Abstract
Spatiotemporal control over the release or activation of biomacromolecules such as siRNA remains a significant challenge. Light-controlled release has gained popularity in recent years; however, a major limitation is that most photoactivable compounds/systems respond only to UV irradiation, but not near-infrared (NIR) light that offers a deeper tissue penetration depth and better biocompatibility. This paper reports a simple NIR-to-UV upconversion nanoparticle (UCNP)-based siRNA nanocarrier for NIR-controlled gene silencing. siRNA is complexed onto a NaYF4:Yb/Tm/Er UCNP through an azobenzene (Azo)–cyclodextrin (CD) host–guest interaction. The UV emission generated by the NIR-activated UCNP effectively triggers the trans-to-cis photoisomerization of azobenzene, thus leading to the release of siRNA due to unmatched host – guest pairs. The UCNP-siRNA complexes are also functionalized with PEG (i.e., UCNP-(CD/Azo)-siRNA/PEG NPs), targeting ligands (i.e., EGFR-specific GE11 peptide), acid-activatable cell-penetrating peptides (i.e., TH peptide), and imaging probes (i.e., Cy5 fluorophore). The UCNP-(CD/Azo)-siRNA/PEG NPs with both GE11 and TH peptides display a high level of cellular uptake and an excellent endosomal/lysosomal escape capability. More importantly, NIR-controlled spatiotemporal knockdown of GFP expression is successfully achieved in both a 2D monolayer cell model and a 3D multicellular tumor spheroid model. Thus, this simple and versatile nanoplatform has great potential for the selective activation or release of various biomacromolecules.
Keywords: Spatiotemporally controlled gene silencing, NIR-controlled release, Upconversion nanoparticle, Host–guest interaction
1. Introduction
Controlled activation or release of biomolecules is crucial in various biological applications [1]. Ample means have been investigated, and light appears to be an ideal trigger for controlling the function of bio-molecules spatiotemporally [2–5]. Light-induced release of payloads can be achieved through a photocaging strategy. It involves caging the molecules of interest by a photo-cleavable molecule or bond, which can then be cleaved by light irradiation and thus release the caged molecules to function [6–8]. Various molecules, such as proteins, peptides, nucleic acids, amino acids, and drugs, have been photocaged and delivered to the cells/animals followed by targeted photolysis at the area of interest, thus enabling the activation of these molecules with a high spatiotemporal resolution [6–11].
The azobenzene molecule is one of the most widely studied photoswitchable agents [12]. Trans-azobenzene has a strong host–guest interaction with cyclodextrin [12,13]. Upon UV light irradiation, azo-benzene molecules are isomerized from their thermodynamically stable trans-state into their metastable cis-state, thereby inhibiting their host–guest interactions with cyclodextrin due to changes in polarity and steric hindrance [14,15]. This unique characteristic provides photoswitchable control over the azobenzene–cyclodextrin host–guest complex, which has been explored for a broad range of applications, including light-responsive hydrogels [16,17], micelles [11], vesicles [18], and other drug delivery vehicles [19]. However, similar to most photocaging systems, the trans-to-cis photoisomerization of azobenzene requires UV light activation, which is harmful to biological systems [13]. In addition, UV light also has a short tissue penetration depth, thereby limiting its in vivo and clinical use [1]. In contrast, near-infrared (NIR) light is capable of deeper tissue penetration than visible or UV light. It is also safer and is expected to cause minimal damage to biological systems [20,21]. To bridge this gap, a nano-transducer capable of converting NIR light to UV light is highly desirable. Although converting high-energy UV light to visible or NIR light can be achieved by various types of materials [22,23], the opposite process of converting low-energy NIR light to visible or UV light is restricted to a few rare earth material systems that show a unique upconversion property [24,25]. Lanthanide-based upconversion nanoparticles (UCNPs) have been extensively studied for various biomedical applications [24,25]. Previous studies have demonstrated that lanthanide-based UCNPs have low toxicity and are considered to be safe for long-term usage in vivo, thus providing a safe approach for use in biological applications [26,27].
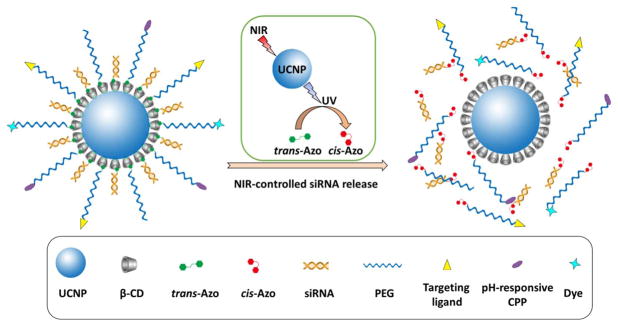
The use of siRNA to silence or interrupt the activity of specific genes by downregulating the expression of their encoding proteins has proven to be a powerful strategy in manipulating genetic functions and cellular phenotypes [28,29]. In this study, we developed a simple NIR-controlled siRNA delivery nanocarrier for the spatiotemporal activation of siRNA in living cells. siRNAs tagged with azobenzene (Azo-siRNAs) were complexed onto the NaYF4:Yb/Tm/Er UCNPs functionalized with β-cyclodextrin (CD) through a host–guest interaction. The NIR-activated UCNP core can emit UV light [30], which can effectively photoisomerize azobenzene from the trans- to cis-state, thus releasing siRNAs due to the unmatched host–guest pairs (Fig. 1(A)). This on-demand NIR-controlled siRNA release could enable precise and spatiotemporal gene silencing. The UCNP-(CD/Azo)-siRNA complex was also functionalized with PEG (i.e., UCNP-(CD/Azo)-siRNA/PEG NPs), providing NPs with good water dispersity, reduced opsonization, and convenient surface modifications (i.e., targeting ligands and imaging probes). In a proof-of-concept study, the NIR-controlled spatiotemporal activation of the UCNP-(CD/Azo)-siRNA/PEG nanoplatform was investigated in EGFR-overexpressing triple-negative breast cancer (TNBC) cells. The EGFR-specific GE11 peptide was conjugated on the surface of the NPs as an active-targeting ligand [31]. In light of the slightly acidic tumor microenvironment, an acid-activated cell-penetrating peptide (CPP), TH peptide (amino acid sequence: AGYLLGHINLHHLAHL(Aib) HHIL) [32], was also decorated onto the surface of the NPs to further enhance the cellular uptake. NPs with both GE11 and TH (i.e., GE11+/TH+ NPs) exhibited a high level of cellular uptake and an excellent endosomal/lysosomal escape capability. More importantly, using siRNA against GFP in GFP-expressing TNBC cells, specific GFP gene silencing was observed in cells exposed to NIR irradiation. Overall, this simple and versatile UCNP-based siRNA nanocarrier system can be used to spatiotemporally control gene regulation for diverse applications. This nanoplatform can also potentially be used to spatiotemporally deliver various types of payloads.
Fig. 1.
An illustration of NIR-triggered trans-to-cis photoisomerization of azobenzene, which subsequently leads to siRNA release from the UCNP-(CD/Azo)-siRNA/PEG NPs.
2. Materials and methods
2.1. Materials
CF3CO2Na, Y(CF3CO2)3, Yb(CF3CO2)3, Tm(CF3CO2)3, and Er (CF3CO2)3 were purchased from Rare Earth Products, Inc. (Beverly, MA). Carboxymethyl-β-cyclodextrin, dimethyl sulfoxide (DMSO), trans-azobenzene-tagged siRNA against GFP (Azo-siRNA), and trans-Azo-siRNA-Cy5.5 were obtained from Sigma-Aldrich (St. Louis, MO). Cy5 was purchased from Lumiprobe Corporation (Hallandale Beach, FL). Maleimide–PEG–NH2 (i.e., Mal-PEG-NH2, Mn = 5 kDa) was obtained from JenKem Technology (Allen, TX, USA). GE11 peptide (amino acid sequence: YHWYGYTPQNVIGGGGC) was purchased from Tufts University Core Facility (Boston, MA). Other reagents were purchased from Thermo Fisher Scientific (Fitchburg, WI).
2.2. Synthesis of β-CD-functionalized NaYF4:Yb3 +/Er3 +/Tm3 + UCNPs (CD-UCNPs)
The amine-functionalized UCNPs (NH2-UCNPs) were first prepared following the same protocol using a thermal decomposition method as published previously [30]. CD-UCNPs were then synthesized through amidization between NH2-UCNP and carboxymethyl-β-cyclodextrin. Briefly, carboxymethyl-β-cyclodextrin (85.3 mg), 1-ethyl-3-(3-di-methylaminopropyl)-carbodiimide (EDC, 13.2 mg), and N-hydroxysuccinimide (NHS, 8.3 mg) were added into the NH2-UCNP solution (5 mg/mL, 4 mL). The pH of the solution was adjusted to 8.5. The reaction was performed at room temperature for 48 h. After dialysis against DI water for 48 h, CD-UCNPs were obtained by lyophilization.
2.3. Synthesis of Mal-PEG-azobenzene (i.e., Mal-PEG-Azo)
Mal-PEG-Azo was prepared by amidization between Mal-PEG-NH2 and 4-(phenylazo) benzoic acid (Fig. S2). Briefly, Mal-PEG-NH2 (20 mg), 4-(phenylazo)benzoic acid (1.81 mg), N,N′-dicyclohexylcarbodiimide (DCC, 2.02 mg), and NHS (0.94 mg) were added to di-chloromethane (3 mL). The reaction was carried out at room temperature for 24 h. After the removal of dicyclohexylurea by filtration, the reaction solution was added dropwise into cold diethyl ether to get the crude product, which was then purified through a repeated precipitation process. The final polymer, Mal-PEG-Azo, was dried under vacuum. mPEG-Azo was prepared following a similar method using mPEG-NH2 instead. The chemical structures of Mal-PEG-Azo and mPEG-Azo were confirmed by 1H NMR as shown in Fig. S2.
2.4. Synthesis of GE11 (or Cy5 or TH)-PEG-Azo
GE11-PEG-Azo was prepared by an Mal-SH reaction between Mal-PEG-Azo and GE11 peptide. Briefly, Mal-PEG-Azo (5 mg), GE11 (1.9 mg), and tris(2-carboxyethyl)phosphine (0.5 mg) were dissolved in dimethylformamide (DMF, 3 mL). The reaction was carried out at room temperature overnight and the solution was then added dropwise into cold diethyl ether to get the crude product, which was then purified through a repeated precipitation process. The final polymer was dried under vacuum. TH-PEG-Azo and Cy5-PEG-Azo were prepared following a similar method using TH peptide (2.9 mg) and Cy5-SH (0.56 mg), respectively. The chemical structures of GE11-PEG-Azo, TH-PEG-Azo, and Cy5-PEG-Azo were confirmed by 1H NMR as shown in Fig. S2.
2.5. Synthesis of UCNP-(CD/Azo)-siRNA/PEG-GE11/TH/Cy5/OCH3
GE11-PEG-Azo (0.5 mg), TH-PEG-Azo (0.5 mg), Cy5-PEG-Azo (0.25 mg), mPEG-Azo (3.75 mg), and siRNA-Azo (2 mg) were dissolved in DI water (5 mL). The resulting solution was added dropwise into the CD-UCNP aqueous solutions (2 mL, 1 mg/mL) for 2 h. The mixture was stirred for another 12 h at 4 °C and then dialyzed against DI water at 4 °C for 48 h. The final product was freeze-dried in a lyophilizer (Labconco, Kansas City, MO) at a temperature of −80 °C under vacuum for two days. Samples were then stored at −20 °C.
2.6. Characterizations
1H NMR spectra of all polymer products were recorded on an AV400 NMR spectrometer in CDCl3. The morphologies of the NPs were studied by transmission electron microscopy (TEM, FEI Tecnai G2 F30 TWIN 300 KV, E.A. Fischione Instruments, Inc.) and dynamic light scattering (DLS, ZetaSizer Nano ZS90, Malvern Instruments). FTIR spectra were recorded on a Bruker Tensor 27 FT-IR spectrometer. The luminescence spectrum of the UCNPs was acquired on a Nanolog FL3-2iHR spectrofluorometer (HORIBA Jobin Yvon, Inc., USA). The siRNA loading level was determined by a NanoDrop™ One/OneC Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, Fitchburg, WI).
2.7. NIR-controlled in vitro siRNA release
In vitro siRNA release studies were performed in a PBS solution at 37 °C. A UCNP-(CD/Azo)-siRNA/PEG NP solution (1 mg/mL, 1 mL) was enclosed in a dialysis tube (molecular weight cut-off 30 kDa), which was placed in 10 mL of PBS. The samples were irradiated with the 980 nm laser at an output power of 0.75 W/cm2 for 10 or 20 min. At certain time points, 3 mL of media were collected and replaced by an equal amount of fresh media. The amount of siRNA in the collected media was quantified by a NanoDrop™ One/OneC Microvolume UV–vis Spectrophotometer.
2.8. Cellular uptake studies in a 2D monolayer cell model
MDA-MB-468 cells were cultured under standard conditions (37 °C/5% CO2/95% humidity) in DMEM medium (Gibco, Gaithersburg, MD) containing 10% fetal bovine serum (Gibco, Gaithersburg, MD) and 1% penicillin/streptomycin (Thermo Fisher, Fitchburg, WI). The cellular uptake of NPs was studied by fluorescence microscopy and flow cytometry. MDA-MB-468 cells were seeded into a 24-well plate (1 × 105 cells/well) and cultured for 24 h before use. Cells were treated with pure medium (control), GE11−/TH+ NPs (i.e., formed by UCNP-(CD/Azo)-siRNA/PEG-GE11/Cy5/OCH3), and GE11+/TH+ NPs (i.e., formed by UCNP-(CD/Azo)-siRNA/PEG-GE11/TH/Cy5/OCH3) at two different pH conditions (i.e., pH = 6.7 and pH = 7.4). After 2 h incubation, cells were washed with PBS, fixed with 4% of paraformaldehyde for 15 min, stained with Hoechst 33342 (Thermo Fisher, Fitchburg, WI) for 15 min, and then imaged under a fluorescence microscope (Nikon, Japan). For flow cytometry studies, cells were treated the same way as described for fluorescence microscopy studies. After 2 h incubation, cells were washed with PBS three times, harvested with 0.25% EDTA-trypsin (Gibco, Gaithersburg, MD), and fixed with 4% of paraformaldehyde for 15 min. Cells were then spun down (130 g for 5 min) and resuspended in 1 mL PBS for testing. Data were acquired with an Attune NxT flow cytometer (Thermo Fisher, Fitchburg, WI) and analyzed with FlowJo 7.6 (Ashland, OR).
2.9. Intracellular trafficking of NPs
MDA-MB-468 cells (5 × 104 cells/well) were seeded in 8-well chamber Nunc™ Lab-Tek™ II chamber slides (Thermo Fisher, Fitchburg, WI) and cultured for 24 h before use. Cells were treated with GE11+/TH+ NPs. After certain time points (i.e., 15, 30, or 120 min), cells were washed with PBS, stained with LysoTracker™ Green and Hoechst 33342 according to the manufacturer’s protocol, and then imaged under a confocal laser scanning microscope (CLSM, Nikon Eclipse Ti inverted microscope equipped with Nikon A1R confocal diode lasers, Japan).
2.10. NIR-controlled gene silencing tests in a 2D monolayer cell model
Gene silencing efficiencies were studied by fluorescence microscopy. In a proof-of-concept study, GFP-siRNA and GFP-expressing MDA-MB-468 (i.e., GFP-MDA-MB-468) cells were used. GFP-MDA-MB-468 cells were seeded in a 96-well plate and cultured for 24 h before use. Cells were treated with various formulations as summarized in Table 1 at siRNA concentration of 40 nM. After 4 h incubation, cells were irradiated with a 980 nm NIR laser at a power density of 0.75 W/cm2 for 10 min. After incubation for a total of 24 h, cells were washed with PBS, fixed with 4% of paraformaldehyde for 15 min, and then imaged under a fluorescence microscope.
Table 1.
Summary of various formulations for gene silencing and cell viability studies.
| With or without NIR laser | |
|---|---|
| 1 | Pure medium pH 7.4 (control pH 7.4) |
| 2 | GE11−/TH+ NPs pH 7.4 |
| 3 | GE11+/TH+ NPs pH 7.4 |
| 4 | RNAiMAX pH 7.4 |
| 5 | Pure medium pH 6.7 (control pH 6.7) |
| 6 | GE11−/TH+ NPs pH 6.7 |
| 7 | GE11+/TH+ NPs pH 6.7 |
| 8 | RNAiMAX pH 6.7 |
To further verify the NIR-controlled gene silencing ability of the UCNP-(CD/Azo)-siRNA/PEG NPs, cells were first treated with GE11+/TH+ NPs for 4 h. Half of the cells in one well of a 96-well plate were irradiated with NIR light, while the other half were covered by sterilized aluminum foil to avoid NIR-light exposure. After another 24 h of incubation, cells were imaged under a fluorescence microscope.
2.11. Cell viability tests in a 2D monolayer cell model
MDA-MB-468 cells were cultured in a 96-well plate and were treated the same way as described in the gene silencing studies. After 24 h of incubation, the cell viabilities (percentage of the control group without 980 nm laser irradiation) were determined using a standard MTT assay and data were collected using a micro-plate reader (GloMax®-Multi Detection System, Promega, Madison, WI).
2.12. NP penetration studies in a 3D multicellular tumor spheroid (MCTS) culture model
GFP-MDA-MB-468 MCTS 3D culture model was formed using a hanging drop method [33,34]. Briefly, GFP-MDA-MB-468 cells were suspended in a growth medium (50 cell/μL) containing 0.24% (v/v) methylcellulose. The drops of the cell suspension (20 μL/drop) were deposited onto a 96-well plate (low attachment, Corning, NY), which was then inverted onto a lid deposited with PBS drops (10 μL/drop). After incubation for 5 days, GFP-MDA-MB-468 MCTSs with a size of ~500 μm were formed. MDA-MB-468 MCTS was treated with GE11−/TH+ NPs and GE11+/TH+ NPs at a pH of 7.4 and a pH of 6.7. After certain time points (i.e., 6 and 24 h), MCTSs were gently rinsed with PBS and stained with Hoechst 33342. Samples were then analyzed using a CLSM.
2.13. NIR-controlled gene silencing tests in a 3D MCTS culture model
Gene silencing efficiencies of the NPs were also studied in a GFP-MDA-MB-468 MCTS model. Cells were treated with various formulations as summarized in Table 1 at an siRNA concentration of 40 nM. After 24 h of incubation, MCTSs were irradiated with a 980 nm laser at a power density of 0.75 W/cm2 for 10 min. After another 24 h of incubation, MCTSs were gently washed with PBS, stained with Hoechst 33342, and then imaged under a CLSM.
2.14. Cell viability tests in a 3D MCTS culture model
MDA-MB-468 MCTSs were treated with various formulations as summarized in Table 1. After 24 h of incubation, MCTSs were irradiated with a 980 nm laser at a power density of 0.75 W/cm2 for 10 min. After another 24 h of incubation, the cell viability in MCTSs was determined by a standard MTT assay. Data were acquired by a GloMax®-Multi Detection System micro-plate reader.
3. Results and discussion
3.1. Synthesis and characterization of UCNP-(CD/Azo)-siRNA/PEG-GE11/TH/Cy5/OCH3 NPs
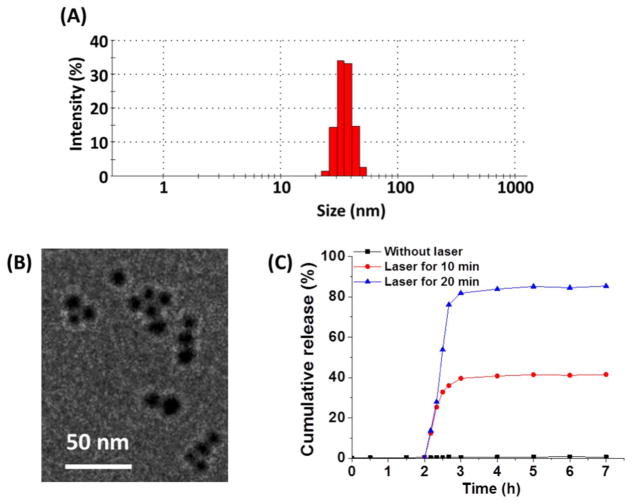
Amine-functionalized NaYF4:Yb/Tm/Er UCNPs (NH2-UCNPs) were prepared using a thermal decomposition method following the same protocol we previously reported [30], which was then converted to β-CD functionalized UCNPs (CD-UCNPs) through amidization. The average size of the CD-UCNPs determined by transmission electron microscopy (TEM) was 15 nm in diameter (Fig. S1 (A)). siRNAs conjugated with azobenzene (Azo-siRNA) were then attached onto the surface of the UCNPs through a CD-Azo host–guest interaction. Fig. S1 (B) shows the luminescence emissions of the CD-UCNPs upon 980 nm excitation. It is noted that the emission around 330–360 nm (UV light) was suitable to photoisomerize azobenzene from the trans- to cis-state, thus releasing the siRNAs (as detailed later). The NPs were also functionalized with PEG-Azo via the same CD-Azo host–guest interaction, thus providing the NPs with good water dispersity as well as reduced opsonization. Moreover, cell/tissue targeting ligands (e.g., the EGFR-specific GE11 peptide in this study; 10% of PEG arms), cell-penetrating peptides (e.g., TH peptide; 10% of PEG arms), and imaging probes (e.g., Cy5 fluorophore; 5% of PEG arms) were conjugated to the distal ends of the PEG arms. The average hydrodynamic diameter of the final UCNP-(CD/Azo)-siRNA/PEG NPs was 38 nm (PDI = 0.21; Fig. 2(A)). The stability of the UCNP-(CD/Azo)-siRNA/PEG NPs were tested by DLS analyses. The sizes of NPs did not change notably after two days in the cell culture media (Fig. S3), demonstrating the excellent stability of the UCNP-(CD/Azo)-siRNA/PEG NPs. The TEM image of the UCNP-(CD/Azo)-siRNA/PEG NPs is shown in Fig. 2(B). The siRNA loading level was 17.3 wt% for GE11+/TH+ NPs and 17.6 wt% for GE11−/TH+ NPs.
Fig. 2.
(A) DLS analyses and (B) TEM image of the UCNP-(CD/Azo)-siRNA/PEG NPs. (C) In vitro siRNA release.
3.2. NIR-controlled siRNA release from UCNP-(CD/Azo)-siRNA/PEG NPs
Due to the strong CD-Azo host–guest interaction, a close proximity between UCNPs and light-responsive guests (Azo-siRNA) is expected [13], leading to higher efficiencies for the photoswitching of azo-benzene under NIR light, and thereby triggering effective siRNA release. To validate this hypothesis, in vitro siRNA release profiles were studied. Azo-siRNA tagged with Cy5.5 (Azo-siRNA-Cy5.5) was used for this experiment. As shown in Fig. 2(C), without NIR light, only a minimal if not zero amount of siRNA (< 0.5%) was released from the UCNP-(CD/Azo)-siRNA/PEG NPs, demonstrating the strong CD-Azo host–guest interaction. In stark contrast, the release of siRNA was clearly observed under NIR light. Moreover, the amount of siRNA being released can be readily controlled by the NIR irradiation time. About 41% and 85% of siRNA were released within 10 and 20 min of NIR irradiation, respectively. This result strongly supports that NIR light is required and essential to controllably release the siRNAs; thus, a spatiotemporal control of gene silencing can be achieved by NIR light. It is worth mentioning that other biomacromolecules (e.g., peptides, microRNAs, ssDNAs, and proteins) can also be conjugated with azo-benzene through either amidization or esterification. Hence, NIR-controlled spatiotemporal release/activation of these biomacromolecules may also be realized using this nanoplatform.
3.3. Cellular uptake behavior in a 2D monolayer cell model
Before the gene silencing efficiency of the nanoplatform was investigated, the in vitro cellular uptake behavior of the NPs was examined. MDA-MB-468 TNBC cells overexpressing EGFR were used in this study. GE11 peptide, a ligand that can efficiently bind to EGFR, was conjugated onto the NPs to achieve their active tumor-targeting capability. Moreover, a cell penetrating peptide, TH [32], which can be activated by acid, was also conjugated onto the NPs to further enhance the cellular uptake of the NPs. Specifically, TH is inactivated at a pH of 7.4 prior to reaching the tumor sites to minimize off-target effects. It can then be activated at an acidic tumor microenvironment (i.e., pH 6.5–7.0) and subsequently promote NP internalization [35,36].
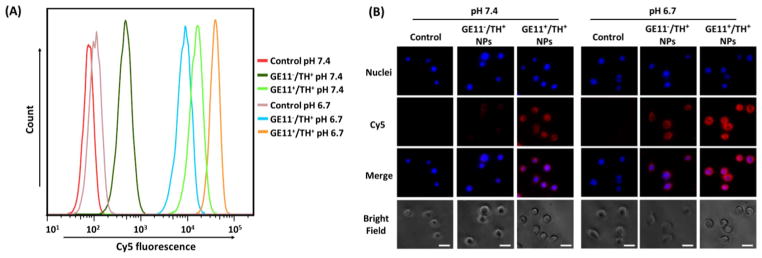
The cellular uptake study was first tested in a 2D monolayer cell model. Cells were treated with GE11−/TH+ and GE11+/TH+ NPs in a normal physiological pH environment (i.e., pH = 7.4) and an acidic pH environment (i.e., pH = 6.7, mimicking a tumor microenvironment). Cy5 fluorophore was conjugated onto the NPs to allow for NP tracking. Fig. 3(A) shows that the Cy5 fluorescence intensities in the cells treated with TH-conjugated NPs (i.e., both GE11−/TH+ and GE11+/TH+ NPs), and the fluorescence at a pH of 6.7 was remarkably higher than at a pH of 7.4, thus strongly indicating that TH peptide enhanced NP internalization in the acidic tumor microenvironment. This is consistent with the fluorescence microscope images shown in Fig. 3(B). Moreover, for both pH conditions, the GE11-conjugated (i.e., GE11+/TH+) NPs displayed a much higher cellular uptake compared to the GE11-lacking (i.e., GE11−/TH+) ones indicated by both flow cytometry results and fluorescence images, demonstrating the active targeting effect of the GE11 peptide. Taken together, a combination of GE11 and TH peptides significantly promoted the internalization of NPs in the tumor micro-environment-mimicking condition.
Fig. 3.
In vitro cellular uptake studies using (A) flow cytometry and (B) fluorescence microscopy in MDA-MB-468 TNBC cells. Cells were treated with pure medium (control), GE11−/ TH+ NPs, and GE11+/TH+ NPs at two different pH conditions (i.e., pH = 6.7 and pH = 7.4) for 2 h. Scale bar: 60 μm.
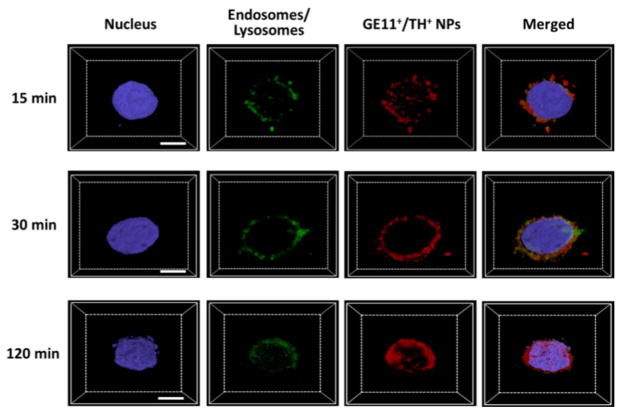
Other than facilitating the internalization of NPs, TH cell penetrating peptide is also considered an endosomolytic peptide for enhancing the endosomal escape of NPs [32]. This is a desirable feature for this application because if the NPs cannot escape from the endosomes efficiently, siRNA will be degraded in the endosomes or lysosomes, and hence hinder the siRNA from functioning in the cytosol [37]. As shown in Fig. 4, after 15 min incubation, NPs co-localized well with endosomes/lysosomes, thus indicating that NPs were trapped in the endosomes/lysosomes. After 30 min incubation, Cy5 fluorescence from NPs partially merged with green fluorescence, demonstrating that a considerable portion of NPs already escaped from the endosomes/lysosomes. At 2 h post-incubation, the extent of co-localization between the red fluorescence (Cy5-tagged NPs) and green fluorescence (i.e., endosomes/lysosomes) significantly decreased. Moreover, the Cy5 fluorescence in the cytoplasm exhibited a more homogeneous distribution pattern, suggesting that most of the NPs escaped from the endosomes/lysosomes.
Fig. 4.
Z-stack CLSM images for the assessment of the endosomal/lysosomal escape of the UCNP-(CD/Azo)-siRNA/ PEG NPs in MDA-MB-468 cells. Scale bar: 10 μm.
3.4. NIR-controlled gene silencing tests in a 2D monolayer cell model
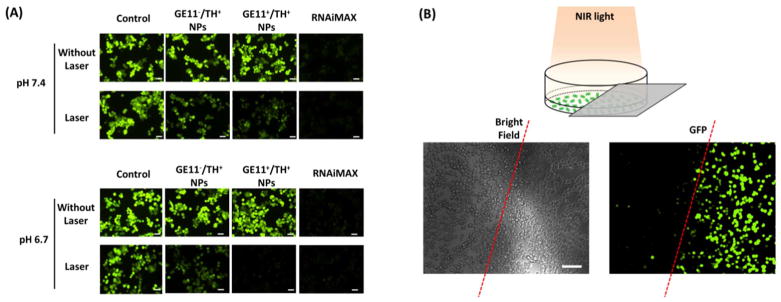
Given that fact that NIR light (980 nm) successfully triggered a rapid release of siRNA from UCNP-(CD/Azo)-PEG NPs and NPs were able to enter the cell cytosol rapidly, the NIR-controlled gene silencing efficiency of the NPs was investigated. In a proof-of-concept study, GFP-siRNA and GFP-MDA-MB-468 cells were used. Cells treated with RNAiMAX were considered a positive control group with a significant reduction of green fluorescence intensities (Fig. 5(A)). However, for cells treated with either GE11−/TH+ or GE11+/TH+ NPs without NIR laser application, no gene silencing effect was observed according to GFP fluorescence intensity. This is in good agreement with the in vitro siRNA release profile showing minimal/no siRNA release from NPs in the absence of NIR light. In stark contrast, NIR laser irradiation (10 min) induced a significant reduction of GFP fluorescence intensity in all NP-treated groups, again demonstrating that NIR light could effectively trigger the release of siRNA to function. More importantly, the GE11+/TH+ NPs at a pH of 6.7 exhibited the best gene silencing efficiency due to their strong cellular uptake capability as discussed above. Notably, longer irradiation time (i.e., 20 min) induced a significantly greater reduction of GFP than shorter irradiation time (i.e., 10 min) (Fig. S4), which is consistent with the siRNA release profile (Fig. 2(C)). To further demonstrate that this technique can control gene silencing with a high spatiotemporal resolution, in the cells incubated with GE11+/TH+ NPs, instead of irradiating all of the cells in a well of a 96-well plate with the NIR laser, the well was half-covered with aluminum foil, meaning only half of the cells in the well were irradiated with the NIR light (Fig. 5(B)). A clear difference was observed between these two areas with the GFP gene greatly silenced in the cells exposed to the NIR light. All of these results indicate that this unique siRNA nanoplatform can precisely regulate gene silencing by selective NIR irradiation. In addition, cell viability assays (Fig. S5) revealed no apparent cytotoxicity associated with these NPs, while the RNAiMAX, a commercially available product, induced a significant amount of cell death (> 20%).
Fig. 5.
(A) NIR-controlled gene silencing tests in a 2D monolayer cell model. siRNA against GFP were used and tested in GFP-expressing MDA-MA-468 TNBC cells. In the laser treatment groups, cells were first incubated with UCNP-(CD/Azo)-siRNA/PEG NPs for 4 h, followed by NIR irradiation for 10 min. Images were taken after 24 h incubation. Scale bar: 100 μm. (B) Spatiotemporally controlled gene silencing under NIR light. Cells half-covered by aluminum foil were irradiated with an NIR laser. Fluorescence images after 24 h show the region-specific down-regulation of GFP in cells. Scale bar: 100 μm.
3.5. NP penetration study in a 3D multicellular tumor spheroid (MCTS) model
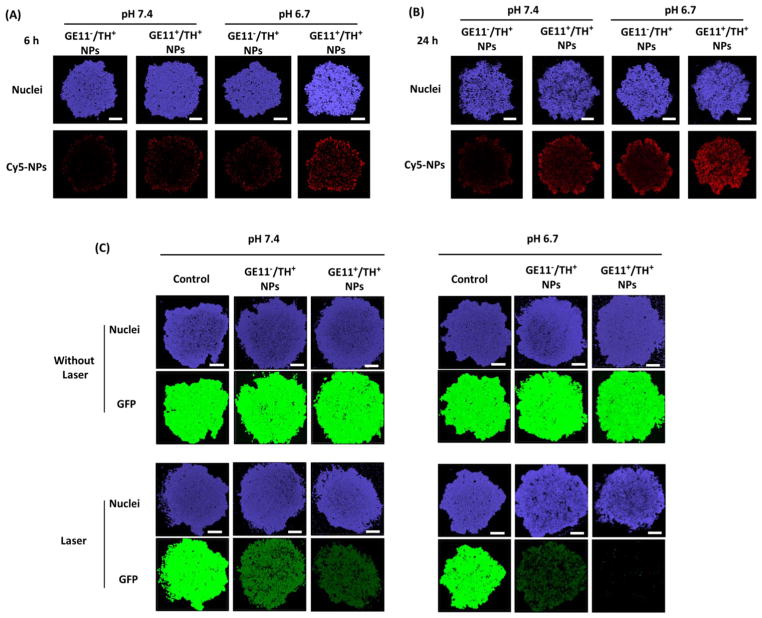
The utility of UCNP-(CD/Azo)-PEG NPs were further investigated in a 3D MCTS model that is considered to be a bridge between 2D monolayer cell models and in vivo models [38,39]. The NP penetration in the MCTS was first examined. MCTSs were treated with GE11−/TH+ and GE11+/TH+ NPs at a pH of 7.4 and a pH of 6.7. As shown in Fig. 6(A), 6 h post-incubation, NPs exhibited limited tumor penetration at a pH of 7.4. Meanwhile, GE11+/TH+ NPs penetrated deeper compared to GE11−/TH+ NPs, thus implying that the targeting ligand (i.e., GE11 peptide) could facilitate NP penetration in tumors [40–42]. In a tumor-mimicking acidic environment (i.e., pH = 6.7), the TH peptide was activated and thus led to enhanced NP penetration depths for both types of NPs [35]. As noted, the GE11+/TH+ NPs at a pH of 6.7 showed the best penetration capability. After 24 h of incubation, a more homogenous NP distribution was observed in the MCTS treated with GE11+/TH+ NPs at a pH of 6.7 (Fig. 6(B)).
Fig. 6.
NP penetration studies in a 3D MCTS model. GFP-MDA-MB-468 MCTSs were treated with Cy5-tagged GE11−/TH+ and GE11+/TH+ NPs for (A) 6 h and (B) 24 h. Scale bar: 100 μm. (C) NIR-controlled gene silencing tests in a 3D MCTS model.
Gene silencing efficiency was then tested in a MCTS 3D model. Consistent with the penetration data, the GE11+/TH+ NPs at a pH of 6.7 exhibited the best gene silencing efficiency under NIR irradiation for 10 min (Fig. 6(C)). Moreover, NPs did not induce any toxicities (Fig. S6), thus warranting further study for in vivo applications.
4. Conclusion
A simple and versatile UCNP-based siRNA nanoplatform was developed for spatiotemporal gene knockdown. siRNA was successfully complexed onto NIR-to-UV UCNPs through an Azo–CD host–guest interaction to from UCNP-(Azo/CD)-siRNA complexes that were further functionalized with PEG. The NIR-activated UCNPs precisely controlled the release of siRNAs from the UCNP-(Azo/CD)-PEG NPs due to the trans-to-cis photoisomerization of azobenzene, thereby inhibiting the host–guest interaction. GE11 and TH peptides, which were conjugated onto the surface of the NPs, facilitated the cellular uptake and endosomal/lysosomal escape of the NPs. Moreover, a spatiotemporal gene knockdown controlled by NIR light was observed in both a 2D monolayer cell model and a 3D MCTS model. We anticipate that this simple nanoplatform can also control the release/activation of various bio-macromolecules (e.g., peptides, microRNAs, ssDNAs, proteins, etc.).
Supplementary Material
Acknowledgments
This project was financially supported by NIH grants (K25CA166178 and R21CA196653).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2017.12.028.
References
- 1.Jayakumar MKG, Idris NM, Zhang Y. Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers. Proc Natl Acad Sci. 2012;109:8483–8488. doi: 10.1073/pnas.1114551109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat Genet. 2001;28:317. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 4.Fomina N, Sankaranarayanan J, Almutairi A. Photochemical mechanisms of light-triggered release from nanocarriers. Adv Drug Deliv Rev. 2012;64:1005–1020. doi: 10.1016/j.addr.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Li J, Wu D, Qiu Z, Zhang Y. Chemistry and biological applications of photo-labile organic molecules. Chem Soc Rev. 2010;39:464–473. doi: 10.1039/b901255a. [DOI] [PubMed] [Google Scholar]
- 6.Young DD, Deiters A. Photochemical control of biological processes. Org Biomol Chem. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 7.Shah S, Rangarajan S, Friedman SH. Light-activated RNA interference. Angew Chem. 2005;117:1352–1356. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 8.Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. Targeting expression with light using caged DNA. J Biol Chem. 1999;274:20895–20900. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 9.Peng K, Tomatsu I, Kros A. Light controlled protein release from a supramolecular hydrogel. Chem Commun. 2010;46:4094–4096. doi: 10.1039/c002565h. [DOI] [PubMed] [Google Scholar]
- 10.Braun GB, Pallaoro A, Wu G, Missirlis D, Zasadzinski JA, Tirrell M, Reich NO. Laser-activated gene silencing via gold nanoshell–siRNA conjugates. ACS Nano. 2009;3:2007–2015. doi: 10.1021/nn900469q. [DOI] [PubMed] [Google Scholar]
- 11.Nalluri SKM, Voskuhl J, Bultema JB, Boekema EJ, Ravoo BJ. Light-responsive capture and release of DNA in a ternary supramolecular complex. Angew Chem Int Ed. 2011;50:9747–9751. doi: 10.1002/anie.201103707. [DOI] [PubMed] [Google Scholar]
- 12.Bandara HD, Burdette SC. Photoisomerization in different classes of azobenzene. Chem Soc Rev. 2012;41:1809–1825. doi: 10.1039/c1cs15179g. [DOI] [PubMed] [Google Scholar]
- 13.Möller N, Hellwig T, Stricker L, Engel S, Fallnich C, Ravoo BJ. Near-infrared photoswitching of cyclodextrin–guest complexes using lanthanide-doped LiYF 4 upconversion nanoparticles. Chem Commun. 2017;53:240–243. doi: 10.1039/c6cc08321h. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Ma N, Wang Z, Zhang X. Photocontrolled reversible supramolecular assemblies of an azobenzene-containing surfactant with α-cyclodextrin. Angew Chem Int Ed. 2007;46:2823–2826. doi: 10.1002/anie.200604982. [DOI] [PubMed] [Google Scholar]
- 15.Inoue Y, Kuad P, Okumura Y, Takashima Y, Yamaguchi H, Harada A. Thermal and photochemical switching of conformation of poly (ethylene glycol)-substituted cyclodextrin with an azobenzene group at the chain end. J Am Chem Soc. 2007;129:6396–6397. doi: 10.1021/ja071717q. [DOI] [PubMed] [Google Scholar]
- 16.Zhao YL, Stoddart JF. Azobenzene-based light-responsive hydrogel system. Langmuir. 2009;25:8442–8446. doi: 10.1021/la804316u. [DOI] [PubMed] [Google Scholar]
- 17.Liao X, Chen G, Liu X, Chen W, Chen F, Jiang M. Photoresponsive pseudopo-lyrotaxane hydrogels based on competition of host–guest interactions. Angew Chem. 2010;122:4511–4515. doi: 10.1002/anie.201000141. [DOI] [PubMed] [Google Scholar]
- 18.Nalluri SKM, Ravoo BJ. Light-responsive molecular recognition and adhesion of vesicles. Angew Chem Int Ed. 2010;49:5371–5374. doi: 10.1002/anie.201001442. [DOI] [PubMed] [Google Scholar]
- 19.Samanta A, Stuart MC, Ravoo BJ. Photoresponsive capture and release of lectins in multilamellar complexes. J Am Chem Soc. 2012;134:19909–19914. doi: 10.1021/ja3101837. [DOI] [PubMed] [Google Scholar]
- 20.Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 21.Yan B, Boyer JC, Branda NR, Zhao Y. Near-infrared light-triggered dissociation of block copolymer micelles using upconverting nanoparticles. J Am Chem Soc. 2011;133:19714–19717. doi: 10.1021/ja209793b. [DOI] [PubMed] [Google Scholar]
- 22.Michalet X, Pinaud F, Bentolila L, Tsay J, Doose S, Li J, Sundaresan G, Wu A, Gambhir S, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang ST, Cao L, Luo PG, Lu F, Wang X, Wang H, Meziani MJ, Liu Y, Qi G, Sun YP. Carbon dots for optical imaging in vivo. J Am Chem Soc. 2009;131:11308–11309. doi: 10.1021/ja904843x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Banerjee D, Liu Y, Chen X, Liu X. Upconversion nanoparticles in biological labeling, imaging, and therapy. Analyst. 2010;135:1839–1854. doi: 10.1039/c0an00144a. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Qiu H, Prasad PN, Chen X. Upconversion nanoparticles: design, na-nochemistry, and applications in theranostics. Chem Rev. 2014;114:5161–5214. doi: 10.1021/cr400425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jalil RA, Zhang Y. Biocompatibility of silica coated NaYF 4 upconversion fluorescent nanocrystals. Biomaterials. 2008;29:4122–4128. doi: 10.1016/j.biomaterials.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Xiong L, Yang T, Yang Y, Xu C, Li F. Long-term in vivo biodistribution imaging and toxicity of polyacrylic acid-coated upconversion nanophosphors. Biomaterials. 2010;31:7078–7085. doi: 10.1016/j.biomaterials.2010.05.065. [DOI] [PubMed] [Google Scholar]
- 28.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Wang Y, Xie R, Gong S. Tumor-targeted pH/redox dual-sensitive un-imolecular nanoparticles for efficient siRNA delivery. J Control Release. 2017;259:105–114. doi: 10.1016/j.jconrel.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G, Jaskula-Sztul R, Esquibel CR, Lou I, Zheng Q, Dammalapati A, Harrison A, Eliceiri KW, Tang W, Chen H. Neuroendocrine tumor-targeted up-conversion nanoparticle-based micelles for simultaneous NIR-controlled combination chemotherapy and photodynamic therapy, and fluorescence imaging. Adv Funct Mater. 2017;27 doi: 10.1002/adfm.201604671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkman AM, Chen G, Wang Y, Hedman CJ, Sherer NM, Havighurst TC, Gong S, Xu W. Aminoflavone-loaded EGFR-targeted unimolecular micelle nanoparticles exhibit anti-cancer effects in triple negative breast cancer. Biomaterials. 2016;101:20–31. doi: 10.1016/j.biomaterials.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Song J, Zhang B, Liu L, Wang K, Wang R. Design of acid-activated cell penetrating peptide for delivery of active molecules into cancer cells. Bioconjug Chem. 2011;22:1410–1415. doi: 10.1021/bc200138d. [DOI] [PubMed] [Google Scholar]
- 33.Nagelkerke A, Bussink J, Sweep FCGJ, Span PN. Generation of multicellular tumor spheroids of breast cancer cells: how to go three-dimensional. Anal Biochem. 2013;437:17–19. doi: 10.1016/j.ab.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, Ma B, Wang Y, Xie R, Li C, Dou K, Gong S. CuS-based theranostic micelles for NIR-controlled combination chemotherapy and photothermal therapy and photoacoustic imaging. ACS Appl Mater Interfaces. 2017;9:41700–41711. doi: 10.1021/acsami.7b14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Tang J, Fu L, Ran R, Liu Y, Yuan M, He Q. A pH-responsive α-helical cell penetrating peptide-mediated liposomal delivery system. Biomaterials. 2013;34:7980–7993. doi: 10.1016/j.biomaterials.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Jiang T, Zhang Z, Zhang Y, Lv H, Zhou J, Li C, Hou L, Zhang Q. Dual-functional liposomes based on pH-responsive cell-penetrating peptide and hyaluronic acid for tumor-targeted anticancer drug delivery. Biomaterials. 2012;33:9246–9258. doi: 10.1016/j.biomaterials.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 37.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 38.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 39.Shin CS, Kwak B, Han B, Park K. Development of an in vitro 3D tumor model to study therapeutic efficiency of an anticancer drug. Mol Pharm. 2013;10:2167–2175. doi: 10.1021/mp300595a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao H, Xiong Y, Zhang S, Yang Z, Cao S, Jiang X. RGD and interleukin-13 peptide functionalized nanoparticles for enhanced glioblastoma cells and neo-vasculature dual targeting delivery and elevated tumor penetration. Mol Pharm. 2014;11:1042–1052. doi: 10.1021/mp400751g. [DOI] [PubMed] [Google Scholar]
- 41.Qin L, Wang CZ, Fan HJ, Zhang CJ, Zhang HW, Lv MH, Cui SD. A dual-targeting liposome conjugated with transferrin and arginine-glycine-aspartic acid peptide for glioma-targeting therapy. Oncol Lett. 2014;8:2000–2006. doi: 10.3892/ol.2014.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao H, Yang Z, Zhang S, Pang Z, Liu Q, Jiang X. Study and evaluation of mechanisms of dual targeting drug delivery system with tumor microenvironment assays compared with normal assays. Acta Biomater. 2014;10:858–867. doi: 10.1016/j.actbio.2013.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.