Abstract
Protein S-glutathionylation is an important reversible post-translational modification implicated in redox signaling. Oxidative modifications to protein thiols can alter the activity of metabolic enzymes, transcription factors, kinases, phosphatases, and the function of contractile proteins. However, the extent to which muscle contraction induces oxidative modifications in redox sensitive thiols is not known. The purpose of this study was to determine the targets of S-glutathionylation redox signaling following fatiguing contractions. Anesthetized adult male CB6F1 (BALB/cBy × C57BL/6) mice were subjected to acute fatiguing contractions for 15 min using in vivo stimulations. The right (stimulated) and left (unstimulated) gastrocnemius muscleswere collected 60 min after the last stimulation and processed for redox proteomics assay of S-glutathionylation. Using selective reduction with a glutaredoxin enzyme cocktail and resin-assisted enrichment technique, we quantified the levels of site-specific protein S-glutathionylation at rest and following fatiguing contractions. Redox proteomics revealed over 2200 sites of S-glutathionylation modifications, of which 1290 were significantly increased after fatiguing contractions. Muscle contraction leads to the greatest increase in S-glutathionylation in the mitochondria (1.03%) and the smallest increase in the nucleus (0.47%). Regulatory cysteines were significantly S-glutathionylated on mitochondrial complex I and II, GAPDH, MDH1, ACO2, and mitochondrial complex V among others. Similarly, S-glutathionylation of RYR1, SERCA1, titin, and troponin I2 are known to regulate muscle contractility and were significantly S-glutathionylated after just 15 min of fatiguing contractions. The largest fold changes (> 1.6) in the S-glutathionylated proteome after fatigue occurred on signaling proteins such as 14-3-3 protein gamma and MAP2K4, as well as proteins like SERCA1, and NDUV2 of mitochondrial complex I, at previously unknown glutathionylation sites. These findings highlight the important role of redox control over muscle physiology, metabolism, and the exercise adaptive response. This study lays the groundwork for future investigation into the altered exercise adaptation associated with chronic conditions, such as sarcopenia.
Keywords: Muscle contraction, Post-translational modification, S-glutathionylation, Redox signaling, Skeletal muscle, Thiol redox proteomics
Graphical abstract
Highlights
-
•
A single bout of fatiguing contractions increase muscle protein S-glutathionylation.
-
•
Mitochondrial proteins are sensitive to oxidative modifications following fatigue.
-
•
The glutathionylated proteome includes cysteines of known functional importance.
1. Introduction
Oxidative modifications are implicated in both the pathological damage of oxidative stress and the physiological and adaptive responses to redox signaling. The thiol group on the amino acid cysteine is a major target for oxidative modification of proteins [1]. The reactive nature of the thiol group means that they also play an important regulatory role in protein structure through disulfide bond formation, cofactor binding, and in catalytic activity [2]. Glutathione is the most abundant antioxidant molecule within cells, especially the mitochondria [3], [4]. It is an essential cofactor of glutathione peroxidase, glutathione S-transferase, and glutaredoxin for the scavenging of hydrogen peroxide and the reversal of oxidative modifications to proteins [3]. Reduced or oxidized glutathione can react with oxidized protein thiols or thiolate anions to form protein S-glutathionylation (P-SSG) modifications through several enzymatic and non-enzymatic mechanisms [5]. Glutathione can also react with oxidized derivatives of protein cysteines, such as sulfenic acid (-SOH), thiyl radicals (-S.), or S-nitroso (-SNO), thereby converting these oxidized forms to a more stable modification [6], [7], [8] and preventing further thiol oxidation to sulfinic and sulfonic acid. This is significant because sulfinylation and sulfonylation are largely irreversible modifications with the exception of reduction of 2-Cys PRX sulfinylation by sulfiredoxin [9], [10]. In addition to this protective effect, P-SSG is considered an important type of oxidative modification that regulates transcription, mitochondrial metabolism, apoptosis, and other critical processes [6], [11]. Glutaredoxin is the primary enzyme responsible for the reversal of this modification and returning oxidized cysteines to their original thiol (SH) redox status [12].
Contraction of skeletal muscle results in an immediate and transient increase in oxidants produced by NADPH oxidase and xanthine oxidase [13], [14], [15], [16], [17], [18]. The oxidants produced during exercise interact with redox-sensitive signaling pathways such as P-38/MAPK, NFkB, AMPK, and NRF2, to promote exercise recovery, biogenesis, protein turnover, and cyto-protective and antioxidant responses [13], [19], [20], [21]. In muscle, these oxidants are implicated in the regulation of metabolism, calcium homeostasis and sensitivity of contracting fibers through proteins like SERCA, troponin I, ATP synthase, and mitochondrial complexes [17], [19], [22], [23]. Efforts to reduce the amount of oxidants generated with exercise have occasionally resulted in decreased protection against subsequent oxidative events (ischemia), injury, and adaptive signaling, emphasizing the importance of redox signaling in exercise [13], [24], [25]. In contrast, mitochondrially-targeted antioxidants, such as elamipretide (SS-31) and AAV-mCAT, improve mitochondrial deficits, fatigue resistance, and reduce oxidative stress in mice [26], [27], [28]. In addition, aged mice, where increased mitochondrial oxidant production leads to chronic oxidative stress, have an impaired adaptive response to contraction compared to younger mice [29], [30].
The goal of this study was to determine the effect of fatiguing contractions on the redox sensitive S-glutathionylated proteome in order to provide new insights into fatigue-associated redox signaling involved in the muscle response to exercise. Given the evidence that the contraction-induced oxidant production is primarily from non-mitochondrial sources [18], we hypothesized that mitochondrial proteins would undergo relatively less redox modification than cytoplasmic and sarcolemmal proteins following an acute bout of contraction. In this study, in vivo electrical stimulation of skeletal muscle was used to induce fatiguing contractions in adult CB6F1 mice. Fatigue increased the P-SSG levels of many new and previously reported Cys sites on mitochondrial, sarcomeric, and calcium homeostasis proteins.
2. Materials and methods
2.1. All animal procedures described in this study were approved by the University of Washington IACUC
2.1.1. In-vivo stimulation
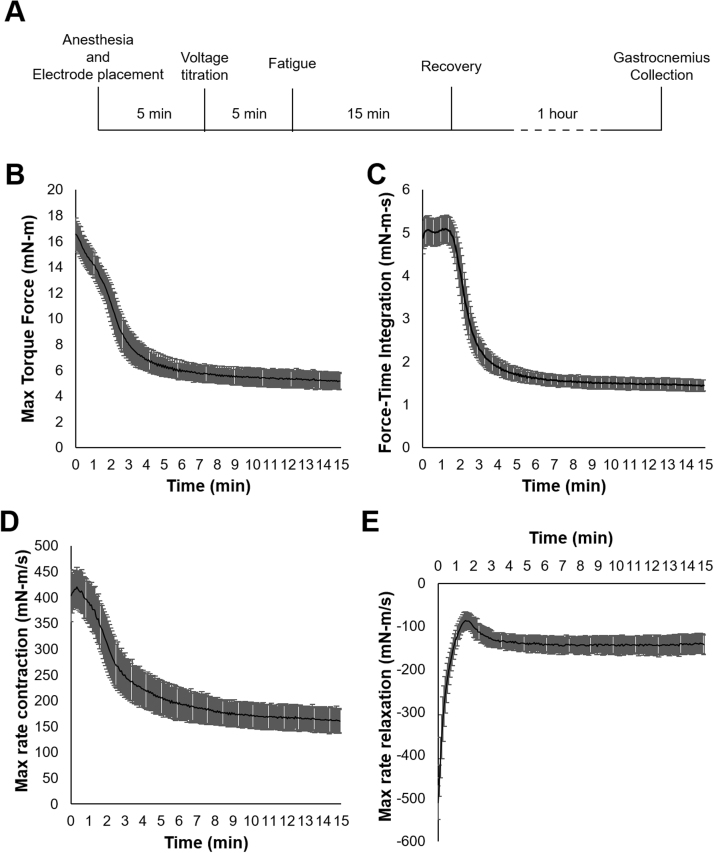
Five littermate male CB6F1(BALB/cBy x C57BL/6) mice at eleven months of age were obtained from National Institute on Aging (NIA) and placed under isofluorane anesthesia just prior to and throughout the duration of the in vivo stimulation and recovery. The right leg of the mouse was secured at the knee and the foot taped to a footplate and force transducer (Aurora Scientific Inc. Ontario). Subdermal stimulation electrodes were placed proximal and distal to the gastrocnemius. Stimulation voltage was optimized to produce maximum twitch force (20–22 V). Muscle was stimulated at 100 Hz for a 300 ms duration every fourth second for 15 min using a Grass stimulator (S88X, Astro-Med, Inc.), and torque force obtained by Aurora DMC software (V5.5), and analyzed by Aurora DMA software (V5.321). The muscle was then allowed to rest for 1 h before the mouse was sacrificed and both the stimulated and unstimulated muscles were frozen in liquid nitrogen and stored at − 80 °C (Fig. 1A). Briefly, maximal force production and the force-time integral for each tetanic stimulation was assessed throughout the 15 min of fatigue (Fig. 1B and C). The maximal rate of contraction and relaxation of each stimulation was measured over the course of fatigue (Fig. 1D, E).
Fig. 1.
In vivo direct muscle stimulation of CB6F1 mouse gastrocnemius to induce fatigue. Five adult (11 month) male CB6F1 mice were anesthetized and subject to a 15 min fatiguing protocol before a 1 h recovery and collection of the left and right gastrocnemius (A). 100 Hz submaximal stimulations for 300 ms every fourth second over fifteen minutes induced fatigue to approximately 30% initial force (B). The prolonged force output of type 2 fibers is observed for up to 2 min with force-time integration analysis (C). The maximum rate of contraction (D) and relaxation (E) during the 15 min of fatigue suggest a shift from fast-twitch to slow-twitch fibers.
2.1.2. Redox proteomics analyses of P-SSG
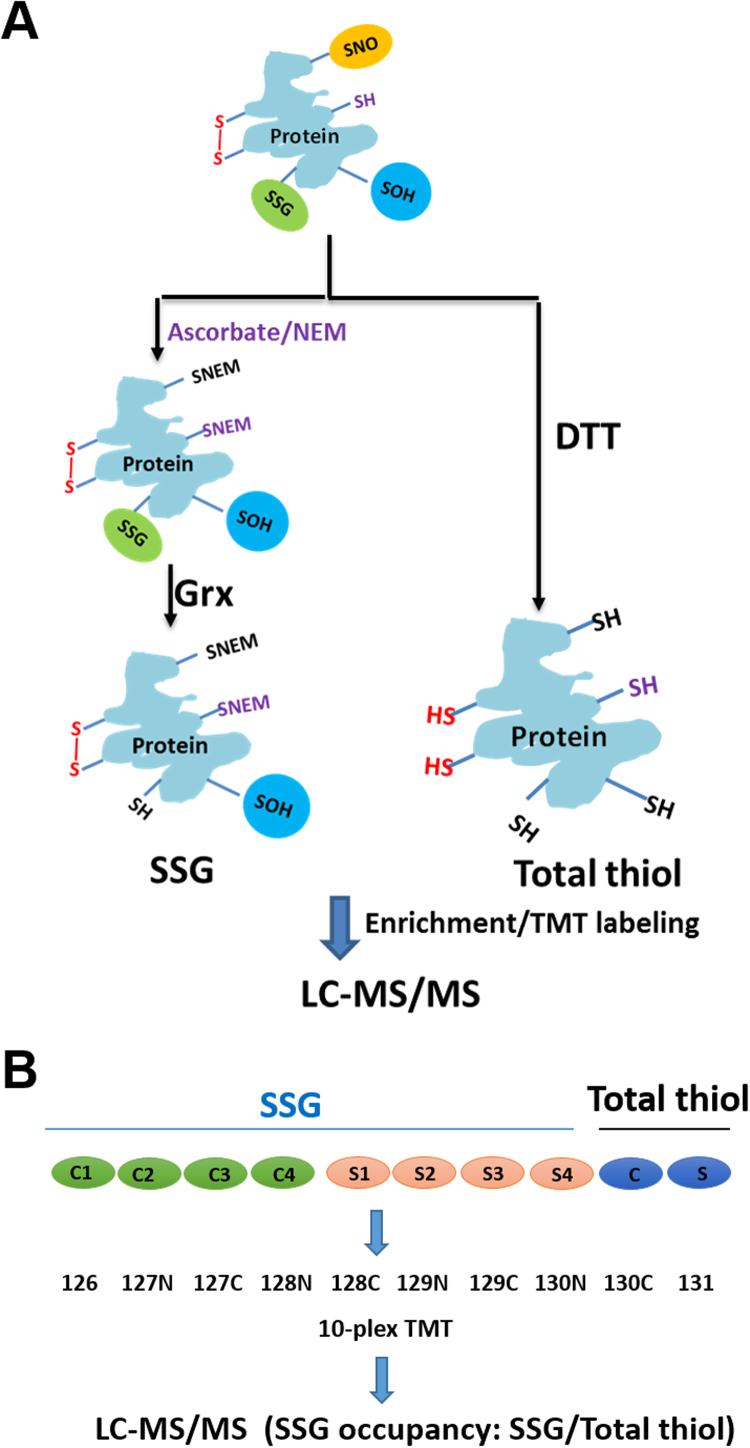
The redox proteomics experiments were performed as previously described for P-SSG [31], [32], [33]. Briefly, frozen mouse gastrocnemius muscles from stimulated and unstimulated legs from four mice were minced while frozen and incubated for 30 min on ice in the dark in the homogenization buffer comprising 250 mM HEPES buffer pH 6.0, 1% v/v SDS, 1% v/v Triton X-100 and 100 mM n-ethyl-maleimide (NEM) to block all free thiols. A small portion of tissues were pooled from the four unstimulated (C) and four stimulated (S) samples, respectively, for total thiol profiling (Fig. 2), in which tissue was incubated in the same homogenization buffer without NEM for thiol blocking. All tissue samples were then homogenized using a hand held homogenizer until completely homogenized. The resulting homogenate was pre-cleared by centrifugation at 14,000 rpm for 10 min at 4 °C. The 8 samples for P-SSG profiling were further incubated with 2 mM sodium ascorbate, 2 µM CuCl, and 1 mM SDS in the dark at 55 °C for selective reduction of nitrosylated cysteine residues and complete alkylation of free thiols. The NEM blocking and ascorbate reduction step was omitted for the two total thiol profiling samples. All 10 samples were then subjected to acetone precipitation overnight for complete removal of all excessive reagents such as detergents or NEM. Precipitated proteins were resuspended/solubilized in 250 mM HEPES buffer pH 7.0 containing 8 M urea and 0.1% SDS. Buffer exchange was performed twice using 250 mM HEPES buffer pH 7.0 containing 8 M urea, resulting in a final volume of 30–40 µL. Protein concentrations were measured by the BCA assay. Approximately 480 µg of the protein solution was diluted to ~ 500 μL by 1 M urea in 25 mM HEPES buffer, pH 7.6 and then subjected to selective reduction of S-glutathionylated thiol residues using GRX1 enzyme cocktail containing 2.5 µg/mL GRX1M (C14S mutant from E. coli), 0.25 mM GSSG, 1 mM NADPH, and 4 U/mL glutathionine reductase [33] for P-SSG or reduced with 20 mM DTT in 25 mM HEPES buffer pH 7.7 containing 2% v/v SDS for 30 min at 37 °C (Fig. 2A).
Fig. 2.
Redox proteomics analysis of protein S-glutathionylation occupancy after fatigue. Sample processing scheme for analyzing SSG occupancy (A). One arm of tissue samples were processed with ascorbate reduction for SNO and NEM blocking for free thiols followed by reduction for SSG through the GRX enzyme cocktail. The other arm of samples for total thiol profiling was directly reduced by DTT without NEM blocking. 10-plex TMT labeling scheme for P-SSG and total thiol profiling (B). 4 replicates of unstimulated (C1–C4) and 4 replicates of stimulated muscles were processed, enriched, and labeled for SSG in parallel. A small portion of tissues for each sample were pooled from unstimulated (C) and stimulated (S), respectively for total thiol measurements. Thiol-containing peptides were eluted from all 10 labeled samples and combined into one final sample for LC-MS/MS analysis. The site-specific P-SSG occupancy was calculated as the ratio of P-SSG over the total thiol for each Cys site for a given protein.
Following the reduction step, free thiol-containing proteins were enriched using Thiopropyl Sepharose 6B resin with 400 µg protein per sample for SSG-channels and 100 µg protein per sample for total thiol [32], [34]. Following on-resin digestion, isobaric labeling with 10-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific) was performed (Fig. 2B). Briefly, 70 µL of anhydrous acetonitrile was added to the manufacturer-provided TMT reagent vials. Forty microliters of 100 mM triethylammonium bicarbonate (TEAB) buffer pH 8.5 and the 70 µL of the TMT reagent solutions were added to the resin containing peptides and the labeling reaction was carried out at room temperature for 1 h. The reaction was stopped by the addition of 8 µL of 5% NH2OH·HCl in 200 mM TEAB buffer for 15 min. The excess TMT reagents were removed by washing five times each with 80% ACN with 0.1% TFA and 25 mM ammonium bicarbonate. The captured, labeled peptides were eluted by DTT as previously described [20].
LC-MS/MS analysis of TMT-labeled cysteine-containing peptides was performed on an orbitrap fusion Lumos mass spectrometer (Thermo Fisher Scientific). A Waters nanoACQUITY UPLC system with a custom packed C18 column (50 cm × 75 µm i.d., Phenomenex Jupiter, 3 µm particle size) and a 3h LC gradient was applied for peptide separation. Full MS spectra were recorded at resolution of 30 K over the range of m/z 400–2000 with an automated gain control (AGC) value of 2 × 105. MS/MS was performed in the data-dependent mode with orbitrap resolution of 30 K, an AGC target value of 1 × 105, a normalized collision energy setting of 30 for high-energy collision dissociation (HCD), a dynamic cycle time of 3 s and a dynamic exclusion time of 60 s were used.
2.1.3. Data Analysis and statistics
LC-MS/MS raw data were converted into dta files using Bioworks Cluster 3.2 (Thermo Fisher Scientific), and MSGF plus algorithm [35] (v9979, released in March 2014) was used to search MS/MS spectra against the mouse protein sequence database (UniProt, released in September 2016). The key search parameters used were 20 ppm tolerance for precursor ion masses, 0.5 Da tolerance for fragment ions, dynamic oxidation of methionine (15.9949 Da), dynamic NEM modification of Cys (125.0477 Da), and static 10-plex TMT modification of lysine and N-termini of peptides (229.1629 Da). Peptides were identified from database searching results applying the following criteria: Q-value < 0.01, and mass measurement error < 10 ppm (± 5 ppm), resulting final FDR < 1% at the unique peptide level. Since NEM blocked original free Cys sites, all glutathionylated Cys residues were identified as un-modified Cys.
For TMT-based relative quantification analysis of P-SSG sites, the intensities of each channel of TMT reporter ions were summed from all spectra corresponding to individual Cys sites and transformed to log2 values. P-SSG occupancy for each Cys site was calculated as the ratio of the average level of P-SSG (n = 4) over total thiol (n = 1 for the pooled sample) in percentage for each of the control or fatigue conditions. A 4-fold load difference between P-SSG channels and total thiol channels was taken into account in the calculation of P-SSG occupancy. Cys sites with significant changes in P-SSG levels were based on two-sample student t-test with permutation based FDR control.
Examination of the dataset by the Core Analysis module of the Ingenuity Pathway Analysis software (IPA, http://www.ingenuity.com/) allowed for the identification of biologically relevant canonical pathways affected by stimulation. IPA core analysis was performed using a FDR cutoff of 0.05 and User Data Set was used as the Reference. Cysteine residues that corresponded to the same protein were consolidated using the maximum of absolute values of log Fold-Change. The significance values for the canonical pathways were calculated by right-tailed Fisher's exact test (p < 0.05). The Cellular Compartment and Biological Process of the top 1290 P-SSG modifications (FDR ≤ 0.05) were identified using DAVID Bioinformatics Resources (https://david.ncifcrf.gov/).
Protein crystal structures were attained from Protein Data Bank (www.rcsb.org) using the accession number 2b05 for 14-3-3 gamma [36], 3ALO for MAP2K4 gamma [37], 5xtb for NDUV2 and NDUV1 [38], and 3tlm for SERCA1 [39]. Figures were constructed using Molsoft software 3.8–6a (www.molsoft.com).
3. Results
Adult mice averaged a submaximal (100 Hz) tetanic force of 16.5 ± 1.3 millinewton-meters (mN-m) (mean ± SD) using the described in vivo stimulation protocol. Isometric fatiguing stimulations reduced the maximal torque force to 31% of initial force over the course of fatigue (Fig. 1B). The force output of the right gastrocnemius declined rapidly in the first three minutes and remained relatively constant for the remainder of the fatigue protocol. The rate of contraction and relaxation also fell precipitously during the first few minutes of the protocol (Fig. 1D, E). This rapid decline in force and kinetics followed by the longer period of sustained force is consistent with the drop-out of Type 2 (IIa, IIb) fast-twitch fibers and the persistence of fatigue-resistant Type 1, slow-twitch fibers [40]. This in vivo electrical stimulation strategy has the benefit of providing a consistent, reproducible muscle stimulus in rodents, in contrast to treadmill protocols, to assess molecular responses to muscle contraction. The in vivo electrical stimulation also leaves one hind limb unstimulated to be used as a control.
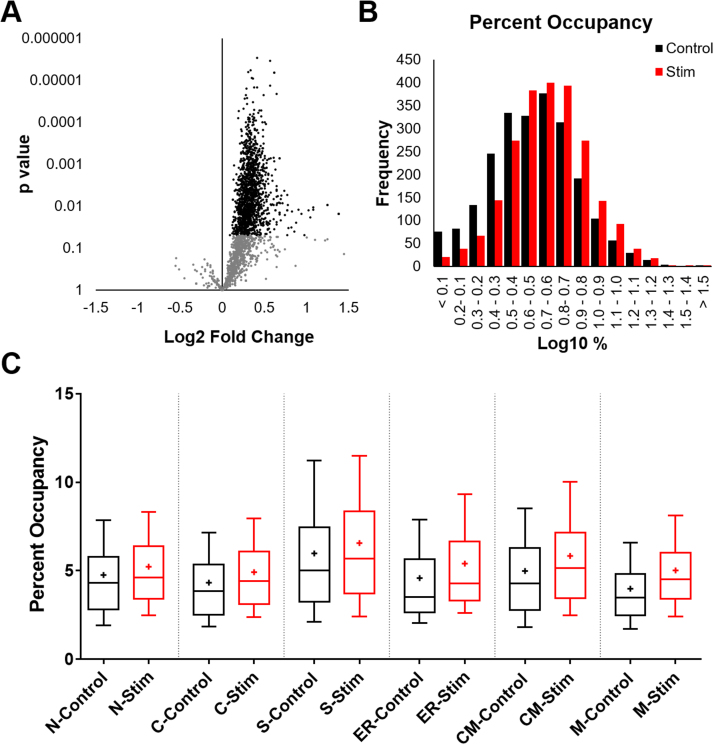
Redox proteomics was performed through parallel processing and enrichment of proteins containing P-SSG and total thiols in parallel samples. A 10-plex TMT labeling strategy was used to detect P-SSG modifications and total thiols (Fig. 2A, B). The 10 TMT-labeled samples were combined into one final sample for LC-MS/MS analysis such that each thiol-containing peptide was identified with 10 reporter ion intensities reflecting the abundance levels of P-SSG and total thiols for each condition. In total, over 2200 Cys sites of P-SSG modifications were identified and quantified across all 10 channels. The levels of P-SSG were significantly increased after fatigue for over half of the sites as illustrated in the volcano plot shown in Fig. 3A. P-SSG occupancy for each Cys site was estimated as the ratio of the average level of P-SSG (n = 4) over total thiol (n = 1 for the pooled sample) in percentage for each condition. As shown, the left unstimulated gastrocnemius had an average P-SSG occupancy of 4.5%, while the right stimulated gastrocnemius showed a 5.22% average occupancy (Fig. 3B). Despite the moderate increase, significant differences in terms of average occupancy were observed in different subcellular compartments. Of the protein Cys sites detected, with some multi-compartmental overlap, 588 sites were annotated as nuclear proteins, 1154 as cytosolic proteins, 333 as secreted proteins, 141 as endoplasmic reticulum proteins, 229 as cell membrane proteins, and 390 as mitochondrial proteins. The average change in P-SSG occupancy was 0.72%; however, mitochondria showed the most significant occupancy shift at an average of 1.03%, and nuclear proteins the lowest occupancy shift at an average of 0.47% (Fig. 3C). Additionally, secreted proteins had the largest baseline occupancy at 5.98%, while mitochondria had the smallest at 3.99%.
Fig. 3.
Protein S-glutathionylation occupancy shift after fatigue. Of the approximately 2200 sites of P-SSG, over half (1290 with FDR adjustment of 0.05) of them had a significant shift in P-SSG level after fatigue as observed in this volcano plot (A). On a log scale, a shift in occupancy of approximately 0.72% is evident between the left (unstimulated) and right (stimulated) leg. (B). Divided into subcellular location (Uniprot defined), the greatest shift in occupancy occurs from right to left, with mitochondria demonstrating a 1.04% shift in occupancy compared to nuclear proteins at 0.47%. The average baseline P-SSG occupancy across all compartments was 4.5% which increased to an average occupancy of 5.22% in the stimulated gastrocnemius. (C). In the 10/90 percentile box and whiskers plot, + indicates the mean of all data points, including outliers, which were not displayed for ease of visualization. N = Nuclear, C = Cytoplasmic, S = Secreted, ER = Endoplasmic/Sarcoplasmic Reticulum, CM = Cell Membrane, M = Mitochondria.
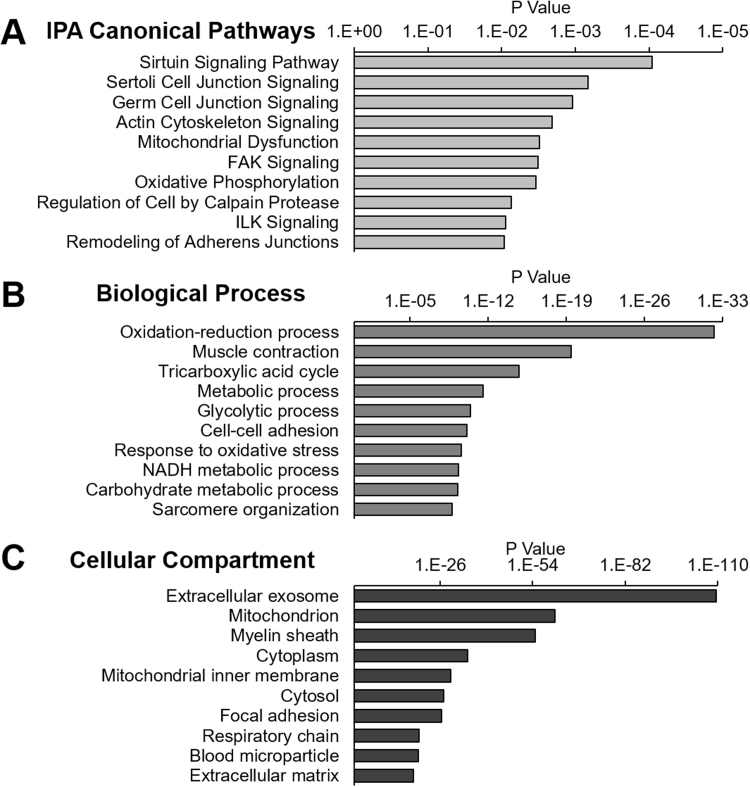
Ingenuity Pathway Analysis (IPA) revealed that the top significantly altered protein targets (p ≤ 0.05) were involved in sirtuin, cell junction, and actin cytoskeleton signaling, as well as mitochondrial dysfunction and oxidative phosphorylation (Fig. 4A). Sirtuin signaling encompassed many proteins associated with mitochondrial complex I and lactate dehydrogenase, among others associated with the regulation of NAD (Supplementary Table 1). S-glutathionylated proteins involved in actin cytoskeleton, cell junction, FAK, and ILK signaling are predominately cytoskeletal and sarcomeric proteins as well as several proteins associate with the MAPK pathway. Although IPA analysis indicates P-SSG of proteins in these pathways was altered by contraction, this analysis does not indicate if the P-SSG modification alters protein activity. The role of these modifications in protein function and redox signaling will require further study.
Fig. 4.
Canonical Pathways, Biological Process, and Cellular Compartment analysis of proteins with significant changes of P-SSG by fatiguing contractions. The top ten Canonical Ingenuity Pathways (IPA) involving the 1290 P-SSG sites with significant changes after fatigue (A). The significance of each pathway was determined by right-tailed Fisher's exact test (p < 0.05). DAVID Bioinformatics Resources identified the Biological Processes (B) and Cellular Compartments (C) associated with the oxidized proteins, ranked by significance.
The significantly changed 1290 P-SSG sites (FDR ≤0.05) were assessed with DAVID Bioinformatics Resources for Biological Process and Cellular Compartment [41], [42]. The Biological Processes of most affected proteins are associated with oxidative and glycolytic metabolism as well as muscle contraction and oxidative stress responses (Fig. 4B, Supplementary Table 2). The compartments to which the most affected proteins were localized include proteins extracellular to the muscle fibers, including exosomes, myelin sheath, blood micro-particle, and extracellular matrix (Fig. 4C, Supplementary Table 3). This is consistent with the observation that NADPH oxidase, a membrane protein, produces oxidants during exercise [18], [24]. Mitochondria, however, was the second most oxidized compartment following fatigue.
Many studies have identified P-SSG sites as modulators of protein activity, several of which are discussed below and outlined in Table 1. However, new analytical approaches, like the ones described here, have allowed for the detection of proteins with novel P-SSG modifications. Some of these proteins were among the most affected by fatigue. We have outlined a selected list of these novel P-SSG sites in Table 2 and Fig. 5.
Table 1.
Known glutathionylation sites associated with altered protein function. Table of P-SSG modifications significantly affected by fatiguing contractions sorted by Excitation/contraction Coupling, Metabolism, Redox Regulation, and Other. Significance determined by p-value (≤ 0.05). Note: Some P-SSG sites were no longer significant with FDR correction.
| Function | Protein | Cysteine Site(s) | Log2 FC | P value | FDR |
|---|---|---|---|---|---|
| EC Coupling | Troponin I, fast skeletal muscle | 134 | 0.5239111 | 0.000948 | 0.009052 |
| Ryanodine receptor 1 | 3636 | 0.300979 | 0.003931 | 0.016153 | |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 349 | 0.2534598 | 0.000531 | 0.007538 | |
| 674 | 0.1620496 | 0.031643 | 0.054613 | ||
| ATP-sensitive inward rectifier potassium channel 11 | 42 | 0.2226592 | 0.039947 | 0.065353 | |
| Metabolism | Glyceraldehyde-3-phosphate dehydrogenase | 150 | 0.2984179 | 0.006478 | 0.020467 |
| Creatine kinase M-type | 283 | 0.4612769 | 0.002407 | 0.012625 | |
| ATP synthase subunit alpha, mitochondrial | 244 | 0.450853 | 0.009337 | 0.025096 | |
| 294 | 0.5469985 | 0.002965 | 0.013841 | ||
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 89 | 0.3130165 | 0.00116 | 0.009595 | |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | 251 | 0.334239 | 0.020124 | 0.039935 | |
| 255 | 0.3191644 | 0.037534 | 0.062227 | ||
| NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | 187 | 0.3499677 | 0.013932 | 0.031497 | |
| 206 | 0.2585537 | 0.011949 | 0.02892 | ||
| Aconitate hydratase, mitochondrial | 385 | 0.2292044 | 0.008289 | 0.023541 | |
| 448 | 0.2875243 | 0.003558 | 0.015214 | ||
| 451 | 0.2168614 | 0.01297 | 0.030181 | ||
| Malate dehydrogenase, cytoplasmic | 137 | 0.2440071 | 0.005084 | 0.018121 | |
| Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial | 560 | 0.3262215 | 0.010501 | 0.026683 | |
| ADP/ATP translocase 1 | 160 | 0.4420761 | 0.012331 | 0.02947 | |
| 257 | 0.3422474 | 0.001147 | 0.009566 | ||
| Peptidyl-prolyl cis-trans isomerase F, mitochondrial (Cyclophilin D) | 202 | 0.2659391 | 0.014669 | 0.032343 | |
| Redox regulation | Peroxiredoxin-1 | 52 | 0.6198489 | 0.035623 | 0.059947 |
| Thioredoxin | 73 | 0.3607812 | 0.013675 | 0.031156 | |
| Thioredoxin, mitochondrial | 90 | 0.3926398 | 0.004439 | 0.016882 | |
| 93 | 0.3926398 | 0.004439 | 0.016882 | ||
| Other | Hemoglobin subunit beta-1 | 94 | 0.3880275 | 0.005266 | 0.018398 |
Table 2.
Novel sites of P-SSG with greatest fold change after stimulation. Table of novel P-SSG modifications significantly affected by fatiguing contractions and with a fold change greater than 1.68. Significance determined by p-value (≤ 0.05). Note: Some P-SSG sites were no longer significant with FDR correction.
| Protein | Cysteine Site(s) | Log2 FC | P value | FDR |
|---|---|---|---|---|
| 14-3-3 protein gamma | 112 | 1.023413 | 0.012581 | 0.029635 |
| 97 | 0.829382 | 0.013471 | 0.030999 | |
| ARL14 effector protein-like | 100 | 1.386748 | 0.015173 | 0.032951 |
| 92 | 1.386748 | 0.015173 | 0.032951 | |
| 98 | 1.386748 | 0.015173 | 0.032951 | |
| Activating signal cointegrator 1 complex subunit 3 | 2004 | 0.824314 | 0.032172 | 0.055351 |
| Dual specificity mitogen-activated protein kinase kinase 4 | 264 | 0.854978 | 0.040535 | 0.066078 |
| Myosin heavy chain 1 | 480 | 0.97775 | 0.048739 | 0.075581 |
| Myosin heavy chain 2b | 523 | 0.942909 | 0.015117 | 0.032951 |
| 541 | 0.907055 | 0.01224 | 0.029316 | |
| Myosin light chain 1/3, skeletal muscle isoform | 62 | 0.781071 | 0.009289 | 0.025048 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | 223 | 1.24754 | 0.009351 | 0.025096 |
| 224 | 1.24754 | 0.009351 | 0.025096 | |
| Phosphomannomutase 2 | 99 | 0.772268 | 0.005397 | 0.01863 |
| Ribosome-binding protein 1 | 1415 | 0.786351 | 0.03373 | 0.05748 |
| Sciellin | 606 | 1.08025 | 0.010851 | 0.027299 |
| 617 | 1.08343 | 0.01201 | 0.02895 | |
| Histone-lysine N-methyltransferase Smyd1 | 332 | 0.775664 | 0.012484 | 0.029529 |
| 335 | 0.775664 | 0.012484 | 0.029529 | |
| Sarcalumenin | 648 | 1.043884 | 0.018395 | 0.037535 |
| Synaptophysin-like protein 2 | 155 | 0.775554 | 0.048596 | 0.075513 |
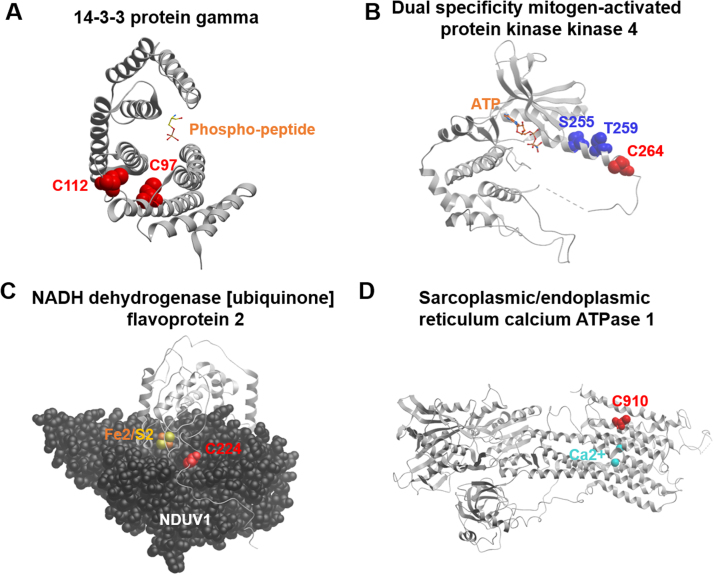
Fig. 5.
Protein structure illustrating sites of novel P-SSG modifications with the greatest fold change after stimulation. Novel P-SSG modifications involved in the regulation of kinase and phosphatase signaling, metabolic pathways, and proteins associated with excitation/contraction coupling. Figure contains significant P-SSG sites with a greater than 1.68 fold change with fatigue. (A) 14-3-3 gamma crystal structure (Protein Data Bank accession number: 2b05) with highlighted P-SSG cysteine residues (red) in relation to the phospho-peptide recognition site. (B) MAP2K4 gamma crystal structure (Protein Data Bank accession number: 3ALO) with highlighted P-SSG cysteine residues (red) and phosphorylation sites (blue). (C) NDUV2 ribbon crystal structure and NDUV1 space-filling crystal structure (Protein Data Bank accession number: 5xtb) with highlighted P-SSG cysteine residues (red) and N1A iron/sulfur cluster. (D) SERCA1 crystal structure (Protein Data Bank accession number: 3tlm) with highlighted P-SSG cysteine residues (red) and calcium (light blue) in the transmembrane domain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Physical activity is one of the most effective interventions for the prevention and treatment of muscle atrophy and promotes beneficial health outcomes in heart disease, diabetes, and many other pathologies [43], [44]. Reactive oxygen/nitrogen species (ROS/RNS) generated during exercise modulate contractile function and participate in the exercise adaptive response through oxidative post-translational modifications of regulatory cysteines in skeletal muscle [9], [14], [19]. Protein S-glutathionylation in particular is a transient and reversible modification that modulates the function and structure of proteins and also prevents the subsequent irreversible oxidation of cysteines [20]. Many of the P-SSG sites identified in muscle are known to alter actin-myosin bridge formation, calcium homeostasis, and metabolism, and contribute to fatigue [40]. The goal of this study was to assess the S-glutathionylated proteome following fatiguing contractions. In addition to the detection of P-SSG sites known to result in physiological effects, proteomic analysis has revealed many novel sites susceptible to S-glutathionylation that may contribute to contraction induced redox signaling.
Compared to other post-translational modifications, such as phosphorylation, the number of known S-glutathionylation sites are small and their roles in regulating adaptive responses in skeletal muscle are less well-characterized. Despite the increasing effort to understand redox dependent signaling in skeletal muscle, the quantitative effect of muscle contraction on the S-glutathionylated proteome has yet to be assessed. Using a thiol-based redox proteomics technique, we detected a significant shift in P-SSG level in mouse gastrocnemius muscle 60 min after 15 min fatiguing contractions (Fig. 3). All detected P-SSG sites were present in both the unstimulated stimulated muscles with an average occupancy of 4.5% for the whole P-SSG proteome in the unstimulated leg that increased to an average occupancy of 5.22% in the stimulated gastrocnemius. This suggests that S-glutathionylation of proteins is a common post-translational modification under basal redox homeostasis; however, a significant shift of homeostasis towards oxidation would lead to higher occupancy of P-SSG. Interestingly, the proteins annotated as secretory proteins or present in the extracellular space have a higher average basal P-SSG occupancy (5.98%), while those in subcellular organelles, like the mitochondria (3.99%), have a lower basal occupancy. Proteins most sensitive to changes in P-SSG after fatiguing contractions (change > 1.4 FC and p ≤ 0.05) began with 1.67% average P-SSG occupancy and increased to 2.52% after fatigue. As previously mentioned, mitochondrial proteins had the most significant shift in occupancy, suggesting they may contribute to ROS production during fatigue or are more susceptible to oxidation (Fig. 3C), despite the evidence indicating that the contraction-induced oxidant production is primarily from non-mitochondrial sources. Indeed, the alkalinity of the mitochondrial matrix is known to promote the ionized thiolate form of cysteine [45].
Discussed below are a few of those P-SSG sites known to result in physiological effects (Table 1). More studies are required to explore the other physiological consequences of the P-SSG sites reported in this study, as well as those sites not previously described (Table 2).
5. Muscle contraction
5.1. Myofibril
Oxidative modification of contractile proteins can have an immediate effect on contraction and may be responsible for some of the rapid changes in contraction and relaxation kinetics observed in the first few minutes of fatigue (Fig. 1C, D). For example, protein S-glutathionylation of Cys134 on troponin I2 is known to increase calcium sensitivity in fast twitch fibers in rats and exercising humans [46], [47], [48]. This modification was increased 1.44 fold (p = 0.0095) in the fatigued muscle compared to its unstimulated control. Post-translational modifications of titin, the largest known mammalian protein, has an increasingly appreciated role in the regulation of myofibril stiffness [49]. While disulfide formation of cryptic cysteines in titin's I-band Ig domains increase titin stiffness, glutathionylation of cysteines exposed during stretch decrease protein folding, thus enhancing elasticity [50], [51]. In this study, over 200 P-SSG modifications were observed on titin in the mouse gastrocnemius, and 50 of them had a significant increase in glutathionylation after fatiguing contractions. Many of these P-SSG sites were in titin's I-band Ig domains.
5.2. E-C coupling
Calcium release and reuptake can also be altered by P-SSG of E-C coupling proteins in response to muscle contraction. P-SSG and S-nitrosylation of ryanodine receptor 1 (RYR) at Cys3636 alter the sensitivity of the receptor to voltage and calcium/calmodulin binding [46], [52], [53], [54]. In this study, P-SSG occupancy of Cys 3636 of RYR-1 was significantly altered after stimulation, but only by 1.23 fold. Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1) P-SSG at Cys674 can disrupt calcium reuptake into the SR, affecting relaxation time [46], [52], [55]. This cysteine is also susceptible to irreversible oxidation, which would permanently impair calcium reuptake, suggesting that P-SSG may serve to protect against oxidative damage from increased oxidant production during muscle contraction [56]. Similarly affecting calcium homeostasis, the activity of ATP-sensitive inward rectifier potassium channel 11 (KCJ11) is reportedly regulated by Cys42 oxidation and is restored by DTT [52], [57]. This cysteine was moderately S-glutathionylated 60 min after fatigue in this study (FDR = 0.065, Log2FC = 0.22).
6. Metabolism
In contrast to our hypothesis, mitochondrial proteins underwent one of the greatest increases in P-SSG following an acute bout of contractions. Mitochondrial and glycolytic metabolism are essential for contraction, energy balance, and regulation of active electrolyte transport in muscle. Mitochondria are a regular source of oxidants in cells [58], [59], though not considered to be a major source during exercise [13], [17], [24]. They contain a robust antioxidant defense with 10–15 mM glutathione and mitochondrial-specific peroxiredoxin (PRX3), superoxide dismutase (MnSOD), thioredoxin-2 (TRX2), GSH peroxidase (GPX4), and glutaredoxin (GRX2), indicating that the redox state of mitochondrial proteins are highly regulated [60]. P-SSG in mitochondria can temporarily regulate the activity of key metabolic enzymes. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key glycolytic enzyme, is inhibited by S-glutathionylation of Cys150, a modification also increased with fatigue in this study [61], [62], [63]. Creatine kinase (M-type) is essential for the production of phosphocreatine and ADP and the reverse reaction, supplying muscle with ATP during contraction. S-glutathionylation at creatine kinase Cys283 was associated with a loss of enzyme activity during ischemia and was identified as redox sensitive through Isotope-Coded Affinity Tag (ICAT) labeling and proteomics [64], [65]. This modification, as well as Cys317 of creatine kinase (mitochondrial, S-Type) was were also observed to be a redox sensitive site in this study and others [66].
6.1. TCA cycle
Within the TCA cycle, malate dehydrogenase (MDH1) Cys137 as well as aconitase (ACO2) at Cys385 were significantly S-glutathionylated with fatigue [66], [67], [68], [69]. When modified, the cysteine on ACO2 is reported to reduce enzyme activity. Like Cys385, Cys448 and Cys451 are also significantly modified with fatigue, and are associated with the 4Fe-4S cluster.
6.2. Electron transport system
Oxidative modifications of protein complexes in the electron transport chain can regulate superoxide and ATP production, and are implicated in the detrimental effects of oxidative stress on bioenergetics [67], [70], [71]. Cys125 and Cys142 of NADH dehydrogenase flavoprotein 1 (NDUFV1) are reported to be S-glutathionylated and possibly form disulfide linkages, but their S-glutathionylation occupancy was not significantly increased under fatiguing conditions [69], [72]. However, Cys187 and 206 were also reported as possible disulfide linkages by Zhang et al., and these were significantly S-glutathionylated with fatigue [72], [73]. Additionally, Cys59 of NDUFB7, Cys463, Cys64, and Cys92 of NDUFS1, and Cys347 of NDUFS2 were among the complex I subunit cysteines to significantly change in P-SSG and were previously reported to be redox sensitive by Danielson et al. [74]. Succinate dehydrogenase (SDHA) S-glutathionylation on Cys89 increases enzyme activity and reduces superoxide production in purified protein and in bovine and ischemic rat hearts [46], [69], [75], [76]. ATP synthase, responsible for utilizing the proton-motive force for ATP production, was S-glutathionylated at Cys244 and 294 of the alpha subunit with fatigue. S-glutathionylation of Cys294, although transiently inhibitory, is shown to protect the enzyme from irreversible oxidation, while S-sulfhydration is reported to improve activity [67], [69], [77], [78].
7. Novel P-SSG sites
Advances in proteomics and enrichment techniques have increased the detection of novel S-glutathionylation sites. We report several new sites of P-SSG on proteins involved in common signaling pathways, metabolic pathways, and proteins associated with excitation/contraction coupling. The sites listed in Table 2 had a minimum fold change of 1.68 and were significantly affected by fatigue (p ≤ 0.05). Briefly, we will describe a few of the most interesting novel P-SSG modifications and speculate on possible downstream effects.
Protein 14-3-3 gamma has been shown to associate with and regulate the activity, localization, and interactions of over a hundred proteins, including phosphatases and kinases like PKA and PKC [79], [80], [81]. These functions are mediated through its specific phosphoserine/phosphothreonine binding activity on target proteins. Two of the most significantly affected S-glutathionylation sites associated with fatigue (C112, C97) reside in or near this phospho-peptide binding domain (Fig. 5A). As these S-glutathionylation sites are previously unreported, it is unknown whether these oxidative modifications affect its function.
Similarly S-glutathionylated after fatiguing contractions, Cys264 is adjacent to the activating phosphosites of S255 and T259 of MAP2K4, a member of the mitogen-activated protein kinase family, and may be a mechanism by which ROS regulates the MAPK pathway (Fig. 5B) [82]. Two cysteines, 223 and 224, on the N1A iron-sulfur cluster of complex I flavoprotein 2 subunit (NDUV2) were among the most sensitive site of glutathionylation following contraction. This protein is believed to act as an antioxidant by capturing one of the two electrons donated to the complex by NADH [83]. Hinchliffe et al. proposes that, in doing so, it prevents the excess reduction of the FMN site in the NDUV1 subunit and the formation of reactive oxygen species. It is possible that the oxidation of the conserved cysteine, Cys224, can form a previously unknown disulfide bond with a similarly oxidized cysteine on the adjacent complex (Cys125 of NDUV1) approximately 5 angstroms away (Fig. 5C), a cysteine already known to participate in disulfide bonds [72]. Further study is required to determine the role of this modification in complex I function with fatigue. Not listed on Table 2, but also among the most significantly S-glutathionylated cysteines (1.64 fold change), was Cys910 of SERCA, the calcium transporter that functions to reuptake calcium into the sarcoplasmic reticulum. Cysteine 910, another previously unreported glutathionylation site, lies in the transmembrane domain next to the glutamic acid residue (Glu908) that assists calcium into the SR (Fig. 5D). Further study is required to determine if this modification might alter calcium uptake and delay muscle relaxation.
There are several potential limitations regarding the current redox proteomics approach. First, there is a possibility of thiol oxidation occurring in the minutes before freezing, during storage, or immediately following the thaw, despite the use of a high concentration of NEM (100 mM) in the first stage of sample processing. Moreover, the indirect nature of P-SSG measurement is considered a surrogate of thiol oxidation since the glutaredoxin reaction does not offer perfect specificity for P-SSG. For example, some protein disulfide bonds or SOH may also be reduced during glutaredoxin treatment and therefore contribute to the P-SSG signal in our analyses [32], [84]. Indeed, we have previously performed several negative controls to evaluate the specificity of reduction where nearly zero background signals were observed when no reducing agents were applied and a minimal signal (estimated to be < 10%) of non-specific reduction was observed for the most proteins when GRX was omitted in the reduction cocktail still containing GSH, NADPH, and glutathione reductase [32]. These data suggests a relative good specificity for the GRX reduction. Finally, the time course of changes in the thiol redox proteome following exercise is not known, and further optimization of post-fatigue analysis would be required to capture the dynamic P-SSG levels.
In conclusion, mapping the S-glutathionylation proteome in muscle has revealed many unreported glutathionylation sites, which are highly susceptible to modification with fatigue. Further study will be required to test the physiological relevance of the novel P-SSG sites through computational means or site-directed mutagenesis [85]. Pathway and cellular compartment analyses implicate P-SSG regulatory control over metabolism, contraction kinetics, and many signal transduction pathways. Nearly all modifications had a relatively small change in glutathionylation (< 2 fold), which suggests that exercise-induced adaptive responses are regulated by relatively subtle changes of in situ P-SSG modifications, that 60 min delay between contraction and biopsy misses the window of peak redox signaling, or downstream adaptive responses require repeated bouts of contraction. On the other hand, some of the P-SSG sites may not play a substantial role in exercise adaptation signaling. However, their identification as redox sensitive may implicate a functional role under more oxidative conditions such as protection from further irreversible modifications. The in vivo stimulation method described in this study provides an effective means of reliably reproducing fatigue and studying redox signaling in stimulated and unstimulated muscle through global quantitative redox proteomics methods for advancing our understanding of redox signaling in health and disease [14], [22], [23]. Thus, the data reported here provide insights into the potential effects on muscle function and adaptive signaling resulting from oxidants produced during exercise.
Funding and conflicts
This study was supported by AFAR BIG, P01AG001751; Glenn Foundation Award; Genetic Approaches to Aging Training Grant (T32AG000057); and Nathan Shock Center of Excellence in the Basic Biology of Aging (P30 AG013280), and NIH grants UC4DK104167, P41GM103493, and U24 DK112349. Some of the experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the DOE under Contract DE-AC05-76RL0 1830. The authors report no conflicts of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.05.011.
Contributor Information
Wei-Jun Qian, Email: Weijun.qian@pnnl.gov.
David J. Marcinek, Email: dmarc@uw.edu.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404(5):902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Go Y.M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribas V., Garcia-Ruiz C., Fernandez-Checa J.C. Glutathione and mitochondria. Front. Pharmacol. 2014;5:151. doi: 10.3389/fphar.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grek C.L. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013;288(37):26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shelton M.D., Chock P.B., Mieyal J.J. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 2005;7(3–4):348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 7.Mieyal J.J. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 2008;10(11):1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalle-Donne I. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 2009;34(2):85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Kramer P.A. The measurement of reversible redox dependent post-translational modifications and their regulation of mitochondrial and skeletal muscle function. Front. Physiol. 2015;6:347. doi: 10.3389/fphys.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee S.G. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. Suppl. 2007;106:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 11.Shelton M.D., Mieyal J.J. Regulation by reversible S-glutathionylation: Molecular targets implicated in inflammatory diseases. Mol. Cells. 2008;25(3):332–346. [PMC free article] [PubMed] [Google Scholar]
- 12.Shelton M.D., Chock P.B., Mieyal J.J. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 2005;7(3–4):348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 13.Wadley G.D. Xanthine oxidase inhibition attenuates skeletal muscle signaling following acute exercise but does not impair mitochondrial adaptations to endurance training. Am. J. Physiol. Endocrinol. Metab. 2013;304(8):E853–E862. doi: 10.1152/ajpendo.00568.2012. [DOI] [PubMed] [Google Scholar]
- 14.Powers S.K., Jackson M.J. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol. Rev. 2008;88(4):1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson M.J. Interactions between reactive oxygen species generated by contractile activity and aging in skeletal muscle? Antioxid. Redox Signal. 2013;19(8):804–812. doi: 10.1089/ars.2013.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasilaki A. Free radical generation by skeletal muscle of adult and old mice: effect of contractile activity. Aging Cell. 2006;5(2):109–117. doi: 10.1111/j.1474-9726.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- 17.Steinbacher P., Eckl P. Impact of oxidative stress on exercising skeletal muscle. Biomolecules. 2015;5(2):356–377. doi: 10.3390/biom5020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakellariou G.K. Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxid. Redox Signal. 2013;18(6):603–621. doi: 10.1089/ars.2012.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri E., Sestili P. Reactive oxygen species in skeletal muscle signaling. J. Signal Transduct. 2012;2012:982794. doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastore A., Piemonte F. S-Glutathionylation signaling in cell biology: progress and prospects. Eur. J. Pharm. Sci. 2012;46(5):279–292. doi: 10.1016/j.ejps.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Webb R. The ability of exercise-associated oxidative stress to trigger redox-sensitive signalling responses. Antioxidants. 2017;6(3) doi: 10.3390/antiox6030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonagh B., Sakellariou G.K., Jackson M.J. Application of redox proteomics to skeletal muscle aging and exercise. Biochem. Soc. Trans. 2014;42(4):965–970. doi: 10.1042/BST20140085. [DOI] [PubMed] [Google Scholar]
- 23.McDonagh B. Differential cysteine labeling and global label-free proteomics reveals an altered metabolic state in skeletal muscle aging. J. Proteome Res. 2014;13(11):5008–5021. doi: 10.1021/pr5006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frasier C.R. Redox-dependent increases in glutathione reductase and exercise preconditioning: role of NADPH oxidase and mitochondria. Cardiovasc. Res. 2013;98(1):47–55. doi: 10.1093/cvr/cvt009. [DOI] [PubMed] [Google Scholar]
- 25.Ristow M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. USA. 2009;106(21):8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel M.P. Mitochondrial-targeted peptide rapidly improves mitochondrial energetics and skeletal muscle performance in aged mice. Aging Cell. 2013;12(5):763–771. doi: 10.1111/acel.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcinek D.J., Siegel M.P. Targeting redox biology to reverse mitochondrial dysfunction. Aging. 2013;5(8):588–589. doi: 10.18632/aging.100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D. Ectopic catalase expression in mitochondria by adeno-associated virus enhances exercise performance in mice. PLoS One. 2009;4(8):e6673. doi: 10.1371/journal.pone.0006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasilaki A. Adaptive responses of mouse skeletal muscle to contractile activity: the effect of age. Mech. Ageing Dev. 2006;127(11):830–839. doi: 10.1016/j.mad.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Ljubicic V. Molecular basis for an attenuated mitochondrial adaptive plasticity in aged skeletal muscle. Aging. 2009;1(9):818–830. doi: 10.18632/aging.100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo J. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 2014;9(1):64–75. doi: 10.1038/nprot.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su D. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2014;67:460–470. doi: 10.1016/j.freeradbiomed.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan J.C. Quantitative profiling of protein s-glutathionylation reveals redox-dependent regulation of macrophage function during nanoparticle-induced oxidative stress. Acs Nano. 2016;10(1):524–538. doi: 10.1021/acsnano.5b05524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su D. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic. Biol. Med. 2014;67:460–470. doi: 10.1016/j.freeradbiomed.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S., Pevzner P.A. MS-GF plus makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014:5. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crystal Structure of 14-3-3 gamma in complex with a phosphoserine peptide. 〈http://www.rcsb.org/structure/2B05〉.
- 37.Matsumoto T. Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state. Biochem. Biophys. Res. Commun. 2010;400(3):369–373. doi: 10.1016/j.bbrc.2010.08.071. [DOI] [PubMed] [Google Scholar]
- 38.Guo R. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell. 2017;170(6):1247–1257. doi: 10.1016/j.cell.2017.07.050. (e12) [DOI] [PubMed] [Google Scholar]
- 39.Sacchetto R. Crystal structure of sarcoplasmic reticulum Ca2+-ATPase (SERCA) from bovine muscle. J. Struct. Biol. 2012;178(1):38–44. doi: 10.1016/j.jsb.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Allen D.G., Lamb G.D., Westerblad H. Skeletal muscle fatigue: cellular mechanisms. Physiol. Rev. 2008;88(1):287–332. doi: 10.1152/physrev.00015.2007. [DOI] [PubMed] [Google Scholar]
- 41.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Warburton D.E., Nicol C.W., Bredin S.S. Health benefits of physical activity: the evidence. CMAJ. 2006;174(6):801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seene T., Kaasik P. Role of exercise therapy in prevention of decline in aging muscle function: glucocorticoid myopathy and unloading. J. Aging Res. 2012;2012:172492. doi: 10.1155/2012/172492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mailloux R.J., Jin X., Willmore W.G. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol. 2014;2:123–139. doi: 10.1016/j.redox.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastore A., Piemonte F. Protein glutathionylation in cardiovascular diseases. Int. J. Mol. Sci. 2013;14(10):20845–20876. doi: 10.3390/ijms141020845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Winter J.M., Ottenheijm C.A. A two-faced cysteine residue modulates skeletal muscle contraction. Focus on "S-nitrosylation and S-glutathionylation of Cys134 on troponin I have opposing competitive actions on Ca(2+) sensitivity in rat fast-twitch muscle fibers. Am. J. Physiol. Cell Physiol. 2017;312(3):C314–C315. doi: 10.1152/ajpcell.00009.2017. [DOI] [PubMed] [Google Scholar]
- 48.Mollica J.P. S-glutathionylation of troponin I (fast) increases contractile apparatus Ca2+ sensitivity in fast-twitch muscle fibres of rats and humans. J. Physiol. 2012;590(6):1443–1463. doi: 10.1113/jphysiol.2011.224535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruger M., Linke W.A. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 2011;286(12):9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alegre-Cebollada J. S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell. 2014;156(6):1235–1246. doi: 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giganti D. Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nat. Commun. 2018;9(1):185. doi: 10.1038/s41467-017-02528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mieyal J.J. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 2008;10(11):1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aracena-Parks P. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J. Biol. Chem. 2006;281(52):40354–40368. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 54.Hidalgo C. Cross talk between Ca2+ and redox signalling cascades in muscle and neurons through the combined activation of ryanodine receptors/Ca2+ release channels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360(1464):2237–2246. doi: 10.1098/rstb.2005.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viner R.I., Williams T.D., Schoneich C. Peroxynitrite modification of protein thiols: oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 1999;38(38):12408–12415. doi: 10.1021/bi9909445. [DOI] [PubMed] [Google Scholar]
- 56.Qin F. Hydrogen peroxide-mediated SERCA cysteine 674 oxidation contributes to impaired cardiac myocyte relaxation in senescent mouse heart. J. Am. Heart Assoc. 2013;2(4):e000184. doi: 10.1161/JAHA.113.000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapp S., Tucker S.J., Ashcroft F.M. Mechanism of ATP-sensitive K channel inhibition by sulfhydryl modification. J. Gen. Physiol. 1998;112(3):325–332. doi: 10.1085/jgp.112.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liemburg-Apers D.C. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch. Toxicol. 2015;89(8):1209–1226. doi: 10.1007/s00204-015-1520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mari M. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 2009;11(11):2685–2700. doi: 10.1089/ars.2009.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barinova K.V. S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase induces formation of C150-C154 intrasubunit disulfide bond in the active site of the enzyme. Biochim. Biophys. Acta. 2017;1861(12):3167–3177. doi: 10.1016/j.bbagen.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 62.Mohr S., Stamler J.S., Brune B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J. Biol. Chem. 1996;271(8):4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 63.Eaton P. Glyceraldehyde phosphate dehydrogenase oxidation during cardiac ischemia and reperfusion. J. Mol. Cell. Cardiol. 2002;34(11):1549–1560. doi: 10.1006/jmcc.2002.2108. [DOI] [PubMed] [Google Scholar]
- 64.Sethuraman M. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 2004;3(6):1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 65.Reddy S. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem. J. 2000;347(Pt 3):821–827. [PMC free article] [PubMed] [Google Scholar]
- 66.Fu C. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J. Proteome Res. 2008;7(9):3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mailloux R.J., Willmore W.G. S-glutathionylation reactions in mitochondrial function and disease. Front. Cell Dev. Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han D. Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione. Biochemistry. 2005;44(36):11986–11996. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- 69.Kumar V. Redox proteomics of thiol proteins in mouse heart during ischemia/reperfusion using ICAT reagents and mass spectrometry. Free Radic. Biol. Med. 2013;58:109–117. doi: 10.1016/j.freeradbiomed.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 70.Mailloux R.J. Glutaredoxin-2 is required to control oxidative phosphorylation in cardiac muscle by mediating deglutathionylation reactions. J. Biol. Chem. 2014;289(21):14812–14828. doi: 10.1074/jbc.M114.550574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mailloux R.J., Treberg J.R. Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 2016;8:110–118. doi: 10.1016/j.redox.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L. Mass spectrometry profiles superoxide-induced intramolecular disulfide in the FMN-binding subunit of mitochondrial Complex I. J. Am. Soc. Mass Spectrom. 2008;19(12):1875–1886. doi: 10.1016/j.jasms.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C.L. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46(19):5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danielson S.R. Quantitative mapping of reversible mitochondrial Complex I cysteine oxidation in a Parkinson disease mouse model. J. Biol. Chem. 2011;286(9):7601–7608. doi: 10.1074/jbc.M110.190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koenitzer J.R. Fatty acid nitroalkenes induce resistance to ischemic cardiac injury by modulating mitochondrial respiration at complex II. Redox Biol. 2016;8:1–10. doi: 10.1016/j.redox.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y.R. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J. Biol. Chem. 2007;282(45):32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 77.Modis K. S-Sulfhydration of ATP synthase by hydrogen sulfide stimulates mitochondrial bioenergetics. Pharmacol. Res. 2016;113(Pt A):116–124. doi: 10.1016/j.phrs.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S.B. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc. Med. 2013;23(1):14–18. doi: 10.1016/j.tcm.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Autieri M.V., Carbone C.J. 14-3-3Gamma interacts with and is phosphorylated by multiple protein kinase C isoforms in PDGF-stimulated human vascular smooth muscle cells. DNA Cell Biol. 1999;18(7):555–564. doi: 10.1089/104454999315105. [DOI] [PubMed] [Google Scholar]
- 80.Dougherty M.K., Morrison D.K. Unlocking the code of 14-3-3. J. Cell Sci. 2004;117(Pt 10):1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 81.Dubois T. Structure and sites of phosphorylation of 14-3-3 protein: role in coordinating signal transduction pathways. J. Protein Chem. 1997;16(5):513–522. doi: 10.1023/a:1026321813463. [DOI] [PubMed] [Google Scholar]
- 82.Son Y. Mitogen-activated protein kinases and reactive oxygen species: how can ROS activate MAPK pathways? J. Signal Transduct. 2011;2011:792639. doi: 10.1155/2011/792639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hinchliffe P., Sazanov L.A. Organization of iron-sulfur clusters in respiratory complex I. Science. 2005;309(5735):771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- 84.Duan J., Gaffrey M.J., Qian W.J. Quantitative proteomic characterization of redox-dependent post-translational modifications on protein cysteines. Mol. Biosyst. 2017;13(5):816–829. doi: 10.1039/c6mb00861e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marino S.M., Gladyshev V.N. Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid. Redox Signal. 2011;15(1):135–146. doi: 10.1089/ars.2010.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material