Abstract
The role of the gut microbiome in animal health has become increasingly evident. Unlike most other insects, honey bees possess a highly conserved and specialized core gut microbiome, which consists of nine bacterial species and is acquired mostly through social transmission. Five of these species are ubiquitous in honey bees and are also present in bumble bees. Recent studies have shown that the bee gut microbiome plays a role in metabolism, immune function, growth and development, and protection against pathogens. Disruption of the gut microbiome has also been shown to have detrimental effects on bee health. Overall, evidence suggests that the gut microbiome plays an important role in bee health and disease.
Introduction
Pathogens make up a small part of the communities of microorganisms associated with animal hosts. The roles of non-pathogenic microbial associates are increasingly appreciated. For example, the human gut microbiome plays a critical role in host physiology, nutrition, development, immune function, behavior, and also protection against pathogenic microorganisms [1]. Honey bees har-bor a specialized gut community, consisting of organisms largely restricted to this niche [2•• ]. The honey bee gut microbiome has some similarities to that of mammals: it is mostly socially transmitted, is largely restricted to guts of its hosts, helps to metabolize dietary carbohydrates, and contributes protection against pathogens. The basic biology of the bee gut microbiome was summarized in a recent review paper [2 ]. This article will focus on recent findings regarding possible roles of the honey bee gut microbiome in protection against disease.
The gut microbiome of corbiculate bees
The guts of honey bee (Apis mellifera) adult workers are dominated by a distinctive set of nine bacterial species (or ‘phylotypes’), largely restricted to the hindgut [2••]. Five of these, Snodgrassella alvi, Gilliamella apicola, two species of Lactobacillus, and a Bifidobacterium species, are ubiquitous and can be found in essentially every adult worker worldwide; these species can be considered as the core gut microbiome. Others (Bartonella apis, Apibacter adven-toris, Frischella perrara, and Acetobacteraceae) are present in guts of many honey bee workers, but sometimes absent. Smaller numbers of bacteria, often representing environmental species, occur in the foregut and midgut [3]. The core species are relatively infrequent in larvae and in adult queens, which contain highly variable communities dominated by environmental bacteria [4, 5]. The mature worker hindgut microbiome is substantial, totaling 108–109 bacterial cells [6] and is established in workers within four days following eclosure, before leaving the hive. Transmission is through a fecal route and facilitated by social interactions and contact with hive surfaces [6]. Each core bacterial species shows a characteristic distribution within the hindgut (Figure 1). S. alvi and G. apicola dominate the ileum region of the hindgut, where S. alvi forms a continuous layer on the lining of the longitudinal folds, and G. apicola occurs on top. F. perrara forms a melanized scab at the pylorus, near the beginning of the ileum. The others, which are Gram positive species, are most abundant in the rectum region of the hindgut.
Figure 1.
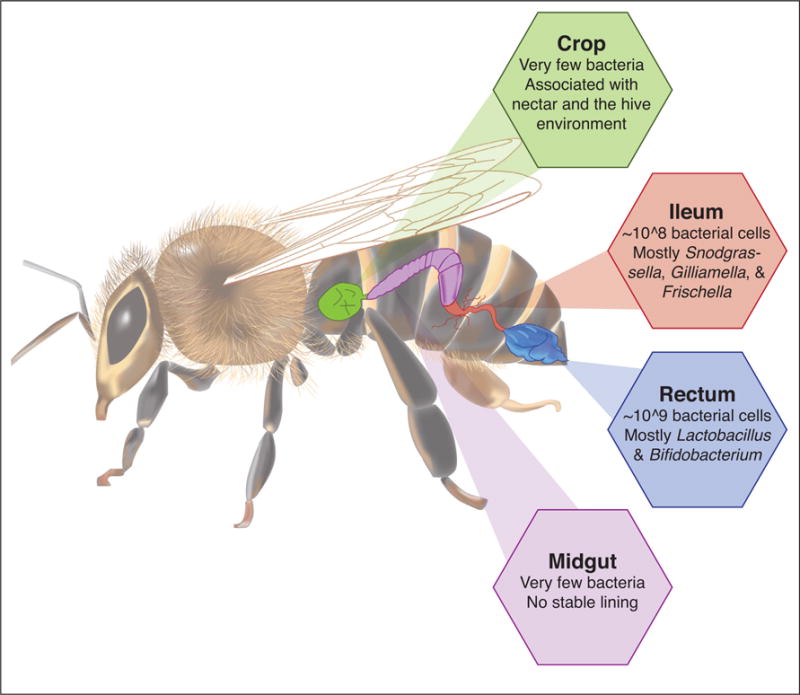
Schematic showing distribution of bacterial communities in the honey bee worker gut. The ileum and rectum are two regions of the hindgut. For detailed overview see Kwong and Moran [2••].
Each of these species exists as multiple strains, even within the gut of a single worker adult [7]. Extensive strain diversity of A. mellifera core gut bacteria, corresponding to different gene repertoires and metabolic capabilities, has been shown for G. apicola [8•], and for the two Lactobacillus clades and Bifidobacterium [9]. Accessory genes (genes present in some strains but not others within a species) include many involved in carbohydrate utilization, as well as genes encoding toxins likely targeted to competing bacteria. Strain level variation is even greater when comparing strains present in Apis versus Bombus, the latter having far fewer genes for using diverse carbohydrates [8•,10].
Other social corbiculate bees, including other honey bees (genus Apis), bumble bees (Bombus), and stingless bees (tribe Meliponini), contain distinct strains of the five core species found in A. mellifera [11••,12]. Phylogenetic analyses of strains from diverse corbiculate bee species suggest that these five bacterial species colonized a common ancestor of the corbiculate clade, about 80 million years ago, and that strains subsequently diversified, with some host lineages acquiring a few additional bacterial types. Based on phylogenetic analyses for S. alvi, G. apicola, and Lactobacillus ‘Firm-5’, related bacterial strains tend to occur within related hosts, with Apis and Bombus strains forming separate clades. All of these bees are social and live in colonies consisting of a queen and workers, enabling transmission of a consistent gut microbiome across generations.
The gut microbiome can be studied experimentally, since core species can be grown in culture and inoculated into bees [13]. If bees are manually removed from the comb at an early pupal stage (before the mouthparts harden) using sterile methods, guts of emerging adults will contain few or no bacteria and lack all core species, enabling experiments in which the gut community is inoculated in a controlled manner (e.g. [10,11••,14••,15••,16•]). Experiments comparing microbiotia-free and inoculated bees have revealed some of the functions of the gut microbiome and its members (Table 1). Controlled inoculations have also shown that strains of S. alvi from Apis cannot inoculate Bombus hosts, and vice versa [11••] but that there is some ability of S. alvi strains to inoculate other host species within Apis [11••] and within Bombus [10]. Strains appear largely host-specific in natural collections, with distinct micro-biomes for each host species, and no evident geographic convergence when bee species co-occur [11••,17].
Table 1.
Experiments comparing microbiota-free bees with inoculated bees
| Bee species | Inoculation | Trait compared | Citation |
|---|---|---|---|
| A. mellifera | S. alvi, G. apicola, and whole community | Immune gene expression, survival rate following E. coli injection | Kwong et al. [30•] |
| A. mellifera | S. alvi | Susceptibility to Lotmaria infection, immune gene expression, vitellogenin expression | Schwarz et al. [29•] |
| A. mellifera | F. perrara | Pylorus scab formation | Engel et al. [16•] |
| A. mellifera | F. perrara | Immune gene expression, melanization response, overall gene expression | Emery et al. [14••] |
| A. mellifera | Whole community | Metabolism, insulin pathway expression, vitellogenin expression, growth | Zheng et al. [15••] |
| A. mellifera | S. alvi | Host specificity | Kwong et al. [10] |
| A. mellifera | S. alvi | Host specificity | Kwong et al. [11•] |
| A. mellifera | Whole community | Routes of colonization | Powell et al. [6] |
| B. terrestris | Whole community | Susceptibility to Crithidia infection | Koch and Schmid-Hempel [39••] |
| B. terrestris | Whole community | Susceptibility to Crithidia infection | Koch and Schmid-Hempel [40] |
| A. mellifera | Whole community | Survival following antibiotic exposure | Raymann et al. [24••] |
Non-core species in the bee microbiome: potential pathogens?
Guts of virtually all honey bee adult workers are dominated by five bacterial species (the core gut microbiome). Worker guts typically also contain low frequencies of other bacteria, which may play important biological roles, through their interactions with other organisms in the gut or through their direct, potentially pathogenic, effects on hosts. Some appear specific to honey bee guts, but are not ubiquitous. For example, Frischella perrara, a relative of G. apicola within the family Orbaceae, is widespread [18]. It colonizes in a distinctive manner in the pylorus region near the junction of the midgut and the hindgut; experiments involving inoculations of microbiota-free bees show that it causes a characteristic brown ‘scab’ [16•], which has been shown to result from stimulation of immune pathways including the melanization response [14••]. This species also causes disordered cell division in gut epithelial cells and produces a complex polyketide molecule that affects cellular replication in human cell lines [19].
Another common non-core bee gut species is, Bartonella apis, a member of a group containing animal pathogens [20•]; B. apis is widespread in honey bee workers, but impacts on hosts are unknown. In a study of associations of microbes with colony collapse symptoms, B. apis was relatively abundant in healthy bees relative to bees from collapsing colonies [21], suggesting the possibility of a positive effect on disease resistance. Likewise, effects on hosts are unknown for Apibacter adventoris, a Bacteroi-detes species sampled repeatedly from bee guts and not elsewhere, but never abundant [22].
Many of the rarer bacterial species in honey bee guts likely represent opportunistic organisms able to invade as pathogens. Commonly sampled groups include species of Enterobacteriaceae, including Hafnia alvi, and species of Enterobacter, Klebsiella, and Serratia. Serratia marcescens strains can be pathogenic, causing sepsis and death [23•]. Strains isolated from hives can cause mortality when administered orally to workers in the laboratory [24••]. Potentially, these Enterobacteriaceae pathogens are under-recognized as causes of bee mortality, since infected bees usually leave the hive to die; they are more likely to accumulate in wintering hives [23•].
Environmental and developmental factors that can alter the bee gut microbiom
In animals generally, gut microbiome composition is influenced by many factors, including diet, stress, immune responses, stress, aging, and exposure to antibiotics. All of these factors appear to affect the bee microbiome (Figure 2). Some evidence suggests that, as workers age and transition to foraging, the microbiome composition shifts slightly [25,26]. Microbiome composition, particularly the relative numbers of S. alvi and G. apicola, can also shift through the season, possibly reflecting changes in diet [27•]. Indeed, poor nutrition has been shown to disrupt the normal gut microbiome, resulting in higher mortality and disease susceptibility [28•]. Disruption of the microbiome (dysbiosis) has many consequences for worker development: such disruption during early adult life affects expression of important developmental genes, including vitellogenin [29•] and is expected to affect immune system function, since the honey bee microbiome stimulates immune pathways [14••,29•,30•]. In turn, honey bee innate immune function has been shown to be compromised by stimulation of cellular stress responses [31•]. Together these findings suggest that dysbiosis may have cascading effects for the ability of bees to respond to environmental stressors such as poor nutrition or temperature stress, and that, conversely, these stressors may impact the microbiome (Figure 2).
Figure 2.
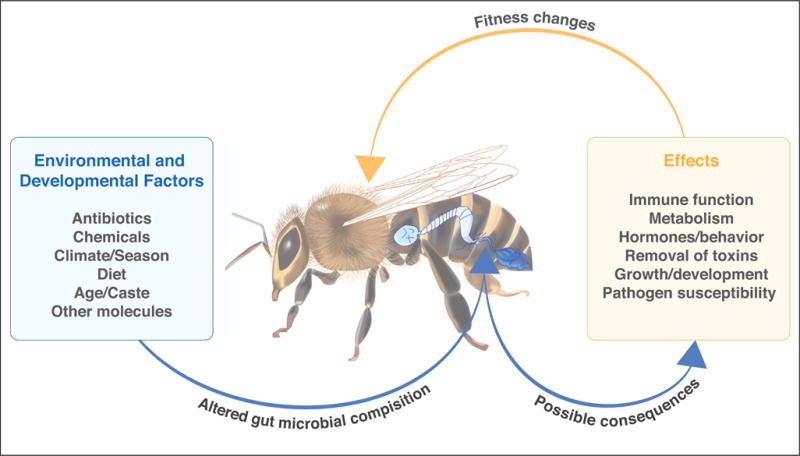
Overview of roles of the gut microbiome in honey bee health.
Honey bees in some regions are routinely exposed to antibiotics used in beekeeping for preventing outbreaks of American or European Foulbrood caused by Paeniba-cillus larvae or Melissococcus plutonius, respectively. Oxy-tetracycline has been used for decades in beekeeping in the USA, and strains of bee gut bacterial species have acquired several tetracycline resistance loci, with frequencies highest in colonies exposed more recently [32]. Tetracycline exposure results in severe gut dysbiosis, with drastic and persistent effects on microbiome size and composition [24••]. The treatment also increases mortality in the hive, potentially due to greater susceptibility to opportunistic pathogens, as observed in the lab [24••]. Certain pesticides have also been shown to impact the honey bee microbiome [33].
While a largely consistent microbiome persists throughout the lifespans of honey bee adult workers, adult queens have a strikingly different microbiome composition, with greater variation among individuals, and consisting of bacteria that are also found in the hive environment [4•,5]. The size of the queen microbiome is highly variable but often smaller than that of workers. The causes of the striking differences in microbiome between queens and workers are not yet known, but may reflect caste-specific differences in immune activities or in physiological conditions within the gut.
A study of microbiome shifts with worker age in the Asian A honey bee, Apis cerana, showed that the core gut bacteria peak in young workers and decline in numbers as workers age [34]; such dramatic shifts have not been reported for A. mellifera, suggesting that microbiome stability over the lifespan differs between species. In bumble bees, shifts with age, stress and exposure to environmental bacteria are even more pronounced, and many individuals exhibit increased frequencies of Enterobacteriaceae and other non-core, and potentially pathogenic, bacterial species, which sometimes dominate in individual bee guts [35–37].
Roles of the bee gut microbiome in nutrition and metabolism
Genomic and metabolic studies on bee gut core species, G. apicola, Lactobacillus species, and Bifidobacterium, indicate capabilities to digest and metabolize a diverse array of plant-produced carbohydrates. In experiments comparing microbiota-free bees and bees possessing a conventional gut microbiome, many physiological effects of the gut microbiome were evident, including a major positive effect on gut size, weight gain following eclosure, insulin and vitellogenin signaling, and sucrose sensitivity [15••]. All of these physiological variables are expected to impact bee health, immune responsiveness, and susceptibility to stress. The gut microbiome has major effects on the profile of short chain fatty acids in the gut and in the hemolymph; for example, butyrate dominates in conventional bees but is entirely absent in microbiota-free bees [15••]. Genomic studies reveal that some core gut bacteria harbor many genes for carbohydrate metabolism [8•,10,38], and different strain compositions possess different abilities to metabolize carbohydrates [8•,9,39••].
Roles of the bee gut microbiome in protection against pathogens
In both honey bees and bumble bees, the gut microbiome has been shown to play some role in protection against pathogen infection (Table 2). In two separate studies, microbiota-free B. terrestis inoculated with the fecal matter of wild-type workers were more resistant to the trypanosomatid gut parasite Crithidia bombi than bees that were not inoculated [40,41]. The protective ability of the microbiome transplant was more strongly influenced by the colony source rather than the bees’ colony of origin, suggesting that different gut microbiome compositions can be more or less protective [41]. These studies did not identify the strains underlying the protection; thus, the specific community members conferring pathogen protection in bumble bees warrants further investigation.
Table 2.
Experiments demonstrating a role of the microbiome in protection against pathogens
| Pathogen | Host | Mode of infection | Protector(s) | Citation |
|---|---|---|---|---|
| Paenibacillus larvae | A. mellifera | Ingested | LAB* mixture | Forsgren et al. [42] |
| Crithidia bombi | B. terrestris | Ingested | Entire community | Koch and Schmid-Hempel [39••] |
| Crithidia bombi | B. terrestris | Ingested | Entire community | Koch and Schmid-Hempel [40] |
| Melissococcus plutonius | A. mellifera | Ingested | LAB* mixture | Vasquez et al. [43] |
| Lotmaria passim | A. mellifera | Ingested | Snodgrassella | Schwarz et al. [29•] |
| Serratia marcescens | A. mellifera | Ingested | Entire community | Raymann et al. [24••] |
| Escherichia coli | A. mellifera | Injected | S. alvi and G. apicola | Kwong et al. [30•] |
| Pathogen | Host | (+) Correlations | (−) Correlations | Citation |
|
| ||||
| Crithidia bombi | B. terrestris | None reported | S. alvi, G. apicola | Koch et al. [41] |
| Crithidia | B. impatiens, B. bimaculatus, B. griseocollis | Alpha 2.2 | G. apicola | Cariveau et al. [36] |
| Nosema | B. impatiens, B. bimaculatus, B. griseocollis | S. alvi | None reported | Cariveau et al. [36] |
| Sacbrood virus | A. cerana | None reported | S. alvi, Lactobacillus | Guo et al. [34] |
| Melissococcus plutonius | A. cerana | None reported | S. alvi, Bifidobacterium | Guo et al. [34] |
| Nosema spp. | A. mellifera | F. perrara | None reported | Maes et al. [28•] |
| Paenibacillus larvae | A. mellifera | None reported | Bifidobacterium, Lactobacillus | Erban et al. [46] |
| Melissococcus plutonius | A. mellifera | G. apicola, F. perrara Bifidobacterium | S. alvi and Lactobacillus | Erban et al. [47] |
A few other studies have indirectly assessed the role the bumble bee microbiome plays in pathogen infection, by correlating the presence of pathogens with the abundance of gut community members (Table 2). For example, B. terrestris infected with Crithidia was shown to possess lower numbers of G. apicola and S. alvi than uninfected individuals [42]. However, in B. impatiens, B. bimaculatus, and B. griseocollis, only the abundance of G. apicola was negatively correlated with the presence of Crithidia, and Parasaccharibacter apium (Alpha 2.2) was positively correlated with Crithidia infection [36]. Correlations between the microbiome composition and Nosema infection have been less consistent, with one study showing a positive correlation with the abundance of S. alvi [36], and other studies finding no differences in microbiome composition between bees infected or not infected with Nosema [40,42]. Although these correlations are interesting, it is not clear if they are the cause or the effect of pathogen infection.
In A. mellifera, several studies provide evidence for a role of the adult gut microbiome in protection against bee pathogens (Table 2). Few studies have examined the protective role of individual members of the core gut microbiome. One study investigated whether colonization with S. alvi, G. apicola, or the whole community could protect against hemolymph infection by E. coli. Bees possessing the entire gut community and, to a smaller extent, bees mono-inoculated with S. alvi or with G. apicola cleared more bacteria from the hemolymph after E. coli injection, and contained more antimicrobial peptide than did microbiome-free bees, suggesting immune priming by these core gut community members [30•]. Immune priming has also been shown for F. perrara, which colonizes locally in the ileum region of the hindgut and stimulates a dramatic increase in production of the antimicrobial peptide apidaecin [14••]. Although our focus is on adult workers, some studies have also examined potential interactions of gut microbiomes with larval pathogens. When larvae are given a sterile sugar diet or a sugar diet spiked with different lactic acid bacteria (LAB) and then exposed to P. larvae or M. plutonius, the LAB cocktails reduced infection by these larval pathogens [43,44]. However, the LAB strains used in these studies were isolated from adult workers crops [45,46], which mostly contain species that inhabit nectar and hive materials and are not part of the core gut microbiome [26].
Dysbiosis of the A. mellifera core gut microbiome can increase susceptibility to pathogens. Treatment with the antibiotic tetracycline, which severely alters the core gut community composition and size, leads to increased infection by the opportunistic pathogen S. marcescens within hives [24••]. Likewise, in laboratory experiments, bees treated with tetracycline were more susceptible to S. marcescens infection than control bees [24••]. Dysbiosis-induced susceptibility to parasite infection also was found for bees pre-inoculated with S. alvi prior to being released into hives [29•]; pre-inoculation perturbs the core micro-biome and increases susceptibility to the protozoan parasite Lotmaria passim [29•].
The presence of various honey bee pathogens of both adult and larval stages sometimes correlates with the presence or abundance of different members of the adult core gut community [28•,34,47,48]. Some of these studies show negative correlations between the presence of pathogens and the relative abundances of core members of the adult gut community (Table 2). However, it is not clear that lower abundance of core gut species is a cause of increased susceptibility to pathogens. In some cases, increased pathogen loads could be due to general perturbation of metabolism or immune responses, with this perturbation also impacting the gut community. Alternatively, dysbiosis may result from pathogen infection. Some experimental studies do support a causative role of core gut species in protection (Table 2), but more experiments are needed to determine the extent to which specific core gut community members may protect against particular pathogens.
Conclusions
Substantial evidence now points to a role of the bee gut microbiome in honey bee and bumble bee health. Most experimental work is based on laboratory studies, while most studies under field conditions are based on correlational analyses only. Thus, experimental studies in apiaries under realistic field conditions are needed. Many infectious diseases of adult bees are caused by organisms that are widespread or ubiquitous in colonies but that occasionally undergo outbreaks causing disease. Potentially these outbreaks are triggered by disruption of the normal microbiome. Active management of the worker gut microbiome could be a tool for improving bee health [49].
Footnotes
This review comes from a themed issue on Parasites/parasitoids/biological control
Edited by Bryony Bonning, Elke Genersch and Annette Bruun Jensen
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–384. doi: 10.1038/nrmicro.2016.43. Comprehensive review of the structure, transmission, and evolution of the gut microbial communities of social bees. The various metabolic functions of core members of the bee gut microbiome are predicted based on genomic culture-based studies. This review also discusses the usefulness of the bee as a model system to study gut microbial communities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Tarpy DF, Mattila HR, Newton ILG. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl Environ Microbiol. 2015;81:3182–3191. doi: 10.1128/AEM.00307-15. Provides a description of the gut microbiome composition of honey bee queens. This study shows that queens possess a specific microbiome that is very different from workers, and that the queen microbiome composition shifts dramatically as queens age. The authors found no correlation between the queen microbiome and the microbiome of workers who cared for them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapheim KM, Rao VD, Yeoman CJ, Wilson BA, White BA, Goldenfeld N, Robinson GE. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera) PLoS ONE. 2015;10:e0123911. doi: 10.1371/journal.pone.0123911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol. 2014;80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio. 2016;7:e01326–e1416. doi: 10.1128/mBio.01326-16. Over 40 strains of G. apicola from honey bees and bumble bees were isolated and sequenced. Although very similar at the 16S rDNA level, strains varied greatly in accessory genes, specifically those involved in sugar utilization. The authors found that G. apicola, has the ability to metabolize mannose, xylose, arabinose, and rhamnose, all which can be toxic to bees. Although sugar metabolism varied among G. apicola strains, every strain from A. mellifera was able to utilize at least one of these sugars. Strains from Bombus species had very limited ability to use different sugars. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SG, Vásquez A. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics. 2015;16:284. doi: 10.1186/s12864-015-1476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A. 2014;111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Jaffé R, Moran NA. Dynamic microbiome evolution in social bees. Sci Adv. 2017;3:e1600513. doi: 10.1126/sciadv.1600513. The authors compared the gut microbiomes of the major groups of eusocial corbiculate bees (honey bees, bumble bees, and stingless bees) from multiple locations on four continents. Different host species have distinct gut communities, but a core corbiculate microbiota was identified, consisting of five core gut bacterial lineages. Most core bacterial strains were specific to their hosts, suggesting co-evolution and diversification with host species. However, some generalist strains were found that could colonize multiple hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol. 2013;22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- 13.Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA. Standard methods for research on Apis mellifera gut symbionts. J Apic Res. 2013;52:1–24. [Google Scholar]
- 14••.Emery O, Schmidt K, Engel P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera) Mol Ecol. 2017;26:2576–2590. doi: 10.1111/mec.14058. Transcriptome changes between bees experimentally colonized with F. perrara and S. alvi and between age-controlled hive bees with and without the scab phenotype were evaluated. F. perrara, but not S. alvi, caused strong activation of the host immune system. F. perrara upregulated the pattern recognition receptors, antimicrobial peptides and transporter genes as well as the melanization cascade, which suggests that the scab phenotype corresponds to a melanization response of the host. [DOI] [PubMed] [Google Scholar]
- 15••.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci U S A. 2017;114:4775–4780. doi: 10.1073/pnas.1701819114. By comparing microbiota-free bees and conventional bees, the authors show that the gut microbiome promotes weight gain of both whole body and the gut in individual honey bees. The gut microbiome was also shown to affect honey bee hormonal signaling, behavior, and gut physicochemical conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Engel P, Bartlett KD, Moran NA. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. mBio. 2015;6:e00193–e215. doi: 10.1128/mBio.00193-15. This study showed that F. perrara causes the characteristic scab phenotype on the gut epithelium of honey bees. Causation was verified through correlations in adult bees between the presence of a pylorus scab and a high abundance of F. perrara, FISH microscopy, and experimental colonization of microbiota-free bees with F. perrara. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell E, Ratnayeke N, Moran NA. Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol Ecol. 2016;25:4461–4471. doi: 10.1111/mec.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel P, Kwong WK, Moran NA. Frischella perrara gen. nov., sp nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int J Syst Evol Microbiol. 2013;63:3646–3651. doi: 10.1099/ijs.0.049569-0. [DOI] [PubMed] [Google Scholar]
- 19.Engel P, Vizcaino MI, Crawford JM. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl Environ Microbiol. 2015;81:1502–1512. doi: 10.1128/AEM.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Kenerová L, Moritz R, Engel P. Bartonella apis sp nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol. 2016;66:414–421. doi: 10.1099/ijsem.0.000736. This report describes the genome sequence of Bartonella apis isolated from the honey bee gut. Bartonella apis sp. nov. was found to have distinct growth characteristics and a number of biochemical properties that distinguish it from other species of the genus Bartonella, therefore representing a new genus. [DOI] [PubMed] [Google Scholar]
- 21.Cornman RS, Tarpy DR, Chen Y, Jeffreys L, Lopez D, Pettis JS, vanEngelsdorp D, Evans JD. Pathogen webs in collapsing honey bee colonies. PLoS ONE. 2012;7:e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong WK, Moran NA. Apibacter adventoris gen. nov., sp nov., a member of the phylum Bacteroidetes isolated from honey bees. Int J Syst Evol Microbiol. 2016;66:1323–1329. doi: 10.1099/ijsem.0.000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Burritt NL, Foss NJ, Neeno-Eckwall EC, Church JO, Hilger AM, Hildebrand JA, Warshauer DM, Perna NT, Burritt JB. Sepsis and hemocyte loss in honey bees (Apis mellifera) infected with Serratia marcescens strain sicaria. PLoS ONE. 2016;11:e0167752. doi: 10.1371/journal.pone.0167752. A strain of S. marcescens was isolated from dead bees in winterkilled hives and from Varroa mites. The S. marcescens strain sicaria (Ss1) was sequenced and characterized. Ss1 was associated with reduced numbers of host-defensive hemocytes, sepsis, and death. These results raise the possibility that opportunistic Serratia strains are important causes of mortality in honey bees, and that stress on workers might increase susceptibility to this mortality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15:e2001861. doi: 10.1371/journal.pbio.2001861. The effects of the antibiotic tetracycline on the gut microbiome of honey bees was evaluated. Tetraycline exposure was shown to severely alter gut microbiome composition, decrease survivorship of bees in the hive and in the lab, and lead to increased susceptibility to the opportunistic pathogen S. marcescens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corby-Harris V, Maes P, Anderson KE. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE. 2014;9:e95056. doi: 10.1371/journal.pone.0095056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson KE, Rodrigues PA, Mott BM, Maes P, Corby-Harris V. Ecological succession in the honey bee gut: shift in Lactobacillus strain dominance during early adult development. Microb Ecol. 2016;71:1008–1019. doi: 10.1007/s00248-015-0716-2. [DOI] [PubMed] [Google Scholar]
- 27•.Ludvigsen J, Rangberg A, Avershina E, Sekelja M, Kreibich C, Amdam G, Rudi K. Shifts in the midgut/pyloric microbiota composition within a honey bee apiary throughout a season. Microbes Environ. 2015;30:235–244. doi: 10.1264/jsme2.ME15019. Authors evaluated how the honey bee midgut/pyloric microbiota changes across seasons. The honey bee midgut/pyloric microbiota composition changed throughout the foraging season. In particular, G. apicola dominated the gut microbiome composition in early in the summer, but S. alvi dominated at the end of summer. In contrast, honey bees maintained a stable microbiota composition during the winter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Maes PW, Rodrigues PA, Oliver R, Mott BM, Anderson KE. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera) Mol Ecol. 2016;25:5439–5450. doi: 10.1111/mec.13862. The authors examined the effect of diet on the gut microbiome composition, mortality, and weight of honey bees. The microbiome composition differed significantly between bees given fresh versus aged pollen. Bees given aged pollen showed decreased thorax weight, increased mortality, and increased numbers of Nosema spores. However, bees given fresh pollen or a fresh substitute showed no difference in microbiome composition, weight, mortaility, or pathogen infection. [DOI] [PubMed] [Google Scholar]
- 29•.Schwarz RS, Moran NA, Evans JD. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci U S A. 2016;113:9345–9350. doi: 10.1073/pnas.1606631113. The effects of early colonization by S. alvi on microbiome composition and susceptibility to the parasite Lotmaria passim were evaluated. Early colonization with S. alvi caused disruption of the gut microbiome, made bees more susceptible to L. passim infection, and altered developmental and detoxification gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Kwong WK, Mancenido AL, Moran NA. Immune system stimulation by the native gut microbiota in honey bees. R Soc Open Sci. 2017;4:170003. doi: 10.1098/rsos.170003. Authors tested whether the gut microbiome of the honey bee can induce expression of antimicrobial peptides (AMPs). They found that up-regulated expression of the AMPs apidaecin and hymenoptaecin in gut tissue when the microbiome is present. In microbiota-free bees, gut apidaecin levels were lower in the gut and the hemolymph than in bees with a normal gut microbiome. Bees inoculated with the normal microbiome showed increased survivorship following injection with E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.McKinstry M, Chung C, Truong H, Johnston BA, Snow JW. The heat shock response and humoral immune response are mutually antagonistic in honey bees. Sci Rep. 2017;7:8850. doi: 10.1038/s41598-017-09159-4. The authors characterized the cellular stress response and the Heat Shock Response (HSR), in honeybees and found that induction of the HSR through heat reduces expression of the antimicrobial peptides hymenoptaecin, defensin 1, and abaecin. Conversely, induction of immune pathways by wounding the bee abdomen resulted in decreased expression of multiple HSR genes, suggesting a mutually antagonistic relationship between the HSR and immune activation in honey bees. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honey bees. mBio. 2012;3:e00377–e412. doi: 10.1128/mBio.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakumanu ML, Reeves AM, Anderson TD, Rodrigues RR, Williams MA. Honey bee gut microbiome is altered by in-hive pesticide exposures. Front Microbiol. 2016;7:1255. doi: 10.3389/fmicb.2016.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Wu J, Chen Y, Evans JD, Dai R, Luo W, Li J. Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J Invertebr Pathol. 2015;127:110–114. doi: 10.1016/j.jip.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Billiet A, Meeus I, Van Nieuwerburgh F, Deforce D, Wa¨ckers F, Smagghe G. Impact of sugar syrup and pollen diet on the bacterial diversity in the gut of indoor-reared bumblebees (Bombus terrestris) Apidologie. 2015;47:548–560. [Google Scholar]
- 36.Cariveau DP, Powell JE, Koch H, Winfree R, Moran NA. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus) ISME J. 2014;8:2369–2379. doi: 10.1038/ismej.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Powell JE, Guo J, Evans JD, Wu J, Williams P, Lin Q, Moran NA, Zhang Z. Two gut community enterotypes recur in diverse bumblebee species. Curr Biol. 2015;25:R652–R653. doi: 10.1016/j.cub.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 38.Lee FJ, Rusch DBDB, Stewart FJ, Mattila HR, Newton ILG. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol. 2015;17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 39••.Ellegaard KM, Engel P. Beyond 16S rRNA community profiling: intra-species diversity in the gut microbiota. Front Microbiol. 2016;7:1475. doi: 10.3389/fmicb.2016.01475. This paper reviews the evolution and functional relevance of strain level diversity in host-associated gut microbial communities. Authors stress importance of intra-species level diversity within gut communities and discuss the limitations of 16S rRNA profiling for evaluating intra-species level diversity. Recent studies that have utilized different approaches to evaluate strain level diversity in humans and honey bees are highlighted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch H, Schmid-Hempel P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett. 2012;10:1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- 42.Koch H, Cisarovsky G, Schmid-Hempel P. Ecological effects on gut bacterial communities in wild bumblebee colonies. J Anim Ecol. 2012;81:1202–1210. doi: 10.1111/j.1365-2656.2012.02004.x. [DOI] [PubMed] [Google Scholar]
- 43.Forsgren E, Olofsson TC, Váasquez A, Fries I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie. 2010;41:99–108. [Google Scholar]
- 44.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS ONE. 2012;7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olofsson TC, Vásquez A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol. 2008;57:356–363. doi: 10.1007/s00284-008-9202-0. [DOI] [PubMed] [Google Scholar]
- 46.Vásquez A, Olofsson TC. The lactic acid bacteria involved in the production of bee pollen and bee bread. J Apic Res. 2009;48:189–195. [Google Scholar]
- 47.Erban T, Ledvinka O, Kamler M, Nesvorna M, Hortova B, Tyl J, Titera D, Markovic M, Hubert J. Honeybee (Apis mellifera)-associated bacterial community affected by American foulbrood: detection of Paenibacillus larvae via microbiome analysis. Sci Rep. 2017;7:5084. doi: 10.1038/s41598-017-05076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erban T, Ledvinka O, Kamler M, Hortova B, Nesvorna M, Tyl J, Titera D, Markovic M, Hubert J. Bacterial community associated with worker honeybees (Apis mellifera) affected by European foulbrood. PeerJ. 2017;5:e3816. doi: 10.7717/peerj.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rangberg A, Diep DB, Rudi K, Amdam GV. Paratransgenesis: an approach to improve colony health and molecular insight in honey bees (Apis mellifera)? Integr Comp Biol. 2012;52:89–99. doi: 10.1093/icb/ics089. [DOI] [PubMed] [Google Scholar]


