ABSTRACT
The cyclodipeptide pulcherriminic acid synthesized by Bacillus licheniformis is an iron chelator that antagonizes certain pathogens by removing iron from the environment. But since the insoluble iron-pulcherriminic acid complex cannot act as an iron carrier as siderophores do, excessive synthesized pulcherriminic acid causes iron starvation for the producer cells. At present, the regulation of pulcherriminic acid synthesis and the mechanism by which B. licheniformis strikes a balance between biocontrol and self-protection from excessive iron removal remain unclear. This study provides insights into the regulatory network and explains the mechanism of pulcherriminic acid biosynthesis. The yvmC-cypX synthetic gene cluster was directly negatively regulated by three regulators: AbrB, YvnA, and YvmB. Within the regulatory network, YvnA expression was repressed not only by AbrB but also by iron-limiting environments, while YvmB expression was repressed by YvnA. The transporter gene yvmA is repressed by YvmB and is required for pulcherriminic acid secretion. The biosynthesis window is determined by the combined concentration of the three regulators in an iron-rich environment. Under iron-limiting conditions, cells close the pulcherriminic acid synthesis pathway by downregulating YvnA expression.
IMPORTANCE The cyclodipeptides are widespread in nature and exhibit a broad variety of biological and pharmacological activities. The cyclodipeptide scaffold is synthesized by nonribosomal peptide synthetases (NRPSs) and cyclodipeptide synthases (CDPSs). At present, it is clear that CDPSs use aminoacyl tRNAs as substrates to synthesize the two peptide bonds, and the pulcherriminic acid synthase YvmC is a member of the eight identified CDPSs. However, little is known about the regulation of cyclodipeptide synthesis and secretion. In this study, we show that AbrB, which is considered to be the main regulator of NRPS-dependent pathways, is also involved in the regulation of CDPS genes. However, AbrB is not the decisive factor for pulcherriminic acid synthesis, as the expression of YvnA determines the fate of pulcherriminic acid synthesis. With this information on how CDPS gene transcription is regulated, a clearer understanding of cyclodipeptide synthesis can be developed for B. licheniformis. Similar approaches may be used to augment our knowledge on CDPSs in other bacteria.
KEYWORDS: Bacillus licheniformis, cyclodipeptide, pulcherriminic acid, iron depletion, AbrB
INTRODUCTION
Iron is an essential micronutrient for bacterial growth and metabolic processes (1). In soil and sediments, the iron concentration stays at a low level under aerobic conditions because its solubility is controlled by stable hydroxides, oxyhydroxides, and oxides (2). For some pathogenic bacteria, iron acquisition is a significant factor for maintaining cell growth and is required for disease manifestations (3–5). To compete for iron in environments where it is limited, a broad array of bacteria secrete siderophores, which are low-molecular-weight compounds that bind Fe(III) with extremely high affinity (2). Siderophore-iron complexes are internalized into cells, and then Fe(III) is reduced to Fe(II) and released from the complexes (2, 6). Consequently, a sessile producer of a potent siderophore might compete at a distance with other microorganisms that have less avid iron uptake systems by depleting iron (7). For example, pyoverdin, a diffusible siderophore produced by fluorescent pseudomonads, acts as an iron carrier for the producer strain by forming ferripyoverdin [Fe(III)-pseudobactin] and inhibiting the growth of bacteria and fungi in iron-limiting environments (8–10). Bacillus licheniformis adopts a similar strategy for competition with other microorganisms through depletion of iron from the environment. Pulcherriminic acid is a cyclodipeptide that has the same iron-chelating group as hydroxamate siderophores and can deplete iron ions by forming insoluble pulcherrimin [Fe(III)-pulcherriminic acid complex] (11, 12). In recent years, pulcherriminic acid was reported to play an important role in the biocontrol of pathogens though competition for iron (13, 14), and it has been considered a biocontrol agent alternative to conventional chemical fungicides for the control of postharvest citrus pathogens.
Pulcherriminic acid structurally belongs to the diketopiperazines (DKPs), a large class of cyclic peptides that exhibit various biological properties (15). The DKP scaffold is mainly accessed by employing the nonribosomal peptide synthetases (NRPSs) and cyclodipeptide synthases (CDPSs) (16, 17). Previous research confirmed that pulcherriminic acid is synthesized by a CDPS in Bacillus species (18, 19). The CDPS YvmC catalyzes leucyl-tRNA to form cyclo-l-leucyl-l-leucyl (cLL), and then cytochrome P450, encoded by cypX, transforms cLL to pulcherriminic acid (12). Transcription of the yvmC-cypX cluster was shown to be negatively regulated by the multiple antibiotic resistance regulator (MarR)-like regulator YvmB, and the yvmB-yvmA cluster is located next to yvmC-cypX in B. subtilis (20). MarR proteins exist widely in bacteria and serve as transcriptional regulators of antibiotic resistance, stress responses, virulence, and catabolism of aromatic compounds (21–23).
Although the mechanisms of tRNA-dependent cyclodipeptide synthase pathways have been well studied, there is little knowledge about their biosynthesis regulation (15). Especially for pulcherriminic acid producer strains, a lack of available iron is one of many environmental challenges for Bacillus species, but insoluble pulcherrimin cannot act as an iron carrier (as siderophores can) for the producer strain, so it is likely that an excessive concentration of pulcherriminic acid would cause iron starvation and damage in producer strains. Therefore, whether a regulatory network balances iron removal for biocontrol and protects the pulcherriminic acid producer strains from excessive iron removal needs to be explored, especially in environments where iron is scarce or where its bioavailability is poor.
NRPSs and CDPSs are the main sources of DKP-containing natural products (4, 15). The NRPS-dependent antibiotics are synthesized mainly in the late exponential and stationary growth phases in Bacillus, as is pulcherriminic acid (24), and the regulation of NRPS-dependent pathways has been well studied. AbrB is one of the main global regulators that determine the timing of processes during the transition from exponential growth to the stationary growth phase, and it is known as the primary regulator of NRPS-dependent pathways in Bacillus (25–27). This information led us to the hypothesis that AbrB is also involved in regulation of CDPS genes. In the present work, due to the lack of understanding about the regulation of CDPS genes and the similarity of the products of CDPSs and NRPSs, AbrB was considered an entry point for analysis of the regulatory network of pulcherriminic acid biosynthesis and secretion in more detail.
The purpose of the present study was to provide insight into the regulatory network of pulcherriminic acid biosynthesis in B. licheniformis. The relationships between AbrB and the yvmC-cypX cluster as well as its neighboring genes, the MarR-like regulator gene yvnA and the transporter gene yvmA, were investigated. We demonstrate that the AbrB/YvnA/YvmB proteins make up a regulatory network for pulcherriminic acid biosynthesis and that YvmA is required for secretion. We also demonstrate that YvnA functions by direct or indirect sensing of the iron concentration in the environment and helps to balance iron removal and protection of the pulcherriminic acid producer strains.
RESULTS
AbrB is a negative transcriptional regulator of pulcherriminic acid biosynthesis.
To gain insights into the relationship between AbrB and pulcherriminic acid biosynthesis, the abrB deletion strain DW2 ΔabrB was constructed from the wild-type strain B. licheniformis DW2. Like its abrB deletion and overexpression derivative strains, DW2 began exponential growth at 6 h and then entered the stationary growth phase at about 26 h. At 36 h, the optical density at 600 nm (OD600) of DW2 ΔabrB was 4.02, which is 64.5% that of DW2 (6.23), suggesting that deletion of the global transcription regulator gene abrB has a negative influence on cell growth. Overexpression of abrB did not have a significant influence on cell density compared to that of DW2. Pulcherriminic acid synthesis in DW2 started at 12 h (Fig. 1A), and the absorbance at 410 nm (A410) reached 1.55 at 36 h. The DW2 ΔabrB strain showed earlier pulcherriminic acid biosynthesis, and the A410 value was 3.21, which was 1.03-fold higher than that for DW2. However, abrB overexpression caused a nonpulcherrimin phenotype. These results indicated that pulcherriminic acid synthesis was probably under negative regulation by AbrB.
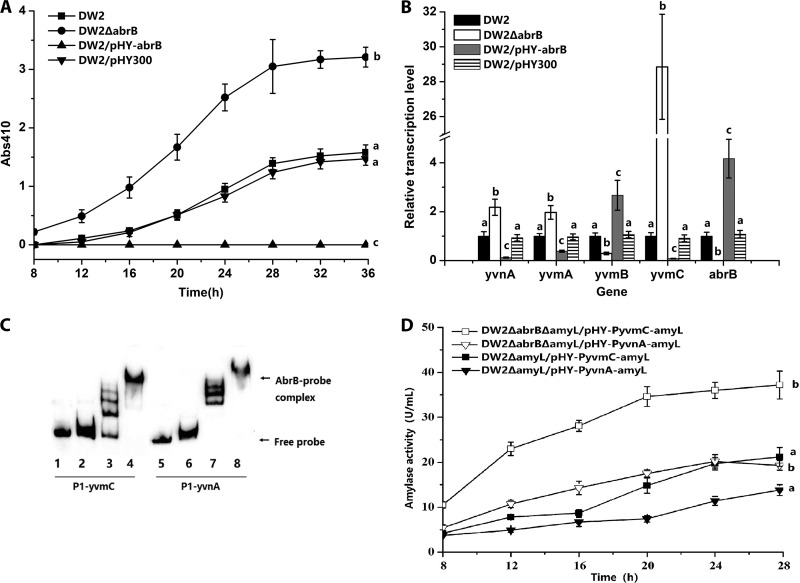
FIG 1.
AbrB negatively regulates pulcherriminic acid biosynthesis by binding to the promoter of yvmC-cypX. (A) Effects of abrB deletion (DW2 ΔabrB) and overexpression (DW2/pHY-abrB) on pulcherriminic acid production. (B) Gene transcription variations in abrB deletion and overexpression strains. The transcription levels of genes in DW2 were set to 1. (C) Binding of the AbrB protein to the promoter regions of yvmC and yvnA. Gel shifts of the labeled 91-bp P1-yvmC and P1-yvnA probes by the AbrB protein are shown. The concentrations of AbrB in lanes 1 to 8 were 0, 1, 2, 5, 0, 1, 2, and 5 ng/μl, respectively. Twenty nanograms of the P1-yvmC (lanes 1 to 4) or P1-yvnA (lanes 5 to 8) probe was added to each lane. The nonspecific competitor poly(dI-dC) was added to the EMSA binding buffer. (D) Expression-level comparisons of PyvmC-amyL and PyvnA-amyL transcriptional fusions. The amyL gene on the chromosome was deleted in DW2 and DW2 ΔabrB for the construction of DW2 ΔamyL and DW2 ΔabrB Δamy, respectively. pHY-PyvmC-amyL was then transformed into DW2 ΔamyL and DW2 ΔabrB ΔamyL to construct DW2 ΔamyL/pHY-PyvmC-amyL and DW2 ΔabrB ΔamyL/pHY-PyvmC-amyL. pHY-PyvnA-amyL was transformed into DW2 ΔamyL and DW2 ΔabrB ΔamyL to construct DW2 ΔamyL/pHY-PyvnA-amyL and DW2 ΔabrB ΔamyL/pHY-PyvnA-amyL.
The influence of abrB on transcription of yvmC, yvmB, yvmA, and yvnA was evaluated by reverse transcription-quantitative PCR (RT-qPCR). The transcription levels of genes in DW2 were set to 1. Consistent with the pulcherriminic acid production levels, the relative transcription level of yvmC increased to 28.85 in DW2 ΔabrB and decreased to 0.08 in DW2/pHY-abrB (Fig. 1B). These results indicated that AbrB controlled pulcherriminic acid biosynthesis by negatively regulating yvmC-cypX transcription. In contrast to that of yvmC, the relative transcription level of yvmB decreased to 0.29 in DW2 ΔabrB and increased to 2.67 in DW2/pHY-abrB. The relative transcription levels of yvmA and yvnA increased to 1.97 and 2.18, respectively, in DW2 ΔabrB and decreased to 0.38 and 0.12, respectively, in DW2/pHY-abrB.
To identify if AbrB directly regulates these genes, binding of the AbrB protein to the promoter regions of yvmC, yvmB, yvmA, and yvnA was investigated with electrophoretic mobility shift assays (EMSAs). Only the P1-yvmC and P1-yvnA probes were retarded due to formation of AbrB-DNA complexes (Fig. 1C). The yvmA and yvmB probes did not interact with the AbrB protein (data not shown). To evaluate the effects of AbrB on promoter activity, transcriptional fusions of the report gene amyL and the promoters PyvmC and PyvnA were constructed, and the expression plasmids pHY-PyvmC-amyL and pHY-PyvnA-amyL were electroporated into DW2 ΔamyL and DW2 ΔabrB ΔamyL, respectively. At 28 h, α-amylase assays with DW2 ΔabrB ΔamyL/pHY-PyvmC-amyL (37.2 U/ml) showed a 75.5% increase in amylase activity compared to that of DW2 ΔamyL/pHY-PyvmC-amyL (21.2 U/ml). DW2 ΔabrB ΔamyL/pHY-PyvnA-amyL (20.2 U/ml) showed a 46.4% increase in amylase activity compared to that of DW2 ΔamyL/pHY-PyvnA-amyL (13.8 U/ml) (Fig. 1D). Collectively, these results confirmed that AbrB directly repressed yvmC-cypX and yvnA expression by binding to promoter regions and influenced yvmA and yvmB transcription indirectly.
YvmB is a negative transcriptional regulator of yvmC and yvmA.
Although YvmB has been reported to be a negative regulator of pulcherriminic acid synthesis in B. subtilis (20), its function in B. licheniformis was still unknown, and whether YvmB participated in the regulation of yvnA and abrB was also not clear. Thus, it was necessary to reexamine the function of YvmB in pulcherriminic acid biosynthesis in B. licheniformis.
yvmB deletion and overexpression did not influence the cell growth of B. licheniformis, but pulcherriminic acid production increased in the yvmB deletion strain DW2 ΔyvmB. The A410 value for DW2 ΔyvmB reached 2.53, which was an increase of 69% compared to that for DW2 (1.55) (Fig. 2A). Pulcherriminic acid was not synthesized by DW2/pHY-yvmB, and further work showed that the relative transcription level of yvmC was 3.42 in DW2 ΔyvmB and 0.13 in DW2/pHY-yvmB (Fig. 2B). Similarly, the relative transcription level of yvmA increased to 2.86 in DW2 ΔyvmB and decreased to 0.11 in DW2/pHY-yvmB. Deletion or overexpression of YvmB had no effect on the transcription of abrB or yvnA.
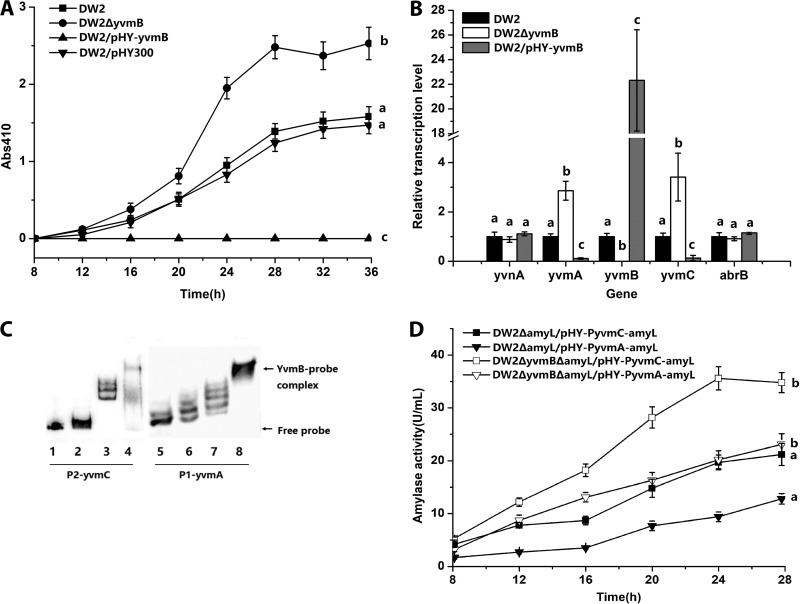
FIG 2.
YvmB represses pulcherriminic acid biosynthesis by binding to the promoter of yvmC-cypX. (A) Effects of yvmB deletion (DW2 ΔyvmB) and overexpression (DW2/pHY-yvmB) on pulcherriminic acid production. (B) Gene transcription variations in yvmB deletion and overexpression strains. (C) Binding of the YvmB protein to the promoter regions of yvmC and yvmA. Gel shifts of the labeled P2-yvmC (87 bp) and P1-yvmA (91 bp) probes by the YvmB protein are shown. The concentrations of YvmB in lanes 1 to 8 were 0, 1, 2, 5, 0, 1, 2, and 5 ng/μl, respectively. Twenty nanograms of the P2-yvmC (lanes 1 to 4) or P1-yvmA (lanes 5 to 8) probe was added to each lane. The nonspecific competitor poly(dI-dC) was added to the EMSA binding buffer. (D) Expression-level comparisons of PyvmC-amyL and PyvmA-amyL transcriptional fusions. The amyL gene on the chromosome was deleted in DW2 ΔyvmB for the construction of DW2 ΔyvmB ΔamyL. pHY-PyvmC-amyL was transformed into DW2 ΔamyL and DW2 ΔyvmB ΔamyL to construct DW2 ΔamyL/pHY-PyvmC-amyL and DW2 ΔyvmB ΔamyL/pHY-PyvmC-amyL. pHY-PyvmA-amyL was transformed into DW2 ΔamyL and DW2 ΔyvmB ΔamyL to construct DW2 ΔamyL/pHY-PyvmA-amyL and DW2 ΔyvmB ΔamyL/pHY-PyvmA-amyL.
EMSAs were performed to investigate protein-DNA interactions between YvmB and promoter probes for yvmC and yvmA. The P2-yvmC probe was retarded in the presence of YvmB (Fig. 2C). Similar to the result for yvmC, interaction of the P1-yvmA probe and the YvmB protein was also confirmed. Transcriptional fusions of the amyL gene and the PyvmA promoter were constructed. The α-amylase activity was 35.6 U/ml in DW2 ΔyvmB ΔamyL/pHY-PyvmC-amyL and 23.1 U/ml in DW2 ΔyvmB ΔamyL/pHY-PyvmA-amyL at 28 h, which was 1.68-fold higher than that of DW2 ΔamyL/pHY-PyvmC-amyL (21.2 U/ml) and 1.80-fold higher than that of DW2 ΔamyL/pHY-PyvmA-amyL (12.8 U/ml) (Fig. 2D). These results confirmed that YvmB represses expression of yvmC and yvmA directly by binding to their promoters in B. licheniformis DW2.
YvnA is a negative transcriptional regulator of yvmC and yvmB.
yvnA deletion and overexpression had no effect on the cell growth of B. licheniformis. Both YvnA deletion and overexpression in DW2 completely abrogated pulcherriminic acid synthesis (Fig. 3A), suggesting that YvnA might have multiple functions in the regulation of pulcherriminic acid synthesis. Transcription levels of yvmC, yvmB, yvmA, and abrB were measured in DW2 ΔyvnA and DW2/pHY-yvnA. Both deletion and overexpression of YvnA repressed yvmC transcription, with relative transcription levels of 0.05 in DW2 ΔyvnA and 0.08 in DW2/pHY-yvnA (Fig. 3B). The relative transcription level of yvmB increased to 4.26 in DW2 ΔyvnA and decreased to 0.12 in DW2/pHY-yvnA. In contrast to yvmB, yvmA was positively regulated by YvnA, with its relative transcription level decreasing to 0.28 in DW2 ΔyvnA and increasing to 2.07 in DW2/pHY-yvnA.
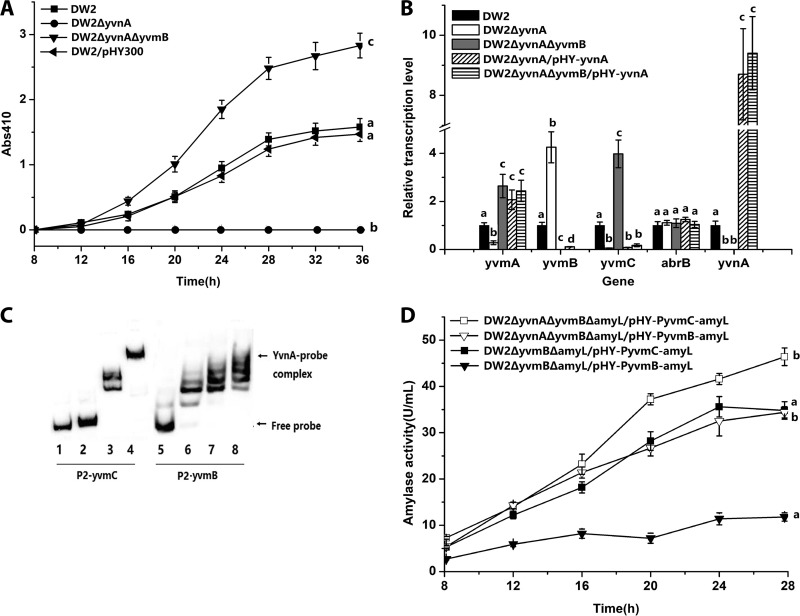
FIG 3.
Influence of YvnA on pulcherriminic acid biosynthesis. (A) Effects of yvnA on pulcherriminic acid production. The DW2 ΔyvnA ΔyvmB strain was set as a control to exclude the effects of yvmB. DW2/pHY-yvnA and DW2 ΔyvnA ΔyvmB/pHY-yvnA did not produce pulcherriminic acid (data not shown). (B) Effects of yvnA deletion and overexpression on gene transcription. (C) Binding of the YvnA protein to the promoter regions of yvmC and yvmB. Gel shifts of the labeled P2-yvmC (87 bp) and P2-yvmB (87 bp) probes by the YvnA protein are shown. The concentrations of YvnA in lanes 1 to 8 were 0, 1, 2, 5, 0, 1, 2, and 5 ng/μl, respectively. Twenty nanograms of the P2-yvmC (lanes 1 to 4) or P2-yvmB (lanes 5 to 8) probe was added to each lane. The nonspecific competitor poly(dI-dC) was added to the EMSA binding buffer. (D) Expression-level comparisons of PyvmC-amyL and PyvmB-amyL transcriptional fusions. The amyL gene on the chromosome was deleted in DW2 ΔyvmB and DW2 ΔyvmB ΔyvnA for the construction of DW2 ΔyvmB ΔamyL and DW2 ΔyvmB ΔyvnA ΔamyL, respectively. pHY-PyvmC-amyL was transformed into DW2 ΔyvmB ΔamyL and DW2 ΔyvmB ΔyvnA ΔamyL to construct DW2 ΔyvmB ΔamyL/pHY-PyvmC-amyL and DW2 ΔyvmB ΔyvnA ΔamyL/pHY-PyvmC-amyL, respectively. pHY-PyvmB-amyL was transformed into DW2 ΔyvmB ΔamyL and DW2 ΔyvmB ΔyvnA ΔamyL to construct DW2 ΔyvmB ΔamyL/pHY-PyvmB-amyL and DW2 ΔyvmB ΔyvnA ΔamyL/pHY-PyvmB-amyL, respectively.
Interactions between YvnA and the promoter regions of yvmC, yvmB, and yvmA were investigated by use of EMSAs. The results showed that the P2-yvmC and P2-yvmB probes were retarded in the presence of YvnA (Fig. 3C), but the probes for the yvmA promoter did not have interactions with the YvnA protein.
To avoid the influence of YvmB, the α-amylase activity assay was performed with DW2 ΔyvmB ΔamyL/pHY-PyvmC-amyL and DW2 ΔyvnA ΔyvmB ΔamyL/pHY-PyvmC-amyL. The results showed that the α-amylase activity of DW2 ΔyvnA ΔyvmB ΔamyL/pHY-PyvmC-amyL (46.3 U/ml) was 1.33-fold higher than that of DW2 ΔyvmB ΔamyL/pHY-PyvmC-amyL (34.8 U/ml) at 28 h (Fig. 3D). In DW2 ΔyvnA ΔamyL/pHY-PyvmB-amyL, with a deletion of yvnA, α-amylase activity increased to 34.4 U/ml at 28 h, which was 1.92-fold higher than that of DW2 ΔamyL/pHY-PyvmB-amyL (11.8 U/ml). These results showed that YvnA repressed yvmC and yvmB by binding directly to promoter regions and that yvnA regulated yvmA indirectly in DW2/pHY-yvnA. On the other hand, DW2 ΔyvnA likely repressed pulcherriminic acid synthesis by upregulating YvmB.
The yvnA and yvmB double-deletion strain DW2 ΔyvnA ΔyvmB was constructed to investigate the function of YvmB in a yvnA deletion strain. The results showed that the requirement for YvnA in pulcherriminic acid production was bypassed by yvmB deficiency, and YvnA overexpression in DW2 ΔyvnA ΔyvmB/pHY-yvnA inhibited pulcherriminic acid biosynthesis (Fig. 3A). RT-qPCR results showed that the relative yvmC transcription level increased to 3.98 in DW2 ΔyvnA ΔyvmB but decreased to 0.16 in DW2 ΔyvnA ΔyvmB/pHY-yvnA (Fig. 3B). These results indicated that YvnA is a negative regulator of yvmC-cypX and yvmB that binds directly to their promoters. YvnA also indirectly regulates the expression of yvmC-cypX and yvmA through repression of yvmB expression.
YvmA is required for pulcherriminic acid secretion.
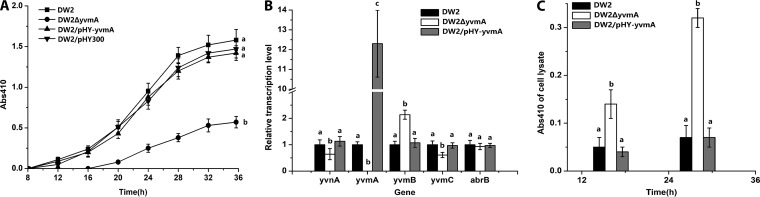
YvmA is a putative major facilitator superfamily (MFS)-like transporter. Our results showed that the yvmA gene is located next to yvmC and negatively regulated by YvmB in B. licheniformis. To investigate the relationship between YvmA and pulcherriminic acid efflux, the deletion strain DW2 ΔyvmA was constructed. YvmA overexpression had no influence on the production of pulcherriminic acid compared to that in DW2/pHY300. However, yvmA deficiency caused a 64% decrease in pulcherriminic acid production, and release of pulcherriminic acid was delayed for about 8 h (Fig. 4A). RT-qPCR results showed that the relative transcription level of yvmB increased to 2.13, that of yvnA decreased to 0.64, and that of yvmC decreased to 0.61 in DW2 ΔyvmA (Fig. 4B). Whether the pulcherriminic acid synthesized before 16 h accumulated in the intracellular space is unknown. Pulcherrimin in the medium was dissolved and removed by use of a 0.1-mmol/liter NaOH solution, and the A410 of cell lysates was measured at 16 and 28 h. The A410 value for DW2 ΔyvmA cell lysate was 0.14 at 16 h and 0.32 at 28 h (Fig. 4C). The A410 value was 0.05 at 16 h and 0.07 at 28 h for DW2, similar to measurements for DW2/pHY-yvmA. These results demonstrate that YvmA is required for secretion of pulcherriminic acid and that disruption of yvmA causes intracellular accumulation of pulcherrimin.
FIG 4.
YvmA is required for secretion of pulcherriminic acid. (A) Effects of yvmA deletion (DW2 ΔyvmA) and overexpression (DW2/pHY-yvmA) on pulcherriminic acid production. (B) Gene transcription variations in yvmA deletion and overexpression strains. The transcription levels of genes in DW2 were set to 1. (C) Effects of yvmA deletion (DW2 ΔyvmA) and overexpression (DW2/pHY-yvmA) on intracellular accumulation of pulcherriminic acid.
Pulcherriminic acid antagonizes fungal growth by iron depletion.
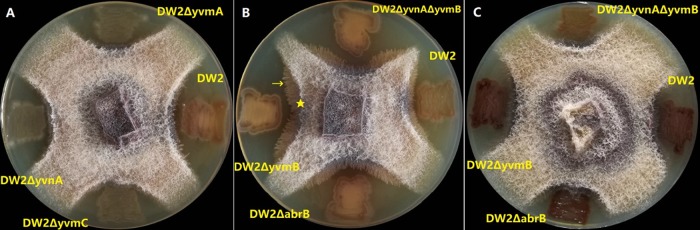
Seven B. licheniformis strains were used to inoculate peptone-dextrose agar (PDA) plates supplemented with no FeCl3 or 120 mg/liter FeCl3. On PDA plates without FeCl3 (Fig. 5), color halos of insoluble pulcherrimin appeared, indicating a background concentration of Fe(III) in the PDA medium. Inhibition zone tests showed that DW2 inhibited growth of Fusarium oxysporum. DW2 ΔyvnA, DW2 ΔyvmA, and the yvmC deletion strain DW2 ΔyvmC, which did not synthesize or secrete less pulcherriminic acid, showed smaller inhibition zones (Table 1). In addition, DW2 ΔabrB, DW2 ΔyvmB, and DW2 ΔyvnA ΔyvmB, which had higher pulcherriminic acid yields, showed larger inhibition zones. These results showed that mainly antagonistic activity was conferred by pulcherriminic acid. But DW2 ΔyvmC and DW2 ΔyvnA still had narrower inhibition zones, suggesting that B. licheniformis has products in addition to pulcherriminic acid that inhibit F. oxysporum growth. On PDA plates with 120 mg/liter FeCl3, the diameters of inhibition zones were smaller than those on plates without FeCl3 and the cells turned dark red, suggesting that pulcherriminic acid antagonizes F. oxysporum growth by iron depletion and that pulcherriminic acid was retained in colonies in the iron-rich environment.
FIG 5.
Inhibition of F. oxysporum by pulcherriminic acid. The asterisk indicates the edge of the inhibition zone. The arrow indicates hyphae growing into the inhibition zone.
TABLE 1.
Formation of pigmented halo and inhibition zone around F. oxysporum on PDA plates
| Strain and FeCl3 presence | Colony color | Width of inhibition zone (mm) | Width of color halo (mm) |
|---|---|---|---|
| No FeCl3 | |||
| DW2 | Pink | 4.5 | 3 |
| DW2 ΔyvmC | White | 1.5 | 0 |
| DW2 ΔyvnA | White | 2.0 | 0 |
| DW2 ΔyvmA | White | 2.0 | 0 |
| DW2 ΔabrB | Pink | 7.5 | 7 |
| DW2 ΔyvmB | Pink | 8.5 | 7.5 |
| DW2 ΔyvnA ΔyvmB | Pink | 8 | 7.5 |
| FeCl3 (120 mg/liter) | |||
| DW2 | Red | 2.5 | 0 |
| DW2 ΔabrB | Dark red | 2 | 0 |
| DW2 ΔyvmB | Dark red | 2 | 0 |
| DW2 ΔyvnA ΔyvmB | Dark red | 2.5 | 0 |
Excessive pulcherriminic acid synthesis damages the producer strain in iron-limiting environments.
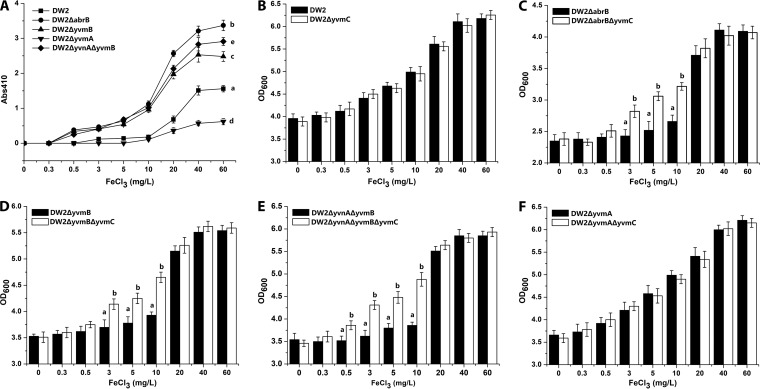
To investigate whether excessive pulcherriminic acid synthesis causes iron starvation for the producer strain, the influence of the iron concentration on pulcherriminic acid yields was examined (Fig. 6A). The B. licheniformis strains were cultivated in media with different final concentrations of FeCl3. In DW2, pulcherriminic acid was synthesized at low levels in the presence of 3 to 10 mg/liter FeCl3 (A410 = 0.11 to 0.18) but rapidly increased in 20 to 40 mg/liter FeCl3 (A410 = 0.24 to 1.56). DW2 ΔabrB, DW2 ΔyvmB, and DW2 ΔyvnA ΔyvmB started pulcherriminic acid synthesis in the presence of a lower FeCl3 concentration (0.5 mg/liter) and showed higher pulcherriminic acid yields in 0.5 to 10 mg/liter FeCl3 (A410 = 0.25 to 1.12) than those of DW2. These results indicated that iron limitation (0.5 to 10 mg/liter FeCl3) repressed pulcherriminic acid synthesis in DW2 to keep its synthetic capability at a low level but that deletion of the regulators relieved the repression. yvmC deletion strains of DW2 ΔabrB, DW2 ΔyvmB, DW2 ΔyvmA, and DW2 ΔyvnA ΔyvmB were then constructed. The cell densities at stationary growth phase (30 h) were examined at different iron concentrations. As shown in Fig. 6B, the cell density of DW2 did not show significant differences from that of DW2 ΔyvmC under all conditions, indicating that pulcherriminic acid produced by DW2 did not cause any iron starvation that affected cell growth. On the other hand, DW2 ΔabrB ΔyvmC had significantly higher cell densities than those of DW2 ΔabrB in 3 to 10 mg/liter FeCl3, but there was no difference in cell densities in an iron-rich environment (20 to 60 mg/liter) (Fig. 6C). Similar situations were also observed with DW2 ΔyvmB versus DW2 ΔyvmB ΔyvmC (Fig. 6D) and DW2 ΔyvnA ΔyvmB versus DW2 ΔyvnA ΔyvmB ΔyvmC (Fig. 6E). DW2 ΔyvmA, which had lower pulcherriminic acid yields than those of DW2, also had cell densities similar to those of DW2 ΔyvmA ΔyvmC at different FeCl3 concentrations (Fig. 6F). These results suggest that in iron-limiting environments, excessive synthesis of pulcherriminic acid depletes iron ions and damages cell growth.
FIG 6.
Influence of pulcherriminic acid yield on cell density. (A) Effect of FeCl3 concentration on the pulcherriminic acid yield of each strain. The A410 of each strain was measured after culture for 36 h. (B to F) Cell densities (OD600) of the strains at different FeCl3 concentrations.
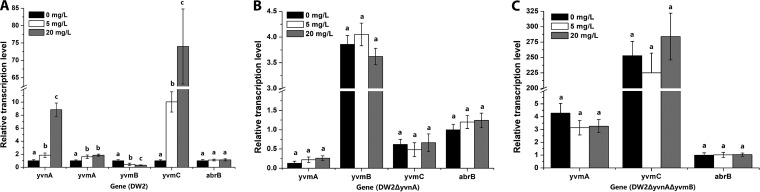
FeCl3 induces expression of YvnA.
Various transcriptional regulators of siderophore biosynthesis function by direct or indirect sensing of the extra- or intracellular presence of Fe/Fe-siderophores (2). To investigate the roles of abrB, yvnA, and yvmB in the sensing of iron, transcription levels of the related genes were examined in the presence of 0 mg/liter, 5 mg/liter, and 20 mg/liter FeCl3. Transcription levels of the genes in DW2 at 0 mg/liter FeCl3 were used as the controls and set to 1. As shown in Fig. 7A, with increasing concentrations of FeCl3, transcription of yvnA, yvmA, and yvmC in DW2 increased to different levels, but in contrast, transcription of yvmB decreased. In addition, the transcription of abrB was not influenced by FeCl3. These results suggested that pulcherriminic acid biosynthesis is indeed induced by an iron-rich environment but still could not confirm which gene was directly induced by iron. In DW2 ΔyvnA, transcription of yvmB did not obviously change with different FeCl3 concentrations (Fig. 7B). In DW2 ΔyvnA ΔyvmB, transcription of yvmA and yvmC also did not show obvious differences at different FeCl3 concentrations after both yvnA and yvmB were deleted (Fig. 7C). These results indicate that expression of abrB, yvmA, yvmB, and yvmC is not directly influenced by FeCl3. YvnA is responsible for direct or indirect iron sensing in the regulatory system.
FIG 7.
Effects of FeCl3 concentration on transcription of genes. The relative transcription levels of the genes in DW2 (A), DW2 ΔyvnA (B), and DW2 ΔyvnA ΔyvmB (C) in media containing 0, 5, and 20 mg/liter FeCl3 are shown. The transcription levels of genes in DW2 in the medium contain 0 mg/liter FeCl3 were set to 1.
DISCUSSION
In neutral or alkaline soils, Fe(III) is poorly soluble, and the total soluble Fe(III) species represent about 10−10 mol/liter at equilibrium with soil iron (7). Competition for iron is proposed to be a mechanism to suppress soilborne pathogens in this environment (28). In a previous study, pathogenic F. oxysporum was efficiently suppressed by siderophores, such as pseudobactin, which is produced by Pseudomonas spp. (29). Our results showed that pulcherriminic acid can function as a biocontrol agent, as do other soluble siderophores, by forming insoluble pulcherrimin. Bacillus-based biological control agents have great potential in integrated pest management systems (30). For Bacillus spp., the global regulator AbrB is known to control a variety of self-protective functions in stressful environments, including production of NRPS-dependent lipopeptide and peptide antibiotics (26, 31), biofilm formation, and root colonization (32–34). In the present study, the regulatory range of AbrB was extended to CDPS-dependent pulcherriminic acid biosynthesis. This helps B. licheniformis to achieve effective iron homeostasis in response to a range of iron stresses.
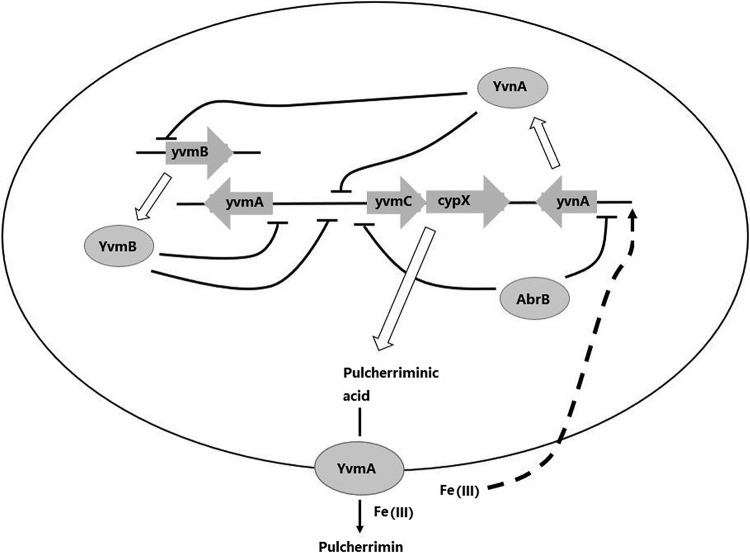
Unlike the case of antibiotic-producing strains, which are self-resistant to excreted antibiotics, efficient secretion of pulcherriminic acid cannot avoid excessive iron removal. To meet the requirements of interspecific competition and self-protection from iron starvation, precise regulation is necessary. Our results show that protection is achieved mainly by modulating the expression levels of the AbrB/YvnA/YvmB system and that the synthesis window is determined by the combined concentration of AbrB, YvnA, and YvmB (Fig. 8). Based on our results, AbrB linked pulcherriminic acid biosynthesis with cell growth status and YvnA had core functions in the regulatory system by sensing the iron concentration (directly or indirectly) and forming two sets of dual-regulation systems, AbrB-YvnA and YvnA-YvmB. Dual-regulation systems are common models for precise regulation of the biosynthesis of a vast number of secondary metabolites. For example, the biosynthesis of subtilosin is under dual regulation by AbrB-Rok (AbrB and Rok repress the subtilosin biosynthesis cluster [sboAX-albABCDEFG], and AbrB represses expression of Rok) in B. subtilis (35–37). In our previous study, due to repression of Spo0A-P, AbrB was expressed at a relatively high level during exponential growth phase and decreased during stationary phase (27). Decreasing expression of AbrB with cell growth led to increased YvnA and, consequently, to decreased YvmB, which is inhibited by YvnA. This result indicated that during exponential growth phase, pulcherriminic acid biosynthesis was mainly repressed by AbrB and YvmB, and YvnA was expressed at a relative low level. When the cells entered stationary phase, YvnA had a higher expression level and acted as the main repressor of yvmC-cypX due to a lower concentration of AbrB. In an iron-rich environment, although yvnA was induced to be expressed fully, which enhanced its repression of yvmC-cypX, it also reduced the YvmB concentration. Therefore, pulcherriminic acid could still be synthesized rapidly.
FIG 8.
Regulatory network for pulcherriminic acid biosynthesis in B. licheniformis.
It is notable that both YvnA deletion and overexpression disrupted pulcherriminic acid biosynthesis. Transcription of yvmC-cypX was inhibited by both excessively low and high activities of YvnA due to the effects on biosynthesis genes and YvmB. This specific phenomenon of dual-regulation systems is called see-saw regulation (38). In iron-limiting environments, cells stop the pulcherriminic acid synthesis pathway completely by downregulating YvnA expression and indirectly increase the YvmB concentration in DW2, giving a result similar to that of deleting the yvnA gene in an iron-rich environment. Therefore, control of yvmC-cypX by the AbrB/YvnA/YvmB regulatory system avoided rapid pulcherriminic acid synthesis and excessive iron ion removal, which can be considered a self-protection mechanism in B. licheniformis DW2.
MFS transporters target a wide spectrum of substrates, including ions, lipids, carbohydrates, amino acids, peptides, nucleosides, and other molecules (39). Sometimes, disruption of an MFS transport gene does not abolish substrate secretion completely. At this point, pulcherriminic acid production has similarities to siderophore secretion. For example, deletion of the MFS-like exporter gene entS results in a 50% reduction of enterobactin secretion in Escherichia coli (40). In B. subtilis, secretion of bacillibactin decreases to about 10% in a strain deficient in the MFS-like transporter gene ymfE compared to the level in the wild-type strain (41). In the present study, the MFS-like protein YvmA was identified as an exporter of pulcherriminic acid, and yvmA deletion resulted in a 64% decrease in pulcherriminic acid secretion compared to that of DW2. Accumulation of intracellular pulcherriminic acid also increased in the yvmA-deficient strain. Based on our results, YvmA is probably not the only pulcherriminic acid exporter in B. licheniformis. The remaining export capacity might be contributed by alternative specific transporters or low-substrate-specificity efflux systems. In previous research, some siderophores were also found to be secreted by multiple transporters. MFS, the resistance, nodulation, and cell division (RND) superfamily, and the ATP-binding cassette (ABC) superfamily are the main classes of siderophore efflux pumps (2). The RND transporters AcrB and AcrD, the multidrug transporter MdtABC (42), and the MFS protein EntS (40) are all involved in enterobactin export in E. coli. In some cases, the single disruption of an exporter gene can cause or not cause accumulation of intracellular siderophores (40, 43, 44). The differences are thought to be unrelated to the export capacity of alternative transporters and the degree of coupling between synthetic and transporter genes (2).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic material.
Strains and plasmids used in this study are listed in Table 2. In general, B. licheniformis strains and their derivatives and E. coli strains were grown on Luria-Bertani (LB) agar plates or in LB broth supplemented with antibiotics (tetracycline, 20 μg/ml; ampicillin, 50 μg/ml; and kanamycin, 20 μg/ml), where necessary, at 37°C. Pulcherriminic acid was synthesized by cultivating cells in 50 ml ME medium (20 g/liter glucose, 12 g/liter sodium citrate, 7 g/liter NH4Cl, 0.5 g/liter K2HPO4, 0.50 g/liter MgSO4, 0.04 g/liter FeCl3, 0.15 g/liter CaCl2·2H2O, 0.01 g/liter MnSO4, pH 7.5) in 250-ml flasks on a rotary shaker (180 rpm) at 37°C for 36 h.
TABLE 2.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Characteristic(s) | Reference or sourcea |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA*1 hsdR17(rK− mK−) phoA supE*44 λ− thi-1 gyrA96 relA*1 | TaKaRa |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | TaKaRa |
| B. licheniformis strains | ||
| DW2 | Wild type (CCTCC M2011344) | CCTCC |
| DW2 ΔabrB | ΔabrB | This study |
| DW2 ΔyvmB | ΔyvmB | This study |
| DW2 ΔyvnA | ΔyvnA | This study |
| DW2 ΔyvmA | ΔyvmA | This study |
| DW2 ΔyvnA ΔyvmB | ΔyvnA ΔyvmB | This study |
| DW2 ΔamyL | ΔamyL | This study |
| DW2 ΔabrB ΔamyL | ΔabrB ΔamyL | This study |
| DW2 ΔyvmB ΔamyL | ΔyvmB ΔamyL | This study |
| DW2 ΔyvnA ΔyvmB ΔamyL | ΔyvnA ΔyvmB ΔamyL | This study |
| DW2 ΔyvmC | ΔyvmC | This study |
| DW2 ΔabrB ΔyvmC | ΔabrB ΔyvmC | This study |
| DW2 ΔyvmB ΔyvmC | ΔyvmB ΔyvmC | This study |
| DW2 ΔyvnA ΔyvmB ΔyvmC | ΔyvnA ΔyvmB ΔyvmC | This study |
| DW2 ΔyvmA ΔyvmC | ΔyvmA ΔyvmC | This study |
| Plasmids | ||
| T2(2)-ori | E. coli-B. licheniformis shuttle vector; oripUC/orits; temperature sensitive; Kanr | Laboratory stock |
| T2(2)-abrB | T2(2)-ori derivative containing homologous arms for abrB knockout | This study |
| T2(2)-yvmB | T2(2)-ori derivative containing homologous arms for yvmB knockout | This study |
| T2(2)-yvnA | T2(2)-ori derivative containing homologous arms for yvnA knockout | This study |
| T2(2)-yvmA | T2(2)-ori derivative containing homologous arms for yvmA knockout | This study |
| T2(2)-amyL | T2(2)-ori derivative containing homologous arms for amyL knockout | This study |
| T2(2)-yvmC | T2(2)-ori derivative containing homologous arms for yvmC knockout | This study |
| pHY-300PLK | E. coli-Bacillus shuttle vector; Ampr (E. coli) Tcr (both E. coli and B. licheniformis) | TaKaRa |
| pHY-abrB | pHY300PLK harboring abrB fused with P43 promoter and amyL terminator | This study |
| pHY-yvmB | pHY300PLK harboring yvmB fused with P43 promoter and amyL terminator | This study |
| pHY-yvnA | pHY300PLK harboring yvnA fused with P43 promoter and amyL terminator | This study |
| pHY-yvmA | pHY300PLK harboring yvmA fused with P43 promoter and amyL terminator | This study |
| pHY-PyvmC-amyL | pHY300PLK harboring amyL fused with PyvmC promoter | This study |
| pHY-PyvmB-amyL | pHY300PLK harboring amyL fused with PyvmB promoter | This study |
| pHY-PyvnA-amyL | pHY300PLK harboring amyL fused with PyvnA promoter | This study |
| pHY-PyvmA-amyL | pHY300PLK harboring amyL fused with PyvmA promoter | This study |
| pET-28a(+) | Protein expression vector in BL21(DE3); Kanr | Novagen |
| pET-28a-abrB | pET-28a(+) harboring abrB gene | This study |
| pET-28a-yvmB | pET-28a(+) harboring yvmB gene | This study |
| pET-28a-yvnA | pET-28a(+) harboring yvnA gene | This study |
CCTCC, China Center for Type Culture Collection.
Analysis of pulcherriminic acid concentration and cell density.
The red pigment produced by B. licheniformis DW2 was shown to be pulcherrimin in our previous research (19). It has three characteristic absorption peaks, at 243, 282, and 410 nm, when dissolved in 2 mol/liter NaOH (19). The absorbance at 410 nm was used to characterize the pulcherriminic acid content (45). Two milliliters of culture broth was centrifuged at 10,000 × g for 2 min. Pellets (containing cells and pulcherrimin) were washed twice with deionized water, resuspended in 0.1 mmol/liter NaOH solution to dissolve pulcherrimin, and centrifuged at 10,000 × g for 2 min, and the supernatant (contain pulcherriminic acid) was removed. The cells were then resuspended in water to measure the cell density (OD600).
Construction of genetically engineered strains and protein purification.
T2(2)-ori was the shuttle plasmid used for construction of the knockout vector for B. licheniformis; this plasmid has a temperature-sensitive replicon. In-frame deletion mutant strains were constructed according to our previously reported method (27). The abrB, yvmB, yvnA, and yvmA overexpression plasmids were constructed based on the shuttle plasmid pHY300PLK. The constitutive P43 promoter and the terminator of the amyL gene were fused with target gene fragments and ligated into pHY300PLK to construct gene expression plasmids. Plasmids were electroporated into B. licheniformis DW2 (46). The α-amylase reporter strains were constructed by fusing the amyL gene to promoter regions of target genes (bp −200 to +21), ligating the fusions with pHY300PLK, and electroporating the constructs into B. licheniformis strains.
The AbrB, YvmB, and YvnA proteins were expressed via the pET-28a(+) plasmid (Novagen, Denmark) in E. coli BL21(DE3). Recombinant E. coli BL21(DE3) strains harboring plasmids were cultured in LB medium at 16°C. At an OD600 of 0.6, protein was induced with 0.3 mmol/liter IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h, and His-tagged proteins were purified using a Ni-nitrilotriacetic acid purification system (GenScript Corporation, USA). Purity and molecular masses of proteins were determined by SDS-PAGE. Concentrations of purified proteins were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA).
Quantitative real-time PCR.
Total RNA was extracted from B. licheniformis cells at mid-log phase (16 h) by use of TRIzol reagent (Invitrogen, USA). DNase I was applied to degrade contaminating DNA from total RNA according to the manufacturer's instructions. RNA concentrations were determined on a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). First-strand cDNA was amplified from 0.5 μg total RNA by use of Revert Aid first-strand cDNA synthesis kits (Thermo Scientific, USA). Reverse transcription-quantitative PCR (RT-qPCR) was performed on a Vii7 real-time PCR system (ABI, USA), using 20-μl reaction mixtures. B. licheniformis DW2 was used as the reference strain, and the 16S rRNA gene was the reference gene used to normalize the data via the threshold cycle (CT) number (ΔCT = CT target gene − CT 16S; ΔΔCT = ΔCT reference strain − ΔCT target strain; gene expression level = 2−ΔΔCT).
Electrophoretic mobility shift assays.
Binding regions for proteins were scanned at the locations from bp −200 to +21 for each promoter. Each 221-bp sequence was divided into three probes, and each probe was amplified from B. licheniformis DW2 chromosomal DNA by use of biotin-labeled primer pairs (Tsingke, China). Probes for bp −70 to +21 were named P1, probes for bp −148 to −61 were named P2, and probes for bp −200 to −137 were named P3. EMSAs were carried out by use of chemiluminescence EMSA kits (Beyotime, China) according to the manufacturer's instructions. Samples were analyzed in 8% native-PAGE gels. Proteins were transblotted to nylon membranes (Beyotime, China) by use of a mini-transblot electrophoresis apparatus (Liuyi, China). Membranes were treated and analyzed by use of an MF-Chemi Bis chemiluminescence imaging system (DNR Bio-Imaging Systems, Israel).
α-Amylase activity assays.
α-Amylase activity assays were performed as described previously (47).
Effect of iron on antagonism.
The antagonistic activity of pulcherriminic acid against F. oxysporum was studied on PDA plates supplemented with 0 mg/liter and 120 mg/liter FeCl3 (30 ml PDA medium per plate). B. licheniformis DW2 and derivative strains were used to inoculate the edges of plates, and F. oxysporum was placed in the center. PDA plates were incubated for 9 days at 30°C.
Statistical analyses.
All samples were analyzed in triplicate. In the figures, statistical differences (P ≤ 0.05) in independent t tests are denoted by letters.
ACKNOWLEDGMENTS
This work was supported by the National Program on Key Basic Research Project (973 Program; grant 2015CB150505) and the Science and Technology Program of Wuhan (grant 20160201010086).
We have no conflicts of interest to declare, and the manuscript was approved for publication by all authors. The work described here is original research that has not been published previously and is not under consideration for publication elsewhere, in whole or in part.
REFERENCES
- 1.Barber MF, Elde NC. 2015. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miethke M, Marahiel M. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 4.Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behnsen J, Raffatellu M. 2016. Siderophores: more than stealing iron. mBio 7:e01906-16. doi: 10.1128/mBio.01906-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat Prod Rep 27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 7.Haas D, Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 8.Kloepper JW, Leong J, Teintze M, Schroth MN. 1980. Pseudomonas siderophores: a mechanism explaining disease-suppressive soils. Cur Microbiol 4:317–320. doi: 10.1007/BF02602840. [DOI] [Google Scholar]
- 9.Kloepper JW, Leong J, Teintze M, Schroth MN. 1980. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885–886. doi: 10.1038/286885a0. [DOI] [Google Scholar]
- 10.Meyer JM, Abdallah MA. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol 107:319–328. doi: 10.1099/00221287-107-2-319. [DOI] [Google Scholar]
- 11.Sipiczki M. 2006. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl Environ Microbiol 72:6716–6724. doi: 10.1128/AEM.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cryle MJ, Bell SG, Schlichting I. 2010. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-l-leucyl-l-leucyl dipeptide oxidase. Biochemistry 49:7282–7296. doi: 10.1021/bi100910y. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman CP, Droby S. 2001. Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst Appl Microbiol 24:395–399. doi: 10.1078/0723-2020-00045. [DOI] [PubMed] [Google Scholar]
- 14.Türkel S, Ener B. 2009. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z Naturforsch C 64:405–410. [DOI] [PubMed] [Google Scholar]
- 15.Belin P, Moutiez M, Lautru S, Seguin J, Pernodet JL, Gondry M. 2012. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat Prod Rep 29:961–979. doi: 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- 16.Giessen TW, Marahiel MA. 2014. The tRNA-dependent biosynthesis of modified cyclic dipeptides. Int J Mol Sci 15:14610–14631. doi: 10.3390/ijms150814610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giessen TW, Marahiel MA. 2015. Rational and combinatorial tailoring of bioactive cyclic dipeptides. Front Microbiol 6:785. doi: 10.3389/fmicb.2015.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang MR, Sternberg D, Behr RK, Sloma A, Berka RM. 2006. Use of transcriptional profiling & bioinformatics to solve production problems: eliminating red pigment production in a Bacillus subtilis strain producing hyaluronic acid. Ind Biotechnol 2:66–74. doi: 10.1089/ind.2006.2.66. [DOI] [Google Scholar]
- 19.Li X, Wang D, Cai D, Zhan Y, Wang Q, Chen S. 2017. Identification and high-level production of pulcherrimin in Bacillus licheniformis DW2. Appl Biochem Biotechnol 183:1323–1335. doi: 10.1007/s12010-017-2500-x. [DOI] [PubMed] [Google Scholar]
- 20.Randazzo P, Aubert-Frambourg A, Guillot A, Auger S. 2016. The MarR-like protein PchR (YvmB) regulates expression of genes involved in pulcherriminic acid biosynthesis and in the initiation of sporulation in Bacillus subtilis. BMC Microbiol 16:190. doi: 10.1186/s12866-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hommais F, Oger-Desfeux C, Van Gijsegem F, Castang S, Ligori S, Expert D, Nasser W, Reverchon S. 2008. PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J Bacteriol 190:7508–7522. doi: 10.1128/JB.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan L, Murray TS, Kazmierczak BI, He C. 2010. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol Microbiol 75:76–91. doi: 10.1111/j.1365-2958.2009.06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol 2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 24.Stein T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 25.Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Breton YL. 2007. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol 189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Choi SK, Kim J, Oh TK, Park SH. 2012. Efficient production of polymyxin in the surrogate host Bacillus subtilis by introducing a foreign ectB gene and disrupting the abrB gene. Appl Environ Microbiol 78:4194–4199. doi: 10.1128/AEM.07912-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Wang Q, Qiu Y, Nomura CT, Li J, Chen S. 2017. Untangling the transcription regulatory network of the bacitracin synthase operon in Bacillus licheniformis DW2. Res Microbiol 168:515–523. doi: 10.1016/j.resmic.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Chet I, Ordentlich A, Shapira R, Oppenheim A. 1991. Mechanisms of biocontrol of soil-borne plant pathogens by Rhizobacteria. Plant Soil 129:85–92. doi: 10.1007/BF00011694. [DOI] [Google Scholar]
- 29.Lemanceau P, Bakker PA, De Kogel WJ, Alabouvette C, Schippers B. 1993. Antagonistic effect of nonpathogenic Fusarium oxysporum Fo47 and pseudobactin 358 upon pathogenic Fusarium oxysporum f. sp. dianthi. Appl Environ Microbiol 59:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen BJ, Zidack NZ, Larson BJ. 2004. The role of Bacillus-based biological control agents in integrated pest management systems: plant diseases. Phytopathology 94:1272. doi: 10.1094/PHYTO.2004.94.11.1272. [DOI] [PubMed] [Google Scholar]
- 31.Duitman EH, Wyczawski D, Boven LG, Venema G, Kuipers OP, Hamoen LW. 2007. Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Appl Environ Microbiol 73:3490–3496. doi: 10.1128/AEM.02751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol 52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng J, Wang Y, Li J, Shen Q, Zhang R. 2013. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl Microbiol Biotechnol 97:8823–8830. doi: 10.1007/s00253-012-4572-4. [DOI] [PubMed] [Google Scholar]
- 35.Albano M, Smits WK, Ho LTY, Kraigher B, Mandicmulec I, Kuipers OP, Dubnau D. 2005. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J Bacteriol 187:2010. doi: 10.1128/JB.187.6.2010-2019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng G, Yan LZ, Vederas JC, Zuber P. 1999. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol 181:7346–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoa TT, Tortosa P, Albano M, Dubnau D. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol Microbiol 43:15–26. doi: 10.1046/j.1365-2958.2002.02727.x. [DOI] [PubMed] [Google Scholar]
- 38.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan N. 2013. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci 38:151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Furrer JL, Sanders DN, Hook-Barnard IG, McIntosh MA. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol Microbiol 44:1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x. [DOI] [PubMed] [Google Scholar]
- 41.Miethke M, Schmidt S, Marahiel MA. 2008. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J Bacteriol 190:5143–5152. doi: 10.1128/JB.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horiyama T, Nishino K. 2014. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. doi: 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu W, Arceneaux JEL, Beggs ML, Byers BR, Eisenach KD, Lundrigan MD. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes. Mol Microbiol 29:629–639. doi: 10.1046/j.1365-2958.1998.00961.x. [DOI] [PubMed] [Google Scholar]
- 44.Brickman TJ, Kang HY, Armstrong SK. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol 183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kupfer DG, Uffen RL, Canale-Parola E. 1967. The role of iron and molecular oxygen in pulcherrimin synthesis by bacteria. Arch Mikrobiol 56:9–21. doi: 10.1007/BF00406050. [DOI] [PubMed] [Google Scholar]
- 46.Xue GP, Johnson JS, Dalrymple BP. 1999. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J Microbiol Methods 34:183–191. doi: 10.1016/S0167-7012(98)00087-6. [DOI] [Google Scholar]
- 47.Cai D, Hao W, He P, Zhu C, Qin W, Wei X, Nomura CT, Chen S. 2017. A novel strategy to improve protein secretion via overexpression of the SppA signal peptide peptidase in Bacillus licheniformis. Microb Cell Fact 16:70. doi: 10.1186/s12934-017-0688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]