Abstract
The vacuolar (H+)-ATPases (V-ATPases) are ATP-dependent proton pumps that acidify intracellular compartments and pump protons across specialized plasma membranes. Proton translocation occurs through the integral V0 domain, which contains five different subunits (a, d, c, c′, and c"). Proton transport is critically dependent on buried acidic residues present in three different proteolipid subunits (c, c′, and c"). Mutations in the 100-kDa subunit a have also influenced activity, but none of these residues has proven to be required absolutely for proton transport. On the basis of previous observations on the F-ATPases, we have investigated the role of two highly conserved arginine residues present in the last two putative transmembrane segments of the yeast V-ATPase a subunit (Vph1p). Substitution of Asn, Glu, or Gln for Arg-735 in TM8 gives a V-ATPase that is fully assembled but is totally devoid of proton transport and ATPase activity. Replacement of Arg-735 by Lys gives a V-ATPase that, although completely inactive for proton transport, retains 24% of wild-type ATPase activity, suggesting a partial uncoupling of proton transport and ATP hydrolysis in this mutant. By contrast, nonconservative mutations of Arg-799 in TM9 lead to both defective assembly of the V-ATPase complex and decreases in activity of the assembled V-ATPase. These results suggest that Arg-735 is absolutely required for proton transport by the V-ATPases and is discussed in the context of a revised model of the topology of the 100-kDa subunit a.
The vacuolar (H+)-ATPases are a family of ATP-dependent proton pumps that are responsible for acidification of a wide variety of intracellular compartments, including endosomes, lysosomes, clathrin-coated vesicles, secretory vesicles, such as synaptic vesicles and chromaffin granules, and the central vacuoles of plants and fungi (1–8). Acidification of these compartments is in turn critical for many cellular processes, including receptor-mediated endocytosis, intracellular targeting, coupled transport of small molecules and ions, and viral entry into cells. V-ATPases in the plasma membrane of such cells as renal intercalated cells, osteoclasts, macrophages, and insect goblet cells also play a role in renal acidification, bone resorption, pH homeostasis, and coupled K+ transport, respectively (9–12).
V-ATPases are composed of two domains (1–8). The peripheral V1 domain, which is responsible for ATP hydrolysis, is composed of nine subunits (subunits A–H) of molecular mass 70–14 kDa. The integral V0 domain, which is responsible for proton translocation, contains five subunits (subunits a, d, c, c′, and c") of molecular mass 100–17 kDa. The V-ATPases are thus structurally similar to the F-ATPases (or ATP synthases) of mitochondria, chloroplasts, and bacteria, which usually function in ATP synthesis (13–18).
The F-ATPases are believed to operate by a rotary mechanism in which ATP hydrolysis in the F1 domain drives rotation of a central γ subunit that connects F1 and F0 (19–21). The γ subunit (together with the ɛ subunit) is in turn linked to a ring of ten proteolipid c subunits in the F0 domain (22–24), such that rotation of γ causes this ring of c subunits to rotate relative to subunit a (also part of the F0 domain; ref. 25). It is this movement of c subunits relative to subunit a that is thought to be directly responsible for proton translocation (14, 15, 26, 27). In particular, a buried carboxyl group near the center of each c subunit is thought to interact transiently with a critical arginine residue (Arg-210) in the penultimate transmembrane helix of subunit a (14, 26, 28, 29).
Unlike the F-ATPases, the V-ATPases contain three different proteolipid subunits (subunits c, c′, and c"), which are homologous both to each other and to the F-ATPase c subunit (30, 31). Subunits c and c′ of the V-ATPase both contain four putative transmembrane helices with a critical acidic residue present in TM4, whereas subunit c" contains five transmembrane helices with the essential glutamic acid residue present to TM3 (31). By contrast, subunit c of the F-ATPases contains just two transmembrane helices, with a critical aspartate residue near the middle of the C-terminal helix (14). The F0 domain contains ten copies of subunit c (22, 32), whereas the V0 domain contains five to six copies of subunits c/c′ and a single copy of subunit c" (33, 34). The difference in the number of buried acidic residues may account for the difference in proton/ATP stoichiometry between the V- and F-ATPases (35).
Although the V-ATPases do not contain a subunit that is obviously homologous to subunit a of the F-ATPases, we have proposed that the 100-kDa subunit of the V-ATPases plays a similar role in proton transport on the basis of mutagenesis studies of the yeast V-ATPase 100-kDa subunit Vph1p (36, 37). Thus, mutagenesis of several buried charged residues near the C terminus of the 100-kDa subunit (including Glu-789) significantly reduce ATP-dependent proton transport by the resultant complex, although none of the residues identified thus far is absolutely essential for proton translocation (36, 37). To determine whether an arginine residue analogous in function to Arg-210 of the F-ATPase a subunit exists in the C-terminal domain of the V-ATPase 100-kDa subunit, site-directed mutagenesis has been performed on two highly conserved arginine residues in the last two putative transmembrane segments of Vph1p. The results have identified Arg-735 as the first 100-kDa subunit residue essential for proton transport.
Experimental Procedures
Materials and Strains.
Zymolyase 100T was obtained from Seikagaku America (Rockville, MD). Concanamycin A was purchased from Fluka. Protease inhibitors were from Roche Molecular Biochemicals. The mouse monoclonal antibody 8B1-F3 against the yeast V-ATPase A subunit and the mouse monoclonal antibody 10D7 against the 100-kDa subunit a were from Molecular Probes. Escherichia coli and yeast culture media were purchased from Difco. Restriction endonucleases, T4 DNA ligase, and other molecular biology reagents were from GIBCO, Promega, and New England Biolabs. ATP, phenylmethylsulfonylfluoride, and most other chemicals were purchased from Sigma. Yeast strain MM112 (MATα Δvph1∷LEU2 Δstv1∷LYS2 his3-(Δ200 leu2 lys2 ura3–52) and plasmid MM322 (VPH1 in pRS316; ref. 38) were used to generate and study the Vph1p mutants. Yeast cells were grown in yeast extract-peptone-dextrose medium or synthetic dropout medium (39).
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed on EcoRI-BamHI fragments of the wild-type VPH1 cDNA plasmid with Altered Sites II in Vitro Mutagenesis Systems (Promega) according to the manufacturer's protocol. Mutations were confirmed by DNA sequencing. Fragments containing the indicated mutations were then substituted back into the vector containing the wild-type VPH1 cDNA. No other mutations were detected in the final product.
Transformation and Selection.
Yeast cells (MM112) lacking functional endogenous Vph1p and Stv1p were transformed by use of the lithium acetate method (40) with the wild-type pRS316-VPH1 as a positive control and the pRS316 vector alone as a negative control. The transformants were selected on uracil minus (Ura−) plates as described (41). Growth phenotypes of the mutants were assessed on yeast extract-peptone-dextrose plates buffered with 50 mM KH2PO4 or 50 mM succinic acid to either pH 7.5 or 5.5.
Detection of the Expression of the 100-kDa Subunit with Various Mutations.
Whole-cell lysates were prepared from overnight cultures in synthetic dropout Ura− medium as described (42), and the proteins were separated by SDS/PAGE on 8% acrylamide gels. The expression of 100-kDa subunit a was detected by Western blotting, using the monoclonal antibody 10D7 against Vph1p (Molecular Probes).
Isolation of Vacuolar Membrane Vesicles and Assessment of V-ATPase Assembly.
Vacuolar membrane vesicles were isolated as described (42). Protein concentrations were measured by the bicinchoninic acid (BCA) protein assay (Pierce). Assembly of the V-ATPase was assessed by measurement of the amount of subunit A relative to the amount of subunit a present on the vacuolar membrane vesicle with the mouse monoclonal antibodies 8B1-F3 against the Vma1p or 10D7 against the Vph1p (both antibodies were obtained from Molecular Probes), followed by horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Blots were developed by using a chemiluminescent detection method obtained from Kirkegaard & Perry Laboratories. Quantitations were done with an IS-1000 digital imaging system (Alpha Innotech, San Leandro, CA).
Detection of V-ATPase Subunits Present on Isolated Vacuolar Membranes.
Vacuolar membranes isolated from each of the mutants were subjected to SDS/PAGE, and Western blots were probed with the mouse monoclonal antibodies 10D7 against the Vph1, 8B1-F3 against the Vma1p, and 13D11-B2 against Vma2p (Molecular Probes). Blots were also probed with rabbit polyclonal antibodies against Vma6p, Vma7p, Vma8p, Vma10p, and Vma13p (all provided by T. Stevens, Univ. of Oregon, Eugene), Vma4p (a gift of D. Klionsky, Univ. of Michigan, Ann Arbor), and Vma5p (a gift of P. Kane, SUNY Upstate Medical Univ., Syracuse). After removal of unbound primary antibodies by washing, blots were incubated with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and developed by using a chemiluminescent detection method obtained from Kirkegaard & Perry Laboratories.
Other Procedures.
ATPase activities were measured by using a coupled spectrophotometric assay (43) with the modification of using 0.35 mM NADH instead of 0.5 mM NADH. ATP-dependent proton transport was measured by fluorescence quenching with the fluorescence probe 9-amino-6-chloro-2-methoxyacridine in transport buffer (25 mM Mes/Tris, pH 7.2/5 mM MgCl2/0.5 mM ATP) as described (44) in the presence or absence of 1 μM concanamycin A, a specific inhibitor of the V-ATPase (45). SDS/PAGE was performed as described by Laemmli (46).
Results
Previous studies of the F-ATPase a subunit have suggested an important role for many charged amino acid residues in the last two transmembrane segments, including Arg-210 and Glu-219 (29, 47–49). Mutagenesis studies of the V-ATPase have similarly implicated several charged residues in the last two transmembrane segments of the 100-kDa subunit a, including Glu-789 in TM9 (36, 37). However, no residue that plays a comparably important role to Arg-210 has been identified. Fig. 1 shows the alignment of amino acid sequences of the last two transmembrane segments of two isoforms of the yeast 100-kDa subunit a (Vph1p and Stv1p), together with three subunit a isoforms identified in mouse (38, 50, 51). Apparently, there are only two arginine residues in this region that are conserved in all a subunit sequences, namely Arg-735 and Arg-799. Arg-735 is predicted to reside near the amino terminus of TM8, whereas Arg-799 is predicted to be near the carboxyl terminus of TM9. There is some sequence similarity between the region containing Glu-789 and Arg-799 of the V-ATPase a subunit and the helix containing Glu-219 and Arg-210 of the F-ATPase a subunit (Fig. 1b). Nevertheless, an evolutionary relationship between these sequences is ruled out because, in the alignment shown, these regions have the opposite orientation (N to C).
Figure 1.
Alignment of the last two transmembrane helices of the 100-kDa subunit a. (a) Alignment of a subunit sequences from yeast (Vph1p, Stv1p) and mouse (a1, a2, a3). Identical residues are indicated by shaded boxes. Vph1p residues mutated in this study (Arg-735, Arg-762, and Arg-799) are indicated by arrowheads. (b) The region of the F-ATPase a subunit that includes the functionally important residues Arg-210, Asn-214, and Glu-219 is aligned with the sequence of Vph1p that includes residues Glu-789 and Arg-799. Note that the orientation of the two sequences is reversed.
To address the role of these residues in V-ATPase function, site-directed mutagenesis was performed on the VPH1 gene encoding one isoform of the a subunit in yeast. The mutant proteins were then expressed in a yeast strain disrupted in both the VPH1 and STV1 genes, as described (36, 37). Arg-735 was changed to Lys, Leu, Ala, Asn, Glu, Gln, and Cys, whereas Arg-799 was changed to Lys, Leu, and Ala. As a control, mutations of an additional arginine residue (Arg-762) that is predicted to reside in the loop between TM8 and TM9, but that is not conserved between species, were also constructed, including R762K, R762L, and R762A.
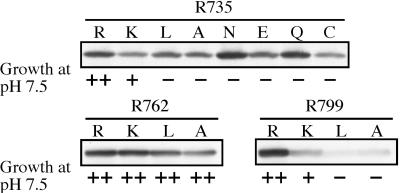
We first determined the effect of each Vph1p mutation on the growth phenotype of the strain expressing the mutant form of the protein. Disruption of V-ATPase function has led to a conditional growth phenotype (vma−) in which cells are able to grow in media buffered to pH 5.5 but not in media buffered to pH 7.5 (52). In general, a mutation must lead to a loss of greater than 80% of wild-type activity to display a vma− phenotype (53, 54). Fig. 2 shows that all of the mutations at Arg-735 and Arg-799 except R735K and R799K cause defective growth at neutral pH, with these latter two mutations showing a partial growth phenotype. By contrast, the mutations at Arg-762 showed normal growth at pH 7.5. These results suggest that mutations at Arg-735 and Arg-799 lead to defective V-ATPase function whereas those at Arg-762 do not.
Figure 2.
Effect of arginine mutations on stability of Vph1p. Whole-cell lysates prepared from cells expressing wild-type or mutant forms of Vph1p were subjected to SDS/PAGE followed by Western blot analysis with the monoclonal antibody 10D7. Also shown are the growth phenotypes of cells transformed with each of the indicated plasmids on yeast extract-peptone-dextrose plates buffered to pH 7.5.
To test whether the mutations introduced caused destabilization of the 100-kDa subunit a, Western blot analysis was performed on whole-cell lysates by using the monoclonal antibody 10D7 specific for Vph1p. As seen in Fig. 2, the mutants at Arg-735 and Arg-762 generally showed levels of Vph1p similar to that observed for the wild type, although some of the mutants (such as R735C and R762A) showed slight reductions in the amount of Vph1p present in whole-cell lysates. By contrast, mutations at Arg-799 caused a dramatic decrease in Vph1p staining, with only the R799K mutation showing significant levels of Vph1p. These results indicate that, of the 13 mutations tested, only R799L and R799A mutations dramatically reduced stability of Vph1p.
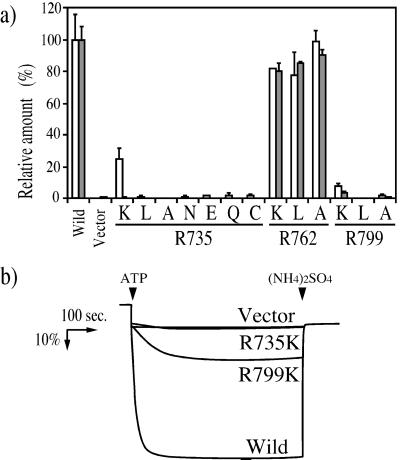
One way in which mutations in Vph1p can lead to loss of V-ATPase function is through disruption of assembly of the complex. To assess the assembly competence of each of the mutants, vacuoles were isolated and Western blotting was performed by using antibodies against subunit A (Vma1p) and subunit a (Vph1p). Defective assembly appears as a reduction in the amount of subunit A present on the vacuolar membrane (55). Fig. 3 Top shows the results of a representative experiment, and Fig. 3 Bottom shows quantitation of the levels of subunit a (open bars) and subunit A (closed bars) for two to three independent vacuole preparations. Subunit A was present at greater than 60% of wild-type levels in the R735K, R735N, R735E, and R735Q mutants, as well as all of the Arg-762 mutants, whereas significantly reduced levels of subunit A were present in the vacuoles from the R735A, R735C, and R799K mutants. By contrast, vacuoles from the R735L, R799L, and R799A mutants were almost completely devoid of subunit A, indicating a dramatic defect in assembly of the V-ATPase in these mutants.
Figure 3.
Effect of arginine mutations in Vph1p on the presence of Vph1p and subunit A on the vacuolar membrane. Vacuolar membranes (1 μg protein) isolated from cells expressing wild-type or mutant forms of Vph1p were subjected to SDS/PAGE followed by Western blot analysis with antibodies against Vph1p and Vma1p. Quantitation of the data were performed by densitometric analysis and the results expressed as the amount of Vph1p or Vma1p present relative to that observed for the wild-type Vph1p. Each bar represents the average of three measurements of two or three independent vacuolar membrane preparations, with the error corresponding to the standard deviation.
We next measured the effects of each of the mutations in Vph1p on the proton transport and concanamycin-sensitive ATPase activity in isolated vacuoles. Fig. 4a shows that all of the mutations at Arg-735 led to a complete loss of proton transport activity and a generally parallel loss of ATPase activity, although the R735 K mutant retained 24% ± 7% of wild-type ATPase activity, suggesting a significant uncoupling of proton transport and ATP hydrolysis in this mutant. Mutations at Arg-799 also led to a significant loss of proton transport and ATPase activity in isolated vacuoles; although, as discussed above, both R799L and R799A showed defective assembly of the complex. The R799K mutant retained 7.5% ± 1.9% of ATPase activity and 3.5% ± 0.8% of proton transport activity. Fig. 4b shows the ATP-dependent 9-amino-6-chloro-2-methoxyacridine quenching observed for vacuoles isolated from cells expressing the wild-type Vph1p, the R799K and R735K mutant proteins, and the vector alone (negative control). Thus, the R735K is completely inactive for proton transport, whereas the R799K mutant retains low but significant proton transport activity. By contrast, all of the mutations at Arg-762 showed nearly wild-type levels of both proton transport and ATPase activity. These results suggest that Arg-735 is absolutely essential for proton transport, whereas Arg-799 may play an important, if not essential, role in activity of the V-ATPase.
Figure 4.
Effect of arginine mutations in Vph1p on ATPase activity and proton transport by the V-ATPase. (a) Vacuolar membranes (3 μg protein) were isolated from cells expressing wild-type or mutant forms of Vph1p, and ATPase activities were measured in the absence or presence of 1 μM concanamycin A as described under Experimental Procedures. Open bars represent the concanamycin-sensitive portion of the ATPase activity. The specific activity of the concanamycin-sensitive ATPase activity of vacuoles containing the wild-typeVph1p is 0.72 μmol/min/mg protein. Approximately 90% of the ATPase activity of wild-type vacuoles was sensitive to concanamycin. ATP-dependent proton transport (solid bars) was measured by using 1 μg protein as the initial rate of ATP-dependent fluorescence quenching as described in Experimental Procedures. ATPase activity and proton transport are expressed relative to wild-type vacuoles (100%), with each value the average of the three measurements on two or three independent vacuole preparations (error bars correspond to the standard deviations). (b) ATP-dependent fluorescence quenching observed by using 3 μg of vacuolar membrane vesicles isolated from cells expressing wild-type Vph1p, vector alone, or the R735K or R799K mutants. ATP (1 mM) to start the reaction or ammonium sulfate (5 mM) to dissipate the proton gradient was added at the arrows.
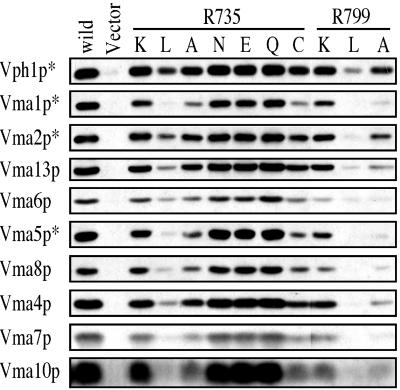
To evaluate further the effect of mutations at Arg-735 and Arg-799 on assembly and stability of the V-ATPase complex, isolated vacuoles were separated by SDS/PAGE and probed by Western blot by using antibodies against all of the V1 subunits (subunits A–H), as well as subunits a and d of the V0 domain. As seen in Fig. 5, the R735K, R735N, R735E, and R735Q mutants all showed normal assembly of the V-ATPase on the vacuolar membrane, whereas the R735A, R735C, and R799K mutants all showed partial loss of V1 and/or V0 subunits from the vacuoles. By contrast, vacuoles from the R735L, R799L, and R799A mutants all showed nearly complete loss of V-ATPase subunits, indicating severely defective assembly. These data are in good agreement with the results shown in Fig. 3. We had reported that the R735K mutant was defective in assembly (58); however, the procedure used to isolate vacuoles in that study differed somewhat from that used in this study (42). Moreover, the assembly competence of the R735K mutant was confirmed by immunoprecipitation of a fully assembled V-ATPase complex by using a monoclonal antibody directed against subunit A (data not shown).
Figure 5.
Effect of Vph1p mutations at Arg-735 and Arg-799 on assembly of the V-ATPase as assessed by Western blot analysis of vacuolar membranes. Vacuolar membranes isolated from cells expressing the wild-type or mutant forms of Vph1p were separated by SDS/PAGE followed by Western blotting, using the subunit-specific antibodies as described in Experimental Procedures. For subunits marked with an asterisk, 1 μg of protein was used; otherwise 10 μg protein was used.
Discussion
Mutagenesis studies have identified several charged residues in the last two transmembrane helices of the F-ATPase a subunit that seem to play some role in proton transport, including Arg-210, Glu-219, and His-245 (29, 47–49). Arg-210 was shown to be absolutely required for ATP-driven proton transport, with even the conservative substitution R210K being completely inactive (29, 48). By contrast, although certain mutations at Glu-219 and His-245 are inactive [such as E219L and H245Y (47, 56)], substitutions at both positions are more tolerated, with mutations such as E219G having more than 50% of wild-type activity (48). These results suggest that, although Arg-210 plays a critical role in ATP-dependent proton transport, Glu-219 and His-245 are in a position to influence proton transport, but are not absolutely essential.
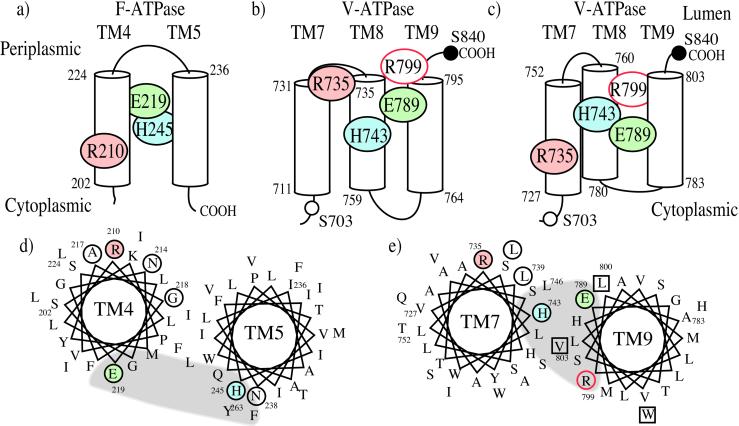
Both Arg-210 and Glu-219 are present in TM4 of the a subunit, and crosslinking studies suggest that the face of TM4 containing Arg-210 is oriented toward the second transmembrane helix of subunit c containing the critical aspartate residue (Asp-61; ref. 57). Arg-210 has been suggested to interact directly with Asp-61 as part of the proton transport cycle (14, 26, 28, 29). A helical structure between Arg-210 and Glu-219 would place Glu-219 on the opposite face of the helix and closer to the extracellular surface of the membrane, as shown in Fig. 6. His-245, which resides on TM5, has been proposed to interact with Glu-219 in TM4, and both residues may contribute to an aqueous access channel that allows protons to gain access to Asp-61 (14, 26, 28, 47, 49).
Figure 6.
Model of the transmembrane topology of the carboxyl-terminal helices of the F-ATPase and V-ATPase a subunits. (a) Proposed transmembrane topology of TM4 and TM5 of the F-ATPase a subunit (14, 59, 60). Putative transmembrane regions are shown as cylinders, with the bordering residue numbers indicated. Colored circles indicate residues either critical for proton transport (Arg-210; refs. 29, 48) or able to influence proton transport (Glu-219 and His-245; refs. 47–49, 56). (b) Location of residues in carboxyl-terminal region of the V-ATPase a subunit that are critical for proton transport (Arg-735; this study) or that influence proton transport [Glu-789, His-735 (36, 37), and Arg-799 (this study)], based on the previously proposed topology of Vph1p, in which Ser-703 (open circle) has a cytoplasmic orientation whereas Ser-840 (solid circle) is luminal (58). (c) Location of important residues in the carboxyl-terminal region of the V-ATPase a subunit by using a revised membrane topology of Vph1p. (d) Proposed helical wheel diagram for TM4 and TM5 of the F-ATPase a subunit (14, 28). Note that Arg-210 and Glu-219 are on opposite sides of the membrane, and that His-245 in TM5 is oriented to interact with Glu-219 on TM4. Arg-210 is available to interact with the carboxyl groups on the ring of c subunits. Other residues whose mutation leads to loss of activity (29) are shown by the open circles. The location of a putative access channel leading to the luminal surface of the protein (28) is indicated by the shaded area. (e) Proposed helical wheel diagram for TM7 and TM9 of the V-ATPase a subunit based on the topological model shown in c. The critical arginine residue is again oriented to interact with the ring of c subunits, and the histidine and glutamic acid residues are shown interacting with each other. The putative access channel (shaded area) is shown closer to the critical arginine residue than in d.
There is no obvious sequence homology between the F-ATPase subunit a and the 100-kDa subunit of the V-ATPase, and these two proteins are structurally quite different. Thus, unlike the F-ATPase a subunit, the 100-kDa subunit possesses a large amino-terminal domain on the cytoplasmic side of the membrane (58). Moreover, topographical studies (58) suggest that the 100-kDa subunit contains nine transmembrane helices instead of the five observed for the F-ATPase a subunit (59, 60). Nevertheless, mutagenesis studies have identified several charged residues in the C-terminal region of the molecule that seem to play some role in ATP-dependent proton transport (36, 37), including Glu-789 (predicted to reside in TM9) and His-743 (predicted to reside in TM8). As for the corresponding residues in the F-ATPase a subunit, neither of these residues is absolutely required for proton transport because even nonconservative substitutions (such as alanine) give 40–60% of wild-type levels of activity. However, several mutations at both Glu-789 and His-743 showed an altered pH profile for activity (36, 37), suggesting that these residues may be in a position to influence proton translocation.
In this study we have investigated the role of two arginine residues in proton transport by the V-ATPases: Arg-735 and Arg-799. Of these two arginine residues, only Arg-735 is absolutely essential for proton transport, with even the conservative R735K mutant being completely inactive in ATP-dependent proton transport. This mutant retained ≈24% of wild-type levels of ATPase activity, suggesting that this mutation leads to uncoupling of proton transport and ATP hydrolysis. The only other V-ATPase mutants displaying such an uncoupled phenotype are those isolated in subunit D (Vma8p; ref. 61), which has been postulated to function as the homologue to the γ subunit in the V-ATPases (62). If Arg-735 interacts with the critical glutamic acid residues on the proteolipid subunits, mutation to lysine may allow movement of the protonated carboxyl groups on subunit c past this point of interaction so that proton transport is no longer coupled to ATP hydrolysis.
In our topographical model of the V-ATPase a subunit (58), Arg-735 would be predicted to reside at the amino-terminal end of TM8, placing it near the luminal surface of the protein (see Fig. 6b). Although this arrangement would place His-743 and Glu-789 at a comparable depth from the luminal surface to Glu-219 and His-245 of the F0 a subunit (Fig. 6a), this would not allow a direct interaction between Arg-735 and the proteolipid carboxyl groups, which are predicted to be located near the center of the membrane (14, 30). An alternate arrangement of helices (shown in Fig. 6c) would place Arg-735 in a position to interact with the c subunit carboxyl groups and still allow Glu-789 and His-743 to interact. In this model, Arg-735 and His-743 still reside on the same helix (TM7), whereas Glu-789 resides on TM9. Both models shown in Fig. 6 b and c are consistent with the available topological data, which indicates that Ser-703 has a cytoplasmic orientation, whereas Ser-840 has a luminal orientation (58). The models differ, however, in the placement of the three putative transmembrane helices (TM7–9). The model in Fig. 6b arranges these helices to optimize overlap with the most hydrophobic residues (711–795; see figure 5 of ref. 58), whereas in Fig. 6c, TM7 begins at residue 727, placing the weakly hydrophobic segment (711) in the cytoplasm. In addition, Fig. 6c includes residues 796–803 (which are also weakly hydrophobic) in TM9. As seen from the helical wheel diagram shown in Fig. 6e, a helical structure between Arg-735 and His-743 would place these residues about 90° apart on TM7. If Glu-789 and His-743 form part of a water-filled channel, such a channel may be somewhat closer to the critical arginine residue for the V-ATPase (Fig. 6e) than for the F-ATPase (Fig. 6d). One possible consequence of this closer proximity of the channel to the critical arginine residue for the V-ATPase may be that the c subunit carboxyl group that is in communication with the luminal surface of the protein spends more of its time interacting with the positively charged arginine residue on subunit a. This increased proximity may prevent access of protons to this carboxyl group from the luminal surface that is necessary for passive proton conductance. In fact, the V0 domain differs from the F0 domain in that V0 does not usually act as a passive proton channel (63).
The other residue suggested to play some role in activity from this study is Arg-799. In the modified topography of subunit a shown in Fig. 6c, Arg-799 is predicted to be located near the luminal surface of TM9. It is conceivable that this residue, like His-743 and Glu-789, might contribute to the aqueous channel that leads to the luminal surface of the protein. An additional arginine residue (Arg-140 on TM3) has also been suggested to contribute to the F-ATPase proton channel from the periplasmic side of the membrane (28). Further work is required to determine the exact topography of the C-terminal region of the a subunit and the packing of the a subunit helices both relative to each other and to the helices of the proteolipid subunits.
Acknowledgments
This work was supported by National Institutes of Health Grant GM34478 (to M.F.) and Charles A. King Trust Fellowship Award (to T.N.).
Abbreviation
- V-ATPase
vacuolar proton-translocating adenosine triphosphatase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stevens T H, Forgac M. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 2.Forgac M. J Biol Chem. 1999;274:12951–12954. doi: 10.1074/jbc.274.19.12951. [DOI] [PubMed] [Google Scholar]
- 3.Bowman E J, Bowman B J. J Exp Biol. 2000;203:97–106. doi: 10.1242/jeb.203.1.97. [DOI] [PubMed] [Google Scholar]
- 4.Kane P M, Parra K J. J Exp Biol. 2000;203:81–87. doi: 10.1242/jeb.203.1.81. [DOI] [PubMed] [Google Scholar]
- 5.Graham L A, Powell B, Stevens T H. J Exp Biol. 2000;203:61–70. doi: 10.1242/jeb.203.1.61. [DOI] [PubMed] [Google Scholar]
- 6.Futai M, Oka T, Sun-Wada G H, Moriyama Y, Kanazawa H, Wada Y. J Exp Biol. 2000;203:107–116. doi: 10.1242/jeb.203.1.107. [DOI] [PubMed] [Google Scholar]
- 7.Nelson N, Perzov N, Cohen A, Padler V, Nelson H. J Exp Biol. 2000;203:89–95. doi: 10.1242/jeb.203.1.89. [DOI] [PubMed] [Google Scholar]
- 8.Sze H, Li X, Palmgren M G. Plant Cell. 1999;4:677–690. doi: 10.1105/tpc.11.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D, Breton S. J Exp Biol. 2000;203:137–145. doi: 10.1242/jeb.203.1.137. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee D, Chakraborty M, Leit M, Neff L, Jamsa-Kellokumpu S, Fuchs R, Baron R. Proc Natl Acad Sci USA. 1992;89:6257–6261. doi: 10.1073/pnas.89.14.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swallow C J, Grinstein S, Rotstein O D. J Biol Chem. 1990;265:7645–7654. [PubMed] [Google Scholar]
- 12.Wieczorek H, Gruber G, Harvey W R, Huss M, Merzendorfer H, Zeiske W. J Exp Biol. 2000;203:127–135. doi: 10.1242/jeb.203.1.127. [DOI] [PubMed] [Google Scholar]
- 13.Weber J, Senior A E. Biochim Biophys Acta. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 14.Fillingame R H, Jiang W, Dmitriev O Y. J Exp Biol. 2000;203:9–17. doi: 10.1242/jeb.203.1.9. [DOI] [PubMed] [Google Scholar]
- 15.Cross R L, Duncan T M. J Bioenerg Biomembr. 1996;28:403–408. doi: 10.1007/BF02113981. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen P L. J Bioenerg Biomembr. 1996;28:389–395. doi: 10.1007/BF02113979. [DOI] [PubMed] [Google Scholar]
- 17.Capaldi R A, Aggeler R, Wilkens S, Gruber G. J Bioenerg Biomembr. 1996;28:397–401. doi: 10.1007/BF02113980. [DOI] [PubMed] [Google Scholar]
- 18.Futai M, Omote H. J Bioenerg Biomembr. 1996;28:409–414. doi: 10.1007/BF02113982. [DOI] [PubMed] [Google Scholar]
- 19.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabbert D, Engelbrecht S, Junge W. Nature (London) 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 21.Noji H, Yasuda R, Yoshida M, Kazuhiko K. Nature (London) 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 22.Stock D, Lelie A G, Walker J E. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Fillingame R H. J Biol Chem. 1995;270:24609–24614. [PubMed] [Google Scholar]
- 24.Schulenberg B, Aggeler R, Murray J, Capaldi R A. J Biol Chem. 1999;274:34233–34237. doi: 10.1074/jbc.274.48.34233. [DOI] [PubMed] [Google Scholar]
- 25.Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Yanagida T, Wada Y, Futai M. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 26.Vik S B, Antonio B J. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 27.Junge W, Sabbert D, Engelbrecht S. Ber Bunsen-Ges Phys Chem. 1996;100:2014–2019. [Google Scholar]
- 28.Vik S B, Long J C, Wada T, Zhang D. Biochim Biophys Acta. 2000;1458:457–466. doi: 10.1016/s0005-2728(00)00094-3. [DOI] [PubMed] [Google Scholar]
- 29.Cain B D, Simoni R D. J Biol Chem. 1989;264:3292–3300. [PubMed] [Google Scholar]
- 30.Mandel M, Moriyama Y, Hulmes J D, Pan Y C, Nelson H, Nelson N. Proc Natl Acad Sci USA. 1988;85:5521–5524. doi: 10.1073/pnas.85.15.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirata R, Graham L A, Takatsuki A, Stevens T H, Anraku Y. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Hermolin J, Fillingame R H. Proc Natl Acad Sci USA. 2001;98:4966–4971. doi: 10.1073/pnas.081424898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arai H, Terres G, Pink S, Forgac M. J Biol Chem. 1988;263:8796–8802. [PubMed] [Google Scholar]
- 34.Powell B, Graham L A, Stevens T H. J Biol Chem. 2000;275:23654–23660. doi: 10.1074/jbc.M004440200. [DOI] [PubMed] [Google Scholar]
- 35.Cross R L, Taiz L. FEBS Lett. 1990;259:227–229. doi: 10.1016/0014-5793(90)80014-a. [DOI] [PubMed] [Google Scholar]
- 36.Leng X H, Manolson M, Liu Q, Forgac M. J Biol Chem. 1996;271:22487–22493. doi: 10.1074/jbc.271.37.22487. [DOI] [PubMed] [Google Scholar]
- 37.Leng X H, Manolson M, Forgac M. J Biol Chem. 1998;273:6717–6723. doi: 10.1074/jbc.273.12.6717. [DOI] [PubMed] [Google Scholar]
- 38.Manolson M F, Wu B, Proteau D, Taillon B E, Roberts B T, Hoyt M A, Jones E W. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- 39.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 40.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short Protocols in Molecular Biology. New York: Wiley; 1992. [Google Scholar]
- 42.Kawasaki-Nishi S, Nishi T, Forgac M. J Biol Chem. 2001;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 43.Forgac M, Cantley L, Wiedenmann B, Altstiel L, Branton D. Proc Natl Acad Sci USA. 1983;80:1300–1303. doi: 10.1073/pnas.80.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y, Forgac M. J Biol Chem. 1992;267:5817–5822. [PubMed] [Google Scholar]
- 45.Drose S, Bindseil K U, Bowman E J, Siebers A, Zeeck A, Altendorf K. Biochemistry. 1993;32:3902–3906. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 46.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Cain B D, Simoni R D. J Biol Chem. 1988;263:6606–6612. [PubMed] [Google Scholar]
- 48.Valiyaveetil F I, Fillingame R H. J Biol Chem. 1997;272:32635–32641. doi: 10.1074/jbc.272.51.32635. [DOI] [PubMed] [Google Scholar]
- 49.Vik S B, Patterson A R, Antonio B J. J Biol Chem. 1998;273:16229–16234. doi: 10.1074/jbc.273.26.16229. [DOI] [PubMed] [Google Scholar]
- 50.Manolson M F, Proteau D, Preston R A, Stenbit A, Roberts B T, Hoyt M, Preuss D, Mulholland J, Botstein D, Jones E W. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- 51.Nishi T, Forgac M. J Biol Chem. 2000;275:6824–6830. doi: 10.1074/jbc.275.10.6824. [DOI] [PubMed] [Google Scholar]
- 52.Nelson H, Nelson N. Proc Natl Acad Sci USA. 1990;87:3503–3507. doi: 10.1073/pnas.87.9.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Kane P M. Biochemistry. 1996;35:10938–10948. doi: 10.1021/bi9608065. [DOI] [PubMed] [Google Scholar]
- 54.MacLeod K J, Vasilyeva E, Baleja J D, Forgac M. J Biol Chem. 1998;273:150–156. doi: 10.1074/jbc.273.1.150. [DOI] [PubMed] [Google Scholar]
- 55.Kane P M, Kuehn M C, Howald-Stevenson I, Stevens T. J Biol Chem. 1992;267:447–454. [PubMed] [Google Scholar]
- 56.Cain B D, Simoni R D. J Biol Chem. 1986;261:10043–10050. [PubMed] [Google Scholar]
- 57.Jiang W, Fillingame R H. Proc Natl Acad Sci USA. 1998;95:6607–6612. doi: 10.1073/pnas.95.12.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leng X H, Nishi T, Forgac M. J Biol Chem. 1999;274:14655–14661. doi: 10.1074/jbc.274.21.14655. [DOI] [PubMed] [Google Scholar]
- 59.Long J C, Wang S, Vik S B. J Biol Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 60.Valiyaveetil F I, Fillingame R H. J Biol Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 61.Xu T, Forgac M. J Biol Chem. 2000;275:22075–22081. doi: 10.1074/jbc.M002983200. [DOI] [PubMed] [Google Scholar]
- 62.Xu T, Vasilyeva E, Forgac M. J Biol Chem. 1999;274:28909–28915. doi: 10.1074/jbc.274.41.28909. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Myers M, Forgac M. J Biol Chem. 1992;267:9773–9778. [PubMed] [Google Scholar]