Abstract
Objective
Expression of microRNAs (miRNAs) in the human placenta is dynamic across gestation, with expression of miRNAs belonging to the C14MC, C19MC and miR-371-3 clusters. Specifically, miRNAs within the C19MC cluster are exclusively expressed in primates with predominant expression in the placenta. Non-human primates can be utilized to study developmental processes of placentation in vivo that cannot be assessed in the human placenta, however, miRNA expression has not been defined in the macaque placenta. Our objective was to profile miRNAs in the macaque placenta, hypothesizing that expression is conserved between the macaque and human placenta.
Methods
Total RNA from first trimester and term macaque placentas (n=4 per group) was analyzed through RNA-sequencing and validated by quantitative real-time PCR (qRT-PCR).
Results
A total of 607 pre-miRNAs previously annotated in the macaque reference database (miRBase21) were detected, and 166 miRNAs were differentially expressed between first trimester and term placentas. A total of 457 unannotated sequences were detected and deemed candidate novel miRNAs by miRDeep2 software. Differential expression was confirmed for six of nine miRNAs evaluated by qRT-PCR. Comparative analysis demonstrated expression of several miRNA orthologs of human pregnancy-associated miRNA clusters in the macaque placenta.
Conclusions
Profiling placental miRNAs of the macaque revealed conserved expression of a number of miRNAs within the C14MC, C19MC and miR-371-3 clusters between the human and macaque. These results establish non-human primates as a model for human placentation and miRNA biology, with the prediction of their functional significance in placental development and function.
Keywords: macaque, placenta, miRNA, gene expression
INTRODUCTION
Nonhuman primates are excellent models for studying the biology of human pregnancy. Although interstitial trophoblast invasion is limited within the decidua compared to human placentation [1, 2], similarities to human placentation in the villous structure, endovascular trophoblast migration and spiral artery remodeling as well as a longer duration of gestation and maturity of young are strengths of the macaque model in comparison with other animal models. Additionally, the immune cell population, endocrine profile, and placental MHC expression are similar between nonhuman primate and human placentas [1, 3].
The central events of implantation and pregnancy are thought to be highly regulated by molecular cues, including the expression of microRNAs (miRNAs). miRNAs are small (18–25 nucleotides) non-coding RNAs that post-transcriptionally regulate gene expression by binding to target mRNAs to repress or degrade the transcripts [4, 5], and act to upregulate transcription through direct and indirect mechanisms [5–7]. miRNAs are involved in diverse cellular processes including embryonic development [8, 9], implantation [10] and pregnancy [11–13]. In humans, there are three miRNA clusters (MCs) highly expressed within the placenta, the C14MC, C19MC and miR-371-3 clusters. The C14MC is expressed from the maternally inherited chromosome and includes 52 miRNAs located within the imprinted DLK-DIO3 domain of human chromosome 14 [11, 13, 14]. The C19MC is expressed from the paternally inherited allele within an imprinted region and contains 46 miRNA genes [13, 15, 16]. The miR-371-3 cluster is adjacent to the C19MC and consists of 3 conserved mammalian miRNAs [13] that are highly expressed in embryonic stem cells with roles in cell cycle regulation [17–19]. The miRNAs of these clusters have roles in placental function and associations with placental pathologies such as preeclampsia [11–13, 20–23]. For instance, miR-519d of the C19MC cluster may indirectly alter extravillous trophoblast migration through regulation of target transcripts CXCL6, NR4A2 and FOXL2 [24]. The C19MC is of particular interest as it is exclusively expressed in primates and predominantly in the placenta [16, 25] with high expression in human trophoblasts [26, 27].
The macaque model has been well defined in terms of placental structure and physiology, however, the expression of miRNAs within the macaque placenta has been essentially unexplored. The objective of this study was to profile miRNA expression within the macaque placenta, hypothesizing that miRNA expression is conserved between macaques and humans. Through a miRNA-sequencing (miR-seq) approach, miRNA expression profiles were defined for first trimester and term macaque placentas. These profiles will be informative for future studies in the macaque model to better understand molecular changes underlying placental developmental processes and pathologies.
MATERIALS AND METHODS
Animals
Rhesus monkeys (Macaca mulatta) were from the colony maintained at the Wisconsin National Primate Research Center. All procedures were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and under the approval of the University of Wisconsin Graduate School Animal Care and Use Committee. A total of 16 placentas were collected from pregnancies terminated in the first trimester (gestation day: 36) or from term clinical cesarean sections (gestation day: ~165).
RNA Extraction
RNA extraction was performed using TRIzol Reagent (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) following protocol recommendations. Total RNA was extracted from 4 first trimester and 4 term placentas and sent to Arraystar Inc. (Rockville, Maryland, USA) to undergo miR-seq.
Library Preparation and miRNA-sequencing
A miR-seq library was prepared for each sample through the following steps: ligation of a 3′-adapter and 5′-adapter, cDNA synthesis, PCR amplification, and extraction and purification of the amplified PCR fragments from a PAGE gel. The sequencing libraries were quantified with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Each sample was diluted to 8 pM and cluster generation was performed on the Illumina cBot using a TruSeq Rapid SR cluster kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina NextSeq 500 using TruSeq Rapid SBS kits (Illumina). All kit protocols were performed following the manufacturer’s instructions.
Raw Data Processing and Differential Expression Analysis
Raw sequences were generated as clean reads by real-time base calling and quality filtering. Reads were trimmed of the 3′-adaptor sequence and reads shorter than 15 nt were discarded. The trimmed reads were then aligned to rhesus macaque pre-miRNA sequences in the miRBase 20 reference database (http://www.mirbase.org), using novoalign software for annotation and generation of miRNA read counts, which were used to estimate the expression level of each miRNA. Expression was normalized as transcripts per million total aligned miRNA reads (TPM). Differential expression was calculated by deriving the fold change of the log2 transformed TPM for each miRNA, and a t-test was performed to determine statistical significance (P< 0.05). All known macaque miRNAs of the pregnancy-associated clusters were analyzed for changes in expression level from first trimester to term. A 2-way ANOVA was performed using GraphPad Prism (version 7) software for all miRNAs in the cluster at different stages of gestation where significance was defined as P < 0.05.
Expression Validation by qRT-PCR
In addition to the 8 sequenced samples, an additional 8 placentas (4 first trimester and 4 term) were extracted for total RNA as described above. Samples were diluted to 80 ng/ul and a spike-in control, C. elegans mature miR-39c, was added to each sample (6.24×105 copies per sample). Total RNA was reverse transcribed using a miRNA II RT kit (Qiagen, Germantown, MD) following the manufacturer’s recommended protocol, utilizing the HiSpec buffer and 920 ng of sample. The cDNA was then diluted 1:5 with RNAse-free water and qRT-PCR reactions were assembled by using a miScript SYBR green PCR kit (Qiagen) combined with a primer specific to the miRNA of interest (see Supplementary File 1 for primer sequences). All reactions were performed in triplicate and a set of no template control reactions were included in parallel for each primer. The qRT-PCR reactions were performed as recommended by the kit protocol and run on a Roche Light Cycler. The spike-in control served as the reference gene for all samples to calculate the ΔCt. Data were analyzed by performing a t-test on the ΔCt values for each miRNA and are graphically represented as the fold change in expression ± the standard error using the 2−ΔΔCt method by Livak and Schmittgen [28].
C14MC, C19MC, and miR-371-3 Cluster Analysis
miRNAs belonging to the human pregnancy associated clusters were analyzed for conservation of expression between human and non-human primate placentas and trends in expression across gestational age. If a human miRNA within one of these clusters was not represented in the macaque dataset, the human miRNA sequence was analyzed by BLAST and miRBase to determine sequence complementarity between the known human miRNA and the rhesus genome or annotated miRNA reference sequences. Sequences conserved in each cluster were assessed for expression changes throughout gestation. Statistical analysis was performed using GraphPad Prism software. A 2-way ANOVA with post-hoc Bonferroni Correction for multiple comparisons was applied to the mean log transformed TPM read count for each miRNA in the cluster at each time point and significance was defined as P < 0.05.
Novel miRNA Candidate Analysis
Unannotated macaque sequences were analyzed by miRDeep2 software for prediction of novel miRNAs. Briefly, a score of the likelihood that a sequence represents a novel miRNA candidate is assigned based on an algorithm that takes into account the number of reads corresponding to the mature and star sequences and also, the relative and absolute stabilities of the sequence structure [29, 30]. To characterize the predicted novel sequences, a BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to align the predicted sequences to the macaque genome (assembly Mmul_8.0.1) and then also to the human genome (assembly GRCh38.p7) if a macaque miRNA annotation did not exist. Additionally, the predicted sequence was searched in miRBase to determine sequence alignments between the macaque and human.
RESULTS
miRNA-Sequencing Analysis of the Rhesus Macaque Placenta
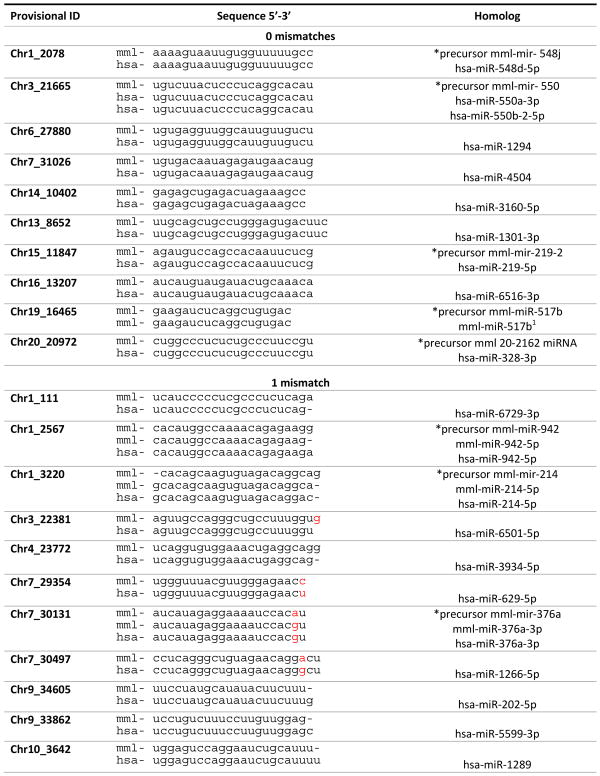
The miRNA expression profiles of 4 first trimester and 4 term rhesus macaque placentas were evaluated by miR-seq analysis. Reads were aligned to annotated rhesus pre-miRNAs within miRBase, where the average number of reads aligned to pre-miRNAs were about 9,476,574 and 8,701,971 for first trimester and term placentas, respectively (Supplementary File 2). A total of 1064 sequences were expressed with at least 1 TPM in one sample, where 607 sequences aligned to known rhesus macaque pre-miRNAs and 457 novel miRNA candidate sequences were predicted through miRDeep2 analysis. Non-supervised hierarchal clustering of all expressed miRNAs revealed segregation of individual placentas based on gestational age with distinct expression patterns between early and late gestation (Figure 1A). A larger proportion of miRNAs were more highly expressed by greater than 2-fold change in first trimester versus term placentas, in distinction from the reverse comparison (Figure 1B).
Figure 1.
miRNA expression in the macaque placenta. A. Unsupervised hierarchical clustering of miRNA expression. Each row represents an expressed miRNA and each column represents a sequenced placenta in which the first trimester placentas (n=4) and the term placentas (n=4) are distinctly clustered. Expression levels increase from green to red. B. Fold change in expression is represented for first trimester compared to term placenta on the y-axis and term compared to first trimester on the x-axis to depict expression levels between placentas of differing gestational age. Each miRNA is represented by a point where red are miRNAs highly expressed in first trimester, green are miRNAs highly expressed in term and gray are miRNAs not differentially expressed.
Differential Expression of miRNAs in First and Term Rhesus Macaque Placentas
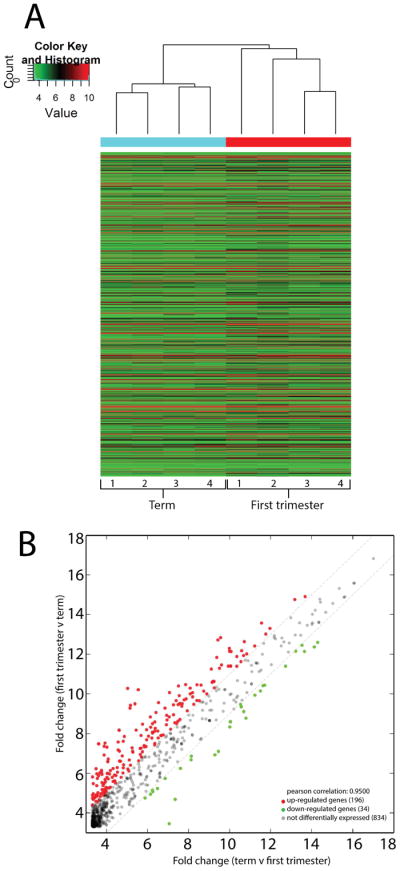
Across gestation, a total of 166 miRNAs were significantly differentially expressed (> 2-fold), where 137 and 29 miRNAs were preferentially expressed in first trimester and term placentas, respectively (Table 1). Supplementary file 3 provides information in regards to the predominant miRNA isoforms expressed, mean fold change in expression between groups, the p-value, and false discovery rate (FDR) for each differentially expressed miRNA. Figure 2 shows the cluster analysis of differentially expressed miRNAs in individual samples (top dendrogram), and in addition, differentially expressed miRNAs clustered in terms of their relative expression levels (left dendrogram). Despite the fact that each of these miRNAs were differentially expressed, most miRNAs segregated as low, moderate or high expression clusters that were maintained across both first trimester and term placentas. Additionally, a subset of six miRNAs were highly abundant as well as differentially expressed (Figure 2, bottom of the heat map).
Table 1.
Differentially expressed miRNAs in macaque placentas.
BLAST novel mature sequence to macaca mulatta
Figure 2.
Hierarchal clustering of differentially expressed miRNAs between first trimester and term placentas. The top dendrogram illustrates the relatedness in expression across placentas, whereas the left dendrogram represents clustering of miRNAs with similar expression levels. The highest levels of expression are indicated in red and the lowest level of expression are indicated in green.
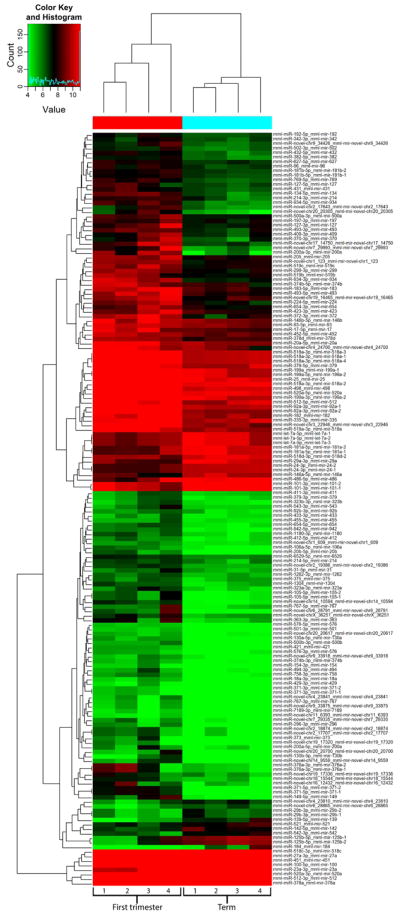
To provide technical validation of the miR-seq differential expression results, expression was evaluated for 9 selected miRNAs by qRT-PCR in sequenced RNA samples (n=4) and in additional biological replicates (n=4) for each first trimester and term placentas. miRNA selected for qRT-PCR analysis included two novel miRNA candidates (Chr19_17320 and Chr19_16465), two C14MC miRNAs (miR-323 and miR-134), two C19MC miRNAs (miR-519a and miR-518c), two miR-371-3 cluster miRNAs (miR-371-5p and miR-372), and an additional miRNA preferentially expressed in term placentas (miR-139). Differential expression was confirmed for Chr19_17320, miR-372, miR-371-5p, miR-519a, miR-518c and miR-139, whereas expression was not significantly different between first trimester and term placentas for Chr19_16456, miR-323 and miR-134 (Figure 3). These results confirm that human pregnancy-related C19MC and miR-371-3 cluster miRNAs are differentially expressed at different stages of gestation in the macaque placenta.
Figure 3.
Gene expression validation by qRT-PCR. A. Fold change in expression between first trimester and term placentas was assessed for nine selected miRNAs. B. The fold change in expression obtained by qRT-PCR and RNA-sequencing with their corresponding P-values. P-values are denoted as follows: * P < 0.05, ** P < 0.01 and *** P < 0.0001.
Conservation of miRNA expression between macaque and human placentas
miRNAs belonging to the human C14MC, C19MC and miR-371-3 placenta-associated clusters were assessed for conservation of expression between the human and macaque placenta. Figure 4 illustrates the annotation of macaque miRNAs representing orthologs of the human placenta-associated clusters, where each gene and species (−5p and/or −3p distinction) is depicted across the chromosome. If a human miRNA was not represented in the macaque dataset, a BLAST of the human pre-miRNA sequence to the macaque genome was performed to determine the existence of a macaque ortholog. The human C14MC is positioned on macaque chromosome 7 and of the 52 miRNAs in the cluster, 45 genes (76 species) were expressed, 4 were not expressed and 3 human miRNAs, mir-1197, mir-300 and mir-655, had no sequence similarities between the macaque and human. Overall, ~88% of all expressed C14MC species had higher expression in first trimester compared to term placentas. A comparison of expression levels across gestation for each miRNA within the cluster by a 2-way ANOVA revealed a significant difference (P <0.0001), suggesting decreased expression of the C14MC through gestation (Figure 4B, Supplementary File 4). The trends in expression for all miRNAs within each cluster are represented in Supplementary File 4.
Figure 4.
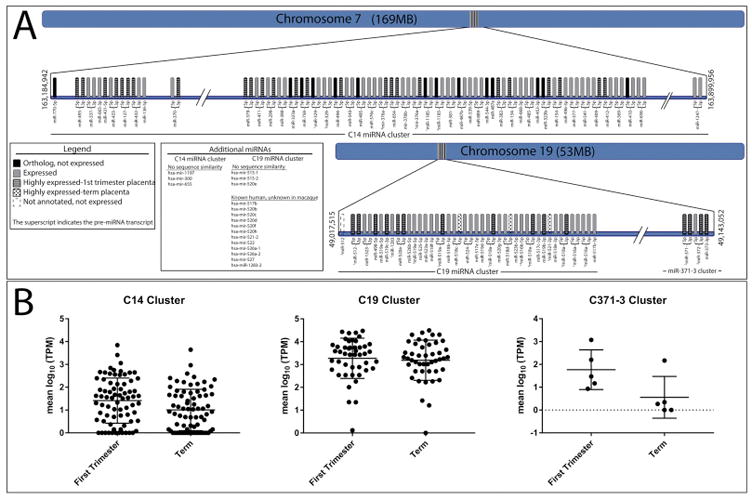
Analysis of the conservation of placenta-associated miRNA clusters between the macaque and human. A. Annotated miRNAs within the C14MC, C19MC and miR-371-3 clusters were assessed in macaque placentas. Comparison in expression between the human and macaque is represented by individual bars along the chromosome designated as either an ortholog but not expressed, expressed, highly expressed in first trimester placenta, highly expressed in term placenta, and not annotated or expressed but is an ortholog. Highly expressed refers to those that are significantly, differentially expressed. B. Expression of miRNAs within placenta-associated clusters across gestation. The mean log10TPM for each miRNA and the overall mean and SE for all miRNAs is represented for each time point. Significantly different (P < 0.0001) expression levels between first trimester and term placentas for the C14MC, C19MC and miR-371-3 cluster are denoted by an asterisk.
Conserved expression of the C19MC was observed in the macaque as 32 of 46 genes were expressed, however, several known human miRNAs within this cluster are not annotated in the macaque (Figure 4A). No sequence similarities were observed between the human and macaque for human pre-miRNA sequences mir-515-1, mir-515-2 and mir-520e. BLAST of human C19MC pre-miRNA sequences to the macaque genome revealed only partial alignments to several annotated macaque C19 miRNAs, and thus a specific ortholog or homolog could not be distinguished. Altogether, 49 C19MC species were expressed in our dataset, ~71% were higher in first trimester. As illustrated in Supplementary File 4, the C19MC miRNA species demonstrate a wide range of expression levels in both the first and third trimesters. A comparison of expression levels between early and late gestation for all C19MC miRNAs revealed a significant difference (P < 0.0001, 2-way ANOVA with post-hoc Bonferrroni Correction) in expression, and supported the apparent trend of higher expression in first trimester placentas. The miR-371-3 cluster had significantly (P < 0.0001) different expression levels from first trimester to term.
Characterization of Novel Macaque miRNA Candidate Sequences
A total of 286 sequences had a miRDeep score greater than 5 and an 87% estimated probability that the miRNA sequence was a true positive call. The pre-miRNA candidate sequences were aligned to the rhesus genome and mature sequences were searched in miRBase. Nine novel candidate sequences aligned with 0 mismatches to known human mature miRNAs, 19 with 1 mismatch, 10 with 2 mismatches and 17 with > 2 mismatches (Table 2). Although sequences with partial alignments could not be fully characterized, we propose that novel rhesus sequences with complete alignments to known human miRNAs may be considered macaque miRNA candidates.
Table 2.
Alignments and characterization of predicted novel macaque miRNA sequences. A comparison of novel macaque sequences were aligned to known human miRNAs identified through NCBI BLAST and miRBase.
| Differentially Expressed Macaque Placenta miRNAs | |||
|---|---|---|---|
| Mature miRNA ID | Fold Change | Mature miRNA ID | Fold Change |
| preferentially expressed in first trimester placentas | |||
| miR-novel_chr7_29993 | 25.273 | miR-431-5p | 3.379 |
| miR-200a-3p | 19.035 | miR-758-3p | 3.377 |
| miR-371-5p (mir-371-2) | 14.551 | miR-novel_chr7_29335 | 3.364 |
| miR-371-5p (mir-371-1) | 14.551 | miR-432-5p | 3.319 |
| miR-149-5p | 11.984 | miR-654-3p | 3.256 |
| miR-376a-3p (mir-376a-2) | 11.704 | miR-500a-3p | 3.203 |
| miR-376a-3p (mir-376a-1) | 11.704 | miR-423-3p | 3.139 |
| miR-novel-chr3_22946 | 9.803 | miR-494-3p | 3.088 |
| miR-novel-chr6_28791 | 9.785 | miR-127-5p | 3.061 |
| miR-519a-3p | 9.701 | miR-1304-5p | 3.018 |
| miR-novel-chr17_14750 | 8.111 | miR-512-3p | 3.001 |
| miR-novel-chr16_12432 | 8.000 | miR-199a-3p | 3.002 |
| miR-934-3p | 7.946 | miR-novel_chr2_19386 | 2.949 |
| miR-novel-chr20_20305 | 7.872 | miR-382-5p | 2.939 |
| miR-372-3p | 7.391 | miR-500b-3p | 2.930 |
| miR-363-3p | 6.894 | miR-498-5p | 2.887 |
| miR-novel-chr20_20700 | 6.644 | miR-323b-3p | 2.869 |
| miR-934-5p | 6.525 | miR-412-5p | 2.859 |
| miR-134-5p | 6.468 | miR-379-5p | 2.752 |
| miR-370-3p | 6.253 | miR-379-3p | 2.682 |
| miR-novel-chr14_10594 | 6.176 | miR-502-3p | 2.675 |
| miR-767-5p | 6.063 | miR-novel_chr9_33875 | 2.667 |
| miR-105-5p (mir-105-2) | 5.731 | miR-novel-chr4_23841 | 2.636 |
| miR-105-5p (mir-105-1) | 5.731 | miR-146b-5p | 2.608 |
| miR-92a-3p | 5.512 | miR-371-3p (mir-371-2) | 2.558 |
| miR-novel-chr14_9559 | 5.241 | miR-371-3p (mir-371-1) | 2.558 |
| miR-335-3p | 4.928 | miR-518a-3p (mir-518a-4) | 2.555 |
| miR-20b-5p | 4.926 | miR-518a-3p (mir-518a-3) | 2.555 |
| miR-224-5p | 4.786 | miR-518a-3p (mir-518a-1) | 2.555 |
| miR-373-3p | 4.776 | miR-18a-3p | 2.532 |
| miR-novel-chr2_17643 | 4.649 | miR-130a-5p | 2.476 |
| miR-493-5p | 4.616 | miR-6529-5p | 2.458 |
| miR-127-3p | 4.542 | miR-374b-3p | 2.458 |
| miR-130b-5p | 4.532 | miR-378d-3p | 2.385 |
| miR-92a-3p | 4.436 | miR-181b-5p | 2.381 |
| miR-novel-chr4_24700 | 4.424 | miR-25-3p | 2.380 |
| miR-296-3p | 4.341 | miR-421-3p | 2.369 |
| miR-novel_chr18_15544 | 4.284 | miR-181b-5p | 2.365 |
| miR-512-5p | 4.182 | miR-767-3p | 2.358 |
| miR-183-5p | 4.124 | miR-31-5p | 2.354 |
| miR-novel_chrx_36251 | 4.079 | miR-520a-3p | 2.350 |
| miR-378a | 4.043 | miR-96-5p | 2.337 |
| miR-409-3p | 3.951 | miR-455-3p | 2.321 |
| miR-323a-3p | 3.939 | miR-411-3p | 2.304 |
| miR-519b -3p | 3.896 | miR-433-3p | 2.294 |
| miR-200a-5p | 3.849 | miR-518a-3p (mir-518a-2) | 2.265 |
| miR-novel_chr2_17707 | 3.848 | miR-627-5p | 2.241 |
| miR-novel_chr2_18874 | 3.848 | miR-1262-3p | 2.235 |
| miR-299-3p | 3.832 | miR-576-3p | 2.222 |
| miR-543-3p | 3.777 | miR-192-5p | 2.198 |
| miR-520a-5p | 3.771 | miR-501-3p | 2.192 |
| miR-374b-5p | 3.771 | miR-novel_chr9_33918 | 2.186 |
| miR-654-5p | 3.763 | miR-452-5p | 2.148 |
| miR-519c-3p | 3.760 | miR-429-3p | 2.133 |
| miR-214-3p | 3.670 | miR-92b-3p | 2.114 |
| miR-205-5p | 3.641 | miR-novel_chr20_20617 | 2.111 |
| miR-novel_chr19_16465 | 3.630 | miR-342-3p | 2.089 |
| miR-182-5p | 3.615 | miR-197-3p | 2.086 |
| miR-novel_chr19_17320 | 3.609 | miR-novel-chr1_609 | 2.080 |
| miR-17-5p | 3.570 | miR-novel_chr11_6393 | 2.075 |
| miR-769-5p | 3.556 | miR-214-5p | 2.068 |
| miR-novel-chr9_34426 | 3.523 | miR-93-5p | 2.052 |
| miR-942-5p | 3.507 | miR-199a-5p (mir-199a-2) | 2.047 |
| miR-novel_chr1_123 | 3.495 | miR-199a-5p (mir-199a-1) | 2.047 |
| miR-493-3p | 3.434 | miR-375-3p | 2.022 |
| miR-20a-5p | 3.428 | miR-576-5p | 2.017 |
| miR-novel-chr19_17336 | 3.423 | miR-7189-3p | 2 |
| miR-106a-5p | 3.415 | miR-154-3p | 2 |
| miR-1180-3p | 3.391 | ||
| preferentially expressed in term placentas | |||
| miR-184-3p | 12.091 | miR-23a-3p | 2.519 |
| miR-139-5p | 6.329 | miR-486-5p | 2.466 |
| miR-125b-5p (mir-125b-2) | 5.130 | miR-521-3p | 2.428 |
| miR-125b-5p (mir-125b-1) | 5.109 | miR-101-3p | 2.412 |
| miR-29a-3p | 4.079 | miR-29b-3p (mir-29b-2) | 2.400 |
| miR-100-5p | 3.382 | miR-29b-3p (mir-29b-1) | 2.393 |
| miR-451-5p | 3.232 | let7a-5p (mir-let-7a-1) | 2.337 |
| miR-146a-5p | 3.210 | let7a-5p (mir-let-7a-2) | 2.328 |
| miR-181a-5p (mir-181a-2) | 2.999 | miR-101-3p | 2.326 |
| miR-181a-5p (mir-181a-1) | 2.999 | let7a-5p (mir-let-7a-3) | 2.310 |
| miR-27a-3p | 2.844 | miR-novel_chr6_28865 | 2.267 |
| miR-24-3p (mir-24-2) | 2.732 | miR-novel_chr4_23810 | 2.178 |
| miR-24-3p (mir-24-1) | 2.721 | miR-518c-3p | 2.103 |
| miR-142-5p | 2.719 | miR-518d-3p | 2.078 |
| miR-542-3p | 2.573 | ||
Blue= C14MC miRNAs, Orange= C19MC miRNAs, Green= miR-371-3 miRNAs. Parenthesis denote the miRNA precursor variant.
DISCUSSION
The expression profile of macaque placental miRNAs was established through a miR-seq approach. Unsupervised clustering of miRNA expression levels in individual placentas revealed distinct profiles for placentas collected in first trimester compared to those collected at term, suggesting that miRNA expression is specific to the stage of pregnancy and consistent across individuals. We predict that study of the macaque placenta miRNAome and their respective target genes will provide a deeper understanding of the molecular basis of physiology and pathophysiology in macaque placental studies.
Profiles of highly expressed placental miRNAs vary across human studies, which may be attributable to gestational age, the placental component analyzed, and/or methods used to assess expression. Luo et al. [31] reported high expression of miR-21, miR-125b and miR-517a in human placentas regardless of gestational age or representative cell types assessed. These miRNAs were also detected in the current study, although miR-148a-3p, miR-26a-5p and miR-516b were the most highly expressed macaque placental miRNAs, suggesting that species-specific differences in miRNA expression may exist. A microarray analysis of human first trimester and term placentas identified 191 differentially expressed genes, of which miRNAs belonging to the miR-17-92, C14MC, C19MC and miR-371-3 clusters were highly expressed in first trimester placenta [32]. miRNAs within these clusters were also detected in first trimester macaque placentas in the present study.
Differential expression of the pregnancy-associated miRNA clusters has been observed in human placentas across gestational age. Luo et al. [31] reported predominant expression of miRNAs from chromosome 19 in whole villous term placenta samples. Similarly, Morales-Prieto et al. [26] reported high expression of C19MC miRNAs in primary term human trophoblasts, whereas members of the C14MC were predominantly expressed in first trimester trophoblasts. The authors suggest that expression of the C14MC decreases and the C19MC increases throughout gestation. In agreement with human studies, ~88 % of the C14MC miRNAs expressed in the macaque placenta decreased from first trimester to term, however, predominant expression of the rhesus C19MC members in term placenta was not observed. This is in concordance with another human study by Gu et al. [32], and differences in C19MC expression patterns could be attributed to the representative placental cell types analyzed.
Genomic sequences of known human and macaque miRNAs were examined in the present study and the results were compared to a previous study conducted by Morales-Prieto et al. [20]. While miRNAs of the miR-371-3 cluster are well conserved across the human and macaque, a subset of the human C14MC and C19MC members were not orthologous in the macaque. Morales Prieto et al. [20] determined that no macaque orthologs exist for hsa-mir-655 of the C14MC or hsa-mir-518a-1, hsa-mir-518a-2 and hsa-mir-520h of the C19MC. In the present study, about 87 % of the human C14MC genes were expressed in the macaque, however, no macaque orthologs could be determined for 3 human C14MC miRNAs, mir-1197, mir-300 and mir-655. In comparison, the homology between the human and macaque C19MC is more complex as 3 human miRNAs have no sequence similarities with the macaque and 11 human miRNAs did not have a macaque ortholog, although a subset partially aligned to the macaque genome. Additionally, the sequence of macaque mir-517b was more closely matched to human mir-517a or mir-519b, hence there are C19MC sequence differences between humans and macaques that warrant further investigation. It has been demonstrated that macaque pre-miRNA and mature miRNA sequences are more dissimilar to human sequences compared to other non-human primates such as chimpanzee or orangutan [33]. Thus, determining orthologs of human miRNAs in the rhesus macaque requires more careful exploration and ultimately functional testing.
In summary, expression profiling of miRNAs in the macaque placenta revealed conserved expression of miRNAs belonging to pregnancy-associated clusters. Given the importance of rhesus macaques in studies to model human pregnancy, further work is needed to improve the annotation and explore both the putative targets and functional roles of C19MC members in the macaque placenta. As miRNAs serve to regulate gene expression in a variety of tissues including the placenta, a greater understanding of the expression profile of the macaque placental miRNAs will enhance interpretation of the biology at the maternal-fetal interface in response to experimental conditions or pathologies in the macaque animal model.
Supplementary Material
Expression of individual miRNAs within placenta-associated clusters where expression is represented by the averaged normalized read count (TPM) for each expressed miRNA within a cluster in first trimester and term placentas. The expression levels for specific miRNAs are connected by a red line if the expression decreases and green line if the expression increases from first trimester to term. Significantly different (P < 0.0001) expression levels between first trimester and term placentas for the C14MC, C19MC and miR-371-3 cluster are denoted by an asterisk.
HIGHLIGHTS.
Macaque placenta miRNA expression changes across placental development.
The human pregnancy-associated miRNA clusters are conserved in the macaque.
Novel predicted macaque miRNA sequences were identified.
Acknowledgments
The authors would like to extend our thanks to the animal care and veterinary staff at the WNPRC for the care of animals throughout pregnancy and for the collection of placenta specimens. We thank undergraduate students Joslin Musick and Megan Murphy for their assistance in analyzing and characterizing novel predicted sequences. This work was supported by NIH grants HD091163 to T.G.G. and P51 OD011106 to the WNPRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carter AM. Comparative studies of placentation and immunology in non-human primates suggest a scenario for the evolution of deep trophoblast invasion and an explanation for human pregnancy disorders. Reproduction. 2011;141(4):391–6. doi: 10.1530/REP-10-0530. [DOI] [PubMed] [Google Scholar]
- 2.Grigsby PL. Animal Models to Study Placental Development and Function throughout Normal and Dysfunctional Human Pregnancy. Semin Reprod Med. 2016;34(1):11–6. doi: 10.1055/s-0035-1570031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golos TG, Bondarenko GI, Dambaeva SV, Breburda EE, Durning M. On the role of placental Major Histocompatibility Complex and decidual leukocytes in implantation and pregnancy success using non-human primate models. Int J Dev Biol. 2010;54(2–3):431–43. doi: 10.1387/ijdb.082797tg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–30. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 7.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 8.Laurent LC. MicroRNAs in embryonic stem cells and early embryonic development. J Cell Mol Med. 2008;12(6a):2181–8. doi: 10.1111/j.1582-4934.2008.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross N, Kropp J, Khatib H. MicroRNA Signaling in Embryo Development. Biology (Basel) 2017;6(3) doi: 10.3390/biology6030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Niu Z, Li Q, Pang RT, Chiu PC, Yeung WS. MicroRNA and Embryo Implantation. Am J Reprod Immunol. 2016;75(3):263–71. doi: 10.1111/aji.12470. [DOI] [PubMed] [Google Scholar]
- 11.Sadovsky Y, Mouillet JF, Ouyang Y, Bayer A, Coyne CB. The Function of TrophomiRs and Other MicroRNAs in the Human Placenta. Cold Spring Harb Perspect Med. 2015;5(8):a023036. doi: 10.1101/cshperspect.a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morales Prieto DM, Markert UR. MicroRNAs in pregnancy. J Reprod Immunol. 2011;88(2):106–11. doi: 10.1016/j.jri.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97(1):51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14(9):1741–8. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37(10):3464–73. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19(18):3566–82. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 17.Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS, Lee JH, Koo KH, Park JW, Kim KS. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28(6):521–7. doi: 10.1007/s10059-009-0158-0. [DOI] [PubMed] [Google Scholar]
- 18.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5(2):351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 20.Morales-Prieto DM, Ospina-Prieto S, Schmidt A, Chaiwangyen W, Markert UR. Elsevier Trophoblast Research Award Lecture: origin, evolution and future of placenta miRNAs. Placenta. 2014;35(Suppl):S39–45. doi: 10.1016/j.placenta.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Hromadnikova I, Kotlabova K, Ondrackova M, Pirkova P, Kestlerova A, Novotna V, Hympanova L, Krofta L. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 2015;34(6):437–57. doi: 10.1089/dna.2014.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hromadnikova I, Kotlabova K, Ivankova K, Krofta L. Expression profile of C19MC microRNAs in placental tissue of patients with preterm prelabor rupture of membranes and spontaneous preterm birth. Mol Med Rep. 2017;16(4):3849–3862. doi: 10.3892/mmr.2017.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu G, Brkic J, Hayder H, Peng C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int J Mol Sci. 2013;14(3):5519–44. doi: 10.3390/ijms14035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knofler M, Sadovsky Y. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology. 2014;155(12):4975–85. doi: 10.1210/en.2014-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 26.Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, Schneider U, Herrmann J, Gruhn B, Markert UR. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725–34. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18(8):417–24. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 31.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, Ohkuchi A, Matsubara S, Takeshita T. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81(4):717–29. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 32.Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol Endocrinol Metab. 2013;304(8):E836–43. doi: 10.1152/ajpendo.00660.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brameier M. Genome-wide comparative analysis of microRNAs in three non-human primates. BMC Res Notes. 2010;3:64. doi: 10.1186/1756-0500-3-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of individual miRNAs within placenta-associated clusters where expression is represented by the averaged normalized read count (TPM) for each expressed miRNA within a cluster in first trimester and term placentas. The expression levels for specific miRNAs are connected by a red line if the expression decreases and green line if the expression increases from first trimester to term. Significantly different (P < 0.0001) expression levels between first trimester and term placentas for the C14MC, C19MC and miR-371-3 cluster are denoted by an asterisk.