Abstract
Exposure to antioxidants and xenobiotics triggers the expression of a myriad of genes encoding antioxidant proteins, detoxifying enzymes, and xenobiotic transporters to offer protection against oxidative stress. This articulated universal mechanism is regulated through the cis-acting elements in an array of Nrf2 target genes called antioxidant response elements (AREs), which play a critical role in redox homeostasis. Though the Keap1/Nrf2/ARE system involves many players, AREs hold the key in transcriptional regulation of cytoprotective genes. ARE-mediated reporter constructs have been widely used, including xenobiotics profiling and Nrf2 activator screening. The complexity of AREs is brought by the presence of other regulatory elements within the AREs. The diversity in the ARE sequences not only bring regulatory selectivity of diverse transcription factors, but also confer functional complexity in the Keap1/Nrf2/ARE pathway. The different transcription factors either homodimerize or heterodimerize to bind the AREs. Depending on the nature of partners, they may activate or suppress the transcription. Attention is required for deeper mechanistic understanding of ARE-mediated gene regulation. The computational methods of identification and analysis of AREs are still in their infancy. Investigations are required to know whether epigenetics mechanism plays a role in the regulation of genes mediated through AREs. The polymorphisms in the AREs leading to oxidative stress related diseases are warranted. A thorough understanding of AREs will pave the way for the development of therapeutic agents against cancer, neurodegenerative, cardiovascular, metabolic and other diseases with oxidative stress.
Abbreviations: 3-MC, 3-methylcholanthrene; Ah, aryl hydrocarbon; AhE, aryl hydrocarbon receptor; AP-1, activator protein-1; ARE, antioxidant response element; ATF4, activating transcription factor 4; β-NF, β-naphthoflavone; β-NF-RE, β-NF-responsive element; β-TRCP, β-transducin repeat-containing protein; B[α]P, benzo[a]pyrene; BACH, broad-complex, tramtrack and bric-a-brac (BTB), and cap'n'collar (CNC) homology proteins; bla, β-lactamase; BTB, broad-complex, tramtrack and bric-a-brac; CAT, chloramphenicol acetyltransferase; ChIP-Seq, chromatin immunoprecipitation sequencing; C-MARE, cyclic AMP responsive element (CRE)-type MARE; CNC-bZIP, cap'n'collar basic-region leucine zipper; CRE, cyclic AMP responsive element; CTD, C-terminal domain; CY1A1, cytochrome P450 1A1; DDI2, DNA-damage inducible 1 homology 2; eGFP, enhanced green fluorescent protein; EpRE, electrophile-responsive element; ER, endoplasmic reticulum; FRET, fluorescence resonance energy transfer; FTL, ferritin light polypeptide; GSH, glutathione; GST, glutathione S-transferase; GSTYa, glutathione S-transferase Ya subunit structural gene; hARE, human ARE; HBE1, human bronchial epithelial 1; HO-1, hemeoxygenase-1; HRD1, hydroxymethylglutaryl reductase degradation protein 1; HRE, heme responsive element; Keap1, Kelch-like ECH-associated protein 1; KO, knock-out; Mafs, musculoaponeurotic fibrosarcoma proteins; MAPT, microtubule-associated protein Tau; MAREs, Maf-recognition elements; MT, metallothionein; Nap, naphthalene; NCBI, national center for biotechnology information; NE, nuclear envelope; NESzip, nuclear export signal co-localized with the leucine zipper domain; NQO1, NAD(P)H quinone dehydrogenase 1; NQR, NAD(P)H:quinone reductase; Nrf1, nuclear factor erythroid 2 (NFE2)-related factor 1; Nrf2, nuclear factor erythroid 2 (NFE2)-related factor 2; Nrf3, Nuclear factor erythroid 2 (NFE2)-related factor 3; NTD, N-terminal domain; PAHs, polyaromatic hydrocarbons; Phe, phenanthrene; PHx, partial hepatectomy; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PMA, phorbol 12-myristate 12-acetate; PMF-1, polyamine-modulated factor-1; PWM, position weight matrix; Pyr, pyrene; ROS, reactive oxygen species; sMAF, small Maf; SNPs, single nucleotide polymorphisms; tBHQ, tert-butylhydroquinone; T-MARE, TRE-type MARE; TPA, tetradecanoyl-phorbol-13-acetate; TRE, TPA-responsive element; VCP, valosin-containing protein; XRE, xenobiotic-responsive element
Keywords: Antioxidant response elements, Antioxidant genes, ARE-reporter constructs, ARE SNPs, Keap1/Nrf2/ARE pathway, Oxidative stress
Highlights
-
•
Antioxidant response elements (ARE) orchestrate the expression of a myriad of genes.
-
•
Since its discovery, numerous experimentally functional AREs have been identified.
-
•
Transcription factors act on the AREs to regulate the cytoprotective genes.
-
•
Applications based on AREs are employed to detect and monitor chemicals/drugs.
-
•
Computational methods and analysis of AREs are still in its infancy.
1. Introduction
The emergence of techniques such as isolation and characterization of genes, construction of reporter genes, and transient transfection of reporter constructs has made it possible to study the regulatory elements of phase II genes. The dynamic expression of phase II genes is regulated by the transcriptional regulatory element called antioxidant response element (ARE). It is a daunting task to recognize and identify the AREs. Though a formidable challenge, scientists have not only identified the AREs, but also characterized these AREs. Many transcription factors interact with these AREs to orchestrate the expression of an array of cytoprotective genes in a spatio-temporal manner. Cells are constantly exposed to various kinds of stressors: chemical, environmental, oxidative, radiation, etc. The defense systems in the cells overcome these stressors through the action of the cis-acting elements of antioxidant, phase II detoxification and xenobiotic responsive genes [1], [2]. AREs are well-established as significant regulators of redox homeostasis and activators of cytoprotection during oxidative stress. The nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2) - the primary transcription factor - acts on these AREs (cis-acting elements), thereby elevating the expression of an array of genes that recuperates the redox insult by chemicals and xenobiotics, which ultimately protects the cells from damage [3], [4]. Kelch-like ECH-associated protein 1 (Keap1) inhibits and holds Nrf2 in the cytoplasm and directs Nrf2 for proteasomal degradation [3]. These findings led to the decoding of the signaling cascade involved in the defense mechanism, called the Keap1/Nrf2/ARE system. As a universal system, the Keap1/Nrf2/ARE pathway regulates the network of genes and offers cytoprotection under various stress conditions. Besides their role in cytoprotection due to oxidative stress, AREs regulate carbohydrate metabolism, cognition, inflammation, iron metabolism, metastasis, NADPH regeneration, lipid metabolism, and tissue remodeling [5]. This review describes the discovery of the AREs, classes of ARE enhancers, computational identifitication and analysis of AREs, transcription factors that act on the AREs, and applications of AREs.
2. Discovery of the ARE
The detoxification of toxic chemicals and xenobiotics happens in a highly regulated manner in three phases namely, phase I, II, and III [6], [7]. Phase I enzymes primarily consist of cytochrome P450 superfamily of enzymes and metabolizes the xenobiotics [8]. Phase II conjugating enzymes include glutathione S-transferases which catalyzes the conjugation of reactive electrophile species with glutathione (GSH) thereby it attenuates the toxic potential of xenobiotics [9]. Phase III transporters eliminate the GSH conjugates and offers protection against deleterious chemicals [10]. The regulation of three phase enzymes plays a chief role to counter environmental toxicants; hence it triggered scientists to unravel the detoxification regulatory mechanism. Xenobiotic metabolites induce the key phase I and phase II enzymes that are essential for the detoxification process [11]. These key enzymes, include GST isoenzymes, epoxide hydrolase and P450 isoenzymes, are expressed in high levels in the hepatocytes of animals treated with xenobiotics [12]. Polycyclic aromatic hydrocarbons (PAHs) such as β-naphthoflavone (β-NF) and 3-methylcholanthrene (3-MC) elicited the inducible expression of glutathione S-transferase Ya subunit structural gene (GSTYa) [13]. Scientists utilized these PAHs to study regulatory regions of GSTYa and other genes.
In the late 1980s, the functional analysis of the rat 5'-flanking sequences of a GSTYa – a phase II enzyme paved the way to dissect out the regulatory elements [14]. This study pioneered the identification of the AREs in many genes since then. The pGTBcat vectors were constructed through the fusion of the 5'-flanking region of GSTYa to the chloramphenicol acetyltransferase (CAT). The different promoter length of pGTBcat vectors - pGTB.7 cat, pGTB1.6 cat, and pGTB4.0 cat were constructed from the genomic subclone pλGTB45-15-5.5. These fusion genes were transiently transfected into rat cell lines (H5–6 and 3MO), mouse cell lines (Hepa1c1c7), and human hepatoma cells (HepG2). The measurement of CAT activity revealed the two regulatory elements between nucleotides −650 and −1550. Of the two cis-acting elements, one is present in human, mouse, and rat cells, and responsive to β-NF in heterologous cells; whereas the other element is found only in human and rat cell lines, and exhibited maximal basal promoter activity in homologous cells. The element responsive to β-NF is found to be functional only in the presence of dioxin receptors. Though this groundbreaking study first identified the two regulatory elements and their approximate location in the promoter, it did not discover the exact cis-acting sequence elements.
2.1. β-NF-responsive element
Three years later, Paulson et al. [15] identified the critical cis-acting sequence elements necessary for both the basal and 3-MCinducible expression of GSTYa in HepG2 cells. Dimethyl sulfate methylation protection footprints identified the presence of a regulatory element between −905 and −885, and these cis-acting elements resembled the xenobiotic-responsive element (XRE) or dioxin-responsive element. This study narrowed down the location of the cis-acting elements identified by the Telakowski-Hopkins team [14]. Rushmore et al. [16] identified three regulatory regions at different locations in the promoter of GSTYa. The first regulatory region identified is similar to the XRE sequence and is present between nucleotides −867 and −857, and controlled the maximum basal expression; the second regulatory region localized to positions −908 to −899 and has identity with the core XRE sequence of the cytochrome P450 1A1 (CYP1A1) gene; the third regulatory region is distinct from that of XRE sequence between −722 and −682 and is responsible for the inducible expression and the basal level expression of both planar aromatic compounds - β-NF and 3-MC. Hence, this element was termed the β-NF-responsive element (β-NF-RE). This study provided the evidence that the regulation of GSTYa is mediated by a regulatory element that is different from that of XRE.
2.2. Antioxidant response element
The further unravelling of this distinct β-NF-RE revealed that the phenolic antioxidant t-butylhydroquinone activated the transcription of GSTYa through this element without the need of aryl hydrocarbon (Ah) receptors [17]. As this β-NF-RE is responsive to phenolic antioxidants, this element is also named as an antioxidant-responsive element (ARE). The hypothesis put forward by Talalay et al. [18] and Prochaska and Talalay [19] that the induction of phase II drug metabolism enzymes by phenolic antioxidants was confirmed with the identification of ARE and its regulation of GSTYa independent of the Ah receptor. The ARE core sequence of GSTYa was identified as 5'-puGTGACNNNGC-3' (N represents any nucleotide) on characterization with deletion and mutational analysis [20]. This ARE core sequence was clearly different from that of the XRE core sequence of CYP1A1. The difference in the core sequences of ARE and XRE reveals the difference in their functional activity. In addition, Rushmore et al. [20] observed that reactive oxygen species (ROS) and oxidants formed through electron reduction and redox cycling of phenolic antioxidants. These ROS and oxidants activated the transcription of GSTYa through the ARE. This is the first study reporting that oxidative stress activates antioxidant genes through the cis-acting regulatory element - ARE - and offered a defense in eukaryotic cells. In the search for trans-acting proteins that bind to the ARE, a protein from within the untreated cells interacted with the ARE and treated cells but did not result in the induction of a new protein [16]. These findings envisioned that a trans-acting protein that is not inducible and constitutively expressed recognizes the ARE. AREs remain the central part of the signal transduction pathway in eukaryotic cells that respond to oxidative stress.
2.3. Electrophile-responsive element
Another group of scientists were characterizing cis-acting sequences of mouse GSTYa in the late 1980s. Daniel et al. [21] reported the two regulatory elements exactly similar as those found in rat GSTYa in the 5'-flanking region of mouse GSTYa. These two regulatory elements were located between −1.6 and −0.2 kb. One is responsible for basal expression, whereas the other is responsible for inducible expression when exposed to β-NF and 3-MC in both homologous (mouse) and heterologous (rat and human) hepatoma cells. The regulation of one of these two regulatory elements of GSTYa corroborated the metabolic cascade model proposed [18], [19]. This particular common element present in nucleotides −754 to −713 responded not only to planar aromatic compounds (β-NF, 3-MC, and Dioxin) but also electrophilic compounds (tert-butylhydroquinone, dimethyl fumarate, and trans-4-phenyl-3-buten-2-one) [22]. As this element responded surprisingly to an electrophilic signal, it was termed an electrophile-responsive element (EpRE). A DNA motif with an inverse repeat 5'-TGGAAAT(GACATTGC)TAATGGT-3' was identified as the EpRE. The trans-acting proteins acting on this cis-element were observed in both the nuclear extracts and the cytosolic fractions from DNaseI footprint and gel mobility-shift assays.
As scientists analyzed the function of the 5'-flanking region of GSTYa during the same period, Bayney et al. [23] investigated the cis-acting sequences regulating NAD(P)H: quinone reductase (NQR) in rat liver - a different antioxidant gene. The findings revealed that cis-acting regulatory elements are responsible for inducible expression of NQR. As observed in the rat and mouse GSTYa, β-NF induced the expression of NQR. In addition to β-NF, Sudan III - an azo dye - induced a 4–5 fold elevation in the expression of NQR. Favreau and Pickett [24] located two regulatory elements: one at position −393 to −352, which shared XRE sequence identity, and another at −434 to −404, which is distinct from that of XRE. This observation is very similar to that of findings of Rushmore and Pickett [17] in rat GSTYa. Both the planar aromatic compounds and phenolic antioxidants regulate the inducible expression of rat NQR through the ARE. As demonstrated in GSTYa, hydrogen peroxide (H2O2) induced the expression of rat NQR through the ARE sequence [25]. The ARE sequence of rat NQR is protected from digestion with the nuclear extracts and the sequence is palindromic (5’-AGTCACAGTGACT-3’). The 3'-end of this ARE sequence showed strong protection from digestion and high homology between the rat GSTYa and human NQR. The determination of specific ARE nucleotide sequences of rat NQR revealed three regions: 3'-flanking region, proximal, and distal half sites [26]. These two half-sites are oriented in a palindromic fashion. All three regions are essential for the high basal expression of rat NQR in HepG2 cells. Mutational analysis of the proximal half-site highlighted the importance of 3 nucleotides in the proximal half-site, 5'-gTGActtgca-3' that are essential for the maximum basal and inducible activity of rat NQR. Many of the features of the rat NQR ARE are similar to that of rat GSTYa ARE. Yet another study identified the ARE in the human NQR located between −470 and −445 [27]. This ARE regulated the expression of NQR in both hepatoma (Hepa-1 and HepG2) cells and non-hepatic (HeLa) cells on exposure to β-NF and 3-(2)-tert-butyl-4-hydroxyanisole. The sequence of the human NQR ARE is 5'-TGAGTCAGC-3'. Within a span of half a decade, groups of scientists discovered the AREs that regulate GSTYa in the mouse and rat, and NQR in the rat and human, and laid the foundation for the discovery of the Keap1-Nrf2-ARE pathway in later years. The experimentally identified functional AREs in human, mice, rat, zebrafish, and cell lines are summarized in this review (Table 1).
Table 1.
The experimentally identified functional AREs in human, mice, rat, zebrafish and cell lines.
| ARE | Location/Position of ARE with respect to + 1 transcription start site | Experimentsa | Gene | Category/Function | Species/Cell lines | References |
|---|---|---|---|---|---|---|
| TGACTCAGC, TGACTAAGC, TTACGAAGC | −3118, −291, −330 | MA | Gclc | Antioxidant | Homo sapiens | [28] |
| TGACTTAGC, TGAGAGGGC | −76, −387 | SDM | Gpx2 | Antioxidant | Homo sapiens | [29] |
| TGAATCAGC, TGCCTCAGC | −3429, −4322 | ChIP, SDM, EMSA | Prdx1 | Antioxidant | Homo sapiens | [30] |
| TGACCGAGC | −349 | ChIP, SDM | Prdx6 | Antioxidant | Homo sapiens | [31] |
| TTACTCAGC | −416 | EMSA | Trx | Antioxidant | Homo sapiens | [32] |
| TGACAAAGC | −301 | EMSA | Txnrd1 | Antioxidant | Homo sapiens | [33] |
| TGACTCAGC | −1565 | RA, SDM | Ftl | Metal binding protein | Homo sapiens | [34] |
| TGACCTGGC | −99 | TTA | Mt1b | Metal binding protein | Homo sapiens | [35] |
| TGACTCAGC | −5522 | EMSA, ChIP | Akr1c2 | Detoxification protein | Homo sapiens | [36] |
| TGACAAAGC | −499 | CL, SA | Mgst1 | Detoxification protein | Homo sapiens | [37] |
| TGACTCAGC | −463 | CAT RA, SA | Nqo1 | Detoxification protein | Homo sapiens | [38] |
| TGACTTGGC | −3296 | EMSA | Ugt1a1 | Detoxification protein | Homo sapiens | [39] |
| TGATTCAGC, TGAGGAGGC, TGACTCAGC, TGACGCAGC, TGACTCGCC, TGAGTGAGC, TGATTCAGC, TGATTCTGC, TGACAAGGC, TGACTCAGC, TGACTCAGC, TGACTTAGC, TGACTCAGC, TGACAGAGC, TGAGGCAGC | −3231, −3995, −4009, −4073, −4186, −5520, −6047, −6080, −7184, −9059, −9088, −9117, −9146, −9579, −11779 | ChIP | Hmox1 | Antioxidant | Homo sapiens | [40] |
| TGCTGAGTC | – | ChIP | MAPT | Microtubule associated protein Tau | Homo sapiens | [41] |
| TGACCTCGC | −2603 to −2629 | EMSA, ChIP | Osx | Transcription factor | Homo sapiens | [121] |
| TGACGTCGC, | −52, −341 | ChIP | Psmb5 | Proteolytic degradation of misfolded proteins | Mus musculus | [42] |
| TGACCAAAC | ||||||
| TGAGTCAGC, | −59, −915, −937 | EMSA, FP | Gst-p1 | Antioxidant | Mus musculus | [43] |
| TGAATCTGG, TGACATCTC | ||||||
| TGACTAAGC, TGACCAAGC | −35, −804 | ChIP | Gsr1 | Antioxidant | Mus musculus | [44] |
| TGAGAAAGC | −440 | SDM | Slc7a11 | Antioxidant | Mus musculus | [45] |
| TGACAAAGC, | −4076, −4023 | RA | Fth1 | Metal binding protein | Mus musculus | [46] |
| TGACTCAGC | ||||||
| TGACTCAGC | −1118 | RA, SDM | Ftl1 | Metal binding protein | Mus musculus | [34] |
| TGACTATGC | −69 | TTA | Mt1 | Metal binding protein | Mus musculus | [35] |
| TGACTCAGC | −214 | TTA | Mt2 | Metal binding protein | Mus musculus | [35] |
| TGACCCAGC | −925 | RA, EMSA | Akr1b3 | Detoxification protein | Mus musculus | [47] |
| TGACAAAGC | −728 | EMSA | Gsta1 | Detoxification protein | Mus musculus | [48] |
| TGACATTGC | −147 | SDM, RA, EMSA | Gsta3 | Detoxification protein | Mus musculus | [49] |
| TGACATAGC | −94 | RA, EMSA | Mrp2 | Excretion pump | Mus musculus | [50] |
| TGAGTCGGC | −435 | SDM, GW, 5’RACE, EMSA | Nqo1 | Detoxification protein | Mus musculus | [51] |
| TGAGCGAGC, TGACTTGGC, TGACCCTGC, TGAGGCTGC, TGACTCAGC, TGACACAGC, CAACTCAGC, TGACTCTGC, TGACTCAGC, TGACTAAGC, TGACTCAGC, TGACTCAGC, TGACTCAGC, TGAGCAAGC | −126, −3318, −3547, −3612, −3990, −4042, −4090, −6069, −7426, −8709, −9734, −9763, −9791, −9878 | EMSA, SDM | Hmox1 | Antioxidant | Mus musculus | [52], [53], [54] |
| TGACATAGC | −784 | EMSA, SDM | Ppargγ | Anti-inflammation | Mus musculus | [55] |
| TGACCCAGC | −983 to −944 | EMSA, RA | Akr1b3 | Catalyzing the reduction of glucose to sorbitol | Mus musculus | [47] |
| GTGGTAGTG | −731 to −722 | RA | Gclc | Antioxidant | Mus musculus | [56] |
| GCAACAGTG | −502 to −493 | RA | Hsp70 | Antioxidant | Mus musculus | [56] |
| TGATCTGGC, TGACATTGC, TGACAAAGC, TGATTTGGC | −913 to −904, −743 to −734, −728 to −719, −148 to −139 | RA | Gst | Antioxidant | Mus musculus | [56] |
| TGACATTGC | −1762 to −1733 | EMSA, ChIP | Osx | Transcription factor | Mus musculus | [121] |
| TGAGTCAGC | −247 | SDM | Srxn1 | Antioxidant | Rattus norvegicus | [57] |
| TGACAAAGC | −696 | MA | Gsta2 | Detoxification protein | Rattus norvegicus | [20] |
| TGACTTGGC | −421 | CAT RA | Nqo1 | Detoxification protein | Rattus norvegicus | [24] |
| TGACGGTGC | −3470 to −3500 | EMSA, ChIP | Osx | Transcription factor | Rattus norvegicus | [121] |
| TGACTCATC, TGACTCATC | – | EMSA | Gstp1 | Antioxidant | Danio rerio | [58] |
| TGACAACGC, TGACTCAGC | −453 to −444, −355 to −346 | RA | Gclc | Antioxidant | Danio rerio | [56] |
| TGACGCGGC | −690 to −681 | RA | Hsp70 | Antioxidant | Danio rerio | [56] |
| TGATAGAGC, GCCACAGTG | −749 to –740, −1331 to −1322 | RA | Gst | Antioxidant | Danio rerio | [56] |
| TGACTCATC | −53 to −45 | EMSA | Gstp1 | Antioxidant | Danio rerio | [58] |
| TGCTGATTC | −86 to −77 | ChIP | Nrf2 | Transcription factor | Adenocarcinomic human alveolar basal epithelial cells | [59] |
| TGACAAAGC, TGACTCTGG | −48 to −62, −46 to −32 | ChIP, EMSA | Trxr1 | Antioxidant | Bovine arterial endothelial cells | [60] |
| TGCTGAGTA | −452 to −420 | EMSA | Trx | Antioxidant | Erythroleukemic cell line | [32] |
| TGACTGTGG | −754 | EMSA, ChIP | Nrf2 | Transcription factor | Murine keratinocyte cell line | [2] |
| TGACATTGC | – | SDM, EMSA, ChIP | Mgsta3 | Antioxidant | Mouse hepatoma Hepa1c1c7 | [49] |
| TGCTGAGTC | – | EMSA | HO-1 | Antioxidant | Immortalized rat proximal tubular epithelial cells | [61] |
| TGAATCAGC, TGCCTCAGC | −536 to −528, −1429 to −1421 | SDM, ChIP | Prx1 | Antioxidant | Human lung cancer cell line | [30] |
| TGACTCAGC | −461 to −455 | CAT RA | Dtd | Anticancer | Human colon adenocarcinoma cell lines | [62] |
| TGACTCAGC | – | CAT RA, EMSA | Nqo1 | Detoxification protein | Human monocyte cell lines | [63] |
| TGCTGTGTC | – | EMSA | HO-1 | Antioxidant | Human breast adenocarcinoma cell line and cervical cancer cell line | [64] |
| TGACTCAGC, TGAGTCAGC | – | EMSA | Bach1 | Transcription factor | Human umbilical vein endothelial cells | [65] |
| TGACTCAGC | – | EMSA | Nqo1 | Antioxidant | Human umbilical vein endothelial cells | [65] |
| TGACAAAGC | −4117 | EMSA | Ferritin H | Antioxidant | Hepatoma cells, Liver hepatoblastoma cell line | [66] |
| TGACTCAGC | – | EMSA | Nrf2 | Transcription factor | Human umbilical vein endothelial cells | [67] |
| TGACATAGC | – | EMSA, RA | Mrp2 | Excretion pump | Mouse hepatoma cell, human hepatoblastoma cell line | [50] |
| TGACATGGC | – | EMSA, RA | Mrp2 | Excretion pump | Mouse hepatoma cell, human hepatoblastoma cell line | [50] |
| TGAGCATGC, TGATCTTTC, TGAGTGAGC, TGACTCCGC, | −152 to −121, −212 to −183, −78 to −49, −62 to −31 | EMSA | Ncb5or | Antioxidant | Mouse embryonic fibroblast | [68] |
| TGACAGAGC, TGGCATTGC, TGACAAAGC | – | EMSA | Gsta5 | Drug metabolizing enzyme | Liver hepatoblastoma cell line | [69] |
| TGACATTGC, TGACAAAGC | – | EMSA | GstA1 | Drug metabolizing enzyme | Liver hepatoblastoma cell line | [69] |
| TGACTCAGC | – | EMSA | Gcsh | Oxidative stress response | Liver hepatoblastoma cell line | [70] |
| TGACTAAGC | – | EMSA | Gcs1 | Oxidative stress response | Liver hepatoblastoma cell line | [70] |
| TGACTCAGC | −312 | SDM, EMSA | Gclm | Antioxidant | Liver hepatoblastoma cell line | [71] |
| TGGCATTGC | – | EMSA | Gsta2 | Antioxidant | Rat hepatocyte derived cell line | [72] |
| TGACAAAGC | – | ChIP | Ferritin H | Antioxidant | Human hepatocarcinoma cell line | [73] |
| TGACCAAGC | – | – | Ets-1 | Transcription factor | Human Ovarian carcinoma cell line | [74] |
| TGACTCAGC | – | EMSA | Bach1 | Transcription factor | Human umbilical | [65] |
The experimental methods employed in the identification of AREs abbreviated in this column are as follows: MA, mutational analysis; ChIP, chromatin immunoprecipitation assay; SDM, site directed mutagenesis; EMSA, electrophoretic mobility shift assay; RA, Reporter gene assay; TTA, transient transfection assay; CL, chromosomal localization; SA, sequence analysis; CAT RA, chloramphenicol acetyltransferase reporter assay; FP, foot printing; GW, genome walking; 5'RACE, rapid amplification of 5' complementary DNA ends.
3. A regulatory element within another regulatory element
It was a daunting task for scientists to clearly define the ARE and its functions due to the presence of another element within the ARE called the activator protein-1 (AP-1) site. The AP-1 site is an element that is transcriptionally activated by tetradecanoyl-phorbol-13-acetate (TPA) and hence called a TPA-responsive element (TRE) [48]. It is also known as phorbol 12-myristate 12-acetate (PMA) response element because it responds to PMA. AP-1 is a transcription factor that acts on the cis-element of human metallothionein IIa promoter [75]. An array of genes carried these TREs as they were discovered from DNase I protection analyses [76]. The consensus sequence of the TRE is defined as TGA(C/G)TCA. The ARE contains a tetranucleotide sequence 5'-TGAC-3', which is a half-site of TRE. Besides phorbol esters, UV light, metals, growth factors, and other stressors activate AP-1, which then binds the TRE. The binding of the transcription factors Fos and Jun to the ARE and its transactivation were ascertained by the presence of a cryptic AP-1 site (TRE) within the ARE [48]. Despite the absence of a consensus TRE sequence within the ARE (EpRE), TPA triggered the response through the interaction between c-Jun/c-Fos and TRE. Like ARE, TRE also activates the transcription of an array of genes on exposure to various enhancers, so both these elements (ARE and TRE) belong to the class of inducible enhancers [77]. The human ARE (hARE) of NQR carries a TRE in 5'-CTAGTGAtgagtcaGCCGTC-3' [27]. The presence of such a TRE within the ARE allows the competition of different transcription factors to bind to both these elements. Such competition encouraged researchers to identify and distinguish the transcription factors that bind to TRE and ARE and the regulation of the gene expression by these two elements. Although AP-1 binds AREs with low affinity, numerous studies differentiated the TRE-mediated regulation from ARE-induced transcription. Nguyen et al. [78] unmasked the essentiality of the GC-3' in the ARE core sequence and distinguished the ARE from the TRE. The close similarity of these two elements discloses that their regulatory proteins are also closely related. The investigations showed that the perfect consensus TRE sequence within the hARE is essential for the basal and xenobiotic inducible expression of human NQR [79]. Both TRE and ARE are present in the 5'-flanking sequences of a number of genes and two or more TRE copies are present within the AREs [80]. Cells that are devoid of AP-1 induced ARE transcription, but in contrast, ARE activators did not mediate TRE transcription [81]. Mutational analysis revealed that two copies rather than a single copy of the TRE are responsive to antioxidants and xenobiotics [79].
Another atypical AP-1 family of transcription factors - musculoaponeurotic fibrosarcoma proteins (Mafs) - binds the 13 bp consensus Maf-recognition elements (MAREs) 5'-TGCTGAG/CTCAGCA-3', which is palindromic in nature. The core MARE region has high similarity to the core ARE region [82]. Based on the type of core regions harbored within the MAREs, MAREs are termed TRE-type MARE (T-MARE) 5'-TGCtgactcaGCA-3' and cyclic AMP responsive element (CRE)-type MARE (C-MARE) 5'-TGCtgacgtcaGCA-3'. Maf, as a homodimer with itself, recognizes the GC dinucleotide of MAREs, and with Nrf2 as a heterodimer, binds the GC-3' dinucleotide of the ARE [83]. A variant of T-MARE (5'-TGCTGACTCATCA-3') interaction with MafG was identified from the crystal structure of MafG. MAREs are found in the regulatory regions of erythroid and megakaryocyte-specific genes [84]. The dimers such as small Maf (sMaf) homodimers and sMaf: Nrf2 heterodimers bind to the ARE [85].
Through sequence conservation and mutation analyses, Inamdar et al. [86] identified a 10 bp consensus motif (T/C)GCT(G/T)(A/T)GTCA called the heme responsive element (HRE). The HRE contains AP-1 heptad and the ARE, though these AREs are not responsive to heme or cadmium. The trans-acting factors that bind to the ARE may also regulate HO-1. Besides heme and cadmium, other inducers may activate the transcription of the HO-1 through the ARE with cap'n'collar basic-region leucine zipper (CNC-bZIP) interfamily heterodimers [87]. The HRE, being a composite ARE, can serve as a binding site for the different bZIP proteins.
4. Four classes of ARE enhancers
The importance of the 5'-end of the core ARE made Wasserman and Fahl [88] extend the ARE to 20-bp by adding nucleotides at both ends and termed the resulting ARE an extended ARE. The consensus extended ARE sequence is 5'-TMAnnRTGAYnnnGCRwwww-3', in which M represents A/C, R - A/G, Y - C/T, W - A/T, and n – A/T/G/C. In this, the trinucleotide at the 5'end is called 'TMA' motif, which is essential for the induction of Gsta1. Through point mutations, it was observed that the minimal length of 16-bp 5'-TMAnnRTGAYnnnGCR-3' was enough for induction of mouse NQO1. The alignment of AREs from different genes revealed the better conservation of core AREs than the extended AREs. The distinct sequence within one ARE that is essential for the function of a particular gene will not be efficient in the other genes. In this context of specific nucleotide and conservation, AREs are grouped into four different classes of enhancers [89]. Class I contains a 16-bp ARE with a TMA motif and AP-1 binding site. e.g. mouse Fth1 cCtccATGACaaaGCA [46]. Class II contains a 16-bp ARE with the presence of a TMA motif and the absence of the AP-1 site. e.g., mouse Gsta1 TAAtgGTGACaaaGCA [48]. Class III (e.g., mouse Mt2 GTGACtcaGCG) and IV (e.g. mouse Mrp2 ATGACataGCA) include 11-bp ARE, excluding the TMA with the presence and absence of the AP-1 site respectively [35], [50]. Classes I and III ARE genes are activated by AP-1 whereas classes I and II ARE genes are induced by Nrf2. Though the classification of ARE enhancers was put forth, it was ambiguous and could not classify all the AREs under these four classes.
5. AREs - Computational methods and analysis
As a result of numerous experimental investigations of promoter sequences of specific genes, functional ARE sequences were identified. Additionally, consensus ARE sequences were proposed for several organisms by utilizing mutational studies. A survey of the literature revealed that there are a few computational studies performed to identify and analyze ARE sequences.
Wang et al. [90] collected a set of experimentally identified ARE sequences from the NCBI literature database and developed a position weight matrix (PWM) statistical model using a set of experimentally identified ARE sequences in the human genome, and identified a set of ARE sequences with functional evidence. By aligning the binding sites, a nucleotide position frequency matrix was created and then converted into a PWM, which is a widely used statistical model that reflects the observed nucleotide frequencies of each nucleotide at each position. Further, a sliding window approach with a search pattern was performed to retrieve sub-sequences with the size of PWM in the human genomic sequence to identify the putative ARE sequences. A sub-sequence which matches the search pattern and also holds the higher PWM score than the lowest PWM score of the experimentally identified functional AREs. In addition, conservation of AREs at the sequence level was identified using phylogenetic footprinting by analyzing the upstream 5-kb regions of a set of orthologous genes from human, mouse, and rat genomes [91]. With the development of an integrated computer system, functional polymorphisms were identified within the AREs in the human genome and checked how these single nucleotide polymorphisms (SNPs) in the AREs bring changes in the expression of the target genes. Further experimental analysis of these SNPs will help us identify individuals who are highly prone to diseases in response to xenobiotics and oxidative stress as a consequence of genetic predisposition.
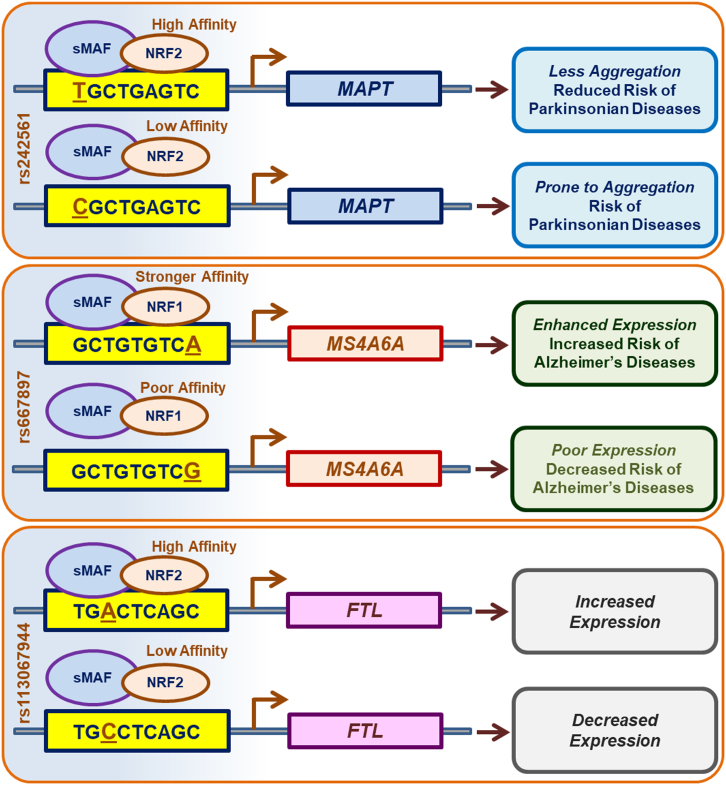
In recent years there have been a few investigations on ARE SNPs (Fig. 3). A functional ARE SNP was identified in the promoter of microtubule-associated protein Tau (MAPT) using computational data analysis with input data from chromatin immunoprecipitation sequencing (ChIP-Seq) and a genome-wide association study (GWAS) [41]. The ARE SNP rs242561 C/T (Cgctgagtc/Tgctgagtc) exhibited differential affinity to Nrf2/sMaf heterodimers. The T allelic variant of rs242561 showed high affinity, whereas the C allelic variant had a low affinity towards Nrf2/sMaf. This strong affinity of Nrf2/sMaf due to the presence of the T allelic variant in the ARE of MAPT will produce MAPT tau protein that is less prone to aggregation due to the inclusion of exon 3 in the MAPT of the tau protein product. Thus, the high affinity enhanced the MAPT transcript and reduced the risk of Parkinsonian diseases. Using an online tool ‘Region Report’, Kuosmanen et al. [92] identified the SNP – rs113067944 in the ARE of the ferritin light polypeptide (FTL) gene promoter. When there is a change in the allele from A to C in the core ARE of TG[A/C]CTCAGC, the Nrf2 has a poor affinity towards the ARE with a C allele and results in decreased FTL transcription. By using an Nrf2 binding prediction model, this study revealed 14 different SNPs in the ARE sequences that had clinical relevance. Yet another GWAS reported the creation of an ARE in the membrane-spanning 4-domain family, subfamily A (MS4A) locus due to an SNP rs667897 [93]. A putative cis-regulatory SNP - GCTGTGTC[A/G] was identified at the MS4A6A locus. The terminal nucleotide change from G to A results in the allele-specific transcription factor binding site. Though both Nrf1 and Nrf2 in association with sMafs displayed the potential to activate the rs667897A, Nrf1 is more effective in the transcriptional activation of the rs667897A cis-SNP. A subset of a population carrying rs667897A is associated with enhanced MS4A6A gene expression and Alzheimer's disease. The ARE SNPs associated disease studies still remain in their infancy.
Fig. 3.
ARE SNPs influence the expression of target genes. The SNPs present in the AREs alter the affinity of the transcription factors towards AREs and modulate the expression of their target genes. The specific allele carried within the AREs may either increase or decrease the risk of disease.
There are several computational tools developed to predict/identify transcription factor binding sites. Transcription factor flexible model overcomes the fixed length motifs prediction of PWMs to predict variable length motifs [94]. One of the recent models - DRAF, adopts machine learning approach using transcription factor binding sequences and physicochemical properties of transcription factor binding domains to predict the binding sites [95]. However, to the best of our knowledge, there is no single method available to predict/identify ARE sequences of query genes and no computational studies were attempted to identify ARE sequences at the genome-level using the organism specific consensus ARE sequences, which are highly needed and thus offer challenges in the computational research field.
6. Transcription factors that interact with AREs
The transcription factors that interact with AREs intrigued scientists for over half a decade, despite the first ARE being identified in the late 1980s. The cryptic TRE (AP-1 binding site) within the ARE not only baffled scientists but also created dispute among them, a few said that AP-1 regulates the expression of detoxification genes through the ARE [48], [96], [97] while others argued that unique proteins bind the ARE, not the AP-1 [98], [99], [100]. A clearer picture emanated from work on mouse F9 embryonal carcinoma cells [81]. Tert-butylhydroquinone (tBHQ) - an inducer - stimulated the ARE site but not the TRE site in F9 cells. Growth hormone reporter constructs carrying ARE elements transfected into F9 cells that lacked TRE-binding proteins triggered the activation of transcription of this reporter gene. Even the TRE consensus sequence was not activated by ARE-activating protein. The transcriptional activation of the most phase II enzymes failed even when ARE sequences are swapped with TRE consensus sequence which eventually revealed that proteins interacts with ARE rather TRE [81]. Other studies evidenced that ARE-binding proteins induce the phase II enzymes through ARE but not with AP-1 [78], [100]. Hence, both the electrophilic and antioxidant inducers mediate the regulation only through ARE-binding proteins.
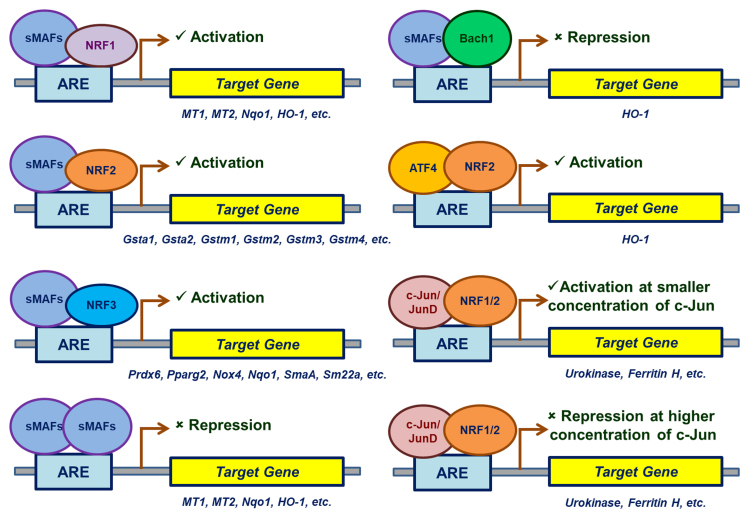
The activation or repression of the target genes is determined by the dimerizing partners of the transcription factors and AREs in the promoter of the target genes (Fig. 1). Of all the transcription factors, Nrfs are the primary transcription factors that interact with AREs. Locus control regions (LCRs) are the evolutionarily conserved regulatory sequences present on the upstream α- and β-globin genes [101]. Sequence-specific transcription factor called nuclear factor erythroid 2 (NF-E2) regulates the expression of these globin genes by binding with LCRs [102], [103]. Besides their regulatory sites in α- and β-globin genes, NF-E2 possess recognition sites in ferrochelatase, heme biosynthesis and porphobilinogen deaminase genes [84], [104]. More erythroid-specific transcription factors were identified that recognizes the LCRs after the identification of NF-E2. Two of them are nuclear factor erythroid 2 (NFE2)-related factor 1 (Nrf1) and nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2) falls under same bZIP transcription factor family [105].
Fig. 1.
Overview of the transcription factors involved in the activation or repression of target gene expression via AREs. Besides the partners in the dimerization, the transcription factors may either activate or repress the target gene expression based on the nature of cell type, electrophilic compound, oxidative stressor, and target gene.
6.1. Nuclear factor erythroid 2 (NFE2)-related factor 1 (Nrf1)
Despite the AREs of NQO1 of human and rat being highly conserved, the interaction of Jun and Fos transcription factors occurred only with the hARE but not with the rat ARE [22], [106]. Venugopal and Jaiswal [107] rightly observed this difference and hypothesized that AP-1 related proteins might interact with AREs. From earlier studies, they observed the similar human tissue-specific pattern of expression of Nrf1/Nrf2 and NQO1 are regulated by the ARE [105], [108], and the binding site was identical to that of hARE [109]. The presence or absence of an absolute AP-1 binding site within the ARE governs the binding of the transacting proteins with the AREs [109]. The hARE regulated CAT transcripts were elevated when Nrf1 and Nrf2 proteins were overexpressed independently and combinatorially in both human hepatoblastoma cells (HepG2) and monkey kidney (Cos1) cells [107]. Yet no significant difference in the levels of CAT transcripts was observed when Nrf1 and Nrf2 were expressed individually or in combination, and hence no heterodimerization of these two transcription factors was obligatory. β-NF and tBHQ also triggered the hARE regulated CAT transcription through Nrf1 and Nrf2. These evidence unraveled the Nrf1/Nrf2 regulation of AREs and pioneered the identification of signal transduction mechanisms in antioxidant and xenobiotic metabolism.
Through a complementation assay in yeast, genes that encode the NF-E2/AP-1 site binding proteins were studied; one of the proteins showed extreme closeness to the NF-E2 and CNC proteins and was designated as Nrf1 [105]. Though it was well-established that Nrf1 binds AREs and transactivates an array of genes [110], their subcellular localization and regulation mechanisms remained elusive for some years. The Nrf1 carries 155 additional amino acids in its N-terminal domain (NTD), which distinguishes it from Nrf2 [111]. Keap1 negatively regulates Nrf2 but not Nrf1. NTD negatively regulates Nrf1 and is responsible for its endoplasmic reticulum (ER) localization. When this NTD of Nrf1 was fused to the N-terminus of Nrf2, the Nrf2 was still regulated by Keap1 but localized to the ER. The localization of Nrf1 to the ER membrane raises the question of nuclear localization and the activation of ARE-driven genes. During ER stress, Nrf1 is cleaved in the ER and is translocated to the nucleus to activate the transcription of its target genes [112]. When residing within the ER, Nrf1 is glycosylated, whereas it is not in the nucleus [113]. The deglycosylated and proteolytically cleaved Nrf1 of different truncated forms make an entry into the nucleus through the nuclear pore and activate the transcription of its target genes on binding with AREs [113], [114]. Scientists are still finding the precise details of subcellular localization of Nrf1 and its functional forms. The affinity for the ARE is dependent on cofactor assembly of Nrf1 in a cell-specific manner [115]. Nrf1 generally spans across the cell membrane as an integral membrane protein with both cytoplasmic and luminal domains [116]. The 65 residues in the N-terminus of Nrf1 are essential for the integration into both the ER and nuclear envelope (NE) membranes. tBHQ activates the NE bound Nrf1 but not by the ER stressors.
The hepatocyte-specific deletion of Nrf1 resulted in liver damage, but no deleterious effects were observed in the Nrf2 knock-out (KO) mice [115]. In addition, metallothionein gene (MT1 and MT2) expression decreased in Nrf1 null mutant mice. MT1 and MT2 carry an ARE in their regulatory region, and Nrf1 exclusively regulates these two genes over Nrf2. An interesting observation disclosed that under acute copper exposure, Nrf2 regulates MT1, whereas under chronic copper exposure, Nrf1 regulates MT1 through the ARE [117]. Furthermore, Nrf1 regulates glutamate-cysteine ligase catalytic/modifier subunits, NQO1, HO-1, and ferritin [115], [118], and thereby defends oxidative stress in vivo [119]. The indispensable role of Nrf1 in development was evidenced from global KO of Nrf1 in mouse embryos leading to lethality [120]. The critical ARE-mediated developmental genes are under the control of Nrf1, and its target genes are constitutively expressed and are essential for development. Nrf1 regulates osterix - a zinc finger transcription factor essential for osteoblast differentiation and bone formation [121]. The osterix promoter carries a conserved ARE that interacts with Nrf1.
Nrf1, similar to that of Nrf2, responds to xenobiotics and thereby binds AREs and activates GCLC, NQO1, HO-1, and ferritin. Instead of Keap1, NTD of Nrf1 inhibits Nrf1 by localizing it into the ER. After the proteolytic cleavage of NTD, Nrf1 translocates into the nucleus. Nrf1 responds to metal toxicity through MT1 and MT2 regulation. Unlike Nrf2, Nrf1 is indispensable for redox homeostasis during development.
6.2. Nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2)
Though Nrf1 regulates ARE-driven target genes, another class of CNC-bZIP transcription factor predominates the regulation of ARE-driven phase II genes during oxidative stress [122], [123] and hence is called a master regulator of stress responses [124]. Unlike Nrf1, the global deletion of Nrf2 produced viable animals, but they were susceptible to stressors [119]. Though Nrf2 was well-known as the master regulator of ARE-dependent genes, it was first discovered as a transcriptional regulator of β-globin genes [108]. Of the several DNA binding proteins, one clone was isolated, characterized, and found to have a bZIP domain with high homology to NF-E2 and was ultimately named Nrf2. Initially, it was hypothesized that Nrf2 was an essential factor for hematopoiesis [125]; later it was evident that Nrf2 induced ARE-mediated genes. Further investigation on Nrf2 discovered the Keap1/Nrf2/ARE pathway, which is a requisite molecular mechanism in the metabolism of electrophiles and oxidants. The resemblance in the sequence motifs between the ARE and NF-E2 binding site led to the elucidation that Nrf2 is an absolute transcription factor that regulates ARE transcription.
The activity of Nrf2 is repressed by a cytoplasmic protein called Keap1 with CNC homology-associated protein 1 [4]. Keap1 serves as a sensor of ARE inducers and thereby regulates Nrf2. The binding of the ARE inducers to the Keap1 liberates Nrf2 from its complex and initiates the ARE response. The Nrf2-/- KO mouse model confirmed that Nrf2 is essential for both the constitutive and inducible expression of ARE-mediated glutathione S-transferase (Gsta1, Gsta2, Gstm1, Gstm2, Gstm3, and Gstm4) genes [126].
Several lines of evidence support that Nrf2 regulates a repertoire of ARE-dependent genes in mammalian cells. In human neuroblastoma IMR-32 cells, microarray analysis revealed that the PI3K-Nrf2-ARE pathway triggers a set of ARE-induced genes on tBHQ treatment [127]. Another in vivo study in the small intestine of wild-type and Nrf2-/- KO mice on exposure to sulforaphane reported that ARE-driven genes are upregulated in wild-type mice by Nrf2 but not in Nrf2 deficient mice [128]. A similar investigation revealed that the Keap1-Nrf2 pathway increases the expression of an array of ARE-mediated genes in the liver of 3H-1,2,-dithiole-3-thione exposed wild-type mice but not in Nrf2-/- KO mice [129]. Hypoxia treatment markedly increased the expression of Nrf2-ARE pathway regulated genes in the lungs of Nrf2+/+ mice than in Nrf2-/- mice [130]. These studies not only revealed that Nrf2 regulates an array of genes, but also indicated the presence of AREs in the promoter of Nrf2 target genes.
Nrf2 exerts multiple functions besides its defense against oxidants and electrophiles [131]. A repertoire of ARE-regulated genes was identified while mining the genome, and these genes regulate processes such as cell proliferation, metabolic reprogramming, and proteasomal protein degradation [42], [132], [133]. The promoter region of Nrf2 itself has AREs, which helps in the amplification of its own transcription during the defense response [134]. In addition to the AREs, the Nrf2 promoter possesses an XRE at −712 bp and two XRE-like motifs upstream of a lone XRE. AhR binds to XRE and regulates the expression of Nrf2 [135]. Likewise, the AhR promoter also carries an ARE at −230 bp to which Nrf2 interacts and activates the AhR expression [136]. The presence of each of these regulatory elements in these two genes - Nrf2 and AhR - confirms the interaction between AhR and Nrf2 signaling pathways. The complexity of these signaling pathways exhibited through different regulatory elements requires further investigation to envision their roles in health and disease.
6.3. Nuclear factor erythroid 2 (NFE2)-related factor 3 (Nrf3)
In search of a novel CNC transcription factor, Kobayashi et al. [137] identified Nrf3 with a high homology bZIP domain to other CNC proteins. The function of Nrf3 still remains elusive since its discovery. This high homology among CNC factors reveals that they all share similar and overlapping regulatory elements. Nrf3 interacts with MARE in the β-globin promoter of chicken, which is also a target for Nrf1 and Nrf2. Nrf3 binding to the ARE and transactivation of its target genes was well-evidenced. The ARE-driven NQO1 expression was triggered by Nrf3 in monkey kidney COS-1 cells [138], but in contrast Nrf3 repressed the NQO1 expression in human hepatoma HepG2 cells [139] and mouse embryonic stem cells [140]. Depending on the cell types, expression levels of Nrf3 and its cofactors, Nrf3 either activates or represses the transcription of their target genes.
Nrf3 contains an ER signal peptide in its NHB1 domain, which targets Nrf3 to the ER and gets activated during ER stress and translocates itself into the nucleus to transactivate ARE-dependent genes [141]. Glycosylation and deglycosylation control the activation of Nrf3. In the ER and Golgi apparatus, Nrf3 gets glycated, and when glycated, Nrf3 becomes a negative regulator [142]. In vitro studies revealed potential target genes of Nrf3 such as PRDX6, PPARG2, NFE2L2, NOX4, NQO1, SMAA, and SM22A but there is hardly any in vivo evidence.
Both Nrf1 and Nrf3 are regulated by the same proteins [143]. The ER-associated degradation ubiquitin ligase hydroxymethylglutaryl reductase degradation protein 1 (HRD1) and valosin-cointaining protein (VCP) degrade Nrf3 in the cytoplasm during normal physiological conditions. Under stress conditions, DNA-damage inducible 1 homology 2 (DDI2) cleavage of Nrf3 activates it and translates it into the nucleus. In the nucleus, β-transducin repeat-containing protein (β-TRCP)-based E3 ubiquitin ligase facilitates the degradation of Nrf3 in the nucleus.
Though all the three Nrfs (Nrf1, Nrf2, and Nrf3) regulates redox homeostasis, Nrf2 reins the regulation of phase II genes during oxidative insult [119]. They all transactivate the same genes yet they transcribe different genes depending on their partners. Besides redox regulation, all three regulates distinct genes that regulate different processes. In competition towards common ARE targets, Nrf2 supersedes Nrf1 complexes in mice during extreme electrophilic or oxidative stress [144]. The subcellular localization of Nrf1/Nrf3 and Nrf2 is ER and cytosol respectively. In humans, elevated expression of Nrf1 was detected in lung, heart, skeletal muscle, kidney and ovary where low expression was observed in brain, thymus, liver, spleen, pancreas, small intestine, colon, prostate, testis and peripheral blood leukocytes [108]. Nrf1 is mandatory for normal growth, development and cytoskeletal organization [120]. In mice, during development Nrf1 is ubiquitously expressed [145]. Nrf1 maintains normal homeostasis whereas Nrf2 responds to metabolic stress [115]. Nrf1 and Nrf2 are expressed ubiquitously albeit at different levels in vertebrate tissues and thus perform cooperative as well as competitive functions. Nrf2 expression is observed in wide range of tissues and even in embryonic stem cells [146]. Elevated expression of Nrf2 was detected in the fetal liver and muscle and in adult lung, muscle and kidney [105], [147]. High levels of Nrf3 expression were observed in the B cell lineage and mammalian placenta [148]. The expression of Nrf3 was well established in the cells and tissues of mouse and human sources [137]. In humans, intermediate and low levels of Nrf3 express in the brain, thymus, lung, heart, pancreas, spleen, colon, kidney, and leukocytes whereas Nrf3 expression was absent in skeletal muscle, ovary, testis and prostate [149]. In contrast, mouse expressed high levels of Nrf3 in brain, thymus, lung, adipose tissue, stomach, placenta, uterus, and testis [150]. Although the expression was observed to be common in some tissues and species, yet the expression and distribution of Nrf1, Nrf2 and Nrf3 display difference in species as well as tissues.
6.4. Small musculoaponeurotic fibrosarcoma proteins (sMafs)
v-maf - an avian musculoaponeurotic fibrosarcoma viral oncogene - codes for a leucine zipper motif that shows similarity with other bZIP proteins [151]. Fujiwara et al., [152] isolated two sMafs, namely MafK and MafF, coding for proteins with MW of ~18 kDa. Another novel Maf-related gene, MafG was reported to have extensive homology with its related genes MafK and MafF [153]. Due to their low MW and small size, these three Mafs (MafK, MafF, and MafG) are grouped into the small Maf (sMaf) family. The sMafs possess two important domains: a basic region responsible for DNA binding and a leucine zipper responsible for dimerization. There is another group of Maf family proteins called the large Mafs - c-Maf, MafA/L-Maf, MafB/KREISLER, and NRL [154], [155]. All these large Mafs differ from the sMafs by having an acidic N-terminal transactivation domain, yet sMafs play an important role in transcriptional regulation [156]. sMafs form homodimers in all possible combinations and bind response elements but lack transcriptional activation domains in all their possible homodimers, and thus function as transcriptional repressors [157]. Depending on the dimerization partners, sMafs activate or repress the transcription of ARE-regulated genes. sMafs can form heterodimers with most CNC family transcription factors: p45 NF-E2, Nrf1, Nrf2, Nrf3, Bach1, and Bach2 [1], [158], [159]. Nrf2-sMaf heterodimers bind an ARE present in the upstream region of MafG and trigger MafG expression on exposure to H2O2 [160], [161]. “RTGACTCAGCA” serves as the binding site for Bach1-, p45-NFE2-, and Nrf2-sMaf heterodimers [162], [163]. The extended GC bases are essential for the sMaf-specific DNA binding [83]. A nuclear export signal co-localized with the leucine zipper domain (NESzip) is present in the Nrf2 [164]. This NESzip is switched-off due to heterodimerization with MafG. NESzip is not exposed to the nuclear export protein Nrf2zip/CRM1, preventing its export and degradation in the cytosol. Thus, MafG retains and stabilizes the Nrf2 within the nucleus, and Nrf2-MafG transcriptionally activates the cytoprotective genes by binding with AREs. ARE regulated genes are activated through Nrf2-sMaf dimers and thus serve as the key components of antioxidant and xenobiotic metabolism. It has been evidenced recently from ChIP-Seq that Nrf2-sMaf dimers bound to a large number of genomic regions and the majority was novel members of the Nrf2 pathway [165].
6.5. Broad-complex, Tramtrack and Bric-a-brac (BTB), and cap'n'collar (CNC) homology proteins (BACH)
Bach - a novel bZIP transcription factor family heterodimerizes with MafK and interacts with AREs. Bach1 and Bach2 are identified to interact with MafK and carried similar BTB and bZIP domains [159]. The presence of BTB and bZIP domains makes them unique from other bZIP and zinc finger family proteins. The BTB domain is essential for both homodimerization and heterodimerization with similar BTB domain proteins. Though they possess similarity in their BTB and bZIP domains, Bach1 and Bach2 with MafK function as an activator and repressor in erythroid cells respectively. Bach1 heterodimerizes with sMafs and they both act together as repressor of MAREs [166]. Bach1 primarily serve as a repressor as it lacks a transactivation domain [167]. Bach1 represses the expression of HO-1 by forming heterodimers with sMafs, and these Bach1/sMafs interact with AREs of HO-1 and make them inaccessible for the activation of Nrf2 [168]. It is well-known that oxidative stress induces HO-1 expression when Bach1 exposes the cryptic AREs. The cysteine residues, particularly C574 of Bach1, act as sensors of redox status; the increased oxidants oxidize this cysteine residue and allow the dissociation of Bach1 from AREs and thus exposes AREs for transactivation by Nrf2 [169]. The redox status regulates the ARE regulated expression of their target genes through Bach1. Bach1 represses gene expression of its target genes in an unstressed state. The presence of a functional intronic ARE at +1411 from the TSS in Bach1 revealed that it is a bona fide target of Nrf2 and thereby establishes a feedback-inhibitory mechanism [65]. The differential expression of ARE-regulated genes is decided by the balance between Nrf2 (activator) and Bach1 (repressor). Antioxidant enzymes are brought back to normal levels due to the repression of ARE-regulated antioxidant genes as Bach1 accumulates within the nucleus in response to antioxidants [170]. Bach2 - a B-cell-specific Bach protein - heterodimerizes with sMafs and negatively regulates MAREs, thereby repressing the expression of immunoglobulin genes in undifferentiated B-cells [171]. With aging, Bach1 expression increases with a concomitant decrease in Nrf2-regulated antioxidant genes. This increase of Bach1 expression as aging progresses has to be investigated to combat age-related oxidative stress diseases.
6.6. Activating transcription factor 4
The hunt for Nrf2 partners that regulate the expression of stress response genes resulted in the discovery of activating transcription factor 4 (ATF4) [172]. The dimerization of ATF4 with Nrf2 was confirmed in mammalian cells through co-immunoprecipitation and two-hybrid assays. When mouse hepatoma cells are exposed to cadmium chloride - a potent inducer of HO-1, the ATF4-Nrf2 dimer binds to the ARE of HO-1 and activates its transcription. The ATF-Nrf2 dimer binds to the AREs depending on the nature of the stress stimulus [173]. During the induction of atherosclerotic vessels by oxidized phospholipids, Nrf2 upregulates ATF4 leading to the increased expression of vascular endothelial growth factor [174]. Nrf2 and ATF4 are the major regulators of electrophilic stress response and unfolded protein response (UPR) respectively. Both ER stress and oxidative stress induce ATF4 [175]. Fisetin - a polyphenol - has the potential to increase not only these transcription factors (Nrf2 and ATF4) but also their respective target genes through distinct mechanisms [176]. These two transcription factors (Nrf2 and ATF4) and their target genes are essential to enhance and maintain GSH levels and thus offer protection against oxidative stress. Cells respond to stressors through a novel convergence point between UPR and electrophilic stress response. There are a limited number of studies exploring the crosstalk between Nrf2 signaling pathway and UPR pathway. The roles of Nrf2 and ATF4 during UPR and oxidative stress require investigation on whether they act synergistically or antagonistically. Further research on the key players will unravel the interconnections within the oxidative and ER stress that can be translated into therapeutics for these stress related diseases.
6.7. Jun proteins
Jun proteins (c-Jun, Jun-B, and Jun-D) heterodimerize with both Nrf1 and Nrf2 and bind the hARE of NQO1 [27], [107]. The expression of NQO1 and GSTYa subunit genes are coordinately induced by Nrf2 and Nrf2 in association with Jun proteins through the cis-acting element - ARE [110]. Environmental factors and extracellular physiological stimuli trigger the three MAP kinase families: ERK, JNK and p38 which in turn regulates the expression of c-Jun [177], [178], [179], [180]. c-Jun controls cellular proliferation, apoptosis and oncogenic transformation [181]. JNK, ERK, and P38 induce c-Jun expression and phosphorylate c-Jun in response to stress stimuli [182]. c-Jun expression gets down-regulated through the inhibition of JNK by p38 MAPK in response to epidermal growth factor. Depending on the nature of the stimuli the levels of c-Jun varies and thus the regulation of downstream processes by c-Jun gets altered. The concentrations of c-Jun determine the positive and negative regulation of hARE-induced CAT expression. Interestingly, with a small increase in the concentration, c-Jun can heterodimerize with Nrf1 and Nrf2 to form positive regulatory complexes (Nrf1+c-Jun and Nrf2+c-Jun). The dimerizing partners of c-Jun determines the regulation of an array of genes [183]. These bind the hARE of CAT and activate the transcription of CAT. In contrast, with an increased concentration, c-Jun complexes with c-Fos and inhibits the positive regulatory factors, thereby suppressing the transcription of CAT. Though Jun-B and Jun-D are found to be as efficient as c-Jun, the Nrf2 in association with Jun-B and Jun-D is found more efficacious than Nrf1. c-Jun regulates the expression of urokinase [184] and γ-GCS [185] in a similar fashion. In human K562 leukemia cells, c-jun along with two other transcription factors - Fra-1 and NF-E2 p45 - regulates the expression of GSTP1 [186]. Ferritin plays a major role in the homeostasis of iron. JunD regulates human ferritin H via the ARE present upstream of ferritin H [73]. In addition to Nrf2, JunD also plays an important role in the defense against oxidative stress. JunD responds to both oxidative stressors and electrophilic xenobiotics. The phosphorylation of c-Jun led to the increased transcriptional activation of its target genes through ARE sequences in human bronchial epithelial (HBE1) cells; but in contrast, phosphorylation is not required for the activation in HepG2 cells [187]. This phosphorylation was well evidenced in HBE1 cells, triggering the increased expression of NQO1, Gclc, and Gclm. This difference in the phosphorylation of c-Jun may not only be cell dependent but also gene dependent. On binding with the ARE, Nrf2/c-Jun enhanced the transcription of HO-1, NQO1, Sod2, and Cat in rat primary astrocytes on exposure to ginsenoside Rh1 [188]. In contrast to c-Jun, p34 phosphorylates JunB independent of JNK and leads to the degradation of JunB rather activation [189], [190]. The homodimerization of JunB and the affinity of JunB dimers to AP-1 site are feeble [191]. Dynamics in the signaling pathways and gene regulation by the Jun proteins is due to the differences in the tissue-specific expression, activation, dimerization and affinity towards the AP-1 site [192]. c-Jun promotes cell proliferation and oncogenesis whereas JunB inhibits cell proliferation through cyclin-dependent kinase inhibitor [193], [194]. JunD and c-Jun share the same DNA binding sites and regulates the same genes such as c-myb or human telomerase reverse transcriptase gene [195]. JunD exhibits antioxidant defense, protects cells against oxidative stress and inhibits angiogenesis in tumor [196]. JNK phosphorylates c-Jun more efficiently than JunD [197]. JunD displays activities antagonistic to that of c-Jun with respect to cell growth and apoptosis [198]. All the three members of Jun protein family display functions that are redundant and distinct.
6.8. Other transcription factors that interact with AREs
c-Fos and Fra proteins form heterodimers with Jun proteins, thereby regulating the expression of genes via AREs [199]. hARE-mediated CAT expression is repressed by c-Fos and Fra1 in HepG2 cells [107]. Jun-Fos and Jun-Fra1 heterodimers exhibited high affinity for hARE. Nrf2/polyamine-modulated factor-1 (PMF-1) complex formation enhances the spermidine/spermine N(1)-acetyltransferase 1 expression on exposure to the anti-tumor agent PX-12 [200].
Organisms’ exposure to countless number of xenobiotics is inevitable. Such exposure generates ROS causing damage to biomolecules and modulates the metabolic process. The cells overcome such deleterious effect through the Keap1-Nrf2-ARE defense pathway. This regulation of antioxidant and detoxification enzymes by Nrf2 was studied [201], [202] and such studies revealed the multitude roles of Nrf2 in the diverse cellular processes and it is the primary player in this complex regulatory pathway. Nrf2 regulates endogenous antioxidant systems through the basal and inducible expression of ARE-regulated genes. Of the discussed transcription factors, Nrf2 is the primary factor that regulates Keap1-Nrf2-ARE pathway. AREs are present in the promoters of transcription factor encoding genes and Nrf2 regulates these genes [132], [161]. Though Nrf1, Nrf2 and Nrf3 regulate common genes yet they exhibit definite functions. sMafs act as transcription repressors in all their possible homodimers. Nrf2-sMaf dimers are the primary components of antioxidant and xenobiotic metabolism. In an unstressed state, Bach1 represses the ARE regulated genes. During ER stress, UPR is activated where ATF4 dimers with Nrf2 and offers protection. Jun family proteins in association with Nrf1 and Nrf2 regulate various cellular processes such as cell proliferation, apoptosis and oncogenesis. The combination of different transcription factors that act upon AREs has still not been resolved completely. However, the constitution differs based on the nature of cell type, electrophilic compound, oxidative stressor, and target gene. Though different homodimers and heterodimers are known to activate or repress the transcription of ARE regulated genes, the binding specificities of these diverse combinations have not been categorized yet. Future research is required to determine how different combination of transcription factors shape under different stress conditions. The spatial and temporal expression of ARE regulated genes are determined by the transcription factors and when this regulation goes awry results in oxidative stress related diseases.
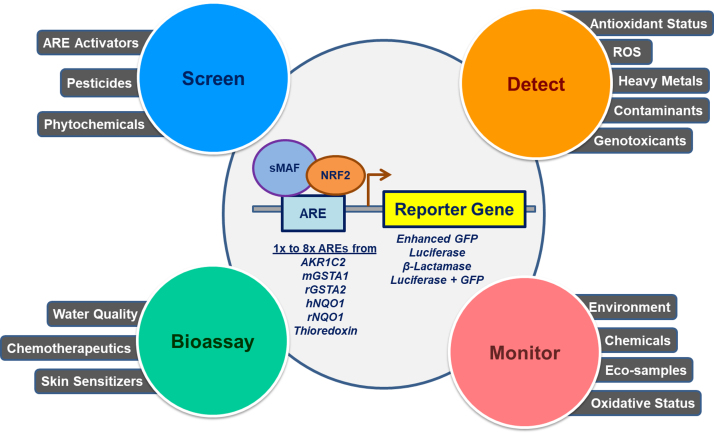
7. Applications based on AREs
The design of ARE-mediated reporter constructs was started in the late 1980s to discover and characterize AREs. Scientists adopted these ARE-based reporter constructs for a wide variety of applications (Fig. 2). The design of the ARE reporter constructs is different for various applications. There is no universal ARE reporter construct for all the assays. The application determines the design of the ARE reporter construct. Based on the assay requirement, the design of the ARE-reporter constructs and nature of the cell lines are chosen. The key components essential for the ARE reporter constructs are one or more copies of the same or different AREs from well-studied antioxidant genes and a reporter gene downstream of the AREs. The reporter gene includes fluorescence protein or an enzyme that converts a substrate into a chromogenic substance or light emitting product. The size of the construct influences the transfection efficiency. If there is more than one ARE in the construct, the size of the construct gets increase which may affect the transfection efficiency. Yet there is no clear evidence on the number of AREs required for a specific application. More research is required for the optimization on the number of AREs and the precise reporter gene that can be employed the most. The delivery method differs for different types of hosts.
Fig. 2.
The different applications based on ARE reporter constructs. The ARE based reporter constructs are used in the bioassay, detection, monitoring, and screening of various compounds/drugs in both in vitro and in vivo models.
7.1. Monitoring samples and ARE activity
A stable ARE-reporter Huh7-1×-ARE-luc was successfully developed to monitor chemicals and environmental samples [203]. This reporter construct has a luciferase gene, which is controlled by an ARE from the human NQO1 promoter. When these hepatoma Huh7 reporter cells are exposed to ARE-activating substances, they exhibit luciferase activity. The Huh7-1×-ARE-luc reporter carries one copy of ARE within the reporter construct and is sensitive enough to monitor oxidative stress in vitro. ARE-reporter cell lines are utilized to monitor chemicals, drugs and environmental samples for their oxidative stress inducing potentials. These cells serve as a rapid and inexpensive tool to monitor samples instead of using animals.
Nanoparticles are well-known to cause oxidative stress responses in vitro [204]. Nrf2-ARE luciferase reporter constructs harbored in HepG2 cells have been successfully employed to assess the induction of oxidative stress response by metal and metal oxide nanoparticles [205]. These cytotoxicity assays will serve to analyze the safety of nanoparticles in the future.
ARE-driven enhanced green fluorescent protein (eGFP) was tethered with nanoparticles [206]. These magnetic nanoparticles were then transfected into adult dog retinal endothelial cells. The transfection was efficient in dividing and migrating cells rather quiescent cells. Under hyperoxia, the ARE-mediated eGFP expression gets activated. This method of ARE-tethered magnetic nanoparticles can be adopted in the treatment of retinopathy of prematurity (ROP). ROS generated during hyperoxic conditions lead to ROP. Replacing the reporter eGFP with therapeutic genes will offer therapeutic approach to ROP. The transcriptional activation of the therapeutic genes relies on the pathogenic status of the cells. During hyperoxic conditions retinal vasculature develops from migrating cells and most of the cells divide and migrate in the developing eye. As the transfection to these magnetic nanoparticles in migrating and dividing cells is more efficient, this method can be employed for the treatment of ROP. A similar strategy using the ARE-GFP/nanoparticle construct was used to monitor tissue oxidative status in vivo [207]. The construct consists of a minimal thymidine kinase promoter with multiple AREs followed by eGFP. Undue ROS lead to mitochondrial oxidative stress and is an early sign of injury in diabetes. The sub-retinal injection of these ARE-GFP/nanoparticle constructs reported the activation of AREs due to oxidative stress in diabetic rat retinal pigment epithelium. These systems adopt the use of nanoparticles for the attachment and delivery of the reporter constructs in in vitro and in vivo models to detect the ROS and oxidative stress. However, these nanoparticles have the potential to generate ROS and cause oxidative stress in both cells and tissues, which is a concern in the use of ARE-GFP/nanoparticle constructs. The detection of ROS was efficiently performed with the generation of nanoscale biosensors by tethering magnetic nanoparticles with the AREs [208].
Using a high throughput screening approach, Shukla et al. [209] profiled environmental chemicals for ARE activators from a U.S. National Toxicology Program with 1340 compounds from a 1408 compound library. Two ARE reporters - ARE (3×) derived from hNQO1 with a downstream beta-lactamase (bla) reporter gene and ARE-luc reporter with 7 consensus AREs that have affinity only for Nrf2 were transfected into HepG2 cells for quantitative high throughput screening of chemicals [209]. Of the 1340 compounds, 30 exhibited ARE activity induced through oxidative stress in both the ARE-bla and ARE-luc HepG2 cells. The AREs in the two reporters are different in terms of activation, ARE-bla activates the ARE through interactions of different transcription factors, while ARE-luc activates the ARE only through Nrf2 during oxidative stress. These two different constructs sort chemicals based on their specific signatures in the activation of AREs and distinguish compounds that mediate ARE activation through Nrf2 and non-Nrf2. This approach is quite novel in the use of AREs to differentiate compounds that exhibit different chemical properties. The advantage of this method is that a large number of compounds can be tested in a short time.
The monitoring of ARE activity in liver regeneration post partial hepatectomy (PHx) was made possible in vivo by using an ARE-driven luciferase reporter gene from the firefly (Photinus pyralis) [210]. Nrf2 activity was measured through in vivo bioluminescence imaging and in vitro luminescence assays. The Nrf2 activity was at its peak on day 3 and became saturated at day 7 after PHx. The ARE-luc mouse serves as an excellent model to study Nrf2-mediated expression, which interacts with important pathways during regeneration. In addition, ARE-driven luciferase reporter assays help us study antioxidant stress responses during post-operative treatments.
7.2. Screening of ARE activators, pesticides and genotoxicants
With the stable transfection of ARE(4×)/TK-GFP reporter constructs into HepG2 cells, the screening of ARE activators was performed from chemical libraries containing small molecules [211]. In a high throughput HepG2 ARE functional assay, the novel small molecule, LAS0811 − 1,2‐dimethoxy‐4,5‐dinitrobenzene was identified as an activator of the ARE. The 10 combinatorial chemical libraries examined consisted of 9400 small molecules with no known ARE-activators. An ARE-driven reporter is not only a powerful tool to discover ARE activators but also a high throughput tool to screen large numbers of small molecules and thus remains as one of the best methods to identify potential agents for cancer chemoprevention yet, the use of these tools in still in its infancy.
A HepG2 cell-based assay was recently developed to measure the change in activity of the ARE signaling pathway [212]. A reporter construct with β-lactamase under the control of ARE was transfected into HepG2 cells. Fluorescence resonance energy transfer (FRET) technology detects the activation of the ARE. CCF4-AM is a FRET reporter substrate, which is turned into a polar CCF4 through hydrolysis by cytoplasmic esterase. Upon cleavage, CCF4 shifts the emission form 530–460 nm as a result of disruption in energy transfer. The cleavage of CCF4 is mediated by the expression of β-lactamase, which is under the control of the ARE. When there is no β-lactamase expression, energy transfer occurs within the CCF4, resulting in the 530 nm emission. This principle was employed for quantitative high-throughput screening of compounds capable of activating the ARE signaling pathway. This assay employed FRET, which is completely different from other ARE-reporter construct assays.
The ARE activating potential of botanical pesticides was tested using primary mixed cultures derived from ARE-human placental alkaline phosphatase reporter mice [213]. An ARE-driven luciferase reporter in human neuroblastoma cells detected the subtoxic concentration of botanical pesticides capable of eliciting the Keap1/Nrf2/ARE pathway. In the testing of compounds in primary cortical neurons and neuroblastoma cells, using ARE-reporter assays, neuroprotective phytochemicals can be identified and characterized. This assay will help us limit the use of potential toxic botanical pesticides in the environment.
The stereospecific properties of the phytochemicals were validated in the activation of EpRE-mediated gene transcription using EpRE(mGstYa)-Lux and EpRE(hNQO1)-Lux reporter constructs [214]. A difference in EpRE mediated transcription activation was observed between planar and non-planar flavonoids. The planar structured flavonoids exhibited luciferase activity, but non-planar flavonoids failed to elicit any luciferase activity. This indicates that these EpRE constructs can be employed to screen molecules based on the planarity and mechanism of activation.
In continuation with the above approaches, a workflow was developed using ARE-bla reporter gene assay models and high-throughput screening data to mechanistically profile compounds that elicit hepatotoxicity [215]. The genotoxicity of the compounds was assessed adopting ARE-luc reporter constructs transfected into HepG2 cells [216]. This Nrf2 mediated ARE-reporter assay identified the genotoxic compounds successfully. The induction of luminescence was lower in the non-genotoxic compounds. The screening of drugs for genotoxicity in the early phase of research can be achieved using ARE-reporter constructs. In this study, two different vectors were tested for the efficient induction of luminescence, one with 1× ARE and the other with 3× AREs. No difference in luminescence was observed from these two constructs. The findings from this study suggest that the use 1× ARE is sufficient for the efficient use of ARE-reporter assays.
7.3. Detecting antioxidant levels and heavy metal contamination
The level of antioxidants plays an important role in the newborn brain; when the levels go down, oxidative stress is caused, leading to injury and inflammation [217]. To know the status in the levels of antioxidants, an EpRE-luc reporter was constructed with two EpREs from the thioredoxin promoter and inserted into a mouse strain. The luminescence activity was high in the first 4 days and declined at day 10. These constructs offer insight into the levels of antioxidants and their influence in development in the newborn. In all the above reporter assays, the reporter protein is induced at the transcriptional level, but the reporter protein product is not stabilized. The OKD48 system was developed to overcome this problem, which successfully monitored oxidative stress in both in vitro (HeLa and HEK293T) and in vivo (C57BL/6 mouse) systems [218]. This OKD48 construct consists of 3xARE in the TK basal promoter along with the Neh2 domain fused to flag luciferase. The Neh2 domain of the hNrf2 binds Keap1 and becomes stabilized. In normal conditions, the OKD48 construct is not induced, and even when transcribed and translated, the OKD48 protein is degraded through the Keap1 system. The 3xARE in the OKD48 construct are activated during oxidative stress and the resulting protein Neh2(Nrf2)-Luciferase is stabilized by the Keap1 protein, thus aiding in the detection of luminescence. The 3xARE present in the construct lack the TRE and are derived from GSTya to prevent the interaction of these AREs with transcription factors other than Nrf2. Hence, this ARE based OKD48 model serves as a reporter system to screen Nrf2 inducers in both in vitro and in vivo settings.
The detection of heavy metal contamination in aquatic environments is still a herculean task using animal models. A study evidenced the detection of heavy metal mercury using a fusion protein (green fluorescent-luciferase) under the control of EpRE constructed into a plasmid pEpRE-luc-GFP [219]. This plasmid construct has one copy of EpRE from the murine Gsta1incorporated into the minimal promoter from the murine MT1. Both the transient and stable transfection of these plasmids into the zebrafish detected the low levels of mercury (0.1–0.2 μM HgCl2) that do not show external signs of toxicity. These whole fish reporter assays are more sensitive than cultured cells. The oxidative stressors in the aquatic environments can successfully be detected by these ARE-mediated plasmid constructs transfected into zebrafish. Yet another novel Nrf2-responsive reporter gene revealed that heavy metal induction of Nrf2 activity is cell specific [220]. Reporters such as ARE-luciferase and AREmut-luciferase constructs were derived from the 7 tandem repeats from hARE. These two reporter constructs were transduced into 5 cell lines - A172, A549, HEK293T, HepG2, and MCF7. Of the ten metals tested (Ag, As, Au, Cd, Co, Cu, Fe, Hg, Pb, and Zn), three metals (As, Cd, and Cu) elicited an Nrf2 response in all the 5 engineered cell lines. This cell specific variation of heavy metal activation of Nrf2 activity may due to a difference in the pathways displayed by different cells. The Nrf2 antioxidant pathway-inducing potential of the polyaromatic hydrocarbons (PAHs) - benzo[a]pyrene (B[α]P), naphthalene (Nap), phenanthrene (Phe), and pyrene (Pyr) - and heavy metal/loid(s) - As, Cd, and Pb was tested both individually and as mixtures using ARE-luc HepG2 cells [221]. Both individual and binary to seven-component mixtures of heavy metal/loid(s) and PAHs elicited the Nrf2 antioxidant pathway. This ARE-luc HepG2 cell model of heavy metal/loid(s) activates the Nrf2 antioxidant pathway. Thus, the ARE-luc HepG2 cell model can be used to test mixtures for oxidative stress response pathways of common contaminants from the environment. The environment contains mixtures of pollutants, instead of detecting the specific components in the mixture, it is helpful to detect whether the components in the mixture elicits oxidative insult which is the major cause of deleterious diseases.
7.4. Bioassays
A water quality bioassay based on oxidative stress response was developed using the AREc32 cell line [222]. Water samples from different sources - effluents, drinking water, surface water, and wastewater, were tested using the AREc32 cell line. While these cells could respond using the oxidative stress response pathway, the concentration of contaminants in water determines whether it activates this defense pathway or induces cytotoxicity in cells. The ARE-luciferase reporter was transfected into HepG2 cells and used to estimate the risk of organic contaminant extracts from drinking water [223]. This method revealed several components of the organic extracts from the drinking water that elicited ARE activity in the ARE-luciferase HepG2 cells individually or in combination. Though there are lots of assays to test the contaminants in the water, they all fail to test whether the contaminants elicit oxidative stress in the system.
The efficacy of cancer chemotherapeutic agents was assessed by the generation of AREc32, a stable MCF-7 breast cancer-reporter cell line [224]. The reporter construct contains 8 copies of ARE derived from the mouse Gsta1 and rat Gsta2. AREc32 cells displayed enhanced luciferase activity on treatment with a few cancer chemotherapeutic agents such as carmustine [1,3-bis (2-chloroethyl)-1-nitrosourea], chlorambucil, cisplatin, and melphalan when the cells are pre-exposed with L-buthionine-S,R-sulfoximine – a GSH depleter. GSTs conjugate electrophilic compounds with GSH and bring about resistance towards anticancer agents. The anticancer agents used are mostly alkylating agents and redox-cycling compounds. Such agents activate Nrf2 mediated ARE-genes. To test whether these anticancer agents will induce ARE-based reporter gene expression, this reporter was constructed. This AREc32 revealed that chemotherapeutic agents triggered Nrf2 activation in a redox-dependent fashion as evidenced from increased luciferase activity. This assay will reveal whether Nrf2 play a role in the response of tumor cells to anticancer agents based on the redox status. In addition, AREc32 cells will aid in optimal concentration of the chemotherapeutic agents that can be utilized for treatment. The use of suboptimal concentration may assist tumor cells to adapt them against chemotherapeutic agents with increased induction of cytoprotective genes.
The skin sensitizing potential of cosmetic products was tested using animals till the ban was imposed during 2013. Then scientists employed in vitro tests with keratinocytes for the toxicological evaluation of skin irritants. Chemicals in the cosmetics react with proteins in the skin and modify them covalently [225]. These covalently modified proteins are recognized by the immune system as non-self antigens and trigger a T-cell mediated immune response. This innate response is similar to that of innate response elicited by pathogenic organisms [226]. Hence, scientists utilized Keap1-Nrf2-ARE defense pathway to identify the skin sensitizing chemicals. Most of the skin sensitizing in vitro test employs the vectors constructed with one or more of the AREs upstream of the luciferase reporter gene. These vectors are transfected into the different types of cell lines to screen and test skin sensitizing chemicals. When the cells are exposed to different cosmetics, they trigger the activation of Nrf2, which then binds AREs in the construct and starts the transcription of the reporter gene. Thus the chemicals that are skin irritants activate reporters in the construct.
In vitro testing of skin sensitizing chemicals was assayed using ARE-regulated quinone reductase and ARE-dependent luciferase activity in Hepa1C1C7 and AREc32 cells respectively [225]. These ARE constructs reported the skin sensitization potential of 102 different chemicals in vitro and classified them into extreme, strong, and moderate allergens. This ARE-based molecular endpoint assay offers advantages over empirical methods such as gene expression analyses (gene-chip and reverse-transcription-PCR), surface marker expression analysis by flow cytometry, and measurement of cytokine levels by enzyme-linked immunosorbent assay as these experiments rely on analytical endpoints. A further advancement was made to match the human skin conditions, where luciferase reporter constructs with one copy of the ARE from human AKR1C2 was stably incorporated into HaCaT keratinocytes to screen skin sensitizers [227]. Yet another study developed a similar construct of luciferase reporter using the ARE of the rat NQO1 and introduced it into human keratinocyte – LuSens [228]. As evidenced from these experiments, the number of copies of AREs determines the sensitivity of the reporters [229]. The use of number of copies of AREs in the design of efficient reporter construct is still a debate which necessitates further research how the number of AREs influence the assays.
The combination of four assays - direct peptide reactivity assay, KeratinoSens assay, MUSST assay, and h-CLAT assay - yielded excellent results in the prediction of potential skin sensitizing chemicals. The KeratinoSens assay in combination with the MUSST assay offered increased accuracy [230]. The validation of chemicals with pairing of assays yielded better screening of compounds.
A novel in vitro photosafety assay was developed to evaluate potential photoallergens and phototoxicants using AREc32 and KeratinoSens reporter cell lines [231]. Chemicals that are photoallergens/photoxicans activate the Keap1-Nrf2-ARE pathway when exposed to UV radiation. With the photo-ARE assay and photo-KeratinoSens, potential photoallergenic and phototoxic chemicals can be screened.
Even the KeratinoSens cell line distinguished Nrf2-independent and -dependent activation through the difference in gene expression on exposure to different skin sensitizers [232]. ARE-luciferase transfected stable keratinocyte cell lines offer an alternative to animal testing for the assessment of the skin sensitizing potential of chemicals [233]. There will be more and more applications based on the ARE-reporter constructs in the coming years.
8. Conclusion and future perspectives
Since the discovery of the first ARE in the late 1980s, more AREs have been identified in a myriad of genes, which led to an understanding of the complexity of regulatory control through various transcription factors in the Keap1/Nrf2/ARE pathway. These transcription factors in different combinations either activate or repress the transcription of their target genes, depending on the heterogeneity in the ARE sequences. Within the past three decades, we have witnessed how ARE-regulated gene expression maintains redox equilibrium through accelerated research. With the advent of ARE-reporter assays, manifold applications were developed using ARE sequences for a range of investigations from environmental chemical profiling through to the screening of Nrf2 activators. Though considerable progress has been made in the mechanistic understanding of ARE-mediated gene regulation, there exist many new questions and gaps that remain to be explored. First, the number of AREs present upstream of cytoprotective genes varies, and studies are required to test whether this difference in the number of AREs determine the activation of its target gene expression in a spatio-temporal manner. Second, most of the experimentally identified AREs carry cytosine in the core ARE sequence, and determining whether the methylation of these cytosine residues plays any role in the gene regulation besides the primary ARE sequence will be a major challenge. Third, for decades, genetic variants in the coding regions of the genome responsible for diseases have been reported. Recently, there are reports that a considerable number of genetic variants that fall into the cis-regulatory region are associated with diseases. Though a few studies reported the presence of SNPs within the AREs leading to the neurodegenerative diseases, more studies are warranted to identify SNPs in the AREs and their predisposition to oxidative stress related diseases. The polymorphisms within the ARE sequences influence the binding of Nrf2 and their target gene regulation. This difference in the binding affinity may result in a differential response during oxidative stress, leading to disease. Fourth, the intrinsic affinity of AREs and their neighboring sequences with histone octamers has not been studied yet. Finally, the deep conserved ARE sequences can be exploited for different applications based on AREs such as detection of environmental pollutants and monitoring of oxidative stress status. In the years to come, research will shed new insights into ARE-regulated gene expression that offers therapeutic interventions in oxidative stress related diseases.
Acknowledgements
Azhwar Raghunath thanks the University Grants Commission – Basic Scientific Research (UGC-BSR), New Delhi, India for its Senior Research Fellowship (UGC-BSR-SRF – No.F.7-25/2007). The authors also thank the UGC-SAP DRS II: F-3-30/2013 and DST FIST: SR/FST/LSI-618/2014, New Delhi, India for their partial financial assistance.
Acknowledgments
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Alan P. Kumar, Email: csiapk@nus.edu.sg.
Gautam Sethi, Email: phcgs@nus.edu.sg.
Ekambaram Perumal, Email: ekas2009@buc.edu.in.
References
- 1.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Kwak M.K., Itoh K., Yamamoto M., Kensler T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motohashi H., Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trend Mol. Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Itoh K., Ishii T., Wakabayashi N., Yamamoto M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 1999;31:319–324. doi: 10.1080/10715769900300881. [DOI] [PubMed] [Google Scholar]
- 5.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Meyer U.A. Overview of enzymes of drug metabolism. J. Pharmacokinet. Biopharm. 1996;24:449–459. doi: 10.1007/BF02353473. [DOI] [PubMed] [Google Scholar]
- 7.Xu C., Li C.Y., Kong A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 8.Lewis D.F. Human cytochromes P450 associated with the phase 1 metabolism of drugs and other xenobiotics: a compilation of substrates and inhibitors of the CYP1, CYP2 and CYP3 families. Curr. Med. Chem. 2003;10:1955–1972. doi: 10.2174/0929867033456855. [DOI] [PubMed] [Google Scholar]
- 9.Strange R.C., Jones P.W., Fryer A.A. Glutathione S-transferase: genetics and role in toxicology. Toxicol. Lett. 2000;112–113:357–363. doi: 10.1016/s0378-4274(99)00230-1. [DOI] [PubMed] [Google Scholar]
- 10.Mizuno N., Niwa T., Yotsumoto Y., Sugiyama Y. Impact of drug transporter studies on drug discovery and development. Pharmacol. Rev. 2003;55:425–461. doi: 10.1124/pr.55.3.1. [DOI] [PubMed] [Google Scholar]
- 11.Wolf C.R., Moll E., Friedberg T., Oesch F., Buchmann A., Kuhlmann W.D., Kunz H.W. Characterization, localization and regulation of a novel phenobarbital-inducible form of cytochrome P450, compared with three further P450-isoenzymes, NADPH P450-reductase, glutathione transferases and microsomal epoxide hydrolase. Carcinogenesis. 1984;5:993–1001. doi: 10.1093/carcin/5.8.993. [DOI] [PubMed] [Google Scholar]
- 12.Paulson K.E., Darnell J.E., Jr, Rushmore T., Pickett C.B. Analysis of the upstream elements of the xenobiotic compound-inducible and positionally regulated glutathione S-transferase Ya gene. Mol. Cell. Biol. 1990;10:1841–1852. doi: 10.1128/mcb.10.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rushmore T.H., King R.G., Paulson K.E., Pickett C.B. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc. Natl. Acad. Sci. USA. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telakowski-Hopkins C.A., King R.G., Pickett C.B. Glutathione Stransferase Ya subunit gene: dentification of regulatory elements required for basal level and inducible expression. Proc. Natl. Acad. Sci. USA. 1988;85:1000–1004. doi: 10.1073/pnas.85.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulson K.E., Darnell J.E., Jr, Rushmore T., Pickett C.B. Analysis of the upstream elements of the xenobiotic compound-inducible and positionally regulated glutathione S-transferase Ya gene. Mol. Cell. Biol. 1990;10:1841–1852. doi: 10.1128/mcb.10.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rushmore T.H., King R.G., Paulson K.E., Pickett C.B. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc. Natl. Acad. Sci. USA. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rushmore T.H., Pickett C.B. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J. BioI. Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 18.Talalay P., De Long M.J., Prochaska H.J. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenensis. Proc. Natl. Acad. Sci. USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prochaska H.J., Talalay P. Regulatory mechanisms of monofunctional and bifunctional anticarcinogenic enzyme inducers in murine liver. Cancer Res. 1988;48:4776–4782. [PubMed] [Google Scholar]
- 20.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. BioI. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 21.Daniel V., Sharon R., Bensimon A. Regulatory elements controlling the basal and drug-inducible expression of glutathione S-transferase Ya subunit gene. DNA. 1989;8:399–408. doi: 10.1089/dna.1.1989.8.399. [DOI] [PubMed] [Google Scholar]
- 22.Friling R.S., Bensimon A., Tichauer Y., Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayney R.M., Morton M.R., Favreau L.V., Pickett C.B. Rat liver NAD(P)H:quinone reductase. regulation of quinone reductase gene expression by planar aromatic compounds and determination of the exon structure of the quinone reductase gene. J. Biol. Chem. 1989;264:21793–21797. [PubMed] [Google Scholar]
- 24.Favreau L.V., Pickett C.B. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J. BioI. Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- 25.Favreau L.V., Pickett C.B. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. characterization of a DNA-protein interaction at the antioxidant responsive element and induction by 12-0-tetradecanoylphorbol 13-acetate. J. BioI. Chem. 1993;268:19875–19881. [PubMed] [Google Scholar]
- 26.Favreau L.V., Pickett C.B. The rat quinone reductase antioxidant response element. Identification of the nucleotide sequence required for basal and inducible activity and detection of antioxidant response element-binding proteins in hepatoma and non-hepatoma cell lines. J. BioI. Chem. 1995;270:24468–24474. doi: 10.1074/jbc.270.41.24468. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Jaiswal A.K. Regulation of human NAD(P)H:quinone oxidoreductase gene. role of API binding site contained within human antioxidant response element. J. BioI. Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 28.Mulcahy R.T., Wartman M.A., Bailey H.H., Gipp J.J. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 29.Banning A., Deubel S., Kluth D., Zhou Z., Brigelius-Flohé R. The GI-GPx gene is a target for Nrf2. Mol. Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y.J., Ahn J.Y., Liang P., Ip C., Zhang Y., Park Y.M. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 31.Chowdhury I., Mo Y., Gao L., Kazi A., Fisher A.B., Feinstein S.I. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic. Biol. Med. 2009;46:146–153. doi: 10.1016/reeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y.C., Masutani H., Yamaguchi Y., Itoh K., Yamamoto M., Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J. Biol. Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 33.Rundlöf A.K., Carlsten M., Arnér E.S. The core promoter of human thioredoxin reductase 1: cloning, transcriptional activity, and Oct-1, Sp1, and Sp3 binding reveal a housekeeping-type promoter for the AU-rich element-regulated gene. J. Biol. Chem. 2001;276:30542–30551. doi: 10.1074/jbc.M101452200. [DOI] [PubMed] [Google Scholar]
- 34.Hintze K.J., Theil E.C. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc. Natl. Acad. Sci. USA. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalton T., Palmiter R.D., Andrews G.K. Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor antioxidant response element and metal response promoter elements. Nucleic Acids Res. 1994;22:5016–5023. doi: 10.1093/nar/22.23.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou H., Du S., Ji Q., Stolz A. Induction of AKR1C2 by phase II inducers: identification of a distal consensus antioxidant response element regulated by NRF2. Mol. Pharmacol. 2006;69:1662–1672. doi: 10.1124/mol.105.019794. [DOI] [PubMed] [Google Scholar]
- 37.Kelner M.J., Bagnell R.D., Montoya M.A., Estes L.A., Forsberg L., Morgenstern R. Structural organization of the microsomal glutathione S-transferase gene (MGST1) on chromosome 12p13.1-13.2. Identification of the correct promoter region and demonstration of transcriptional regulation in response to oxidative stress. J. Biol. Chem. 2000;275:13000–13006. doi: 10.1074/jbc.275.17.13000. [DOI] [PubMed] [Google Scholar]
- 38.Jaiswal A.K. Human NAD(P)H: quinone oxidoreductase (NQO1) gene structure and induction by dioxin. Biochemistry. 1991;30:10647–10653. doi: 10.1021/bi00108a007. [DOI] [PubMed] [Google Scholar]
- 39.Yueh M.F., Tukey R.H. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J. Biol. Chem. 2007;282:8749–8758. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- 40.Reichard J.F., Motz G.T., Puga A. Heme oxygenase-1induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X., Campbell M.R., Lacher S.E., Cho H.Y., Wan M., Crowl C.L. A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of Parkinsonian disorders. Cell Rep. 2016;15:830–842. doi: 10.1016/j.celrep.2016.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwak M.K., Wakabayashi N., Greenlaw J.L., Yamamoto M., Kensler T.W. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikeda H., Serria M.S., Kakizaki I., Hatayama I., Satoh K., Tsuchida S. Activation of mouse Pi-class glutathione S-transferase gene by Nrf2 (NF-E2-related factor 2) and androgen. Biochem. J. 2002;364:563–570. doi: 10.1042/BJ20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harvey C.J., Thimmulappa R.K., Singh A., Blake D.J., Ling G., Wakabayashi N. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki H., Sato H., Kuriyama-Matsumura K., Sato K., Maebara K., Wang H. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 46.Tsuji Y., Ayaki H., Whitman S.P., Morrow C.S., Torti S.V., Torti F.M. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol. Cell Biol. 2000;20:5818–5827. doi: 10.1128/mcb.20.16.5818-5827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishinaka T., Yabe-Nishimura C. Transcription factor Nrf2 regulates promoter activity of mouse aldose reductase (AKR1B3) gene. J. Pharmacol. Sci. 2005;97:43–51. doi: 10.1254/jphs.fp0040404. [DOI] [PubMed] [Google Scholar]
- 48.Friling R.S., Bergelson S., Daniel V. Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc. Natl. Acad. Sci. USA. 1992;89:668–672. doi: 10.1073/pnas.89.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jowsey I.R., Jiang Q., Itoh K., Yamamoto M., Hayes J.D. Expression of the aflatoxin B1-8,9-epoxide-metabolizing murine glutathione S-transferase A3 subunit is regulated by the Nrf2 transcription factor through an antioxidant response element. Mol. Pharmacol. 2003;64:1018–1028. doi: 10.1124/mol.64.5.1018. [DOI] [PubMed] [Google Scholar]
- 50.Vollrath V., Wielandt A.M., Iruretagoyena M., Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem. J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H: quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alam J. Multiple elements within the 5' distal enhancer of the mouse heme oxygenase-1 gene mediate induction by heavy metals. J. Biol. Chem. 1994;269:25049–25056. [PubMed] [Google Scholar]
- 53.Gong P., Hu B., Stewart D., Ellerbe M., Figueroa Y.G., Blank V. Cobalt induces heme oxygenase- 1 expression by a hypoxia-inducible factor-independent mechanism in Chinese hamster ovary cells: regulation by Nrf2 and MafG transcription factors. J. Biol. Chem. 2001;276:27018–27025. doi: 10.1074/jbc.M103658200. [DOI] [PubMed] [Google Scholar]
- 54.Gong P., Stewart D., Hu B., Li N., Cook J., Nel A. Activation of the mouse heme oxygenase-1 gene by 15-deoxy-Delta(12,14)-prostaglandin J(2) is mediated by the stress response elements and transcription factor Nrf2. Antioxid. Redox Signal. 2002;4:249–257. doi: 10.1089/152308602753666307. [DOI] [PubMed] [Google Scholar]
- 55.Cho H.Y., Gladwell W., Wang X., Chorley B., Bell D., Reddy S.P. Nrf2-regulated PPAR{gamma} expression is critical to protection against acute lung injury in mice. Am. J. Respir. Crit. Care Med. 2010;182:170–182. doi: 10.1164/rccm.200907-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Almeida D.V., Nornberg B.F., Geracitano L.A., Barros D.M., Monserrat J.M., Marins L.F. Induction of phase II enzymes and hsp70 genes by copper sulfate through the electrophile-responsive element (EpRE): insights obtained from a transgenic zebrafish model carrying an orthologous EpRE sequence of mammalian origin. Fish Physiol. Biochem. 2010;36:347–353. doi: 10.1007/s10695-008-9299-x. [DOI] [PubMed] [Google Scholar]
- 57.Soriano F.X., Léveillé F., Papadia S., Higgins L.G., Varley J., Baxter P. Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J. Neurochem. 2008;107:533–543. doi: 10.1111/j.1471-4159.2008.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki T., Takagi Y., Osanai H., Li L., Takeuchi M., Katoh Y. Pi class glutathione S-transferase genes are regulated by Nrf2 through an evolutionarily conserved regulatory element in zebrafish. Biochem. J. 2005;388:65–73. doi: 10.1042/BJ20041860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yaekashiwa M., Wang L.H. Nrf2 regulates thromboxane synthase gene expression in human lung cells. DNA Cell. Biol. 2003;22:479–487. doi: 10.1089/10445490360708883. [DOI] [PubMed] [Google Scholar]
- 60.Sakurai A., Nishimoto M., Himeno S., Imura N., Tsujimoto M., Kunimoto M. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J. Cell. Physiol. 2005;203:529–537. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 61.Alam J., Killeen E., Gong P., Naquin R., Hu B., Stewart D. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am. J. Physiol. Ren. Physiol. 2003;284:F743–F752. doi: 10.1152/ajprenal.00376.2002. [DOI] [PubMed] [Google Scholar]
- 62.Yao K.S., Hageboutros A., Ford P., O’Dwyer P.J. Involvement of activator protein-1 and nuclear factor-kappaB transcription factors in the control of the DT-diaphorase expression induced by mitomycin C treatment. Mol. Pharmocol. 1997;51:422–430. [PubMed] [Google Scholar]
- 63.Ng D., Kokot N., Hiura T., Faris M., Saxon A., Nel A. Macrophage activation by polycyclic aromatic hydrocarbons: evidence for the involvement of stress-activated protein kinases, activator protein-1, and antioxidant response elements. J. Immunol. 1998;161:942–951. [PubMed] [Google Scholar]
- 64.Alam J., Wicks C., Stewart D., Gong P., Touchard C., Otterbein S. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J. Biol. Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 65.Jyrkkanen H.K., Kuosmanen S., Heinaniemi M., Laitinen H., Kansanen E., Mella-Aho E. Novel insights into the regulation of antioxidant-response-element mediated gene expression by electrophiles: induction of the transcriptional repressor BACH1 by Nrf2. Biochem. J. 2011;440:167–174. doi: 10.1042/BJ20110526. [DOI] [PubMed] [Google Scholar]
- 66.Pietsch E.C., Chan J.Y., Torti F.M., Torti S.V. Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. J. Biol. Chem. 2002;278:2361–2369. doi: 10.1074/jbc.M210664200. [DOI] [PubMed] [Google Scholar]
- 67.Hsieh C.Y., Hsiao H.Y., Wu W.Y., Liu C.A., Tsai Y.C., Chao Y.J. Regulation of shear-induced nuclear translocation of the Nrf2 transcription factor in endothelial cells. J. Biomed. Sci. 2009;16:12. doi: 10.1186/1423-0127-16-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larade K., Jiang Z.G., Dejam A., Zhu H., Bunn H.F. The reductase NCB5OR is responsive to the redox status in beta-cells and is not involved in the ER stress response. Biochem. J. 2007;404:467–476. doi: 10.1042/BJ20061859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miao W., Hu L., Kandouz M., Batist G. Oltipraz is a bifunctional inducer activating both phase I and phase II drug-metabolizing enzymes via the xenobiotic responsive element. Mol. Pharmacol. 2003;64:346–354. doi: 10.1124/mol.64.2.346. [DOI] [PubMed] [Google Scholar]
- 70.Wild A.C., Moinova H.R., Mulcahy R.T. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 71.Erickson A.M., Nevarea Z., Gipp J.J., Mulcahy R.T. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. Revision of the ARE consensus sequence. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 72.Park E.Y., Cho I.J., Kim S.G. Transactivation of the PPAR-responsive enhancer module in chemopreventive glutathione S-transferase gene by the peroxisome proliferator-activated receptor-gamma and retinoid X receptor heterodimer. Cancer Res. 2004;64:3701–3713. doi: 10.1158/0008-5472.CAN-03-3924. [DOI] [PubMed] [Google Scholar]
- 73.Tsuji Y. Jun D activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson L.A., Gemin A., Espiritu R., Singh G. ets-1 is transcriptionally up-regulated by H2O2 via an antioxidant response element. FASEB J. 2005;19:2085–2087. doi: 10.1096/fj.05-4401fje. [DOI] [PubMed] [Google Scholar]
- 75.Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 76.Angel P., Imagawa M., Chiu R., Stein B., Imbra R.J., Rahmsdorf H.J. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 77.Pinkus R., Weiner L.M., Daniel V. Role of oxidants and antioxidants in the induction of AP-1, NF-kappaB, and glutathione S-transferase gene expression. J. Biol. Chem. 1996;271:13422–13429. doi: 10.1074/jbc.271.23.13422. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen T., Rushmore T.H., Pickett C.B. Transcriptional regulation of a rat liver glutathione S-transferase Ya subunit gene. analysis of the antioxidant response element and its activation by the phorbol ester 12-O-tetradecanoylphorbol- 13-acetate. J. Biol. Chem. 1994;269:13656–13662. [PubMed] [Google Scholar]
- 79.Xie T., Belinsky M., Xu Y., Jaiswal A.K. ARE- and TRE-mediated regulation of gene expression. response to xenobiotics and antioxidants. J. BioI. Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]
- 80.Prestera T., Holtzclaw W.D., Zhang Y., Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc. Natl. Acad. Sci. USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prestera T., Talalay P. Electrophile and antioxidant regulation of enzymes that detoxify carcinogens. Proc. Natl. Acad. Sci. USA. 1995;92:8965–8969. doi: 10.1073/pnas.92.19.8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kataoka K., Noda M., Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurokawa H., Motohashi H., Sueno S., Kimura M., Takagawa H., Kanno Y. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell. Biol. 2009;29:6232–6244. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mignotte V., Eleouet J.F., Raich N., Romeo P.H. Cis and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc. Natl. Acad. Sci. USA. 1989;86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Motohashi H., O’Connor T., Katsuoka F., Engel D.J., Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 86.Inamdar N.M., Ahn Y.I., Alam J. The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem. Biophys. Res. Commun. 1996;221:570–576. doi: 10.1006/bbrc.1996.0637. [DOI] [PubMed] [Google Scholar]
- 87.Prestera T., Talalay P., Alam J., Ahn Y.I., Lee P.J., Choi A.M. Parallel induction of heme oxygenase-1 and chemoprotective phase 2 enzymes by electrophiles and antioxidants: regulation by upstream antioxidant-responsive elements (ARE) Mol. Med. 1995;1:827–837. [PMC free article] [PubMed] [Google Scholar]
- 88.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 90.Wang X., Tomso D.J., Chorley B.N., Cho H.Y., Cheung V.G., Kleeberger S.R. Identification of polymorphic antioxidant response elements in the human genome. Hum. Mol. Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sandelin A., Wasserman W.W. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 92.Kuosmanen S.M., Viitala S., Laitinen T., Perakyla M., Polonen P., Kansanen E. The effects of sequence variation on genome-wide NRF2 binding--New target genes and regulatory SNPs. Nucleic Acids Res. 2016;44:1760–1775. doi: 10.1093/nar/gkw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lacher S.E., Alazizi A., Wang X., Bell D.A., Pique-Regi R., Luca F. A hypermorphic antioxidant response element is associated with increased MS4A6A expression and Alzheimer's disease. Redox Biol. 2017;14:686–695. doi: 10.1016/j.redox.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mathelier A., Wasserman W.W. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013;9:e1003214. doi: 10.1371/journal.pcbi.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.A.M. Khamis, O. Motwalli, R. Oliva, B.R. Jankovic, Y.A. Medvedeva, H. Ashoor, M. Essack, X. Gao, V.B. Bajic, A novel method for improved accuracy of transcription factor binding site prediction, Nucleic Acids Res. DOI: 10.1093/nar/gky237. [DOI] [PMC free article] [PubMed]
- 96.Bergelson S., Daniel V. Cooperative interaction between Ets and AP-1 transcription factors regulates induction of glutathione S-transferase Ya gene expression. Biochem. Biophys. Res. Commun. 1994;200:290–297. doi: 10.1006/bbrc.1994.1447. [DOI] [PubMed] [Google Scholar]
- 97.Pinkus R., Weiner L.M., Daniel V. Role of quinone-mediated generation of hydroxyl radicals in the induction of glutathione S-transferase gene expression. Biochemistry. 1995;34:81–88. doi: 10.1021/bi00001a010. [DOI] [PubMed] [Google Scholar]
- 98.Nguyen T., Pickett C.B. Regulation of rat glutathione-S-transferase Ya subunit gene expression: DNA-protein interaction at the antioxidant response element. J. Biol. Chem. 1992;267:13535–13539. [PubMed] [Google Scholar]
- 99.Wang B., Williamson G. Detection of a nuclear protein which binds specifically to the antioxidant responsive element (ARE) of the human NAD(P)H: quinone oxidoreductase gene. Biochim. Biophys. Acta. 1994;1219:645–652. doi: 10.1016/0167-4781(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 100.Yoshioka K., Deng T., Cavigelli M., Karin M. Antitumor promotion by phenolic antioxidants: inhibition of AP-1 activity through induction of Fra expression. Proc. Natl. Acad. Sci. USA. 1995;92:4972–4976. doi: 10.1073/pnas.92.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Orkin S.H. Globin gene regulation and switching: circa 1990. Cell. 1990;63:665–672. doi: 10.1016/0092-8674(90)90133-y. [DOI] [PubMed] [Google Scholar]
- 102.Mignotte V., Wall L., deBoer E., Grosveld F., Romeo P.H. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 1989;17:37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ney P.A., Sorrentino B.P., Lowrey C.H., Nienhuis A.W. Inducibility of the HS II enhancer depends on binding of an erythroid specific nuclear protein. Nucleic Acids Res. 1990;18:6011–6017. doi: 10.1093/nar/18.20.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taketani S., Inazawa J., Nakahashi Y., Abe T., Tokunaga T. Structure of the human ferrochelatase gene. Exon/intron gene organization and location of the gene to chromosome 18. Eur. J. Biochem. 1992;205:217–222. doi: 10.1111/j.1432-1033.1992.tb16771.x. [DOI] [PubMed] [Google Scholar]
- 105.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rushmore T.H., Pickett C.B. Glutathione S-transferases, structure, regulation, and therapeutic implications. J. Biol. Chem. 1993;268:11475–11478. [PubMed] [Google Scholar]
- 107.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H: quinone oxidoreductase1 gene. PNAS USA. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chan J.Y., Han X.L., Kan Y.W. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jaiswal A.K. Antioxidant response element. Biochem. Pharmacol. 1994;48:439–444. doi: 10.1016/0006-2952(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 110.Venugopal R., Jaiswal A.K. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Y., Crouch D.H., Yamamoto M., Hayes J.D. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 2006;399:373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang W., Chan J.Y. Nrf1 is targeted to the endoplasmic reticulum by an N-terminal transmembrane domain. inhibition of nuclear translocation and transacting function. J. Biol. Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y., Lucocq J.M., Yamamoto M., Hayes J.D. The N-terminal homology box 1 (NHB1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also enable its asparagine-glycosylation. Biochem. J. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sha Z., Goldberg A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits andp97. Curr. Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohtsuji M., Katsuoka F., Kobayashi A., Aburatani H., Hayes J.D., Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y., Lucocq J.M., Hayes J.D. The Nrf1 CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem. J. 2009;418:293–310. doi: 10.1042/BJ20081575. [DOI] [PubMed] [Google Scholar]
- 117.Song M.O., Mattie M.D., Lee C.H., Freedman J.H. The role of Nrf1 and Nrf2 in the regulation of copper-responsive transcription. Exp. Cell Res. 2014;322:39–50. doi: 10.1016/j.yexcr.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang H., Magilnick N., Lee C., Kalmaz D., Ou X., Chan J.Y. Nrf1 and Nrf2 regulate rat glutamate–cysteine ligase catalytic subunit transcription indirectly via NF-κB and AP-1. Mol. Cell. Biol. 2005;25:5933–5946. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leung L., Kwong M., Hou S., Lee C., Chan J.Y. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 120.Chan J.Y., Kwong M., Lu R., Chang J., Wang B., Yen T.S. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xing W., Singgih A., Kapoor A., Alarcon C.M., Baylink D.J., Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J. Biol. Chem. 2007;282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- 122.Sykiotis G.P., Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwak M.K., Kensler T.W. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 2011;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Higgins L.G., Kelleher M.O., Eggleston I.M., Itoh K., Yamamoto M., Hayes J.D. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol. 2009;237:267–280. doi: 10.1016/j.taap.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 125.Martin F., van Deursen J.M., Shivdasani R.A., Jackson C.W., Torutman A.G., Ney P.A. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood. 1998;91:3459–3466. [PubMed] [Google Scholar]
- 126.Chanas S.A., Jiang Q., McMahon M., McWalter G.K., McLellan L.I., Elcombe C.R. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li J., Lee J.M., Johnson J.A. Microarray analysis reveals an antioxidant responsive element-driven gene set involved in conferring protection from an oxidative stress-induced apoptosis in IMR-32 cells. J. Biol. Chem. 2002;277:388–394. doi: 10.1074/jbc.M109380200. [DOI] [PubMed] [Google Scholar]
- 128.Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamoto M., Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 129.Kwak M.K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T.W. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 130.Cho H.Y., Reddy S.P., Debiase A., Yamamoto M., Kleeberger S.R. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic. Biol. Med. 2005;38:325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 131.Ma Q., He X. Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol. Rev. 2012;64:1055–1081. doi: 10.1124/pr.110.004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 134.Nguyen T., Sherratt P.J., Pickett C.B. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 135.Miao W., Hu L., Scrivens P.J., Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J. Biol. Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 136.Shin S., Wakabayashi N., Misra V., Biswal S., Lee G.H., Agoston E.S. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol. Cell. Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kobayashi A., Ito E., Toki T., Kogame K., Takahashi S., Igarashi K. Molecular cloning and functional characterization of a new Cap'n' collar family transcription factor Nrf3. J. Biol. Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Y., Kobayashi A., Yamamoto M., Hayes J.D. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J. Biol. Chem. 2009;284:3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- 139.Sankaranarayanan K., Jaiswal A.K. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H: quinone oxidoreductase1 gene. J. Biol. Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 140.Pepe A.E., Xiao Q., Zampetaki A., Zhang Z., Kobayashi A., Hu Y. Crucial role of nrf3 in smooth muscle cell differentiation from stem cells. Circ. Res. 2010;106:870–879. doi: 10.1161/CIRCRESAHA.109.211417. [DOI] [PubMed] [Google Scholar]
- 141.Nouhi Z., Chevillard G., Derjuga A., Blank V. Endoplasmic reticulum association and N-linked glycosylation of the human Nrf3 transcription factor. FEBS Lett. 2007;581:5401–5406. doi: 10.1016/j.febslet.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y., Kobayashi A., Yamamoto M., Hayes J.D. The Nrf3 transcription factor is a membrane-bound trans-activator domain. Oncogene. 2009;8:2371–2380. [Google Scholar]
- 143.Chowdhury A.M.M.A., Katoh H., Hatanaka A., Iwanari H., Nakamura N., Hamakubo T. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci. Rep. 2017;7:12494. doi: 10.1038/s41598-017-12675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsujita T., Peirce V V., Baird L., Matsuyama Y., Takaku M., Walsh S.V. Transcription factor Nrf1 negatively regulates the cystine/glutamate transporter and lipid-metabolizing enzymes. Mol. Cell Biol. 2014;34:3800–3816. doi: 10.1128/MCB.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Murphy P., Kolsto A. Expression of the bZIP transcription factor TCF11 and its potential dimerization partners during development. Mech. Dev. 2000;97:141–146. doi: 10.1016/s0925-4773(00)00413-5. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Y., Xiang Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 2016;472:961–1000. doi: 10.1042/BJ20151182. [DOI] [PubMed] [Google Scholar]
- 147.Chan K., Lu R., Chang J.C., Kan Y.W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chenais B., Derjuga A., Massrieh W., Red-Horse K., Bellingard V., Fisher S.J. Functional and placental expression analysis of the human NRF3 transcription factor. Mol. Endocrinol. 2005;19:125–137. doi: 10.1210/me.2003-0379. [DOI] [PubMed] [Google Scholar]
- 149.Derjuga A., Gourley T.S., Holm T.M., Heng H.H., Shivdasani R.A., Ahmed R. Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Mol. Cell Biol. 2004;24:3286–3294. doi: 10.1128/MCB.24.8.3286-3294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chevillard G., Nouhi Z., Anna D., Paquet M., Blank V. Nrf3-deficient mice are not protected against acute lung and adipose tissue damages induced by butylated hydroxytoluene. FEBS Lett. 2010;584:923–928. doi: 10.1016/j.febslet.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 151.Nishizawa M., Kataoka K., Goto N., Fujiwara K.T., Kawai S. v-maf, a viral oncogene that encodes a “leucing zipper” motif. Proc. Natl. Acad. Sci. USA. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fujiwara K.T., Kataoka K., Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- 153.Kataoka K., Igarashi K., Itoh K., Fujiwara K.T., Noda M., Yamamoto M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Benkhelifa S., Provot S., Lecoq O., Pouponnot C., Calothy G., Felder-Schmittbuhl M.P. mafA, a novel member of the Maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene. 1998;17:247–254. doi: 10.1038/sj.onc.1201898. [DOI] [PubMed] [Google Scholar]
- 155.Yang Y., Cvekl A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einst. J. Biol. Med. 2007;23:2–11. doi: 10.23861/ejbm20072347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 157.Motohashi H., Katsuoka F., Shavit J.A., Engel J.D., Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–876. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 158.Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 159.Oyake T., Itoh K., Motohashi H., Hayashi N., Hoshino H., Nishizawa M. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Crawford D.R., Leahy K.P., Wang Y., Schools G.P., Kochheiser J.C., Davies K.J. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic. Biol. Med. 1996;21:521–525. doi: 10.1016/0891-5849(96)00160-8. [DOI] [PubMed] [Google Scholar]
- 161.Katsuoka F., Motohashi H., Engel J.D., Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J. Biol. Chem. 2005;280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- 162.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Warnatz H.J., Schmidt D., Manke T., Piccini I., Sultan M., Borodina, D T. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem. 2011;286:23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li W., Yu S., Liu T., Kim J.H., Blank V., Li H. Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim. Biophys. Acta. 1783;2008:1847–1856. doi: 10.1016/j.bbamcr.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F. Identification of novel NRF2-regulated genes by ChIP-seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Igarashi K., Hoshino H., Muto A., Suwabe N., Nishikawa S., Nakauchi H. Multivalent DNA binding complex generated by small maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 168.Sun J., Hoshino H., Takaku K., Nakajima O., Muto A., Suzuki H. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ishikawa M., Numazawa S., Yoshida T. Redox regulation of the transcriptional repressor Bach1. Free Radic. Biol. Med. 2005;38:1344–1352. doi: 10.1016/j.freeradbiomed.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 170.Dhakshinamoorthy S., Jain A.K., Bloom D.A., Jaiswal A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H: quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 171.Muto A., Hoshino H., Madisen L., Yanai N., Obinata M., Karasuyama H. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3' enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He C.H., Gong P., Hu B., Stewart D., Choi M.E., Choi A.M. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 173.Rossler O.G., Thiel G. Specificity of stress-responsive transcription factors Nrf2, ATF4, and AP-1. J. Cell Biochem. 2017;118:127–140. doi: 10.1002/jcb.25619. [DOI] [PubMed] [Google Scholar]
- 174.Afonyushkin T., Oskolkova O.V., Philippova M., Resink T.J., Erne P., Binder B.R. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler. Thromb. Vasc. Biol. 2010;30:1007–1013. doi: 10.1161/ATVBAHA.110.204354. [DOI] [PubMed] [Google Scholar]
- 175.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ehren J.L., Maher P. Concurrent regulation of the transcription factors Nrf2 and ATF4 mediates the enhancement of glutathione levels by the flavonoid fisetin. Biochem. Pharmacol. 2013;85:1816–1826. doi: 10.1016/j.bcp.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 177.Quantin B., Breathnach R. Epidermal growth factor stimulates transcription of the c-jun proto-oncogene in rat fibroblasts. Nature. 1988;334:538–539. doi: 10.1038/334538a0. [DOI] [PubMed] [Google Scholar]
- 178.Brenner D.A., O'Hara M., Angel P., Chojkier M., Karin M. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature. 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 179.Gille H., Strahl T., Shaw P.E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 180.Whitmarsh A.J., Yang S.H., Su M.S., Sharrocks A.D., Davis R.J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wisdom R., Johnson R.S., Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kayahara M., Wang X., Tournier C. Selective regulation of c-jun gene expression by mitogen-activated protein kinases via the 12-o-tetradecanoylphorbol-13-acetate- responsive element and myocyte enhancer factor 2 binding sites. Mol. Cell Biol. 2005;25:3784–3792. doi: 10.1128/MCB.25.9.3784-3792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van Dam H., Huguier S., Kooistra K., Baguet J., Vial E., van der Eb A.J., Herrlich P., Angel P., Castellazzi M. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 1998;12:1227–1239. doi: 10.1101/gad.12.8.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.De Cesare D., Vallone D., Caracciolo A., Sassone-Corsi P., Nerlov C., Verde P. Heterodimerization of c-jun with ATF-2 and c-fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 185.Jeyapaul J., Jaiswal A.K. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 2000;59:1433–1439. doi: 10.1016/s0006-2952(00)00256-2. [DOI] [PubMed] [Google Scholar]
- 186.Borde-Chiche P., Diederich M., Morceau F., Wellman M., Dicato M. Phorbol ester responsiveness of the glutathione S-transferase P1 gene promoter involves an inducible c jun binding in human K562 leukemia cells. Leuk. Res. 2001;25:241–247. doi: 10.1016/s0145-2126(00)00118-1. [DOI] [PubMed] [Google Scholar]
- 187.Levy S., Jaiswal A.K., Forman H.J. The role of c-Jun phosphorylation in EpRE activation of phase II genes. Free Radic. Biol. Med. 2009;47:1172–1179. doi: 10.1016/j.freeradbiomed.2009.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jung J.S., Lee S.Y., Kim D.H., Kim H.S. Protopanaxatriol ginsenoside Rh1 upregulates phase II antioxidant enzyme gene expression in rat primary astrocytes: involvement of MAP kinases and Nrf2/ARE signaling. Biomol. Ther. 2016;24:33–39. doi: 10.4062/biomolther.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bakiri L., Lallemand D., Bossy-Wetzel E., Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fuchs S.Y., Xie B., Adler V., Fried V.A., Davis R.J., Ronai Z. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J. Biol. Chem. 1997;272 doi: 10.1074/jbc.272.51.32163. (32163-31268) [DOI] [PubMed] [Google Scholar]
- 191.Deng T., Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 192.Hsu J.C., Cressman D.E., Taub R. Promoter-specific trans-activation and inhibition mediated by JunB. Cancer Res. 1993;53:3789–3794. [PubMed] [Google Scholar]
- 193.Passegue E., Wagner E.F. JunB suppresses cell proliferation by transcriptional activation ofp16(INK4a) expression. EMBO J. 2000;19:2969–2979. doi: 10.1093/emboj/19.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Konishi N., Shimada K., Nakamura M., Ishida E., Ota I., Tanaka N. Function of JunB in transient amplifying cell senescence and progression of human prostate cancer. Clin. Cancer Res. 2008;14:4408–4416. doi: 10.1158/1078-0432.CCR-07-4120. [DOI] [PubMed] [Google Scholar]
- 195.Kuhlmann A.S., Villaudy J., Gazzolo L., Castellazzi M., Mesnard J.M., Dodon M. Duc. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT) Retrovirology. 2007;4:92. doi: 10.1186/1742-4690-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gerald D., Berra E., Frapart Y.M., Chan D.A., Giaccia A.J., Mansuy D. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 197.Kallunki T., Deng T., Hibi M., Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 198.Weitzman J.B., Fiette L., Matsuo K., Yaniv M. JunD protects cells from p53-dependent senescence and apoptosis. Mol. Cell. 2000;6:1109–1119. doi: 10.1016/s1097-2765(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 199.Angel P., Karin M. The role of Jun, Fos and the AP-1complex in cell- proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 200.Kim Y.H., Coon A., Baker A.F., Powis G. Antitumor agent PX-12 inhibits HIF-1a protein levels through an Nrf2/PMF-1-mediated increase in spermidine/spermine acetyl transferase. Cancer Chemother. Pharmacol. 2011;68:405–413. doi: 10.1007/s00280-010-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nguyen T., Nioi P., Pickett C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nguyen T., Sherratt P.J., Huang H.C., Yang C.S., Pickett C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 203.Motahari P., Sadeghizadeh M., Behmanesh M., Sabri S., Zolghadr F. Generation of stable ARE-driven reporter system for monitoring oxidative stress. Daru. 2015;23:38. doi: 10.1186/s40199-015-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sharma A., Gorey B., Casey A. In vitro comparative cytotoxicity study of aminated polystyrene, zinc oxide and silver nanoparticles on a cervical cancer cell line. Drug Chem. Toxicol. 2018;23:1–15. doi: 10.1080/01480545.2018.1424181. [DOI] [PubMed] [Google Scholar]
- 205.Prasad R.Y., McGee J.K., Killius M.G., Suarez D.A., Blackman C.F., DeMarini D.M. Investigating oxidative stress and inflammatory responses elicited by silver nanoparticles using high-throughput reporter genes in HepG2 cells: effect of size, surface coating, and intracellular uptake. Toxicol. Vitr. 2013;27:2013–2021. doi: 10.1016/j.tiv.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 206.Prow T., Grebe R., Merges C., Smith J., McLeod D., Leary J. Nanoparticle tethered antioxidant response element as a biosensor for oxygen induced toxicity in retinal endothelial cells. Mol. Vis. 2006;12:616–625. [PubMed] [Google Scholar]
- 207.Prow T.W., Bhutto I., Grebe R., Uno K., Merges C., McLeod D.S. Nanoparticle-delivered biosensor for reactive oxygen species in diabetes. Vision. Res. 2008;48:478–485. doi: 10.1016/j.visres.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Prow T.W., Sundh D., Lutty G.A. Nanoscale biosensor for detection of reactive oxygen species. Methods Mol. Biol. 2013;1028:3–14. doi: 10.1007/978-1-62703-475-3_1. [DOI] [PubMed] [Google Scholar]
- 209.Shukla S.J., Huang R., Simmons S.O., Tice R.R., Witt K.L., Vanieer D. Profiling environmental chemicals for activity in the antioxidant response element signaling pathway using a high throughput screening approach. Environ. Health Prespect. 2012;120:1150–1156. doi: 10.1289/ehp.1104709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Alizai P.H., Bertram L., Kroy D., Kummer J., Andert A., Neumann U.P. Expression of VEGFR-2 during liver regeneration after partial hepatectomy in a bioluminescence mouse model. Eur. Surg. Res. 2017;58:330–340. doi: 10.1159/000479628. [DOI] [PubMed] [Google Scholar]
- 211.Zhu M., Baek H., Liu R., Song A., Lam K., Lau D. LAS0811: from combinatorial chemistry to activation of antioxidant response element. J. Biomed. Biotechnol. 2009;2009:420194. doi: 10.1155/2009/420194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhao J., Shukla S.J., Xia M. Cell-based assay for identifying the modulators of antioxidant response element signaling pathway. Methods Mol. Biol. 2016;1473:55–62. doi: 10.1007/978-1-4939-6346-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Son T.G., Camandola S., Arumugam T.V., Cutler R.G., Telljohann R.S., Mughal M.R. Plumbagin, a novel Nrf2/ARE activator, protects against cerebral ischemia. J. Neurochem. 2010;112 doi: 10.1111/j.1471-4159.2009.06552.x. (1316-132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Boerboom A.M., Vermeulen M., van der Woude H., Bremer B.I., Lee-Hilz Y.Y., Kampman E. Newly constructed stable reporter cell lines for mechanistic studies on electrophile-responsive element-mediated gene expression reveal a role for flavonoid planarity. Biochem. Pharmacol. 2006;72:217–226. doi: 10.1016/j.bcp.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 215.Kim M.T., Huang R., Sedykh A., Wang W., Xia M., Zhu H. Mechanism profiling of hepatotoxicity caused by oxidative stress using antioxidant response element reporter gene assay models and big data. Environ. Health Perspect. 2016;124:634–641. doi: 10.1289/ehp.1509763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Westerink W.M., Stevenson J.C., Horbach G.J., Schoonen W.G. The development of RAD51C, Cystatin A, p53 and Nrf2 luciferase-reporter assays in metabolically competent HepG2 cells for the assessment of mechanism-based genotoxicity and of oxidative stress in the early research phase of drug development. Mutat. Res. 2010;696:21–40. doi: 10.1016/j.mrgentox.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 217.Dohlen G., Odland H.H., Carlsen H., Blomhoff R., Thaulow E., Saugstad O.D. Antioxidant activity in the newborn brain: a luciferase mouse model. Neonatology. 2008;93:125–131. doi: 10.1159/000107777. [DOI] [PubMed] [Google Scholar]
- 218.Oikawa D., Akai R., Tokuda M., Iwawaki T. A transgenic mouse model for monitoring oxidative stress. Sci. Rep. 2012;2:229. doi: 10.1038/srep00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kusik B.W., Carvan M.J., 3rd, Udvadia A.J. Detection of mercury in aquatic environments using EPRE reporter zebrafish. Mar. Biotechnol. 2008;10:750–757. doi: 10.1007/s10126-008-9113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Simmons S.O., Fan C.Y., Yeoman K., Wakefield J., Ramabhadran R. NRF2 oxidative stress induced by heavy metals is cell type dependent. Curr. Chem. Genom. 2011;5:1–12. doi: 10.2174/1875397301105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Muthusamy S., Peng C., Ng J.C. Effect of multi-component mixtures of polyaromatic hydrocarbons and heaby metal/loid(s) on Nrf2-antioxidant response element (ARE) pathway in ARE reporter-HepG2 cells. Toxicol. Res. 2016;5:1160–1171. doi: 10.1039/c6tx00024j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Escher B.I., Dutt M., Maylin E., Tang J.Y., Toze S., Wolf C.R. Water quality assessment using the AREc32 reporter gene assay indicative of the oxidative stress response pathway. J. Environ. Monit. 2012;14:2877–2885. doi: 10.1039/c2em30506b. [DOI] [PubMed] [Google Scholar]
- 223.Wang S., Zhang H., Zheng W., Wang X., Anderson M.E., Pi J. Organic extract contaminants from drinking water activate Nrf2-mediated antioxidant response in a human cell line. Environ. Sci. Technol. 2013;47:4768–4777. doi: 10.1021/es305133k. [DOI] [PubMed] [Google Scholar]
- 224.Wang X.X.J., Hayes J.D., Wolf C.R. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 225.Natsch A., Emter R. Skin sensitizers induce antioxidant response element dependent genes: application to the in vitro testing of the sensitization potential of chemicals. Toxicol. Sci. 2008;102:110–119. doi: 10.1093/toxsci/kfm259. [DOI] [PubMed] [Google Scholar]
- 226.Freudenberg M.A., Esser P.R., Jakob T., Galanos C., Martin S.F. Innate and adaptive immune responses in contact dermatitis: analogy with infections. G. Ital. Dermatol. Venereol. 2009;144:173–185. [PubMed] [Google Scholar]
- 227.Emter R., Ellis G., Natsch A. Performance of a novel keratinocyte-based reporter cell line to screen skin sensitizers in vitro. Toxicol. Appl. Pharmacol. 2010;245:281–290. doi: 10.1016/j.taap.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 228.Ramirez T., Mehling A., Kolle S.N., Wruck C.J., Teubner W., Eltze T. LuSens: a keratinocyte based ARE reporter gene assay for use in integrated testing strategies for skin sensitization hazard identification. Toxicol. Vitr. 2014;28:1482–1897. doi: 10.1016/j.tiv.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 229.Zhu M., Chapman W.G., Oberley M.J., Wasserman W.W., Fahl W.E. Polymorphic electrophile response elements in the mouse glutathione S-transferase GSTa1 gene that confer increased induction. Cancer Lett. 2001;164:113–118. doi: 10.1016/s0304-3835(00)00664-9. [DOI] [PubMed] [Google Scholar]
- 230.Bauch C., Kolle S.N., Fabian E., Pachel C., Ramirez T., Wiench B. Intralaboratory validation of four in vitro assays for the prediction of the skin senstizing potential of chemicals. Toxicol. Vitr. 2011;25:1162–1168. doi: 10.1016/j.tiv.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 231.Tsujita-Inoue K., Hirota M., Atobe T., Ashikaga T., Tokura Y., Kouzuki J. Development of novel in vitro photosafety assays focused on the Keap1-Nrf2-ARE pathway. J. Appl. Toxicol. 2016;36:956–968. doi: 10.1002/jat.3234. [DOI] [PubMed] [Google Scholar]
- 232.Emter R., van der Veen J.W., Adamson G., Ezendam J., van Loveren H., Natsch A. Gene expression changes induced by skin sensitizers in the KeratinoSensTM cell line: discriminating Nrf-dependent and Nrf-independent events. Toxicol. Vitr. 2013;27:2225–2232. doi: 10.1016/j.tiv.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 233.Andreas N., Caroline B., Leslie F., Frank G., Kimberly N., Allison H. The intra- and inter-laboratory reproducibility and predictivity of the KeratinoSens assay to predict skin sensitizers in vitro: results of a ring-study in five laboratories. Toxicol. Vitr. 2011;25:733–744. doi: 10.1016/j.tiv.2010.12.014. [DOI] [PubMed] [Google Scholar]