Abstract
The β-class carbonic anhydrase (CA, EC 4.2.1.1) from the pathogenic bacterium Burkholderia pseudomallei, BpsCAβ, that is responsible for the tropical disease melioidosis was investigated for its activation with natural and non-natural amino acids and amines. Previously, the γ-CA from this bacterium has been investigated with the same library of 19 amines/amino acids, which show very potent activating effects on both enzymes. The most effective BpsCAβ activators were L- and D-DOPA, L- and D-Trp, L-Tyr, 4-amino-L-Phe, histamine, dopamine, serotonin, 2-pyridyl-methylamine, 1-(2-aminoethyl)-piperazine and L-adrenaline with KAs of 0.9–27 nM. Less effective activators were D-His, L- and D-Phe, D-Tyr, 2-(2-aminoethyl)pyridine and 4-(2-aminoethyl)-morpholine with KAs of 73 nM–3.42 µM. The activation of CAs from bacteria, such as BpsCAγ/β, has not been considered previously for possible biomedical applications. It would be of interest to perform studies in which bacteria are cultivated in the presence of CA activators, which may contribute to understanding processes connected with the virulence and colonization of the host by pathogenic bacteria.
Keywords: Carbonic anhydrase, metalloenzymes, pathogens, activators, Burkholderia pseudomallei
Introduction
Carbonic anhydrases (CAs, EC 4.2.1.1) are a superfamily of ubiquitous metalloenzymes with the catalytically active form represented by a metal hydroxide derivative acting as a potent nucleophile1–15. They catalyze a simple but physiologically relevant reaction in which carbon dioxide is reversibly hydrated to bicarbonate and protons2,5,6,13,15. CAs are grouped in seven genetically distinct families, named α-, β-, γ-, δ-, ζ-, η- and ɵ-CAs, and share a relatively low similarity for both sequences and three-dimensional structures5,6,8,13,15–17. α-CAs are normally monomers and rarely dimers; β-CAs are dimers, tetramers or octamers; γ-CAs are trimers18–21. Interesting, ζ-CA has three slightly different active sites on the same polypeptide chain5,8,15,16,22–25. X-ray crystal structures of δ-, η- and θ-CAs are not available to date. The catalytic active sites of α-, β-, δ-, η- and, perhaps θ-CAs contain a Zn(II) ion. γ-CAs are Fe(II) enzymes but they are also active coordinating Zn(II) or Co(II) ions, whereas ζ-CAs bind Cd(II) or Zn(II) within their active site, being cambialistic enzymes1,5,6,15,16,26,27. The metal ion from the CA active site is coordinated by three His residues in the α-, γ-, δ- and probably the θ-classes: by one His and two Cys residues in β- and ζ-CAs or by two His and one Gln residues in the η-class, with the fourth ligand being a water molecule/hydroxide ion acting as nucleophile in the catalytic cycle of the enzyme18,19,21,28–35.
CAs are involved in many crucial physiologic and pathologic processes connected to pH regulation, secretion of electrolytes, biosynthetic processes, photosynthesis, tumorigenesis, etc. The existence in bacteria of genes encoding for CAs from at least one of the α-, β- and γ-classes suggests that these enzymes are essential for the bacterial physiology. In fact, it has been demonstrated that CAs are involved in the transport and supply of CO2 or HCO3−, pH regulation, acclimatization of the pathogen within the stomach, induction of the cholera toxin expression, pathogenicity and/or the growth of the microorganism, and, at least in Escherichia coli, in the cyanate degradation16,36–44. Generally, CAs hydrate carbon dioxide at a very high rate, with pseudo first order kinetic constants as high15,16 as 104–106 s –1. The rate-determining step of the entire catalytic process for all CA genetic families is likely the formation of the metal hydroxide species of the enzyme, via the transfer of a proton from the metal-coordinated water molecule to the surrounding solvent to form the nucleophilic form of the enzyme5,6,13,15,16. As a consequence, in all CAs, a proton shuttle residue is present which controls the transfer of the proton from the active site to the protein surface45,46. For α-CAs, a His residue (His64, hCA I numbering system) plays the role of proton shuttle, whereas for other genetic families, the nature and the role of the proton shuttle are less well understood25–31. Thus, the manipulation of the proton shuttle within the CAs active sites is crucial to the function of these enzymes and explains the efficacy and prominence of the catalytic processes in which these enzymes participate45,47. A multitude of physiologically active compounds such as biogenic amines (histamine, serotonin, and catecholamines), amino acids, oligopeptides or small proteins were shown to act as efficient carbonic anhydrase activators (CAAs)47. CAAs may be useful in the treatment of Alzheimer’s disease, in aging, in achieving spatial learning and memory therapy45,46. Indeed, the action of CAAs can be mediated by extracellular signal-regulated kinase (ERK) pathways in a critical step for memory formation, within the cortex and the hippocampus, which are two brain areas involved in memory processing and rich in various CA isoforms46.
Whereas bacterial CA inhibitors (CAIs) were extensively studied, leading to a detailed understanding of the catalytic and inhibition mechanisms, only a few studies are available on the bacterial CAAs. Recently, our groups described the biochemical properties of a β- and γ-CA from the pathogenic bacterium Burkholderia pseudomallei, which is responsible for the tropical disease melioidosis48–51. These enzymes, called BpsCAβ and BpsCAγ, showed high catalytic activity for the physiologic CO2 hydration reaction to bicarbonate and protons (kcat 105 s−1)48–51. Moreover, the study of the inhibition profiles with the classical CA inhibitors (sulfonamides and anions) revealed an interesting structure–activity relationship for the interaction of these enzymes with the inhibitors48–51. We also investigated the activation profiles of BpsCAγ with a series of natural and non-natural amino acids and aromatic/heterocyclic amines52. Here, we report the effects the aforementioned CAAs on the activity of BpsCAβ, which has not yet been investigated for its activation profile. We also compare the effects of these classes of these CAAs on the β- and γ-class enzymes from this bacterium.
Materials and methods
Gene identification and cloning
The identification of the genes encoding B. pseudomallei β-CA (BpsCAβ) and γ-CA (BpsCAγ) was performed as described by Del Prete et al.50. Briefly, the β-CA gene with the accession number WP_004189176.1 and the γ-CA gene (accession number: WP_038762492.1) from Burkholderia pseudomallei were identified running the “BLAST” program, using the nucleotide sequences of known bacterial β-CAs or γ-CAs as query sequence. The GeneArt Company (Invitrogen, Carlsbad, CA), specializing in gene synthesis, designed the synthetic BpsCAβ (BpsCAβ-DNA) and BpsCAγ (BpsCAγ-DNA) genes encoding for the β- and γ-CAs and containing four base-pair sequences (CACC) necessary for directional cloning at the 5′ end of the BpsCAβ and BpsCAγ genes. The recovered BpsCAβ and BpsCAγ genes and the linearised expression vector (pET-100/D-TOPO) were ligated by T4 DNA ligase to form the expression vector pET-100/BpsCAβ or pET-100/BpsCAγ.
Expression and purification
BL21-CodonPlus(DE3)-RIPL competent cells (Agilent, Palo Alto, CA) were transformed with pET-100/BpsCAβ or pET-100/BpsCAγ, grown at 37 °C, and induced with 1 mM IPTG. After 30 min, ZnSO4 (0.5 mM) was added to the culture medium and cells were grown for an additional 3 h. Subsequently, cells were harvested and re-suspended in the following buffer: 50 mM Tris/HCl, pH 8.0, 0.5 mM PMSF and 1 mM benzamidine. Cells were then disrupted by sonication at 4 °C. After centrifugation at 12,000× g for 45 min, the supernatant was incubated with His Select HF nickel affinity gel resin (Sigma, St. Louis, MO) equilibrated in lysis buffer for 30 min. Following centrifugation at 2000× g, the resin was washed in buffer (50 mM Tris/HCl, pH 8.3, 500 mM KCl, 20 mM imidazole). The protein was eluted with the wash buffer containing 300 mM imidazole. The collected fractions were dialyzed against 50 mM Tris/HCl, pH 8.3. At this stage of purification, the proteins were at least 95% pure and the obtained recovery was of about 2 mg of the recombinant proteins.
Carbonic anhydrase activity assay and determination of the activation constant
An applied photophysics stopped-flow instrument was used for assaying the CA catalysed CO2 hydration activity53. Phenol red (at a concentration of 0.2 mM) was used as an indicator, working at the absorbance maximum of 557 nm, with 10 mM TRIS (pH 8.3) as buffer, 0.1 M Na2SO4 (for maintaining constant ionic strength), following the CA-catalysed CO2 hydration reaction for a period of 10 s at 25 °C. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and activation constants. For each activator, at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The unactivated rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of activators 1–19 (10 mM) were prepared in distilled-deionized water and dilutions up to 0.01 nM were done thereafter with distilled-deionized water. Activator and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E-A complex. The activation constant (KA), defined similarly with the inhibition constant KI5–8, can be obtained by considering the classical Michaelis–Menten equation (Equation (1)), which has been fitted by non-linear least squares by using PRISM 3:
| (1) |
where [A]f is the free concentration of activator.
Working at substrate concentrations considerably lower than KM ([S] ≪ KM), and considering that [A]f can be represented in the form of the total concentration of the enzyme ([E]t) and activator ([A]t), the obtained competitive steady-state equation for determining the activation constant is given by Equation (2)45,47,54,55:
| (2) |
where v0 represents the initial velocity of the enzyme-catalyzed reaction in the absence of activator45,47,54,55.
Results and discussion
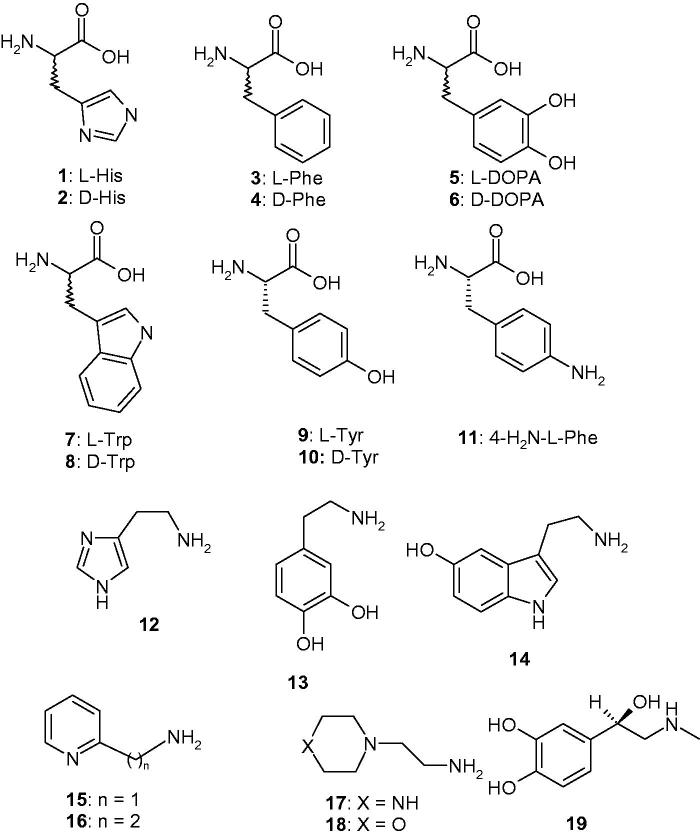
The activators 1–19 were included in this study (Figure 1), as they were employed for investigations as CAAs against many classes of CAs, including the bacterial BpsCAγ52. Both natural and non-natural amino acids and amines are among the investigated compounds (Figure 1)53.
Figure 1.
Chemical structures of compounds 1–19 investigated as CAAs in the present paper.
L-Tyr (at 10 μM concentration) is a very effective CAA for all enzymes considered here, i.e. hCA I, II and BpsCAγ/β, that significantly enhances the kcat values for each enzyme, whereas KM remains unchanged (Table 1). This situation has been observed for all CAAs investigated so far54–56. L-Tyr is a nanomolar activator for the α-class enzymes (hCA I and II) and BpsCAβ with KAs in the range of 3–20 nM and a submicromolar activator for BpsγCA (Table 2). Owing to the very high efficacy as a BpsCAβ activator, L-Tyr increased the kinetic constant of this enzyme by a factor of 19.3 compared with the unactivated rate. This is one of the highest kinetic effects observed so far for an activator of these enzymes.
Table 1.
Activation of human carbonic anhydrase (hCA) isozymes I, II and BpsCAγ/β with L-Tyr, at 25 °C, for the CO2 hydration reaction53.
| kcatd | KMd | (kcat)L-Tyre | KAf (μM) | |
|---|---|---|---|---|
| Isozyme | (s−1) | (mM) | (s−1) | L-Tyr |
| hCA Ia | 2.0 × 105 | 4.0 | 13.9 × 105 | 0.020 |
| hCA IIa | 1.4 × 106 | 9.3 | 12.8 × 106 | 0.011 |
| BpsCAγb | 5.3 × 105 | 21.2 | 13.8 × 105 | 0.200 |
| BpsCAβc | 1.6 × 105 | 4.7 | 3.10 × 106 | 0.003 |
Human recombinant isozymes, from Ref.7.
Bacterial recombinant enzyme, from Ref.52.
Bacterial recombinant enzyme, this work.
Observed catalytic rate without activator. KM values in the presence and the absence of activators were the same for the various CAs (data not shown).
Observed catalytic rate in the presence of 10 μM activator.
The activation constant (KA) for each enzyme was obtained by fitting the observed catalytic enhancements as a function of the activator concentration53. Mean from at least three determinations by a stopped-flow, CO2 hydrase method. Standard errors were in the range of 5–10% of the reported values (data not shown).
Table 2.
Activation constants of hCA I, hCA II and the bacterial CAs BpsCAγ/β with amino acids and amines 1–19. Data for hCA I, II and BpsCAγ are from Refs.25,52.
| KA (μM)d | |||||
|---|---|---|---|---|---|
| No. | Compound | hCA Ia | hCA IIa | BpsγCAb | BpsCAβc |
| 1 | L-His | 0.03 | 10.9 | 24.7 | 31.6 |
| 2 | D-His | 0.09 | 43 | 0.086 | 0.98 |
| 3 | L-Phe | 0.07 | 0.013 | 1.73 | 3.42 |
| 4 | D-Phe | 86 | 0.035 | 0.13 | 0.075 |
| 5 | L-DOPA | 3.1 | 11.4 | 0.072 | 0.009 |
| 6 | D-DOPA | 4.9 | 7.8 | 0.98 | 0.007 |
| 7 | L-Trp | 44 | 27 | 0.43 | 0.002 |
| 8 | D-Trp | 41 | 12 | 0.052 | 0.001 |
| 9 | L-Tyr | 0.02 | 0.011 | 0.20 | 0.003 |
| 10 | D-Tyr | nt | nt | 32.8 | 1.89 |
| 11 | 4-H2N-L-Phe | 0.24 | 0.15 | 0.009 | 0.0009 |
| 12 | Histamine | 2.1 | 125 | 0.12 | 0.012 |
| 13 | Dopamine | 13.5 | 9.2 | 0.014 | 0.006 |
| 14 | Serotonin | 45 | 50 | 0.10 | 0.027 |
| 15 | 2-Pyridyl-methylamine | 26 | 34 | 2.36 | 0.016 |
| 16 | 2-(2-Aminoethyl)pyridine | 13 | 15 | 0.034 | 0.94 |
| 17 | 1-(2-Aminoethyl)-piperazine | 7.4 | 2.3 | 0.018 | 0.004 |
| 18 | 4-(2-Aminoethyl)-morpholine 0.14 | 0.19 | 0.015 | 0.073 | |
| 19 | L-Adrenaline | 0.09 | 96 | 0.019 | 0.002 |
Human recombinant isozymes, stopped flow CO2 hydrase assay method25.
From Ref.52, stopped flow CO2 hydrase assay method.
This work.
Mean from three determinations by a stopped-flow, CO2 hydrase method14. Standard errors were in the range of 5–10% of the reported values (data not shown); nt: not tested.
In Table 2, the CAA profiles of the amino acids and amines 1–19 that were measured for two human (α-class) and two bacterial (γ-class) CAs (i.e. four diverse enzymes) are shown. The CAA profiles for hCA I, hCA II and BpsγCA were reported previously25,52. The amino acids/amines 1–19 are effective activators of BpsCAβ (Table 2). In fact, these amino acids and amines show activation constants ranging between 0.9 nM and 31.6 µM, leading to a very interesting structure-activity relationship, as outlined below:
A large number of the investigated amino acids and amines showed extremely effective activating properties against BpsCAβ, with activation constants in the subnanomolar – low nanomolar range, more precisely of 0.9–27 nM. They include L- and D-DOPA, L- and D-Trp, L-Tyr, 4-amino-L-Phe, histamine, dopamine, serotonin, 2-pyridyl-methylamine, 1-(2-aminoethyl)-piperazine and L-adrenaline. 4-Amino-L-Phe 11 was the most effective, subnanomolar activator, followed by D-Trp, L-Trp, L-Tyr and L-adrenaline which showed KAs in the range of 1-3 nM. Although many of these derivatives also showed effective BpsCAγ activating properties,52 their effects on the β-class enzyme are more potent, as observed by comparing the KAs of these compounds against the two pathogenic enzymes. Furthermore, in many cases, these compounds were more effective in selectively activating the bacterial versus the human α-class CAs (Table 2). Again, the D-amino acid derivatives were more effective BpsCAβ activators compared with their L-enantiomer, except for Tyr, for which the L-enantiomer was a better activator compared with the D-enantiomer. Small changes in the scaffold (e.g. the presence of an amino moiety as in 11 or two OH groups as in DOPA) lead to an enhanced effect compared to the parent L-/D-Phe scaffold. In fact, D-Phe is a rather effective activator (KA of 75 nM) whereas the L-enantiomer is a weak, micromolar activator with a KA of 3.42 µM.
Compounds with medium CAA potency for BpsCAβ were D-His, L- and D-Phe, D-Tyr, 2-(2-aminoethyl)pyridine and 4-(2-Aminoethyl)-morpholine, which have KAs ranging between 73 nM and 3.42 µM (Table 2). Again small differences in the scaffold of the activator lead to significant differences in activity. For example, the two amines 15 and 16 only differ by an extra CH2 moiety. However, 16 is 58.7 times less effective as a CAA compared with 15. Compounds 17 and 18 are also structurally similar, with an oxygen atom in the ring of 17 being replaced by an NH group in 18. However, the two compounds show a highly different action on BpsCAβ: the piperazine 17 was 18.2 times more effective as a CAA compared with the morpholine 18.
The least effective activator was L-His, with a KA of 31.6 µM (Table 2). It may be observed that the D-enantiomer 2 was a much more effective activator (32.2 times) compared with the L-enantiomer 1.
Conclusions
The present study evidenced that many natural and non-natural amino acids and amines show very potent activating effects on both CAs present in this pathogenic bacterium. Among them are L- and D-DOPA, L- and D-Trp, L-Tyr, 4-amino-L-Phe, histamine, dopamine, serotonin, 2-pyridyl-methylamine, 1-(2-aminoethyl)-piperazine and L-adrenaline, which showed activation constants ranging between 0.9 and 27 nM. Less effective activators were D-His, L- and D-Phe, D-Tyr, 2-(2-aminoethyl)pyridine and 4-(2-aminoethyl)-morpholine, which showed KAs ranging between 73 nM and 3.42 µM. The activation of CAs in bacteria, such as BpsCAγ/β, has not been previously considered until now for possible biomedical applications. In fact, no growth studies of these bacteria in media which are enriched in CAAs have been reported in the literature. It would be of interest to perform such studies which may bring new light in understanding processes connected with the virulence and colonization of the host by such bacteria, which are responsible for a rather difficult to treat disease, melioidosis56. Moreover, biogenic CAAs may be useful as potential biomarkers for diagnosis of such pathogen born diseases to direct treatment and prevent sepsis.
Funding Statement
This research was financed in part by a Distinguished Scientist Fellowship Programme (DSFP) of King Saud University, Saudi Arabia and the Australian Research Council (DP160102681).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Supuran CT, Capasso C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63. [DOI] [PubMed] [Google Scholar]
- 2.Del Prete S, De Luca V, Supuran CT, Capasso C.. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–4. [DOI] [PubMed] [Google Scholar]
- 3.De Simone G, Di Fiore A, Capasso C, Supuran CT.. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9. [DOI] [PubMed] [Google Scholar]
- 4.De Luca V, Del Prete S, Supuran CT, Capasso C.. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82. [DOI] [PubMed] [Google Scholar]
- 5.Capasso C, Supuran CT.. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32. [DOI] [PubMed] [Google Scholar]
- 6.Capasso C, Supuran CT.. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704. [DOI] [PubMed] [Google Scholar]
- 7.Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. . Inhibition of human carbonic anhydrase isozymes I, II, IX and XII with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. J Enzyme Inhib Med Chem 2015;30:52–6. [DOI] [PubMed] [Google Scholar]
- 8.Kikutani S, Nakajima K, Nagasato C, et al. . Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A 2016;113:9828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Prete S, Vullo D, De Luca V, et al. . Cloning, expression, purification and sulfonamide inhibition profile of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem Lett 2016;26:4184–90. [DOI] [PubMed] [Google Scholar]
- 10.Del Prete SD, Luca VD, Simone G, et al. . Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;1–6: 4184–90. [DOI] [PubMed] [Google Scholar]
- 11.Del Prete S, Vullo D, Fisher GM, et al. . Discovery of a new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum – the eta-carbonic anhydrases. Bioorg Med Chem Lett 2014;24:4389–96. [DOI] [PubMed] [Google Scholar]
- 12.Supuran CT, Capasso C.. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J Enzyme Inhib Med Chem 2016;31:1254–60. [DOI] [PubMed] [Google Scholar]
- 13.Ozensoy Guler O, Capasso C, Supuran CT.. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem 2016;31:689–94. [DOI] [PubMed] [Google Scholar]
- 14.Del Prete S, Vullo D, De Luca V, et al. . Sulfonamide inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:1115–20. [DOI] [PubMed] [Google Scholar]
- 15.Capasso C, Supuran CT.. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr Top Med Chem 2016;16:2359–68. [DOI] [PubMed] [Google Scholar]
- 16.Capasso C, Supuran CT.. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9. [DOI] [PubMed] [Google Scholar]
- 17.Del Prete S, Vullo D, De Luca V, et al. . Biochemical characterization of the delta-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem 2014;29:906–11. [DOI] [PubMed] [Google Scholar]
- 18.De Simone G, Monti SM, Alterio V, et al. . Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6. [DOI] [PubMed] [Google Scholar]
- 19.Di Fiore A, Capasso C, De Luca V, et al. . Ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9. [DOI] [PubMed] [Google Scholar]
- 20.Lomelino CL, Mahon BP, McKenna R, et al. . Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111. Bioorg Med Chem 2016;24:976–81. [DOI] [PubMed] [Google Scholar]
- 21.Ferraroni M, Del Prete S, Vullo D, et al. . Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56. [DOI] [PubMed] [Google Scholar]
- 22.Supuran CT.Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32. [DOI] [PubMed] [Google Scholar]
- 23.Buzas GM, Supuran CT.. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932–2015). J Enzyme Inhib Med Chem 2016;31:527–33. [DOI] [PubMed] [Google Scholar]
- 24.Carta F, Supuran CT, Scozzafava A.. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med Chem 2014;6:1149–65. [DOI] [PubMed] [Google Scholar]
- 25.Supuran CT.Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81. [DOI] [PubMed] [Google Scholar]
- 26.Capasso C, Supuran CT.. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87. [DOI] [PubMed] [Google Scholar]
- 27.Alterio V, Langella E, De Simone G, Monti SM.. Cadmium-containing carbonic anhydrase CDCA1 in marine diatom Thalassiosira weissflogii. Mar Drugs 2015;13:1688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinard MA, Lotlikar SR, Boone CD, et al. . Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem 2015;23:4831–8. [DOI] [PubMed] [Google Scholar]
- 29.Zolnowska B, Slawinski J, Pogorzelska A, et al. . Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N'-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;71:135–47. [DOI] [PubMed] [Google Scholar]
- 30.De Luca L, Ferro S, Damiano FM, et al. . Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Eur J Med Chem 2014;71:105–11. [DOI] [PubMed] [Google Scholar]
- 31.Supuran CT.Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72. [DOI] [PubMed] [Google Scholar]
- 32.Supuran CT. Carbonic anhydrases-an overview. Curr Pharm Des 2008;14:603–14. [DOI] [PubMed] [Google Scholar]
- 33.Supuran CT.Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt A, Mahon BP, Cruzeiro VW, et al. . Structure–activity relationships of benzenesulfonamide-based inhibitors towards carbonic anhydrase isoform specificity. Chembiochem 2017;18:213–22. [DOI] [PubMed] [Google Scholar]
- 35.Alterio V, Langella E, Viparelli F, et al. . Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie 2012;94:1232–41. [DOI] [PubMed] [Google Scholar]
- 36.Cobaxin M, Martinez H, Ayala G, et al. . Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb Pathog 2014;66:5–13. [DOI] [PubMed] [Google Scholar]
- 37.Abuaita BH, Withey JH.. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 2009;77:4111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimori I, Onishi S, Takeuchi H, Supuran CT.. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des 2008;14:622–30. [DOI] [PubMed] [Google Scholar]
- 39.Morishita S, Nishimori I, Minakuchi T, et al. . Cloning, polymorphism, and inhibition of beta-carbonic anhydrase of Helicobacter pylori. J Gastroenterol 2008;43:849–57. [DOI] [PubMed] [Google Scholar]
- 40.Joseph P, Ouahrani-Bettache S, Montero JL, et al. . A new beta-carbonic anhydrase from Brucella suis, its cloning, characterization, and inhibition with sulfonamides and sulfamates, leading to impaired pathogen growth. Bioorg Med Chem 2011;19:1172–8. [DOI] [PubMed] [Google Scholar]
- 41.Modak JK, Liu YC, Machuca MA, et al. . Structural basis for the inhibition of Helicobacter pylori alpha-carbonic anhydrase by sulfonamides. PLoS One 2015;10:e0127149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimori I, Vullo D, Minakuchi T, et al. . Carbonic anhydrase inhibitors: cloning and sulfonamide inhibition studies of a carboxyterminal truncated alpha-carbonic anhydrase from Helicobacter pylori. Bioorg Med Chem Lett 2006;16:2182–8. [DOI] [PubMed] [Google Scholar]
- 43.Vullo D, De Luca V, Del Prete S, et al. . Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg Med Chem Lett 2016;26:1253–9. [DOI] [PubMed] [Google Scholar]
- 44.Del Prete S, Isik S, Vullo D, et al. . DNA cloning, characterization, and inhibition studies of an alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J Med Chem 2012;55:10742–8. [DOI] [PubMed] [Google Scholar]
- 45.Supuran CT.Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med Chem 2011;3:1165–80. [DOI] [PubMed] [Google Scholar]
- 46.Supuran CT.How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60. [DOI] [PubMed] [Google Scholar]
- 47.Supuran CT.Carbonic anhydrases: from biomedical applications of the inhibitors and activators to biotechnological use for CO(2) capture. J Enzyme Inhib Med Chem 2013;28:229–30. [DOI] [PubMed] [Google Scholar]
- 48.Del Prete S, Vullo D, di Fonzo P, et al. . Comparison of the anion inhibition profiles of the beta- and gamma-carbonic anhydrases from the pathogenic bacterium Burkholderia pseudomallei. Bioorg Med Chem 2017;25:2010–15. [DOI] [PubMed] [Google Scholar]
- 49.Del Prete S, Vullo D, Di Fonzo P, et al. . Sulfonamide inhibition profile of the gamma-carbonic anhydrase identified in the genome of the pathogenic bacterium Burkholderia pseudomallei the etiological agent responsible of melioidosis. Bioorg Med Chem Lett 2017;27:490–5. [DOI] [PubMed] [Google Scholar]
- 50.Del Prete S, Vullo D, Di Fonzo P, et al. . Anion inhibition profiles of the gamma-carbonic anhydrase from the pathogenic bacterium Burkholderia pseudomallei responsible of melioidosis and highly drug resistant to common antibiotics. Bioorg Med Chem 2017;25:575–80. [DOI] [PubMed] [Google Scholar]
- 51.Vullo D, Del Prete S, Di Fonzo P, et al. . Comparison of the sulfonamide inhibition profiles of the beta- and gamma-carbonic anhydrases from the pathogenic bacterium Burkholderia pseudomallei. Molecules 2017;22:E421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vullo D, Del Prete S, Osman SM, et al. . Burkholderia pseudomallei gamma-carbonic anhydrase is strongly activated by amino acids and amines. Bioorg Med Chem Lett 2017;27:77–80. [DOI] [PubMed] [Google Scholar]
- 53.Khalifah RG.The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73. [PubMed] [Google Scholar]
- 54.Licsandru E, Tanc M, Kocsis I, et al. . A class of carbonic anhydrase I – selective activators. J Enzyme Inhib Med Chem 2017;32:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vullo D, Del Prete S, Capasso C, Supuran CT.. Carbonic anhydrase activators: activation of the beta-carbonic anhydrase from Malassezia globosa with amines and amino acids. Bioorg Med Chem Lett 2016;26:1381–5. [DOI] [PubMed] [Google Scholar]
- 56.Capasso C, Supuran CT.. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704. [DOI] [PubMed] [Google Scholar]