Abstract
The HSF and FOXO families of transcription factors play evolutionarily conserved roles in stress resistance and lifespan. In humans, the rs2802292 G-allele at FOXO3 locus has been associated with longevity in all human populations tested; moreover, its copy number correlated with reduced frequency of age-related diseases in centenarians. At the molecular level, the intronic rs2802292 G-allele correlated with increased expression of FOXO3, suggesting that FOXO3 intron 2 may represent a regulatory region. Here we show that the 90-bp sequence around the intronic single nucleotide polymorphism rs2802292 has enhancer functions, and that the rs2802292 G-allele creates a novel HSE binding site for HSF1, which induces FOXO3 expression in response to diverse stress stimuli. At the molecular level, HSF1 mediates the occurrence of a promoter–enhancer interaction at FOXO3 locus involving the 5′UTR and the rs2802292 region. These data were confirmed in various cellular models including human HAP1 isogenic cell lines (G/T). Our functional studies highlighted the importance of the HSF1-FOXO3-SOD2/CAT/GADD45A cascade in cellular stress response and survival by promoting ROS detoxification, redox balance and DNA repair. Our findings suggest the existence of an HSF1-FOXO3 axis in human cells that could be involved in stress response pathways functionally regulating lifespan and disease susceptibility.
INTRODUCTION
Cellular homeostasis is dependent upon the interaction between genes and the environment, and relies on the coordinated regulation of interlinked molecular processes including stress resistance. The ability to resist to cellular stress has been associated with good health and longevity and requires the concerted action of pathways involved in antioxidant defense, heat protection, protein degradation, glycogen storage, DNA repair and metabolism. These pathways ultimately lead to the activation of stress resistance transcription factors, such as HSF1 and FOXO3 (1,2), which in turn regulate interconnected gene expression networks that are evolutionarily conserved from yeast to mammals (3,4).
HSF1 coordinates the transcription of stress-induced genes to ensure adaptation of eukaryotic cells and organisms to environmental changes. Although its pivotal role in cellular homeostasis has been mainly linked to the transactivation of genes encoding heat shock proteins, recent genome-wide studies have revealed that HSF1 can reprogram gene expression more extensively, being involved both in physiological processes, including metabolism and aging, and pathological conditions, such as neurodegenerative diseases and cancer. Specifically, HSF1 plays a key role in the regulation of core cellular processes in cells under stress – where it controls genes encoding for transcription factors, cell-cycle regulators and components of the translational apparatus – and allows for reactivation of the cellular machinery upon restoration of conditions compatible with proliferation (5). Indeed, it has been shown that HSF1 regulates cellular response to oxidative stress, hypoxia and metabolic imbalance (6–9), and is required together with FOXO3 to sustain lifespan extension in model organisms (4).
FOXO3 belongs to the FOXO family of transcription factors, which share the FHRE DNA consensus sequence. FOXO3 controls the expression of a complex network of genes regulating proliferation, survival, metabolism and autophagy, ultimately leading to tumor suppression, nutrient sensing and stress resistance in several cell types and tissues. In multicellular organisms, FOXO proteins have been involved in the regulation of response to oxidative stress, starvation and caloric restriction with the final effect of increasing lifespan and prevent aging-related diseases, such as diabetes and cancer (10).
The human FOXO3 gene comprises four exons and three introns, but only exon 2 and 3 are actively transcribed and translated into FOXO3 protein (see Figure 1A). Intriguingly, intron 2 is very large (101 625 bp) and contains a p53 binding site which is able to activate FOXO3 expression in response to DNA damage (11). Furthermore, several single nucleotide polymorphisms (SNPs) were characterized in this intron and used for association studies with longevity and age-related diseases in humans. Importantly, all the previously reported FOXO3 coding variants failed to show any role in longevity (12). Of note, among non-coding SNPs the rs2802292 minor allele (G), located at intron 2, has been shown to be strongly associated with long-lived subjects in all human populations tested (13–20) (collectively 5746 subjects >90 years of age and 6554 controls – see Supplementary Table S1), and its copy number correlated well with healthy aging phenotypes and lower prevalence of age-related diseases (13). We recently showed that an inverse correlation exists between rs2802292 G-allele copy number and cancer risk in patients with hamartomatous polyposis syndromes that are caused by mutations in STK11 (Peutz-Jeghers syndrome) or PTEN (PTEN hamartoma tumor syndrome) (21), two genes whose protein products are proximate upstream regulators of FOXO3 (10). At the molecular level, preliminary data seem to indicate that the intronic rs2802292 G-allele correlates with increased basal expression of FOXO3 (21,22), suggesting the hypothesis that FOXO3 intron 2 may act as a regulatory region.
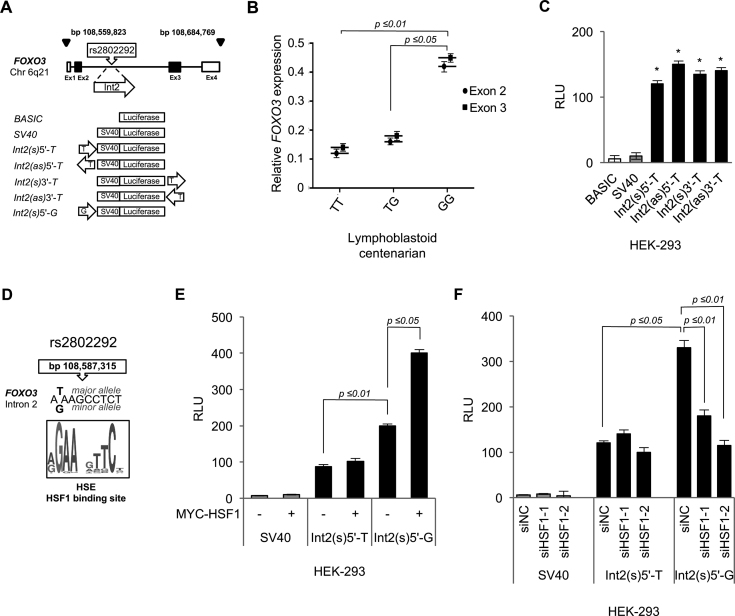
Figure 1.
The rs2802292 locus has enhancer properties and the presence of the G-allele creates a unique binding site for HSF1, which is required for transcriptional activation. (A) Top: structure of the human FOXO3 gene. The four exons are shown as white (non-coding) or dark (coding) boxes. Bottom: schematic representation of the luciferase constructs generated. (B) Expression levels of FOXO3 exon 2 and 3, as measured by quantitative PCR (qPCR) in cell lines established from centenarians that were homozygous for the rs2802292 T-allele (TT, n = 3), heterozygous (TG, n = 3) or homozygous for the rs2802292 G-allele (GG, n = 3). (C) Luciferase reporter assays (n = 3) measuring the enhancer activity of the indicated constructs in HEK-293 cells. Along the y-axis, relative luciferase units (RLU) are normalized to Renilla luciferase signal. (D) HSF1 recognition motif identified in the rs2802292 SNP minor G-allele. (E) Enhancer activity of the indicated constructs in the presence or absence of exogenous MYC-tagged-HSF1, as assessed by luciferase reporter assays (n = 3) in HEK-293 cells. (F) Luciferase reporter assays (n = 3) measuring the effect of HSF1 silencing by siRNAs on the enhancer activity of the indicated constructs. siNC, siRNA negative control. Two different siRNAs for HSF1 (siHSF1-1 and siHSF1-2) were tested. The presented results are representative of at least three independent experiments. P-values were derived from t-tests: *P ≤ 0.05.
Here, we show that the 90-bp sequence around the intronic SNP rs2802292 at FOXO3 locus has enhancer properties, and that HSF1 binding to this region through a novel HSE binding site created by the rs2802292 minor G-allele promotes expression of FOXO3 in response to diverse stress stimuli. Molecular analyses carried out in various established cell lines, primary cultures and human near-haploid HAP1 isogenic cells (G/T) generated by the CRISPR/Cas9 system for genome editing revealed that HSF1 mediates the occurrence of physical and functional interactions between the 5′UTR and the rs2802292 region at FOXO3 locus thereby activating their enhancer functions. At last, we performed functional studies showing that the identified regulatory intronic region is involved in cellular stress response and lifespan regulation.
MATERIALS AND METHODS
Cell lines, primary culture of biopsies and reagents
HEK-293 and OVCAR-3 cells (both from ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, 11966025), HAP1 FOXO3 SNP rs2802292 (G to T) and HAP1 parental (G) cell lines (from Invitrogen GeneArt Engineered Cell Models Collection, ID HZGHK481 and ID C631, respectively) were grown in Iscove's Modified Dulbecco's medium (IMDM) (Gibco, 12440-053), supplemented with 10% Fetal Bovine Serum (FBS) (Gibco, 25030081), 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco, 15140122), 2 mM L-glutamine (HEK-293) (Gibco, 25030081) and 1% non-essential amino acids in Medium Essential Medium (MEM) solution (HEK-293) (Gibco, 11140035).
Lymphoblastoid cell lines and primary dermal fibroblasts are archival samples from the UOC Laboratory of Medical Genetics at the University of Bari ‘Aldo Moro’. Lymphoblastoid cell lines were generated by transformation with Epstein–Barr virus (23) of peripheral B lymphocytes derived from centenarian donors (≥100 years). Primary dermal fibroblasts were obtained from skin biopsies of healthy donors (24). All donors signed a regular informed consent for research purposes approved by the local Ethics Committee (AOU Policlinico, 70124 Bari, Italy) in compliance with international and national data protection laws.
Lymphoblastoid cell lines and human primary dermal fibroblasts were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, 11875-093) supplemented with 10% FBS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. All cell lines were maintained at 37°C and 5% CO2 avoiding confluence at any time. Human primary fibroblasts were used for experiments at the second passage. KRIBB11 (Calbiochem, 385570) was purchased from Merck. H2O2 (H1009) and Menadione (M2518) were purchased from Sigma.
CRISPR/Cas9-mediated genome editing
HAP1 FOXO3 SNP rs2802292 (G to T) cells were generated by a GeneArt Engineered Cell Model service based on the near-haploid human cell line HAP1 using CRISPR/Cas9-mediated genome editing. Briefly, a double strand break is introduced by targeting the CRISPR/Cas9 to a specific site, in the presence of a suitably designed homologous donor sequence. gRNAs and oligo donor sequences used for engineering are available from the authors upon request.
Genotyping
Genomic DNA from cell lines and primary fibroblasts was extracted using QIAsymphony SP/AS instruments (QIAGEN) according to the manufacturer’s protocol and quantified on a NanoDROP 2000 spectrophotometer (Thermo Scientific). Polymerase chain reactions (PCRs) were carried out in 25 μl reaction mixtures containing 50 ng of genomic DNA, 1× PCR buffer (Tris–HCl, (NH4)2SO4, 15 mM MgCl2; pH 8.7), 200 μM dNTPs (Thermo Fisher Scientific, R0181) and 0.5 U Taq DNA Polimerase Recombinant (Thermo Fisher Scientific, EP0405) and the following primers (10 pmol each): FOXO3 rs2802292g/t Fw and FOXO3 rs2802292g/t Rv. PCR amplification cycles were carried out at 95°C for 15 min followed by 29 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min and extension at 72°C for 1 min, and by a final extension at 72°C for 10 min on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems). A total of 5 μl of the amplified products were loaded onto 2% Agarose Standard Low EEO (Sigma, A9539) in 0.5× Tris-Borate-EDTA (TBE) (Serva, 39320.01) and visualized using GelRed™ (Biotium, 41003). Sequencing products were purified by use of the DyeEx™ 2.0 Spin Kit (QIAGEN, 63206) and sequenced on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Primer sequences are listed in ‘Additional Material’.
Quantitative real-time PCR
Total RNAs were extracted using TRI Reagent (Sigma, 93289). Samples were treated with DNase-1 (Ambion, AM2222) and retro-transcribed using the Superscript Vilo Master Mix (Invitrogen, 11755-050). PCRs were carried out using the SsoAdvanced Universal SYBR Green PCR Super Mix (Biorad, 172–5272) on a CFX Connect™ Real-Time PCR Detection System (Bio-Rad). Relative quantification was done using the ΔΔCt method. Primer sequences are listed in ‘Additional Material’.
Immunoblotting
Immunoblots were carried out as previously described (25). A total of 20 μg of protein extracts from each sample were denatured in 4× Laemmli sample buffer (Biorad, 161–0747) and loaded into a Sodium dodecyl sulphate-polyacrylamide gel for immunoblot analysis. Immunoblots were performed using anti-ACTB (Sigma, A2066), anti-HSF1 (Cell Signaling, 4356), anti-MYC Tag (Cell Signaling, 2278), anti-HSPA1A (Cell Signaling, 4872) and anti-FOXO3 (Cell Signaling, 2497). Immunoblots were developed with the ECL-plus chemiluminescence reagent (GE Healthcare, RPN2232) according to the manufacturer’s instructions.
Cell transfection and RNA interference
Transient transfection experiments were performed using Lipofectamine 3000 (Thermo Fisher Scientific, L3000075) according to the manufacturer’s instructions. A total of 500 ng of plasmid were used for transfection experiments. All cell lines were transfected at 80% confluence and cultured for an additional 48 h to express the plasmids before performing the experiments.
For RNA interference, cells were transfected with 5 nM siRNAs directed against HSF1 (siHSF1-1, Ambion, validated oligos AM16704; siHSF1-2, Stealth, D1016853) using Lipofectamine 3000. On-TARGET-plus control 5 nM siRNAs (siNC, Thermo Scientific, D-001810-10-20) were used as control sequences. siRNA sequences are listed in ‘Additional Material’.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed using the MAGnify Chromatin Immunoprecipitation System (Thermo Fisher Scientific, 49–2024) as previously described (26). IgG antibodies were included in the kit and anti-HSF1 (Abcam ChIP Grade, AB2923), anti-MYC-tag (Cell Signaling, 2278), anti-histone H3-acetyl K27 (Abcam ChIP Grade, AB4729), anti-histone H3-mono methyl K4 (Abcam ChIP Grade, AB8895) antibodies were used according to the manufacturer’s instructions. The set of primers used for ChIP allows amplification of target regions including HSF1 binding sites at intron 2 and at the 5′UTR region of the locus. FOXO3 exon 3 region was used as a control (sequences are listed in ‘Additional Material’).
ChIP-loop assay
The ChIP-loop assay was carried out by combining the ChIP procedure as previously described with the Chromosome Conformation Capture (3C) procedure as performed by Gondor et al. (27). Briefly, chromatin was cross-linked with paraformaldehyde 1% (Sigma, P6148) and digested with Csp6I (Fermentas, ERO211) for the 3C step. Cross-linked chromatin complexes were enriched by performing a ChIP with anti-HSF1 antibodies (Abcam, ab2923). Immunoprecipitated complexes were resuspended in T4 ligase buffer with 400 U of T4 DNA Ligase (NEB, MO202L). Ligation mixes were subjected to reverse cross-link at 65°C overnight. Circular DNA was isolated with exonuclease treatment, then purified with Qiagen PCR purification kit (Qiagen, 28106) according to the manufacturer’s instructions and eluted with 50 μl of 0.3× TE buffer containing 10 ng of Escherichia coli DNA. Ligated DNA was amplified with Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, F530S) by performing 1 cycle at 98°C for 30 s and 35 cycles at 98°C for 10 s, 60°C for 30 s and 72°C for 30 s, followed by 5 min at 72°C. We used five different couples of primers to analyze five possible ligation products. Upon Csp6I digestion and ligation, we expected to obtain specific PCR products using primers matching fragments that could be in close proximity. Primer sequences are listed in ‘Additional Material’.
Cloning
We constructed luciferase reporter plasmids by cloning 90 nucleotides located around the SNP into the pGL3-Promoter Vector (Promega) upstream to the SV40 promoter (Int2(s)5′-T, Int2(as)5′-T, Int2(s)5′-G) or downstream to the Luciferase gene (Int2(s)3′-T, Int2(as)3′-T). Sequences were cloned in the antisense and sense orientation relative to the SV40 promoter. For the Int2(s)5′-T, Int2(as)5′-T, Int(s)5′-G plasmids, we introduced a KnpI site at the 5′-end and a XhoI site at the 3′-end of the 90 nucleotide sequences, after which we inserted them into a vector that was digested with KnpI and XhoI restriction enzymes (NEB, R0142S and R0146L, respectively). For the Int2(s)3′-T, Int2(as)3′-T plasmids, we introduced a SalI site at the 5′-end and a BamHI site at the 3′-end of the 90 nucleotide sequences, after which we inserted them into a vector that was digested with SalI and BamHI restriction enzymes (NEB, R0138L and R0136L, respectively). The pGL3 plasmid without the enhancer region (BASIC) and with the SV40 promoter alone (SV40) were used as controls. A pcDNA-MYC-HSF1 mammalian expression clone of human HSF1 and the pcDNA, PGL3 basic, pRL and FHRE-Luc plasmids were all purchased from Addgene. Sequences used for cloning are listed in ‘Additional Material’.
Luciferase assay
Cells were transfected into a 24-well plate with either the empty vector (pcDNA) and/or the pcDNA-MYC-HSF1 expression constructs. After 24 h, cells were transfected with the luciferase reporter plasmid. About 20–24 h after transfection, cells were washed with phosphate-buffered saline and the medium was replaced with 100 μl Passive Lysis 1× (Promega, E1910) according to the manufacturer’s instructions. Firefly and Renilla luminescence were measured with SPECTRO star Omega (BMG Labtech). Firefly luciferase/Renilla luciferase ratios were calculated for each well.
Assays for intracellular ROS levels
Intracellular Reactive Oxygen Species (ROS) levels were evaluated with the Image-iT Live Green Reactive Oxygen Species Detection Kit (Invitrogen, I36007) according to the manufacturer’s instructions. Briefly, cells were stained with 5-(and-6)-carboxy-2′,7′dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA) for 30 min and analyzed for intracellular ROS levels under an Axio Observer Z1 fluorescence microscope (Zeiss). Zen Image software (Zeiss) was used for quantitative analysis of fluorescent intensity. Background taken just outside the cells was subtracted from each image.
Cellular glutathione assay
The GSH/GSSG-Glo assay (Promega, V6611) was used to measure the change in the redox state of the cells due to oxidants, which are downstream metabolites of O2−. The assay was performed according to the manufacturer’s instructions.
DNA damage assay
The DNA Damage Quantification Kit (Abnova, KA0784) was used to measure the level of apurinic/apyrimidinic (AP) sites, an indicator of DNA lesion and repair against chemical damage and cell aging. The assay was performed according to the manufacturer’s instructions.
Cell proliferation assay
Colony formation assays were performed as previously described (28). Briefly, cells were seeded and grown in 60 mm dishes 24 h before treatment with different glucose concentrations. The medium in the dishes was changed every 3 days. Cells were then fixed and stained using Coomassie Brilliant Blue (Biorad, 161–0400). Percent cell growth inhibition at each glucose concentration was quantified by densitometric evaluation using Image J software. For proliferation assays, cells were seeded and grown into 96-well plates 24 h before treatment. Sulphorhodamine B (S1402, Sigma) assay (SRB) was carried out as described (29). Briefly, at the end of incubation, cultures were fixed with 10% trichloroacetic acid and stained with SRB at 0.4% (w/v) in 1% acetic acid for 30 min. Unbound SRB was washed out with 1% acetic acid and SRB-bound cells were solubilized with 10 mM Trizma base (Sigma, T1503). The absorbance was read at a wavelength of 540 nm using a microplate reader (BMG Labtech Spectrostar Omega). The survival fraction was calculated as the ratio of SRB absorbance of treated cells to SRB absorbance of control cells.
Statistical analysis
The statistical significance of the results was analyzed using Student’s t-tail test, and a *P-value < 0.05, **P < 0.005 and ***P < 0.001 were considered statistically significant.
RESULTS
The rs2802292 locus has enhancer properties and the presence of the G-allele creates a unique binding site for HSF1, which is required for transcriptional activation
To get insight into the role of the rs2802292 G-allele in the regulation of FOXO3, we first sequenced it in 14 lymphoblastoid cell lines established from centenarians and found that four of them were homozygous for the rs2802292 T-allele (TT), 7 were heterozygous (TG) and three were homozygous for the rs2802292 G-allele (GG) (Supplementary Table S2), thus reflecting the SNP distribution previously reported in long-lived subjects (13). The intronic localization of the rs2802292 SNP prompted us to ascertain whether it could affect regulation of FOXO3 splicing or expression of alternative starting sites at exon 3. Quantitative RNA analysis failed to show significant differences in the relative expression ratio of exon 2 to exon 3 in the three different genotypes (Figure 1B), indicating that the minor allele (G) does not influence the assembly of FOXO3 mRNA nor does it induce an alternative transcription start site. Importantly, the copy number of the minor G-allele correlated with increased levels of both exon 2 and exon 3 mRNAs, thus confirming a transcriptional regulatory role for this region, as previously hypothesized (21,22) (Figure 1B).
To explore the possibility that the rs2802292 region could regulate FOXO3 expression, we selected a 90-bp sequence in intron 2 (bp 108 587 283 to 108 587 372, referred to as Int2) located around the SNP major allele (T) and cloned it upstream (5′) to a PGL3-luciferase vector containing the SV40 promoter sequence (Figure 1A). Based on an in silico analysis, this sequence comprises transcription factor response elements for SP1, GATA1 and ESR1 (Supplementary Table S3). Preliminary analysis of the transcriptional ability of the Int2(s)5′-T construct revealed that this sequence could be a regulatory region (Figure 1C). Thus, we also cloned this fragment in the antisense (as) orientation upstream (5′) to the luciferase gene (Int2(as)5′-T), and in the sense (Int2(s)3′-T) and antisense (Int2(as)3′-T) orientations downstream (3′) to the luciferase gene (Figure 1A). All these vectors were able to potently activate transcription in HEK-293 cells, indicating that this region of intron 2 has enhancer properties (Figure 1C).
To deepen our understanding of the functional significance of the G to T change at genomic position 108 587 315 in the rs2802292 SNP, we carried out an in silico analysis on this sequence to search for additional transcription factor response elements and found that the presence of a G creates a unique binding site for HSF1 (Figure 1D). This finding prompted us to test the activity of the construct containing a G (Int2(s)5′-G) instead of a T (Int2(s)5′-T) at position corresponding to bp 108 587 315 (Figure 1A). Intriguingly, the Int2(s)5′-G variant significantly enhanced transcriptional activity compared with the Int2(s)5′-T vector both in basal conditions and upon HSF1 overexpression (Figure 1E and Supplementary Figure S1A). Moreover, genetic silencing of HSF1 by two different siRNAs reduced the activity of the Int2(s)5′-G variant to the levels observed with the Int2(s)5′-T vector, confirming that endogenous HSF1 is required for enhanced transcriptional activation (Figure 1F and Supplementary Figure S1B).
HSF1 binds to the genomic rs2802292 enhancer region containing the SNP minor G-allele and induces expression of FOXO3 in a stress-dependent manner
Consistent with the role of HSF1 in stress resistance, the Int2(s)5′-G vector was significantly activated by H2O2-oxidative stress and heat shock in an HSF1-dependent manner (Figure 2A and B; Supplementary Figure S2). Indeed, the Int2(s)5′-T vector failed to respond to either type of stress, and genetic silencing (Figure 2A and Supplementary Figure S2A) or pharmacological inhibition (Figure 2B and Supplementary Figure S2B) of HSF1 abrogated stress-dependent Int2(s)5′-G transcriptional activation.
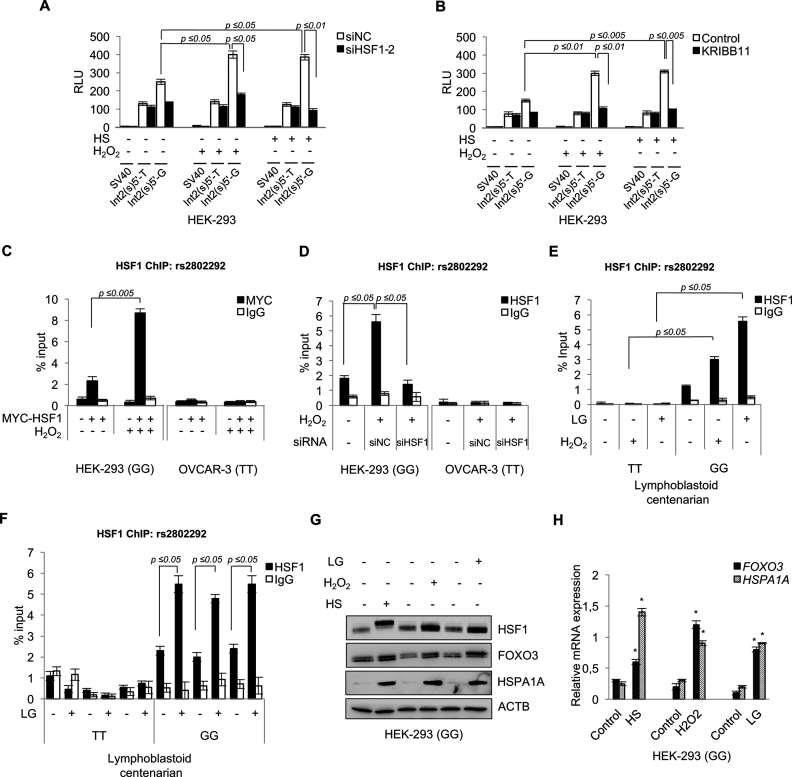
Figure 2.
HSF1 binds to the genomic rs2802292 SNP region in cells homozygous for the rs2802292 G-allele and induces the expression of FOXO3 in a stress-dependent manner. Luciferase reporter assays (n = 3) measuring the effect of HSF1 silencing by siRNA (siHSF1-2) (A) or HSF1 chemical inhibition (1 h KRIBB11, 10 μM) (B) on the enhancer activity of the rs2802292 SNP region upon hydrogen peroxide (1 h H2O2, 100 μM) or heat shock (HS) (2 h at 43°C) treatment. Cells (HEK-293) were collected and analyzed after 2 h of recovery from the indicated treatment. (C) ChIP assays (n = 3) of HSF1 occupancy of the genomic rs2802292 SNP region in HEK-293 cells homozygous for the rs2802292 G-allele (GG) in the presence or absence of exogenous MYC-tagged-HSF1 with and without H2O2 treatment (1 h, 100 μM). (D) ChIP assays (n = 3) testing the effect of HSF1 silencing by siRNA (siHSF1: siHSF1-2) on HSF1 binding to the genomic rs2802292 SNP region in HEK-293 cells homozygous for the rs2802292 G-allele (GG) and OVCAR-3 cells homozygous for the rs2802292 T-allele (TT) with and without H2O2 treatment (1 h, 100 μM). (E) ChIP analysis (n = 3) of HSF1 occupancy of the genomic rs2802292 SNP region in homozygous GG and TT cells originated from centenarians upon oxidative (1 h H2O2, 100 μM) and glucose restriction (LG) (10 h, 0.75 mM) stress stimuli. (F) ChIP analysis measuring the effect of LG (10 h, 0.75 mM) on HSF1 occupancy of the genomic rs2802292 SNP region in additional homozygous GG (n = 3) and TT (n = 3) cells originated from centenarians. (C–F) IgGs were used as an immunoprecipitation control. Expression levels of FOXO3, HSF1 and HSPA1A protein (G) and FOXO3 and HSPA1A transcript (H) were assessed by immunoblot and qPCR (n = 3), respectively, in HEK-293 cells homozygous for the rs2802292 G-allele subjected to stress conditions by heat shock (HS) (2 h at 43°C), hydrogen peroxide (1 h H2O2, 100 μM) and LG (24 h, 0.75 mM). P-values were derived from t-tests: *P ≤ 0.05.
ChIP analysis revealed that both exogenous MYC-tagged-HSF1 and endogenous HSF1 could bind to the genomic rs2802292 SNP region in HEK-293 cells homozygous for the rs2802292 G-allele (GG) (Figure 2C and D), but not in OVCAR-3 cells homozygous for the rs2802292 T-allele (TT) (Figure 2C and D). Moreover, binding of HSF1 to the genomic rs2802292 SNP region was strongly induced by oxidative stress in HEK-293 GG cells, and prevented by genetic depletion of HSF1 (Figure 2C and D). Similar results were obtained in GG cells, but not in TT cells, derived from centenarians (Figure 2E). Furthermore, stress conditions induced by glucose restriction (LG) (30) were also able to significantly induce HSF1 recruitment at the G-allele in cells derived from centenarians (Figure 2E). Importantly, the effect of LG was observed in all GG cells (n = 3), but not in TT cells (n = 3) (Figure 2F). We next performed a molecular analysis of homozygous GG HEK-293 cells subjected to cellular stress. Heat shock, H2O2 and LG were all able to activate HSF1 (Figure 2G) and to induce expression of the heat shock protein HSPA1A and of FOXO3 both at the protein and the mRNA level (Figure 2G and H).
Collectively, these results show that cells homozygous for the minor G-allele (GG) respond to oxidative stress, heat shock and glucose restriction by activating HSF1, which in turn binds to the rs2802292 SNP region at the unique HSE site and induces transcription of FOXO3.
HSF1 mediates the occurrence of a promoter–enhancer interaction at FOXO3 locus involving the 5′UTR and the rs2802292 region
Enhancers are distal sequences of DNA that can burst transcription of target genes by acting in cis, and their distance from target promoters may vary from 100 bp to Mb on the same chromosome (31). These sequences can lie upstream or downstream of the target gene and even in the gene body (introns), independently of orientation (32). Furthermore, enhancer–promoter looping has been shown to be required for gene activation (33), and chromatin of active enhancers is ‘marked' by the combination of H3K4me1/H3K27ac histone marks (34). Thus, we speculated that the rs2802292-containing enhancer region could functionally and physically interact with the FOXO3 promoter under stress conditions in an HSF1-dependent manner. To test this hypothesis, we first performed an in silico analysis of the 5′UTR region of FOXO3 locus, which revealed the presence of a putative HSF1 consensus motif at genomic positions 108 553 298–108 553 314 (Figure 3A). This finding prompted us to ascertain whether HSF1 could be recruited at this element while concurrently occupying the rs2802292 enhancer region in a stress-dependent manner. Our data show that, upon induction of oxidative stress, HSF1 could efficiently bind to both regions of FOXO3 locus in HEK-293 cells (GG) and in human dermal primary fibroblasts that were homozygous (GG) for the minor G-allele of the rs2802292 SNP (Figure 3B and E). Of note, we failed to detect HSF1 binding at the 5′UTR locus in human primary fibroblasts that were homozygous (TT) for the major T-allele, even after H2O2 treatment (Figure 3B and E). This suggests that HSF1 may bind to these regions in a coordinate manner, i.e. it can occupy its binding site at the 5′UTR only in cells where it is bound to the rs2802292 SNP region, which requires the presence of the minor G-allele.
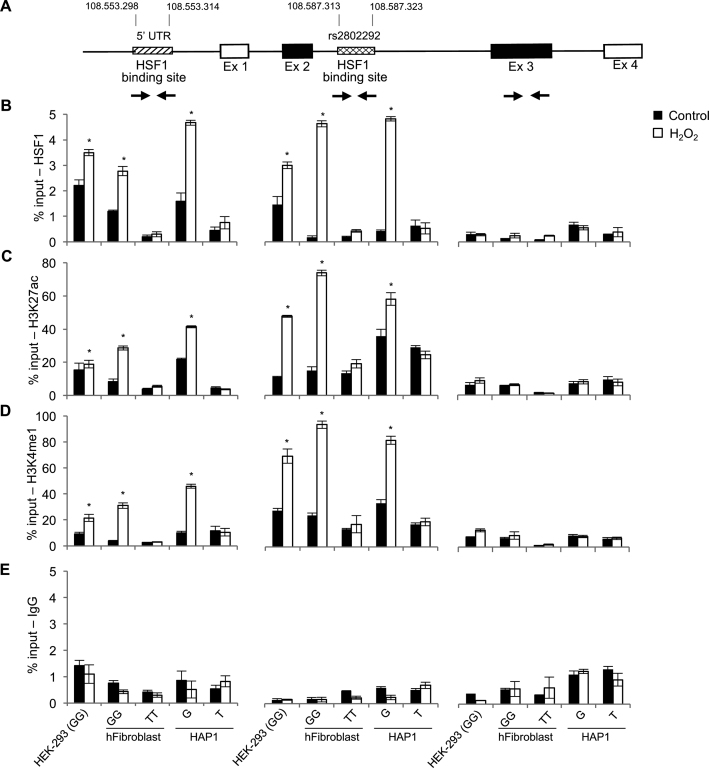
Figure 3.
HSF1 binds to both the 5′UTR promoter and the rs2802292 SNP enhancer region at FOXO3 locus only in cells containing the minor G-allele under oxidative stress. (A) Schematic representation of FOXO3 locus revealing the presence of a putative HSF1 consensus motif in the 5′UTR region at genomic positions 108 553 298–108 553 314. (B–E) ChIP experiments (n = 3) performed on HEK-293 cells homozygous for the rs2802292 G-allele (GG), homozygous GG and TT cells originated from human dermal primary fibroblasts and HAP1 FOXO3 SNP rs2802292 (G to T) and HAP1 parental (G) cells cultured in normal conditions and under oxidative stress (1 h H2O2, 100 μM) showing HSF1 occupancy of the 5′UTR region and of the genomic rs2802292 SNP region (B) and the involvement of activating epigenetic modifications such as acetylation of lysine 27 (H3K27ac) (C) and monomethylation of lysine 4 of histone H3 (H3K4me1) (D) on the 5′UTR and the rs2802292 region in the presence of the minor G-allele, but not in TT cells. IgGs were used as an immunoprecipitation control (E). Exon 3 region was used as a control. P-values were derived from t-tests: *P ≤ 0.05.
Further characterization of these cells indicated that, upon induction of oxidative stress, the rs2802292 SNP region is marked by activating epigenetic modifications typical of active enhancers involving acetylation of lysine 27 (H3K27ac) and monomethylation of lysine 4 of histone H3 (H3K4me1) only in the presence of the minor G-allele, and not in TT cells (Figure 3C–E). These data corroborate the hypothesis that the 5′UTR and the rs2802292 region of FOXO3 locus may act in cis in stressed cells carrying the minor G-allele.
Moreover, to address the role of the minor G-allele in chromatin structural organization and regulation, we first employed the CRISPR/Cas9 system for genome editing to generate human near-haploid HAP1 isogenic cell lines: HAP1 FOXO3 SNP rs2802292 (G to T) and HAP1 parental (G) cells (Supplementary Figure S3). Then, we performed a ChIP analysis of these cells subjected or not to oxidative stress. We found that HSF1 could bind to the rs2802292 region and the 5′UTR site only in HAP1 parental cells, due to the presence of the G, in a H2O2-dependent manner (Figure 3B and E). Consistently, also in this isogenic cell system, H2O2 induced the H3K27ac and H3K4me1 epigenetic modifications typical of active enhancers only in the presence of the minor G-allele (Figure 3C–E). These results suggest that HSF1 may mediate the occurrence of physical and functional interactions between the 5′UTR and the rs2802292 region at FOXO3 locus thereby activating their enhancer functions.
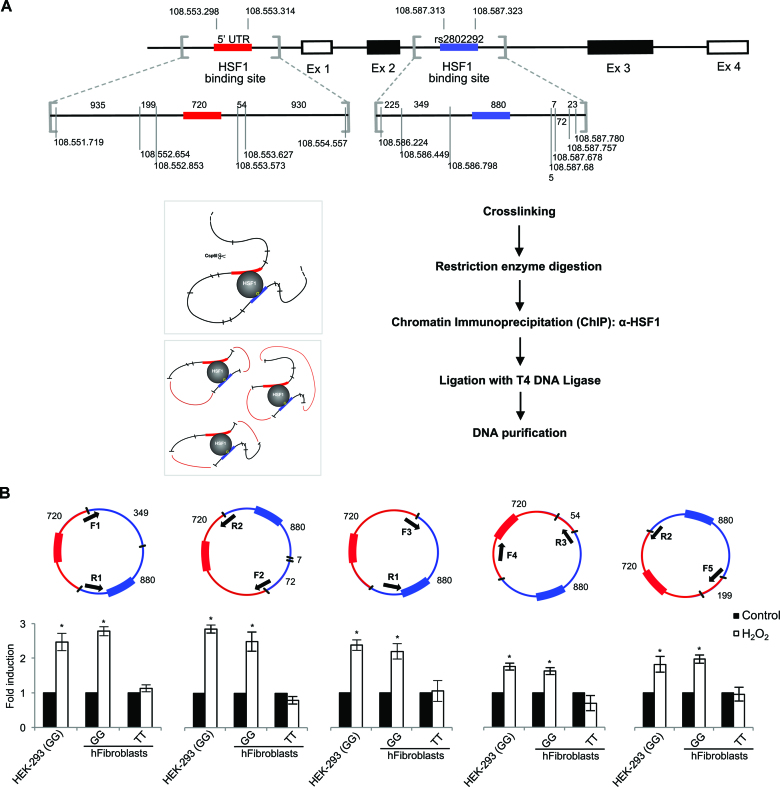
To further confirm this observation, we performed a ChIP-loop analysis of HEK-293 cells and primary human fibroblasts (GG and TT). H2O2-stressed cells were treated with formaldehyde to obtain cross-linked chromatin, after which DNA/protein complexes were first digested with the Csp6I restriction enzyme and then selected with anti-HSF1 antibodies; at last, ligation of chromatin fragments was carried out with the T4 DNA Ligase (Figure 4A). Purified DNA was analyzed by PCR with primers specific for the various possible combinations of ligated chromatin fragments (Figure 4B). Our data revealed that HSF1 complexes mediated the occurrence of a promoter–enhancer interaction involving the 5′UTR and the rs2802292 SNP region in cells homozygous for the minor G-allele (GG) that were subjected to oxidative stress (Figure 4B).
Figure 4.
HSF1 mediates the occurrence of a promoter–enhancer interaction at FOXO3 locus involving the 5′UTR and the rs2802292 region. (A) Top: physical map of the human FOXO3 gene. The scheme shows the Csp6I restriction enzyme sites flanking the baits (red for the 5′UTR and blue for the rs2802292 region). Bottom: schematic representation of the 3C and ChIP-loop assay. Crosslinked chromatin was digested with Csp6I and immunoprecipitated with anti-HSF1. The immunoprecipitated samples were diluted in a ligation buffer and ligated with the T4 DNA Ligase. After reversing the crosslinks, the ligated DNA was purified and amplified by PCR with various combinations of primers as indicated in (B). (B) This strategy allows the amplification of sequences ligated to the bait in circular DNA. The arrows indicate the positions of the primers within the bait sequence. Five different couples of primers were designed to analyze the five possible ligation products. Purified DNA was analyzed by PCR with primers specific for the various possible combinations of chromatin fragments. The values are the results of the densitometric analysis and are expressed as fold induction. HEK-293 cells and primary human fibroblasts (GG, n = 3; TT, n = 3) were collected after induction of oxidative stress (1 h H2O2, 100 μM). The presented results are representative of three independent experiments. P-values were derived from t-tests: *P ≤ 0.05.
Functional studies highlight the importance of the identified regulatory intronic region in stress response and lifespan regulation
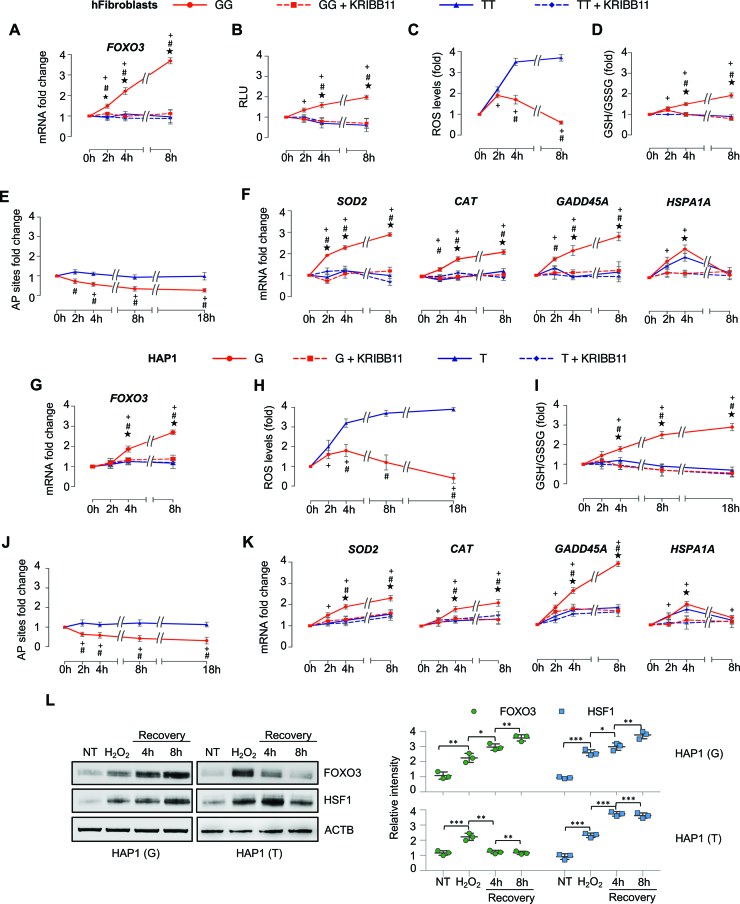
To determine the impact of the rs2802292 genotype in stress resistance phenotypes of human primary dermal fibroblasts, we treated homozygous GG and TT cells with H2O2 (or menadione, a potent oxidative stress inducer) (35) and characterized their oxidative stress response at different time points during recovery. We first measured the expression of FOXO3 in these cells and found that its mRNA levels were significantly increased in an HSF1- and time-dependent manner only in GG cells (Figure 5A and Supplementary Figure S4A). Then, we evaluated FOXO3-dependent transcription in these fibroblasts by transfection of a FOXO3 responsive vector (FHRE-Luc). Our data showed that stressed GG cells were able to induce significantly higher levels of FOXO3 transcriptional activity than TT fibroblasts in an HSF1-dependent manner (Figure 5B). These findings were confirmed by analysis of the expression of FOXO3 endogenous target genes involved in stress resistance, such as SOD2, CAT, GADD45A, CCND1, RBL2, BCL2L11 and BCL6. Indeed, stressed GG cells expressed higher mRNA levels of FOXO3 and its target genes in an HSF1-dependent manner, while TT fibroblasts did not (Figure 5F and Supplementary Figure S4B). Of note, no significant differences were detected between GG and TT cells when the expression of HSPA1A was analyzed, thus confirming that the G/T change could influence only FOXO3 transcription and not other HSF1 target genes (Figure 5F and Supplementary Figure S4A). The observed stress-dependent upregulation of SOD2 and CAT, two major antioxidant enzymes involved in ROS detoxification, prompted us to measure ROS levels in GG versus TT stressed primary fibroblasts. Our results revealed that GG cells could very efficiently lower ROS amounts after stress induction, while TT cells could not (Figure 5C). Consistently, GG fibroblasts showed a better overall response to oxidative stress than TT cells and this response was HSF1-dependent as shown by the ratio between reduced glutathione (GSH) and oxidized glutathione (GSSG) (Figure 5D). Indeed, after H2O2 withdrawal, GG cells exhibited a significant antioxidant activity lasting for up to 8 h, which was reduced to levels comparable to TT cells upon pharmacological inhibition of HSF1 (Figure 5D). Moreover, inhibition of HSF1 did not show a significant effect on TT-cell response (Figure 5D). At last, we quantified DNA damage by measuring the level of AP sites, an indicator of DNA lesion and repair against chemical damage and cell aging, showing that GG cells displayed a significantly better DNA repair response (AP site reduction) than TT cells (Figure 5E).
Figure 5.
The rs2802292 minor G-allele has a key role in the stress resistance phenotype of human cells. (A–F) Experiments performed on human primary dermal fibroblasts homozygous for the rs2802292 G-allele (GG, n = 3) or for the rs2802292 T-allele (TT, n = 3) treated with hydrogen peroxide (1 h H2O2, 100 μM) in the presence (where indicated) or absence of an HSF1 inhibitor (KRIBB11, 10 μM). Cells were collected and analyzed at the indicated time points during recovery. (A) Expression levels of FOXO3 as measured by qPCR. (B) Evaluation of FOXO3-dependent transcription by transfection of a FOXO3 responsive vector (FHRE-Luc). Data were obtained by a luciferase reporter assay. (C) Assays performed to measure intracellular ROS levels. (D) Cellular glutathione assay to measure oxidative stress response. The GSH/GSSG ratio indicates the redox status of the cells and is used as a marker for oxidative stress. (E) DNA damage quantification assay to measure the level of DNA lesion and repair. AP sites were used as an indicator of DNA damage. (F) Expression levels of FOXO3 target genes involved in stress resistance (SOD2, CAT, GADD45A, HSPA1A) as measured by qPCR. (G–L) Experiments performed on HAP1 (T, n = 3) and HAP1 (G, n = 3) cells treated with hydrogen peroxide (1 h H2O2, 100 μM) in the presence (where indicated) or absence of an HSF1 inhibitor (KRIBB11, 10 μM). Cells were collected and analyzed at the indicated time points during recovery. (G) Expression levels of FOXO3 as measured by qPCR. (H) Assays performed to measure intracellular ROS levels. (I) Cellular glutathione assay to measure oxidative stress response. (J) DNA damage quantification assay to measure the level of DNA lesion and repair. (K) Expression levels of FOXO3 target genes involved in stress resistance (SOD2, CAT, GADD45A, HSPA1A) as measured by qPCR. (L) Immunoblot analysis (left panel) and densitometric analysis after normalization against the loading control (right panel) of FOXO3 and HSF1 protein expression levels in HAP1 (T) and HAP1 (G) cells at the indicated time points during recovery from treatment with hydrogen peroxide (1 h H2O2, 100 μM). All experiments were repeated three to six times. P-values were derived from t-tests: ★P ≤ 0.05: GG relative to GG+KRIBB11. #P≤ 0.05: GG relative to TT. +P ≤ 0.05: GG relative to 0 h time point. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001.
Then, to further confirm the role of the minor G-allele in oxidative stress response, we employed the HAP1 isogenic cell lines described above. We found that only parental (G) cells could robustly activate FOXO3 mRNA expression upon H2O2 exposure (Figure 5G). Moreover, consistent with our previous observations, the presence of the minor G-allele promoted greater ROS detoxification (Figure 5H), antioxidant activity (GSH/GSSG ratio) (Figure 5I) and DNA damage repair (measured by AP site reduction) in cells subjected to oxidative stress (Figure 5J). These results were corroborated by the upregulation of FOXO3 target genes involved in stress resistance (SOD2, CAT) and DNA repair (GADD45A) in an HSF1-dependent manner (Figure 5K) and specificity was ensured by the analysis of HSPA1A expression which was independent of the G/T rs2802292 status (Figure 5K). At last, we performed a protein analysis of HAP1 FOXO3 SNP rs2802292 (G to T) and HAP1 parental (G) cells subjected to H2O2 treatment. Oxidative stress activated HSF1 in both cell lines, but a robust and persistent expression of FOXO3 could only be observed in HAP1 parental (G) cells (Figure 5L).
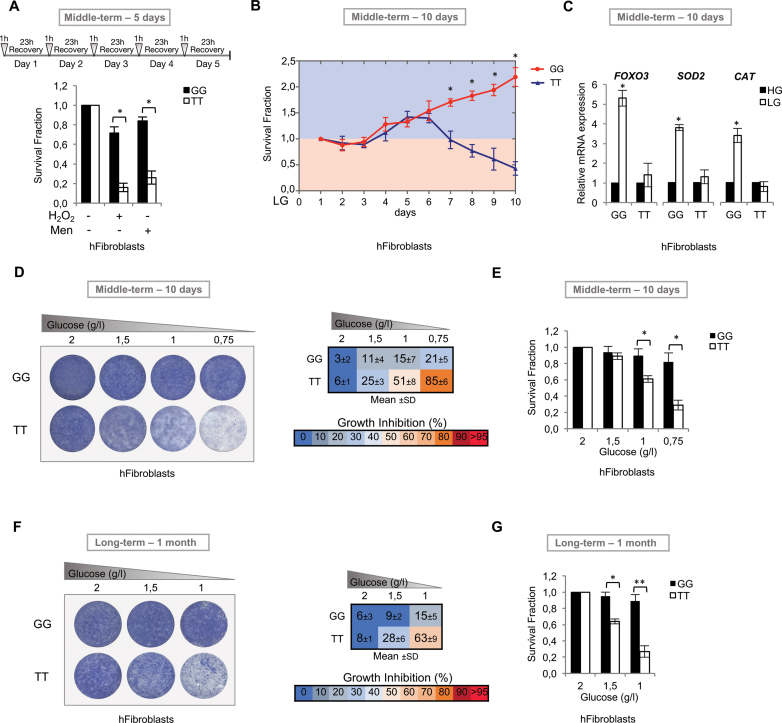
These data were further supported by the observation that human GG fibroblasts showed a better survival rate than TT cells under oxidative stress conditions in short-term cultures only in the presence of active HSF1 (Supplementary Figure S5). Thus, we decided to extend these findings by subjecting human primary dermal fibroblasts (GG versus TT) to different types of chronic stress. First, we treated GG and TT cells with a 1 h pulse of H2O2 or menadione followed by 23 h of recovery and repeated this scheme for up to 5 days. Primary human GG fibroblast survival fractions remained significantly high (around 70–80%), while TT cell survival fractions eventually declined (Figure 6A). Then, we cultured cells under glucose restriction (LG). As shown in Figure 6B, fibroblasts homozygous for the minor G-allele were able to survive and even proliferate in LG conditions, while TT cells underwent death. Molecular analysis performed in these cells confirmed LG-dependent upregulation of FOXO3 and its target genes involved in stress resistance (SOD2, CAT) only in GG cells (Figure 6C). Moreover we cultured these cells for up to 10 days (Figure 6D and E) or 1 month (Figure 6F and G) under different glucose concentrations (2, 1.5, 1, 0.75 g/l) and measured both the percentage of growth inhibition (Figure 6D and F) and the survival fraction (Figure 6E and G). Collectively, middle-term (10 days) and long-term (1 month) culture experiments unequivocally showed that human primary GG fibroblasts can survive and grow in glucose shortage conditions, while TT-cell survival was dependent upon glucose concentration (Figure 6).
Figure 6.
The rs2802292 minor G-allele is involved in lifespan regulation of human cells. (A) Survival fraction of human primary dermal fibroblasts (GG, n = 3; TT, n = 3). Cells were treated with a pulse of hydrogen peroxide (1 h, 100 μM) or menadione (1 h, 20 μM) followed by 23 h of recovery for up to 5 days and analyzed at the indicated time points. The effect was determined using the SRB assay. (B) Survival fraction of human primary dermal fibroblasts (GG, n = 3; TT, n = 3). Cells were cultured in low glucose (LG, 0.75 g/l) and analyzed at the indicated time points. The effect was determined using the SRB assay. (C) Expression levels of FOXO3 and FOXO3 target genes involved in stress resistance (SOD2, CAT) as measured by qPCR in human primary dermal fibroblasts (GG, n = 3; TT, n = 3) in middle-term (10 days) culture experiments under high (HG, 2 g/l) or LG (0.75 g/l) conditions. (D and E) Middle-term (10 days) and (F and G) long-term (1 month) culture experiments on human primary dermal fibroblasts (GG, n = 3; TT, n = 3) grown under the indicated glucose concentrations. The effect on cell survival is dependent on glucose concentration as shown by colony formation assays (D and F) and by determination of the survival fraction using the SRB assay (E and G). Cell growth percent inhibition at each glucose concentration is presented in D and F. All experiments were performed in triplicate and repeated at least three times. Data are expressed as the increase or decrease percentage with respect to controls (HG). P-values were derived from t-tests: *P ≤ 0.05; **P ≤ 0.005.
Taken together, these results show that GG cells display a better response to oxidative and metabolic stress than TT cells, resulting in increased survival under glucose restriction in both middle-term and long-term cultures.
DISCUSSION
Aging is characterized by the accumulation of cellular and genetic damage throughout life (36). This time-dependent process is modulated at least in part by exposure to stress and by the resulting effects on gene expression. Evidence gathered from fundamental science studies has revealed that exposure to certain low-intensity or short-term stressors can be beneficial and promote longevity as it enhances the activity of molecular chaperones and other recovery mechanisms thereby boosting the stress response of the cell. Inversely, prolonged or high-intensity stressor exposure is detrimental and reduces lifespan as it causes increases in oxidative stress, inflammation, stress hormones and insulin and can overcome compensatory mechanisms (37).
The use of model organisms has uncovered the existence of evolutionarily conserved pathways related to aging, and highlighted that genes regulating endocrine signaling, stress responses and metabolism can all promote increased lifespan. Moreover, these studies showed that various age-related diseases are affected by genes involved in longevity pathways thus suggesting that a link exists between aging and disease susceptibility (2). Overall, these findings indicate that longevity is regulated by a complex genetic network governing stress resistance and genomic stability, and FOXO3 is an important player in this process (38).
Indeed, the minor G-allele of FOXO3 SNP rs2802292 is strongly associated with longevity in humans (13–19), and its copy number showed a positive correlation with reduced age-related disease susceptibility (13). Furthermore a recent large, prospective cohort study revealed that the strong survival advantage seen in carriers of the rs2802292 minor G-allele is associated with a mortality risk reduction for coronary heart disease (CHD). The observation that allelic variation in FOXO3 affects this risk may have important clinical implications as CHD is the major cause of death in industrialized countries (39,40). Emerging evidence strongly suggests that CHD is a manifestation of a chronic inflammatory response associated with systemic metabolic perturbations leading to cellular and endoplasmic reticulum stress due to oxidative stress, hypoxia and glucose deprivation (41–45). Of note, FOXO3 activity might counteract all these stressors at both the cellular and systemic level. Indeed, FOXO3 regulates glucose homeostasis and lipid metabolism (46–50) in the liver-muscle axis and is an evolutionarily conserved major sensor of endoplasmic reticulum stress (51,52).
Overall, our results show for the first time that different types of cellular stresses induce the recruitment of HSF1 on a unique DNA binding site that is created by the presence of a G at the FOXO3 intronic SNP rs2802292. This region has enhancer properties and its activation triggers the upregulation of FOXO3. These data are in agreement with a recent work conducted by Donlon and colleagues, in which they provided evidence that, when stimulated by oxidative stress, the FOXO3 locus could activate several cis-regulating elements located at intron 2. Moreover, FOXO3 became physically connected, through chromatin looping, with 46 other genes on chromosome 6 thus forming a larger functional unit that could represent a chromatin domain defining an aging hub (53). In addition, two intronic FOXO3 SNPs, one located at intron 2 (rs12206094) and the other at intron 3 (rs4946935), were recently associated with increased FOXO3 expression and greater transactivation activity in a luciferase assay (54).
The stress- and HSF1-dependent increase in FOXO3 expression levels activates its antioxidant and DNA repair transcriptional program leading to increased tolerance to stress in short-, middle- and long-term cultures. These findings shed light on the role of FOXO3 rs2802292 minor G-allele in long-lived subjects and might significantly further our understanding of the molecular mechanisms involved in healthy aging and reduced susceptibility to age-related diseases, in particular CHD, diabetes and obesity. Moreover, we believe that the rs2802292 minor G-allele may serve as an important predictive marker for prognosis and therapy response in several conditions triggering oxidative stress response in cells and tissues.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Francesco Paolo Jori for his helpful discussion during the preparation of the manuscript and editorial assistance.
Authors’ contribution: C.S. designed the research and wrote the manuscript with the contribution of V.G.; V.G., G.F., P.S., A.P., T.T., M.L.S., C.F., R.B., R.L., D.C.L., F.C.S. and N.R. conducted the biological and computational assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC) ‘Investigator Grant 2014′ [IG15696 to C.S., in part]; Italian Ministry of Health ‘GIOVANI RICERCATORI GRANT 2011-2012′ [GR-2011-02351968 to C.S.]; Italian Ministry of Health ‘RICERCA FINALIZZATA GRANT 2011-2012′ [RF-2011-02352088 to C.S.]. Funding for open access charge: AIRC [IG15696].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fontana L., Partridge L., Longo V.D.. Extending healthy life span-from yeast to humans. Science. 2010; 328:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005; 120:449–460. [DOI] [PubMed] [Google Scholar]
- 3. Longo V.D., Fabrizio P.. Regulation of longevity and stress resistance: a molecular strategy conserved from yeast to humans. Cell. Mol. Life Sci. 2002; 59:903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsu A.L., Murphy C.T., Kenyon C.. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003; 300:1142–1145. [DOI] [PubMed] [Google Scholar]
- 5. Vihervaara A., Sistonen L. HSF1 at a glance. J. Cell Sci. 2014; 127:261–266. [DOI] [PubMed] [Google Scholar]
- 6. Akerfelt M., Morimoto R.I., Sistonen L.. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell. Biol. 2010; 11:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martindale J.L., Holbrook N.J.. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 2002; 192:1–15. [DOI] [PubMed] [Google Scholar]
- 8. Fulda S., Gorman A.M., Hori O., Samali A.. Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010; 2010:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morimoto R.I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998; 12:3788–3796. [DOI] [PubMed] [Google Scholar]
- 10. Chiacchiera F., Simone C.. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010; 9:1091–1096. [DOI] [PubMed] [Google Scholar]
- 11. Renaul V.M., Thekkat P.U., Hoang K.L., White J.L., Brady C.A., Kenzelmann Broz D., Venturelli O.S., Johnson T.M., Oskoui P.R., Xuan Z. et al. The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene. 2011; 30:3207–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donlon T.A., Curb J.D., He Q., Grove J.S., Masaki K.H., Rodriguez B., Elliott A., Willcox D.C., Willcox B.J.. FOXO3 gene variants and human aging: coding variants may not be key players. J. Gerontol. A Biol. Sci. Med. Sci. 2012; 67:1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willcox B.J., Donlon T.A., He Q., Chen R., Grove J.S., Yano K., Masaki K.H., Willcox D.C., Rodriguez B., Curb J.D.. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:13987–13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anselmi C.V., Malovini A., Roncarati R., Novelli V., Villa F., Condorelli G., Bellazzi R., Puca A.A.. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009; 12:95–104. [DOI] [PubMed] [Google Scholar]
- 15. Flachsbart F., Caliebe A., Kleindorp R., Blanché H., von Eller-Eberstein H., Nikolaus S., Schreiber S., Nebel A.. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2700–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., Wang W.J., Cao H., Lu J., Wu C., Hu F.Y., Guo J., Zhao L., Yang F., Zhang Y.X. et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum. Mol. Genet. 2009; 18:4897–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pawlikowska L., Hu D., Huntsman S., Sung A., Chu C., Chen J., Joyner A.H., Schork N.J., Hsueh W.C., Reiner A.P. et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009; 8:460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soerensen M., Dato S., Christensen K., McGue M., Stevnsner T., Bohr V.A., Christiansen L.. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010; 9:1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun L., Hu C., Zheng C., Qian Y., Liang Q., Lv Z., Huang Z., Qi K., Gong H., Zhang Z. et al. FOXO3 variants are beneficial for longevity in Southern Chinese living in the Red River Basin: a case-control study and meta-analysis. Sci. Rep. 2015; 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soerensen M., Nygaard M., Dato S., Stevnsner T., Bohr V.A., Christensen K., Christensen L.. Association study of FOXO3A SNPs and aging phenotypes in Danish oldest-old individuals. Aging Cell. 2015; 14:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forte G., Grossi V., Celestini V., Lucisano G., Scardapane M., Varvara D., Patruno M., Bagnulo R., Loconte D., Giunti L. et al. Characterization of the rs2802292 SNP identifies FOXO3A as a modifier locus predicting cancer risk in patients with PJS and PHTS hamartomatous polyposis syndromes. BMC Cancer. 2014; 14:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banasik K., Ribel-Madsen R., Gjesing A.P., Wegner L., Andersson A., Poulsen P., Borglykke A., Witte D.R., Pedersen O., Hansen T. et al. The FOXO3A rs2802292 G-allele associates with improved peripheral and hepatic insulin sensitivity and increased skeletal muscle-FOXO3A mRNA expression in twins. J. Clin. Endocrinol. Metab. 2011; 96:E119–E124. [DOI] [PubMed] [Google Scholar]
- 23. Hui-Yuen J., McAllister S., Koganti S., Hill E., Bhaduri-McIntosh S.. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. Vis. Exp. 2011; 57:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loconte D.C., Grossi V., Bozzao C., Forte G., Bagnulo R., Stella A., Lastella P., Cutrone M., Benedicenti F., Susca F.C. et al. Molecular and functional characterization of three different postzygotic mutations in PIK3CA-Related overgrowth spectrum (PROS) patients: Effects on PI3K/AKT/mTOR signaling and sensitivity to PIK3 inhibitors. PLoS One. 2015; 27:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiacchiera F., Simone C.. Signal-dependent control of autophagy-related gene expression. Methods Enzymol. 2009; 453:305–324. [DOI] [PubMed] [Google Scholar]
- 26. Peserico A., Germani A., Sanese P., Barbosa A.J., Di Virgilio V., Fittipaldi R., Fabini E., Bertucci C., Varchi G., Moyer M.P. et al. A SMYD3 small-molecule inhibitor impairing cancer cell growth. J. Cell. Physiol. 2015; 230:2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gondor A., Rougier C., Ohlsson R.. High-resolution circular chromosome conformation capture assay. Nat. Protoc. 2008; 3:303–313. [DOI] [PubMed] [Google Scholar]
- 28. Germani A., Matrone A., Grossi V., Peserico A., Sanese P., Liuzzi M., Palermo R., Murzilli S., Campese A.F., Ingravallo G. et al. Targeted therapy against chemoresistant colorectal cancers: Inhibition of p38α modulates the effect of cisplatin in vitro and in vivo through the tumor suppressor FoxO3A. Cancer Lett. 2014; 344:110–118. [DOI] [PubMed] [Google Scholar]
- 29. Holford J., Sharp S.Y., Murrer B.A., Abrams M., Kelland L.R.. In vitro circumvention of cisplatin resistance by the novel sterically hindered platinum complex AMD473. Br. J. Cancer. 1998; 77:366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peserico A., Chiacchiera F., Grossi V., Matrone A., Latorre D., Simonatto M., Fusella A., Ryall J.G., Finley L.W., Haigis M.C. et al. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell. Mol. Life Sci. 2013; 70:2015–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Williamson I., Hill R.E., Bickmore W.A.. Enhancers: from developmental genetics to the genetics of common human disease. Dev. Cell. 2011; 21:17–19. [DOI] [PubMed] [Google Scholar]
- 32. Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G.. Enhancers: five essential questions. Nat. Rev. Genet. 2013; 14:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng W., Bickmore W., Dean A., Nobrega M.A., Bejerano G.. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y. et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004; 303:2011–2015. [DOI] [PubMed] [Google Scholar]
- 36. López-Otín C., Blasco M.A., Partridge L., Serrano M, Kroemer G.. The hallmarks of aging. Cell. 2013; 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Epel E.S., Lithgow G.J.. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2014; 69:S10–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levine M.E., Crimmins E.M.. A genetic network associated with stress resistance, longevity, and cancer in humans. J. Gerontol. A Biol. Sci. Med. Sci. 2016; 71:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Willcox B.J., Tranah G.J., Chen R., Morris B.J., Masaki K.H., He Q., Willcox D.C., Allsopp R.C., Moisyadi S., Poon L.W. et al. The FoxO3 gene and cause-specific mortality. Aging Cell. 2016; 15:617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Willcox B.J., Morris B.J., Tranah G.J., Chen R., Masaki K.H., He Q., Willcox D.C., Allsopp R.C., Moisyadi S., Gerschenson M. et al. Longevity-associated FOXO3 genotype and its impact on coronary artery disease mortality in japanese, whites, and blacks: a prospective study of three american populations. J. Gerontol. A Biol. Sci. Med. Sci. 2017; 72:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kathryn E., Wellen G.S., Hotamisligil. Inflammation, stress, and diabetes. J. Clin. Invest. 2005; 115:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minamino T., Kitakaze M.. ER stress in cardiovascular disease. J. Mol. Cell. Cardiol. 2010; 48:1105–1110. [DOI] [PubMed] [Google Scholar]
- 43. Mozzini C., Cominacini L., Garbin U., Fratta Pasini A.M.. Endoplasmic reticulum stress, NRF2 signalling and cardiovascular diseases in a nutshell. Curr. Atheroscler. Rep. 2017; 10:1–7. [DOI] [PubMed] [Google Scholar]
- 44. Luo T., Kim J.K., Chen B., Abdel-Latif A., Kitakaze M., Yan L.. Attenuation of ER stress prevents post-infarction-induced cardiac rupture and remodeling by modulating both cardiac apoptosis and fibrosis. Chem. Biol. Interact. 2015; 225:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang M.Y., Wang Y.B., Han B., Yang B., Qiang Y.W., Zhang Y., Wang Z., Huang X., Liu J. et al. Activation of aldehyde dehydrogenase 2 slows down the progression of atherosclerosis via attenuation of ER stress and apoptosis in smooth muscle cells. Acta Pharmacol. Sin. 2017; 39:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greer E.L., Oskoui P.R., Banko M.R., Maniar J.M., Gygi M.P., Gygi S.P., Brunet A.. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007; 282:30107–30119. [DOI] [PubMed] [Google Scholar]
- 47. Lützner N., Kalbacher H., Krones-Herzig A., Rösl F.. FOXO3 is a glucocorticoid receptor target and regulates LKB1 and its own expression based on cellular AMP levels via a positive autoregulatory loop. PLoS One. 2012; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeo H., Lyssiotis C.A., Zhang Y., Ying H., Asara J.M., Cantley L.C., Paik J.H.. FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 2013; 32:2589–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang K., Li L., Qi Y., Zhu X., Gan B., DePinho R.A., Averitt T., Guo S.. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012; 153:631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee S., Dong H.H.. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017; 233:R67–R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Safra M., Fickentscher R., Levi-Ferber M., Danino Y.M., Haviv-Chesner A., Hansen M., Juven-Gershon T., Weiss M., Henis-Korenblit S.. The FOXO transcription factor DAF-16 bypasses ire-1 requirement to promote endoplasmic reticulum homeostasis. Cell. Metab. 2014; 20:870–881. [DOI] [PubMed] [Google Scholar]
- 52. Haeusler R.A., Hartil K., Vaitheesvaran B., Arrieta-Cruz I., Knight C.M., Cook J.R., Kammoun H.L., Febbraio M.A., Gutierrez-Juarez R., Kurland I.J., Accili D.. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat. Commun. 2014; 5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Donlon T.A., Morris B.J., Chen R., Masaki K.H., Allsopp R.C., Willcox D.C., Elliott A., Willcox B.J.. FOXO3 longevity interactome on chromosome 6. Aging Cell. 2017; 16:1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Flachsbart F., Dose J., Gentschew L., Geismann C., Caliebe A., Knecht C., Nygaard M., Badarinarayan N., ElSharawy A., May S. et al. Identification and characterization of two functional variants in the human longevity gene FOXO3. Nat. Commun. 2017; 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.