Abstract
The ETS family of transcription factors has been repeatedly implicated in tumorigenesis. In prostate cancer, ETS family members such as ERG, ETV1, ETV4 and ETV5 are frequently overexpressed due to chromosomal translocations, but the molecular mechanisms by which they promote prostate tumorigenesis remain largely undefined. Here we show that ETS family members such as ERG and ETV1 directly repress the expression of the checkpoint kinase 1 (CHK1), a key DNA Damage Response (DDR) cell cycle regulator essential for the maintenance of genome integrity. Critically, we find that ERG expression correlates with CHK1 downregulation in human patients, and demonstrate that Chk1 heterozygosity promotes the progression of high-grade prostatic intraepithelial neoplasia (HGPIN) into prostatic invasive carcinoma (CaP) in Pten+/− mice. Importantly, CHK1 downregulation sensitizes prostate tumor cells to etoposide but not to docetaxel treatment. Thus, we identify CHK1 as a key functional target of the ETS proto-oncogenic family with important therapeutic implications.
INTRODUCTION
The complex network of pathways known as DDR (for DNA Damage Response) orchestrate the cellular response to damaged DNA (1, 2). One consequence of activation of these pathways is the induction of cell-cycle checkpoints that halt proliferation in order to repair the damaged DNA, or, if the damage is beyond repair, trigger apoptosis (3). Significant numbers of studies have demonstrated that derangement of the normal function of members of the DDR leads to unrepaired or mis-repaired DNA, and consequent genomic instability, which is often a prelude to cancer (1, 3).
CHK1 is a key regulator of the DDR best known for its involvement in various cell-cycle checkpoints and its monitoring of DNA integrity during replication (4–11). Loss of heterozygosity of 11q24, where CHK1 is located, have been reported along with frame-shift mutations in the CHK1 gene in genetically unstable stomach, colorectal, breast, colon and endometrial cancers (12–16), while overexpression of PPM1D in breast cancer and other tumor types has been proposed to contribute to tumorigenesis by inappropriate inactivation of CHK1 (17). Variations of CHK1 mRNA and protein levels have been detected in lymphoid neoplasms in the absence of gene mutations, deletions or promoter hypermethylation, suggesting the possibility of transcriptional and/or post-transcriptional downregulation mechanisms (18).
ERG is overexpressed in 50% of human prostate cancers as a result of chromosomal rearrangements involving mainly the TMPRSS2 and ERG genes (3,23–27). Several studies have to some extent defined possible mechanisms by which ERG may promote prostate tumorigenesis (19–25). We have previously shown that overexpression of ERG in combination with Pten heterozygosity in the mouse prostate converts Pten haploinsufficiency-dependent HGPIN to invasive adenocarcinoma through the up-regulation of gene involved in cell migration such as ADAMTS1 and CXCR4 (19). Microarray analysis has likewise demonstrated that a significant number of genes involved in various pathways are deregulated by excess amounts of ERG (19–25), however, the relationship of these genes to ERG-dependent oncogenesis is still unclear and their in vivo genetic validation remains to be addressed.
RESULTS
CHK1 is downregulated upon ERG or ETV1 induction
As most of the transcription factors, ERG functions as an activator or a repressor on different target genes. Accordingly, microarray analysis from HEK-293 cells overexpressing ERG (19) revealed downregulated expression of a number of interesting genes, including CHK1 (Supplementary Fig. S1A), which may represent a potential new link between ERG, DNA replication stress, DDR, and tumorigenesis.
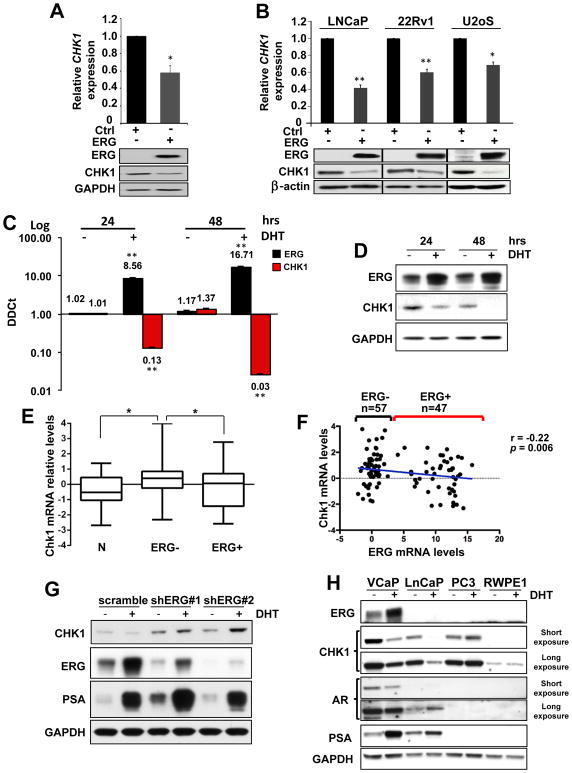
CHK1 downregulation was further confirmed using quantitative RT-PCR (RT-qPCR) and Western blot analysis in HEK-293 (Fig. 1A and Supplementary Fig. S1B) and in additional cell lines overexpressing exogenous ERG (Fig. 1B and Supplementary Fig. S1C). To test the ability of ERG to downregulate CHK1 endogenously, we treated VCaP cells (a prostate cell line characterized by the endogenous TMPRSS2-ERG translocation) with 10 nM dihydrotestosterone (DHT) for 24 and 48 hours. RT-qPCR (Fig. 1C) and Western blot (Fig. 1D) data clearly showed a progressive downregulation of CHK1 with increasing amounts of ERG, while CHK2 levels remained unchanged (Supplementary Fig. S1D). Analysis of a publicly available expression profile dataset of VCaP cells (either untreated or treated with 10 nM DHT for 16 hours) confirmed CHK1 down-regulation upon ERG induction (Supplementary Fig. S1E) (26). Importantly, expression profile data analysis of ERG negative (ERG-, n=57) and ERG positive (ERG+, n=47) human low Gleason untreated primary CaP presented in the Taylor dataset (27), clearly shows a statistically significant reduction of CHK1 transcript in ERG+ compared to ERG- CaP (Fig. 1E), that inversely correlates with the amount of ERG transcript (Fig. 1F).
Figure 1. CHK1 downregulation upon ERG overexpression.
A, RT-qPCR and Western blot analyses showing reduction in CHK1 levels in response to overexpression of ERG in 293 cells. B, CHK1 mRNA and protein downregulation 24 hours following overexpression of ERG in the indicated cell lines. C, Increasing amount of ERG transcript in the VCaP cells inversely correlates with CHK1 mRNA levels after 24 and 48 hours of DHT (10 nM) treatment. D, ERG and CHK1 protein levels in samples described in C. E, Box plot showing mean, standard deviation and range of z-scores of CHK1 expression in normal prostate tissue (N), ERG positive (ERG+) or ERG negative (ERG-) prostate tumors. P < 0.05 calculated using two-tailed t-test. F, Dot plot of CHK1 mRNA expression versus ERG mRNA expression. Each dot represents an individual patient. A blue line shows the linear regression of the data. The slope of the line is significantly non-zero (r=−0.022, p=0.006). Error bars in bar graphs indicate mean±s.d. G, Western blot analysis of ERG, CHK1, and PSA in VCaP cell line transduced with two independent shRNAs targeting ERG and treated for 24 hours with 10 nM DHT. VCaP cells transduced with a scramble shRNA sequence were used as control. H, Western blot analysis of ERG, CHK1, AR, PSA, in VCaP, LNCaP, PC3, RWPE1 prostate cell lines after 24 hours of DHT (10 nM) treatment. GAPDH was used as loading control. Error bars indicate mean±s.d. Experiments were performed in triplicate, data were analyzed using unpaired t-test. Values of P < 0.05 were considered statistically significant (*p<0.05, **p<0.01).
Due to the many caveats to be considered in analyzing most of publicly available expression profile datasets, in order to further investigate the status and a possible correlation of ERG and CHK1 in human CaP, we created a highly controlled tissue micro array (TMA) data set with matched clinical information, including 130 biopsied specimens from prostatectomized human primary CaP, and stained serial sections for ERG or CHK1. ERG and CHK1 staining were scored independently on 89 specimens as percentage of tumor cells expressing the marker. The histopathological analysis defined a significant inverse correlation between ERG and CHK1 expression in human primary CaP (Supplementary Fig. S1F), fully confirming the expression profile analysis (Fig. 1E–F).
Since ERG expression in human CaP depends by the androgen receptor (AR) activity, we overexpressed AR in HEK-293 either untreated or treated with 10 nM DHT for 24 hours, in order to exclude a possible direct role of AR in CHK1 transcriptional repression. No sizable differences in CHK1 levels were detected between control and AR overexpressing cells (Supplementary Fig. S2A). Next, we silenced ERG in VCaP cells untreated or treated with 10 nM DHT for 24 hours. As shown in Figure 1G, ERG knockdown consistently increased CHK1 levels in untreated conditions (Fig. 1G line 1 compared to 3, and 5) while, unexpectedly, DHT treatment increased CHK1 amount in ERG silenced VCaP cells instead of decreasing it (Fig. 1G line 1–2 compared to 3–4, and 5–6). Finally, we treated a panel of AR proficient or deficient prostate cell lines (VCaP, LnCaP, RWPE1, 22RV1, and PC3) with 10 nM DHT for 24 hours (Fig. 1H and Supplementary Fig. S2B). As expected, VCaP cells showed a robust induction of ERG and a consequent downregulation of CHK1 (Fig. 1H). Importantly, CHK1 levels remained unchanged in ERG-negative AR-positive RWPE1 and 22RV1 (Fig. 1H and Supplementary Fig. S2B), as well as in ERG-negative AR-null PC3 prostate cell lines (Fig. 1H).
Intriguingly, however, CHK1 downregulation was observed at protein and RNA level in ERG-negative AR-positive LnCaP cell line (Fig. 1H and Supplementary Fig. S2C). This caught our attention because LnCaP cells are characterized by the translocation of ETV1 gene into an intronic sequence of the MIPOL1 locus at 14q13.3–14q21.1 (28). ETV1 is an additional member of the oncogenic ETS transcription factors family that is overexpressed in approximately 15% of human CaP patients who are TMPRSS2-ERG negative (29). As in the case of ERG, ETV1 up-regulation is the consequence of gene translocation in a genetic locus regulated by the androgen receptor and its induction after DHT treatment is clearly evident by RT-qPCR and by Western blot after proteasome inhibition (Supplementary Fig. S2C–D) (28, 30–32). Since ETV1 and ERG share the same DNA binding site on the promoter of their target genes, they can potentially regulate a common oncogenic signature in human CaP (19, 23, 26, 30). Accordingly, ETV1 overexpression in HEK-293 and PC3 cell lines robustly downregulated CHK1 (Supplementary Fig. 1B–C), while ETV1 knockdown in LnCaP was sufficient to increase CHK1 amount (Supplementary Fig. S2E–F). Finally, analysis of a publicly available expression profile dataset of RWPE1 cells overexpressing ETV1 confirmed CHK1 down-regulation (Supplementary Fig. S2G) (26). Notably, while ERG overexpression in LnCaP was sufficient to downregulate CHK1 (Fig. 1B and Supplementary Fig. S2H), ETV1 induction by DHT treatment did not further decrease CHK1 levels in LnCaP overexpressing ERG (Supplementary Fig. S2H), thus suggesting a possible redundancy between ERG and ETV1 in CHK1 downregulation.
Overall these results suggest CHK1 as a new transcriptional direct target of both ERG and ETV1 oncogenic activity in human prostate cancer.
ERG directly binds and transcriptionally represses CHK1 promoter
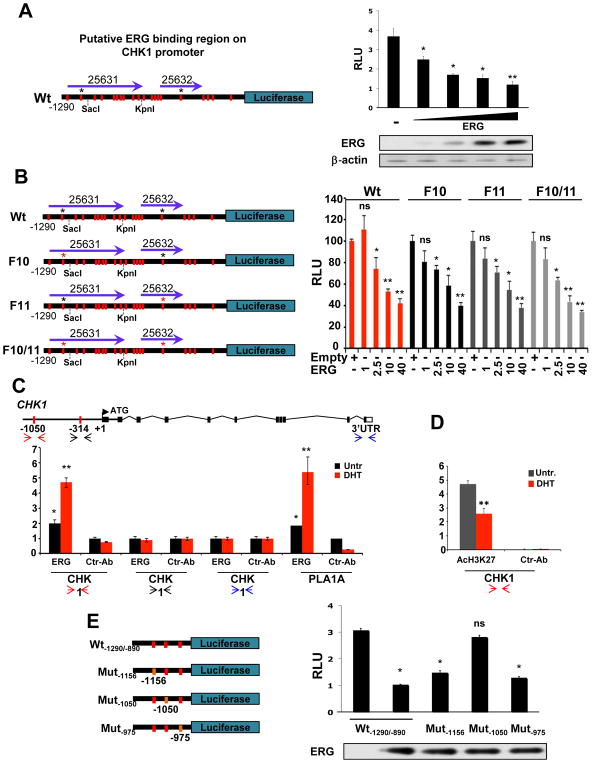
To identify the ERG binding site/s on CHK1 promoter, we took advantage of a recently published ChIP-seq dataset mapping, genome-wide, the ERG binding sites and the putative ERG regulated genes in VCaP cells (21). Supporting our thesis, this dataset lists two different sequences belonging to the CHK1 promoter, both immediately upstream the ATG (ERG 25631, ERG 25632, Supplementary Fig. S3A) (33), that prompted us to clone and study the first 1290 nucleotides of the regulatory region of the human CHK1 gene (Fig. 2A and Supplementary Fig. S3B). A luciferase assay in HEK-293T clearly showed the ability of ERG to repress CHK1 promoter in a dose dependent manner (Fig. 2A). Through an in silico analysis, we identified a total of 20 core motif consensus DNA-binding sites GGA(A/T) for ETS factors spread along the 1290 bp sequence of CHK1 promoter, suggesting 20 putative ERG binding sites (red bars in Fig. 2A, highlighted in red in Supplementary Fig. S3B). Among them, according to Nhili and colleagues’ study (34), only two (positions −1050bp and −314 bp) were characterized by strong flanking sequences (black asterisks in Fig. 2A). A mutational strategy transforming ACC*GGAT*GCC at position −1050 bp in ACC*GT*GCC (F10 mutant) and GCC*GGAA*AGC at position −314 bp in GCC*AA*AGC (F11 mutant) was used to eliminate the two putative ERG binding sites both individually (F10 and F11 mutants) or in combination (F10/11 mutant) (Fig. 2B, red asterisk indicates the mutagenized site). In a luciferase assay in HEK-293T, however, ERG overexpression still repressed in a dose dependent manner the two single mutants (F10 and F11) as well as the double mutant (F10/11) similar to the wild type promoter of CHK1 (Fig. 2B).
Figure 2. ERG binds CHK1 promoter and represses its transcription.
A, On the left, luciferase reporter assay construct showing the 20 core consensus sequences GGA(A/T) for ERG binding (red bars). Asterisks indicate the most probable ERG binding sites according to Nihil and colleagues (34). Purple arrows indicate the two regions of CHK1 promoter identified by Yu and colleagues in a ChIP-seq analysis for ERG (21). On the right, ERG represses CHK1 regulatory region activity in a dose-dependent manner. ERG expression levels are shown in the bottom panel. B, On the left, mutational scheme of the luciferase constructs F10, F11, and F10/11 with mutagenized ERG binding sites indicated by red asterisks. On the right, dose dependent ERG repression of both wild type and mutant CHK1 regulatory region. C, ChIP assay from VCaP prostate cell lines both untreated and treated with DHT (10 nM) for 24 hours, showing levels of direct binding of endogenous ERG in specific sites of the promoter region of human CHK1. PLA1A promoter was used as positive control (23). Error bars indicate mean±s.d.. D, ChIP assay from VCaP prostate cell lines both untreated and treated with DHT (10 nM) for 24 hours, showing levels of AcH3K27 in the promoter region of human CHK1 corresponding to −1050 bp described in C. E, On the left, scheme of wild type Wt-1290/-890, Mut-1156, Mut-1050, and Mut-975 luciferase constructs. Core consensus sequences GGA(A/T) for ERG binding are indicated by red bars, mutant sites are in orange. On the right, ERG-dependent downregulation of basal reporter activity is abolished only when the core consensus motif GGAT at position −1050 bp is mutated to GT. Transfected ERG levels are shown below the bar graph. Experiments were performed in triplicate, data were analyzed using unpaired t-test. Values of P < 0.05 were considered statistically significant (*p<0.05, **p<0.01).
Although results obtained by luciferase assay seemed to disprove the prediction, further evidence indicating the distal part of the −1290 bp CHK1 promoter sequence as a potential important region for ERG repressive activity were obtained by a chromatin immunoprecipitation (ChIP) assay analysis in VCaP cells either untreated or treated for 24 hours with 10 nM DHT. Different sets of primers were designed to amplify respectively the promoter regions around the putative ERG binding sites at −1050 bp (two-headed red arrows), −314 bp (two-headed black arrows) and in the 3′-UTR (two-headed blue arrows) of CHK1 gene (Fig. 2C). PLA1A was used as a positive control for the ChIP assay (23). Notably, ChIP results fully confirmed the ability of ERG to bind the region of CHK1 promoter around the putative ERG binding site at −1050 bp in normal growth conditions (DMEM plus 10% FBS) (Fig. 2C, black bars), and, consistently, showed an increase in the amount of ERG binding after its induction triggered by 24 hours DHT treatment (Fig. 2C, red bars). On the other hand, the region of CHK1 promoter around the putative ERG binding site at −314 bp and the one in the 3′-UTR were not enriched in the ERG ChIP compared to control (Fig. 2C). Finally, in line with the repressive activity of ERG on CHK1 promoter, ChIP assay for AcH3K27 showed a significant decrease in the acetylation of lysine 27 of histone 3 after 24 hours of DHT treatment (Fig. 2D). Conclusive evidence for the functional role of −1050 bp ERG binding site in the transcriptional regulation of CHK1 promoter was obtained from a luciferase assay with the region of CHK1 promoter included from −1290 bp to −890 bp (Wt-1290/-890) (Fig. 2E). Indeed, while ERG overexpression significantly repressed the basal reporter activity of both wild type Wt-1290/-890, and mutant constructs Mut-1156 and Mut-975 (where core motifs of ERG consensus DNA-binding sites located at −1156 bp “GGAT” and at −975 bp “GGAA”, were mutated to “GT” and “GA” respectively) (Fig. 2E), a robust suppression of the downregulation of the reporter activity was evident when the ETS binding DNA core motif “GGAT” located at position −1050 bp was mutated to “GT” (Fig. 2E).
Overall, these results strongly suggest that the ERG binding site at position −1050 bp of CHK1 promoter is functional for CHK1 transcriptional repression. However, generation of two additional luciferase constructs progressively loosing the distal part of CHK1 promoter (SacI and KpnI in Supplementary Fig. S3C), demonstrated the ability of ERG to still repress CHK1 transcription in the absence of the −1050 bp binding site (Supplementary Fig. S3C), thereby suggesting the existence of other ERG consensus sequence among the 20 predicted that can be probably used by ERG to repress CHK1 promoter.
Lastly, in order to further exclude a potential contribution of AR in CHK1 transcriptional regulation, we analyzed the luciferase activity of the wild type CHK1 promoter construct (Wt) in HEK-293 overexpressing AR, ERG, or an empty vector as control. AR reporter construct (4xARE) was used as positive control for AR activity. As shown in Supplementary Figure S3D, AR robustly induced the transcription of 4xARE in a dose dependent manner while it did not affect the basal transcription of CHK1 promoter. An interesting question that remains to be addressed, however, regards the possible role of ERG and AR in the transcriptional regulation of CHK1 through their binding in the 3′-UTR region of the gene. Indeed, the ERG binding site identified by Yu and colleagues (21) on the 3′-UTR of CHK1 (ERG 25633, Supplementary Fig. S3A) falls in the same region where two independent datasets mapped an AR binding site (Supplementary Fig. 3A), even though the ERG site was not enriched in our ChiP assay (21, 35).
Chk1 and Pten compound haploinsufficiency triggers the transition between high-grade prostate intraepithelial neoplasia to invasive prostate carcinoma
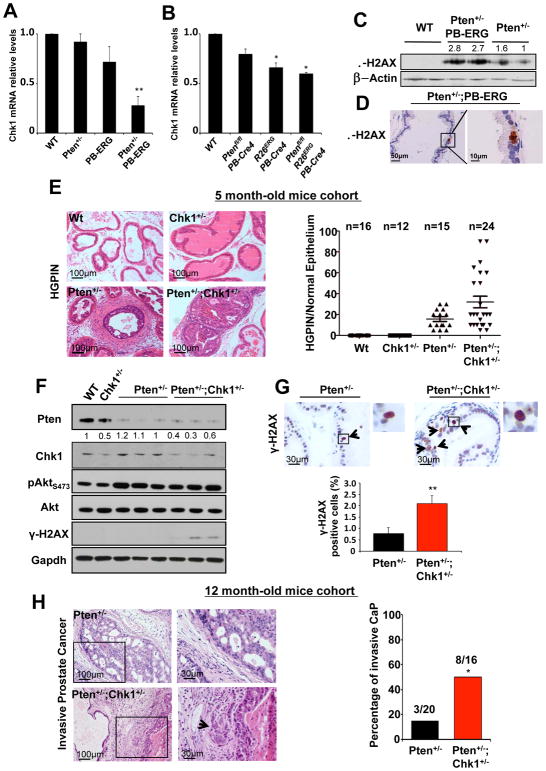
TMPRSS2-ERG fusion and the consequent oncogenic activity of ERG are early events in human CaP, and are frequently accompanied by heterozygous loss of PTEN (19, 27). In order to assess the in vivo relevance of our observations, we took advantage of the Pten+/−;PB-ERG mouse model (19, 25). Our previous study demonstrated that Pten+/−;PB-ERG mice (in which ERG is specifically expressed in the epithelial prostate cells through the androgen receptor-responsive mouse Probasin (PB) promoter) develop invasive adenocarcinoma with 100% penetrance by the age of 6 months, whereas Pten+/− mouse prostates are characterized by HGPIN at this age with no signs of stroma invasion (19, 36). The function of CHK1 as a guardian of the DNA integrity and replication forks number and stability becomes particularly important in a proliferative state in which cells are rapidly replicating their DNA. Bearing in mind our in vitro data, we therefore hypothesized that one of the critical mechanisms by which ERG promotes progression to a more aggressive stage of prostate cancer in Pten+/−;PB-ERG mice is through downregulation of Chk1 levels. To test this hypothesis, we examined Chk1 mRNA amounts in prostates from 7-week old wild-type, Pten+/−, PB-ERG, and Pten+/−;PB-ERG mice. RT-qPCR analysis revealed that the relative levels of Chk1 mRNA were decreased in the prostates of PB-ERG and Pten+/−;PB-ERG mice compared to those of age-matched Pten+/− or wild-type mice (Fig. 3A). Chk1 downregulation upon ERG induction was also observed in the prostate of R26ERG;PB-Cre4, and Ptenflox/flox;R26ERG;PB-Cre4 mouse models recently published by Sawyers’ group (25) (Fig. 3B). Interestingly, in both cases down regulation of Chk1 was more pronounced when overexpression of ERG was combined with loss of Pten (Fig. 3A–B). Although this data might simply reflect the relative amount of Chk1 mRNA belonging to the contaminating ERG negative stroma component of the prostate (sizable in PB-ERG and R26ERG;PB-Cre4 normal prostates, while minimal in Pten+/−;PB-ERG and Ptenflox/flox;R26ERG;PB-Cre4 prostate epithelial tumors), a recent publication describes ERG transcriptional function positively modulated by AKT activation (32). Transient knock down of PTEN, however, did not affect CHK1 levels in VCaP cells (Supplementary Fig. S4A).
Figure 3. Chk1 heterozygosity accelerates Pten-heterozygous driven prostate cancer.
A, Relative expression of Chk1 mRNA in prostates of wild type, Pten+/−, PB-ERG, Pten+/−;PB-ERG and B, wild type, Ptenfl/fl;PB-Cre4, R26ERG;PB-Cre4, and Ptenfl/fl;R26ERG;PB-Cre4 conditional mouse models. C, Immunoblotting showing increased levels of γ-H2AX in prostates of Pten+/−;PB-ERG mice. D, Immunohistochemistry staining for γ-H2AX confirms the presence of DNA damage in Pten+/−;PB-ERG prostate epithelial cells. E, Prostate sections of 5-month old wild type, Chk1+/−, Pten+/− and Pten+/−;Chk1+/− mice were stained with haematoxylin and eosin for histopathological studies. Incidence and grade of HGPIN are reported in the dot plot. F, Western blot analyses of Pten, Chk1, Akt, pSer473Akt, γ-H2AX in wild type, Chk1+/−, Pten+/− and Pten+/−;Chk1+/− mouse prostates. Gapdh was used as loading control. G, γ-H2AX staining in Pten+/− and Pten+/−;Chk1+/− mouse prostates. Percentage of positive cells is shown in the graph (n=3 mice per genotype on a total of 10,000 cells analyzed). H, Sections from prostates of Pten+/− and Pten+/−;Chk1+/− mice stained with haematoxylin and eosin. High magnifications of the region inside the black squares are shown on the right. Arrowheads indicate points of stroma invasion. Histopathological results describing the incidence of invasive prostate cancer in the Pten+/− (n=20) and Pten+/−;Chk1+/− (n=16) cohorts of mice are shown in the graph (Fisher’s Test, p=0.033). Data were analyzed using unpaired t-test. Values of P < 0.05 were considered statistically significant (*p<0.05, **p<0.01).
Given the high frequency of concomitant PTEN-loss and TMPRSS2-ERG translocation in human primary prostate cancer, in order to determine whether over-expression of ERG also lead to DNA damage response in vivo in a PTEN deficient context as previously demonstrated in vitro in prostate cell lines (23, 37), the status of γ-H2AX was investigated in immunoblot and immunohistochemistry analyses using prostates samples of wild type, Pten+/−, and Pten+/−;PB-ERG mice. Increased levels of γ-H2AX was detected in all genetically modified mice, although it was most prominently found in Pten+/−;PB-ERG mice (Fig. 3C–D and Supplementary Fig. S4B). Impairment of CHK1 function has been described in PTEN-null contexts as a consequence of AKT phosphorylation of CHK1 on S280 promoting a partial delocalization of CHK1 from the nucleus to the cytoplasm (10). In order to understand if this was also the case in prostate lesions developed by Pten+/− mice, we fractionated the cytosolic cellular component of wild type and Pten+/− mouse prostates and analyzed Chk1 by western blot. As shown in Supplementary Figure S4C, the amount of the cytosolic fraction of Chk1 compared to the total, resulted comparable in wild type and Pten+/− mouse prostates, likely suggesting the requirement of a marked AKT activation, exclusive of a PTEN-null condition, for the cytosolic delocalization of CHK1 or otherwise the species/context specificity of the AKT dependent phosphorylation of CHK1.
These results indicate that downregulation of Chk1 is a possible mechanism by which overexpression of ERG contributes to prostate cancer progression. However, it is interesting to note that CHK1 was always downregulated while never completely repressed in the Pten+/−;PB-ERG and in Ptenfl/fl;R26ERG;PB-Cre4 mouse prostates, as well as in human CaP and in human prostate cell lines endogenously (VCaP, LnCaP) or exogenously (HEK-293, LnCaP, 22RV1, PC3, U2oS, or RWPE1) overexpressing ERG or ETV1, respectively (Fig. 1–2, and Supplementary Fig. S1 and S2).
If downregulation of CHK1 is an important downstream mediator of ERG-dependent prostate oncogenesis, we would expect Pten+/−;Chk1+/− mice to develop a more aggressive prostate phenotype than their age-matched Pten+/− littermates. To investigate this possibility, we generated a Pten+/−;Chk1+/− cohort of mice by crossing Pten+/− mice (38) with Chk1+/− mice (39–41). Pten+/−;Chk1+/− male compound mutants behave and age in a similar manner to their Pten+/− littermates (Supplementary Fig. S4D–G) although they are generally characterized by an exacerbation of classical Pten+/− phenotypes such as benign lymphoproliferation, spleen enlargement and pheochromocytoma (38) (Supplementary Fig. S4F, G). As for the Pten+/− mice (38), the same phenotypes were far more pronounced in the Pten+/−;Chk1+/− female compound mutant cohort, which dictated euthanization in some cases (Supplementary Fig. S4E). To study the possible oncogenic role of Chk1 downregulation in Pten heterozygous mouse prostates, a cohort of age-matched Pten+/−;Chk1+/−(n=24), Pten+/− (n=15), Chk1+/− (n=12), and wild-type (n=16) mice were sacrificed and characterized histopathologically at 5 months of age, while another large cohort was analyzed at 12 months (see below). At 5 months, 8 out of 24 Pten+/−;Chk1+/− mouse prostates showed a robust increase of high grade prostatic intraepithelial neoplasia (HGPIN >40%) in the tissue of at least one of the three lobes (Fig. 3E). The remaining 16 Pten+/−;Chk1+/− mice were characterized by HGPIN in approximately 20% of their prostate tissue, a condition fully comparable to the prostate phenotype found in the 14 out of 15 age-matched Pten+/− mice (Fig. 3E). All wild type and Chk1+/− prostates were normal. Importantly, Western blot analysis of wild type, Pten+/−, Chk1+/−, and Pten+/−;Chk1+/− mouse prostates showed the presence of γ-H2AX only in the Pten+/−;Chk1+/− prostates (Fig. 3F), while no substantial differences were detected in Akt activation (Fig. 3F). We further proved the increased amount of DNA damage response in Pten/Chk1 double heterozygous mouse prostatic epithelial cells by performing IHC analysis for γ-H2AX and phospho-53BP1 on prostate sections from Pten+/− (n=3) and Pten+/−,Chk1+/− (n=3) mice. Prostate epithelial cells presenting regular nuclear morphology accompanied by γ-H2AX or phospho-53BP1 nuclear dots staining were counted. As shown in Figure 3G and Supplementary 4H, the number of prostate epithelial cells showing evidence of DNA damage response was two times higher in Pten+/−;Chk1+/− than Pten+/− prostates, thereby validating our hypothesis. These results suggest that by reducing the levels of CHK1, ERG can favor genomic instability by increasing the presence of unrepaired DNA damage, and, stochastically, accelerating tumor progression in PTEN deficient human prostate cancer.
As previously reported (36), 12-month-old Pten+/− mice rarely develop invasive prostatic adenocarcinomas. Accordingly, in order to determine whether Chk1 downregulation in a Pten heterozygous context was sufficient to transform the Pten+/− driven HGPIN into invasive prostate carcinoma, as it is the case for ERG (19), a further cohort of age-matched Pten+/−;Chk1+/−(n=16), Pten+/− (n=22), Chk1+/− (n=16), and wild-type (n=15) mice were sacrificed at 12 months of age. Indeed, histopathology analysis carried out on serial sections of the three prostate lobes of all the mice unveiled the presence of invasive prostate carcinoma in 8 of the 16 (50%) Pten+/−;Chk1+/− mice, while only 3 of the 20 (15%) Pten+/− showed tumor epithelial cells invading the surrounding stroma (Fig. 3H; Fisher Test p=0.033). All the wild type and Chk1+/− mice were characterized by normal prostate epithelium with sporadic signs of hyperplasia.
Overall, these results strongly support the hypothesis that ETS family members might favor tumorigenesis by lowering CHK1 levels, thus promoting DNA damage accumulation, aberrant G2/S cell cycle check point, and, in turn, genomic instability (6, 10, 42, 43).
Lowering of CHK1 levels in prostate cancer cells bearing functional p53 enhances sensitivity to etoposide, but not to docetaxel
Affecting more than half of the patients, TMPRSS2-ERG translocation is one of the most studied oncogenic lesions in CaP. Although a growing body of evidence describes ERG overexpression involved in different aspects of prostate tumor progression, its correlation with specific clinical and pathological parameters is still matter of debate (44), while very little effort has been devoted, until now, to translate the knowledge acquired on the oncogenic activity of ERG in rationalized therapeutic strategies.
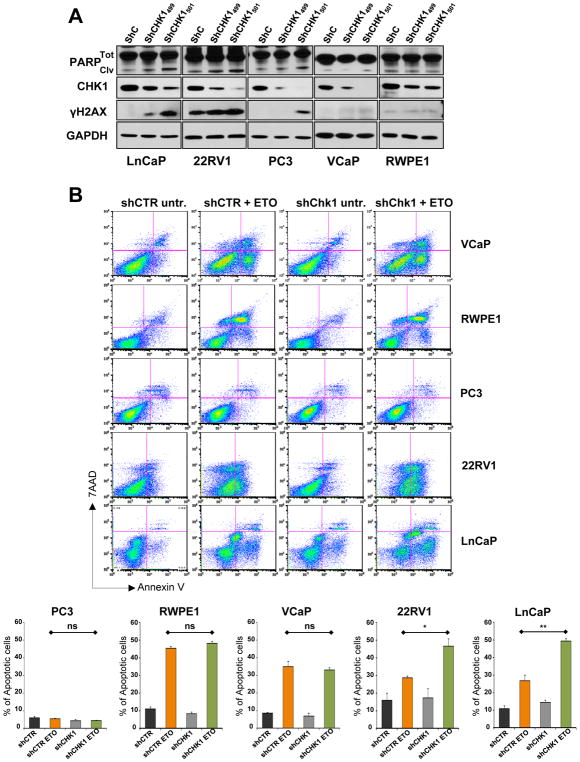
Intriguingly, we frequently found fragmented nuclei in cells overexpressing high levels of exogenous ERG (Supplementary Fig. S5A), while immunoblot analysis showed high levels of p53, its phosphorylation on serine 15, and the up-regulation of p21, suggesting the presence of extensive DNA damage in these cells and the expected p53 response (45, 46) (Supplementary Fig. S5B). Consistent with these results, excess amounts of ERG prevented colony formation in LNCap and 22Rv1 bearing a functional p53 pathway (see below), but not in PC3 cells completely lacking p53 expression (Supplementary Fig. S5C). Therefore, in order to study: 1. the possible effect of lowering CHK1 levels in different human CaP cell lines and 2. if this downregulation might influence sensitivity to specific drug treatments, we stabilized a panel of human immortalized and CaP cell lines (RWPE-1, VCaP, LnCaP, 22RV1, and PC3) with two different shRNAs (shCHK1499 and shCHK1501) targeting CHK1 transcript with different efficacy (Fig. 4A). In all four CaP cell lines (LnCaP, 22RV1, PC3, and VCaP), with the exclusion of the immortalized RWPE1, shCHK1501 robustly reduced CHK1 levels (Fig. 4A). Consistent with the literature (47–50), however, the extremely low levels of CHK1 in shCHK1501 stable LnCaP, 22RV1, PC3, and VCaP cell lines caused an excessive accumulation of unrepaired DNA damage (γH2AX in Fig. 4A) and, in turn, a persistent apoptotic response (cleaved PARP in Fig. 4A) that made it impossible to maintain the stable clones for more than few passages. On the other hand, the lower efficacy of shCHK1499 in targeting CHK1 (Fig. 4A) permitted the generation and propagation of stable clones for all of them. Since ATR/CHK1 pathway is primarily activated by replicative stress during S/G2 phases of cell cycle, and CHK1 activity is essential to genomic stability by maintaining replication fork integrity (42), suppressing inappropriate firing of late or cryptic DNA replication origins (43), and promoting homologous recombination DNA damage repair (11), we tested whether reduction of CHK1 levels might be detrimental for prostate tumor cells when treated with agents directly targeting the replication machinery (9, 51). ShControl and shCHK1499 stable RWPE-1, VCaP, LnCaP, 22RV1, and PC3 prostate cell lines were treated with 20 μM etoposide for 48 hours. Untreated cells served as the control. AnnexinV/7-AAD flow cytometric analysis defined PC3 cell line as completely resistant to etoposide treatment, while VCaP, LnCaP, 22RV1, and RWPE1 showed a robust apoptotic response (Fig. 4B, upper and lower quadrants on the right in the plots; orange and green bars in the graphs). RWPE1 was the most sensitive to etoposide among the prostate cell lines tested, probably as consequence of the fact that RWPE1 are immortalized normal prostate epithelial cells and not tumor cells. CHK1 downregulation, however, did not change the amount of apoptotic RWPE1 cells after etoposide treatment (Fig. 4B). VCaP, 22RV1, and LnCaP showed comparable sensitivity to etoposide (orange bars), CHK1 downregulation, however, significantly increased the percentage of apoptotic cells in LnCaP and 22RV1 while it did not affect the response to etoposide in VCaP (Fig. 4B, quadrants on the right in the plot; red and green bars in the graphs). The differential sensitivity to etoposide after CHK1 knockdown of LnCaP and 22RV1 compared to VCaP cells might be reconciled by the status of p53 among these three different prostate cell lines. Indeed, while LnCaP cells have both p53 alleles wild type and 22RV1 one allele wild type and one expressing a mutant form of p53 (Q331R) mildly affecting p53 activity, VCaP are characterized by a dysfunctional p53 pathway expressing a classical hot spot mutation of p53 (R248W) (52).
Figure 4. CHK1 levels dictate etoposide sensitivity in CaP cell lines bearing functional p53 pathway.
A, Western blot analysis comparing the levels of PARP (total and cleaved), CHK1, and γ-H2AX in shControl, shCHK1499, and shCHK1501, stable LnCaP, 22RV1, PC3, VCaP, and RWPE1 prostate cell lines. GAPDH was used as loading control. B, FACS analysis of AnnexinV/7-AAD staining showing the amount of apoptotic cells in shControl and shCHK1499 VCaP, RWPE1, PC3, 22RV1, and LnCaP stable cell lines treated with 20 μM of etoposide for 48 hours. Experiments were performed in triplicate, data were analyzed using unpaired t-test. Values of P < 0.05 were considered statistically significant (*p<0.05, **p<0.01).
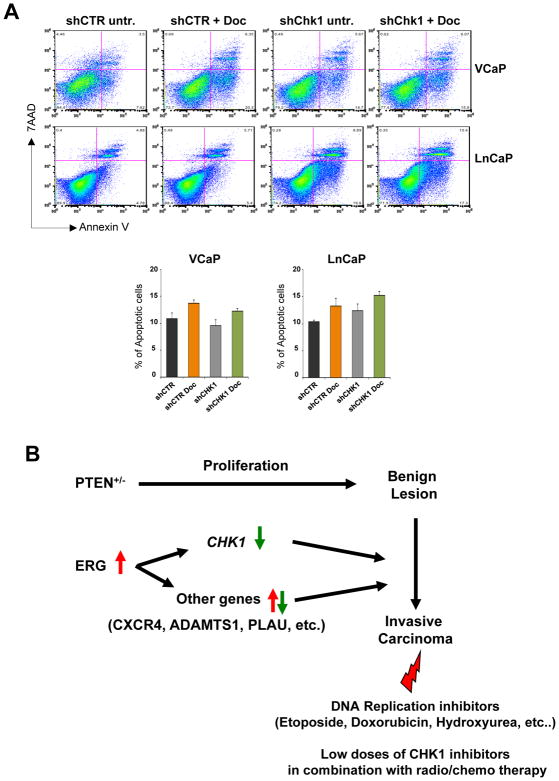
Finally, to understand if CHK1 levels might influence sensitivity to other types of drug, we treated shControl and shCHK1499 stable LnCaP and VCaP cells with docetaxel, a widely used standard-of-care therapy for prostate cancer. As shown in figure 5A, downregulation of CHK1 did not sensitize either LnCaP or VCaP cells to docetaxel treatment.
Figure 5. CHK1 down regulation does not improve docetaxel efficacy.
A, FACS analysis of AnnexinV/7-AAD staining showing the amount of apoptotic cells in shControl and shCHK1499 VCaP and LnCaP stable cell lines treated with 0.2 nM of docetaxel for 48 hours. Experiments were performed in triplicate, data were analyzed using unpaired t-test. Values of P < 0.05 were considered statistically significant (*p<0.05, **p<0.01). B, Model summarizing the cooperative contribution of PTEN heterozygosity and ERG expression to prostate cancer progression and the potential sensitivity of these tumors to specific drug regimens.
Overall these results strongly support the concept that reduction of CHK1 levels renders prostate tumor cells bearing functional p53 specifically sensitive to genotoxic agents targeting the DNA replication machinery and causing stalling of replication forks such as etoposide.
DISCUSSION
ETS family members have been implicated in tumors of various histologies, including leukemia, sarcoma and prostate cancer (53–55). In particular, ERG and ETV1 have been found translocated and ectopically expressed in a large proportion of human prostate cancer (60%), a tumor type that still represents the second leading cause of cancer-related deaths in males in the USA (22, 27, 55, 56). Thus defining the molecular mechanisms underlying the contribution of ETS family members to tumorigenesis is of great relevance, as it may also offer new opportunities for therapeutic intervention. Ectopic and aberrant overexpression of ETS proteins may conceivably mediate their proto-oncogenic role in tumorigenesis through the mis-expression of their transcriptional target genes. The data presented here suggest that CHK1 may represent such a critical gene. ERG most likely does not initiate tumorigenesis in the prostate, since overexpression of ERG alone does not affect proliferation in a post-mitotic tissue such as the prostate gland (19, 25, 57). Nonetheless, expression of ERG in a Pten heterozygous or null background causes a more aggressive stage of cancer, suggesting that the genes deregulated by ERG become functionally relevant when cells are proliferating (19, 25, 57). In accordance with this notion, ERG overexpression in human patients is more frequently observed in conjunction with mutations that are known to trigger proliferation, such as partial PTEN loss (19, 57, 58). Recently, Brenner and colleagues have associated ERG or ETV1 overexpression in prostate cell lines with increased numbers of γ-H2AX nuclear foci, which offers evidence of DNA damage in these cells (23). They have also described ERG in a multiproteic complex with PARP1 and DNAPK, and have shown that this complex regulates Erg activity (23). However, the mechanism responsible for the increased DNA damage remained elusive. Our findings provide a compelling transcriptional mechanism explaining how ERG and other ETS family members could cause genomic instability. We propose that ERG overexpression may facilitate the persistency of mild levels of DNA-damage through the downregulation of CHK1. Reductions in CHK1 levels have been shown to cause DNA damage, most likely due to impairment of the monitoring of DNA integrity during replication (5–7, 11, 43, 59–62). Thus it is conceivable that ERG-dependent DNA damage and consequent genomic instability might be due at least in part to reduced CHK1 levels and, consequently, an impaired cell-cycle check-point process during cellular proliferation. This particular condition, when associated with a hyper-proliferative context such as the one triggered by PTEN-loss, may dictate increased genomic instability and the acquisition of further genetic lesions, which can drive prostate tumor progression and resistance to treatments (Fig. 5B).
Importantly, our study also offers straightforward therapeutic implications. Inactivating CHK1 function has been given serious consideration as a way to abrogate the cell-cycle checkpoints believed to account for the survival of cancer cells in response to chemo- and radiotherapy (63–68), and a number of CHK1 inhibitors are currently in clinical trials. Our data suggest that tumors harboring elevated levels of ERG, or other ETS family members, may be more sensitive to specific classes of drugs targeting the replication machinery than, for instance, to agents such as taxanes hitting microtubules polymerization, in a p53 functional context. Finally, CHK1 inhibitors, due to already reduced levels of CHK1, may be administered at very low doses in ETS positive tumors and therefore would be extremely effective in a synthetically lethal approach in combination with radiation/chemotherapy and PARP inhibitors (69).
METHODS
In vitro experiments
HEK-293, VCaP, LnCaP, 22RV1, PC3, RWPE1, and U2oS human cell lines were purchased from the ATCC. Cells were tested and authenticated by ATCC (DNA fingerprinting, karyotyping, and morphology), and additionally by Western blot and qRT-PCR for specific markers in our laboratory. Cells were cultured in DMEM or RPMI medium (as indicated by the ATCC) supplemented with 10% FBS and tested for mycoplasma contamination every month. For DHT treatments, the regular medium was replaced with regular medium supplemented with 10 nM DHT. Lipofectamine 2000 (Invitrogen) was used as transfection reagent.
Microarray analysis
Human prostate cancer data set has been previously described (27). Briefly, copy number data was generated on Agilent 244KaCGH arrays and mRNA expression data was obtained on Affymetrix Human Exon 1.0 ST arrays. The complete genomes dataset and analytic methods is reported separately (27). Specimens were classified as harboring ERG aberrant expression with a z-score greater than 4.0. This correlated with ERG genetic re-arrangments as identified by CGH analysis. CHK1 levels (z-score) were analyzed according to ERG aberrant expression status and a t-test was used to compare the mean z-score between tumors with ERG aberrant expression and those without. Publicly available expression data GSE39388 (26), GSE21032 (27), GSE14595 (19), GSE14097 (21), GSE46799 (25), were obtained from GEO database. Differential expression and statistical significance have been evaluated by Bioconductor limma package. Values of p<0.05 were considered statistically significant.
Generation of plasmid vectors
The pCMV-SPORT6-ERG (human) was purchased by Life Technologies (MHS6278-202759378). ERG cDNA was recovered by digestion with XhoI/SalI from pCMV-SPORT6-ERG and cloned in pBabe-puro by using SalI site. Human ETV1 cDNA was cloned into EcoRI site in pBabe-3xFlag-puro. ERG target sequence in pLKO were: 5′-CGACATCCTTCTCTCACAT-3′ (ERG-shRNA#1), and 5′-GATGATGTTGATAAAGCCTTA-3′ (ERG-shRNA#2). ShRNAs targeting human CHK1 were purchased from the Thermo Fisher Scientific (RHS4533-EG1111). pLKO-shScramble (#1864) was purchased from Adgene and used as control. Smart pool siRNA for PTEN was purchased from Dharmacon (siGENOME siPTEN M-003023-02-0005). pCDNA3.1-AR was kindly provided by Dr Balk (Beth Israel Deaconess Medical Center of Boston). The wild type (Wt) construct used in luciferase reporter activity was generated by amplifying 1290 bp upstream translation initiation site of CHK1 using the following primers: CCGGACGCGTGCACCACGAGTACCGCACTCTGAGG and CCGGAGATCTGACTCCACCGAGCACCTCGG. Sequences in bold indicate recognition sites for MluI and BglII, respectively. This PCR fragment was sub-cloned into the MluI and BglII sites of the pGL2-enhancer luciferase reporter vector (Promega). The mutants forms of this construct were generated using the following primers and their reverse complementary oligos in site-directed mutagenesis (Strategene) PCR reactions: for mutant F10: AAATCTCTTCAGCCAGTCTCTCCCCGACTGCAAAG; for mutant F11: CTTCCCCAGTCGTTCGCCAAAGCATTTGTCTCCC; for mutant Mut-1156: GCCCGCAGCCCCGCCTGAGCGCAGCGCGGTCGG; for mutant Mut-1050: AAATCTCTTCAGCCAGTCTCTCCCCGACTGCAAAG; and for mutant Mut-975: GTCGAGCCTCACACCGTGCCACTTCATATTTGG. The mutated sites are shown in bold.
Chromatin Immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) analysis was performed accordingly to the Simple ChIP Enzymatic Chromatin IP Kit (9003 Cell Signaling Technology). Briefly, 5 X 107 VCaP cells per condition were fixed with 1% formaldehyde for 10 min at room temperature. Relief of crosslinking was performed by adding 125mM glycine for 5 minutes at room temperature. Cells were washed twice with cold PBS 1X plus protease inhibitor cocktail and then were lysed with cold Buffer A. Collected nuclei were re-suspended in cold buffer B and then treated with 7 ul of micrococcal nuclease for 30 minutes at 37°C. Digested chromatin was sonicated and purified accordingly to the manufacture’s instruction. 100 ug of chromatin was incubated with the following Antibodies: 3ug of ERG (C-17 X; Santa Cruz Biotechnology), 3ug of H3 (provided by the kit), 3ug of AcH3K27 (Abcam ab4729) and 3ug of IgG (provided by the kit). The primers used to amplify by RT-qPCR the regions containing the putative consensus DNA-binding sites of ERG in the CHK1 promoter were as following: red arrowheads, forward 5′-CCAGCAGCGCTCGAGCACC-3′, and reverse 5′-AGACAGCGCGCGTTCCCGTG-3′; black arrowheads, forward 5′-CAGGCGTTTTCTGCCCCATAC-3′, and reverse 5′-CTAAACCTGACCATTTTGTCG-3′; blue arrowheads 5′-CATTTCCACTTGAATGTCTAGTAG-3′, and reverse 5′-CAGTGTCTTTACTGAGGCAATTAC-3′. The primers used to amplify the regions containing the characterized consensus DNA-binding sites of ERG in the PLA1A were as following: forward 5′-TGGCCACCCAGAGATGCAGGA-3′, and reverse 5′-ACACACTGTCCCTCTTTGAGCCA-3′.
Quantitative RT-PCR analysis
RNA was prepared from human cell lines and mouse prostates using RNeasy Mini Kit (Qiagen) followed by cDNA synthesis using the Super Script III First-Strand Synthesis system (18080-051, Invitrogen), or the iScript cDNA synthesis kit (170-8890, Bio-Rad). Taqman analysis was performed through Biopolymers Facility at Harvard Medical School using probes Mm00432485_m1 for mouse Chk1 and Mm00446953_m1 for mouse Gusb, Hs00967506_m1 for human CHK1, Hs00171666_m1 for human ERG, Hs00951941_m1 for human ETV1, Hs99999908_m1 for human GUSB, Hs00984230_m1 for human Beta-2 microglobulin. Mouse Gusb and human GUSB and Beta-2 microglobulin were used as a control and results were normalized against them.
Western blot, immunofluorescence and immunohistochemistry
For western blot, cell and tissue lysates were prepared with 150-RIPA buffer and protease inhibitor cocktail (Roche). The following antibodies were used for western blotting: rabbit polyclonal anti-ERG (2805-1; 1:1000, Epitomics), rabbit polyclonal anti-ETV1 (ab81086; 1:1000, Abcam), mouse monoclonal anti-CHK1 (2G1D5; 1:1000, Cell Signaling Technology), rabbit polyclonal anti CHK2 (2662; 1:1000, Cell Signaling Technology), mouse monoclonal anti-TP53 (DO1, 1:1000, Santa Cruz Biotechnology), rabbit polyclonal anti-CDKN1A (C-19; 1:1000, Santa Cruz Biotechnology), rabbit polyclonal anti-AR (PG21; 1:1000, Millipore), rabbit polyclonal anti-PSA (k92110R; 1:1000, Meridian Life Science), rabbit monoclonal anti-γ-H2AX (9718S; 1:1000, Cell Signaling Technology), rabbit polyclonal anti-Lamin β1 (ab16048; 1:5000, Abcam) mouse polyclonal anti-β-Actin (A5316; 1:5000, Sigma-Aldrich), rabbit polyclonal anti-Pten (138G6; 1:1000, Cell Signaling Technology), rabbit polyclonal anti-phospho-Akt(S473) (9271S; 1:1000, Cell Signaling Technology), rabbit polyclonal anti-Akt (9272S; 1:1000, Cell Signaling Technology), rabbit polyclonal anti-GAPDH (14C10; 1:6000, Cell Signaling Technology), mouse monoclonal anti-FLAG (M2; 1:3000, Sigma Aldrich). For immunohistochemistry (IHC), tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Mouse prostate sections were stained for γ-H2AX (9718S; 1:250, Cell Signaling Technology) and phospho-53BP1 (ab82550; 1:500, Abcam). Human prostate TMA sections were stained for CHK1 ab40866; 1:250, EP691Y Abcam) and ERG (ab92513, 1:250, EPR3864 Abcam). For immunofluorescence (IF), cells were fixed in paraformaldehyde 4% in PBS 1x for half an hour, washed in PBS1x three times, treated with Glycine 0.1 M in PBS 1x for 1 hour, and permeabilized with 0.1% Tryton in PBS 1x for 15 minutes. Rabbit polyclonal anti-γ-H2AX (9718S; 1:500, Cell Signaling Technology), and mouse monoclonal anti-ERG (C-1, 1:500, Santa Cruz) were used in PBS 1x.
Stable cell lines
Stable cell lines were generated by transduction of the indicated shRNAs. 12 hours after infection, cells were washed and puromycin (2 μg/mL) was added to fresh media. Selection was maintained for 2 weeks after transduction before cells were used for experiments. Stable clones were maintained under puromycin selection (0.5 μg/mL). Stable cell lines were tested and authenticated by Western blot and qRT-PCR for specific markers in our laboratory.
AnnexinV/7-AAD FACS Analysis
PC3, LnCaP, 22RV1, VCaP, and RWPE1 shControl and shCHK1499 stable cells lines were plated in 6 well/multiwells (0.2x106 PC3 and 0.5x106 LnCaP, 22RV1, VCaP, and RWPE1). 24 hours later, 20 μM of etoposide or 0.2 nM of docetaxel was added to the medium. Cells were collected 48 hours later and processed following manufacturer’s instructions (Thermo Fisher Scientific, BDB559763).
Pten+/−, Pten+/−;PB-ERG and Pten+/−;Chk1 mutant mice
Pten+/− (C57BL/6) and Pten+/−;PB-ERG (C57BL/6) mice were generated as previously described (19). Chk1+/− (C57BL/6) mice were obtained from the Jackson Laboratory following permission given by Dr. Stephen Elledge. Pten+/−;Chk1+/− mice were generated by crossing Pten+/− and Chk1+/− mice. All mouse work was done in accordance with our IACUC protocol.
Cytosolic fractionation
12 months old wild type (n=2) and Pten+/− (n=2) mice were sacrificed and prostates were immediately extracted and processed for cytosolic fractionation following manufacturer’s instructions (Pierce Biotechnology 78833).
B and T cells staining
Mice were sacrificed at the age of 5 months and the axillary, cervical and inguinal lymph nodes were surgically excised. Organs were forced through a nylon screen to make single cell suspensions and cells were washed and re-suspended in PBS with 2% FBS. Thereafter, cells were stained with fluorochrome-labeled anti-CD3 PE and anti-B220 FITC (Biolegend). Analysis was performed with LRS II (BD bioscience).
Histopathology
Murine prostate tissues were harvested and fixed in formalin overnight, washed in PBS, and dehydrated in Et-OH. Paraffin embedding and H&E staining were performed by the Histology Core Facility at the Beth Israel Deaconess Cancer Center. Parts of the tissues were used for proteins and RNAs extraction. Histopathological analysis of human prostate TMA was performed by the Pathology Histology Core Facility at the Icahn School of Medicine of Mount Sinai.
Supplementary Material
SIGNIFICANCE.
Genetic translocation and aberrant expression of ETS family members is a common event in different types of human tumors. Here we show that through the transcriptional repression of CHK1, ETS factors may favor DNA damage accumulation and consequent genetic instability in proliferating cells. Importantly, our findings provide a rationale for testing DNA replication inhibitor agents in ETS positive p53 proficient tumors.
Acknowledgments
The help and advice of all members of the Pandolfi lab is gratefully acknowledged. We thank Thomas Garvey for insightful editing, and Kaitlyn Webster for excellent technical assistance. We also thank all the staff at the Animal and Histology Core facilities at BIDMC for all their help. R.T. has been granted leave of absence from the Department of Oncology, Università degli Studi di Torino (Italy).
Grant Support
This work was supported by the NIH grant R01 CA082328 to P.P.P. AL has been supported in part by a fellowship from the Istituto Toscano Tumori (ITT, Italy).
Footnotes
Disclosure of Potential Conflict of Interest
The authors declare no competing financial interests.
Author’s Contributions
Conception and design: A. Lunardi, S. Varmeh, P.P. Pandolfi.
Development of methodology: A. Lunardi, S. Varmeh, R. Taulli, U. Ala, B.S. Carver, R.M. Hobbs, J. Guarniero, M. Chen, V. Quarantotti, K.A. Webster, C. Ng, A.H. Berger, C. Nardella, L. Poliseno, S. Signoretti.
Acquisition of data: A. Lunardi, S. Varmeh, R. Taulli, J. Guarniero, M. Chen, V. Quarantotti, K.A. Webster, C. Ng, N. Seitzer, A.H. Berger, C. Nardella, L. Poliseno, S. Signoretti.
Analysis and interpretation of data: A. Lunardi, S. Varmeh, U. Ala, B.S. Carver, A.H. Berger, C. Nardella, L. Poliseno, M. Castillo-Martin, R. Montironi, C. Cordon-Cardo, S. Signoretti, P.P. Pandolfi.
Writing, review, and/or revision of the manuscript: A. Lunardi, S. Varmeh, P.P. Pandolfi.
Administrative, technical, or material support: A. Lunardi, S. Varmeh, R. Taulli, U. Ala, B.S. Carver, R.M. Hobbs, J. Guarniero, M. Chen, V. Quarantotti, K.A. Webster, C. Ng, A.H. Berger, C. Nardella, L. Poliseno, S. Signoretti.
Study supervision: A. Lunardi, S. Varmeh, P.P. Pandolfi.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. doi: 10.1038/nature08467. Epub 2009/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular cell. 2007;28(5):739–45. doi: 10.1016/j.molcel.2007.11.015. Epub 2007/12/18. [DOI] [PubMed] [Google Scholar]
- 3.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–23. doi: 10.1038/nature03097. Epub 2004/11/19. [DOI] [PubMed] [Google Scholar]
- 4.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Molecular oncology. 2011;5(4):368–73. doi: 10.1016/j.molonc.2011.07.002. Epub 2011/08/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enders GH. Expanded roles for Chk1 in genome maintenance. The Journal of biological chemistry. 2008;283(26):17749–52. doi: 10.1074/jbc.R800021200. Epub 2008/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer cell. 2004;6(1):45–59. doi: 10.1016/j.ccr.2004.06.015. Epub 2004/07/21. [DOI] [PubMed] [Google Scholar]
- 7.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer cell. 2003;3(5):421–9. doi: 10.1016/s1535-6108(03)00110-7. Epub 2003/06/05. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277(5331):1497–501. doi: 10.1126/science.277.5331.1497. Epub 1997/09/05. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Molecular biology of the cell. 2006;17(1):402–12. doi: 10.1091/mbc.E05-07-0594. Epub 2005/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer cell. 2005;7(2):193–204. doi: 10.1016/j.ccr.2005.01.009. Epub 2005/02/16. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nature cell biology. 2005;7(2):195–201. doi: 10.1038/ncb1212. Epub 2005/01/25. [DOI] [PubMed] [Google Scholar]
- 12.Vassileva V, Millar A, Briollais L, Chapman W, Bapat B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer research. 2002;62(14):4095–9. Epub 2002/07/19. [PubMed] [Google Scholar]
- 13.Gentile M, Ahnstrom M, Schon F, Wingren S. Candidate tumour suppressor genes at 11q23–q24 in breast cancer: evidence of alterations in PIG8, a gene involved in p53-induced apoptosis. Oncogene. 2001;20(53):7753–60. doi: 10.1038/sj.onc.1204993. Epub 2001/12/26. [DOI] [PubMed] [Google Scholar]
- 14.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer research. 2001;61(21):7727–30. Epub 2001/11/03. [PubMed] [Google Scholar]
- 15.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes, chromosomes & cancer. 1999;26(2):176–80. Epub 1999/09/01. [PubMed] [Google Scholar]
- 16.Connolly KC, Gabra H, Millwater CJ, Taylor KJ, Rabiasz GJ, Watson JE, et al. Identification of a region of frequent loss of heterozygosity at 11q24 in colorectal cancer. Cancer research. 1999;59(12):2806–9. Epub 1999/06/26. [PubMed] [Google Scholar]
- 17.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes & development. 2005;19(10):1162–74. doi: 10.1101/gad.1291305. Epub 2005/05/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tort F, Hernandez S, Bea S, Camacho E, Fernandez V, Esteller M, et al. Checkpoint kinase 1 (CHK1) protein and mRNA expression is downregulated in aggressive variants of human lymphoid neoplasms. Leukemia. 2005;19(1):112–7. doi: 10.1038/sj.leu.2403571. Epub 2004/11/05. [DOI] [PubMed] [Google Scholar]
- 19.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nature genetics. 2009;41(5):619–24. doi: 10.1038/ng.370. Epub 2009/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer research. 2008;68(20):8516–24. doi: 10.1158/0008-5472.CAN-08-1147. Epub 2008/10/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. Epub 2010/05/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–88. doi: 10.1593/neo.07822. Epub 2008/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer cell. 2011;19(5):664–78. doi: 10.1016/j.ccr.2011.04.010. Epub 2011/05/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai C, Wang H, He HH, Chen S, He L, Ma F, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. The Journal of clinical investigation. 2013;123(3):1109–22. doi: 10.1172/JCI66666. Epub 2013/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nature medicine. 2013 doi: 10.1038/nm.3216. Epub 2013/07/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes & development. 2013;27(6):683–98. doi: 10.1101/gad.211011.112. Epub 2013/03/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. Epub 2010/06/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39(1):41–51. doi: 10.1038/ng1935. Epub 2006/12/19. [DOI] [PubMed] [Google Scholar]
- 29.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nature reviews Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. Epub 2008/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai C, Hsieh CL, Omwancha J, Zheng Z, Chen SY, Baert JL, et al. ETV1 is a novel androgen receptor-regulated gene that mediates prostate cancer cell invasion. Mol Endocrinol. 2007;21(8):1835–46. doi: 10.1210/me.2006-0480. Epub 2007/05/17. [DOI] [PubMed] [Google Scholar]
- 31.Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474(7351):403–6. doi: 10.1038/nature10005. Epub 2011/05/17. [DOI] [PubMed] [Google Scholar]
- 32.Selvaraj N, Budka JA, Ferris MW, Jerde TJ, Hollenhorst PC. Prostate cancer ETS rearrangements switch a cell migration gene expression program from RAS/ERK to PI3K/AKT regulation. Molecular cancer. 2014;13:61. doi: 10.1186/1476-4598-13-61. Epub 2014/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic acids research. 2013;41(Database issue):D177–87. doi: 10.1093/nar/gks1060. Epub 2012/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nhili R, Peixoto P, Depauw S, Flajollet S, Dezitter X, Munde MM, et al. Targeting the DNA-binding activity of the human ERG transcription factor using new heterocyclic dithiophene diamidines. Nucleic acids research. 2013;41(1):125–38. doi: 10.1093/nar/gks971. Epub 2012/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. The EMBO journal. 2010;29(13):2147–60. doi: 10.1038/emboj.2010.106. Epub 2010/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS biology. 2003;1(3):E59. doi: 10.1371/journal.pbio.0000059. Epub 2003/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee P, Choudhary GS, Sharma A, Singh K, Heston WD, Ciezki J, et al. PARP inhibition sensitizes to low dose-rate radiation TMPRSS2-ERG fusion gene-expressing and PTEN-deficient prostate cancer cells. PloS one. 2013;8(4):e60408. doi: 10.1371/journal.pone.0060408. Epub 2013/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nature genetics. 1998;19(4):348–55. doi: 10.1038/1235. Epub 1998/08/11. [DOI] [PubMed] [Google Scholar]
- 39.Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, Hakem R, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(10):3805–10. doi: 10.1073/pnas.0611584104. Epub 2007/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes & development. 2000;14(12):1439–47. Epub 2000/06/20. [PMC free article] [PubMed] [Google Scholar]
- 41.Boles NC, Peddibhotla S, Chen AJ, Goodell MA, Rosen JM. Chk1 haploinsufficiency results in anemia and defective erythropoiesis. PloS one. 2010;5(1):e8581. doi: 10.1371/journal.pone.0008581. Epub 2010/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zachos G, Rainey MD, Gillespie DA. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Molecular and cellular biology. 2005;25(2):563–74. doi: 10.1128/MCB.25.2.563-574.2005. Epub 2005/01/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. The EMBO journal. 2007;26(11):2719–31. doi: 10.1038/sj.emboj.7601714. Epub 2007/05/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasi Tandefelt D, Boormans J, Hermans K, Trapman J. ETS fusion genes in prostate cancer. Endocrine-related cancer. 2014;21(3):R143–52. doi: 10.1530/ERC-13-0390. Epub 2014/03/25. [DOI] [PubMed] [Google Scholar]
- 45.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–7. doi: 10.1126/science.281.5383.1674. Epub 1998/09/11. [DOI] [PubMed] [Google Scholar]
- 46.Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nature reviews Molecular cell biology. 2012;13(9):579–90. doi: 10.1038/nrm3420. Epub 2012/08/24. [DOI] [PubMed] [Google Scholar]
- 47.Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell death and differentiation. 2010;17(6):901–11. doi: 10.1038/cdd.2010.35. Epub 2010/04/10. [DOI] [PubMed] [Google Scholar]
- 48.Wu GS, El-Diery WS. p53 and chemosensitivity. Nature medicine. 1996;2(3):255–6. doi: 10.1038/nm0396-255a. Epub 1996/03/01. [DOI] [PubMed] [Google Scholar]
- 49.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer research. 1991;51(23 Pt 1):6304–11. Epub 1991/12/01. [PubMed] [Google Scholar]
- 50.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–25. doi: 10.1016/0092-8674(93)90500-p. Epub 1993/11/19. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez R, Gagou ME, Meuth M. Apoptosis induced by replication inhibitors in Chk1-depleted cells is dependent upon the helicase cofactor Cdc45. Cell death and differentiation. 2008;15(5):889–98. doi: 10.1038/cdd.2008.4. Epub 2008/02/02. [DOI] [PubMed] [Google Scholar]
- 52.Dinnen RD, Drew L, Petrylak DP, Mao Y, Cassai N, Szmulewicz J, et al. Activation of targeted necrosis by a p53 peptide: a novel death pathway that circumvents apoptotic resistance. The Journal of biological chemistry. 2007;282(37):26675–86. doi: 10.1074/jbc.M701864200. Epub 2007/07/20. [DOI] [PubMed] [Google Scholar]
- 53.Peeters P, Raynaud SD, Cools J, Wlodarska I, Grosgeorge J, Philip P, et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90(7):2535–40. Epub 1997/11/05. [PubMed] [Google Scholar]
- 54.Ida K, Kobayashi S, Taki T, Hanada R, Bessho F, Yamamori S, et al. EWS-FLI-1 and EWS-ERG chimeric mRNAs in Ewing’s sarcoma and primitive neuroectodermal tumor. International journal of cancer Journal international du cancer. 1995;63(4):500–4. doi: 10.1002/ijc.2910630407. Epub 1995/11/15. [DOI] [PubMed] [Google Scholar]
- 55.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. Epub 2005/10/29. [DOI] [PubMed] [Google Scholar]
- 56.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26(18):2667–73. doi: 10.1038/sj.onc.1210070. Epub 2006/10/18. [DOI] [PubMed] [Google Scholar]
- 57.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nature genetics. 2009;41(5):524–6. doi: 10.1038/ng.371. Epub 2009/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU international. 2011;107(3):477–85. doi: 10.1111/j.1464-410X.2010.09470.x. Epub 2010/07/02. [DOI] [PubMed] [Google Scholar]
- 59.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Molecular and cellular biology. 2005;25(9):3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. Epub 2005/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6(22):2760–7. doi: 10.4161/cc.6.22.4932. Epub 2007/11/08. [DOI] [PubMed] [Google Scholar]
- 61.Gagou ME, Zuazua-Villar P, Meuth M. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Molecular biology of the cell. 2010;21(5):739–52. doi: 10.1091/mbc.E09-07-0618. Epub 2010/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & development. 2000;14(12):1448–59. Epub 2000/06/20. [PMC free article] [PubMed] [Google Scholar]
- 63.Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Molecular interventions. 2011;11(2):133–40. doi: 10.1124/mi.11.2.11. Epub 2011/05/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engelke C, Parsels L, Qian Y, Zhang Q, Karnak D, Robertson J, et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor, MK8776. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-12-3748. Epub 2013/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai Y, Chen S, Kmieciak M, Zhou L, Lin H, Pei XY, et al. The Novel Chk1 Inhibitor MK-8776 Sensitizes Human Leukemia Cells to HDAC Inhibitors by Targeting the Intra-S Checkpoint and DNA Replication and Repair. Molecular cancer therapeutics. 2013;12(6):878–89. doi: 10.1158/1535-7163.MCT-12-0902. Epub 2013/03/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G, et al. CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(20):5650–61. doi: 10.1158/1078-0432.CCR-12-1322. Epub 2012/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tse AN, Rendahl KG, Sheikh T, Cheema H, Aardalen K, Embry M, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(2 Pt 1):591–602. doi: 10.1158/1078-0432.CCR-06-1424. Epub 2007/01/27. [DOI] [PubMed] [Google Scholar]
- 68.Ashwell S, Zabludoff S. DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(13):4032–7. doi: 10.1158/1078-0432.CCR-07-5138. Epub 2008/07/03. [DOI] [PubMed] [Google Scholar]
- 69.Maugeri-Sacca M, Bartucci M, De Maria R. Checkpoint kinase 1 inhibitors for potentiating systemic anticancer therapy. Cancer treatment reviews. 2013;39(5):525–33. doi: 10.1016/j.ctrv.2012.10.007. Epub 2012/12/05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.