Abstract
Rationale: Genetic factors are involved in acute respiratory distress syndrome (ARDS) susceptibility. Identification of novel candidate genes associated with increased risk and severity will improve our understanding of ARDS pathophysiology and enhance efforts to develop novel preventive and therapeutic approaches.
Objectives: To identify genetic susceptibility targets for ARDS.
Methods: A genome-wide association study was performed on 232 African American patients with ARDS and 162 at-risk control subjects. The Identify Candidate Causal SNPs and Pathways platform was used to infer the association of known gene sets with the top prioritized intragenic SNPs. Preclinical validation of SELPLG (selectin P ligand gene) was performed using mouse models of LPS- and ventilator-induced lung injury. Exonic variation within SELPLG distinguishing patients with ARDS from sepsis control subjects was confirmed in an independent cohort.
Measurements and Main Results: Pathway prioritization analysis identified a nonsynonymous coding SNP (rs2228315) within SELPLG, encoding P-selectin glycoprotein ligand 1, to be associated with increased susceptibility. In an independent cohort, two exonic SELPLG SNPs were significantly associated with ARDS susceptibility. Additional support for SELPLG as an ARDS candidate gene was derived from preclinical ARDS models where SELPLG gene expression in lung tissues was significantly increased in both ventilator-induced (twofold increase) and LPS-induced (5.7-fold increase) murine lung injury models compared with controls. Furthermore, Selplg−/− mice exhibited significantly reduced LPS-induced inflammatory lung injury compared with wild-type C57/B6 mice. Finally, an antibody that neutralizes P-selectin glycoprotein ligand 1 significantly attenuated LPS-induced lung inflammation.
Conclusions: These findings identify SELPLG as a novel ARDS susceptibility gene among individuals of European and African descent.
Keywords: acute respiratory distress syndrome, genome-wide association study, selectin P ligand gene
At a Glance Commentary
Scientific Knowledge on the Subject
An important aspect of the clinical management of acute respiratory distress syndrome (ARDS) is early recognition to prevent further lung injury during mechanical ventilatory support. Despite advances in these preventive strategies aimed at minimizing lung injury among at-risk patients, there are currently no useful genetic or nongenetic biomarkers to identify those who are more likely to develop ARDS or to explain the observed health disparities. There is a need to identify both genetic and nongenetic markers of increased ARDS susceptibility.
What This Study Adds to the Field
This genome-wide association study with preclinical validation is the first to identify SELPLG (selectin P ligand gene) as a novel ARDS susceptibility gene. The SELPLG gene and its encoded protein, P-selectin glycoprotein ligand 1, are potentially novel therapeutic targets for reducing ARDS pathobiology.
Acute respiratory distress syndrome (ARDS) affects more than 200,000 individuals annually in the United States and, despite improvements in overall care over the past 2 decades, the mortality rate remains at roughly 30–40% (1–4). Current therapeutic options to improve ARDS survival consist solely of a preventive strategy using low Vts and distending pressures during mechanical ventilatory support to prevent ventilator-induced lung injury (VILI) (5). Other potentially beneficial adjunctive strategies include prone positioning to improve oxygenation, neuromuscular blockade to prevent patient–ventilator dyssynchrony, inhaled vasodilators to improve oxygenation and unload the right ventricle, extracorporeal membrane oxygenation to improve gas exchange, and fluid management to reduce pulmonary edema (6–10). An important aspect of ARDS management is early recognition to prevent further lung injury during mechanical ventilatory support (11). At present, there are no clinically useful biomarkers to identify the less than 10% of patients with well-recognized precipitating conditions, such as pneumonia, septic shock, or severe trauma, who will eventually develop the syndrome.
Racial and ethnic differences in susceptibility to and mortality from ARDS (12–16) support the increasingly recognized role of genetic factors in ARDS susceptibility (17–27), and highlight the importance of identifying genetic biomarkers of ARDS susceptibility in at-risk individuals to further improve risk stratification (28). We have previously identified several novel genetic ARDS biomarkers and therapeutic targets, including NAMPT (nicotinamide phosphoribosyl transferase), which encodes for a Toll-like receptor 4–binding cytozyme (17, 24), MYLK (encoding MLCK [myosin light-chain kinase]) (19, 26, 29), GADD45a (growth arrest and DNA damage–inducible gene) (30), and sphingosine 1-phosphate receptor 3 (25).
In the current study, we identified three exonic SNPs (from two independent cohorts) within SELPLG (selectin P ligand gene; encoding PSGL-1 [P-selectin glycoprotein ligand 1]) that associate with ARDS susceptibility in at least one population. The selectin pathway is critically involved in the transmigration of leukocytes from the vascular lumen to sites of inflammation (31–33). In preclinical ARDS models, SELPLG expression in lung tissues was significantly increased for both VILI and LPS-induced (LPS) murine models compared with controls. LPS-induced inflammatory lung injury in genetically engineered Selplg−/− mice was significantly reduced compared with wild-type C57/B6 mice. Finally, an inhibitory PSGL-1 antibody significantly attenuated lung inflammation and injury produced by either LPS or VILI. These findings indicate that SELPLG is a novel ARDS susceptibility gene among individuals of European and African descent. Some of the results of these studies have been previously reported in the form of an abstract (34).
Methods
Genome-Wide Association Analysis of African American Patients with ARDS
The discovery population was a collection of well-phenotyped patients with ARDS and DNA samples from multiple sites, including patients enrolled in the FACTT (Fluid and Catheter Treatment Trial) study (10), and data from investigators at the University of Chicago, University of Illinois, University of Washington, University of Pennsylvania, Harvard University, and the University of Tennessee. Race was self- or proxy-reported. All patients with ARDS met the diagnostic criteria per the American–European Consensus Conference (35) or the Berlin definition (36). A detailed description of the methods used to identify ARDS cases in the discovery cohort is included in the online supplement. The clinical characteristics of study participants are presented in Table 1. The control subjects were self-identified African American patients with sepsis or other ARDS risk factors admitted to the ICU at the University of Chicago and the University of Washington under the direct care of experienced intensive care specialists. The Institutional Review Board of the University of Arizona approved the protocol for this genome-wide association study (GWAS) analysis.
Table 1.
Patient Characteristics
| Characteristics | ARDS (n = 232) | Non-ARDS (n = 162) |
|---|---|---|
| Cohort (n) | ||
| FACTT | 80 | 0 |
| U. Chicago | 23 | 57 |
| Harvard | 26 | 0 |
| U. Tennessee | 4 | 0 |
| U. Washington | 96 | 102 |
| U. Illinois | 3 | 3 |
| Female, % | 89 | 63 |
| Male, % | 143 | 99 |
| Age, yr, mean (SD) | 51 (15) | 55 (17) |
| Comorbidities, % | ||
| Sepsis | 61 | 69 |
| Pneumonia | 21 | 19 |
| APACHE II, mean (SD) | 22 (6) | 25 (7) |
| APACHE III, mean (SD) | 88 (34) | 54 (22) |
| ICU LOS, d, mean (SD) | 14 (15) | 7 (9) |
| HOSP LOS, d, mean (SD) | 22 (21) | 16 (14) |
Definition of abbreviations: APACHE = Acute Physiology and Chronic Health Evaluation; ARDS = acute respiratory distress syndrome; FACTT = Fluid and Catheter Treatment Trial; HOSP = hospital; LOS = length of stay; U. = University of.
APACHE scores: 37 APACHE II and 99 APACHE III scores in the control group; 48 APACHE II and 203 APACHE III scores in the ARDS group. APACHE III scores were significantly different between the control and ARDS groups (P < 0.05).
We investigated the genetic variants underlying the development of ARDS (cases) among patients with sepsis or other ARDS risk factors (control subjects) through the analysis of data on 1,428,996 SNPs from 394 individuals (232 cases and 162 ICU control subjects) of African American descent. Specifically, ’ the genotypes of subjects with ARDS for 2,129,301 SNPs with the Affymetrix PanAFR array were assayed. Genotyping was followed by several quality control analyses. First, we discarded samples with: 1) excessive number of missing genotypes (≥5%); 2) high degree of identity by descent estimate, proportional identity by descent greater than 0.20; and 3) sex and race/ethnicity discordance between recorded clinical data and genomic data. Patient genomic ancestry was assessed using data from HapMap3 (37) by principle component analysis (PCA) (see plots in Figure E1 in the online supplement). SNPs with minor allele frequency less than 5% and Hardy-Weinberg P values less than 0.001 were discarded. SNPs with genotyping call rate greater than 95% and a mean genotyping call rate that was 99.5% were included.
Biological Pathway Analysis of GWAS Findings
We used the Identify Candidate Causal SNPs and Pathways (ICSNPathway) analysis (38) to infer the previously annotated biological gene sets that are associated with the host genes of the GWAS-based prioritized SNPs. ICSNPathway screens the complete list of GWAS-based nominal SNP P values based on their annotated functional consequence, such as gain or loss of stop codon. In addition, ICSNPathway includes other genetic variants according to their ethnic-specific linkage disequilibrium (LD) patterns, thus increasing the list of potential causal variants. It then maps each of the SNPs to its nearest host gene that is additionally defined based on its distinctive flanking genomic distances. In the case of multiple assigned SNPs, only the SNP with the smallest GWAS nominal P value for each gene of interest is selected. Next, ICSNPathway analysis uses gene set enrichment analysis and permutation of the SNP labels to infer the candidate pathways according to their corrected P values for multiple testing.
Our analysis was performed for SNPs across various GWAS nominal P value thresholds, ranging from 10−8 to 10−3. Host genes of intragenic SNPs were defined at varying flanking distances of genes, specifically as within, 5 KB, 20 KB, and 100 KB up/downstream of each human gene based on GRCh38.p2 assembly (https://www.ncbi.nlm.nih.gov/assembly/gcf_000001405.28/). We corrected for LD (r2 > 0.8) among SNPs according to the specific ethnicity to which the samples belonged (African Ancestry in Southwest U.S.A. parameter in ICSNPathway). We used publicly available gene sets, namely v3.0 of level 4 of Gene Ontology Molecular Functions and biological processes (39, 40) (http://www.broadinstitute.org/gsea/msigdb/index.jsp), KEGG (41) (http://www.genome.jp/kegg/pathway.html), and BIOCARTA (http://www. biocarta.com/genes/index.asp), and inferred the candidate pathways at ICSNPathway false discovery rate (FDR; ICSNPathway) less than 0.05.
The Molecular Epidemiology of Sepsis in the ICU Cohort
The MESSI (Molecular Epidemiology of Sepsis in the ICU) cohort at the University of Pennsylvania has been described previously (42, 43). Patients were eligible if they were admitted to the ICU with sepsis (44, 45) and excluded if an alternative diagnosis explained SIRS criteria, for declining life support on admission, or for lack of informed consent. Clinical data were abstracted from the electronic medical record. All chest imaging studies obtained during the first 6 days (46) were interpreted by trained physician investigators, as described previously (47, 48). ARDS was adjudicated in accordance with Berlin criteria requiring that chest radiograph and oxygenation criteria be met on the same calendar day while invasively ventilated (36, 43). Mortality was determined at 30 days. Source of sepsis was adjudicated by critical care physician investigators. Genomic DNA was extracted from whole blood using the QIAamp DNA Mini kit (Qiagen) and assayed with the Affymetrix Axiom TxArray v.1, a genome-wide platform comprised of approximately 780,000 markers (49), which were used to identify genetic ancestry and 8 genotypes in the SELPLG locus (Chr12:109017264–109028341).
Models of LPS-induced and Ventilator-induced Murine Lung Injury
Male C57BL/6J mice aged 8–12 weeks (Jackson Laboratories) were used for all experiments. The Animal Care and Use Committee at the University of Arizona approved all animal care procedures and experiments. Mice were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited institution in autoclaved micro-isolator cages with free access to food and water. Mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg) given by intraperitoneal injection and then intubated with a 20-Ga angiocatheter. For the LPS-induced lung injury, mice were intratracheally injected with LPS (Escherichia coli 0127:B8, 1 mg/kg; Sigma). For VILI models, mice were connected to Inspira mechanical ventilator (Harvard Apparatus) using a Vt of 40 ml/kg, respiratory rate of 65 breaths/min, and positive end-expiratory pressure of 0 cm H2O for 4 hours. Spontaneously breathing control animals were anesthetized, given intravenous phosphate-buffered saline (PBS), and allowed to breathe spontaneously for 4 hours. In specific experiments, Selplg knockout mice (Selplg−/−) were obtained from Jackson Laboratories (catalog no. 006336) and maintained under the same conditions as wild-type mice.
Murine VILI and LPS-induced SELPLG Gene Expression
The transcriptomic analyses for both VILI- and LPS-exposed mice were based on the data generated by Affymetrix Mouse Genome 430 2.0 Array (National Center for Biotechnology Information Gene Expression Omnibus [GEO] dataset ID: GSE9368 and GSE9314). The GeneChip Robust Multiarray Averaging method was used to summarize the genome-wide gene expression levels (50, 51). Only the probe sets present (determined by function “mas5calls” in the Bioconductor “affy” package) in all the samples of at least one group were retained. The robust multiarray average function in the “affy” package of Bioconductor was used to summarize the expression level of each probe set. The significance analysis of microarrays algorithm was used to identify the genes deregulated by VILI and LPS based on log2-transformed gene expression levels (52). FDR was controlled using the q value method (53).
PSGL-1 Inhibition and Lung Inflammation in a Mouse Model of ARDS
C57BL/6J mice were pretreated with PSGL-1 antibody (CD162, 30-min pretreatment, intravenous injection, 1 mg/kg) before LPS challenge (1 mg/kg, intratracheal instillation, 18 h) or high Vt ventilation (40 ml/kg, 4 h). BAL and lung tissue were collected for further lung injury analysis.
BAL Analysis
At the termination of each animal experiment, BAL fluid was collected by instilling 1 ml of Hank’s balanced salt solution (Invitrogen) intratracheally, followed by slow recovery of the fluid as we have previously described (54). Cells were recovered from the collected BAL fluid by centrifugation (500 × g, 20 min, 4°C) and counted using an automated cell counter (TC20; Bio-Rad). A blinded investigator performed manual differential cell counts (to elucidate the number of BAL neutrophils) on cytospin samples (500 cells/sample) (Shandon, Cytospin 4, Thermo Fisher Scientific) after staining with Kwik Diff Stain (Thermo Scientific). The supernatant was used to measure total protein concentration (Pierce BCA protein assay kit; Thermo Scientific). The BAL fluid supernatant was centrifuged again (16,500 × g, 10 min, 4°C) and stored at −80°C for further analysis.
Lung Histopathology
To assess alterations in the lung tissue morphology, lungs were fixed in 10% formalin, embedded in paraffin, sectioned, mounted onto slides, and stained with hematoxylin and eosin (H&E). These sections were examined under microscope and histologically evaluated with established lung injury criteria as we previously reported (54).
Statistical Analysis
Continuous data were compared using nonparametric methods and categorical data by chi-square test. Where applicable, standard one-way ANOVA was used and groups were compared using the Newman-Keuls test. Differences between groups were considered statistically significant when P was less than 0.05. Logistic regression analysis with adjustment for sepsis status, age, sex, and population stratification (the first principal component) was used in an additive genetic model. Imputation of 200-kb region around the SELPLG was performed using Michigan Imputation Server (55). After the imputation, SNPs with poor imputation quality (R2 < 0.3, minor allele frequency 0.1%, Hardy-Weinberg equilibrium P < 0.00001) were removed. In the GWAS analysis, a Bonferroni correction for multiple testing (n = 1,428,996 SNPs) was used as the genome-wide threshold for significance (α = 3.50 × 10−8). We also examined the associations between three missense mutations on the SELPLG exon 3 and ARDS using logistic regression assuming an additive model of genetic risk and adjusting for genetic ancestry the MESSI cohort. Models were performed separately for subjects genetically of European ancestry (EA) and African ancestry (AA). Subjects of other ancestry were excluded due to low numbers. Analysis was performed using Golden Helix SNP and Variation Suite and PLINK v1.07 (56).
Results
GWAS of the Discovery Population
Table 1 shows the association of ARDS with various clinical covariates. When compared with ICU control subjects, the participants with ARDS in our study exhibited significantly higher mortality and longer hospital and ICU length of stay (Table 1). No SNP satisfied the conservative genome-wide threshold of Bonferroni correction (P < 3.50 × 10−8; Table E1). The genomic inflation factor based on median chi-square was estimated at 1.02, showing no indication of genomic inflation (Figure E2, Q–Q plot).
Biological Pathway Analysis
We then employed the ICSNPathway analysis (38) to evaluate the association between the GWAS-based prioritized genomic loci and previously known gene sets (e.g., Gene Ontology [40], KEGG [41]). At a nominal GWAS P value less than 10−4, we prioritized a single intronic SNP, rs2115740 (Table 2; ICSNPathway false discovery rate [FDR] = 0.021; GWAS nominal P = 6.5 × 10−5; odds ratio [OR] = 1.78; 95% confidence interval [CI] = 1.33–2.38). rs2115740 is in LD (r2 = 0.83) with two other deleterious nonsynonymous coding SNPs, namely rs2275687 and rs1126627 (LD r2 = 1 between them), which reside within the HEATR1 (HEAT repeat containing 1) gene. HEATR1 is annotated to the Gene Ontology term “Ribosome biogenesis and assembly” (GO:0042254).
Table 2.
Characteristics of the Acute Respiratory Distress Syndrome–associated SNPs and Associated Gene Sets
| GWAS-associated Results |
ICSNPathway-associated Results |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −log10(Nominal GWAS P Value Threshold) | Chromosome (Coordinate) | Assayed SNP in GWAS | Reference/Alternative Alleles | GWAS Nominal P Value of Assayed SNP | Minor Allele Frequency (1000 Genomes) |
Functional Class/eQTL SNPs (Yes/No) | Associated Gene | Candidate Causal Pathway | FDR-corrected P Value of Candidate Causal Pathway Prioritized with ICSNPathway Method Given Various Distances Flanking the Gene of Interest |
||||
| African American | European | Within | 5 KB | 20 KB | 100 KB | ||||||||
| 4 | 1 (236,739,416) | rs2115740 | T/C | 6.53 × 10−5 | 0.4836 | 0.2992 | Intronic (yes) | HEATR1 | GO:0042254: ribosome biogenesis and assembly | 0.021 | 0.007 | 0.074 | 0.028 |
| 3 | 12 (109,017,898) | rs2228315 | T/C | 2.74 × 10−4 | 0.2951 | 0.06759 | Nonsynonymous coding, altering methionine to isoleucine (yes) | SELPLG | GO:0050901: leukocyte tethering or rolling | 0.01 | 0.01 | 0.004 | 0.006 |
| BIOCARTA pathway: granulocytes pathway | 0.069 | 0.182 | 0.013 | 0.024 | |||||||||
| 10 (71,176,066) | rs61732394 | T/C | 6.24 × 10−4 | 0.08197 | <0.0001 | Nonsynonymous coding, altering aspartic acid to glycine (no) | TACR2 | GO:0008528: peptide receptor activity, G protein coupled | 0.025 | 0.102 | 0.062 | 0.002 | |
| GO:0042277: peptide binding | 0.029 | 0.205 | 0.02 | 0.002 | |||||||||
| GO:0042923: neurotransmitter binding | 0.824 | 0.217 | 0.004 | 0.005 | |||||||||
| GO:0042923: neuropeptide binding | 0.183 | 0.205 | 0.011 | 0.002 | |||||||||
| GO:0008188: neuropeptide receptor activity | 0.073 | 0.048 | 0.011 | 0.022 | |||||||||
Definition of abbreviations: eQTL = expression quantitative trait loci; FDR = false discovery rate; GWAS = genome-wide association study; ICSNPathway = Identify Candidate Causal SNPs and Pathways.
To consider a greater number of SNPs, the GWAS nominal P value threshold was increased to less than 10−3, which led to identification of two additional nonsynonymous coding SNPs at ICSNPathway FDR less than 0.05 (Table 2). rs2228315 (GWAS nominal P = 2.7 × 10−4; OR = 1.88; 95% CI = 1.32–2.68) resides within the SELPLG and alters Methionine to Isoleucine. rs61732394 (GWAS nominal P = 6.2 × 10−4; OR = 3.01; 95% CI = 1.51–6.08) changes aspartic acid to glycine within tachykinin receptor 2 (TACR2). The SELPLG-prioritized pathway was Leukocyte Tethering or Rolling (GO:0050901, ICSNPathway FDR = 0.01; Table 2).
At various flanking distances of the three genes up to 100 kb, only the SELPLG-associated pathway, GO:0050901, remained significant (ICSNPathway FDR = 0.01, 0.004, and 0.006, respectively, at 5, 20, and 100 kb; Table 2).
Fine Mapping and Validation of SELPLG Gene Using the MESSI Cohort
To explore the role of SELPLG, we performed imputation of 200-kb region around the gene, and 1,855 SNPs were successfully imputed. The most significant SNP was rs2228315, which is located on the exon 3 (Figure E4). Because this SNP causes a missense mutation, we examined if this SNP and other missense mutations in the same exon were associated with ARDS in two independent datasets, African Americans in the discovery cohort and MESSI cohort. The MESSI cohort has 1,041 patients, 440 with ARDS and 601 sepsis control subjects. There were 362 patients of AA and 607 of EA (Tables E3–E5). Three nonsynonymous mutations were successfully genotyped or imputed in both studies. The rs2228315 minor allele T increases odds of ARDS in the discovery cohort (OR = 2.06; 95% CI = 1.42–2.99) and EAs from the MESSI (OR = 1.41; 95% CI = 0.89–2.24), but the association was not significant in the MESSI (Table 3). rs61729674 was associated with decreased risk of ARDS in both the discovery population and subjects of EA in the MESSI cohort (OR = 0.66 [95% CI = 0.46–0.96] and OR = 0.10 [95% CI = 0.01–0.75], respectively). rs73009 was also associated with decreased risk of ARDS in subjects of EA from the MESSI cohort (OR = 0.21; 95% CI = 0.05–0.96). In the meta-analysis of three nonsynonymous mutations in African Americans from the discovery population and the MESSI cohort, rs2228315 remained significant (OR = 1.38; 95% CI = 1.10–1.72).
Table 3.
Three Missense Mutations in Exon 3 on the SELPLG Gene
| SNP | Position | Allele | Residue Change | Discovery Population: AA (n = 394) |
MESSI Cohort |
Meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA (n = 362) |
EA (n = 607) |
|||||||||||||
| MAF | OR (95% CI) | P Value | MAF | OR (95% CI) | P Value | MAF | OR (95% CI) | P Value | OR (95% CI) | P Value | ||||
| rs7300972 | 109017264 | C/T | Met→Val | 0.24 | 0.76 (0.55–1.05) | 0.11 | 0.27 | 1.28 (0.91–1.79) | 0.43 | 0.01 | 0.21 (0.05–0.96) | 0.05 | 0.95 (0.75–1.20) | 0.65 |
| rs61729674 | 109017266 | A/G | Ser→Phe | 0.18 | 0.66 (0.46–0.96) | 0.03 | 0.19 | 1.12 (0.80–1.78) | 0.15 | 0.01 | 0.10 (0.01–0.74) | 0.02 | 0.86 (0.67–1.11) | 0.24 |
| rs2228315 | 109017898 | T/G | Met→Ile | 0.31 | 2.06 (1.42–2.99) | 1.5 × 10−4 | 0.27 | 0.97 (0.69–1.37) | 0.36 | 0.07 | 1.41 (0.89–2.24 | 0.14 | 1.38 (1.10–1.72) | 0.005 |
Definition of abbreviations: AA = African ancestry; CI = confidence interval; EA = European ancestry; Ile = isoleucine; MAF = minor allele frequency; MESSI = Molecular Epidemiology of Sepsis in the Intensive Care Unit; Met = methionine; OR = odds ratio; Phe = phenylalanine; SELPLG = selectin P ligand gene; Ser = serine; Val = valine.
Results of the discovery population are based on imputed data (allelic association was examined with logic regression adjusting for age, sex, sepsis, and three principal components using PLINK). For the MESSI cohort, the logistic regression was used assuming an additive genetic model and adjusting for one (European) or two (African) principal components.
Increased SELPLG Gene Expression in VILI and LPS-induced Mouse Models of Lung Injury
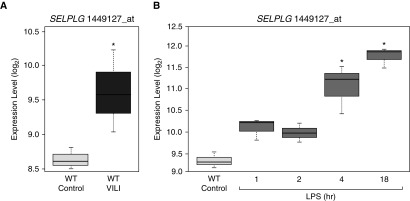
Preclinical validation of SELPLG and PSGL-1 in ARDS pathobiology was next assessed in preclinical mouse models of lung injury. Bioinformatic analysis of SELPLG gene expression in mouse lung tissues from VILI and LPS-induced lung injury preclinical models showed consistent and significant SELPLG upregulation (Figure 1). When compared with controls, VILI-exposed mice exhibited significantly higher SELPLG gene expression levels (Figure 1A, from GEO dataset GSE14525). Similarly, when compared with controls, LPS-induced lung injury resulted in significantly increased SELPLG gene expression, especially after 4 hours and 18 hours (Figure 1B, from GEO dataset GSE9314). Therefore, SELPLG expression was observed to be upregulated in both VILI- and LPS-exposed mouse lungs with a FDR less than 5%, respectively.
Figure 1.
Increased SELPLG (selectin P ligand) expression levels in ventilator-induced lung injury (VILI) and LPS models of preclinical murine lung injury. Affymetrix Mouse Genome 430 2.0 Arrays were used to profile whole-genome gene expression levels in both (A) VILI and (B) LPS models, including SELPLG. The robust multiarray average function in the “affy” package of Bioconductor was used to summarize the expression level of each probe set. SELPLG expression is markedly increased in both preclinical murine models of acute respiratory distress syndrome. Box-and-whisker plots show median (line inside the box), first and third quartiles (bottom and top of box), and 1.5 × interquartile range (whiskers). *P < 0.05 compared with control. WT = wild type.
Effect of PSGL-1 Inhibition on VILI and LPS-induced Lung Injury in Mouse Models
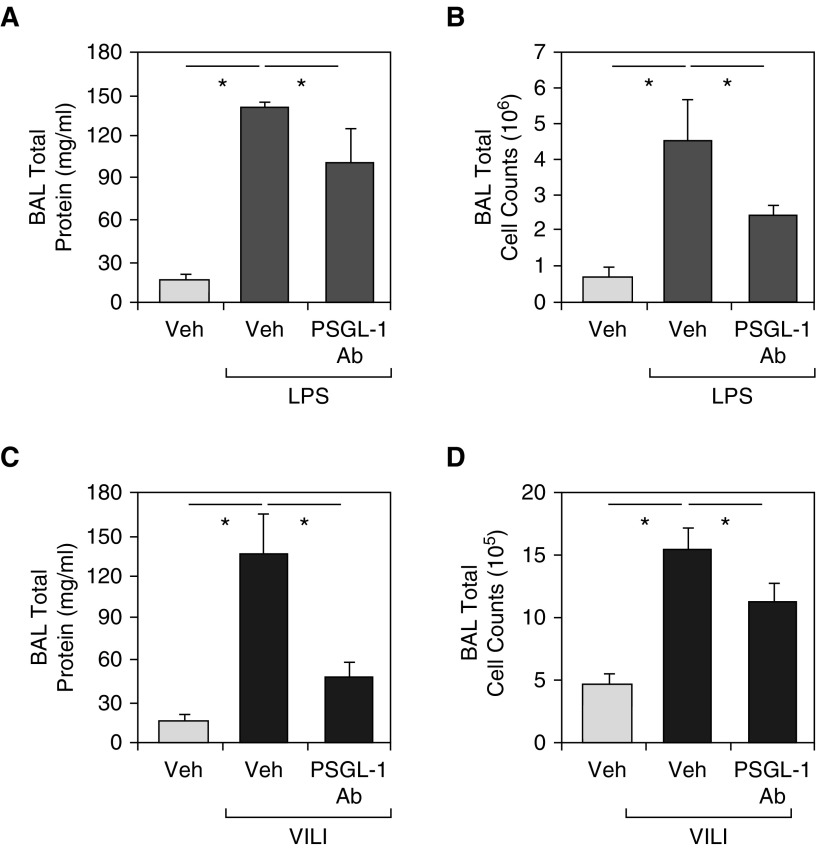
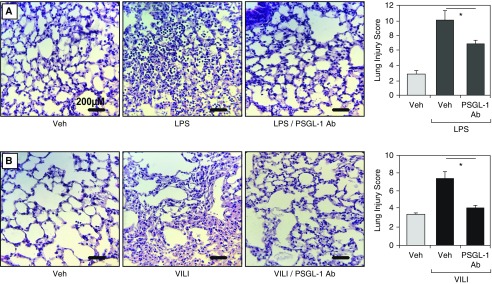
We next examined the therapeutic efficacy of PSGL-1 inhibition (via a neutralizing polyclonal antibody, CD162) on VILI and LPS-induced lung injury. The addition of the PSGL-1 antibody simultaneously with LPS challenge resulted in significant attenuation in LPS-induced BAL total protein increases (P < 0.01; Figure 2A) and BAL cell count increases (P < 0.01; Figure 2B). PSGL-1 also reduced LPS-induced BAL increase in polymorphonuclear cells (PMNs) by 10%. Similar lung injury protection with PSGL-1 antibody pretreatment was observed in VILI-exposed mice, with significant reductions in BAL total protein levels (P < 0.01; Figure 2C) and BAL total cell counts (8% reduction of PMNs; P < 0.01; Figure 2D). In both LPS-induced and VILI models, inflammatory cell number, thickness of alveolar walls in the lung tissues (H&E stained), and lung injury scores were decreased in the PSGL-1 antibody–pretreated group compared with the PBS-treated mice (Figures 3A and 3B).
Figure 2.
Effects of a neutralizing PSGL-1 (P-selectin glycoprotein ligand 1) antibody on ventilator-induced lung injury (VILI) and LPS-induced murine lung inflammation. When compared with spontaneously breathing control mice (Veh), exposure to VILI (40 ml/kg, 4 h) or LPS (1 mg/kg, 18 h) in C57/B6 mice produces robust increases in (A and C) total BAL protein levels and (B and D) total BAL cell counts. (A and B) In vivo PSGL-1 inhibition with a neutralizing antibody, CD162 (intravenous injection, 1 mg/kg) attenuates LPS-mediated lung injury. Compared with mice treated with phosphate-buffered saline vehicle, PSGL-1 antibody–pretreated mice had significant reductions in BAL total protein levels (A) and BAL total cell counts (B) when exposed to LPS-induced lung injury. (C and D) In vivo PSGL-1 inhibition attenuates VILI with significant reductions in BAL total protein levels (C) and BAL total cell counts (D). These studies confirm that PSGL-1 contributes to VILI and LPS-induced lung injury and pathobiology. Results are expressed as mean (±SEM); n = 3–5 per condition; *P < 0.01. Ab = antibody.
Figure 3.
Effects of a neutralizing PSGL-1 (P-selectin glycoprotein ligand 1) antibody on ventilator-induced lung injury (VILI) and LPS-induced histological indices of lung injury. (A) In vivo PSGL-1 inhibition with a neutralizing antibody (CD162, intravenous injection, 1 mg/kg) attenuates LPS-mediated lung injury. Compared with mice treated with phosphate-buffered saline (PBS) vehicle (Veh), PSGL-1 antibody–pretreated mice exhibited fewer inflammatory cells, reduced alveolar wall thickness, and significantly lower histology injury scores. (B) In vivo PSGL-1 inhibition with CD162 also attenuates ventilator-mediated lung injury, with fewer inflammatory cells, reduced alveolar wall thickness, and significantly lower lung injury scores compared with PBS vehicle mice. Results are expressed as mean (±SEM); n = 3–5 per condition; *P < 0.01. Ab = antibody.
Effect of LPS-induced Lung Injury in SELPLG Knockout Mice
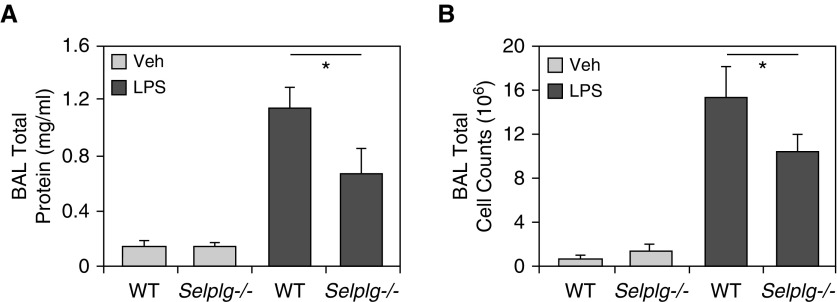
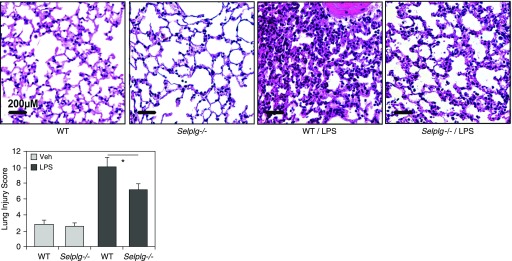
Mice with targeted SELPLG deletion, Selplg−/−, exhibited significantly less cellular inflammation in response to LPS-induced injury than wild-type C57/B6 mice (Figure 4). Compared with C57/B6 mice, Selplg−/− mice demonstrated significant reductions in BAL total protein levels (P < 0.05; Figure 4A) and BAL total cell counts (10% reduction of PMNs; P ≤ 0.05; Figure 4B) when exposed to LPS-induced lung injury. LPS produces robust increases in total BAL protein levels and total leukocyte counts in wild-type C57/B6 mice, which is significantly attenuated in Selplg−/− mice. Similarly, when compared with C57/B6 mice, Selplg−/− mice exhibited fewer inflammatory cells, less alveolar wall thickness (H&E stained), and significantly lower lung injury scores (Figure 5).
Figure 4.
LPS-induced lung injury is attenuated in Selplg−/− mice. Wild-type (WT) C57/B6 mice or Selplg−/− mice were challenged with LPS (1 mg/kg, intratracheal instillation, 18 h). LPS produces robust increases in (A) total BAL protein levels and (B) total leukocyte counts in WT C57/B6 mice. Both LPS-induced increases in BAL protein levels and total leukocyte counts were significantly attenuated in Selplg−/− mice. Results are expressed as mean (±SEM); n = 3 per condition; *P < 0.05. Selplg = selectin P ligand; Veh = vehicle.
Figure 5.
LPS-induced histological indices of lung injury are attenuated in Selplg−/− mice. LPS produces robust increases in number of inflammatory cells, thickness of alveolar walls in the lung tissues (hematoxylin and eosin stained), and histology scores in wild-type C57/B6 mice. LPS-induced increases in number of inflammatory cells, thickness of alveolar walls, and lung injury scores were significantly attenuated in Selplg−/− mice. Results are expressed as mean (±SEM); n = 3 per condition; *P < 0.05. Selplg = selectin P ligand; Veh = vehicle; WT = wild type.
Discussion
To our knowledge, this is the first report implicating SELPLG as a candidate gene involved in ARDS susceptibility in subjects with sepsis. To support our findings, we confirmed that: 1) SELPLG gene expression was significantly increased in VILI and LPS-induced mouse models of lung injury; 2) antibody neutralization of PSGL-1, the SELPLG-encoded protein, attenuated experimental VILI and LPS-induced lung injury; and 3) SELPLG−/− mice are protected against LPS-induced lung injury. We identified nonsynonymous coding variants in SELPLG that associate with differential ARDS risk in populations of varied ancestry and ARDS predisposition. Our group has previously used similar translational systems biology approaches to identify and validate several ARDS and VILI susceptibility candidate genes (17, 19, 24, 27, 29, 30, 57–59).
Although the genetic associations warrant further replication, the strong biological plausibility and preclinical validation strongly support an important role of SELPLG in ARDS susceptibility. The selectins are a versatile family of three calcium-dependent, type I transmembrane glycoproteins (E-selectin [CD62E], L-selectin [CD62L], and P-selectin [CD62P]) that mediate leukocyte tethering and rolling interactions with activated endothelial cells, an early and important part of the innate immune inflammatory response (33, 60–62). Among the many candidate ligands for selectins, only the PSGL-1, the SELPLG-encoded protein product, has been extensively characterized at the molecular, cellular, and functional levels (62). SELPLG is located on the long arm of chromosome 12 (12q24.110) and PSGL-1 spans 412 amino acids. The interaction between the selectins and their specific ligands demonstrates much redundancy in terms of ligand recognition and functional overlap. However, PSGL-1 is well recognized as the most important ligand for both L-selectin and P-selectin (62), with several studies supporting the critical role of PSGL-1 in mediating the early phase of neutrophil rolling onto the activated endothelium (63). Furthermore, PSGL-1–deficient mice demonstrate impaired leukocyte tethering to E-selectin (64). Our mechanistic validation supports a critical role of PSGL-1 in the acute inflammatory response, as mice pretreated with the PSGL-1–neutralizing antibody (CD162) exhibit significantly reduced lung inflammation and injury in response to LPS and VILI. Similarly, when compared with wild-type C57/B6 mice, SELPLG−/− demonstrated significantly attenuated LPS-induced lung inflammation and injury. These results support the finding that increased SELPLG expression and PSGL-1 activity may increase susceptibility to ARDS.
The lack of a genome-wide signal using a Bonferroni correction increases the potential for a false-positive result in our study. Because the effect size and significance of the GWAS signal is modest, we do not believe these SNPs will be useful as a genetic predictor of ARDS. However, our pathway prioritization strategy, gene expression profiles, and animal models provide strong evidence for the role of SELPLG in ARDS. The most significant contribution of our data is likely to be mechanistic insight into ARDS pathology rather than identification of genetic predictors of ARDS susceptibility. A general limitation of using conventional GWAS statistics is that they require large cohorts for studying traits of complex inheritance with small effect size. Such cohorts, though readily obtainable in some common disorders, are not available for uncommon, complex diseases, such as ARDS. Selecting the optimal analytical framework to discover the genetic inheritance of infrequent, complex disorders thus remains an open challenge. Over the last decade, pathway-level analytical methods have been shown in small cohorts to prioritize a signal that would otherwise be considered underpowered by conventional GWAS approaches (65). We have previously demonstrated in a historical rollback validation that SNPs could be unveiled by mining legacy-underpowered SNPs in GWAS cohorts jointly with protein interactions in adult-onset diabetes and systemic lupus (66). Here, we prioritized intragenic SNPs using external knowledge of pathways by the ICSN method (38), which was confirmed to discover overlooked SNPs in three schizophrenia GWASs (67), and to identify common inheritance between lupus and rheumatoid arthritis (68).
We did not observe a similar association between rs2228315 and ARDS in the MESSI population of either ancestry. In EA, there was a signal for increased ARDS risk that was not statistically significant, and this may represent a relative lack of power, given the lower allele frequency in EA compared with AA individuals. It is possible that the association identified in the discovery population is a false positive, although we believe that this is unlikely, given the statistical robustness with several sensitivity analyses and our animal data. We believe the observation of disparate results by race are due to the substantial differences in minor allele frequencies observed rather than different underlying genetic effects between race groups. Such inconsistent associations between race groups are common in complex disease association studies (65–68). Our observation of an inconsistent association among different race groups also suggests the limited use of these SNPs as a predictive biomarker. Furthermore, the identification of rare exonic, nonsynonymous SNPs in European MESSI subjects that also associate with ARDS risk supports the potential for SLEPLG to be an ARDS risk–modifying gene.
Another limitation is the lack of knowledge of the functional consequences of the SNPs identified to be associated with ARDS susceptibility. Interestingly, the observed protective role of the rs7300972 and rs61729674 could be due to a deleterious impact on the amino acid profile of SELPLG, which contrasts with the putative activating effect of rs2228315 (69). However, further evaluation of this speculation, which is in agreement with the observed decreased inflammatory responses in SELPLG knockout and PSGL-1 antibody–pretreated mice in our study demands further work. Our future studies will extend to identification of promoter and coding SNPs that underlie this association. Next, we will investigate the functional role of the exonic and promoter SNPs. For example, for the nonsynonymous coding SNP (rs2228315, Met62Ile), we hypothesize that the mutation from Met to Ile on position 62 might lead to a changed binding affinity of PSGL-1 to its receptor P-selectin, as Met62Ile is in close proximity to a key amino acid, Thr57, which determines PSGL-1 binding affinity (70). The altered lipophilicity of Met/Ile might lead to a change on the localized structure near Thr57, which could impose a key functional outcome on P-selectin binding and neutrophil rolling.
In summary, our genome-wide genetic approaches, novel bioinformatic strategies, and preclinical validation studies have identified SELPLG as a novel ARDS-susceptibility gene and a potentially novel therapeutic target in ARDS pathobiology.
Footnotes
Supported by NIH/NHLBI grants P01HL126609 (J.G.N.G.), R01HL91889 (J.G.N.G.), and HL122474 (N.J.M.); R01HL060710 (D.C.C.); and National Library of Medicine (NLM) grant K22LM008308 (D.C.C.).
Author Contributions: Conception and design of the work, the analysis and interpretation of data for the work, the drafting and revision of the manuscript, and approval of final version to be published—C.B., N.P., T.W., Y.A.L., and J.G.N.G.; conception and design of the work, the analysis and interpretation of data for the work, critical revision of key intellectual content, and approval of final version to be published—S.S., K.B., N.C., T.Z., C.L.K., X.S., and R.A.K.; conception and design of the work, drafting and revision of the manuscript, and approval of final version to be published—S.M.C.; collection and analysis of data, revision of the manuscript, and approval of the final version to be published—T.K.J., J.P.R., and N.J.M.; collection of data and assistance with processing and manuscript revision—J.D.C., D.C.C., C.R.Y., J.H.K., M.G.-G., M.M.W., and G.U.M.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201705-0961OC on February 9, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Rubenfeld GD, Herridge MS. Epidemiology and outcomes of acute lung injury. Chest. 2007;131:554–562. doi: 10.1378/chest.06-1976. [DOI] [PubMed] [Google Scholar]
- 2.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, et al. Incidence and outcomes of acute lung injury. N Engl J Med. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 3.Zambon M, Vincent JL. Mortality rates for patients with acute lung injury/ARDS have decreased over time. Chest. 2008;133:1120–1127. doi: 10.1378/chest.07-2134. [DOI] [PubMed] [Google Scholar]
- 4.Erickson SE, Martin GS, Davis JL, Matthay MA, Eisner MD NIH NHLBI ARDS Network. Recent trends in acute lung injury mortality: 1996–2005. Crit Care Med. 2009;37:1574–1579. doi: 10.1097/CCM.0b013e31819fefdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brower RG, Rubenfeld GD. Lung-protective ventilation strategies in acute lung injury. Crit Care Med. 2003;31(4 S) uppl:S312–S316. doi: 10.1097/01.CCM.0000057909.18362.F6. [DOI] [PubMed] [Google Scholar]
- 6.Guérin C, Reignier J, Richard J-C, Beuret P, Gacouin A, Boulain T, et al. PROSEVA Study Group. Prone positioning in severe acute respiratory distress syndrome. N Engl J Med. 2013;368:2159–2168. doi: 10.1056/NEJMoa1214103. [DOI] [PubMed] [Google Scholar]
- 7.Papazian L, Forel J-M, Gacouin A, Penot-Ragon C, Perrin G, Loundou A, et al. ACURASYS Study Investigators. Neuromuscular blockers in early acute respiratory distress syndrome. N Engl J Med. 2010;363:1107–1116. doi: 10.1056/NEJMoa1005372. [DOI] [PubMed] [Google Scholar]
- 8.Afshari A, Brok J, Møller AM, Wetterslev J. Inhaled nitric oxide for acute respiratory distress syndrome (ARDS) and acute lung injury in children and adults. Cochrane Database Syst Rev. 2010;(7):CD002787. doi: 10.1002/14651858.CD002787.pub2. [DOI] [PubMed] [Google Scholar]
- 9.Peek GJ, Mugford M, Tiruvoipati R, Wilson A, Allen E, Thalanany MM, et al. CESAR trial collaboration. Efficacy and economic assessment of conventional ventilatory support versus extracorporeal membrane oxygenation for severe adult respiratory failure (CESAR): a multicentre randomised controlled trial. Lancet. 2009;374:1351–1363. doi: 10.1016/S0140-6736(09)61069-2. [DOI] [PubMed] [Google Scholar]
- 10.Wiedemann HP, Wheeler AP, Bernard GR, Thompson BT, Hayden D, deBoisblanc B, et al. National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network. Comparison of two fluid-management strategies in acute lung injury. N Engl J Med. 2006;354:2564–2575. doi: 10.1056/NEJMoa062200. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfuss D, Saumon G. Ventilator-induced lung injury: lessons from experimental studies. Am J Respir Crit Care Med. 1998;157:294–323. doi: 10.1164/ajrccm.157.1.9604014. [DOI] [PubMed] [Google Scholar]
- 12.Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979–1996) Crit Care Med. 2002;30:1679–1685. doi: 10.1097/00003246-200208000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Cooke CR, Erickson SE, Eisner MD, Martin GS. Trends in the incidence of noncardiogenic acute respiratory failure: the role of race. Crit Care Med. 2012;40:1532–1538. doi: 10.1097/CCM.0b013e31824518f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson SE, Shlipak MG, Martin GS, Wheeler AP, Ancukiewicz M, Matthay MA, et al. National Institutes of Health National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome Network. Racial and ethnic disparities in mortality from acute lung injury. Crit Care Med. 2009;37:1–6. doi: 10.1097/CCM.0b013e31819292ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemos-Filho LB, Mikkelsen ME, Martin GS, Dabbagh O, Adesanya A, Gentile N, et al. US Critical Illness and Injury Trials Group: Lung Injury Prevention Study Investigators (USCIITG-LIPS) Sex, race, and the development of acute lung injury. Chest. 2013;143:901–909. doi: 10.1378/chest.12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown LM, Kallet RH, Matthay MA, Dicker RA. The influence of race on the development of acute lung injury in trauma patients. Am J Surg. 2011;201:486–491. doi: 10.1016/j.amjsurg.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, et al. Pre–B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005;171:361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 18.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, et al. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L468–L477. doi: 10.1152/ajplung.00109.2005. [DOI] [PubMed] [Google Scholar]
- 19.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006;34:487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc. 2005;2:188–194. doi: 10.1513/pats.200501-005AC. [DOI] [PubMed] [Google Scholar]
- 21.Flores C, Ma SF, Maresso K, Ahmed O, Garcia JG. Genomics of acute lung injury. Semin Respir Crit Care Med. 2006;27:389–395. doi: 10.1055/s-2006-948292. [DOI] [PubMed] [Google Scholar]
- 22.Gao L, Flores C, Fan-Ma S, Miller EJ, Moitra J, Moreno L, et al. Macrophage migration inhibitory factor in acute lung injury: expression, biomarker, and associations. Transl Res. 2007;150:18–29. doi: 10.1016/j.trsl.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamp R, Sun X, Garcia JG. Making genomics functional: deciphering the genetics of acute lung injury. Proc Am Thorac Soc. 2008;5:348–353. doi: 10.1513/pats.200709-152DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Elangovan VR, Mapes B, Camp SM, Sammani S, Saadat L, et al. The NAMPT promoter is regulated by mechanical stress, signal transducer and activator of transcription 5, and acute respiratory distress syndrome–associated genetic variants. Am J Respir Cell Mol Biol. 2014;51:660–667. doi: 10.1165/rcmb.2014-0117OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Ma SF, Wade MS, Acosta-Herrera M, Villar J, Pino-Yanes M, et al. Functional promoter variants in sphingosine 1-phosphate receptor 3 associate with susceptibility to sepsis-associated acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol. 2013;305:L467–L477. doi: 10.1152/ajplung.00010.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascarenhas JB, Tchourbanov AY, Fan H, Danilov SM, Wang T, Garcia JG. Mechanical stress and single nucleotide variants regulate alternative splicing of the MYLK gene. Am J Respir Cell Mol Biol. 2017;56:29–37. doi: 10.1165/rcmb.2016-0053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathew B, Jacobson JR, Siegler JH, Moitra J, Blasco M, Xie L, et al. Role of migratory inhibition factor in age-related susceptibility to radiation lung injury via NF-E2–related factor-2 and antioxidant regulation. Am J Respir Cell Mol Biol. 2013;49:269–278. doi: 10.1165/rcmb.2012-0291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia JG, Sznajder JI. Healthcare disparities in patients with acute respiratory distress syndrome: toward equity. Am J Respir Crit Care Med. 2013;188:631–632. doi: 10.1164/rccm.201307-1394ED. [DOI] [PubMed] [Google Scholar]
- 29.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, et al. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 30.Meyer NJ, Huang Y, Singleton PA, Sammani S, Moitra J, Evenoski CL, et al. GADD45a is a novel candidate gene in inflammatory lung injury via influences on Akt signaling. FASEB J. 2009;23:1325–1337. doi: 10.1096/fj.08-119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Furie BC, Furie B. The biology of P-selectin glycoprotein ligand-1: its role as a selectin counterreceptor in leukocyte–endothelial and leukocyte–platelet interaction. Thromb Haemost. 1999;81:1–7. [PubMed] [Google Scholar]
- 32.Donnelly SC, Haslett C, Dransfield I, Robertson CE, Carter DC, Ross JA, et al. Role of selectins in development of adult respiratory distress syndrome. Lancet. 1994;344:215–219. doi: 10.1016/s0140-6736(94)92995-5. [DOI] [PubMed] [Google Scholar]
- 33.Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. 2007;11:1473–1491. doi: 10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bime C, Pouladi N, Batai K, Zhou T, Kempf C, Sun X, et al. GWAS association of the SELPLG gene with acute respiratory distress syndrome susceptibility in African-Americans: preclinical validation. [abstract] Am J Respir Crit Care Med. 2017;195:A4784. [Google Scholar]
- 35.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, et al. The American-European Consensus Conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 36.Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, et al. ARDS Definition Task Force. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 37.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, et al. International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K, Chang S, Cui S, Guo L, Zhang L, Wang J. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Res. 2011;39:W437–W443. doi: 10.1093/nar/gkr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reilly JP, Anderson BJ, Mangalmurti NS, Nguyen TD, Holena DN, Wu Q, et al. The ABO histo-blood group and AKI in critically ill patients with trauma or sepsis. Clin J Am Soc Nephrol. 2015;10:1911–1920. doi: 10.2215/CJN.12201214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reilly JP, Meyer NJ, Shashaty MG, Feng R, Lanken PN, Gallop R, et al. ABO blood type A is associated with increased risk of acute respiratory distress syndrome in Caucasians following both major trauma and severe sepsis Chest 2014145753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, et al. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20:864–874. [Google Scholar]
- 46.Gajic O, Dabbagh O, Park PK, Adesanya A, Chang SY, Hou P, et al. U.S. Critical Illness and Injury Trials Group: Lung Injury Prevention Study Investigators (USCIITG-LIPS) Early identification of patients at risk of acute lung injury: evaluation of lung injury prediction score in a multicenter cohort study. Am J Respir Crit Care Med. 2011;183:462–470. doi: 10.1164/rccm.201004-0549OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah CV, Lanken PN, Localio AR, Gallop R, Bellamy S, Ma SF, et al. An alternative method of acute lung injury classification for use in observational studies. Chest. 2010;138:1054–1061. doi: 10.1378/chest.09-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer NJ, Feng R, Li M, Zhao Y, Sheu CC, Tejera P, et al. IL1RN coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL-1 receptor antagonist. Am J Respir Crit Care Med. 2013;187:950–959. doi: 10.1164/rccm.201208-1501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li YR, van Setten J, Verma SS, Lu Y, Holmes MV, Gao H, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med. 2015;7:90. doi: 10.1186/s13073-015-0211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 51.McCall MN, Almudevar A. Affymetrix GeneChip microarray preprocessing for multivariate analyses. Brief Bioinform. 2012;13:536–546. doi: 10.1093/bib/bbr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor J, Tibshirani R, Efron B. The ‘miss rate’ for the analysis of gene expression data. Biostatistics. 2005;6:111–117. doi: 10.1093/biostatistics/kxh021. [DOI] [PubMed] [Google Scholar]
- 54.Sammani S, Moreno-Vinasco L, Mirzapoiazova T, Singleton PA, Chiang ET, Evenoski CL, et al. Differential effects of sphingosine 1-phosphate receptors on airway and vascular barrier function in the murine lung. Am J Respir Cell Mol Biol. 2010;43:394–402. doi: 10.1165/rcmb.2009-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mirzapoiazova T, Moitra J, Moreno-Vinasco L, Sammani S, Turner JR, Chiang ET, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol. 2011;44:40–52. doi: 10.1165/rcmb.2009-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camp SM, Ceco E, Evenoski CL, Danilov SM, Zhou T, Chiang ET, et al. Unique Toll-like receptor 4 activation by NAMPT/PBEF induces NFκB signaling and inflammatory lung injury. Sci Rep. 2015;5:13135. doi: 10.1038/srep13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia JG. Genomic investigations into acute inflammatory lung injury. Proc Am Thorac Soc. 2011;8:167–172. doi: 10.1513/pats.201101-002MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kansas GS. Selectins and their ligands: current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 61.Kyriakides C, Favuzza J, Wang Y, Austen WG, Jr, Moore FD, Jr, Hechtman HB. Recombinant soluble P-selectin glycoprotein ligand 1 moderates local and remote injuries following experimental lower-torso ischaemia. Br J Surg. 2001;88:825–830. doi: 10.1046/j.0007-1323.2001.01795.x. [DOI] [PubMed] [Google Scholar]
- 62.Ley K. The role of selectins in inflammation and disease. Trends Mol Med. 2003;9:263–268. doi: 10.1016/s1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Hirata T, Croce K, Merrill-Skoloff G, Tchernychev B, Williams E, et al. Targeted gene disruption demonstrates that P-selectin glycoprotein ligand 1 (PSGL-1) is required for P-selectin–mediated but not E-selectin–mediated neutrophil rolling and migration. J Exp Med. 1999;190:1769–1782. doi: 10.1084/jem.190.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia L, Sperandio M, Yago T, McDaniel JM, Cummings RD, Pearson-White S, et al. P-selectin glycoprotein ligand-1–deficient mice have impaired leukocyte tethering to E-selectin under flow. J Clin Invest. 2002;109:939–950. doi: 10.1172/JCI14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30:390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee Y, Li H, Li J, Rebman E, Achour I, Regan KE, et al. Network models of genome-wide association studies uncover the topological centrality of protein interactions in complex diseases. J Am Med Inform Assoc. 2013;20:619–629. doi: 10.1136/amiajnl-2012-001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernández-Cuervo H, Fanous AH, et al. Molecular Genetics of Schizophrenia Consortium. Uncovering the hidden risk architecture of the schizophrenias: confirmation in three independent genome-wide association studies. Am J Psychiatry. 2015;172:139–153. doi: 10.1176/appi.ajp.2014.14040435. [DOI] [PubMed] [Google Scholar]
- 68.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39:10627–10635. doi: 10.1007/s11033-012-1952-x. [DOI] [PubMed] [Google Scholar]
- 69.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 70.Bernimoulin MP, Zeng XL, Abbal C, Giraud S, Martinez M, Michielin O, et al. Molecular basis of leukocyte rolling on PSGL-1: predominant role of core-2 O-glycans and of tyrosine sulfate residue 51. J Biol Chem. 2003;278:37–47. doi: 10.1074/jbc.M204360200. [DOI] [PubMed] [Google Scholar]