Abstract
The recent completion of the deletion of essentially all of the ORFs in yeast is an important new resource for identifying the phenotypes of unknown genes. Each ORF is replaced with a cassette containing unique tag sequences that allow rapid parallel analysis of strains in a pool by using hybridization to a high-density oligonucleotide array. We examined the utility of this system to identify genes conferring resistance to UV irradiation by using a pool of 4,627 individual homozygous deletion strains (representing deletions of all nonessential genes). We identified most of the nonessential genes previously shown to be involved in nucleotide excision repair, in cell cycle checkpoints, in homologous recombination, and in postreplication repair after UV damage. We also identified and individually confirmed, by replacing the genes, three new genes, to our knowledge not previously reported to confer UV sensitivity when deleted. Two of these newly identified genes have human orthologs associated with cancer, demonstrating the potential of this system to uncover human genes affecting sensitivity to DNA-damaging agents and genes potentially involved in cancer formation.
One of the major challenges of the postsequencing phase of the Human Genome Project is assignment of biological function to the identified genes. A key resource for this is sequence and functional data from model organisms, of which the budding yeast Saccharomyces cerevisiae is one of the more important. Its usefulness as a model organism rests on its facile genetic manipulation and the large degree of homology between genes in yeast and in humans (1), with some 30% of known genes involved in human disease having yeast orthologs. These human homologs often allow functional complementation of phenotypes in yeast with cDNA clones from human cells (2–4).
A powerful method for determining gene function is analysis of the phenotype of mutants lacking the gene. To facilitate such analyses, an international consortium has undertaken the systematic deletion of all of the ≈6,200 known ORFs of yeast by using a PCR-mediated gene deletion strategy (5–7). In addition to a selectable marker, two molecular barcodes, or “tags,” unique 20-base oligonucleotide sequences, are in the replacement cassette. The tags allow pooling of deletion strains and growth in a single culture. These tags, after PCR amplification, can be detected by hybridization to the corresponding complementary sequence on a high-density oligonucleotide array, thus enabling the relative abundance of each tag and hence the abundance of each deletion strain, to be determined (8). This approach, termed here “parallel deletion analysis,” has previously been used with yeast to identify genes required for growth rate in rich and minimal media by using a pool of ≈500 deletion strains (7) and to identify new virulence genes in Salmonella typhimurium (9).
Here we examine the ability of the recently completed set of 4,627 homozygote deletion strains to detect genes conferring resistance to cell killing by a DNA-damaging agent. We chose UV irradiation as the DNA-damaging agent for two reasons. First, the effects of UV irradiation have been exhaustively studied over the past 30 years, and researchers have identified many yeast genes, the mutation or deletion of which produces sensitivity to UV (10). These previously identified genes provide a solid means to check the efficacy of the screen. Second, we chose UV irradiation because many of the yeast genes involved in DNA repair or damage-response checkpoints have human orthologs, the mutation of which produces sensitivity to cancer induction (11).
We treated pools of 4,627 homozygote deletion strains (representing replacement of all of the nonessential genes) with mild doses of UVC and UVB to identify genes conferring UV resistance (i.e., producing UV sensitivity when deleted). As our goal was to find genes affecting UV cell killing, whereas the hybridization assay measures the combination of slowed cell growth and cell survival after treatment, we also tested the sensitivity of individual deletion strains by using the classic colony-forming assay to measure cell killing directly. We show that the results from the parallel deletion analysis with this set of strains (i) are highly reproducible, (ii) allowed the identification of most of the known UV-sensitive deletions in the pool, and (iii) identified three genes not previously reported to be UV sensitive. Importantly, two of the identified genes conferring UV sensitivity when deleted have human orthologs, CaSm and AF9, both of which have been implicated in cancer. We therefore conclude that parallel analysis by oligonucleotide arrays of this pool of yeast deletion strains provides a powerful tool for genetic analysis of the mechanisms of cell killing by cytotoxic agents, as well as a means to identify new genes potentially involved in the etiology of cancer.
Materials and Methods
Yeast Strains.
Genotypes of the parental yeast strain BY4743 and construction of the homozygous diploid deletion strains have been described previously (7). Information is also available in the Saccharomyces Genome Deletion Project web site (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). All of these completed strains can be obtained from Research Genetics (Huntsville, AL) or EUROSCARF (Frankfurt, Germany). The homozygous diploid deletion pool was generated as described (7). In the present study, we used a pool of 4,627 strains representing homozygous deletion of essentially all nonessential genes.
UV Irradiation.
We performed clonogenic survival experiments with the parental strain, BY4743, to determine doses of UVC and UVB that cause ≈50% cell killing. For UVC, this dose was 200 J/m2, whereas for UVB it was 6,400 J/m2. For experiments with the pool, aliquots of the deletion pool representing ≈104 cells of each of the individual strains were grown in yeast extract/peptone/dextrose medium (YPD) in an orbital shaker (New Brunswick Scientific) at 30°C and 300 rpm to midexponential phase (OD600 nm 0.5–1.0). Cells were pelleted (2,000 × g for 3 min), resuspended in ice-cold PBS at 107 cells/ml, dispersed at less than 1-mm depth in glass Petri dishes, and immediately UV irradiated. Mock irradiated cells were treated identically in parallel. For UVC treatments, we irradiated dishes in a UVC light box (Stratalinker, 254 nm, Stratagene). For UVB treatments, the dishes were irradiated beneath a UVB lamp (FS20T12, National Biological, Twinsburg, OH, peak emission 315 nm, with less than 0.6% of the emission with a UVC component of less than 280 nm). The irradiated or control cells were then pelleted and inoculated into prewarmed YPD at OD600 = 0.05 (106 cells/ml). Before reaching OD600 = 1.0, the cultures were back diluted into fresh YPD at a 1:20 dilution to maintain exponential growth. Cultures were harvested and genomic DNA extracted 18 h after treatment. In preliminary experiments with known UV-sensitive strains, we established that this time period was sufficient for sensitive strains to become depleted by a factor of 100–1,000 relative to wild-type cells after the test dose of 200 J/m2. We performed three identical experiments with both UVC and UVB to determine the reproducibility of the system.
PCR Amplification, Microarray Hybridization, and Data Acquisition.
PCR amplification, microarray hybridization, and data acquisition were as described (7). Briefly, after isolation of genomic DNA from the treated and untreated pools, the isolated DNA was used as template in two PCR reactions that amplify the two tags (UPTAG and DOWNTAG) from each strain in the pool by using biotinylated PCR primers complementary to common regions in the transplacement cassette. For both the treated and untreated pool, we combined the PCR products with oligonucleotides complementary to nontag regions of the PCR product, heat denatured the mixtures, and hybridized them to purpose-built oligonucleotide microarrays (DNA TAG3, Affymetrix, Santa Clara, CA) for 16 h at 42°C. After staining with streptavidin–phycoerythrin (Molecular Probes), arrays were scanned at an emission wavelength of 560 nm by using an Affymetrix GeneChip Scanner (Affymetrix). The hybridization intensities for each of the array elements were determined by using Affymetrix genechip software.
Data Analysis.
For the analysis of strain prevalence in the pool, each strain was represented by four values of signal intensity (sense and antisense array elements for each UPTAG and DOWNTAG). These four values were averaged to give a single value for each strain present in the pool. To assess the degree of sensitivity to UV radiation treatment, a ratio of treated/untreated signal for each strain was calculated. Those strains with lower signals in the treated compared with the untreated pool had a low ratio (sensitive strains), whereas those with similar signals from treated and untreated had ratios close to 1 (unaffected strains). The ratios for each of the 4,627 strains were calculated for UVC and UVB treatments for three separate experiments. To identify those strains that were significantly sensitive to the treatment in the three experiments, we used a recently developed statistical package, sam (significance analysis of microarrays methodology) (12). sam assigns a score to each strain in the pool on the basis of the ratio of treated to control relative to the standard deviation of repeated measurements. For strains with scores greater than an adjustable threshold, sam uses permutations of the repeated measurements to estimate the percentage of strains identified by chance, the false discovery rate (FDR). We chose the most stringent criterion of the FDR equal to zero, which produced 31 strains identified as UV sensitive. However, the absolute order of sensitivities of the strains by using this method can only be regarded as approximate because the ratios of treated/control depend on the absolute values of the hybridization signals in the control samples, as is apparent from Fig. 1.
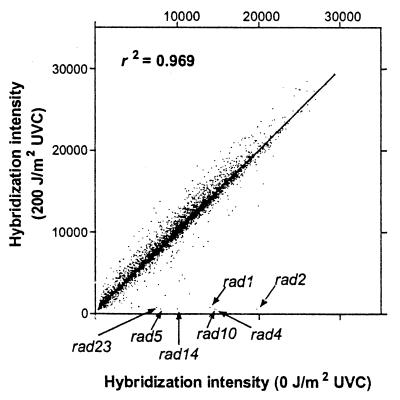
Figure 1.
Scatterplot showing hybridization intensities of the 4,627 deletion strains in the untreated pool (0 J/m2 UVC) versus the treated pool (200 J/m2 UVC). Data for most strains fall close to the line of equivalence, whereas UV-sensitive strains fall near the x axis. Seven known UV sensitive strains are shown with arrows.
Related to the above is the problem of strains in the pool with a hybridization value not very different from background. The 31 strains identified by the sam analysis had ratios of treated/control of from 0.04 (deletion of RAD2) to 0.65 (deletion of YHR134W). Thus to be identified as sensitive in the pool, a strain would need to have a hybridization signal in the control sample of at least 1/0.65 times background levels. To identify the background signal (presumably arising from cross hybridization of irrelevant sequences), we calculated the average hybridization signal for the strains not in the pool (the TAG3 chip has complementary oligonucleotide sequences for the designed tags of all genes irrespective of whether they are essential). These signals varied from 400 to 601 with a mean of 487. Therefore, under the conditions of our experiments, we would not be able to detect the sensitivity to UV of any strains with control hybridization signals of 487 × 1/0.65 or 749. There were 540 (11.7%) of the strains with hybridization signals less than this value in the untreated pool at the time of harvest.
Confirmation of Mutant Phenotype by Clonogenic Assay.
The microarray analysis allowed us to identify individual deletion strains in the pool whose response to UV treatment resulted in a reduction of signal intensity compared with controls. In theory, this could have occurred as a result of either a reduction in growth rate or cell survival after UV. To determine whether the signal reduction corresponded to UV sensitivity leading to cell death, we performed clonogenic survival experiments on several of the strains with reduced ratios of treated/controls. Briefly, midlogarithmic phase cultures were diluted in PBS, and various cell dilutions were spread on replicate 100-mm YPD plates and treated with different doses of UVC radiation. Surviving colonies were counted after incubation for 2–5 days at 30°C and compared with survival of the parental line, BY4743.
Reintroduction of Deleted ORF.
To determine whether the sensitivity of individual deletion strains was because of the absence of the deleted ORF, we reintroduced the ORF by means of an inducible expression vector. We PCR amplified ORFs from BY4743 genomic DNA by using primers which included hemagglutin epitope tags on the 3′ end of the encoded ORFs and HindIII and XhoI restriction sites.
THR1 sense 5′-CTCTCGAAGCTTATGGTTCGTGCCTTCAAAA-3′, THR1 antisense 5′-GTGAGCCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGGTATTGCTGTTCGCGCTA-3′; LSM1 sense 5′-CTCTCGAAGCTTATGTCTGCAAATAGCAAGGACAG-3′, LSM1 antisense, 5′-GTGAGCCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGGTAGTACATGTCAGATTTATGG-3′, YAF9 sense 5′- CTCTCGAAGCTTATGGCTCCGA CAATAAGCAA-3′, YAF9 antisense 5′-GTGAGCCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGGTAACTTCCGTTAATGGCTTCTTG-3′; RAD18 sense, 5′-CTCTCGAAGCTTATGGACCACCAAATAACCACTG-3′, RAD18 antisense, 5′-GTGAGCCTCGAGCTAAGCGTAGTCTGGGACGTCGTATGGGTAATTGTTACCGGGTGGGTCT-3′.
PCR reactions were carried out in a final volume of 50 μl containing genomic DNA (200 ng) by using Vent DNA polymerase (New England Biolabs) according to the manufacturer's instructions. The PCR products were cloned into the galactose-inducible and uracil-selectable expression vector, pMyr (Stratagene), and the constructs expanded in Escherichia coli (DH5α, Life Technologies, Rockville, MD) and sequenced to ensure correct ORF insertion. The yeast deletion strains were transfected by using the lithium acetate method (13). The empty vector was also cloned into parallel cultures to serve as controls. Transformed cells were selected and grown in synthetic media without uracil with 2% dextrose. These were subsequently grown in synthetic media minus uracil with 2% galactose (SMG) for 4 h to allow for production of the protein before UVC exposure and clonogenic survival assay on SMG plates. Western blot analysis by using an antihemagglutin tag antibody (Covance, Cumberland, VA) was used to confirm protein production.
Results
Signal Intensities from Individual Strains.
From hybridization of the PCR products to the oligonucleotide array, we obtained signal intensities from the sense and antisense tags for both the UPTAG and DOWNTAG for each deletion strain in the pool. From these four, a mean signal intensity was calculated for each strain and the data averaged from three experiments. The mean signal intensity ranged from a background level of 386 to 29,354 for the 4,627 strains in the pool. Fig. 1 shows the distribution of these pooled signals from the control and UVC-treated cultures for all 4,627 strains from the three experiments, as well as the position of seven known radiosensitive deletion strains.
Calculation of Sensitivity.
Once mean signal intensities for each strain and treatment were obtained, a ratio of treated/untreated was calculated for each strain in each experiment from which a geometric mean for that strain was calculated. A ratio of 1 suggests the treatment had no effect on the strain, whereas a low ratio suggests a growth deficit. Ratios above 1 were rare and represented experimental variation. This variation was confirmed by statistical analysis by using the sam methodology (12) (Fig. 2). Table 1 shows the list of 31 deletion strains identified by sam method to be significantly sensitive to UVC with the expectation of no false positives (see Materials and Methods), together with the ranking of each strain in the UVC and UVB experiments.
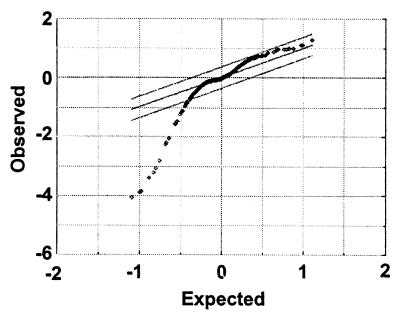
Figure 2.
sam analysis of the UVC data set. sam computes a statistic for each strain measuring the strength of the relationship between the sensitivity ratios in the three repeat experiments. Each point represents an individual strain. The observed values are the paired t statistics for the signal intensities of strains in the treated and control pools. The expected values are the t statistic obtained when the paired data are randomly permuted 100 times. The region defined by the outer parallel lines is set by the investigator and varies the likely number of false positives. The lines shown define the strains with significant sensitivity to UV (outside the lines with no estimated false positives). The 31 strains considered sensitive to UVC are listed in Table 1 along with their ranking in the UVB experiments.
Table 1.
Ranking of the 31 UV-sensitive deletion strains
| UVC rank | Gene name | Reported UVC sensitivity | UVC rank, experiment 1 | UVC rank, experiment 2 | UVC rank, experiment 3 | Protein function | UVB rank |
|---|---|---|---|---|---|---|---|
| 1 | RAD2 | High | 1 | 1 | 1 | Endonuclease involved in NER | 1 |
| 2 | RAD4 | High | 3 | 2 | 2 | DNA damage binding involved in NER | 3 |
| 3 | RAD10 | High | 2 | 3 | 3 | DNA endonuclease involved in NER | 2 |
| 4 | RAD14 | High | 4 | 4 | 6 | DNA damage-binding protein involved in NER | 4 |
| 5 | RAD1 | High | 5 | 5 | 4 | DNA endonuclease involved in NER | 5 |
| 6 | RAD5 | High | 6 | 6 | 5 | DNA helicase involved in postreplication repair | 6 |
| 7 | RAD23 | Moderate | 7 | 7 | 7 | DNA damage binding involved in NER | 8 |
| 8 | MUS81 | Moderate | 8 | 8 | 8 | DNA repair; interacts with Rad54p | 7 |
| 9 | MMS2 | Moderate | 9 | 10 | 13 | Postreplication repair; complex with Ubc13p–Rad5p | 10 |
| 10 | MMS4 | None | 10 | 12 | 10 | Repairs alkylating agent damage | 9 |
| 11 | RAD7 | Moderate | 14 | 9 | 9 | DNA-dependent ATPase | 14 |
| 12 | YBR099C | Unknown | 11 | 11 | 11 | Hypothetical ORF on opposite strand to MMS4 | 11 |
| 13 | RAD59 | Mild | 13 | 13 | 12 | RAD52 epistasis group; involved in dsb repair | 12 |
| 14 | THR1 | Unknown | 12 | 18 | 18 | Homoserine kinase; involved in threonine biosynthesis | 28 |
| 15 | RAD9 | High | 16 | 14 | 20 | Involved in cell cycle checkpoints after DNA damage | 22 |
| 16 | DUN1 | Mild | 15 | 16 | 22 | Protein kinase required for induction of DNA repair genes | 18 |
| 17 | MEC3 | Moderate | 18 | 15 | 15 | DNA damage checkpoint; complex with Rad17p and Ddc1p | 23 |
| 18 | RAD57 | Mild | 26 | 17 | 21 | RAD52 epistasis group; forms dimer with Rad55p | 20 |
| 19 | SAE2 | Mild | 29 | 24 | 19 | Initiation of meiotic recombination and DNA repair | 21 |
| 20 | RAD54 | Mild | 20 | 20 | 37 | DNA repair; interacts with Rad51p and Mus81p | 15 |
| 21 | RAD18 | High | 17 | 39 | 39 | Postreplication repair; partner of Rad6p | 13 |
| 22 | SRS2 | High | 19 | 33 | 35 | DNA helicase involved in DNA repair | 17 |
| 23 | RAD24 | Moderate | 34 | 25 | 34 | Involved in G2 checkpoint after DNA damage | 32 |
| 24 | LSM1 | Unknown | 33 | 22 | 52 | RNA-binding protein involved in mRNA decay | 41 |
| 25 | RAD55 | Mild | 28 | 30 | 36 | RAD52 epistasis group; interacts with Rad51p and Rad57p | 19 |
| 26 | SNF2 | Unknown | 22 | 27 | 114 | Component of swi-snf global transcription activator complex | 30 |
| 27 | YAF9 | Unknown | 21 | 34 | 83 | Protein with similarity to human AF-9 protein | 43 |
| 28 | RAD17 | Moderate | 36 | 49 | 56 | DNA damage checkpoint required for G2 arrest | 45 |
| 29 | REV7 | Moderate | 44 | 28 | 81 | DNA polymerase ζ, required for translesion synthesis | 37 |
| 30 | RAD51 | Mild | 23 | 66 | 156 | RAD52 epistasis group; involved in dsb repair | 24 |
| 31 | YHR134W | Unknown | 684 | 26 | 16 | Protein of unknown function | 31 |
Identification of significant UV sensitivity was performed by sam analysis by using the ratios of hybridization signals in the treated/control pools and their variance obtained in three replicate experiments. Shown also are the sensitivities to UV irradiation categorized from the primary literature as high (DMF > 3.0), moderate (DMF = 1.5–3.0), and mild (DMF < 1.5), where DMF is the dose modifying factor (ratios of doses of wild-type and mutant at equisurvival) at 10% cell survival. Also shown are the individual sensitivity rankings in the three replicate UVC experiments and the sensitivity ranking from three replicate experiments with UVB. All references to the primary data and the known function of the proteins are given in the Proteome database (http://www.proteome.com/databases/index.html).
Similarity Between UVC and UVB Sensitive Strains.
Considering the pool was comprised of 4,627 individual strains, there was remarkable consistency in the reproducibility of identifying UVC sensitive strains between individual experiments and between the UVC and UVB treatments (Table 1). sam analysis identified 31 UVC- and 30 UVB-sensitive strains from the pool of 4,627 strains (Table 1, Fig. 2). This analysis showed the top 10 UVC-sensitive strains were also in the top 10 UVB strains. Twenty-seven of the 31 UVC strains were also in the 30 sam UVB-sensitive strain list.
Identification of Known UV Sensitivity Strains.
The 31 UVC-sensitive strains in Table 1 comprised 25 strains previously reported to have UV sensitivity and six with no prior reports of sensitivity (see Discussion). The strains known to have high UV sensitivity were near the top of the sensitivity rankings, and conversely the mild sensitivity strains were nearer the bottom of the list. The known sensitive strains were deleted for genes involved in nucleotide excision repair (NER), postreplication repair, recombinational repair, or in DNA cell cycle checkpoint functions. NER is the major pathway used to repair UV-induced lesions, and NER-defective strains comprised the top five in the sensitivity rankings.
Identification and Confirmation of Unknown UV Sensitivity Genes.
Six strains that had no previous reported UV sensitivity were identified as significantly sensitive to UVC in our screen. We obtained the individual strains for all six and examined them for UVC sensitivity by clonogenic assay. Two of the six (snf2Δ/snf2Δ and YHR134WΔ/YHR134WΔ) (Table 1, rankings 26 and 31) showed only a marginal increase in cell killing compared with the parental strain, whereas the other four showed marked sensitivity. That these two strains were not sensitive to cell killing suggests that they either experienced more growth delay or had a slower growth rate than the average strain after UV treatment.
One of the four unknown strains that displayed substantial UVC sensitivity was ybr099cΔ/ybr099cΔ (Table 1, ranking 12). This is a hypothetical ORF that is located on the opposite coding strand to the known UV sensitivity gene MMS4. Thus, deletion of YBR099C would result in disruption also of the MMS4 ORF and hence UV sensitivity. The remaining three strains not previously identified as UV sensitive were thr1Δ/thr1Δ, lsm1Δ/lms1Δ and yaf9Δ/yaf9Δ (Table 1, rankings 14, 24, and 27). Fig. 3A shows UVC radiation survival curves of these three along with the parental strain BY4743 for comparison. Also shown for comparison (Fig. 3B) are survival curves for three strains of known UV sensitivity, two with reportedly moderate sensitivity (mus81Δ/mus81Δ, and mms2Δ/mms2Δ) and one with high sensitivity (rad18Δ/rad18Δ) (Table 1, ranking 21). On the basis of a comparison between the known and unknown stains, the three newly identified strains would be classified as moderately sensitive to UVC.
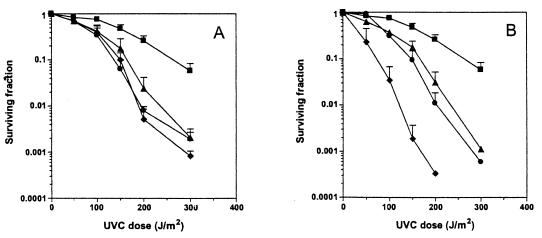
Figure 3.
(A) UVC radiation survival curves of the parental strain, BY4743, and three strains not previously known to be UV sensitive. BY4743, filled squares; lsm1Δ/lsm1Δ, filled triangle; thr1Δ/thr1Δ, filled circle; yaf9Δ/yaf9Δ, filled diamond. (B) UVC radiation survival curves of the parental strain, BY4743, and three known sensitive strains. BY4743, filled squares; mus81Δ/mus81Δ, filled triangle; mms2Δ/mms2Δ, filled circle; rad18Δ/rad18Δ, filled diamond.
To confirm that the sensitivities of the strains were the result of deletion of the ORF and not of other genetic alterations, we reintroduced and expressed the gene products in a plasmid-based inducible expression vector. In each case, we confirmed the expression of the relevant protein by Western blot by using an antihemagglutin antibody before testing for UV sensitivity. As a negative control, we introduced the expression plasmid without the ORF insert. Fig. 4 shows the data for survival to 200 J/m2 of the three newly identified UV sensitive strains with introduction of the empty vector or with the vector containing the relevant ORF. As a positive control, we also performed the same procedure with the known UV sensitive strain rad18Δ/rad18Δ. The data show that in each case, the introduced ORF corrected the sensitive phenotype, restoring survival to that of the wild-type strain BY4743. Thus the sensitivity of each strain can be attributed to deletion of the corresponding gene.
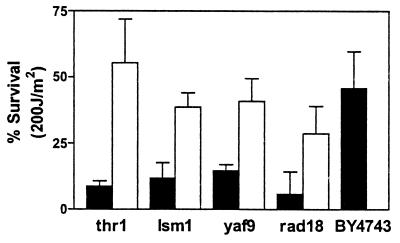
Figure 4.
Reintroduction of deleted ORF abrogates UV sensitivity. We transformed four strains, thr1Δ/thr1Δ, lsm1Δ/lsm1Δ, yaf9Δ/yaf9Δ, and rad18Δ/rad18Δ, with vector alone (black bars) or vector containing the deleted ORF with a galactose inducible promoter (white bars) and tested survival to a single dose of 200 J/m2 UVC. The pooled results of three experiments are shown with the standard deviation.
Discussion
The present study was undertaken to assess the ability of the complete genome set of homozygous yeast deletion strains to discover genes conferring sensitivity to a cytotoxic agent. Previous screens for such genes conferring sensitivity when deleted required replica plating, with the subsequent nontrivial identification of the mutated or deleted gene. The power of the present pool of deletion mutants is that each deleted gene is replaced by two tags that uniquely identify the strain by hybridization to high-density oligonucleotide arrays containing the complementary sequences to each of the tags. In a previous report with this system using a 500-strain pool, Winzeler et al. demonstrated that some 40% showed quantitative growth defects in either rich or minimal medium by using hybridization to a first generation oligonucleotide array, and that the system identified all of the strains in the pool known to be essential for growth in minimal medium (7).
Here we show that the system has considerably more power than previously demonstrated. First, we show that it can semiquantitatively rank the sensitivities to killing by an acute exposure to a DNA-damaging agent. Further, we demonstrate that this comparison of cell killing can be done with the final pool of 4,627 strains representing deletion of all nonessential genes in yeast. We chose UV radiation as the DNA-damaging agent because of numerous studies by investigators over the past 30 years that have identified many genes in yeast that, when mutated or deleted, give rise to sensitivity (10). These genes can be broadly grouped into three classes representing NER (14), postreplication repair (15), including DNA damage checkpoint genes (16), and recombinational repair (17).
We used three criteria to determine the utility of parallel deletion analysis with the present pool: (i) the reproducibility of the results; (ii) its ability to discover previously known genes whose deletion confers sensitivity to UV, and (iii) its ability to discover previously unknown genes affecting UV sensitivity. The deletion pool performed well on all three criteria. In relation to reproducibility, three replicate experiments with UVC and three replicate experiments with a higher dose of the less damaging UVB produced essentially the same results. In fact, the UVC experiments identified the same strains in the top eight most sensitive in each of the three experiments, and the same strains were in the top eight most sensitive to UVB. Analysis of the data with a new statistical method for analyzing microarray data (12) identified 31 strains as significantly sensitive after UVC and 30 for UVB, with the list of strains being very similar for the two types of irradiation. In addition to the close similarity of replicate experiments, the similarity of the data for UVB and UVC is a further test of the reproducibility of the system because the cytotoxic lesions introduced by these two wavelengths are very similar (18).
The second criterion was for the system to correctly identify known UV-sensitive genes. From a search of the Proteome database (http://www.proteome.com/databases/), the primary literature, and recent reviews (10, 14–16), we identified 53 genes that have been reported to confer UV sensitivity when mutated or deleted in any strain background. Of these 53, 13 are essential genes and therefore not in the pool (all members of the TFIIH transcription factor, as well as RFA1, RFC1, POL30, CDC9, CDC7, MEC1, and RAD53). Of the remaining 40, eight strains had hybridization signals too close to background (less than 759 hybridization signal in the untreated pool) to be detected as sensitive (mre11Δ/mre11Δ, xrs2Δ/xrs2Δ, rad50Δ/rad50Δ, rad52Δ/rad52Δ, met18Δ/met18Δ, rad6Δ/rad6Δ, ubc13Δ/ubc13Δ, and ddc1Δ/ddc1Δ) because of slow growth or other reasons. We identified as sensitive 24 of the remaining 32 by using the stringent criterion of no false positives in the sam analysis (Table 1). These involved genes involved in NER, postreplication repair, DNA damage checkpoints, and homologous recombination. The latter included members of the rad52 epistasis group RAD51, RAD54, RAD55, RAD57, RAD59 (Table 1 rankings 30, 20, 25, 18, and 13). The absence of the rad52 homozygous deletion strain is explained by the fact that this strain had a signal intensity in the untreated sample that was too low for it to be detected as sensitive.
There were, however, eight strains that we did not identify as statistically significantly sensitive to UV by this “no false positives” criterion: those with deletions in REV1, REV3, RAD16, RAD30, CAC2, RFL2, UMP1, and MSI1. These eight strains have been reported to confer mild to moderate UV sensitivity when mutated (19–22). However, with the exception of the strains with deletions in CAC2, RFL2, and UMP1, these were in the top 90 (top 2%) of the 4,627 strains in the pool in sensitivity rank. It is likely that the statistical criterion that we used was too stringent to identify these strains and/or there could be differences in UV sensitivity depending on strain background.
Twenty-four of the 31 strains in Table 1 that we identified as sensitive have been reported previously to be sensitive to UV. One other strain, mms4Δ/mms4Δ, although previously reported not to be UV sensitive (23), is likely to be sensitive, because mms4p has been shown to physically interact with mus81p (24), which we identified as conferring UV sensitivity when deleted. The sensitivity of mms4Δ/mms4Δ is also suggested by the fact that UV sensitivity is produced by deletion of YBRO99C, an ORF on the opposite strand and partially overlapping that of MMS4 (SGD database, http://genome-www.stanford.edu/Saccharomyces). Thus 26 of the 31 strains in Table 1 have either been previously reported to be UV sensitive or are likely to be so.
To test the third criterion, namely whether parallel deletion analysis could identify previously unreported genes conferring UV sensitivity, we found that three of the five previously unreported UV sensitive strains were sensitive to cell killing by testing the individual strains for clonogenic survival following UV irradiation. However, this sensitivity does not necessarily implicate these particular genes for two potential reasons. First, there could have been an error in strain construction so that the particular bar-code tag could have been assigned to these genes by mistake. Second, it has been reported that some 8% of these deletion strains have other changes in the genome in addition to elimination of the ORF (25). To test for these possibilities, we expressed the relevant gene in the deletion strain to determine whether this expression would complement the radiation-sensitive phenotype. In all three instances, we showed that replacing the gene restored the wild-type resistance to the sensitive strain (Fig. 4), strongly implicating that gene in UV sensitivity. A possible reason that we have identified genes not previously found in screens for UV sensitivity is that the present study was conducted with diploids, whereas most other mutants were identified in screens with haploids because radiation-sensitive mutants are recessive. Homozygous deletions in diploid cells allow mutants in homologous recombination to be more easily identified.
Two of the three newly identified UV sensitivity genes have human orthologs. The LSM1 gene codes for a protein of the Sm class of RNA-binding proteins involved in control of mRNA decapping and decay (26). The gene is 68% similar and orthologous to a novel human gene of unknown function, CaSm, the product of which has been found to be elevated in pancreatic cancer and in several pancreatic cancer cell lines (27). The YAF9 gene codes for a protein whose molecular function and biological processes are unknown. However, its human ortholog, AF9, is also involved in cancer. AF9 is the partner gene of MLL in a 9:11 translocation associated with de novo acute myeologenous leukemia (28). The third newly identified UV-sensitive gene, THR1, has no known human homolog. The gene codes for homoserine kinase, the first step in the threonine biosynthesis pathway in yeast. The UV sensitivity of the thr1null mutant raises the possibility, among others, that threonine biosynthesis may be involved in UV damage signaling through threonine phosphorylation.
Because of the connection between cellular sensitivity to DNA damage agents and the induction of human cancer, identification of novel genes affecting the sensitivity of yeast to DNA damage has wider applicability than finding human homologs involved in the same process. Many familial cancer susceptibility syndromes are the result of mutations in genes that promote DNA repair or induce cell cycle checkpoints after DNA damage (11). Such genes include the XP family, ATM, TP53, BRCA1, BRCA2, NBS1, WRN, BLM, and hRAD51. Yeast strains mutant in homologs of these genes are sensitive to cell killing by DNA damaging agents, particularly to UV and ionizing radiation (10, 29). The association with cancer of the orthologs of the newly identified UV-sensitive genes, CaSm and AF9, strengthens this hypothesis of an association of DNA damage sensitivity with cancer and provides leads to two new genes potentially involved in cancer predisposition.
In summary, we have tested the recently completed pool of yeast deletion strains by hybridization to an oligonucleotide array for its ability to identify genes, the absence of which confers sensitivity to UV radiation. In addition to identifying known mutants in nucleotide excision repair, cell cycle checkpoints, and postreplication repair, we identified three genes previously unknown to confer resistance. Two of these genes have human orthologs, AF9 and CaSm, both of which have been implicated in human cancer. We conclude that this pool of yeast deletion strains is a powerful tool for discovery of genes not only affecting the sensitivity of cells to cytotoxic agents and hence the mechanism of action of such agents, but also human genes potentially involved in cancer predisposition.
Acknowledgments
We are grateful for help from R. Tibshirani with statistical analysis and N. Denko with the galactose expression system. This work was supported by National Institutes of Health Grants R01 CA15201 and P01 CA67166 (to J.M.B.), H600198 (to R.W.D.), by a grant from Aventis (to R.W.D.), and by Australian National Health and Medical Research Council Fellowship 997034 (to G.W.B.).
Abbreviations
- YPD
yeast extract/peptone/dextrose medium
- sam
significance analysis of microarrays methodology
- NER
nucleotide excision repair
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Foury F. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 2.Schild D, Brake A J, Kiefer M C, Young D, Barr P J. Proc Natl Acad Sci USA. 1990;87:2916–2920. doi: 10.1073/pnas.87.8.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plon S E, Leppig K A, Do H N, Groudine M. Proc Natl Acad Sci USA. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glerum D M, Tzagoloff A. Proc Natl Acad Sci USA. 1994;91:8452–8456. doi: 10.1073/pnas.91.18.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier K O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 7.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Davis R W, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 8.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 9.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 10.Game J C. Mutat Res. 2000;451:277–293. doi: 10.1016/s0027-5107(00)00055-5. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Kinzler K W. The Genetic Basis of Human Cancer. New York: McGraw–Hill; 1998. [Google Scholar]
- 12.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. . (First Published April 17, 2001; 10.1073/pnas.091062498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gietz R D, Schiestl R H. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 14.Prakash S, Prakash L. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 15.Xiao W, Chow B L, Broomfield S, Hanna M. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox B, Game J. Mutat Res. 1974;26:257–264. doi: 10.1016/s0027-5107(74)80023-0. [DOI] [PubMed] [Google Scholar]
- 18.Kuluncsics Z, Perdiz D, Brulay E, Muel B, Sage E. J Photochem Photobiol B. 1999;49:71–80. doi: 10.1016/S1011-1344(99)00034-2. [DOI] [PubMed] [Google Scholar]
- 19.Rajpal D K, Wu X, Wang Z. Mutat Res. 2000;461:133–143. doi: 10.1016/s0921-8777(00)00047-1. [DOI] [PubMed] [Google Scholar]
- 20.Bang D D, Verhage R, Goosen N, Brouwer J, van de Putte P. Nucleic Acids Res. 1992;20:3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 22.Mieczkowski P, Dajewski W, Podlaska A, Skoneczna A, Ciesla Z, Sledziewska-Gojska E. Curr Genet. 2000;38:53–59. doi: 10.1007/s002940000136. [DOI] [PubMed] [Google Scholar]
- 23.Xiao W, Chow B L, Milo C N. Mol Gen Genet. 1998;257:614–623. doi: 10.1007/s004380050689. [DOI] [PubMed] [Google Scholar]
- 24.Mullen J R, Kaliraman V, Ibrahim S S, Brill S J. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes T R, Roberts C J, Dai H, Jones A R, Meyer M R, Slade D, Burchard J, Dow S, Ward T R, Kidd M J, Friend S H, Marton M J. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 26.Tharun S, He W, Mayes A E, Lennertz P, Beggs J D, Parker R. Nature (London) 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 27.Schweinfest C W, Graber M W, Chapman J M, Papas T S, Baron P L, Watson D K. Cancer Res. 1997;57:2961–2965. [PubMed] [Google Scholar]
- 28.Strissel P L, Strick R, Tomek R J, Roe B A, Rowley J D, Zeleznik-Le N J. Hum Mol Genet. 2000;9:1671–1679. doi: 10.1093/hmg/9.11.1671. [DOI] [PubMed] [Google Scholar]
- 29.Sung P, Trujillo K M, Van Komen S. Mutat Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]