Abstract
Acquired point mutations of pre-mRNA splicing factors recur among cancers, leukemias, and related neoplasms. Several studies have established that somatic mutations of a U2AF1 subunit, which normally recognizes 3′ splice site junctions, recur among myelodysplastic syndromes. The U2AF2 splicing factor recognizes polypyrimidine signals that precede most 3′ splice sites as a heterodimer with U2AF1. In contrast with the well-studied U2AF1 subunit, descriptions of cancer-relevant U2AF2 mutations and their structural relationships are currently lacking. Here, we survey databases of cancer-associated mutations and identify recurring missense mutations in the U2AF2 gene. We determine ultrahigh structures of the U2AF2 RNA recognition motifs (RRM1 and RRM2) at 1.1 Å resolutions and map the structural locations of the mutated U2AF2 residues. Comparison with prior, lower resolution structures of the tandem U2AF2 RRMs in the RNA-bound and apo-states reveals clusters of cancer-associated mutations at either the U2AF2 RRM–RNA or apo-RRM1–RRM2 interfaces. Although the role of U2AF2 mutations in malignant transformation remains uncertain, our results show that cancer-associated mutations correlate with functionally-important surfaces of the U2AF2 splicing factor.
Graphical abstract
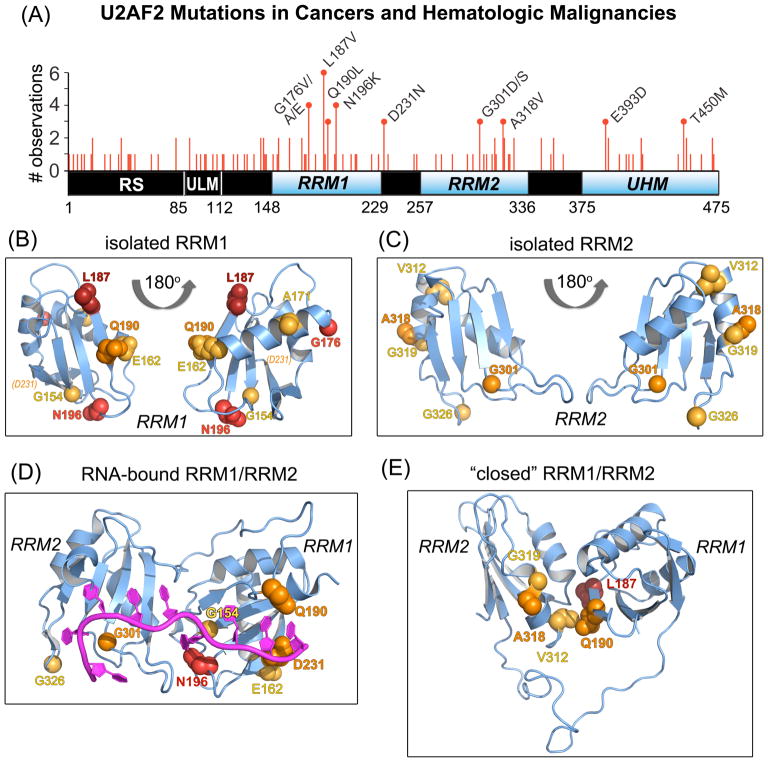
Next-generation sequencing of hematologic malignancies discovered recurrent somatic mutations of genes encoding pre-mRNA splicing factors U2AF1, ZRSR2, SRSF2, or SF3B1 (reviewed in 1). In an essential early step of splicing, U2AF1 recognizes consensus 3′ splice site signals as a heterodimer with a larger subunit, U2AF22. Whole-exome sequencing of myelodysplasias also identified somatic mutations in U2AF23, albeit at lower frequency. To explore the extent of U2AF2 mutations among a range of human malignancies, we queried The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), and Catalogue of Somatic Mutations in Cancer (COSMIC) using the cBi-oportal4, 5 and COSMIC interfaces6 (Supporting Methods). Most catalogued U2AF2 defects were missense mutations (~70%) and 22% of the U2AF2 residues were affected by at least one mutation (Table S1). Nine U2AF2 amino acid changes recurred in at least three patient samples. The recurrent U2AF2 mutations were concentrated in the globular domains (RRM1, RRM2, and UHM; two-sided p-value 0.01 from Fisher’s exact test) (Figure 1A). In particular, an L187V amino acid change in RRM1 was observed in six patient samples (p <0.01), all of which have hematologic malignancies including acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML) and myelodysplastic syndrome (MDS). We also identified “recurring” U2AF2 mutations (observed in three or more patient samples) in lung adenocarcinoma (e.g. G176V/E and Q190L in RRM1), as for U2AF1 mutations7, as well as in cancers of the liver, breast, and colon, among other malignancies (Table S1).
Figure 1.
U2AF2 missense mutations in neoplasms mapped on RRM structures. (A) Non-redundant U2AF2 missense mutations in cancer genomes mapped on the protein domains. Line heights are proportional to the number of independent observations for each mutation summed among independent patient samples. Amino acids that are changed in three or more samples are labeled. RNA recognition motifs (RRM1 and RRM2) are blue and other domains black. Residue numbers of domain boundaries are given below the schematic diagram. RS, arginine-serine-rich; ULM, U2AF ligand motif; UHM, U2AF homology motif. (B–C) Crystal structures of U2AF2 RRM1 (B) and RRM2 (C) at 1.07 and 1.11 Å resolutions. Views are rotated 180° about the y-axis. An affected residue (D231) located beyond the C-terminus of the crystallized construct (P229) is indicated in italicized parentheses. (D) The RNA-bound, “open” U2AF2 conformation (PDB ID 5EV4). (E) A typical “closed” U2AF2 conformation in the absence of RNA (PDB ID 2YH0). For clarity, a single representative of the NMR/PRE-fitted ensemble is shown in (E). Affected residues in (B–E) are space-filling atoms colored by the number of non-redundant mutations: 5, firebrick; 4, red; 3, orange; 2, yellow-orange.
Atomic resolution structures provide three-dimensional maps of mutated residues and could potentially aide drug design. Two central RNA recognition motifs of U2AF2 (RRM1 and RRM2) recognize a polypyrimidine (Py) tract preceding the 3′ splice site8. A C-terminal U2AF2 homology motif (UHM) concurrently recognizes an upstream branch point sequence as a complex with Splicing Factor-1 (SF1)9, and the U2AF2 N-terminus positions U2AF1 to recognize a downstream “AG” dinucleotide10, 11. Subsequently, U2AF2 recruits SF3B1 to the assembling spliceosome and dissociates prior to the first catalytic step of splicing12.
Major U2AF2 conformations in the absence of RNA include “closed” and “open” structures that have been characterized by NMR, small-angle X-ray scattering, and single molecule Förster resonance energy transfer13–16. The “open” U2AF2 conformation is selectively stabilized by RNA binding and has been characterized at high resolution by X-ray crystallography15. Despite structures of the RNA-bound U2AF2 RRM1/RRM2 at up to 1.50 Å resolutions15, 17, 18, the apo-RRM1 at 1.47 Å resolution19, and NMR/PRE-based models of the linked RRMs13, to date ultra-high resolution structures of the U2AF2 RRMs are lacking. Indeed, only three prior RRM structures are known at <1.1 Å resolutions: splicing factor hnRNP A1, nuclear transport protein Nup53, or transcription terminator Seb1 (PDB IDs 1L3K, 5HB7, 5MDU).
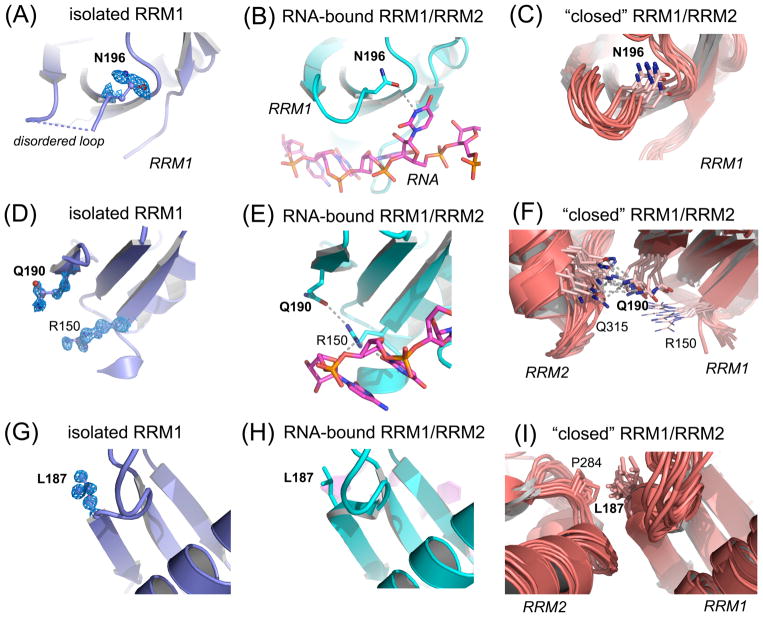
To address this knowledge gap, we determined crystal structures of U2AF2 RRM1 and RRM2 at 1.1 Å resolutions (Figure 1B–C, Supporting Methods, Table S2). The overall conformations of the separate U2AF2 RRMs are similar in the presence and absence of RNA (RMSD 0.34/0.44 Å for 64/51 Cα-atoms of RNA-bound RRM1/RRM2 in PDB ID 5EV4). RNA-binding alters the side chain conformers and displaces ordered water molecules from the RRM surfaces (Figure S1–S2). The RRM folds of the NMR ensemble also match the crystal structures (RMSD 0.69/0.73 Å for 72/59 Cα-atoms of apo-RRM1/RRM2 in PDB ID 2YH0), consistent with starting models for NMR/PRE refinement13 derived from a U2AF2 crystal structure18. The new structures show the recurrently mutated RRM residues at atomic resolution (e.g. Figure 2A,D,G). Three hotspot residues (G176, G301, A318) remain similar among all structures. In contrast, the Q190 and N196 hotspots change conformation to bind RNA (Figure 2A–F). As discussed below, the NMR model further suggests that RRM1 Q190 and L187 contribute inter-RRM contacts of the “closed” U2AF2 conformation (Figure 2F, G–I).
Figure 2.
U2AF2 hotspot residues show local structural changes between the isolated RRM1 (slate) and the linked, tandem U2AF2 RRMs, either a crystal structure bound to a Py tract oligonucleotide (cyan protein and magenta oligonucleotide, PDB ID 5EV4) or an NMR/PRE-based model in the apo-state (salmon, PDB ID 2YH0). Structures were superimposed by matching Cα-atoms and are shown from identical viewpoints. Feature enhanced maps (FEM) contoured at 1σ level are shown for new RRM1 structure. (A) N196 is poorly ordered and precedes a disordered loop of the isolated RRM1 structure (dashed blue line). (B) This loop folds for RNA binding; N196 forms a well-defined hydrogen bond (dashed gray line) with the rU6 base. (C) N196 is qualitatively similar in the “closed” NMR model. (D) Q190 is well-defined in the FEM map of high resolution apo-RRM1 despite lack of contacts. (E) In the oligonucleotide-bound structure, Q190 switches conformation to accept a hydrogen bond from the RNA-bound R150. (F) RRM1 Q190 mediates a hydrogen bond with RRM2 Q315 in the NMR model of the tandem U2AF2 RRMs in the “closed” conformation. (G) The FEM electron density for L187 of the isolated RRM1 structure is well-defined and remains similar in the RNA-bound state (H). The L187 position differs in the “closed” U2AF2 model (I), in which it makes a hydrophobic contact with P284 in RRM2.
We next comprehensively mapped the recurrent, cancer-associated mutations on the U2AF2 structures (Figure 1B–E, Figure S3). Nearly all U2AF2 hotspots localize at RNA and protein interfaces. Most often, the cancer-associated U2AF2 mutations recur at the RRM1 interface with nucleotides in the 3′ half of the Py tract (Figure 1D, Figure 2B,E). An N196 residue that is mutated to lysine in four AML patients recognizes the sixth uracil base (U6 of PDB ID 5EV4) (Figure 2B). A Q190 residue that is mutated to leucine in two CLL samples and one lung adenocarcinoma, interacts with the terminal nucleotide of the bound Py tract (C9) (Figure 2E). The C9 base also is recognized by D231, which is mutated to asparagine in three cancer samples, including squamous cell carcinoma, pediatric high-grade glioma, and Wilms’ tumor. Mutations at the RNA interface of RRM2, which drives U2AF2–RNA binding17, are less common than in RRM1. A glycine in RRM2 (G301) is mutated to aspartate in patients with colorectal or prostate carcinomas or to serine in a papillary renal cell carcinoma, and stacks with the first bound uridine (U1). Such U2AF2 mutations may subtly alter its RNA binding, by analogy with the molecular consequences of the more prevalent U2AF1 mutations20–24. Accordingly, a G301I mutation reduces U2AF2–Py tract binding affinity by five-fold18.
The most frequently mutated U2AF2 residue, L187 in RRM1, is solvent-exposed in the “open”, RNA-bound conformation of U2AF2. In contrast, the “closed” conformation of U2AF2 in the absence of RNA13, 16 buries L187 at the interface of the two RRMs (Figure 1E, Figure 2I). Likewise, Q190 in RRM1 and A318 in RRM2 which is mutated to valine in two colon cancer samples and one stomach cancer, are embedded near L187 at the “closed” U2AF2 RRM1/RRM2 interface (Figure 1E, Figure 2F). In particular, the buried hydrophobic surface area of the Q190L mutation is expected to subtly alter the equilibrium between the “closed” (unbound) and “open” (RNA binding-competent) U2AF2 conformations. Since the “closed” apo-conformation partially obscures the RNA binding surface of U2AF2 compared to the “open” RNA-bound U2AF2, mutation-induced differences in the conformational ensemble of U2AF2 would in turn influence its association with splice site RNAs and possibly other downstream effectors.
Outside of the RRMs, both of two recurrent mutations of the U2AF2 UHM (T450M and E393D) affect residues at its SF1 interface25, 26 (Figure S3). U2AF2 T450, which is replaced by methionine in two cases of liver cancer and one of bile duct cancer, packs against an SF1 linker connecting the U2AF ligand motif (ULM) and a coiled-coil domain. T450 is strongly predicted as a kinase substrate by the NetPhos server27. Accordingly, phosphorylation regulates the SF1 ULM and coiled-coil regions28–30. At a distinct SF1–U2AF2 interface (Figure S3), a U2AF2 E393 residue is mutated to aspartate in AML, MDS, and colon cancer samples. Although the charge of E393D remains negative, E393 mutations can disrupt SF1 association31.
None of the U2AF2 hotspots fall within known interfaces with U2AF110, 32. However, the U2AF2 RRM1 abuts the U2AF1 binding site. It remains possible that cancer-associated mutations of U2AF2, or conversely the more common mutations of U2AF1, could influence the opposing subunit. Already, U2AF2 binding the U2AF1 UHM has been shown to promote the “open” as opposed to “closed” U2AF2 conformation16. The consequences of U2AF2 (or U2AF1) mutations remain to be tested in the context of the U2AF1 zinc knuckles, which are hotspots of MDS mutations3, 33.
As we find for U2AF2, the MDS-relevant mutational hotspots of SRSF2, SF3b1, and U2AF1 localize at RNA-binding interfaces of these proteins (reviewed in 34). Future studies are needed to resolve the potential roles of recurrent U2AF2 mutations in the transformation of normal cells to cancers. In the interim, we conclude that recurrent mutations in hematologic malignancies mark key molecular interfaces for gene expression.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by grants from the NIH (R01 GM070503) and the Edward P. Evans Foundation to C.L.K. Data generated by the TCGA Research Network: http://cancergenome.nih.gov/ contributed to the results reported here. CHESS is supported by NSF award DMR-0936384 and NIH/NIGMS award GM103485.
We thank S. Henderson and A. Patel for contributions to RRM1 structural refinement and RRM2 crystallization.
ABBREVIATIONS
- AML
acute myeloid leukemia
- CMML
chronic myelomonocytic leukemia
- MDS
myelodysplastic syndrome
- Py
polypyrimidine
- PRE
paramagnetic resonance enhancement
- RRM
RNA recognition motif
- snRNP
small nuclear ribonucleoprotein
- UHM
U2AF homology motif
- ULM
U2AF ligand motif
Footnotes
Accession Codes
The coordinates for the RRM1 and RRM2 structures are available as Protein Data Bank entries 5W0G and 5W0H.
The Supporting Information is available free of charge on the ACS Publications website.
Detailed materials and methods, tables of U2AF2 mutations and crystallographic statistics, electron density showing side chain conformations and ordered water molecules (PDF)
References
- 1.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamore PD, Green MR. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 4.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamore PD, Patton JG, Green MR. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 9.Abovich N, Rosbash M. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 10.Kielkopf CL, Rodionova NA, Green MR, Burley SK. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida H, Park SY, Oda T, Akiyoshi T, Sato M, Shirouzu M, Tsuda K, Kuwasako K, Unzai S, Muto Y, Urano T, Obayashi E. Genes Dev. 2015;29:1649–1660. doi: 10.1101/gad.267104.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett M, Michaud S, Kingston J, Reed R. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 13.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins JL, Laird KM, Kielkopf CL. Biochemistry. 2012;51:5223–5225. doi: 10.1021/bi300277t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal AA, Salsi E, Chatrikhi R, Henderson S, Jenkins JL, Green MR, Ermolenko DN, Kielkopf CL. Nat Commun. 2016;7:10950. doi: 10.1038/ncomms10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voith von Voithenberg L, Sanchez-Rico C, Kang HS, Madl T, Zanier K, Barth A, Warner LR, Sattler M, Lamb DC. Proc Natl Acad Sci U S A. 2016;113:E7169–E7175. doi: 10.1073/pnas.1605873113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins JL, Agrawal AA, Gupta A, Green MR, Kielkopf CL. Nucleic Acids Res. 2013;41:3859–3873. doi: 10.1093/nar/gkt046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thickman KR, Sickmier EA, Kielkopf CL. J Mol Biol. 2007;366:703–710. doi: 10.1016/j.jmb.2006.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, Makishima H. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, Lawrence MS, DeLuca DS, Getz G, Meyerson M. PLoS One. 2014;9:e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, Kielkopf CL, Ley TJ, Walter MJ, Graubert TA. Leukemia. 2015;29:909–917. doi: 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fei DL, Motowski H, Chatrikhi R, Prasad S, Yu J, Gao S, Kielkopf CL, Bradley RK, Varmus H. PLoS Genet. 2016;12:e1006384. doi: 10.1371/journal.pgen.1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. Genome Res. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Maucuer A, Gupta A, Manceau V, Thickman KR, Bauer WJ, Kennedy SD, Wedekind JE, Green MR, Kielkopf CL. Structure. 2013;21:197–208. doi: 10.1016/j.str.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Madl T, Bagdiul I, Kern T, Kang HS, Zou P, Mausbacher N, Sieber SA, Kramer A, Sattler M. Nucleic Acids Res. 2013;41:1343–1354. doi: 10.1093/nar/gks1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blom N, Gammeltoft S, Brunak S. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, Robinson PJ. EMBO J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manceau V, Swenson MC, Le Caer JP, Sobel A, Kielkopf CL, Maucuer A. FEBS J. 2006;273:577–587. doi: 10.1111/j.1742-4658.2005.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatrikhi R, Wang W, Gupta A, Loerch S, Maucuer A, Kielkopf CL. Biophys J. 2016;111:2570–2586. doi: 10.1016/j.bpj.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Mol Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H, Park SY, Oda T, Akiyoshi T, Sato M, Shirouzu M, Tsuda K, Kuwasako K, Unzai S, Muto Y, Urano T, Obayashi E. Genes Dev. 2015;29:1649–1660. doi: 10.1101/gad.267104.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, McLellan MD, Dooling DJ, Abbott RM, Fulton RS, Schmidt H, Kalicki-Veizer J, O’Laughlin M, Grillot M, Baty J, Heath S, Frater JL, Nasim T, Link DC, Tomasson MH, Westervelt P, DiPersio JF, Mardis ER, Ley TJ, Wilson RK, Walter MJ. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins JL, Kielkopf CL. Trends Genet. 2017;33:336–348. doi: 10.1016/j.tig.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.