Abstract
Secondary bacterial coinfections following influenza virus pose a serious threat to human health. Therefore, it is of significant clinical relevance to understand the immunological causes of this increased susceptibility. Influenza-induced alterations in alveolar macrophages have been shown to be a major underlying cause of the increased susceptibility to bacterial superinfection. However, the mechanisms responsible for this remain under debate, specifically, whether alveolar macrophages are depleted in response to influenza infection or are maintained post-infection, but with disrupted phagocytic activity. The data presented here resolves this issue by showing that either mechanism can differentially occur in individual mouse strains. BALB/c mice exhibited a dramatic IFN-γ-dependent reduction in levels of alveolar macrophages following infection with influenza A, whereas alveolar macrophage levels in C57Bl/6 mice were maintained throughout the course of influenza infection, although the cells displayed an altered phenotype, namely an upregulation in CD11b expression. These strain differences were observed regardless of whether infection was performed with low or high doses of influenza virus. Furthermore, infection with either the H1N1 A/California/04/2009 (CA04) or H1N1 A/PR8/1934 (PR8) virus strain yielded similar results. Regardless of alveolar macrophage viability, both BALB/c and C57Bl/6 mice showed a high level of susceptibility to post-influenza bacterial infection. These findings resolve the apparent inconsistencies in the literature, identify mouse strain-dependent differences in the alveolar macrophage response to influenza infection, and ultimately, may facilitate translation of the mouse model to clinical application.
Keywords: Viral-bacterial synergy, immunosuppression, mouse genetic background, alveolar macrophages
Introduction
Influenza A virus is one of the leading causes of respiratory infections in the United States. Complications involving secondary infections with bacterial pathogens, such as Staphylococcus aureus and Streptococcus pneumoniae, significantly exacerbate the risk of severe disease resulting in increased rates of hospitalization and death (1). This is especially true for young, elderly or immunocompromised individuals. Furthermore, it is estimated that >95% of the deaths that occurred during the 1918 pandemic were due to post-influenza bacterial infection in the lungs (2). Similarly, roughly half of hospitalized patients during the 1957 and 2009 influenza pandemics presented with bacterial pneumonia (3, 4). Influenza-induced inflammation results in epithelial damage, enhanced exposure of bacterial binding sites, and especially, suppression of innate lung immune responses, all of which are believed to promote bacterial coinfection (5).
Influenza-bacterial synergy has been extensively studied in both mice and humans. Evidence suggests that influenza infection compromises both the host immune response and lung barrier function to promote increased susceptibility to bacterial superinfection. Alveolar macrophage (AM) dysregulation has been particularly identified as an underlying cause of this phenomenon. AMs are crucial for the elimination of bacterial and viral pathogens from the pulmonary space and restriction of this activity has been shown to worsen disease outcome (6–10). Located in the air spaces of the alveoli, AMs have direct contact with environmental stimuli and form the first line of defense against pulmonary pathogens through phagocytosis and bacterial killing. It is commonly accepted that influenza-induced suppression of AMs reduces bacterial clearance and significantly enhances susceptibility to post-influenza bacterial pneumonia. However, the underlying mechanism is controversial (8, 10). A prominent theory suggests that influenza infection promotes AM depletion, consequently resulting in an increased vulnerability to secondary infections (10–13). Conversely, multiple studies have demonstrated the sustained presence of AMs throughout influenza infection, but with reduced phagocytic function (8, 14–16). Nonetheless, the concept that AMs are depleted as a result of influenza infection remains pervasive in the literature (11–13, 17). Inconsistencies in experimental approach have hindered efforts to resolve the conflicting models. Thus, whether AM depletion or altered function is primarily responsible for enhanced mortality in influenza-pneumococcal coinfection has yet to be determined. Importantly, the opposing results presented in the literature compromise the validity of the consequent conclusions and potential translation to human application. To address these issues, we utilized a protocol that monitored AM maintenance post-influenza using the fluorescent and relatively long-lived phagocytic cell labeling dye, PKH26 (8, 10, 14). We further compared multiple experimental conditions that were not consistent between contradictory reports (8, 10), including the potential influence of mouse strain (BALB/c vs. C57BL/6), viral dose (10 vs. 100 PFU) and virus strain (PR8 vs CA04) to clarify experimental factors which could influence the response of AMs to influenza virus infection.
Our results demonstrate that AM expression in post-influenza infected mice was largely dependent on mouse genetic background. Influenza infection of BALB/c mice resulted in a dramatic decline in AM numbers; however, AMs from infected C57Bl/6 mice were not depleted, but exhibited an altered phenotype. Specifically, C57Bl/6 AMs upregulated expression of CD11b, a marker uncommon to naïve resident AMs. Regardless of AM depletion or persistence, both BALB/c and C57Bl/6 mice showed similar levels of susceptibility to bacterial superinfection. These studies resolve a major enigma in the field and provide clarification of mouse strain-related differences in the host response to influenza infection. Furthermore, our results substantiate the accuracy of previously reported conclusions. Understanding the diverse modes of influenza-induced suppression of AMs will be essential in designing future studies and therapies in mice and humans, respectively.
Materials and Methods
Mouse infection models
6–8-week-old female BALB/cAnNCrl and C57Bl/6NCrl mice were purchased from Charles River Laboratories (Wilmington, MA). BALB/cJ wild type (WT) and IFN-γ−/− mice were obtained from Jackson Laboratories (Bar Harbor, ME). All mice were maintained under specific pathogen-free conditions in the Animal Research Facility at Albany Medical College. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Albany Medical College (Protocol Number 11-04004).
H1N1 A/California/04/2009 (CA04) influenza virus was obtained from the Centers for Disease Control and Prevention (Atlanta, GA) and mouse-adapted using previously described methods (18). H1N1 A/Puerto Rico/8/34 (PR8) influenza virus was purchased from Charles River Laboratories. Stock viruses were propagated in the allantoic cavities of 10-day-old embryonated chicken eggs for 48 h at 37°C, and aliquots were stored at 80°C until used. Titers of virus were determined by plaque assays on Madin-Darby canine kidney (MDCK) cell monolayers. The A66.1 S. pneumoniae strain, was cultured at 37°C in Todd-Hewitt broth until mid-log phase (using BSL-2 conditions), washed and resuspended in fresh broth containing 15% glycerol, and stored at −70°C until use. Anaesthetized mice were challenged with sub-lethal doses (10 or 100 PFU) of H1N1 CA04 or PR8 influenza virus via intranasal (i.n.) instillation. For bacterial coinfection, anaesthetized mice were i.n. inoculated with 500 CFU of S. pneumoniae A66.1 eight days post-influenza infection. Survival and weight loss were monitored daily.
Plaque Assay and Bacterial Burden
On day 8 after influenza infection, viral levels in the bronchoalveolar lavage (BAL) and lungs of infected mice were determined by plaque assays on MDCK cell monolayers. Bacterial burdens in the lungs and BAL were measured by sacrificing infected mice at 24 h post-bacterial i.n. challenge and plating serial 10-fold dilutions of each sample onto blood agar plates.
Isolation of lung cells
To collect bronchoalveolar lavage (BAL), mouse lungs were lavaged four times with 1 ml of PBS/EDTA (0.1 mM) and the samples were centrifuged to obtain cells for flow cytometry. Whole lungs were perfused, harvested and placed in a PBS buffer containing DNAse I (150 μg/ml; Roche, Basel, Switzerland) and collagenase D (1 mg/ml; Roche) for 45 min at 37°C. Following digestion, lung tissue was passed through a 40μm filter and blood cells were subsequently removed by incubation in ACK buffer. Single cell suspensions were analyzed using flow cytometry.
Flow cytometric analysis
BAL cells and lung single cell suspensions were first incubated with anti-mouse FcγIII/II receptor mAb (clone 2.4G2) for 15 min. The Fc receptor-blocked cells were further stained with Fixable Viability Dye (eBioscience, Waltham, MA) to label dead cells. Cells were stained for surface markers using the following mAbs: anti-CD11b (clone M1/70; eBioscience), anti-Ly6G (clone 1A8; Biolegend, San Diego, CA), anti-CD11c (clone HLC; BD Biosciences, San Jose, CA), anti-F4/80 (clone BM8; Biolegend), anti-Siglec F (clone E50-2440; BD Bioscience), MHC Class II (clone M5/114.15.2; Biolegend), CD24 (clone M1/69; Biolegend), CD64 (clone X54-57.1; Biolegend), Ly6C (clone HK1.4; Biolegend). Stained cells were analyzed on a FACSCanto (BD Biosciences) and the data were analyzed using FlowJo software (Tree Star Inc. Ashland, OR).
In vivo labeling of lung-resident phagocytes
Resident macrophages were fluorescently labeled as previously described (8). Briefly, we i.n. administered 100 ul of 10 uM PKH26 phagocytic cell labeling dye (Sigma, St. Louis, MO) 1 week prior to influenza virus infection. Lung and BAL cells were evaluated using flow cytometry on days 7 and 9 post-influenza infection.
Depletion of lung macrophages with clodronate
Anesthetized C57Bl/6 mice were given 100 ul of clodronate- or PBS-containing liposomes (FormuMax, Sunnyvale, CA) administered by either the i.n. or intravenous (i.v.) route on days 7 and 8 post-influenza infection. Lung and BAL cells were evaluated the following day for depletion of AMs and interstitial macrophages (IMs) by flow cytometry.
Statistics
All statistical calculations were performed using GraphPad Prism (LaJolla, CA). Tests between two groups used a two-tailed Mann-Whitney U test. Tests between multiple groups used two-way ANOVA with Bonferroni’s multiple comparisons. Survival curves were evaluated by log-rank Mantel-cox test and weight loss data were analyzed by using a two-tailed Mann-Whitney U test.
Results
Influenza A virus infection severely diminishes AM numbers in BALB/c mice
The impact of influenza virus infection on the presence and activity of AMs has been thoroughly studied, with different groups reporting conflicting results. Some previous data suggest that AM populations are depleted in response to influenza infection (10). However contradictory evidence indicates that AMs are maintained following infection, albeit phagocytic function is reduced (8, 9, 19–21). We reasoned that these apparent inconsistencies might be attributed to differences in experimental design, i.e., viral dose/strain or mouse genetic background. Furthermore, discrepancies in AM identification and immunophenotyping strategies could further account for contrasting conclusions. To clarify these issues, we employed a careful flow cytometry gating technique to differentiate AMs in the lung and BAL (Supplemental Fig. 1A). First, cells were selected based on size and granularity, followed by the exclusion of doublets, dead cells and neutrophils from the analysis. As previously described, AMs were identified as cells with high expression of CD11c and positive for F4/80 (CD11chiF4/80+) (Supplemental Fig. 1A) (22). In addition, CD11chiF4/80+ cells exhibit high levels of Siglec F expression, further characterizing these cells as AMs (Supplemental Fig. 1B) (23). We also utilized a phagocytic cell labeling dye (PKH26) to monitor the persistence of resident AMs following influenza virus infection. We administered 10 uM of PKH26 dye i.n. one week prior to either high (100 PFU) or low dose (10 PFU) CA04 virus infection. Finally, we evaluated the maintenance of AMs following influenza virus infection in both BALB/c and C57Bl/6 mice since studies reporting macrophage depletion utilized BALB/c mice while the other reports used C57Bl/6 mice.
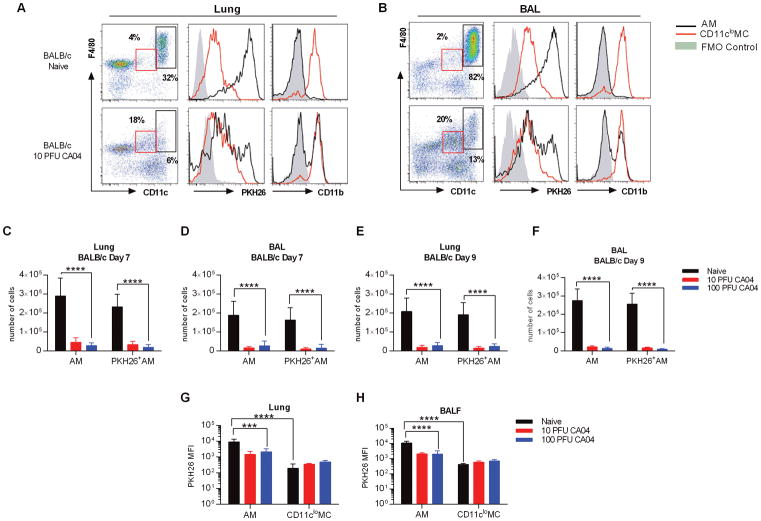
As expected, AMs represented the majority of cells found in the BAL of naïve BALB/c mice (8), while the percentage of AMs in the lungs was less frequent, but still substantial (Fig. 1A, 1B). Prior studies demonstrated that PKH26 labeling of pulmonary macrophages remained stable for up to thirty days (10), and in agreement with this, we found that AMs from naïve animals exhibited robust PKH26 labeling two weeks after PKH26 administration (Fig. 1A, 1B). Also, consistent with published data (24, 25), CD11clo myeloid cells (MCs) expressed CD11b while CD11chi AMs from naïve animals had minimal expression of CD11b (Fig. 1A, 1B). Following CA04 virus infection, we observed a major reduction in the number of total AMs, as well as the number of PKH26+ AMs, in both lung and BAL of BALB/c mice, regardless of infection dose (Fig. 1A–F). The numbers of AMs were reduced to similar levels at both days 7 and 9 post-infection (Fig. 1A–F). Siglec F expression was reduced in CD11chigh cells following infection, indicating a decline in the AM population (Supplemental Fig. 1B). Furthermore, the mean fluorescence intensity (MFI) of PKH26 in AMs from influenza-infected BALB/c mice was diminished and exhibited a bimodal pattern of expression, which suggested that only a portion of the CD11chi cells detected were present at the time of PKH26 i.n. delivery (Fig. 1A, 1B, 1G, 1H). Of note, many of the remaining CD11chi cells upregulated expression of CD11b, in response to CA04 virus infection (Fig. 1A, 1B). Concomitant with diminished AM numbers, there was a notable increase in a distinct population of cells that exhibited comparable expression of F4/80, but displayed levels of CD11c lower than that of AMs (Fig. 1A, 1B and Supplemental Fig. 2A, 2B). These CD11cloF4/80+ cells also presented with high levels of CD11b, but did not express Siglec F (Fig. 1A, 1B and Supplemental Fig. 1B), resulting in a surface expression profile previously used to define interstitial macrophages (IMs) (10, 22, 23), cells that have been reported to proliferate in response to influenza virus infection (10). Moreover, in naïve BALB/c mice, the MFI of PKH26 was significantly lower in CD11cloF4/80+ cells when compared to AMs (Fig. 1G, 1H), again in agreement with the attributes previously described for IMs (10). Thus, we concluded that resident AMs could be identified using CD11c and Siglec F in combination with the PKH26 label. Our data show that AMs were depleted in BALB/c mice following CA04 virus infection, but there was a substantial increase in the number of CD11clo MCs, which may represent the IM population. Furthermore, our analyses confirmed that AMs can be discriminated from lung myeloid populations based on the differential expression of CD11c and PKH26 MFI, in combination with Siglec F.
Figure 1.
Influenza A virus infection induces depletion of AMs in BALB/c mice. (A) Flow cytometry analysis of AMs and IMs in the lung (A) and BAL (B) of naive and CA04-infected BALB/c mice 7 days post-infection. Representative plots show frequency and expression of PKH26 and CD11b on AMs (CD11chiF4/80+) and CD11cloMCs (CD11cloF4/80+). Fluorescence minus one (FMO) control represents negative control as solid gray peak. (C–D) Total numbers of AMs and PKH26+AMs in the lung (C) and BAL (D) of BALB/c mice on day 7 post-CA04 infection. (E–F) Total numbers of AMs and PKH26+AMs in the lung (E) and BAL (F) of BALB/c mice on day 9 post-CA04 infection. Statistical analyses were performed by two-way ANOVA; *** p<0.001; **** p<0.0001. (G–H) MFI of PKH26 on AMs and IMs in the lung (G) and BAL (H) of naïve and influenza-infected BALB/c mice. Statistical analyses were performed by Mann-Whitney U test; ** p<0.01. Data shown are representative of 2 independent experiments with 5 mice per group.
AMs from C57Bl/6 mice are resistant to influenza-induced depletion
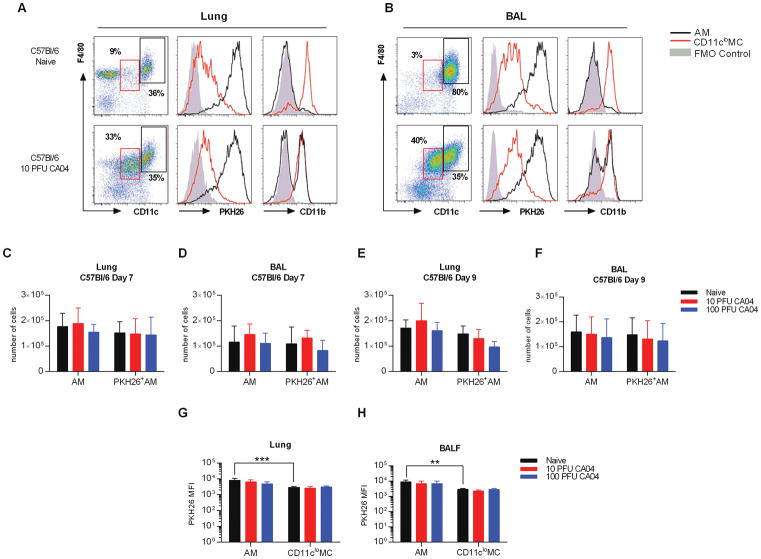
Similar to the results of Ghoneim et al. (10), we found that AMs in BALB/c mice were depleted following influenza virus infection, regardless of infection dose. In contrast, we and others reported no apparent effect of influenza on C57Bl/6 AM cell numbers (8, 14). Considering our results with C57Bl/6 mice and those of others using BALB/c mice, we further evaluated AM persistence in influenza virus-infected C57Bl/6 mice using strategies similar to those described above. Like naïve BALB/c mice, AMs represented the majority of cells detected in the lungs and BAL of naïve C57Bl/6 mice. These cells were strongly stained with PKH26, but displayed very low levels of CD11b (Fig. 2A, 2B). Furthermore, the PKH26 MFI was again higher in AMs compared to CD11clo MCs (Fig. 2G, 2H). In addition, AMs and CD11clo MCs exhibited high and low levels of Siglec F, respectively (Supplemental Fig. 1C). However, unlike BALB/c mice, the numbers of AMs, including PKH26+ AMs, in CA04 virus-infected C57Bl/6 mice remained at levels comparable to those of naïve animals at both day 7 and day 9 post-infection (Fig. 2C–F). This was observed in mice infected with either 10 or 100 PFU of influenza virus (Fig. 2C–F). Furthermore, resident AMs presented with increased CD11b expression following CA04 virus challenge, while the level of PKH26 remained unchanged (Fig. 2A, 2B, 2G, 2H), suggesting that resident AMs present at the time of PKH26 inoculation were sustained post-infection, albeit with an altered phenotype. Siglec F levels were similarly maintained following infection, confirming that resident AMs persisted in C57Bl/6 mice (Supplemental Fig. 1C). In contrast, CD11clo MCs exhibited high levels of CD11b surface expression in both naïve and CA04-infected animals (Fig. 2A, 2B). Furthermore, influenza virus infection induced significant expansion of the total CD11cloF4/80+ cell population (Fig. 2A, 2B and Supplemental Fig. 2C, 2D). Our analysis further revealed that CD11clo F4/80+CD11b+ cells in naïve mice consisted of several different myeloid populations, including IMs, dendritic cells (DCs), and Ly6C+ and Ly6C− monocytes (MO) (23). However, only the IM population (CD11cloF4/80+CD11b+ CD64+MHC II+CD24−) was significantly increased in response to influenza infection (Supplemental Fig. 3A–D), supporting the conclusion that expansion of CD11clo MCs was due to an increase in IMs. Taken together, these data indicate that in C57Bl/6 mice, AMs are more resilient following virus infection compared to AMs from BALB/c mice. Resident AMs in C57Bl/6 mice are maintained post-infection, exhibit similar PKH26 fluorescence as naïve animals, but express higher levels of CD11b. Thus, AMs are not depleted following infection of C57Bl/6 mice, but do present with an altered phenotype.
Figure 2.
Lung resident AMs are maintained in post-influenza infected C57Bl/6 mice. (A) Flow cytometry analysis of AMs and IMs macrophages in the lung (A) and BAL (B) of naive and CA04-infected C57Bl/6 mice 7 days post-infection. Representative plots show frequency and expression of PKH26 and CD11b on AMs (CD11chiF4/80+) and CD11cloMCs (CD11cloF4/80+). Fluorescence minus one (FMO) control represents negative control as solid gray peak. (C–D) Total numbers of AMs and PKH26+AMs in the lung (C) and BAL (D) of C57Bl/6 mice on day 7 post-CA04 infection. (E–F) Total numbers of AMs and PKH26+AMs in the lung (E) and BAL (F) of C57Bl/6 mice on day 9 post-CA04 infection. Statistical analyses were performed by two-way ANOVA; ** p<0.01; *** p<0.001. (G–H) MFI of PKH26 on AMs and IMs in the lung (G) and BAL (H) of naïve and influenza-infected C57Bl/6 mice. Statistical analyses were performed by Mann-Whitney U test; ** p<0.01. Data shown are representative of 2 independent experiments with 5 mice per group.
Ghoneim et al. (10) further reported that both the CA04 and PR8 virus infections resulted in AM depletion in BALB/c mice. We wondered whether the PR8 viral strain, which is known to induce robust inflammation and cellular apoptosis in infected hosts (26, 27), would reduce AM numbers more effectively in C57Bl/6 mice than the less inflammatory CA04 influenza virus. We therefore investigated the persistence of AMs in C57Bl/6 mice following infection with PR8 influenza virus and discovered that both the total number of AMs and the numbers of PKH26+ AMs were maintained in the lungs and BAL of C57Bl/6 mice (Supplemental Fig. 4A–D). These data confirmed that AMs from C57Bl/6 mice are not depleted following influenza virus infection, regardless of viral dose or strain, and further support the concept that AM depletion in response to influenza infection is highly dependent on mouse genetic background.
C57Bl/6 resident AMs are maintained post-influenza and not derived from the CD11clo population
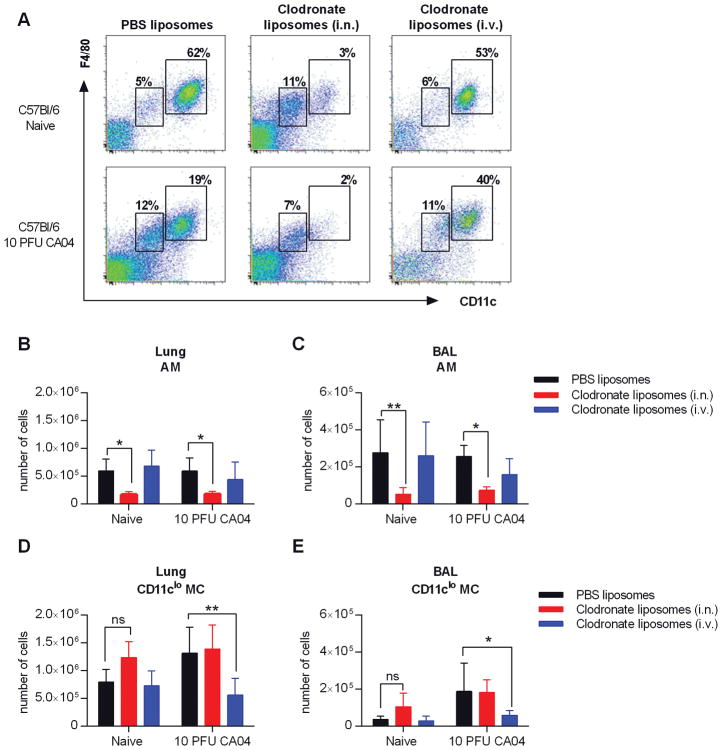
We have shown that AMs in C57Bl/6 mice are maintained post-influenza infection, but upregulate CD11b expression, a marker typically expressed by monocytes and macrophages (22, 23). We further demonstrated that influenza infection results in increased number of IMs. Reconstitution of AM populations following cell ablation has been found to occur via in situ differentiation of monocyte-derived IMs (28–30). Therefore, IMs may contribute to the presence of CD11b+ AMs in influenza-infected C57Bl/6 mice. In this case, a reduction in the IM population would presumably result in decreased AM numbers due to a deficiency in the available precursors (22). Evidence further suggests that i.n. or i.v. administration of clodronate-containing liposomes selectively depletes AMs and IMs, respectively (22). We exploited these procedures to investigate whether AMs in influenza-infected C57Bl/6 mice are derived from lung IMs and therefore would be vulnerable to clodronate-mediated depletion following i.v. injection (22). Naïve and CA04-infected C57Bl/6 mice received liposomal clodronate by either i.n. instillation or i.v. injection on days 7 and 8 post-influenza infection. On Day 9, cells were harvested from the lung and BAL to evaluate levels of AMs and IMs using flow cytometry (Fig. 3A). In both naïve and CA04-infected mice, AMs (CD11chiF4/80+) were depleted after i.n. clodronate treatment (Fig. 3B, 3C). However, consistent with previously published data (22), CD11cloMCs (CD11cloF4/80+) cells, which include the IM population, were maintained (Fig. 3D, 3E and Supplemental Figure 3A–D). Of note, i.n. clodronate treatment modestly increased the number of CD11cloMCs in naïve mice, most likely due to cell recruitment or proliferation in response to AM depletion (28–30). (Fig. 3D, 3E). Conversely, i.v. injection of liposomal clodronate dramatically reduced the numbers of IM-containing CD11cloMCs in CA04-infected mice, but did not significantly deplete AMs in either naïve or infected animals (Fig. 3B–E). Interestingly, we did not observe diminished numbers of CD11cloF4/80+ cells in naïve, i.v. clodronate-treated mice (Fig. 3D, 3E). These data indicate that the AM population in infected C57Bl/6 mice is not derived from the IM pool. Furthermore, these results confirm that resident AMs are not depleted following influenza infection of C57Bl/6 mice.
Figure 3.
AMs are reduced in C57Bl/6 mice following i.n., but not i.v., administration of clodronate liposomes. (A) Naïve and CA04 infected mice were treated with clodronate- or PBS-containing liposomes by i.n. or i.v. injection on days 7 and 8 post-influenza infection. Representative dot plots show the gating strategy for AMs and CD11cloMCs in the BAL of naïve and CA04-infected C57Bl/6 mice on day 9 following CA04 infection. (B–E) Total numbers of AMs (B–C) and IMs (D–E) in the lung (B and D) and BAL (C and E). Data shown are representative of 2 independent experiments with 5 mice per group. Statistical analyses were performed by two-way ANOVA; * p<0.05; ** p<0.01.
IFN-γ promotes AM depletion in BALB/c mice
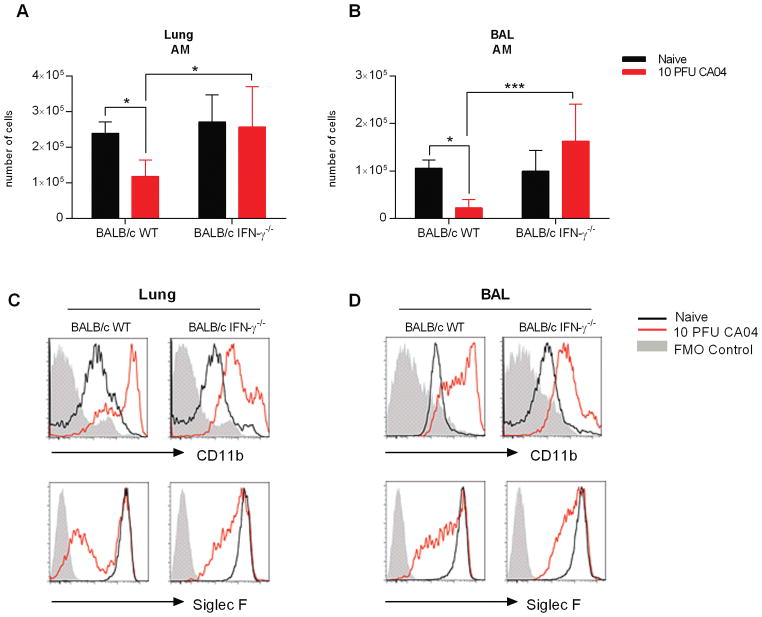
Influenza virus infection substantially diminished the AM pool in BALB/c, but not in C57Bl/6 mice. Earlier reports indicated that C57Bl/6 mice are nonetheless highly susceptible to bacterial outgrowth during post-influenza pneumococcal infection, due to diminished AM phagocytosis that results from increased IFN-γ expression in the infected lung (8, 19, 31). Interestingly, neutralization of IFN-γ promotes survival and bacterial clearance in both C57Bl/6 and BALB/c mice (8). Considering these findings, we suspected that IFN-γ might also promote depletion of BALB/c AMs following influenza virus infection. Using flow cytometry, we evaluated AM numbers in the lungs and BAL of naïve and CA04-infected BALB/c wild type (WT) and IFN-γ−/− mice 9 days after infection. Consistent with the data presented above, the AM population was reduced in response to influenza infection of BALB/c WT mice (Fig. 4A, 4B). Furthermore, CD11chiF4/80+ cells presented with increased CD11b expression, but Siglec F levels were diminished (Fig. 4C and D). In contrast, the numbers of AMs in influenza-infected BALB/c IFN-γ−/− mice was comparable to those in naïve animals (Fig. 4A, 4B). In addition, although IFN-γ−/− CD11chiF4/80+ cells upregulated CD11b in response to influenza infection, the levels of expression were considerably reduced when compared to WT animals (Fig 4C and D). Furthermore, Siglec F levels were better maintained in the absence of IFN-γ (Fig 4C and D). The data demonstrate for the first time that IFN-γ promotes AM cell death in influenza-infected BALB/c mice. Moreover, absence of IFN-γ restricts CD11b upregulation in AMs.
Figure 4.
IFN-γ deficiency prevents AM depletion in influenza-infected BALB/c mice. (A and B) Total number of AMs in the lung (A) and BAL (B) of BALB/c WT and IFN-γ−/− mice on Day 9 after infection with 10 PFU of the CA04 virus. (C and D) Representative plots show frequency and expression of Siglec F and CD11b on AMs in the lungs (C) and BAL (D) of naïve and influenza infected BALB/c WT and IFN-γ−/− mice. Data shown are representative of 2 independent experiments with 5 mice per group. Statistical analyses were performed by two-way ANOVA; * p<0.05; ** p<0.01.
Influenza-pneumococcal coinfection enhances mortality in both BALB/c and C57Bl/6 mice
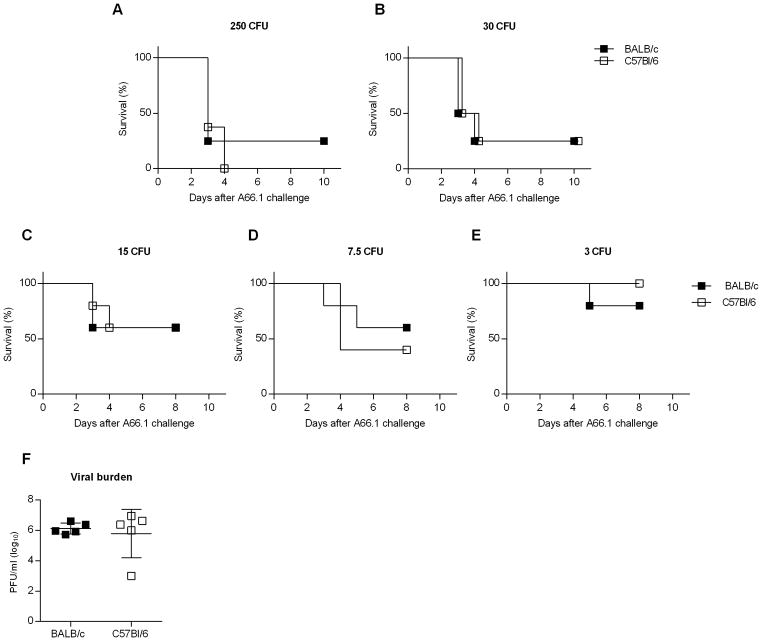
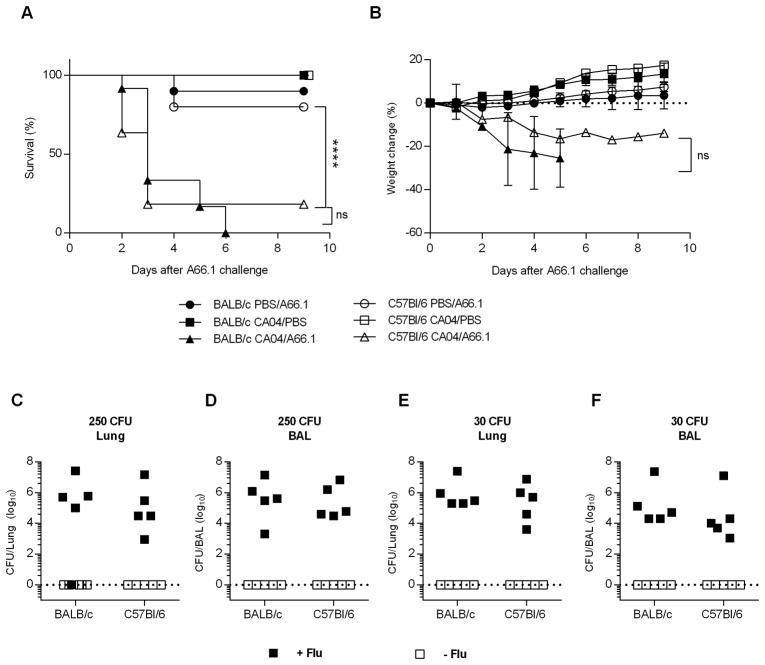
Enhanced susceptibility to bacterial infection is a common complication of influenza, and can lead to severe disease and death. There are multiple pathways underlying the pathogenesis of bacterial superinfections, including viral-induced lung damage and impaired bacterial clearance through suppression of host innate immunity (4, 8–10, 20, 32). Our data support the idea that influenza-induced alterations in AM expression/function are mouse strain dependent. Specifically, influenza infection results in either AM depletion or a reduced phagocytic activity, in BALB/c and C57Bl/6 mice, respectively, either of which could contribute to reduced bacterial clearance following pneumococcal infection. We therefore questioned whether post-influenza infected BALB/c and C57Bl/6 mice would exhibit similar high levels of susceptibility to bacterial co-infection. To examine this, BALB/c and C57Bl/6 mice were infected with a sublethal dose (10 PFU) of CA04 virus and eight days later, challenged i.n. with a sublethal dose of S. pneumoniae strain A66.1 (500 CFU). Both strains of mice rapidly lost weight when co-infected and exhibited enhanced mortality compared to mice infected with CA04 virus or A66.1 alone (Fig. 5A, 5B). Furthermore, post-influenza infected mice presented with high bacterial titers in the lung and BALF 24 h following secondary bacterial infection (Fig. 5C–F). In contrast, mice infected with A66.1 alone showed minimal bacterial burden (Fig. 5C–F). The results showed comparable bacterial replication in co-infected BALB/c and C57Bl/6 mice, indicating that both strains of mice were deficient in bacterial clearance, regardless of the presence or absence of AMs. Consistent with high bacterial titers, BALB/c and C57Bl/6 mice were highly susceptible to co-infection when inoculated with challenge doses as low as 30 CFU (Fig. 6A, 6B). Inoculum doses less than 30 CFU revealed that the dose-response to secondary bacterial infection was similar in BALB/c and C57Bl/6 mice (Figure 6C–E). We also challenged mice with a sterile, filtered inoculum to identify whether non-viable bacterial particles or debris enhanced susceptibility. Our results demonstrated that mice given filtered inoculum exhibited a similar level of susceptibility as PBS-challenged control mice (data not shown). These data taken together indicate that influenza infection enhances susceptibility to bacterial superinfection in both C57Bl/6 and BALB/c mice, due to inhibition of bacterial clearance. The results further demonstrate that mice sufficient in AM populations (i.e., C57BL/6 mice) are highly susceptible to bacterial superinfection, suggesting that AM depletion alone does not promote enhanced susceptibility to secondary bacterial infection.
Figure 5.
BALB/c and C57Bl/6 mice are highly susceptible to synergistic viral-bacterial coinfection. BALB/c and C57Bl/6 mice were challenged with PBS or 10 PFU of CA04 virus. At day 8 post-influenza infection, mice were inoculated i.n. with PBS or 500 CFU of S. pneumoniae A66.1. (A and B) Survival (A) and weight loss (B) were monitored daily for 10 days following S. pneumoniae A66.1 infection. Survival data were analyzed using the log-rank Mantel-cox test and weight loss data were analyzed by the Mann-Whitney U test; **** p<0.0001; ns = not significant. Data shown were pooled from 2 independent experiments with 8 mice per group. (C–F) BALB/c and C57Bl/6 were i.n. inoculated with 250 CFU (C and D) or 30 CFU (E and F) of S. pneumoniae A66.1 eight days after influenza infection. Bacterial burden was evaluated in total lung (C and E) and BAL (D and F) 24 h post-bacterial i.n. challenge. Data shown were pooled from 2 independent experiments with 5 mice per group.
Figure 6.
BALB/c and C57Bl/6 both exhibit similar mortality to influenza-pneumococcal infection superinfection, even at low bacterial challenge doses. BALB/c and C57Bl/6 mice were challenged with PBS or 10 PFU of CA04 virus. At day 8 post-influenza infection, the mice were inoculated i.n. with 250 (A), 30 (B), 15 (C), 7.5 (D) or 3 (E) CFU of S. pneumoniae A66.1. Survival was monitored daily for 10 days following S. pneumoniae A66.1 infection. Data shown were pooled from 2 independent experiments with 8 mice per group. (F) Viral titers were measured in whole lung homogenate of BALB/c and C57Bl/6 mice at day 8 post-influenza infection. Data shown are representative of 2 independent experiments with 5 mice per group.
Of note, viral burden at the time of bacterial infection (Day 8) was similar between both strains of mice (Fig. 6F). Our analyses demonstrated that BALB/c and C57Bl/6 mice exhibited similar susceptibility to lethal secondary bacterial infection following influenza.
Discussion
Our results demonstrate that influenza-induced suppression of AMs occurs via different mechanisms depending upon the mouse genetic background. We established that AMs from BALB/c mice are readily depleted in response to influenza infection, but C57Bl/6 AMs are not depleted yet do show phenotypic changes. Our data are indirectly supported by studies among various independent laboratories, but have yet to be described in a directly comparative analysis. Of note, BALB/c and C57Bl/6 mice are both highly susceptible to post-influenza bacterial infection, suggesting that either AM depletion or restriction of AM-mediated phagocytosis can diminish immunity against bacterial pathogens. These results resolve an existing conundrum in the literature by showing mouse strain-related differences in response to influenza infection.
In recent years, influenza-associated alterations to the host’s innate defenses have been identified as a major underlying cause of increased susceptibility to bacterial superinfection. AM-mediated microbial clearance, which is essential for the elimination of bacteria from the airways, is suppressed post-influenza infection (10, 19, 20). However, the responsible molecular mechanisms have been only poorly understood. Specifically, it is unclear whether influenza infection promotes AM depletion or disrupts bacterial recognition and uptake without reducing AM numbers (8, 10, 14, 20). Conflicting evidence has been obtained in support of both mechanisms, differences presumably related to nuances in experimental design, including viral dose/strain and the timing of infection and analysis (5, 13). However, the precise cause of these differences has yet to be described. Moreover, data in support of AM maintenance was questioned due to insufficient data points and inadequate AM identification (10). Our findings now explain the inconsistencies that are described in the literature and demonstrate that AM viability following influenza infection is dependent on mouse genetic background. We found that AMs were depleted in BALB/c mice in response to influenza infection, while infected C57Bl/6 mice retained the resident AM population, regardless of viral dose/strain. These results are corroborated by several reports demonstrating depletion of AMs in BALB/c mice (10) and a sustained presence of AMs in C57Bl/6 mice (8, 14–16). Using PKH26 to label cells, Janssen et al. (14) found that resident of C57Bl/6 AMs are resistant to apoptosis following influenza infection. Indeed, C57Bl/6 AMs, which can live for up to 8 months at steady state (33), persist for more than two weeks following influenza infection (14).
Several reports agree that the presence of AM is crucial for resistance against influenza infection in both C57Bl/6 and BALB/c mice (7, 34–37). Deletion of AM prior to influenza infection (either through genetic disruption or clodronate treatment) is detrimental and results in increased morbidity and respiratory failure (7, 34). In contrast, administration of GM-CSF promotes AM proliferation and protects against damage-associated mortality in influenza-infected mice (36, 37). In these studies, alterations in the AM population occurred prior to influenza infection. However, to our knowledge, our study is the first to investigate mouse strain-dependent responses of AMs in post-influenza infected BALB/c and C57Bl/6 mice. Consistent with our data, AM are also depleted in BALB/c mice in response to gamma herpes virus (38). Interestingly, AMs are not depleted in response to RSV in neonatal BALB/c mice (39). However, these results have yet to be investigated in C57Bl/6 mice. Whether mouse genetic background influences AM response in a virus-specific or age-dependent manner thus remains to be determined.
Similar to the data presented here and elsewhere (10), the peak of AM depletion in BALB/c mice occurs at day 7 post-influenza infection, when less than 20% of the initial AM population remains. A recent report modeled the dynamic relationship between AM depletion and the minimum infection dose that is required to inhibit bacterial clearance (17). Using available data to depict influenza-pneumococcal coinfection kinetics, the authors predicted that as the number AMs diminish during the course of influenza infection, the inoculum size necessary to support bacterial outgrowth also decreases. Thus, the authors predicted that a single bacterium would be sufficient to promote bacterial invasion at this time point (17). Our current findings in BALB/c mice further support this claim, given that following influenza infection low dose bacterial challenge (30 CFU) was sufficient to establish rapid bacterial outgrowth and thus enhanced mortality in these mice. However, our data also demonstrate that AM depletion in C57Bl/6 mice was negligible and even though resident AMs persisted in C57Bl/6 mice, the animals were still highly susceptible to S. pneumoniae following influenza infection. Moreover, clearance of S. pneumoniae was similarly inhibited in the presence of C57Bl/6 AMs, indicating that that AM depletion is not directly responsible for bacterial superinfection-associated mortality in C57Bl/6 mice. Nevertheless, influenza-induced alterations to AM function are also known to compromise the host’s ability to control bacterial growth (5, 13). IFN-γ production during the recovery phase of influenza downregulates the expression of the MARCO class A scavenger receptor, resulting in reduced bacterial uptake and clearance, and consequently, increased mortality during coinfection with influenza virus and S. pneumoniae (8). Furthermore, suppression of NADPH oxidase following influenza infection reduces killing activity in AMs and neutrophils, leaving mice highly vulnerable to subsequent S. aureus infections (9). In addition, resolution of inflammation can require AM inhibition, which occurs via cell-to-cell contact with respiratory epithelial cells through CD200-CD200R interactions, as well as negative regulation by TGFβ and IL-10 (40–42). Although our results do not rule out alternative possibilities, the data clearly support the concept that AM dysfunction, and not depletion, is responsible for the increased mortality of C57Bl/6 mice infected with both influenza virus and S. pneumoniae.
The mechanism(s) responsible for differential responses of AMs following influenza in BALB/c and C57Bl/6 mice are largely unknown. There is a considerable amount of data that supports the fact that C57Bl/6 and BALB/c mice exhibit disparate immune responses to infectious diseases (43, 44). In C57Bl/6 mice, T cell-mediated responses are sometimes preferentially skewed toward the Th1 lineage, while BALB/c mice are believed to be somewhat Th2 dominant (45). Similarly, M1 (classically activated) or M2 (alternatively activated) macrophage phenotypes are preferentially exhibited in C57BL/6 and BALB/c mice, respectively (46). Moreover, these differences have been largely attributed to the differential expression of key genes that influence functional characteristics of immune cells such as reduced STAT4 expression and increased prostaglandin E2 production in BALB/c mice, both of which limit IFN-γ production (47, 48).
Genetic factors also influence susceptibility to infection in BALB/c and C57Bl/6 mice. C57Bl/6 mice are naturally resistant to infections with intracellular pathogens such as Leishmania major and Yersinia enterocolitica, whereas BALB/c mice are susceptible to both pathogens (49, 50). BALB/c mice are in turn, resistant to parasitic infection with Toxoplasma gondii as well as diet-induced obesity and atherosclerosis (51, 52). BALB/c mice also exhibit enhanced responses to allergens (53). Overall susceptibility to PR8 (H1N1) influenza virus is comparable between BALB/c and C57Bl/6 mice, although C57Bl/6 mice lose more weight in the early stages of infection (54). In addition, C57Bl/6 mice are more susceptible to avian influenza A/H7N9 virus infection, as well as infection with pandemic H1N1, due to reduced viral clearance, coupled with enhanced lung injury and impaired tissue regeneration (55, 56). Increased resistance of BALB/c mice in this case was associated with elevated cytokine production, including IL-4, IL-10, TNFα and IFN-γ, suggesting that the Th1/Th2 dichotomy is dependent on immunological context (55, 56).
The role of IFN-γ during influenza infection is controversial, but is generally thought to have minimal influence on viral burden (57). However, emerging evidence indicates that IFN-γ can play diverse immunoregulatory roles during the recovery phase of influenza infection including suppression of virus-specific CD8+ T cell responses, inhibition of influenza-induced lung inflammation and reduced tissue repair following lung injury (58–60). IFN-γ is similarly important for the resolution of AM-mediated inflammation following influenza infection in both C57Bl/6 and BALB/c mice (8). Production of IFN-γ by infiltrating T cells downregulates the expression of the MARCO phagocytic receptor, which in turn, reduces AM phagocytosis and increased susceptibility to bacterial colonization in post-influenza infected mice. Furthermore, IFN-γ deficiency in influenza-infected C57Bl/6 mice restores MARCO expression on AMs and consequently AM-mediated phagocytosis, which enhances survival following bacterial superinfection. Interestingly, our data showed that AM depletion was rescued in IFN-γ−/− BALB/c mice, which is a novel finding and is consistent with the fact that IFN-γ deficiency improves bacterial clearance and disease outcome in this mouse strain (8). In addition, we now demonstrate that AMs upregulate CD11b following influenza infection, which is modestly blocked in the absence of IFN-γ, suggesting possible co-regulation. AMs from C57BL/6 mice express high amounts of MARCO, however BALB/c AMs preferentially express alternative scavenger receptors such as CD36, which could potentially account for differential responses to IFN-γ (61). BALB/c mice produce elevated levels of IFN-γ compared to C57Bl/6 mice during influenza infection, suggesting that AM depletion in response to IFN-γ may be dose dependent. IFN-γ, along with TNFα, are known to enhance macrophage apoptosis and nitric oxide production to promote bacterial clearance (62). Furthermore, enhanced levels of TNFα correlate with an increase in AM apoptosis following exposure to bleomycin (63). IFN-γ further promotes apoptosis of FAS ligand expressing AMs through upregulated expression of FAS on T lymphocytes (64–66). Additional investigation will be required to determine the precise molecular mechanism responsible for IFN-γ-mediated depletion of AMs in BALB/c mice.
In our analysis of pulmonary macrophage expression, we used differential expression of CD11c to discriminate between AMs and lung myeloid cells. Recent evidence suggests that in addition to high levels of CD11c (22), AMs exhibit increased PKH26 MFI (10), which is further supported by our data. AMs are the predominant CD11chi expressing cell in the lung compartment of naïve animals (23); therefore, we can reason that PKH26-labeled CD11chi cells characterize resident AMs in post-influenza infected mice. In addition, we demonstrated that AMs from infected C57Bl/6 mice were phenotypically altered and expressed increased CD11b. The sustained expression of Siglec F further confirms the stability of the resident AM population in C57Bl/6 mice, albeit with an altered phenotype. Misharin et al. utilized a comprehensive flow cytometric panel to identify myeloid cells in the lung, and excluded CD11b+ cells in their analysis of AMs (23). Furthermore, Ghoneim et al. reported that CD11b expression is a sufficient marker to differentiate between IM and AM populations (10). Although this is a useful distinction for naïve animals, substantial evidence indicates that resident AMs upregulate CD11b during inflammation (14, 24, 25), which in turn, suggests a need for more precise immunophenotyping protocols to assess expression of lung macrophage populations in influenza-infected animals. Multiple investigators have reported that CD11b+ macrophages in the lung represent recruited cells (10, 28). However, we observed maintenance of the PKH26 label in CD11b+AMs following infection of C57Bl/6 mice, suggesting that these AMs were present in the lung preceding the influenza infection. In addition, recruited macrophages express low levels of CD11c compared to resident AMs (14). Alternatively, Ghoneim et al. proposed that AMs present in the lung following influenza infection are derived from the monocyte-derived IM pool (10, 28–30). We now provide evidence that in the absence of IMs (resulting from i.v. clodronate administration), AM populations were still intact in influenza-infected C57Bl/6 mice. Therefore, we conclude that our current gating strategy, based on differential expression of CD11c and PKH26 MFI, in combination with Siglec F, appropriately identifies resident AMs. Moreover, inclusion of CD11b+ cells identifies a subpopulation that may have been overlooked based on previous gating strategies (10, 23). Taken together, our data indicate that C57Bl/6 AMs are maintained during influenza infection, but exhibit phenotypic characteristics different from naïve AMs.
In summary, we present conclusive data demonstrating that the AM response to influenza infection varies in different inbred mouse strains. We confirmed that influenza infection induces robust AM depletion in BALB/c mice, and furthermore, these cells fail to repopulate the lungs by day 9 post-infection. In contrast, C57BL/6 mice maintain the presence of resident AMs, as evidenced by sustained levels of PKH26. We further demonstrated that resident AMs in C57Bl/6 mice can upregulate expression of CD11b, a phenotypic profile which may have resulted in their exclusion from previous analyses. Both macrophage depletion in BALB/c mice and phenotypic alterations in C57Bl/6 AMs were dependent upon IFN-γ (8), and both mouse strains were highly susceptible to secondary bacterial infections. These data substantiate previous results, while identifying mouse strain-related differences in regard to the response of AMs to influenza infection This novel information will be of crucial importance for choosing appropriate mouse models for future experiments. Interrogating distinct mechanisms of influenza-induced suppression of AMs could also prove beneficial for ultimate prevention and/or treatment of viral-bacterial synergy.
Supplementary Material
Acknowledgments
We thank Sharon Salmon, Sean Roberts and Dr. Don Steiner for valuable advice and technical assistance. We also acknowledge Dr. Douglas Cohn, Ms. Victoria Boppert, and animal facility staff at Albany Medical College for the care of the mice.
Footnotes
This work was supported by NIH grants RO1 AI75312 and RO1 HL140496 to D.W.M.
Author contributions
D. Califano, conducted the experiments and analyzed the data. Y. Furuya provided technical assistance. D. Califano and D.W. Metzger conceived and designed the experiments. D. Califano and D.W. Metzger wrote the paper.
Disclosure
The authors declare no competing financial interests
References
- 1.Falsey AR, Becker KL, Swinburne AJ, Nylen ES, Formica MA, Hennessey PA, Criddle MM, Peterson DR, Baran A, Walsh EE. Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis. 2013;208:432–441. doi: 10.1093/infdis/jit190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Memoli MJ, Morens DM, Taubenberger JK. Pandemic and seasonal influenza: therapeutic challenges. Drug Discov Today. 2008;13:590–595. doi: 10.1016/j.drudis.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louria DB, Blumenfeld HL, Ellis JT, Kilbourne ED, Rogers DE. Studies on influenza in the pandemic of 1957–1958. II. Pulmonary complications of influenza. J Clin Invest. 1959;38:213–265. doi: 10.1172/JCI103791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill JR, Sheng ZM, Ely SF, Guinee DG, Beasley MB, Suh J, Deshpande C, Mollura DJ, Morens DM, Bray M, Travis WD, Taubenberger JK. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–243. doi: 10.5858/134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzger DW, Sun K. Immune dysfunction and bacterial coinfections following influenza. J Immunol. 2013;191:2047–2052. doi: 10.4049/jimmunol.1301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dockrell DH, Marriott HM, Prince LR, Ridger VC, Ince PG, Hellewell PG, Whyte MK. Alveolar macrophage apoptosis contributes to pneumococcal clearance in a resolving model of pulmonary infection. J Immunol. 2003;171:5380–5388. doi: 10.4049/jimmunol.171.10.5380. [DOI] [PubMed] [Google Scholar]
- 7.Schneider C, Nobs SP, Heer AK, Kurrer M, Klinke G, van Rooijen N, Vogel J, Kopf M. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 2014;10:e1004053. doi: 10.1371/journal.ppat.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 9.Sun K, Metzger DW. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. J Immunol. 2014;192:3301–3307. doi: 10.4049/jimmunol.1303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol. 2013;191:1250–1259. doi: 10.4049/jimmunol.1300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 12.Robinson KM, Kolls JK, Alcorn JF. The immunology of influenza virus-associated bacterial pneumonia. Curr Opin Immunol. 2015;34:59–67. doi: 10.1016/j.coi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rynda-Apple A, Robinson KM, Alcorn JF. Influenza and Bacterial Superinfection: Illuminating the Immunologic Mechanisms of Disease. Infect Immun. 2015;83:3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, Henson PM. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am J Respir Crit Care Med. 2011;184:547–560. doi: 10.1164/rccm.201011-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo SU, Kwon HJ, Ko HJ, Byun YH, Seong BL, Uematsu S, Akira S, Kweon MN. Type I interferon signaling regulates Ly6C(hi) monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 2011;7:e1001304. doi: 10.1371/journal.ppat.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 17.Smith AM, Smith AP. A Critical, Nonlinear Threshold Dictates Bacterial Invasion and Initial Kinetics During Influenza. Sci Rep. 2016;6:38703. doi: 10.1038/srep38703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun K, Ye J, Perez DR, Metzger DW. Seasonal FluMist vaccination induces cross-reactive T cell immunity against H1N1 (2009) influenza and secondary bacterial infections. J Immunol. 2011;186:987–993. doi: 10.4049/jimmunol.1002664. [DOI] [PubMed] [Google Scholar]
- 19.Hang do TT, Choi EJ, Song JY, Kim SE, Kwak J, Shin YK. Differential effect of prior influenza infection on alveolar macrophage phagocytosis of Staphylococcus aureus and Escherichia coli: involvement of interferon-gamma production. Microbiol Immunol. 2011;55:751–759. doi: 10.1111/j.1348-0421.2011.00383.x. [DOI] [PubMed] [Google Scholar]
- 20.Warshauer D, Goldstein E, Akers T, Lippert W, Kim M. Effect of influenza viral infection on the ingestion and killing of bacteria by alveolar macrophages. Am Rev Respir Dis. 1977;115:269–277. doi: 10.1164/arrd.1977.115.2.269. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson CLJ, akab GJ. Pulmonary antibacterial defenses during mild and severe influenza virus infection. Infect Immun. 1990;58:2809–2814. doi: 10.1128/iai.58.9.2809-2814.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaynagetdinov R, Sherrill TP, Kendall PL, Segal BH, Weller KP, Tighe RM, Blackwell TS. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol. 2013;49:180–189. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan M, Li WC, Vlahos R, Maxwell MJ, Anderson GP, Hibbs ML. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J Immunol. 2012;189:946–955. doi: 10.4049/jimmunol.1200660. [DOI] [PubMed] [Google Scholar]
- 25.Duan M, Steinfort DP, Smallwood D, Hew M, Chen W, Ernst M, Irving LB, Anderson GP, Hibbs ML. CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs. Mucosal Immunol. 2016;9:550–563. doi: 10.1038/mi.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 27.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landsman L, Jung S. Lung macrophages serve as obligatory intermediate between blood monocytes and alveolar macrophages. J Immunol. 2007;179:3488–3494. doi: 10.4049/jimmunol.179.6.3488. [DOI] [PubMed] [Google Scholar]
- 29.Maus UA, Janzen S, Wall G, Srivastava M, Blackwell TS, Christman JW, Seeger W, Welte T, Lohmeyer J. Resident alveolar macrophages are replaced by recruited monocytes in response to endotoxin-induced lung inflammation. Am J Respir Cell Mol Biol. 2006;35:227–235. doi: 10.1165/rcmb.2005-0241OC. [DOI] [PubMed] [Google Scholar]
- 30.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen CI, Anekalla KR, Joshi N, Williams KJN, Abdala-Valencia H, Yacoub TJ, Chi M, Chiu S, Gonzalez-Gonzalez FJ, Gates K, Lam AP, Nicholson TT, Homan PJ, Soberanes S, Dominguez S, Morgan VK, Saber R, Shaffer A, Hinchcliff M, Marshall SA, Bharat A, Berdnikovs S, Bhorade SM, Bartom ET, Morimoto RI, Balch WE, Sznajder JI, Chandel NS, Mutlu GM, Jain M, Gottardi CJ, Singer BD, Ridge KM, Bagheri N, Shilatifard A, Budinger GRS, Perlman H. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breslow-Deckman JM, Mattingly CM, Birket SE, Hoskins SN, Ho TN, Garvy BA, Feola DJ. Linezolid decreases susceptibility to secondary bacterial pneumonia postinfluenza infection in mice through its effects on IFN-gamma. J Immunol. 2013;191:1792–1799. doi: 10.4049/jimmunol.1300180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B, Robinson KM, McHugh KJ, Scheller EV, Mandalapu S, Chen C, Di YP, Clay ME, Enelow RI, Dubin PJ, Alcorn JF. Influenza-induced type I interferon enhances susceptibility to gram-negative and gram-positive bacterial pneumonia in mice. Am J Physiol Lung Cell Mol Physiol. 2015;309:L158–167. doi: 10.1152/ajplung.00338.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy J, Summer R, Wilson AA, Kotton DN, Fine A. The prolonged life-span of alveolar macrophages. Am J Respir Cell Mol Biol. 2008;38:380–385. doi: 10.1165/rcmb.2007-0224RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardani A, Boulton A, Kim TS, Braciale TJ. Alveolar Macrophages Prevent Lethal Influenza Pneumonia By Inhibiting Infection Of Type-1 Alveolar Epithelial Cells. PLoS Pathog. 2017;13:e1006140. doi: 10.1371/journal.ppat.1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He W, Chen CJ, Mullarkey CE, Hamilton JR, Wong CK, Leon PE, Uccellini MB, Chromikova V, Henry C, Hoffman KW, Lim JK, Wilson PC, Miller MS, Krammer F, Palese P, Tan GS. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat Commun. 2017;8:846. doi: 10.1038/s41467-017-00928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam R, Hillberry Z, Chen H, Feng Y, Fletcher K, Neuenschwander P, Shams H. Delivery of GM-CSF to Protect against Influenza Pneumonia. PLoS One. 2015;10:e0124593. doi: 10.1371/journal.pone.0124593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang FF, Barnes PF, Feng Y, Donis R, Chroneos ZC, Idell S, Allen T, Perez DR, Whitsett JA, Dunussi-Joannopoulos K, Shams H. GM-CSF in the lung protects against lethal influenza infection. Am J Respir Crit Care Med. 2011;184:259–268. doi: 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machiels B, Dourcy M, Xiao X, Javaux J, Mesnil C, Sabatel C, Desmecht D, Lallemand F, Martinive P, Hammad H, Guilliams M, Dewals B, Vanderplasschen A, Lambrecht BN, Bureau F, Gillet L. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat Immunol. 2017;18:1310–1320. doi: 10.1038/ni.3857. [DOI] [PubMed] [Google Scholar]
- 39.Eichinger KM, Egana L, Orend JG, Resetar E, Anderson KB, Patel R, Empey KM. Alveolar macrophages support interferon gamma-mediated viral clearance in RSV-infected neonatal mice. Respir Res. 2015;16:122. doi: 10.1186/s12931-015-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snelgrove RJ, Goulding J, Didierlaurent AM, Lyonga D, Vekaria S, Edwards L, Gwyer E, Sedgwick JD, Barclay AN, Hussell T. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat Immunol. 2008;9:1074–1083. doi: 10.1038/ni.1637. [DOI] [PubMed] [Google Scholar]
- 41.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, Rifkin DB, Sheppard D. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 42.Lim S, Caramori G, Tomita K, Jazrawi E, Oates T, Chung KF, Barnes PJ, Adcock IM. Differential expression of IL-10 receptor by epithelial cells and alveolar macrophages. Allergy. 2004;59:505–514. doi: 10.1111/j.1398-9995.2004.00455.x. [DOI] [PubMed] [Google Scholar]
- 43.Sellers RS, Clifford CB, Treuting PM, Brayton C. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol. 2012;49:32–43. doi: 10.1177/0300985811429314. [DOI] [PubMed] [Google Scholar]
- 44.Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 45.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–2095. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. [Google Scholar]
- 47.Kuroda E, Kito T, Yamashita U. Reduced expression of STAT4 and IFN-gamma in macrophages from BALB/c mice. J Immunol. 2002;168:5477–5482. doi: 10.4049/jimmunol.168.11.5477. [DOI] [PubMed] [Google Scholar]
- 48.Kuroda E, Yamashita U. Mechanisms of enhanced macrophage-mediated prostaglandin E2 production and its suppressive role in Th1 activation in Th2-dominant BALB/c mice. J Immunol. 2003;170:757–764. doi: 10.4049/jimmunol.170.2.757. [DOI] [PubMed] [Google Scholar]
- 49.Sacks D, Noben-Trauth N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat Rev Immunol. 2002;2:845–858. doi: 10.1038/nri933. [DOI] [PubMed] [Google Scholar]
- 50.Autenrieth IB, Beer M, Bohn E, Kaufmann SH, Heesemann J. Immune responses to Yersinia enterocolitica in susceptible BALB/c and resistant C57BL/6 mice: an essential role for gamma interferon. Infect Immun. 1994;62:2590–2599. doi: 10.1128/iai.62.6.2590-2599.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen SJ, Zhang YX, Huang SG, Lu FL. Galectins expressed differently in genetically susceptible C57BL/6 and resistant BALB/c mice during acute ocular Toxoplasma gondii infection. Parasitology. 2017;144:1064–1072. doi: 10.1017/S0031182017000270. [DOI] [PubMed] [Google Scholar]
- 52.Friedman G, Ben-Yehuda A, Dabach Y, Hollander G, Babaey S, Ben-Naim M, Stein O, Stein Y. Macrophage cholesterol metabolism, apolipoprotein E, and scavenger receptor AI/II mRNA in atherosclerosis-susceptible and -resistant mice. Arterioscler Thromb Vasc Biol. 2000;20:2459–2464. doi: 10.1161/01.atv.20.11.2459. [DOI] [PubMed] [Google Scholar]
- 53.Gueders MM, Paulissen G, Crahay C, Quesada-Calvo F, Hacha J, Van Hove C, Tournoy K, Louis R, Foidart JM, Noel A, Cataldo DD. Mouse models of asthma: a comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm Res. 2009;58:845–854. doi: 10.1007/s00011-009-0054-2. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava B, Blazejewska P, Hessmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K. Host genetic background strongly influences the response to influenza a virus infections. PLoS One. 2009;4:e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao G, Liu C, Kou Z, Gao T, Pan T, Wu X, Yu H, Guo Y, Zeng Y, Du L, Jiang S, Sun S, Zhou Y. Differences in the pathogenicity and inflammatory responses induced by avian influenza A/H7N9 virus infection in BALB/c and C57BL/6 mouse models. PLoS One. 2014;9:e92987. doi: 10.1371/journal.pone.0092987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otte A, Sauter M, Alleva L, Baumgarte S, Klingel K, Gabriel G. Differential host determinants contribute to the pathogenesis of 2009 pandemic H1N1 and human H5N1 influenza A viruses in experimental mouse models. Am J Pathol. 2011;179:230–239. doi: 10.1016/j.ajpath.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price GE, Gaszewska-Mastarlarz A, Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prabhu N, Ho AW, Wong KH, Hutchinson PE, Chua YL, Kandasamy M, Lee DC, Sivasankar B, Kemeny DM. Gamma interferon regulates contraction of the influenza virus-specific CD8 T cell response and limits the size of the memory population. J Virol. 2013;87:12510–12522. doi: 10.1128/JVI.01776-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiley JA, Cerwenka A, Harkema JR, Dutton RW, Harmsen AG. Production of interferon-gamma by influenza hemagglutinin-specific CD8 effector T cells influences the development of pulmonary immunopathology. Am J Pathol. 2001;158:119–130. doi: 10.1016/s0002-9440(10)63950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Califano D, Furuya Y, Roberts S, Avram D, McKenzie ANJ, Metzger DW. IFN-gamma increases susceptibility to influenza A infection through suppression of group II innate lymphoid cells. Mucosal Immunol. 2017 doi: 10.1038/mi.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamilton RF, Jr, Thakur SA, Mayfair JK, Holian A. MARCO mediates silica uptake and toxicity in alveolar macrophages from C57BL/6 mice. J Biol Chem. 2006;281:34218–34226. doi: 10.1074/jbc.M605229200. [DOI] [PubMed] [Google Scholar]
- 62.Herbst S, Schaible UE, Schneider BE. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS One. 2011;6:e19105. doi: 10.1371/journal.pone.0019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortiz LA, Moroz K, Liu JY, Hoyle GW, Hammond T, Hamilton RF, Holian A, Banks W, Brody AR, Friedman M. Alveolar macrophage apoptosis and TNF-alpha, but not p53, expression correlate with murine response to bleomycin. Am J Physiol. 1998;275:L1208–1218. doi: 10.1152/ajplung.1998.275.6.L1208. [DOI] [PubMed] [Google Scholar]
- 64.Kearns MT, Barthel L, Bednarek JM, Yunt ZX, Henson PM, Janssen WJ. Fas ligand-expressing lymphocytes enhance alveolar macrophage apoptosis in the resolution of acute pulmonary inflammation. Am J Physiol Lung Cell Mol Physiol. 2014;307:L62–70. doi: 10.1152/ajplung.00273.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martins GA, Vieira LQ, Cunha FQ, Silva JS. Gamma interferon modulates CD95 (Fas) and CD95 ligand (Fas-L) expression and nitric oxide-induced apoptosis during the acute phase of Trypanosoma cruzi infection: a possible role in immune response control. Infect Immun. 1999;67:3864–3871. doi: 10.1128/iai.67.8.3864-3871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shustov A, Nguyen P, Finkelman F, Elkon KB, Via CS. Differential expression of Fas and Fas ligand in acute and chronic graft-versus-host disease: up-regulation of Fas and Fas ligand requires CD8+ T cell activation and IFN-gamma production. J Immunol. 1998;161:2848–2855. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.