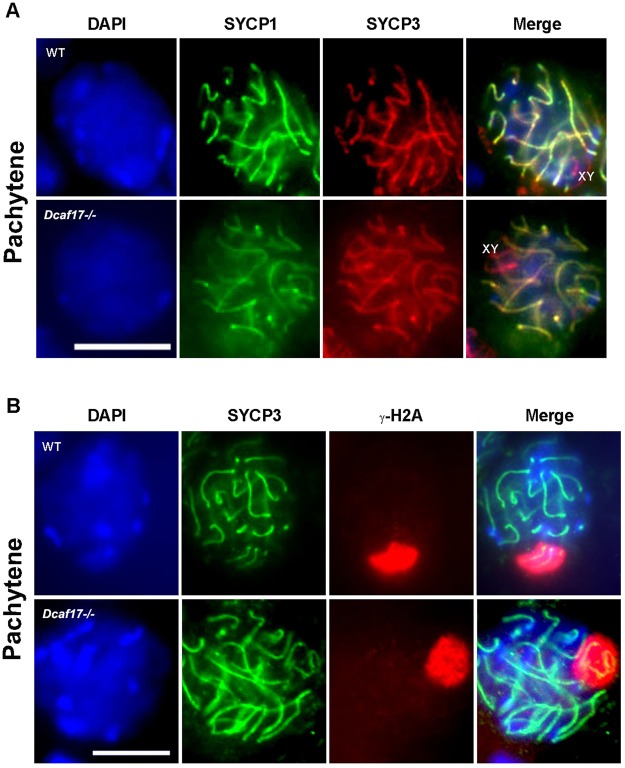
To determine whether the Dcaf17 mutant spermatocytes progress properly through meiosis, we analyzed the expression and localization patterns of meiotic markers by immunolabeling of squashed preparation from WT and Dcaf17−/− adult mice testes. We used antibodies against axial element, SYCP3 (also called SCP3) and transverse filaments, SYCP1 (also called SCP1) to examine the formation of synaptonemal complex and phosphorylated histone H2AFX (γH2AX) antibody to detect DNA double strand breaks (DSBs) before the synaptonemal complex is fully established and the unsynapsed X and Y chromosomes during the process of meiosis34–36. The labeling patterns for SYCP3 and SYCP1 proteins, in Dcaf17−/− spermatocytes were indistinguishable from that in WT littermate spermatocytes (Fig. 9A). Thus synapsis formation appears to be normal in Dcaf17 mutant germ cells. During meiosis, DNA DSBs occur in early prophase and are then repaired as germ cells reach the pachytene stage. To examine the effect of Dcaf17 disruption on DNA DSBs and XY body formation during meiosis, double immunostaining of SYCP3 and γH2AX was performed. During leptotene and early zygotene, γH2AX spreads in the nucleus in response to DNA DSBs. In mid-pachytene to late diplotene spermatocytes, γH2AX was located on the sex chromosomes as a result of meiotic sex chromosome inactivation. The localization of γH2AX was identical in leptotene/zygotene and pachytene of both Dcaf17 mutants and WT mice (Fig. 9B), suggesting that similar to WT spermatocytes, DSBs occurred and were repaired in the Dcaf17 mutant spermatocytes. Together, our data suggests that DCAF17 does not play significant role in meiotic events during spermatogenesis.
Figure 9.
Immunofluorescence analysis of meiotic spermatocytes of 8 weeks old WT and Dcaf17−/− testes. Immunostaining of the synaptonemal complex with SYCP1 (green) and SYCP3 (red) antibodies in pachytene spermatocytes of WT and Dcaf17 mutant mice (Panel A). Immunofluorescnece staining of γ-H2AX (red) and the lateral element of the synaptonemal complex, SYCP3 (Green) in pachytene spermatocytes of WT and Dcaf17−/− mice (Panel B). Spermatocytes with synaptic X and Y chromosomes (green) around which γ-H2AX (red) was normally distributed in WT and Dcaf17−/− strains. Nucleus is counter stained with DAPI (blue). Magnification 1000X. Scale bar: 10 µm.
Defective manchette formation and nuclear compaction during spermiogenesis in Dcaf17−/− mice
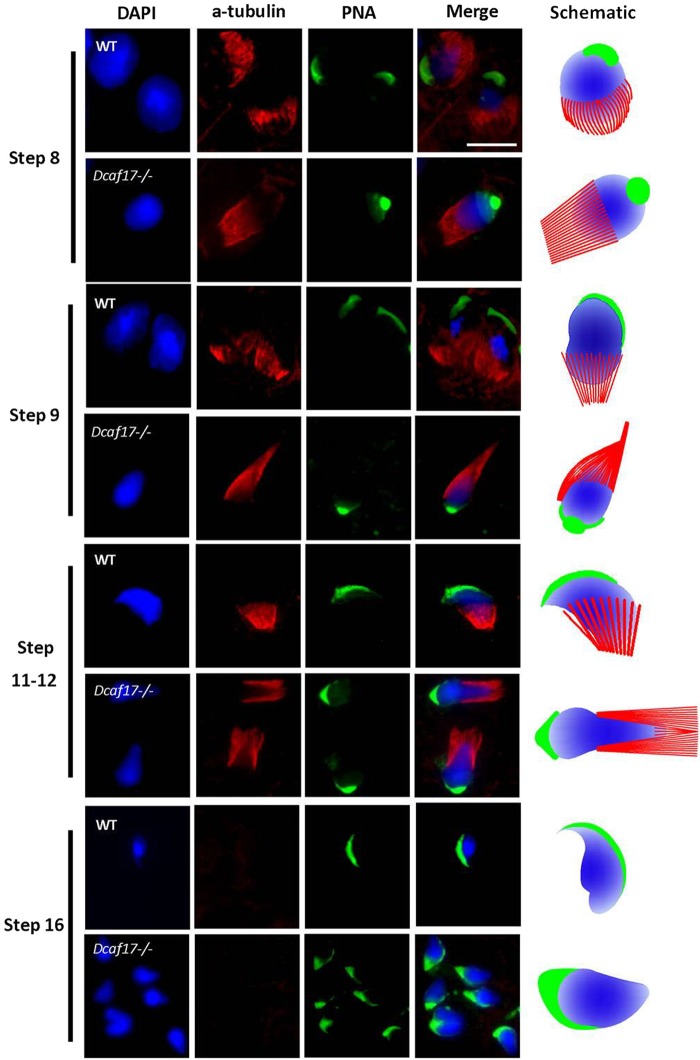
Disruption of Dcaf17 in mice caused male infertility due to abnormality in sperm morphogenesis. Aberrations in the sperm head and flagellar structures are often associated with malformation of the manchette, a transient microtubule structure that appears around the nucleus of elongating spermatid and plays crucial roles both in head shaping and cargo transport to the tail region in developing spermatids10,37–40. The timing of manchette formation is very precise and coincides with the process of nuclear shaping and flagellum development. The sperm head and tail deformities observed in Dcaf17 mutant mice might arise from defects in the formation or function of manchette. To examine abnormalities in manchette formation, acrosome morphogenesis and nuclear elongation during spermiogenesis in the Dcaf17 KO testis, immunofluorescence staining was performed on WT and Dcaf17−/− testes squash preparations using anti α-tubulin and FITC conjugated PNA lectin antibodies and DAPI to identify manchette, acrosome and nucleus, respectively. No significant morphological differences were observed between early round spermatids from spermiogenesis steps 1 to 7 of WT and Dcaf17 mutant mice (Data not shown). In WT testis, the manchette formation begins at step 8 of spermiogenesis, when elongation of the round spermatid starts by nucleus polarization and change of nucleus shape from spherical to slightly elongated (Fig. 10). During the normal manchette biogenesis, the microtubule bundles assemble at the perinuclear ring surrounding posterior region of early elongating spermatid nucleus (Fig. 10)10,40. As the spermiogenesis progresses, the perinuclear ring and manchette move towards distal part of the sperm head forming a slanted conical-shaped manchette (Fig. 10). The manchette moves toward the tail neck region and disappears by the end of spermiogenesis. While assembly of manchettes in Dcaf17−/− testis appeared to start at the correct time, their organization and nuclear elongation were affected (Fig. 10). In the Dcaf17 mutant testis, abnormal manchette organization and nuclear elongation started from step 8 onwards and continued throughout the subsequent processes of spermiogenesis (Fig. 10). The manchettes of Dcaf17−/− spermatids were more elongated and showed abnormal distal movement along the nuclear surface (Fig. 10). Throughout the elongation phase of Dcaf17−/− spermatids, the perinuclear ring of the manchette continued to narrow that resulted in an unusual bulb-like nucleus structure with an enlarged spherical anterior region and a long tapered posterior region (Fig. 10).
Figure 10.
Manchette and acrosome formation based on α-tubulin and PNA immunofluorescence staining of testes squash preparations from WT and Dcaf17−/− mice. Testes squash preparations from WT and Dcaf17−/− testes from adult mice were stained using antibodies against α-tubulin (red) and lectin PNA (green) to analyze manchette and acrosome formation, respectively. Nucleus was stained with DAPI (blue). Representative fluorescence images of microtubule (red), acrosome (green) and nucleus (blue) staining and corresponding merge images of post-meiotic male germ cells at different steps of spermiogenesis are shown for both the genotypes (WT and Dcaf17−/−) at similar stages. Different steps of sperimioenesis are depicted on the basis of nuclear and acrosome staining. WT post-meiotic germ cells show normal microtubule bundles (manchette) assembly (red), acrosome morphogenesis (green) and nuclear (blue) elongation during spermiogenesis. In the Dcaf17 mutant spermiogenic cells, the organization of microtubule bundles (red) around the nucleus (blue) is disrupted resulting in abnormal manchette formation, defective nuclear (blue) condensation and elongation, and abnormal acrosome (green) morphogenesis. Ectopic microtubules, defective acrosome and abnormal nuclear head shaping are noticeable in Dcaf17−/− post-meiotic germ cells. Magnification 1000X. Scale bar: 10 µm. Schematic representations of microtubule bundles (red) assembly, acrosome (green) formation in relation to the nucleus (blue) shaping during spermiogenesis in wild-type and Dcaf17 KO spermatids are shown.
Co-immunostaining of WT and Dcaf17 mutant testes squash preparations by PNA-FITC to stain acrosome revealed that there were no significant differences in acrosome biogenesis during Golgi phase (steps 1–3) and cap phase (steps 4–7) of spermiogenesis between two genotypes, which is consistent with PAS staining of testes sections (Fig. 7). During the acrosome phase (steps 8–12) in WT testis, when nuclear condensation and elongation take place and manchette formed around the posterior region of nucleus, the acrosomal cap further elongated and produced characteristic crescent-shaped acrosome. However, the Dcaf17−/− spermatids during acrosome phase showed impaired acrosome morphogenesis that was manifested by failure of acrosome cap extension (Fig. 10).
To characterize the abnormalities in Dcaf17−/− spermiogenesis at the ultrastructural level, TEM analyses of testes from WT and Dcaf17 mutant mice were carried out. No obvious differences in the ultrastructure of spermatids at Golgi phase and cap phase (steps 1–7) were observed between WT and Dcaf17 mutant mice (Fig. 11A,B,G,H,M,N). However, the elongating and elongated spermatids from Dcaf17−/− testis were most affected and showed various abnormalities in acrosome, nucleus, perinuclear ring and manchette. While elongating and elongated spermatids in WT seminiferous tubules showed typical flattened and elongated nuclei, the Dcaf17−/− spermatids at corresponding developmental steps showed abnormally shaped nuclei (Fig. 11C–F,I–L,O–R). The WT elongating and elongated spermatids showed normal appearing condensed chromatin (Fig. 11D–F). In contrast, chromatin compaction was defective in the elongating and elongated spermatids of Dcaf17−/− mice (Fig. 11J–L,P–R). In differentiating WT spermatids, the acrosome is attached to nuclear envelop symmetrically covering the anterior part of the nucleus (Fig. 11B–F). Although the acrosome in most of the Dcaf17−/− elongating spermatids was attached to the nuclear envelope, its distribution was asymmetric and aligned the deformed nucleus (Fig. 11I–L,O–R). The manchette in normal spermatids appeared as a parallel arrangement of microtubule bundles extending from a perinuclear ring into a distal cytoplasm and attached to posterior nuclear membrane (Fig. 11C–E). In Dcaf17−/− spermatids, the microtubule bundles of manchette was severely disorganized (Fig. 11I–L,O–R). Furthermore, Dcaf17−/− spermatids displayed abnormal positioning of the perinuclear ring (Fig. 11I–L,O–Q). During normal spermiogenesis, the manchette moves toward the tail neck region around steps 13–14 and gradually removed in subsequent steps10 (Fig. 11C–F). Interestingly, disruption of Dcaf17 resulted in delayed removal of manchette and retention of residual cytoplasm (Fig. 11L,R). Collectively, these data suggests the role of DCAF17 in microtubule manchette organization during spermiogenesis.
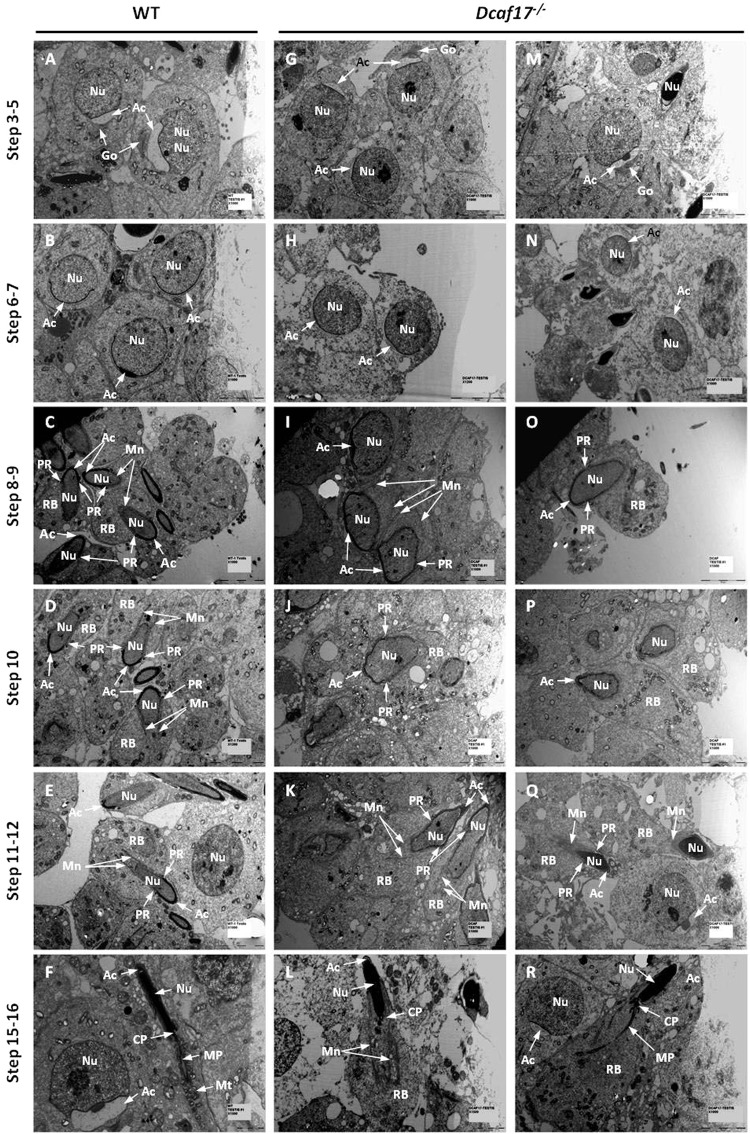
Figure 11.
Testicular ultrastructure of WT and Dcaf17−/− adult mice by transmission electron microscopy (TEM). Representative TEM images of 8 weeks old WT (A–F) and Dcaf17−/− (G–R) mice testes ultrathin sections displaying different steps of spermiogenesis. Spermatids in step 3–5 (A,G,M), step 6–7 (B,H,N), step 8–9 (C,I,O), step 10 (D,J,P), step 11–12 (E,K,Q) and step 15–16 (F,L,R) are represented. In WT (A) and Dcaf17−/− (G,M) testes, round spermatids during steps 3–5(A,G,M) show acrosome (Ac) attached with spherical nucleus (Nu) at one end. Some round spermatids show Golgi (Go) apparatus located close to acrosome (Ac). During steps 6 and 7 of spermiogenesis in WT (B) and Dcaf17−/− (H,N) testes, the acrosome (Ac) flattens and grows to form a cap covering approximately half of the nuclear (Nu) surface. During steps 8–12 of WT spermiogenesis, the spermatid nucleus (Nu) starts to elongate and the acrosome (Ac) extends along the nuclear envelope (C,D,E). The manchette (Mn) in WT elongating spermatids (C,D,E) extends from perinuclear ring (PR) into residual body (RB) and attached to posterior region of nuclear membrane. The Dcaf17−/− elongating spermatids in steps 8–12 (I–K and O–Q) show abnormal nuclear elongation and asymmetric extension of the acrosome (Ac) that is associated with distorted nucleus (Nu). The perinuclear ring (PR) in the elongated spermatids of Dcaf17−/− mutant (I–K and O–Q) is abnormally positioned and microtubule bundles of manchette (Mn) are severely disorganized along the posterior surface of the deformed nucleus (Nu). WT elongated spermatids in steps 15–16 show elongated nucleus (Nu), a midpiece (MP) region containing uniformly distributed mitochondria (Mt) and connecting piece (CP) joining the midpiece region of the tail to the head of maturing spermatozoa. The manchette (Mn) and residual body (RB) gradually disappear during the maturation steps 15–16 in WT. Elongated spermatids at steps 15–16 in Dcaf17−/− testis (L,R) show oval shaped nucleus (Nu), abnormal manchette (Mn) (L) and residual body (RB). Scale bars: (A–E), (G–K) and (M–R) 5 µm; F and L 2 µm.
Discussion
Spermiogenesis is a complex metamorphosis process of male germ cell where the round haploid spermatids undergo a series of remarkable cytoskeletal and nuclear changes to generate mature elongated spermatozoa. Multiple and tightly regulated cellular and molecular events are orchestrated in the generation of normal mature spermatozoa6,8,41. Disruption in underlying mechanisms could cause abnormal sperm formation, which often leads to male infertility8,10. In the present study, we demonstrated that Dcaf17 gene is highly expressed in testis with low level of expression in other organs like brain, liver, pancreas and skin. Targeted disruption of the Dcaf17 gene revealed that it is not essential for normal embryonic development and resulted in male infertility without other identifiable anatomical or behavioral abnormalities. Despite normal female KO mice fertility, the male infertility phenotype of the Dcaf17 KO mice is attributed to the severe malformation and disorganization of specialized structures of the spermatozoa.
E3 ubiquitin ligases play essential roles in regulating every stage of spermatogenesis starting from spermatogonia to differentiated spermatozoa12,15,42. Large number of E3 ligases have been identified that are expressed during mouse spermatogenesis with many of them are shown to be expressed highly or specifically in the testis43. Similarly, Cul4A and Cul4B have been shown to express highly in mouse testis23,25. The CUL4A and CUL4B exhibited complementary expression patterns in adult mouse testis, where CUL4A primarily expressed in spermatocytes during meiosis, while CUL4B was highly expressed in Sertoli cells, spermatogonia and spermatids24. Differential expression patterns of CUL4A and CUL4B in testis at different space and time, highlight key, non-overlapping roles for each protein in testicular development and spermatogenesis. Our findings showed predominant abundance of Dcaf17 mRNA levels in adult mouse testis, implying a crucial role of DCAF17 in testicular function. Further, the Dcaf17 transcripts were gradually increased with the mouse age from low at neonatal age to highest at puberty and remained constant. At 5 dpp, when there were mainly Sertoli cells and spermatogonia within the testis, the Dcaf17 mRNA levels were low. Low levels of Dcaf17 transcripts at neonatal age suggest that DCAF17 may not have crucial role during early mitotic phase of spermatogenesis. However, this need to be confirm by experimental analysis. High expression of Dcaf17 gene at the age of puberty may be due to a pivotal role of DCAF17 during later stages of spermatogenesis.
Male infertility phenotype observed in Dcaf17 KO mice can be due to defective reproductive physiology, sex organ development or sexual behavior. Normal mating behavior of Dcaf17 mutant mice and comparable anatomical structure, size and weight of Dcaf17−/− testes suggested that the infertility is not due to defective reproductive physiology, sex organ development or sexual behavior. However, the infertility phenotype in Dcaf17 KO mice is caused mainly because of the defective germ cell development as shown by testicular histology, sperm analyses and TEM. The onset of spermatogenic defect in Dcaf17−/− mice occurred at 14 days after birth when pachytene spermatocytes occur. Early disruption of spermatogenesis in the Dcaf17−/− testis was evidenced by premature sloughing of spermatogenic cells from seminiferous epithelium into the lumen of seminiferous tubules. The TUNEL data indicated that Dcaf17 deficiency caused germ cell apoptosis that is exacerbated with animal age and resulted in accumulation of sloughed premature germ cells within the KO mice epididymides.
The most remarkable phenotype observed in Dcaf17−/− mice was an acute defect in post-meiotic germ cell development, resulting in increased number of dysmorphic and immotile sperm. During the normal spermiogenesis process, the round haploid spermatids undergo a series of extensive morphological changes to generate mature functional spermatozoa. The major changes during this process include acrosome biogenesis, acroplaxome and manchette formation, chromatin condensation, nucleus elongation, and assembly and elongation of flagellum8,44. In the final step of spermiation, residual cytoplasm is eliminated, specialized intercellular adhesion structures are removed and the spermatid is detached from the Sertoli cell and released into the lumen of the seminiferous tubule2,8. Defects in these processes lead to a lack of functional spermatozoa, which is a major cause of male infertility in the human population45,46. We showed that Dcaf17−/− mice produced malformed spermatozoa due to defective spermiogenesis. This defect was histologically characterized by defective elongating and elongated spermatids and degenerative germ cells that were observed in the lumen of seminiferous tubules and epididymides. Aberrant sperm head and flagellar structures are often attributed to malformed manchette development during spermiogenesis8,10,40. Alpha-tubulin localization at different stages of spermatid development in Dcaf17−/− testis revealed abnormal microtubule assembly, suggesting the role of DCAF17 in manchette formation. Further, electron microscopic analysis of testis ultrathin sections confirmed the aberrant manchette formation during Dcaf17−/− spermiogenesis. In addition to manchette malformation, the ultrastructural studies also showed defects in nucleus shaping, chromatin condensation and asymmetric localization of acrosome. Along with the prematurely degenerated round sloughed germ cells, the cauda epididymis lumen of Dcaf17−/− mice showed very low number of malformed globular shaped sperm which were rarely motile. Immunofluorescence staining of micotchondria, acrosome and nucleus of Dcaf17−/− cauda epididymal sperm showed ectopic localization of mitochondrial sheath and acrosome with less condensed nucleus.
In several studies, it was shown that the CUL4 ubiquitin ligases (CUL4A and CUL4B) play essential roles in male germ cell development. Both Cul4A and Cul4B mutant male mice were infertile22–25. Although CUL4A and CUL4B share sequence and structural similarities, both the proteins play diverse roles in spermatogenesis23–25,47. Disruption of Cul4A in mice resulted in male infertility due to abnormal meiotic progression, whereas disruption of Cul4B in mice caused male infertility because of aberrant post-meiotic sperm development22–25. Macroscopic features of Cul4A KO mice testes showed two fold reductions in the size and the weight of the Cul4A−/− testis compared to that of wild type25. Whereas, Cul4B KO mice had no significant effect on the size and weight of the testis23. Similar to Cul4B KO mice, deletion of Dcaf17 did not affect the testicular size and weight, indicating that disruption of Dcaf17 in mice do not have major effect on the development of testis at macroscopic level. Drastic reduction in sperm production and sperm motility due to defective spermatogenesis have been shown in both the, Cul4A and Cul4B KO mice23,25. Comparable to Cul4A and Cul4B KO mice, deletion of Dcaf17 also disrupted normal spermatogenesis causing significant reduction in sperm number and motility. Epididymal histology of Cul4A and Cul4B KO mice showed to have very few sperm with mostly abnormal morphology. In addition to few sperm, epididymis lumen of Cul4A and Cul4B KO mice have also been shown to contain degenerated cells23,25. Likewise, histological examination of Dcaf17−/− epdidymis in this study also showed that the lumen of Dcaf17 KO epididymis contained very few, mostly abnormal sperm along with round sloughed cells of varying sizes. Deletion of Dcaf17 in mice resulted in formation of vacuoles in the seminiferous epithelium, significant increase in apoptotic cells, remarkable reduction in elongated spermatids and spermatozoa, and release of degenerated germ cells in the lumen of the seminiferous tubules. Similar testicular phenotypes have also been reported in the testes of Cul4A and Cul4B KO mice, independently23,25. Investigation of sperm development in Cul4A deficient mice by two independent groups revealed that CUL4A is required for the regulation of meiotic progression through controlling meiotic recombination and double stranded break24,25. Deletion of Cul4A in mice have been shown to disrupt meiosis process in the testis, causing considerable reduction in the number of round spermatids25. Unlike Cul4A KO mice, the Cul4B KO mice testis showed significant decrease in elongating and elongated spermatids due to increased apoptosis during cap phase and acrosome phase23. The Cul4A and Cul4B KO mice have been shown to have severe defects in acrosome formation, mitochondrial sheath and chromatin condensation during sperm development at ultrastructural level22–24. Equally, fluorescence microscopic and ultrastructural analysis of Dcaf17 KO testis and spermatozoa showed severe defects in normal acrosome formation, manchette organization, chromatin condensation and nuclear elongation during spermiogenesis. The severe infertility phenotype observed in Dcaf17 KO male mice implicates indispensible function(s) of DCAF17 proteins during normal sperm development.
DCAF17 is a member of a DCAF family proteins that function as receptor protein for different Cullin4-based E3 ligase complexes and provides substrate specificity to the E3 ligase complex for protein ubiquitination27. Ubiquitination plays very important roles in regulation of different aspects of normal sperm development and testicular functions15,48–50. The CUL4A and CUL4B function as a scaffold protein for Cullin4-based E3 ligase complex assembly26. Due to the comparable similarities in the testicular phenotypes of Cul4A, Cul4B and Dcaf17 KO mice, we speculate that the function(s) of CUL4A, CUL4B and DCAF17 proteins may be interlinked for the regulation of normal spermatogenesis via ubiquitin mediated protein homeostasis. However, it is needed to be confirmed by appropriate experimental analyses.
In conclusion, our studies on Dcaf17 KO mice successfully characterized the male infertility phenotype caused due to Dcaf17 deletion and demonstrated the importance of DCAF17 protein in maintaining normal spermiogenesis. It has also provided an ideal model to further investigate the molecular mechanisms of DCAF17, E3 ligase complex components and their target proteins in sperm development and male fertility.
Materials and Methods
Animals
Conditional Dcaf17 knockout (KO) mice were generated by inGenious Targeting Laboratory, Inc. (Ronkonkoma, NY, USA). The CMV-Cre mice (B6.C-Tg(CMV-cre)1Cgn/J) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Mice were housed in the animal facility at King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia, under controlled conditions with 12 hours light/dark cycles and free access to feed and water. All the experiments and animal procedures were approved by the Animal Care and Use Committee of the King Faisal Specialist Hospital and Research Centre (Protocol RAC# 210-0006), and executed according to institutional and international guidelines for the care of experimental animals.
Targeted disruption of Dcaf17 gene
Disruption of Dcaf17 gene in the mouse genome was carried out using gene targeting approach. Briefly, a 10.46 kb fragment containing exon 4 of Dcaf17 gene from Dcaf17-containing C57BL/6 BAC clone (RP23: 359E23) was subcloned to construct the targeting vector (pSP72 backbone). The FRT/loxP flanked Neo cassette was inserted upstream of exon 4 in intron 3–4 in an opposite direction in relation to the target gene. The single loxP site was inserted downstream of the exon 4 in intron 4–5 sequence. The target region was 930 bp long and included exon 4 of Dcaf17 gene. The region was designed to have a short homology arm (SA) extending 1.4 kb upstream of the LoxP/FRT-flanked pGK-gb2 Neo cassette and a long homology arm (LA) extending 8.06 kb to the 3′-end of the single lox P located downstream of the exon 4. The targeting construct was linearized by NotI restriction digestion and transfected into the BA1 (C57BL/6 × 129/SvEv) mouse embryonic stem cells (mESCs). The targeted mESC clones were selected for G418 antibiotic resistance. Positive mESC clones were screened by PCR using A1 or A2 forward primers and UNI reverse primer to confirm the short homology arm integration (Figs 2A and S2B). The PCR-positive mESC clones were further confirmed by Southern blotting (Fig. S2C) using the specific probes shown in the Figures S2A and B. Targeted mESC clones containing floxed exon 4 of Dcaf17 gene were microinjected into C57BL/6 blastocysts to produce chimeric mice. Chimeras with a high percentage agouti coat color were mated to wild-type C57BL/6 mice to generate F1 heterozygous offspring (Dcaf17wt/fl)). The Dcaf17wt/fl mice were back-crossed to generate homozygous mice for Dcaf17fl/fl, on a C57BL/6 background. The breeding was continued to establish the founder line. Genomic DNA from tail biopsies of offspring was genotyped for the presence of the Dcaf17fl/fl allele by PCR using LOX1 and LAN1 primers, and sequenced by SDL2 and LOX1 primers to confirm LoxP site retention (Figs 2A and S2B). The Dcaf17fl/fl floxed mice were mated with CMV-Cre mice (B6.C-Tg(CMV-cre)1Cgn/J) to remove the neomycin cassette and Dcaf17 exon 4. Cre-mediated deletion of Dcaf17 exon 4 was confirmed by performing PCR using WSS-F and WSS-R primers on tail DNA samples collected from the F1 pups as shown in Fig. 2B. As a further confirmation, RNA was extracted from tail snips and RT-PCR with primers Dcaf17-3F and Dcaf17-5R was used to discriminate between the various genotypes (Fig. 2C). Dcaf17+/− mice were crossed to generate Dcaf17−/− homozygous mutants.
Genomic DNA extraction and PCR Genotyping
Genomic DNA was isolated from tail biopsies using Gentra Puregene Mouse Tail Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. Genotype analysis, to confirm excision of Dcaf17 exon 4, was performed by routine PCR. The primers used for genotyping are listed in Table 2. PCR conditions were 94 °C for 15 minutes, followed by 35 cycles at 94 °C for 30 seconds, 58 °C for 30 seconds, and 72 °C for 1 minute, followed by a 10 minute extension at 72 °C. The PCR products were separated on 1% agarose gel prepared in 1X TAE (Tris-acetate-EDTA) buffer (40 mM Tris acetate and 1 mM EDTA) containing ethidium bromide at the final concentration of 0.5 µg/ml. Agarose gel electrophoresis was carried our using Owl Horizontal Electrophoresis running chamber (Thermo Scientific, USA) and PowerPac 300 (BIO-RAD Laboratories, Hercules, CA, USA) power supply. Agarose gel images were taken using ImageQuant LAS 4000 (GE Healthcare Life Sciences, Pittsburgh,PA, USA) gel imaging system under standard UV exposure of 1/8 seconds. Adobe Photoshop software (Adobe Inc., San Jose, CA, USA) and Microsoft PowerPoint software were used to assemble images into figures. No post-acquisition modifications were made to the original images.
Table 2.
List of primers used in this study.
| Primer Name | Primer Sequence (5′-3′) | Purpose |
|---|---|---|
| Sp6 | GAGTGCACCATATGGACATATTGTC | Sequencing targeting vector |
| T7 | TAATGCAGGTTAACCTGGCTTATCG | Sequencing targeting vector |
| N1 | TGCGAGGCCAGAGGCCACTTGTGTAG | Sequencing targeting vector |
| N2 | TTCCTCGTGCTTTACGGTATCG | Sequencing targeting vector |
| CARF3 | TGCTCTAGTTGAAGACTTGAGG | Sequencing targeting vector |
| A2 | ACA CGT TCA TGT ATG AGC CCA G | ESC screening |
| A1 | ACACTCTGAGGTAGTTTATAGTCG | ESC screening |
| UNI | AGCGCATCGCCTTCTATCGCCTTC | ESC screening |
| LOX1 | TTGCATACTCACATCCATTTGG | F1 and floxed mice genotyping/LoxP site retention |
| SDL2 | TGCTCTAGTTGAAGACTTGAGG | F1 and floxed mice genotyping/LoxP site retention |
| LAN1 | CCAGAGGCCACTTGTGTAGC | F1 genotyping/LoxP site retention |
| WSS-F | TTGCTCCTCTTTCCCTTCTG | Dcaf17 KO mice genotyping |
| WSS-R | CAAATTGCATACTCACATCCATT | Dcaf17 KO mice genotyping |
| Dcaf17-3F | GTTGCATCAGAGCCAAGAAA | RT-PCR |
| Dcaf17-5R | TTGTTCTGAGCCGACTTCAC | RT-PCR |
| Dcaf17-F | GTTGTACCTCGCAGTGTTCC | qRT-PCR |
| Dcaf17-R | GATGGCTTGGAAGCTGTAGA | qRT-PCR |
| 18 s rRNA-F | CTCTTTCGAGGCCCTGTAAT | qRT-PCR |
| 18 s rRNA-R | CTCCCAAGATCCAACTACGA | qRT-PCR |
Mating behavior and fertility test
Sexually mature (8–10 weeks old) male mice (n = 3 to 5) of each genotypes, WT and Dcaf17−/−, were mated individually with sexually mature (8–10 weeks old) female mice of WT or Dcaf17−/− genotype (n = 3 to 5) in a controlled breeding experiment for the period of 2–6 months. Females were observed for the presence of vaginal plugs and signs of pregnancy. Litter numbers and pups produced by contorlled breeding were recorded.
Sperm count, motility and morphology
The cauda epididymides from 8 weeks and 8 months old WT and Dcaf17−/− mice (n = 3) were dissected and collected in 1 ml of EmbryoMax Human Tubal Fluid (HTF) medium (Millipore, MA, USA) pre warmed at 37 °C. The collected cauda epididymides were incised using fine scissors and sperms were allowed to swim out by incubating for 15–30 minutes at 37 °C under 5% CO2. Ten μl of sperm suspension medium was transferred to the hemocytometer and the total number of sperm was determined using a brightfield microscope (MICROMASTER, Fisher Scientific, Waltham, MA, USA). The total numbers of sperm were counted and the average was calculated from 5 microscopic fields per caudal epididymis. Sperm motility was evaluated immediately by placing a 10 μl drop of sperm suspension on hemocytometer and counting the sperm under a brightfield at 400× magnification. Sperm motility presented as percentage of motile sperm of total sperm number.
To study the sperm morphology, sperm smears were prepared by pipetting 20 µl of cauda epididymides sperm suspension on glass slides. After drying the slide, the sperm smear was fixed with 2% paraformaldehyde (PFA) by incubating at room temperature for 10–15 minutes, washed three times with 1X PBS and stained with Diff-Quick stain (Shandon™ Kwik-Diff™ Stains, ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Diff-Quick stained slides were observed under optical microscope (Olympus IX71, Olympus, Center Valley, PA, USA). Digital images were captured using OLYMPUS DP72 microscope camera (Olympus, Center Valley, PA, USA) and OLYMPUS cellSens Entry 1.6 imaging software (Olympus, Center Valley, PA, USA). Adobe Photoshop software (Adobe Inc., San Jose, CA, USA) and Microsoft PowerPoint software were used to assemble images into figures. No post-acquisition modifications were made to the original images.
Histology
For the evaluation of spermatogenesis, histological analyses were performed on the testes of WT and Dcaf17 KO mice at different postnatal ages (5, 14, 23, 32, 42 and 56 days) as well as on epididymides of sexually mature (8 weeks old) mice (n = 3). The tissues were collected and directly fixed in 4% paraformaldehyde (PFA) or in Bouin’s fixative for overnight at 4 °C. Fixed tissues were washed with distilled water and dehydrated in a series of 70–100% ethanol solutions. Dehydrated tissues were embedded in paraffin and 5 µm thick sections were prepared on glass slides using microtome (Leica RM2155, Leica Biosystems, Buffalo Grove, IL, USA). Paraffin embedded tissue sections were deparaffinized, rehydrated and stained with hematoxylin and eosin (H&E) or periodic acid-Schiff (PAS) staining according to standard protocols51,52. Stained testis and epididymis sections were observed at room temperature using Olympus BX53 (Olympus, Center Valley, PA, USA) optical microscope. Digital images were captured using OLYMPUS DP72 microscope camera (Olympus, Center Valley, PA, USA) and OLYMPUS cellSens Entry 1.6 imaging software (Olympus, Center Valley, PA, USA). Adobe Photoshop software (Adobe Inc., San Jose, CA, USA) and Microsoft PowerPoint software were used to assemble images into figures. No post-acquisition modifications were made to the original images.
Testes squash preparation
Squash preparation of testicular samples was performed as previously described53. Briefly, the testes of adult WT and Dcaf17−/− mice were dissected and decapsulated by removing tunica albuginea. Seminiferous tubules of decapsulated testis were dispersed and fixed in freshly prepared 2% formaldehyde in phosphate-buffered saline (PBS) containing 0.05% Triton X-100 (Sigma, St. Louis, MO, USA) for 10 minutes at room temperature. Pieces (~1 mm) of seminiferous tubules were placed in a drop (~15 µl) of fixing solution on a clean glass slide. The pieces were gently minced with fine tip tweezers and were covered with glass cover slip. The cells were squashed by applying external pressure on coverslip. The slides were frozen by immersing in liquid nitrogen. The slides were immediately used for immunostaining or stored at −80 °C until required.
Immunofluorescence
Evaluation of defects in sperm head and mid piece was carried out using MitoTracker, Peanut agglutinin (PNA) and 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) staining as described previously24. Briefly, the cauda epididymides were dissected from sexually mature (8–10 weeks old) WT and Dcaf17−/− mice (n = 3 for each genotype) and sperm were allowed to swim out as described before. Aliquots of sperm suspension were transferred to 1.5 ml centrifuge tubes and centrifuged at 500 × g for 10 minutes at room temperature. The HTF medium was removed and the sperm were incubated with MitoTracker solution (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) at 37 °C for 30 minutes. Sperm were collected by centrifugation and then fixed with 4% PFA, permeabilized in 0.2% Triton X-100 (sigma-Aldrich, St. Louis, MO, USA) and a smear was prepared. Slides were then incubated with PNA-FITCI (GeneTex, Irvine, CA, USA) (1:1000 dilution in PBS) at room temperature for 45 minutes. Slides were washed and mounted with VECTASHIELD mounting medium containing DAPI (1.5 µg/ml) (Vector Laboratories, Burlingame, CA, USA).
For immunostaining of testes squash preparations, the slides were hydrated and washed in PBS. After washing in PBS, the slides were boiled for 20 min in 1× DakoCytomation Target Retrival Solution (pH 6.0) (Dako, Carpinteria, CA, USA) using a microwave oven for antigen unmasking. The slides were then washed three times in PBS for 5 min each and blocked for non-specific antibody binding by incubating with 10% normal goat serum (Gibco, ThermoFisher Scientific, Waltham, MA, USA) in PBS for 1 h at room temperature. After blocking, the slides were washed with PBS and incubated overnight at 4 °C with appropriate primary antibodies diluted in 0.5% BSA and 0.1% Triton X100 in PBS. The following antibodies were used: mouse monoclonal anti-SYCP3 (97672, Abcam, Cambridge, MA, USA) and rabbit polyclonal anti-SYCP1 (15090, Abcam, Cambridge, MA, USA) at 1:200 dilution; rabbit polyclonal phospho-Histon H2A.x (ser139) antibody (2577, Cell Signaling, Danvers, MA, USA) at 1:500 dilution or mouse monoclonal alpha-tubulin (DM1A) (3873, Cell signaling, Danvers, MA, USA) at 1:1000 dilution. Following the primary antibody incubation, the slides were washed with PBS (3 times 5 minutes each) and were further incubated in dark with appropriate secondary antibodies at room temperature for 1 hour. All secondary antibodies; FITC conjugated goat anti-rabbit-IgG (SC-2012), rodamin conjugated goat anti-rabbit-IgG (SC-2091), FITC conjugated goat anti-mouse-IgG (SC-2010), or rodamin conjugated goat anti-mouse-IgG (SC-2092) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and used at 1:300 dilution. After secondary antibody incubation, slides were washed in PBS, and mounted in VECTASHIELD mounting medium with DAPI (1.5 µg/ml) (Vector Laboratories, Inc., Burlingame, CA, USA). For sperm and testes immunofluorescence analyses, three mice of each genotype were used. From each mouse, three slides were prepared. All the samples were observed at room temperature using a Zeiss Axio imager Z2 epifluorescence microscope (Carl-Zeiss, Oberkochen, Germany) and digital images were captured with an Axiocam MRm microscope camera (Carl-Zeiss, Oberkochen, Germany) and AxioVision imaging software ZEN2011. Adobe Photoshop software (Adobe Inc., San Jose, CA, USA) and Microsoft PowerPoint software were used to assemble images into figures. No post-acquisition modifications were made to the original images.
TUNEL assay
Terminal deoxynucleotidyl transferase (TdT) mediated dUTP-biotin nicked end-labeling (TUNEL) assay was performed by using In Situ Cell Death Detection kit (Roche Diagnostics, Basel, Switzerland) to detect DNA fragmentation in apoptotic cells according to manufacturer’s recommended protocol. Briefly, testis paraffin sections from 8 weeks and 8 months old WT and Dcaf17−/− mice testes were deparaffinized with xylene, rehydrated in dilution series of ethanol, and permeabilized by incubating the sections with proteinase K (Qiagen, Hilden, Germany) solution (20 µg/ml in 10 mM Tris-HCL, pH 7.4) for 15–30 minutes at room temperature. Following permeabilization, the tissue sections were washed twice in PBS and incubated with TUNEL reaction mixture for 60 minutes at 37 °C in the dark. Positive controls were treated with 3 U/ml Dnase I (Roche Diagnostics, Basel, Switzerland) solution (prepared in 50 mM Tris-HCL, pH 7.5, 10 mM MgCl2 and 1 mg/ml BSA) for 10 minutes before TUNEL labeling, while negative controls were treated with labeling solution without TdT enzyme. After TUNEL mixture incubation, sections were washed three times (5 minutes each) with PBS, and mounted in VECTASHIELD mounting medium (Vector Laboratories, Inc., Burlingame, CA, USA). Three serial testicular sections (5 µm) each from three different mice of each WT and Dcaf17−/− genotypes were used for TUNEL assay. All the samples were observed under 20× objective at room temperature using a Zeiss Axio imager Z2 epifluorescence microscope (Carl-Zeiss, Oberkochen, Germany) and digital images were captured with an Axiocam MRm microscope camera (Carl-Zeiss, Oberkochen, Germany) and AxioVision imaging software ZEN2011. All the fluorescence images were acquired under the same exposure time (190.5 ms). The average number of TUNEL-positive cells per tubule was determined by analyzing 100 seminiferous tubules per animal. Adobe Photoshop software (Adobe Inc., San Jose, CA) and Microsoft PowerPoint software were used to assemble images into figures. No post-acquisition modifications were made to the original images.
Total RNA extraction and reverse transcription
To determine Dcaf17 mRNA levels in different tissues and during testis development, adult WT mouse tissues (brain, liver, pancreas, skin and testis) and 5, 14, 23, 32, 42 and 56 days postpartum (dpp) testes were collected and snap frozen immediately by immersing in liquid nitrogen. Total RNA was extracted using QIAGEN RNeasy mini kit and then treated with DNase I according to the manufacturer’s protocol (Qiagen, Valencia, CA, USA). The RNA quality was checked by visualizing intact 28S and 18S RNA bands on agarose gel-electrophoresis using ethidium bromide staining. Quantification of total RNA was carried out using NanoDrop 2000c spectrophotometer (Thermo Scientific, ThermoFisher Scientific, Waltham, MA, USA). Reverse transcription was performed on 500 ng total RNA from each tissue using Superscript III Frist Strand Synthesis system (Life Technologies, ThermoFisher Scientific, Waltham, MA, USA) as per manufacturer’s protocol.
Quantitative Real Time PCR (qRT-PCR)
The qRT-PCR was performed using 5 µl of 1:5 diluted cDNA products as templates from different tissues in a 20 µl of total reaction mixture for each cDNA sample. The qPCR reaction mixture contained 10ul, SYBER Green Supermix (1725271, BIO-RAD Laboratories, Hercules, CA, USA) and 1 µl of 100 nM of each forward and reverse primer. For each sample, a parallel reaction was done using 18 s rRNA primers as endogenous control. Quantitative RT-PCR was carried out using C1000 Touch thermal cycler (CFX96 Real-Time system, BIO-RAD Laboratories, Hercules, CA, USA). Reaction steps were 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30 sec, primer annealing at 58 °C for 30 sec, and primer extension at 72 °C for 30 sec, followed by melting curve analysis (55 to 95 °C; in 0.5 °C increments) to verify specificity of amplicons. All samples were analyzed in triplicate and 18 s rRNA was used as internal control for data normalization, which were analyzed by the 2−ΔCt method and reported as fold change. Primers used for this experiment are listed in Table 2.
Transmission electron microscopy (TEM)
For the ultra-structural analysis, testes and epididymides of WT and Dcaf17−/− adult mice were collected, cut roughly into 1 mm cubes and immediately fixed in 2.5% glutaraldehyde solution prepared in 0.1 M PBS (pH 7.4) for 1 hour at room temperature. After glutaraldehyde fixation, tissues were rinsed twice 5 minutes each with 0.1 M PBS and post-fixed in 1% Osmium Tetroxide (OsO4) for 1 hour at room temperature. After the OsO4 treatment, tissues were washed with distilled water twice for 5 minutes each and pre-stained in 2% uranyl acetate for 30 minutes at room temperature. Tissues were then washed with distilled water for 5 minutes, dehydrated in a series of gradient acetone and embedded in Epon resin (Poly/BED 812). Semi-thin sections (0.5–0.7 µm) were cut using LEICA EM UC6 ultra microtome (Leica Biosystems, Buffalo Grove, IL, USA), stained with toluidine blue and inspected under the bright field microscope. The relevant areas were chosen and ultrathin (50–70 nm) sections were prepared with a diamond knife using LEICA EM UC6 ultra microtome. Sections were picked up on Gilder hexagonal mesh copper grids (G200HH, TED PELLA, INC., Redding, CA, USA) and stained with uranyl acetate followed by lead citrate. Ultrathin sections were examined with JEM-1230 JEOL transmission electron microscope (JEOL USA Inc., Peabody, MA, USA) operated at 60 kV. Digital images were acquired using KeenView Soft Imaging System camera (ResAlta Research Technologies, Golden, CO, USA) and iTEM imaging software (ResAlta Research Technologies, Golden, CO, USA).
Statistical Analysis
All data from sperm count, motility, TUNEL assay and qRT-PCR were analyzed using Graphpad prism 5 software and presented as the mean ± standard error of the mean (SEM). All the experiments were performed in triplicate and were repeated three times. For each experiment, 3 to 5 animals were used. A one-way ANOVA or two-Ways ANOVA followed by post hoc Bonferroni’s multiple or Tukey’s multiple comparisons tests were performed to compare the data. A P-value < 0.05 was considered statistically significant.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Electronic supplementary material
Acknowledgements
We would like to thank Mrs. Trupti Mistry for preparing schematic diagrams for Figure 10 of the manuscript. This research work (RAC # 210-0006) was approved and funded by King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia.
Author Contributions
Ab.As., F.S.A. and B.V.M. designed experiments; As.Al., B.V.M., Ha.A.Ah., R.A. and An.M.Al. performed experiments; As.Al., B.V.M. and Ab.As. wrote the manuscript; As.Al., Ab.As. and B.V.M prepared figures; Ha.A.Am., A.P. and Ab.As. supervised the research work; All authors reviewed the manuscript and approved it for publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Asmaa Ali and Bhavesh V. Mistry contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-29836-2.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27379-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/1/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
- 1.Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Genetics of Male Infertility. Current urology reports. 2016;17:70. doi: 10.1007/s11934-016-0627-x. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reproductive biology and endocrinology: RB&E. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh K, Jaiswal D. Human male infertility: a complex multifactorial phenotype. Reproductive sciences. 2011;18:418–425. doi: 10.1177/1933719111398148. [DOI] [PubMed] [Google Scholar]
- 4.O’Flynn O’Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertility and sterility. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 5.Krausz C. Male infertility: pathogenesis and clinical diagnosis. Best practice & research. Clinical endocrinology & metabolism. 2011;25:271–285. doi: 10.1016/j.beem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 6.de Kretser DM, Loveland KL, Meinhardt A, Simorangkir D, Wreford N. Spermatogenesis. Human reproduction. 1998;13(Suppl 1):1–8. doi: 10.1093/humrep/13.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 7.Roosen-Runge EC. The process of spermatogenesis in mammals. Biological reviews of the Cambridge Philosophical Society. 1962;37:343–377. doi: 10.1111/j.1469-185X.1962.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 8.O’Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014;4:e979623. doi: 10.4161/21565562.2014.979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leblond CP, Clermont Y. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. The American journal of anatomy. 1952;90:167–215. doi: 10.1002/aja.1000900202. [DOI] [PubMed] [Google Scholar]
- 10.Lehti MS, Sironen A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction. 2016;151:R43–54. doi: 10.1530/REP-15-0310. [DOI] [PubMed] [Google Scholar]
- 11.Hou CC, Yang WX. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Molecular biology reports. 2013;40:3213–3230. doi: 10.1007/s11033-012-2397-y. [DOI] [PubMed] [Google Scholar]
- 12.Sutovsky P. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone. Microscopy research and technique. 2003;61:88–102. doi: 10.1002/jemt.10319. [DOI] [PubMed] [Google Scholar]
- 13.Baarends WM, van der Laan R, Grootegoed JA. Specific aspects of the ubiquitin system in spermatogenesis. Journal of endocrinological investigation. 2000;23:597–604. doi: 10.1007/BF03343782. [DOI] [PubMed] [Google Scholar]
- 14.Gao, J. et al. The CUL4-DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. eLife4, 10.7554/eLife.07539 (2015). [DOI] [PMC free article] [PubMed]
- 15.Bose R, Manku G, Culty M, Wing SS. Ubiquitin-proteasome system in spermatogenesis. Advances in experimental medicine and biology. 2014;759:181–213. doi: 10.1007/978-1-4939-0817-2_9. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Ruiz JC, Chun KT. CUL-4A is critical for early embryonic development. Molecular and cellular biology. 2002;22:4997–5005. doi: 10.1128/MCB.22.14.4997-5005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. Journal of biosciences. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 19.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nature structural & molecular biology. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 20.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays in biochemistry. 2005;41:15–30. doi: 10.1042/bse0410015. [DOI] [PubMed] [Google Scholar]
- 21.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochimica et biophysica acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Yin Y, et al. Cell Autonomous and Nonautonomous Function of CUL4B in Mouse Spermatogenesis. The Journal of biological chemistry. 2016;291:6923–6935. doi: 10.1074/jbc.M115.699660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin CY, et al. Human X-linked Intellectual Disability Factor CUL4B Is Required for Post-meiotic Sperm Development and Male Fertility. Scientific reports. 2016;6:20227. doi: 10.1038/srep20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Developmental biology. 2011;356:51–62. doi: 10.1016/j.ydbio.2011.05.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopanja D, et al. Cul4A is essential for spermatogenesis and male fertility. Developmental biology. 2011;352:278–287. doi: 10.1016/j.ydbio.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends in biochemical sciences. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Molecular cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 29.Alazami AM, et al. Mutations in C2orf37, encoding a nucleolar protein, cause hypogonadism, alopecia, diabetes mellitus, mental retardation, and extrapyramidal syndrome. American journal of human genetics. 2008;83:684–691. doi: 10.1016/j.ajhg.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agopiantz M, et al. Endocrine disorders in Woodhouse-Sakati syndrome: a systematic review of the literature. Journal of endocrinological investigation. 2014;37:1–7. doi: 10.1007/s40618-013-0001-5. [DOI] [PubMed] [Google Scholar]
- 31.Woodhouse NJ, Sakati NA. A syndrome of hypogonadism, alopecia, diabetes mellitus, mental retardation, deafness, and ECG abnormalities. Journal of medical genetics. 1983;20:216–219. doi: 10.1136/jmg.20.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juneja SC, van Deursen JM. A mouse model of familial oligoasthenoteratozoospermia. Human reproduction. 2005;20:881–893. doi: 10.1093/humrep/deh719. [DOI] [PubMed] [Google Scholar]
- 33.Oehninger S. Pathophysiology of oligoasthenoteratozoospermia: are we improving in the diagnosis? Reproductive biomedicine online. 2003;7:433–439. doi: 10.1016/S1472-6483(10)61887-1. [DOI] [PubMed] [Google Scholar]
- 34.de Vries F. A.T. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes & Development. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Capetillo Oscar, et al. H2AX Is Required for Chromatin Remodeling and Inactivation of Sex Chromosomes in Male Mouse Meiosis. Developmental Cell. 2003;4:497–508. doi: 10.1016/S1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 36.Yuan Li, et al. The Murine SCP3 Gene Is Required for Synaptonemal Complex Assembly, Chromosome Synapsis, and Male Fertility. Molecular Cell. 2000;5:73–83. doi: 10.1016/S1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 37.Kierszenbaum AL. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Molecular reproduction and development. 2002;63:1–4. doi: 10.1002/mrd.10179. [DOI] [PubMed] [Google Scholar]
- 38.Li W, et al. A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development. 2015;142:921–930. doi: 10.1242/dev.119834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nozawa YI, Yao E, Gacayan R, Xu SM, Chuang PT. Mammalian Fused is essential for sperm head shaping and periaxonemal structure formation during spermatogenesis. Developmental biology. 2014;388:170–180. doi: 10.1016/j.ydbio.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kierszenbaum AL, Tres LL. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Archives of histology and cytology. 2004;67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- 41.Jan SZ, et al. Molecular control of rodent spermatogenesis. Biochimica et biophysica acta. 2012;1822:1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Kanatsu-Shinohara M, Onoyama I, Nakayama KI, Shinohara T. Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8826–8831. doi: 10.1073/pnas.1401837111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou X, et al. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC genomics. 2012;13:495. doi: 10.1186/1471-2164-13-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Donnell L, Nicholls PK, O’Bryan MK, McLachlan RI, Stanton PG. Spermiation: The process of sperm release. Spermatogenesis. 2011;1:14–35. doi: 10.4161/spmg.1.1.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Molecular and cellular endocrinology. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavallini G. Male idiopathic oligoasthenoteratozoospermia. Asian journal of andrology. 2006;8:143–157. doi: 10.1111/j.1745-7262.2006.00123.x. [DOI] [PubMed] [Google Scholar]
- 47.Hannah J, Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene. 2015;573:33–45. doi: 10.1016/j.gene.2015.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng K, Liang X, Huang S, Xu W. The role of histone ubiquitination during spermatogenesis. BioMed research international. 2014;2014:870695. doi: 10.1155/2014/870695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richburg JH, Myers JL, Bratton SB. The role of E3 ligases in the ubiquitin-dependent regulation of spermatogenesis. Seminars in cell & developmental biology. 2014;30:27–35. doi: 10.1016/j.semcdb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura N. Ubiquitination regulates the morphogenesis and function of sperm organelles. Cells. 2013;2:732–750. doi: 10.3390/cells2040732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardiff RD, Miller CH, Munn RJ. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harbor protocols. 2014;2014:655–658. doi: 10.1101/pdb.prot073411. [DOI] [PubMed] [Google Scholar]
- 52.Clermont Y, Leblond CP. Spermiogenesis of man, monkey, ram and other mammals as shown by the periodic acid-Schiff technique. The American journal of anatomy. 1955;96:229–253. doi: 10.1002/aja.1000960203. [DOI] [PubMed] [Google Scholar]
- 53.Page J, Suja JA, Santos JL, Rufas JS. Squash procedure for protein immunolocalization in meiotic cells. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 1998;6:639–642. doi: 10.1023/A:1009209628300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.