Abstract
Controlled macrophage differentiation and activation in the initiation and resolution of inflammation is crucial for averting progression to chronic inflammatory and autoimmune diseases. Here we show a negative feedback mechanism for proinflammatory IFN-γ activation of macrophages driven by macrophage-associated matrix metalloproteinase 12 (MMP12). Through C-terminal truncation of IFN-γ at 135Glu↓Leu136 the IFN-γ receptor-binding site was efficiently removed thereby reducing JAK-STAT1 signaling and IFN-γ activation of proinflammatory macrophages. In acute peritonitis this signature was absent in Mmp12–/– mice and recapitulated in Mmp12+/+ mice treated with a MMP12-specific inhibitor. Similarly, loss-of-MMP12 increases IFN-γ–dependent proinflammatory markers and iNOS+/MHC class II+ macrophage accumulation with worse lymphadenopathy, arthritic synovitis and lupus glomerulonephritis. In active human systemic lupus erythematosus, MMP12 levels were lower and IFN-γ higher compared to treated patients or healthy individuals. Hence, macrophage proteolytic truncation of IFN-γ attenuates classical activation of macrophages as a prelude for resolving inflammation.
IFN-γ is central in inflammatory pathogenesis, response to infection and autoimmune diseases. Here the authors show that MMP12 expression is reduced in patients with SLE and that MMP12 post-translationally truncates IFN-y, inhibiting its function and affecting pathogenesis of mouse models of peritonitis, SLE and rheumatoid arthritis.
Introduction
In the initiation and resolution of inflammation the transition of macrophage populations from proinflammatory to immunosuppressive is orchestrated by coordinated multiple stimuli1, 2. Proinflammatory macrophages are classically activated by interferon-γ (IFN-γ) secreted from type 1 T helper (Th1) cells and natural killer (NK) cells, which also blocks Th2 cell proliferation3–5. IFN-γ receptor (IFNGR) engagement rapidly induces janus kinase (JAK)-signal transducer and activator of transcription 1 (STAT1) phosphorylation leading to IFN-γ-response gene transcription that increases phagocytosis, inducible nitric oxide synthase (iNOS) and radical oxygen species (ROS) levels1, 4. In the resolution of inflammation, alternatively activated immunosuppressive macrophages are induced by Th2 cytokines including interleukin 4 (IL-4), IL-13, IL-10, TGF-β, and immune complexes3, 5. An underappreciated regulatory mechanism of cytokines is post-translational truncation, as has been shown for IFN-α6 and long known for chemokines7, 8. Indeed, most chemokines are cleaved in their N-termini or C-termini by matrix metalloproteinases (MMPs), which can inactivate9, activate10, 11, convert to antagonists12, 13, or switch receptor specificity14 of chemokines. Thus, cytokine transcript analyses alone may misinform biological data interpretation.
The mechanisms underlying the inappropriate balance of Th1 to Th2 cells, and macrophage phenotypes in chronic inflammation and autoimmunity are incompletely understood. Perturbed upregulation of IFN-γ can lead to autoimmune diseases, such as rheumatoid arthritis, lupus nephritis, and systemic lupus erythematosus (SLE), in which elevated Th1 versus Th2 cell populations increase IFN-γ levels and the activation and number of destructive proinflammatory macrophages15–18. In a mouse genetic knockout of macrophage-associated MMP12 (metalloelastase)19, 20 the balance between Th1 and Th2-induced macrophage populations was swayed toward a Th1 signature in chronic experimental autoimmune encephalomyelitis20. This switch was associated with higher levels of IFN-γ activity, but the mechanism was unknown.
MMP12 cleaves substrates important for macrophage migration21, 22, for example, elastin, fibronectin, laminin, entactin, type I and IV collagens, and proteoglycan core proteins22–24. Mmp12–/– mice are relatively healthy and, consistent with the extracellular matrix substrates of MMP12, have a defect in elastinolytic activity and a reduction in macrophage invasion to penetrate basement membranes, and a decrease in IL-1322, 25. MMP12 also has antimicrobial activity26 and a function in antiviral immunity by cleavage of IFN-α, but not IFN-β6, and regulates neutrophil influx via chemokine processing23, 27.
Conflicting reports suggest IFN-γ has dichotomous time-dependent activity and actions in autoimmunity that is not well understood4, 16. We hypothesized that MMP12 may contribute to the temporal regulation of IFN-γ activity by proteolytic processing. Here we show an inverse correlation between mRNA levels of IFN-γ and macrophage MMP12 in SLE patients and upon treatment. We characterize inactivation of IFN-γ by C-terminal proteolytic processing that contributes to attenuation of proinflammatory macrophage activation in acute inflammation. This negative feedback mechanism is driven by MMP12 secretion by proinflammatory IFN-γ-activated macrophages, with elevated MMP12 expression in IL-4-activated immunosuppressive macrophages reinforcing inflammation resolution. Contrary to the detrimental functions classically ascribed to MMPs in inflammation, low-MMP12 levels may be a risk factor underlying excessive proinflammatory IFN-γ macrophage activation in disease.
Results
Negative association of MMP12 with human lupus
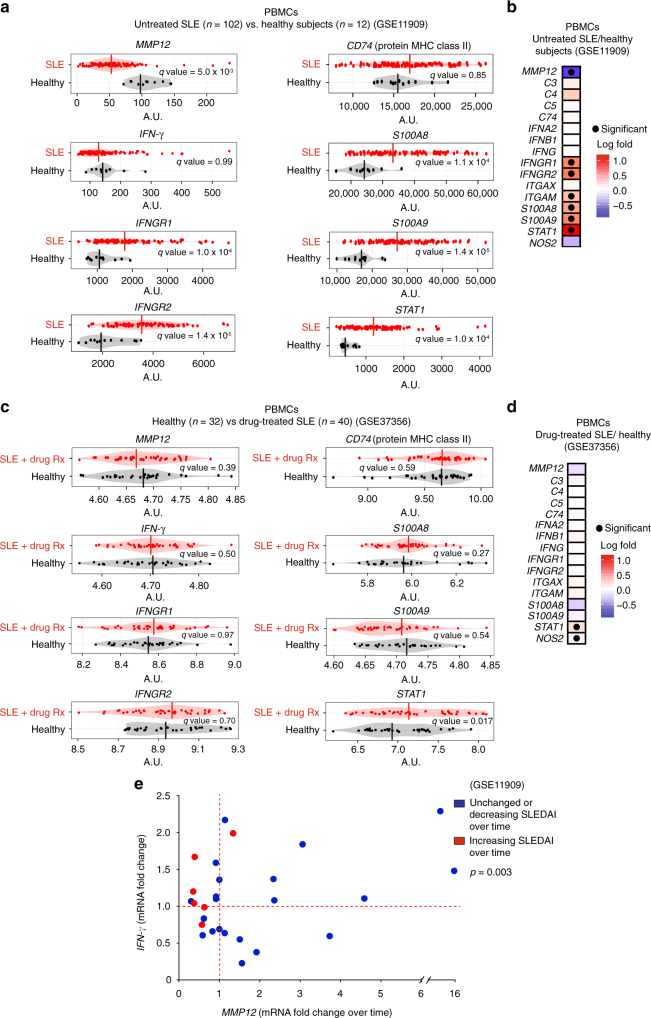
To examine the association of MMP12 expression in human autoimmune disease, we analyzed two human SLE peripheral blood mononuclear cell (PBMC) transcriptome datasets. In the GSE1190928 study, untreated active SLE was associated with significantly reduced MMP12 mRNA (q = 5.0 × 10−3) (n = 102) compared with healthy subjects (n = 12) (Fig. 1a). IFN-γ response genes were correspondingly significantly higher than in healthy controls: IFNGR1 (q = 1 × 10−4), IFGNR2 (q = 1 × 10−5), ITGAM (q = 3 × 10−2), S100A8 (q = 1 × 10−4), S100A9 (q = 1 × 10−4), and STAT1 (q = 1 × 10−4) (Fig. 1b; Supplementary Fig. 1). In a different cohort of stabilized SLE patients that had been successfully treated with corticosteroid, hydroxychloroquine, and/or immunosuppressants (n = 40) (GSE37356)15, the MMP12 levels were comparable (q = 0.39) to those of healthy controls (n = 32) (Fig. 1c), with no significant differences observed in the levels of IFN-γ response genes IFNGR1 (q = 0.97), IFGNR2 (q = 0.70), ITGAM (q = 0.41), S100A8 (q = 0.27) and S100A9 (q = 0.54), or IFNG (q = 0.50) (Fig. 1c; Supplementary Fig. 2).
Fig. 1.
Human PBMC MMP12 and IFN-γ response gene mRNAs in SLE. a PMBC mRNA levels and b quantification of MMP12, C3, C4, C5, CD74, IFNA2, IFNB1, IFNG, IFNGR1, IFNGR2, ITGAM, ITGAX S100A8, S100A9, STAT1, and NOS2 mRNAs in SLE patients (n = 102) or healthy control subjects (n = 12) in the GSE11909 transcript dataset28. Vertical bars are the mean values in all bean plots. c Comparison of these mRNA levels in PBMCs of drug-treated SLE patients (n = 40) or healthy control subjects (n = 32) in the GSE37356 mRNA dataset15. Statistical significance in b and d was determined by calculating the q-values in a and in c. e Longitudinal comparison (31 time points) in 20 individual subjects of the fold changes of MMP12 and IFNG PBMC mRNA levels upon clinical deterioration (increasing SLEDAI in individual patients over time, n = 7 time points), or upon clinical improvement (unchanged or decreasing SLEDAI in individual patients over time, n = 24 time points). Patient information (ethnicity, gender, age, SLEDAI and drug treatment or not) from GSE11909 is in Supplementary Tables S4-S528. Statistical significance was determined by a two-tailed paired Student’s t-test: p = 3 × 10−3
Associated with increasing systemic lupus erythematosus disease activity index (SLEDAI) and clinical deterioration of SLE patients over time was increased IFN-γ associated with significantly decreased MMP12 mRNA levels (two-tailed paired Student’s t-test: p = 6 × 10−3) (GSE11909, n = 114)28 (Fig. 1e, Supplementary Fig. 3). Conversely, in patients showing clinical improvement with decreasing SLEDAI over time, MMP12 mRNA was markedly increased up to ~16-fold (two-tailed paired Student’s t-test: p = 3 × 10−2) (Fig. 1e, Supplementary Fig. 3). In sum, our reanalysis of human clinical sample transcript data revealed that low levels of MMP12 were associated with autoimmune disease and elevated IFN-γ signature genes. Thus, MMP12 may exert clinically significant protective roles that are deficient in the development of SLE. The return of higher MMP12 levels upon treatment and resolution of clinical symptoms are consistent with this hypothesis.
C-terminal truncation of IFN-γ
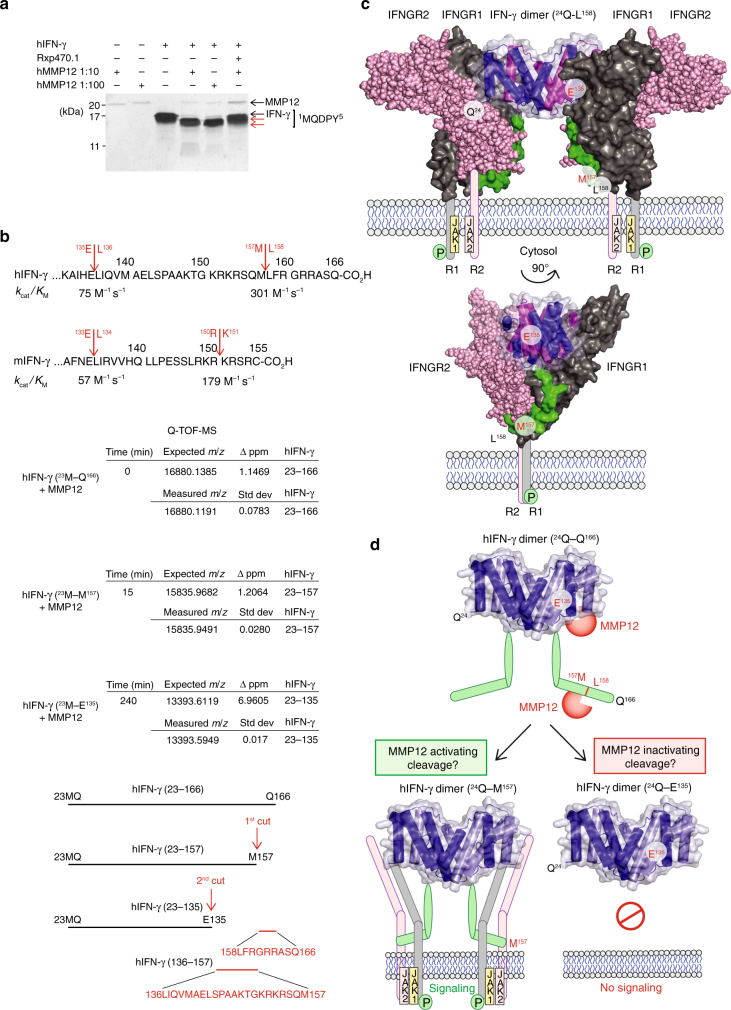
We hypothesized that MMP12 cleaves IFN-γ and attenuates classical activation of macrophages in vivo. Enzyme kinetic analyses revealed that human and mouse IFN-γ were efficiently cleaved proximal to the C-terminus by human and mouse MMP12, respectively (Fig. 2a; Supplementary Fig. 4a). IFN-γ cleavage was prevented by the MMP12-specific inhibitor, Rxp470.16, 29, 30. The first cleavage product of human IFN-γ was apparent within 15 min of co-incubation, consistent with a kcat/KM (1) of 301 M−1 s−1 (Fig. 2b). A second MMP12 cleavage rapidly followed (kcat/KM = 75 M−1 s−1 (1)) and both products displayed an intact N-terminus (Met23 of the recombinant protein) as shown by Edman microsequencing (Fig. 2a). High-mass accuracy top down mass spectrometry identified the two cleavage sites in human IFN-γ at 157Met↓Leu158 and 135Glu↓Leu136 (Fig. 2b, Supplementary Figs. 4b and 5a–c). The 135Glu↓Leu136 scissile bond is conserved in human, monkey, mouse, and rat (N/HE↓LIR/QV) suggestive of a conserved function (Supplementary Fig. 4b). In contrast, MMP12 did not cleave the immunosuppressive macrophage alternative activator, IL-4 (Supplementary Fig. 6).
Fig. 2.
MMP12 cleaves IFN-γ removing the IFN-γ receptor-binding site. a Silver stained 15% SDS-PAGE analysis of in vitro cleavage of human (h) IFN-γ by 10 or 100 ng hMMP12 catalytic domain at 1:10 and 1:100 enzyme to substrate ratios, incubated over 18 h at 37 °C. Revealing C-terminal cleavage, N-terminal sequencing identified an intact N-terminus commencing at 1MQDPY both in IFN-γ and in the two cleavage products (red arrows). The MMP12-specific inhibitor, Rxp470.1, blocked IFN-γ cleavage and autocatalytic cleavage of MMP12 resulting in stabilized MMP12 protein levels over 18 h. Molecular weight marker positions are shown. b Q-TOF-MS analysis of IFN-γ cleavage reaction products revealed C-terminal cleavage first between 157Met↓Leu158 and then at 135Glu↓Leu136 (see Supplementary Fig. 4). The kcat/KM values calculated for each cleavage event in human and mouse (m) IFN-γ are shown. c Based on the crystal structures of the IFN-γ homodimer (pdb entry: 1HIG)33, 51, a secondary complex consisting of the IFN-γ dimer, two high-affinity IFN-γ receptor 1 molecules (IFNGR1(pdb entry: 1FG9); 29Val-Ser241; dark gray), and two low affinity IFN-γ receptor 2 chains (IFNGR2 (pdb entry: 1FYH); 30Leu-Thr237; light pink) were modeled. The IFN-γ C-terminal peptide (135Glu–158Leu) responsible for IFN-γ receptor interaction and signaling was modeled (green). The transmembrane peptide and JAK1/2 are shown in cartoon form. d Frontal view of the structured region of the IFN-γ homodimer with the C-terminal non-structured flexible region (from 146Ala to Gln166) cartooned in green. The two MMP12 cleavage sites are shown: 157Met↓Leu158 and 135Glu↓Leu136
IFNGRs are comprising two ligand binding IFNGR1 chains and two signal-transducing IFNGR2 chains3, 31. We related the cleavage sites in the IFN-γ dimer to the IFNGR chain structures using in silico structural analysis (Fig. 2c). The IFN-γ non-structured C-terminal strand was modeled from Ala146 since it is absent from the crystal structure32, 33. Our analysis predicted that the strand remaining after cleavage at 157Met↓Leu158 would retain binding to both homodimers of IFNGR1 and IFNGR2, consistent with activation34, 35 (Fig. 2d). However, both cleavage products were consistently identified together at all time points by mass spectrometry indicating rapid processive cleavage leading to the final shorter truncated product ending at Glu135 (Fig. 2b and Supplementary Fig. 5a–c). As the loss-of-IFNGR2 binding is predicted by truncation at 135Glu↓Leu136, this suggests the net effect of MMP12 is loss-of-IFN-γ signaling.
Reduced IFN-γ activation of macrophages by MMP12 in vivo
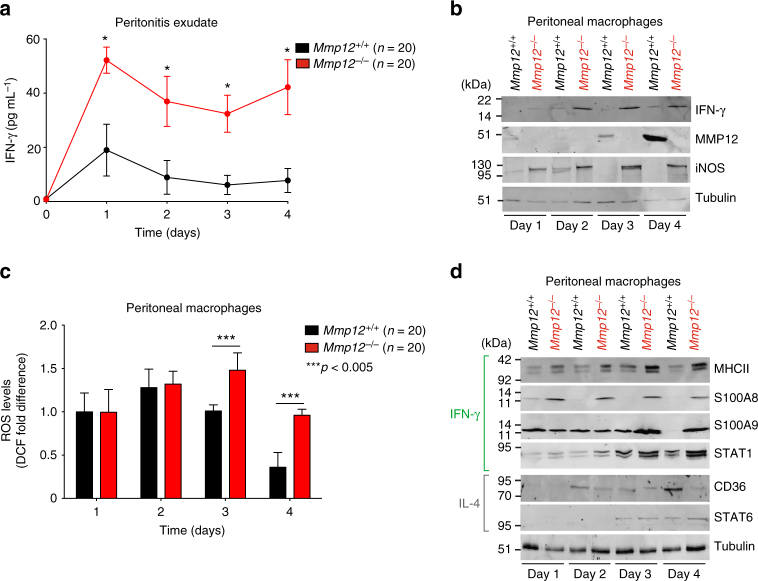
We next examined IFN-γ levels in acute thioglycollate-induced peritonitis from 0 to 4 days in Mmp12+/+ B10.RIII (n = 20) and Mmp12–/– B10.RIII (n = 20) mice (n = 4, for each genotype for each day). Consistent with the initiation of IFN-γ clearance by MMP12 in the Mmp12+/+ mice, ELISA analyses revealed that IFN-γ protein was ~3-fold higher in the peritonitis exudate at days 1 to 4 in the Mmp12–/– mice (Fig. 3a). This difference was also found in peritoneal macrophage lysates by western blotting (Fig. 3b) and was associated with increasing amounts of MMP12 protein over time in the Mmp12+/+ B10.RIII mice (Fig. 3b). The IFN-γ-activated macrophage response markers5, iNOS (Fig. 3b) with attendant ROS production (Fig. 3c), were also up at day 2 and then later declined at days 3 and 4 in the Mmp12+/+ B10.RIII peritoneal macrophages. However, these levels were always greater in the Mmp12–/– B10.RIII primary macrophages, with iNOS and ROS continuing to significantly increase over time; whereas, iNOS protein was undetectable in the Mmp12+/+ B10.RIII macrophages at days 3 and 4.
Fig. 3.
MMP12 decreases IFN-γ-activated macrophage markers in acute peritonitis. a ELISA of IFN-γ protein levels in peritoneal exudate of male Mmp12+/+ B10.RIII (n = 20) and Mmp12–/– B10.RIII (n = 20) mice at days 0–4 after induction of peritonitis (n = 4 for each genotype for each time point, N = 2) expressed as the mean ± s.d. There was no IFN-γ quantified in healthy peritoneum in the absence of inflammation on day 0. Statistical significance was determined by two-tailed unpaired Student’s t-test: *p < 0.05. b 10% SDS-PAGE western blot analysis of IFN-γ, MMP12, and iNOS proteins in primary peritoneal macrophages harvested daily from Mmp12+/+ (n = 20) and Mmp12–/– (n = 20) B10.RIII mice (N = 2). Tubulin, loading control. c Cellular ROS levels in primary peritoneal macrophages were quantified by calculating the mean fluorescence intensity after treatment with 2,-7-dichlorofluorescein diacetate (DCF) (n = 20 for each genotype, n = 4 for each time point, N = 2). Data were normalized to day 1 Mmp12+/+ B10.RIII macrophages and expressed as fold differences. Error bars, s.d. Statistical significance was determined by a two-tailed unpaired Student’s t-test: ***p < 0.005. d 10% SDS-PAGE western blot analysis of markers characteristic for macrophage activation by IFN-γ (MHCII, S100A8, and S100A9) and STAT1, or by IL-4 (CD36) and STAT6 in Mmp12+/+ and Mmp12–/– B10.RIII mouse macrophages harvested daily (n = 20 for each genotype, n = 4 for each time point, N = 2). Tubulin, loading control. Molecular weight marker positions in all blots are as shown
As no one marker is indicative of either proinflammatory classically activated or immunosuppressive alternatively activated macrophages, we supported the iNOS findings in peritonitis by a spectrum of other markers associated with IFN-γ activation. Thus, STAT1, MHCII, S100A8, and S100A9 were also at higher levels in primary Mmp12–/– B10.RIII macrophages compared with Mmp12+/+ B10.RIII cells, particularly at the later time points (Fig. 3d). In contrast, Mmp12+/+ B10.RIII mice displayed higher levels of CD36 on day 4, which is a marker of IL-4 alternative activation. Thus, Mmp12–/– B10.RIII mice displayed elevated IFN-γ protein and prolonged classical macrophage activation in acute inflammation compared with Mmp12+/+ B10.RIII mice. In other words, the absence of MMP12 shifted peritoneal macrophage populations to one enriched with IFN-γ-classically activated proinflammatory macrophages.
IFN-γ cleavage reduces STAT1 and iNOS during phagocytosis
IFN-γ activates JAK-STAT1 signaling with phosphorylation of STAT1 at Tyr701 leading to IFN-γ response gene transcription occurring within 15–30 min and a characteristic upregulation of total STAT1 protein expression after 18–24 h that is highly correlated with IFN-γ activation of macrophages3, 36. To examine the effect of MMP12 cleavage on IFN-γ signaling, we treated mouse RAW264.7 cells lines for 15 min or 24 h with full-length mouse IFN-γ, or with mouse MMP12-cleaved mouse IFN-γ terminating at Glu133 that was mass spectrometry confirmed (Fig. 4a, b). We also treated PMA-matured THP-1 human monocytes with human IFN-γ or with human MMP12-cleaved human IFN-γ terminating at Glu135 (mass spectrometry confirmed) (Fig. 4c, d). In both macrophage cell lines, IFN-γ led to the characteristic phosphorylation of STAT1 at Tyr701 after 15 min with subsequent upregulation of total STAT1 protein at 24 h (Supplementary Fig. 7a, b). Tyr701 phosphorylation was associated with an increase in iNOS protein (Fig. 4b, d). In contrast, incubation with MMP12-truncated IFN-γ neither induced STAT1 phosphorylation nor increased total STAT1 or iNOS proteins in either cell line.
Fig. 4.
MMP12 reduces IFN-γ signaling and responses in macrophages. a Mouse RAW264.7 cells were treated for 15 min or 24 h with PBS, 20 ng/mL mouse IFN-γ, 20 ng/mL mouse IFN-γ pre-incubated with 2 ng/mL mouse MMP12 (37 °C, 18 h), 2 ng/mL mouse MMP12 alone, or 30 ng/mL mouse IL-4 (n = 4, N = 2). After 10% SDS-PAGE, cell lysates were western blotted for pSTAT1-Y701 and STAT1 proteins (Supplementary Fig. 7b). b Western blot of iNOS protein in RAW264.7 cells treated as above for 24 h (n = 4, N = 2). ROS levels were quantified by calculating the mean fluorescence intensity after treatment with 2-,7-dichlorofluorescein diacetate (DCF)(n = 4, N = 2). Data were normalized to PBS-treated cells and expressed as fold differences. Error bars denote s.d. Statistical significance was determined by a two-tailed unpaired Student’s t-test: *p < 0.05; **p < 0.01; ***p < 0.005. c, d Western blotting, quantification, and statistical analyses of pSTAT1-Y701, STAT1, and iNOS protein in human THP-1 cells as described for a and b (n = 4, N = 2) (see also Supplementary Fig. 7a). Tubulin and molecular weight marker positions are shown. e Representative images and f phagocytic index of THP-1 macrophages incubated for 24 h with PBS, 30 ng/mL IL-4, 20 ng/mL IFN-γ, or 20 ng/mL IFN-γ pretreated with 2 ng/mL human MMP12 (37 °C, 18 h), or 2 ng/mL MMP12 alone, and then incubated with serum-coated fluorescent 2-μm microparticles. Scale bars, 20 μm. The phagocytic index was quantified from the number of beads per cell (5–30 cells per field) in each of 20 fields (n = 3, N = 2). Data were normalized to PBS-treated THP-1 macrophages and the means expressed as fold differences. Error bars, s.d. Statistical significance was determined by a two-tailed unpaired Student’s t-test: ***p < 0.005. g MMP12 mRNA analysis of PMA-matured THP-1 cells treated with PBS, IFN-γ, or IL-4 (n = 3, N =2). A-values were normalized to IFN-γ-induced THP-1 cell mean values and expressed as fold differences. Error bars, s.d. Statistical significance was determined by a two-tailed unpaired Student’s t-test: NS not significant difference; ***p = 1 × 10−5
An important response to macrophage classical activation by IFN-γ is elevated phagocytosis with associated induction of microbicidal activity wrought by ROS3 that is generated by elevated iNOS5. In a fluorescent bead phagocytosis assay with differentiated THP-1 cells, PBS-treated and IL-4-activated macrophages showed low levels of phagocytosis (Fig. 4e, f). Phagocytosis was increased 4.9-fold in IFN-γ-activated THP-1 cells, but when IFN-γ was cleaved by MMP12, the cells retained the lower phagocytic activity of non-activated macrophages. These functional data are consistent with the elevation in iNOS protein levels in both IFN-γ-activated RAW264.7 and THP-1 cells compared to PBS-treated or IL-4-alternately activated macrophages, as well as a corresponding increase in cellular ROS (Fig. 4b, d). In contrast, MMP12 cleavage of IFN-γ reduced the increase in the IFN-γ activation markers iNOS and ROS.
As expected, IL-4 treatment of RAW264.7 and THP-1 cells did not affect STAT1 protein phosphorylation (Fig. 4a, c), but did trigger the characteristic Tyr641 phosphorylation of STAT637 (Supplementary Fig. 7c, d). Thus, IFN-γ signaling was reduced by MMP12 cleavage of IFN-γ at 135Glu↓Leu136, which in turn reduced IFN-γ-activated macrophage polarization, whereas IL-4-activated macrophage polarization was unaffected by MMP12. Finally, we analyzed MMP12 mRNA expression in PMA-matured THP-1 cells (M0) that were treated with IFN-γ, IL-4, or PBS vehicle (n = 3, N = 2). PBS controls or IFN-γ-activated cells expressed MMP12 mRNA that were measured with microarray A-values of 9.62 and 8.99, respectively, which were not significantly different (two-tailed paired Student’s t-test: p = 0.09). In contrast, IL-4-activated THP-1 cells expressed 3.2-fold more MMP12 mRNA with an A-value of 10.73 and that was significantly different from the PBS controls and IFN-γ-activated THP-1 cells (two-tailed paired Student’s t-test: p = 1 × 10−5) (Fig. 4g). Hence, these cellular readouts support an IFN-γ function-inactivating cleavage by MMP12 to lessen IFN-γ-activation of macrophages, but not that of IL-4 macrophage activation. Indeed, IL-4 activation led to elevated MMP12 expression, which may act in trans in mixed cell populations to further reduce IFN-γ macrophage activation.
Mmp12 loss is associated with elevated IFN-γ markers in arthritis
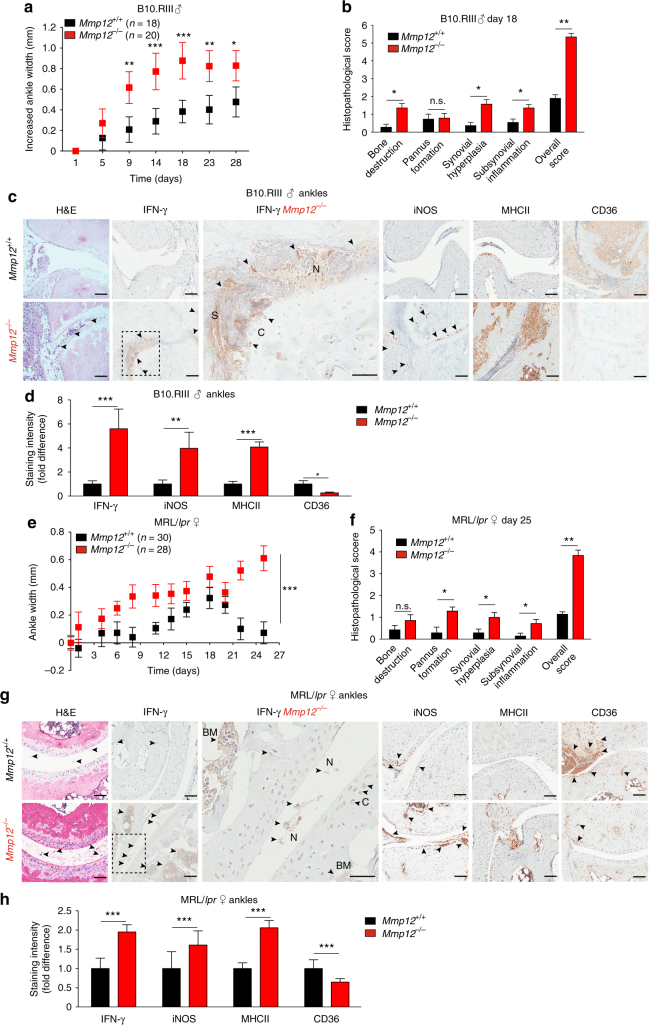
We found that genetic deletion of MMP12 increased the severity of inflammatory arthritis in mice. Twenty-eight days after induction of collagen-induced arthritis, male Mmp12–/– B10.RIII mice (n = 20) exhibited more severe arthritis (Fig. 5a, b), with increased ankle width and histopathological scores compared with male Mmp12+/+ B10.RIII mice (n = 18). The Mmp12–/– B10.RIII mice also had higher levels of IFN-γ and the IFN-γ-activated macrophage markers iNOS and MHCII, with concomitant near absent CD36 staining (Fig. 5c, d). Neutrophils and neutrophil extracellular traps (NETs) in the synovial space23 further indicated an acute IFN-γ-driven proinflammatory response38.
Fig. 5.
Altered macrophage markers in rheumatoid arthritis in Mmp12–/– mice. a Hind ankle widths of Mmp12+/+ and Mmp12–/– male B10.RIII mice (n = 18 and 20, respectively, for each time point) after onset of collagen-induced arthritis (day 0), means ± s.e.m. Mann–Whitney t-test: *p < 0.05, **p < 0.01, ***p < 0.005. b Histopathology of hind ankles after H&E staining (day 18) Mmp12+/+ (n = 3) and Mmp12–/– (n = 3). Bone destruction (p < 6 × 10−5), pannus formation (NS not significant), synovial hyperplasia (p < 3 × 10−5), and subsynovial inflammation (p < 7 × 10−4) were quantified as a histopathological score (p < 9 × 10−4, means ± s.d.), two-tailed unpaired Student’s t-test: *p < 0.05, **p < 0.01. c Representative images of H&E, IFN-γ, iNOS, MHCII, and CD36 immunostaining of hind ankle joints of Mmp12+/+ (n = 3) and Mmp12–/– (n = 3) mice. Here and in g: N NETs; C cartilage; S synovial space; arrowheads, high antibody or H&E staining; scale bars, 100 μm. d Immunostaining quantification of Mmp12+/+ versus Mmp12–/– male B10.RIII mice hind ankle joints for IFN-γ (p < 2 × 10−5), iNOS (p < 1 × 10−3), MHCII (p < 2 × 10−5), and CD36 (p < 2 × 10−2) (means ± s.d.), two-tailed unpaired Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.005. e Ninety-day-female MRL/lpr mice were injected with CFA (day 0). Hind ankle size was measured in Mmp12+/+and Mmp12–/– MRL/lpr mice (n = 30 and 28, respectively, for each time point) (means ± s.d.), two-tailed unpaired Student’s t-test: ***p < 0.005. f Histopathology of H&E stained-hind ankle joints of Mmp12+/+ (n = 3) and Mmp12–/– (n = 3) MRL/lpr (day 25). Bone destruction (n.s.), pannus formation (p < 0.01), synovial hyperplasia (p < 3 × 10−2), and subsynovial inflammation (p < 3 × 10−2) were quantified as the histopathological score (p < 6 × 10−3) (means ± s.d.), two-tailed unpaired Student’s t-test: *p < 0.05, **p < 0.01. g Representative images of H&E, IFN-γ, iNOS, MHCII, and CD36 immunostaining of hind ankle joints of Mmp12+/+ and Mmp12–/– MRL/lpr female mice. BM bone marrow. h Immunostaining intensities of Mmp12+/+ and Mmp12–/– mouse hind ankle joints for IFN-γ (p < 2 × 10−5), iNOS (p < 1 × 10−3), MHCII (p < 4 × 10−4), and CD36 (p < 4 × 10−4) (means ± s.d.), two-tailed unpaired Student’s t-test: ***p < 0.005
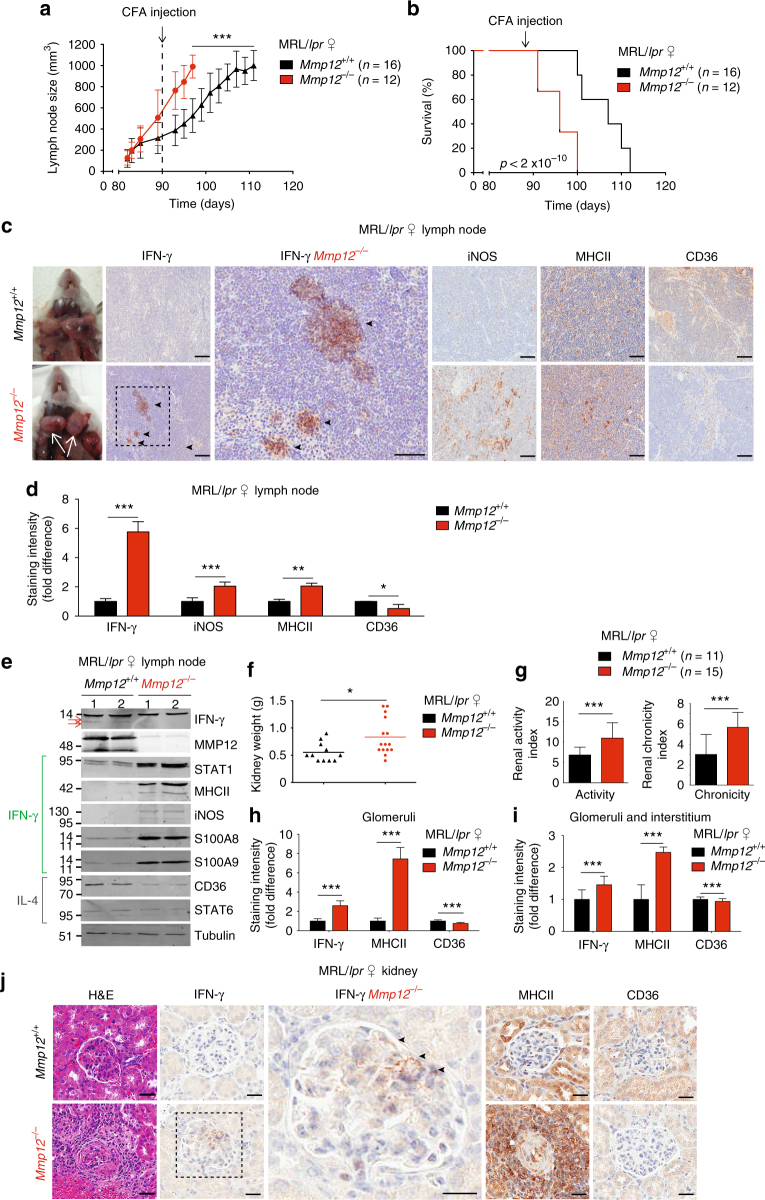
We confirmed these findings in female mice on another background. The autoimmune MRL/lpr mouse strain39 spontaneously develops systemic autoimmunity with severe lymphadenopathy, arthritis and glomerulonephritis. In our substrain, we found it was necessary to initiate autoimmune arthritis earlier with Complete Freund’s adjuvant (CFA), otherwise gross lymphadenopathy necessitated euthanasia too soon to derive meaningful insights into the development of severe arthritis. As CFA stimulates the hyperproduction of IFN-γ, expression of MHC class II genes, and Th1/classical activation of macrophages39, 40 we reasoned this strain was suited to assess the effects of MMP12 on IFN-γ-driven disease and macrophages in vivo.
In female Mmp12–/– MRL/lpr mice, we saw similar associations as found in the B10.RIII mice. Hind ankle joints at baseline were normal with no evidence of disease as quantified by a 4-point histopathological grading scheme41, including no difference in paw edema in both the Mmp12+/+ (n = 30) and Mmp12–/– (n = 28) female MRL/lpr mice (Supplementary Fig. 8b). With loss-of-MMP12 there was an increased incidence of arthritis as compared to Mmp12+/+ MRL/lpr mice (Supplementary Fig. 8a). In the acute disease phase immediately following CFA induction, the Mmp12–/– MRL/lpr mice displayed a faster onset of arthritis that was detectable at day 6 by significant and more severe ankle swelling than Mmp12+/+ MRL/lpr mice, that continued to exacerbate to day 25 (Fig. 5e). In contrast, in Mmp12+/+ MRL/lpr mice, ankle edema started to decrease at day 19 and continued to fall to day 25—at which time the Mmp12–/– MRL/lpr mice showed larger pannus formation, more synovial hyperplasia and subsynovial inflammation (Supplementary Fig. 8c) with a histopathological score of 3.8 in comparison to 1.1 for Mmp12+/+ MRL/lpr mice (Fig. 5f). In quantifying differences in IFN-γ-activated macrophage proteins at day 25 we found the Mmp12–/– MRL/lpr ankles had elevated IFN-γ, as well as elevated iNOS and MHCII classical activation marker proteins, reflecting sustained IFN-γ-stimulated macrophage responses in vivo (Fig. 5g, h). This indicated that without MMP12, MRL/lpr mice experienced a loss-of-control of macrophage activation and inflammation that led to a sustained disease flare.
Elevated IFN-γ activation in Mmp12–/– mouse models of lupus
Elevated levels of IFN-γ and STAT1 signaling are implicated in chronic lymphoid-driven human diseases, such as SLE36, 39, 42 and in mouse models of SLE, including the MRL/lpr strain39, 40. The signature lymphoproliferative response in female MRL/lpr mice, as evidenced by enlarged lymph nodes, was seen by day 90 in female Mmp12–/– (n = 18) and Mmp12+/+ (n = 38) mice (Fig. 6a, Supplementary Fig. 9a, b, N = 2) and later for males (n = 13 each for Mmp12–/– and Mmp12+/+ male mice, Supplementary Fig. 9a). Consistent with the MMP12-dependent differences in lymph node size apparent by day 90 in the female mice, CFA synchronization further expanded the differential increase in lymphadenopathy manifest in the Mmp12–/– MRL/lpr females (n = 18, N = 2) over that seen for the female Mmp12+/+ MRL/lpr mice (n = 38, N = 2) (Fig. 6a, Supplementary Fig. 9b), supporting a role for MMP12 in dampening acute inflammatory responses.
Fig. 6.
Mmp12–/– mouse mortality and IFN-γ macrophages numbers in SLE. a Following CFA injection, the size of the superficial cervical lymph nodes of Mmp12+/+ and Mmp12–/– MRL/lpr (n = 16 and 12, respectively, for each time point) female mice were measured and expressed as means ± s.d. Two-tailed unpaired Student’s t-test: ***p < 0.005. b Kaplan–Meier curves showing mortality rates of female Mmp12+/+ MRL/lpr (n = 16) and Mmp12–/– (n = 12) mice. Two-tailed unpaired Student’s t-test: p < 2 × 10−10, (see Supplementary Fig. 9c, d for survival data). c Representative images of superficial cervical lymph nodes (white arrows) immunostained for IFN-γ, iNOS, MHCII, and CD36 from Mmp12+/+ (n = 3) and Mmp12–/– (n = 3) female MRL/lpr mice at their humane end points (Mmp12+/+ (day 112) and Mmp12–/– (day 98)). Scale bars, 100 μm. d Quantification of immunostaining intensities of Mmp12+/+ (n = 3) versus Mmp12–/– (n = 3) MRL/lpr mice superficial cervical lymph nodes for IFN-γ (p < 9 × 10−6), iNOS (p < 2 × 10−7), MHCII (p < 1 × 10−2), and CD36 (p < 2 × 10−2) expressed as the mean ± s.d. Two-tailed unpaired Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.005. e Superficial cervical lymph nodes were analyzed by western blotting for macrophages markers of IFN-γ activation (iNOS, MHCII, S100A8, and S100A9) and STAT1, as well as for IL-4 induced CD36 and STAT6. Two biological replicates analyses from Mmp12+/+ (n = 8) and Mmp12–/– (n = 8) MRL/lpr mice, are shown. Red arrows indicate lower molecular forms of IFN-γ. f Mean kidney weights and g mean histological scores of activity and chronicity indexes from Mmp12+/+ (n = 11) and Mmp12–/– MRL/lpr (n = 15) mice were measured at their humane end points. Error bars, s.d. Two-tailed unpaired Student’s t-test: *p < 0.05, ***p < 0.005. Quantification of immunostaining in h glomeruli and i in glomeruli and interstitium of Mmp12+/+ (n = 3) and Mmp12–/– MRL/lpr (n = 3) mice at their humane end points for IFN-γ (p < 2 × 10−12), MHCII (p < 7 × 10−17), and CD36 (p < 2 × 10−6). Quantification is expressed as means ± s.d. Two-tailed unpaired Student’s t-test: ***p < 0.005. j Representative images of H&E, IFN-γ, MHCII, and CD36 immunostaining of the renal glomeruli sections of Mmp12+/+ and Mmp12–/– female MRL/lpr mice; arrowheads, strong IFN-γ staining. Scale bars, 100 μm
As in human autoimmune disease, where there is significantly increased prevalence in females versus males43, female mice in the absence of CFA had a shortened survival time than males (112 days versus 131 days) before lymphadenopathy led to humane endpoint euthanasia (Fig. 6b, Supplementary Fig. 9c-e, N = 2). Protein levels of IFN-γ and the IFN-γ-activated macrophage markers iNOS and MHCII were significantly increased in the superficial cervical lymph nodes of the female Mmp12–/– MRL/lpr mice compared with female Mmp12+/+ MRL/lpr mice (day 98) (Fig. 6c, d). As immunohistochemistry is only semi-quantitative we confirmed the elevation of these IFN-γ classical activation macrophage proteins, plus STAT1, S100A8, and S100A9, in lymph node extracts of the knockout versus the wild-type Mmp12 MRL/lpr mice (Fig. 6e). Of note, we detected lower molecular weight forms of IFN-γ in Mmp12+/+ but not Mmp12-/- mice (Fig. 6e), suggestive of in vivo processing of IFN-γ. Thus, the lack of MMP12 was associated with a shift in macrophage populations to one retaining ongoing IFN-γ-activation in vivo.
In association with the enlarged cervical lymph nodes in Mmp12–/– MRL/lpr female mice (Fig. 6a), we found a relative decrease in CD68+ cells consistent with previous reports22, 23 and elevated inflammatory markers e.g., increased neutrophil myeloperoxidase, TUNEL and CD31+ cells (Supplementary Fig. 10b). Although, spleen weights in both sexes were unchanged with or without MMP12 (Supplementary Fig. 9f) and there were no significant differences in the number of circulating leukocytes and monocytes in Mmp12+/+ (n = 15) and Mmp12–/– (n = 27) MRL/lpr female mice (Supplementary Fig. 10a), we observed increased kidney weights in Mmp12–/– MRL/lpr female mice (n = 15) versus Mmp12+/+ MRL/lpr littermates (n = 11) (Fig. 6f). Histological scores of Activity Index and Chronicity Index44, 45 were significantly elevated in Mmp12–/– MRL/lpr female mice (n = 15) versus Mmp12+/+ MRL/lpr littermates (n = 11) (Fig. 6g), indicating both worse acute lupus nephritis activity and resultant chronicity. The concentration of MHCII and IFN-γ proteins were significantly elevated in Mmp12–/– kidney glomeruli compared with Mmp12+/+ littermates, reflecting a shift in the Mmp12–/– MRL/lpr macrophage population in vivo (Fig. 6h–j). Overall, in the MRL/lpr model of SLE, Mmp12–/– mice exhibited sustained IFN-γ signaling favoring ongoing proinflammatory responses versus Mmp12+/+ MRL/lpr mice, which exhibited normal macrophage phenotypic succession to alternatively activated CD36+ macrophages with time.
C-terminal cleavage of IFN-γ is reduced in lupus nephritis
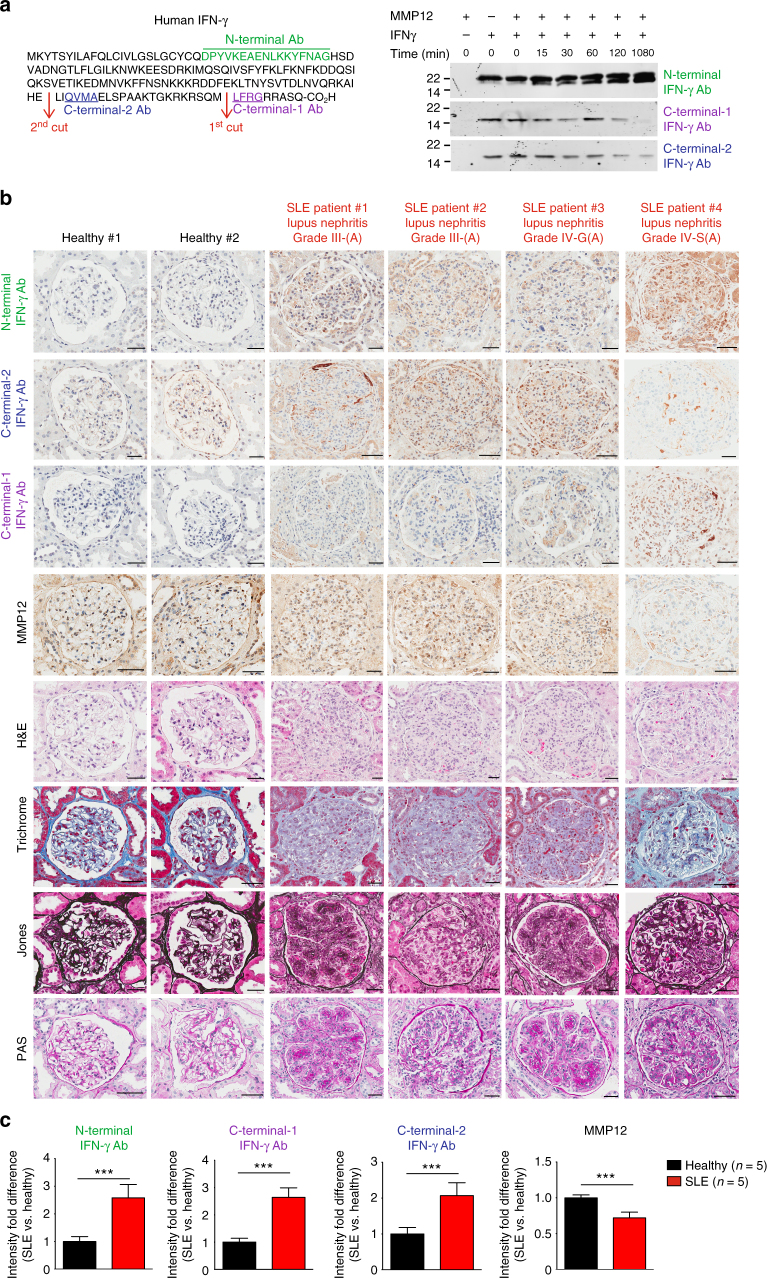
We next analyzed MMP12 and IFN-γ protein levels and the IFN-γ cleavage status in patients with lupus nephritis (Supplementary Table 1). To do so we raised and affinity-purified three antibodies to human IFN-γ to determine its activation status in vivo (Fig. 7a). To profile total (active plus inactive) IFN-γ protein, we raised an antibody against the N-terminal IFN-γ peptide, 25DPYVKEAENLKKYFNAG41. To identify full-length IFN-γ protein, we raised antibodies against the C-terminal sequence 158LFRG161 (C-terminal-1 antibody) distal to the first cleavage site and therefore only present in non-cleaved IFN-γ. By recognizing a sequence (QVMA) between the 1st and 2nd MMP12 cleavage sites, the C-terminal-2 antibody discriminated between the two C-terminal MMP12-truncated forms. So, whereas positive staining for the N-terminal antibody identified all 3 forms of IFN-γ, including the inactive MMP12-truncated IFN-γ 23–135, the C-terminal-2 antibody confirmed the presence of both the active IFN-γ proteoforms past position 135. As the human and murine IFN-γ sequences diverge significantly in the C-terminus (Supplementary Fig. 4b), the anti-human IFN-γ antibodies did not cross react with mouse IFN-γ.
Fig. 7.
IFN-γ epitope antibody staining of human lupus nephritis biopsies. a Left, amino acid sequences (underlined) of human IFN-γ peptides used to raise and affinity-purify rabbit anti–N-terminal, C-terminal-1, and C-terminal-2 IFN-γ epitope antibodies. Right, western blot analysis of human IFN-γ after time-dependent cleavage to 1080 min by human MMP12. Note: disappearance of the C-terminal epitopes as MMP12 cleavage proceeds. Molecular weight marker positions in all blots are as shown. b Immunostaining of human kidney biopsies using anti–N-terminal, C-terminal-2, C-terminal-1, and MMP12 antibodies; and staining with hematoxylin and eosin (H&E), Trichrome, Jones, and PAS of kidney biopsies of healthy (n = 5), lupus nephritis at Grades III-(A) (n = 3) and IV-(A) (n = 2) as diagnosed in Supplementary Fig. 11. Scale bar, 100 μm. Original magnification, ×400. c Quantification of immunostaining intensities in healthy (n = 5) and lupus nephritis (n = 5) kidney biopsies (additional data are included in Supplementary Fig. 11) and expressed as means ± s.d. Statistical significance was determined by a two-tailed unpaired Student’s t-test: ***p < 0.005
After a time-course incubation of IFN-γ with MMP12 for up to 1080 min the presence of a C-terminal cleaved form of IFN-γ was apparent using the N-terminal antibody, even as soon as 15 min (Fig. 7a), confirming the kinetic data of efficient cleavage. The C-terminal-2 antibody will recognize both full-length and IFN-γ cleaved at 157Met↓Leu158. Decreasing immunoreactivity for both C-terminal epitopes over time in vitro confirmed cleavage removal of the short C-terminal peptides (Fig. 7a). Although cleavage at 135Glu↓Leu136 can occur first, the loss-of-C-terminal-1 immunoreactivity with a weak C-terminal-2 band still present at 1080 min indicated that 157Met↓Leu158 cleavage can also precede 135Glu↓Leu136 cleavage. Therefore, the prominent lower immunoreactive band detected by the strong N-terminal antibody is likely the 135Glu↓Leu136 cleaved inactive IFN-γ proteoform.
With these antibodies, we analyzed IFN-γ cleavage status in five healthy subjects and five active (A) patients with SLE Stage III-(A) (n = 3) or Stage IV-(A) (n = 2) lupus nephritis (Fig. 7b, Supplementary Fig. 12 and Supplementary Table 1). As compared to the normal biopsies, the SLE biopsies displayed a significant fold-increase in total glomerular IFN-γ, as shown by the anti–N-terminal antibody (Fig. 7c). This included significant increases in full-length IFN-γ (23–166), as revealed by immunoreactivity against the N-terminal and C-terminal-1 antibodies, and significant increases in active IFN-γ in the SLE patients, as revealed by positive immunostaining of both anti–C-terminal antibodies. Indeed, the significant immunostaining by the anti–C-terminal antibodies was associated with lower MMP12 immunostaining, supporting the hypothesis of reduced MMP12–mediated IFN-γ cleavage in SLE vs. normal kidney.
MMP12 modulation of STAT1 signaling in primary macrophages
To mechanistically explore the in vivo consequences of loss-of-MMP12 on pSTAT1-Y701 signaling by IFN-γ, we induced peritonitis in Mmp12+/+ B10.RIII mice (n = 5) and Mmp12–/– B10.RIII mice (n = 4). At day 4, we collected peritoneal macrophages and stimulated the primary macrophages with IFN-γ. In the Mmp12+/+ cells, we observed the characteristic pulse in pSTAT1-Y701 levels that decayed after ~30 min (Fig. 8a). In contrast, upon IFN-γ treatment primary macrophages from the Mmp12–/– mice displayed prolonged activation of pSTAT1-Y701 phosphorylation to 60 min and beyond (Fig. 8b, c).
Fig. 8.
Prolonged IFN-γ signaling in Mmp12–/– primary peritoneal macrophages. Western blot analysis for pSTAT1-Y701 and STAT1 in primary peritoneal macrophages harvested from individual (a, e) Mmp12+/+ B10.RIII (n = 5 for each time point) and (b, d) Mmp12–/– B10.RIII (n = 4 for each time point) mice 4 days after induction of peritonitis. Cells were treated with 20 ng/mL of IFN-γ for 0–1080 min. c Ratios of pSTAT1-Y701 to STAT1 protein levels were determined after densitometry quantification of the western blots. The data are expressed as fold differences in the ratio of the means for Mmp12+/+ (n = 5 for each time point) and Mmp12–/– (n = 4 for each time point) B10.RIII mice. d Rescue of Mmp12–/– peritoneal macrophages with recombinant mouse MMP12 protein (1:100) incubated for the times shown (n = 4 for each time point). e Mmp12+/+ B10.RIII macrophages were incubated for 30 min with 100 nm specific MMP12 inhibitor Rxp470.1 before addition of 20 ng/mL IFN-γ (n = 4 for each time point). f Western blot analysis of IFN-γ, MMP12, iNOS, MHCII, S100A8, S100A9, STAT1, CD36, and STAT6 proteins in primary peritoneal macrophages harvested from Mmp12+/+ (n = 4) and Mmp12–/– (n = 4) B10.RIII mice at day 4 post-intraperitoneal injection with vehicle; and Mmp12+/+ B10.RIII mice treated daily with 5 mg/kg Rxp470.1 (n = 4) for 4 days during the induction of peritonitis. Actin or tubulin loading controls and molecular weight marker positions in all blots are as shown
We then performed gain- and loss-of-function experiments. Treatment with recombinant mouse MMP12 of four individual cultures of Mmp12–/– B10.RIII primary macrophages harvested from four mice (Fig. 8d) terminated the sustained activation of pSTAT1-Y701 seen in the vehicle control Mmp12–/– cells without exogenous MMP12 (Fig. 8b). Hence adding MMP12 rescued the temporal pattern of pSTAT1-Y701 phosphorylation to that resembling Mmp12+/+ B10.RIII (n = 5) primary peritoneal macrophages, which also decayed at ~30 min (Fig. 8a). Similarly, treatment of primary Mmp12+/+ B10.RIII macrophage cultures with the selective MMP12 inhibitor, Rxp470.1 (Fig. 8e), extended the activation of pSTAT1-Y701 and recapitulated the Mmp12–/– B10.RIII primary cell responses (Fig. 8b). Hence, a genetic lack of MMP12 or the specific inhibition of MMP12 prolonged STAT1 signaling, thereby providing an explanation for the IFN-γ-classically activated macrophage dominated profile of Mmp12–/– B10.RIII mice in vivo.
Finally, to directly link MMP12 catalytic activity with the controlled normal succession of IFN-γ-classically activated macrophage populations to alternately activated macrophages in vivo, we injected Mmp12+/+ B10.RIII mice with 5 mg/kg MMP12-specific inhibitor (Rxp470.1) (n = 4) or vehicle (n = 4) daily for 4 days during the development of peritonitis. Although MMP12 is predominantly a macrophage-associated protease21, 22, other cells also express the enzyme6, which systemic administration of Rxp470.1 will block. In primary macrophages of the vehicle controls harvested at day 4 we found that the cell population was enriched for macrophages with high CD36 expression (indicative of the alternative macrophage activation pathway), an absence of iNOS and MHCII proteins, and low levels of other IFN-γ-classically activated macrophage signature proteins (Fig. 8f). This represents the normal temporal evolution of macrophage phenotypes in acute inflammation. In contrast, treatment of the Mmp12+/+ B10.RIII mice (n = 4) for 4 days with Rxp470.1 shifted the day 4 peritoneal macrophage population to one retaining the early proinflammatory IFN-γ-classically activated macrophage characteristics of Mmp12+/+ B10.RIII at day 2 (Fig. 3b, d) or Mmp12–/– B10.RIII mice at day 4 (Figs. 8f and 3b, d). Thus, IFN-γ, iNOS, MHCII, S100A8, S100A9, and STAT1 proteins were elevated in peritoneal macrophages harvested from the Rxp470.1-treated Mmp12+/+ mice compared with vehicle-treated Mmp12+/+ mice at day 4 and the profile now matched that of the Mmp12–/– mice at day 4. Notably, this loss-of-controlled evolution in macrophage populations over time was also a feature of Mmp12–/– B10.RIII arthritis at 28 days (Fig. 5) and lymphadenopathy in MRL/lpr mice at 96 days (Fig. 6e). Thus, injection of a specific MMP12 inhibitor in vivo in this loss-of- function experiment directly linked MMP12 catalytic activity with the normal controlled shift in proinflammatory to immunosuppressive macrophage cell phenotype ratios occurring in vivo.
Discussion
It is important to recognize all levels of control of key cytokines contributing to the transition from proinflammatory to immunosuppressive macrophage dominated inflammation, and therefore disease risk, susceptibility and resolution. IFN-γ is one of the main effectors in autoimmune diseases, such as SLE, rheumatoid arthritis, and multiple sclerosis19, 46. However, the role of post-translational modification of IFN-γ in these diseases has not been previously recognized. We demonstrated that MMP12, primarily a monocyte-macrophage protease21, 22, downregulates IFN-γ activity by precise C-terminal cleavage at an evolutionary conserved site that prevents phosphorylation of STAT1-Tyr701, and thus IFN-γ signaling and classical activation of macrophages. Initial MMP12 cleavage then sets the stage for IFN-γ clearance in vivo that reduces total IFN-γ levels over time. By forming a post-translational negative feedback mechanism, we found that the normal succession from IFN-γ-classically activated proinflammatory to alternately activated macrophages was dampened when MMP12 was genetically deficient or therapeutically inhibited, especially in acute inflammation, but also in chronic inflammatory autoimmune models experiencing disease flares. Thus, there was a clear shift in inflammatory cell infiltrates to populations enriched for IFN-γ-activated macrophages with attendant tissue pathology in Mmp12 knockout mice in three different models of inflammation, in both male and female mice, and on two different genetic backgrounds.
We observed an important role for the MMP12/IFN-γ processing axis in acute inflammation in models of peritonitis and in the acute collagen-induced model of arthritis. The role of MMP12 was also apparent in MRL/lpr mouse models of arthritis and lupus, especially after CFA hyperinduction of IFN-γ, where differences in its activity would be more manifest depending upon MMP12 initiated inactivation and clearance. This suggests an important role of MMP12 in dampening disease flares. We further showed the significance of dysregulated IFN-γ in human disease by demonstrating that reduced MMP12 mRNA levels were correlated with systemic lupus erythematosus clinical deterioration over time, and also demonstrated increased active IFN-γ in glomeruli of patients suffering from lupus nephritis when compared to healthy individuals, highlighting a potential diagnostic and therapeutic path. Adding to mechanisms in regulation of macrophage activation and to other anti-inflammatory activities of MMP1223, including C-terminal cleavage and inactivation of IFN-α6 and antibacterial roles26, IFN-γ C-terminal truncation provides a new level of post-translational negative feedback control in the evolution of macrophage populations. Our present work further challenges the classical detrimental roles ascribed to other MMPs that are elevated in SLE47, 48.
Numerous pathways and cell types are important in the pathogenesis of chronic inflammation and autoimmune diseases. Multifactorial control of normal macrophage polarization leads to many possible break points that can fail, increasing the risk of chronic inflammation and autoimmunity. Such complexity renders diagnosis and treatment challenging. We were able to translate our findings identifying proteolytic control of IFN-γ activity and IFN-γ activation of macrophages to human autoimmune disease. By analyzing previous data, we found that SLE diagnosed by multiple clinical parameters, including elevated anti-double stranded DNA antibody levels, was associated with lower PBMC MMP12 mRNA levels than in healthy subjects. Further, with clinical improvement upon treatment during the chronic inflammatory phase of the disease, MMP12 mRNA expression increased to near healthy control levels. In a different study in which clinical responses after treatment of 40 SLE patients were examined, we found a strong association in the reduction of IFN-γ response gene expression that coincided with elevated expression of MMP12 mRNA and IL-4 alternatively activated macrophage markers. We found that MMP12 normalization occurs after instituting treatment and precedes clinical improvement, whereas patients whose MMP12 levels are not affected appeared not to improve. Although this clinical association does not prove a mechanistic link for MMP12 in protection from autoimmunity in human disease, it supports the findings from our animal model studies.
The temporal integration of opposing cytokine signals important in differentiation fate decisions for IFN-γ classical activation versus IL-4 activation and other alternate macrophage activation pathways, and the spectrum of cell populations occurring in between, is a multi-faceted, highly nuanced immune mechanism that underpins the development of chronic inflammation and autoimmunity and leads to acute clinical relapses. As MMP12 expression is higher in IL-4 activated macrophages than in IFN-γ-activated cells, MMP12 expression by alternatively activated macrophages may amplify in trans the reduction in IFN-γ activity by IFN-γ-activated macrophage MMP12 to reinforce the decrease in proinflammatory cells. Conversely, low-MMP12 activity favors chronic disease progression and so may be a risk factor for SLE, and potentially other autoimmune diseases. Our findings also further emphasize that using genomic or transcriptomic data alone risks overlooking the key importance of post-translational modifications of immune regulatory and autoimmune signature cytokines. The present data establishing the proteolytic regulation of IFN-γ activity in acute inflammation, including disease flares of chronic disease, suggest a more generally applicable mechanism not limited to SLE or lupus nephritis, but also potentially at play in other IFN-γ-linked phenotypes. Having identified that MMP12 expression provides a benefit in mouse models of IFN-γ-driven inflammatory diseases, it will be important to prospectively evaluate MMP12 activity in a larger cohort of patients with diseases such as SLE. If activity is significantly lower in a large patient cohort with active disease, it will provide crucial information that potentially may point to new therapeutic modalities that restore normal cytokine cellular control to augment the production of MMP12.
Methods
MMP12 cleavage assays of IFN-γ
Recombinant human and mouse MMP12, expressed and purified as described49, were incubated with 10 or 100 ng human or mouse MMP12, 37 °C in assay buffer (100 mM Tris-HCl, 50 mM NaCl, 5 mM CaCl2, pH 8.0) at enzyme/substrate ratios from 1:10 to 1:100. IFN-γ cleavage was determined by silver staining of 15% Tris-Tricine gels and western blots. To identify cleavage sites, cleaved peptides were analyzed by the Edman sequencing, liquid chromatography tandem–mass spectrometry (LC–MS/MS), and MALDI-TOF MS on a Applied Biosystems Voyager DE STR Mass Spectrometer as described50. The specific MMP12 inhibitor RXP470.16, 30 was a control in the cleavage assays. The kcat/KM values were quantified according to (1) from IFN-γ fragment generation over time as described50:
| 1 |
Top down mass spectrometry
IFN-γ and MMP12 cleavage fragments were analyzed by top down liquid chromatography tandem–mass spectrometry (LC–MS/MS). After cleavage (15 min or 240 min), samples were first purified by solid phase extraction on Empore Octyl C8 disks (3M Empore) packed as a C18 StageTips. Tips were activated with 50% acetonitrile, equilibrated with 0.1% formic acid, samples were acidified, loaded and then washed with 2 column volumes of 0.1% formic acid, and eluted in 50% acetonitrile, 0.1% formic acid. Eluates were directly infused into an Impact II high resolution, high-mass accuracy quadrupole time-of-flight (QTOF) system using a CaptiveSpray ion source (Bruker Daltonics) at a flow rate of 3 µL/min, ionized with 4.5 kV, 0.3 bar nebulizer gas pressure, heated to 180 °C and analyzed in positive ion mode. MS1 spectra were acquired in the mass range of 110–5000 m/z at a scan rate of 1 Hz. Following acquisition, a composite MS1 spectrum was generated by summing all MS1 spectra during acquisition time using Compass DataAnalysis v4.3 (Bruker Daltonics). Spectra were deconvoluted by mMass and used to determine the mass of peptide and protein ions. Default mMass baseline correction and smoothing parameters were used.
In silico structural analysis
Based on the crystal structures of the IFN-γ dimer (pdb entry 1HIG51 [10.2210/pdb1HIG/pdb]) and IFN-γ receptor complexes (1FG9 [10.2210/pdb1FG9/pdb]32 and 1FYH [10.2210/pdb1FYH/pdb]33) we modeled a secondary complex consisting of (i) the IFNG dimer (UniProt P01579; Gln24-Leu158), (ii) two high-affinity IFNG receptor 1 molecules (IFNGR1; P15260; Val29-Ser241), and (iii) two low affinity IFNG receptor 2 chains (IFNGR2; P38484; Leu30-Thr237). Missing loops were resolved using ModLoop52 and truncated side chains were rebuilt using Dunbrack’s backbone-dependent rotamer library53 within Chimera (15264254). The C-terminal IFNG sequence (Pro145-Leu158) was based on published biochemical information34, 54 and our data in this manuscript using the Rosetta FlexPepDock web server55 and Modeller56 within Chimera57, and manually evaluated. Molecular graphics figures were made using the molecular visualization system PyMOL (The PyMOL Molecular Graphics System, Version 1.7.1.3, Schrödinger, LLC)58.
Mice
MMP12–deficient mice on the C57BL/6 × 129Sv/Ev background were generated as described by Shipley et al.22 with Mmp12 exon 2-disrupted embryonic stem cells injected into C57BL/6J blastocysts. After the chimeras were crossed to C57BL/6J mice they were backcrossed for 10 generations. As the C57BL/6J strain is considered a low inflammation strain in these disease models, we then backcrossed these homozygous mice for 8 generations onto the MRL/MpJ-Faslpr/J (MRL/lpr) strain (catalog no. substrain 000485; The Jackson Laboratory). Thereafter, the Mmp12+/+ MRL/lpr and Mmp12–/– MRLlpr male and female mice, housed in ventilated racks, were maintained as homozygous lines, leading to an autoimmune disease mouse model after ~90 days that can be synchronized between mice upon injection of CFA for severe arthritic and kidney disease development41 before gross lymphadenopathy reached humane end points as determined by the UBC Animal Care Committee.
To analyze collagen-induced arthritis, we backcrossed Mmp12–/– mice on the 129Sv/Ev background for seven generations onto the B10.RIII-H2r H2-T18b background (The Jackson Laboratory, stock #00045723). Genotyping was performed by the HotShot DNA isolation followed by polymerase chain reaction with Mmp12 primers (forward) and (reverse) and a primer specific for the neomycin insert (reverse). The PCR products were 1100 bp for Mmp12+/+ mice and 550 bp for Mmp12–/– mice. The UBC Animal Care Committee approved all mouse breeding and animal experimental procedures and, established and supervised humane end points.
Thioglycollate-induced sterile peritonitis
At day 90, male and female mice were injected intraperitoneal with 0.5 mL 4% Brewers thioglycollate broth (Sigma-Aldrich, St. Louis, MO) to establish acute sterile peritonitis. Mice were excluded from further studies if obvious adhesions were observed between the peritoneum and skin or if marked increases were observed in the thickness of the peritoneum. Mice were killed at day 0, 1, 2, 3, or 4 and 5 mL of phosphate buffered saline (PBS) was administered via intraperitoneal injection to collect infiltrating cells and peritoneal exudate. Cells were plated for 4–18 h in RPMI 1640 medium, 10% cosmic calf serum and non-essential amino acids. For in vivo MMP12 inhibition, we administered 5 mg/kg Rxp470.1 intraperitoneal once per day for 4 days in Mmp12+/+ B10.RIII (n = 4) and Mmp12–/– B10.RIII (n = 4) mice and collected the peritonitis fluid and cells by PBS lavage on day 4.
Cells
Human THP-1 monocytic cells (ATCC, Manassas, Virginia, USA), mouse RAW264.7 macrophages (ATCC, Manassas, Virginia, USA) and primary mouse peritoneal macrophages were cultured in 10% fetal bovine serum (v/v) in RPMI. All cell lines were tested and found to be mycoplasma negative.
Macrophage activation
Human THP-1 cells (5 × 106 cells in 10 mL) were treated with phorbol 12-myristate 13-acetate (PMA) (100 ng/mL) for 24 h to induce cell attachment and differentiation. THP-1 cells and mouse RAW264.7 cells were activated with 20 ng/mL of IFN-γ (human #300-02, PeproTech, or mouse #315-05, Rocky Hill, NJ, respectively), or 30 ng/mL IL-4 (human #200-04 PeproTech, or mouse #214-14, Rocky Hill, NJ, respectively).
Quantification of intracellular radical oxygen species
Cellular ROS levels were determined by incubating 1 × 105 cells with 5 μg/mL 2,-7-dichlorofluorescein diacetate (DCF-DA, Sigma-Aldrich) for 15 min at 37 °C (n = 4, N = 2). Cells were washed twice with Hank’s complete balanced saline solution (HBSS) and harvested after trypsinization, and the fluorescence (excitation 488 nm, emission 525 nm) was measured using a POLARstar optima (BMG Labtech, Durham, NC). Statistical analysis was determined by a two-tailed unpaired Student’s t-test.
Phagocytosis assay
THP-1 cells (5 × 106 cells in 10 mL) were stimulated with 200 ng/mL PMA for 24 h to induce cell attachment and differentiation and then the medium was changed. A 2.69% Fluoresbrite® 2-μm microparticle suspension (12 μL) (Polysciences, Inc., Warrington, PA) was incubated in 175 μL normal human serum (Complement Technology Inc., Tyler, TX) for 30 min at 37 °C before addition to the PMA-activated cells for 1 h. Phagocytosis was stopped by adding 2 mL of ice-cold HBSS. The bead solution was removed, and the cells were washed twice with HBSS, then harvested by trypsinization and washed. The phagocytosis index was determined in each experiment (n = 3 repeats per experiment, and the experiment itself performed twice, N = 2) by counting the number of beads per cell in 20 separate fields containing 5–30 cells/image for each repeat. Statistical analysis was determined by a two-tailed unpaired Student’s t-test.
Antibodies
Anti-Mouse CD11c FITC (clone N418) was from BioLegend and used at a concentration of 1 μg/mL × 106 cells. Polyclonal rabbit anti-STAT1 (#9172), pSTAT1-Y701 (#9167), STAT6 (#9362), and pSTAT6-Y641 (#9361) were used at a dilution of 1/1000 and were from Cell Signaling. Polyclonal rabbit anti-iNOS (ab15323), anti-IFN-γ (EPR1108, ab133566), anti-MHC Class II (ab55152), and anti-CD36 (ab137320) were used at a dilution of 1/1000 and were from Abcam. Polyclonal goat anti-S100A8 (AF3059) and anti-S100A9 (AF2065) were used at a dilution of 1/1000 and were from R&D Systems. Polyclonal rabbit anti-MMP12 catalytic domain antibody and anti-hinge domain antibody (Triple Point Biologics) were used at a dilution of 1/1000. Anti-mouse myeloperoxidase (MPO) from Hycult (HM1051) were used at a dilution of 1/100 on tissue sections.
Raising and affinity purification of IFN-γ epitope antibodies
Three IFN-γ peptides (GenScript, Piscataway, NJ): DPYVKEAENLKKYFNAG-GC; C-GGLFRG, and CRGG-QVMA (Arg was added for solubility), all synthesized with either a G or GG added as a flexible spacer to a cysteine to enable conjugation to succinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate/Keyhole Limpet Hemocyanin (Sigma-Aldrich, St-Louis, MO). The conjugates were emulsified in CFA and injected in New Zealand white rabbits. Antibody expression was boosted with peptide in incomplete Freund’s adjuvant at 3 weeks. Serum antibody titers to the respective peptides were quantified by ELISA. SulfoLink Immobilization (Pierce Biotechnology, Rockford, IL) was used to conjugate the peptides to affinity-purify antibodies from rabbit sera. Peptides (5 mg) were dissolved in 2 mL of coupling buffer (50 mM Tris, 5 mM EDTA, pH 8.5), reduced with 25 mM TCEP, and incubated for 30 min. The sulfhydryl-containing peptides were coupled to 3 mL SulfoLink columns by mixing with the SulfoLink resin for 15 min and incubated for 2 h. Non-specific binding sites were blocked with 50 mM cysteine for 1 h. The columns were then equilibrated with 50 mM Tris, 5 mM EDTA, pH 8.5 before affinity purification of anti-peptide antibodies from rabbit sera. After column washes, high-affinity antibodies were eluted using 0.1 M glycine, pH 2.5. Antibody specificity was verified by ELISA and western blotting to IFN-γ and MMP12-cleaved IFN-γ. Western blotting of human kidney extracts was used to assess specificity and non-specific binding before translation to ELISA assays.
ELISA
Plates were coated with 5 μg/mL of each of the 3 IFN-γ peptides in Vollers buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) overnight and then blocked with 1% BSA in PBS for 1 h. The wells were rinsed three times with PBS-0.05% Tween 20 and 100 μL of a serial dilution of antibodies (1:102–1:109) were added and incubated at room temperature for 2 h. The wells were rinsed with PBS-Tween 20 (0.05%) before 100 μL of 1:1000 goat anti-rabbit HRP antibody (Bio-Rad Laboratories, Hercules, CA) was added and incubated for 1 h. Wells were aspirated and incubated in the dark with SIGMAFAST™ OPD HRP substrate (Sigma-Aldrich, St-Louis, MO) until visible coloration was seen. At this time 50 μL of 1 M acetic acid was added to stop the reaction and the absorbance was measured at 450 nm on a POLARstar optima microplate reader (BMG Labtech, Durham, NC).
Collagen-induced arthritis in B10.RIII mice
Collagen-induced arthritis was induced by injection with type II collagen (4 mg/mL) at two inguinal sites in 49-day-old-male Mmp12–/– (n = 20) and Mmp12+/+ mice (n = 18) all on the B10.RIII background. Day 0 was defined as 21 days after injection and was typically the date of onset of arthritis. At 18 and 28 days later, the mice were killed and the hind ankles were dissected and prepared for immunostaining and histological staining by hematoxylin and eosin (H&E) and toluidine blue to visualize cartilage proteoglycan23. NETs were previously characterized in collagen-induced arthritis by a variety of stains, including hematoxylin and eosin in combination23 or alone59, 60, the histological appearance and distribution of which are consistent with the NETs identified here by hematoxylin and eosin.
Arthritis in MRL/lpr mice
Spontaneous arthritis and lupus nephritis are a characteristic of the MRL/lpr strain. To ensure reasonably severe and consistent disease stage between animals before excessive lymphadenopathy necessitated animal euthanasia it was necessary to synchronize the initiation of systemic autoimmunity by CFA earlier than spontaneous disease was manifest. We injected 0.05 mL of a 1:1 emulsion of CFA (Sigma) in water intradermal at two inguinal sites in 90-day-old mice41. The width of both hind ankles was measured on three consecutive days before injection and every 2–3 days for 25 days following injection using electronic digital calipers (Marathon). Baseline ankle width was calculated as the average of the pre-injection measurements. Arthritis was deemed present with a 10% increase in average ankle width. Spleen weights were determined of female Mmp12–/– MRL/lpr (n = 22) and Mmp12+/+ MRL/lpr (n = 27) mice with or without CFA injection. Statistical analysis was performed with Prism 5 (GraphPad Software) using two-way analysis of variance with Bonferroni post-tests.
Immunostaining of biopsy sections
Mouse ankle, lymph node and kidney, and human kidney biopsies were dissected and formalin-fixed for 16 h. Hind ankles were decalcified in 10% formic acid for 72 h. Biopsies were then dehydrated through graded ethanol series and paraffin-embedded. Tissue sections (3–5 μm) were deparaffinized, rehydrated through an ethanol series ending in water and stained with hematoxylin and eosin, toluidine blue, trichrome, Jones, or Periodic acid-Schiff (PAS). For immunostaining, tissue sections were rehydrated, quenched to remove endogenous peroxidase, blocked in 1/10 normal goat serum for 1 h and then incubated with antibody in Tris-buffered saline. Primary antibodies and species appropriate secondary antibodies were used as performed by Wax-it Histology Services (Vancouver, BC, Canada).
Mouse glomerulonephritis activity and chronicity index scoring
The means of the activity and chronicity scores from Mmp12+/+ MRL/lpr (n = 11) and Mmp12–/– MRL/lpr (n =15) mice were calculated, for each biopsy, by analysis of 10 components divided into two scoring systems: Renal activity index (1—cellular proliferation, 2—leukocyte infiltration, 3—fibrinoid necrosis or karyorrhexis, 4—cellular crescents, 5—hyaline thrombi, wire loops and 6—mononuclear cell infiltration), and Renal chronicity index (7—glomerular sclerosis, 8—fibrous crescents, 9—interstitial fibrosis, 10—tubular atrophy)61, 62. Each individual component was scored 0 (for normal), 1, 2, or 3 (for severe abnormality, 3 being the worse). In calculating the activity index, fibrinoid necrosis and cellular crescents were weighed by a factor of 2. The renal activity index scale ranges from 0 to 24 points and the renal chronicity index scale from 0 to 12.
For immunostaining analyses of mouse kidney glomeruli, and glomeruli plus the interstitium a minimum of 10 different fields, with each field having a minimum of 5 glomeruli were examined and quantified for each antibody, for each of these two separate analyses (N = 2). Three mice per group (Mmp12+/+ (n = 3) and Mmp12–/– MRL/lpr (n = 3)) were analyzed separately for the glomeruli and the glomeruli and interstitium staining quantifications.
Mouse whole-blood leukocyte and monocyte counts
Whole blood was collected from mice in EDTA-collection tubes and analyzed on a Siemens Advia 120 Hematology System (Oakville, ON, Canada) with the Perox method. The whole blood of each mouse (Mmp12+/+ (n = 15) and Mmp12–/– (n = 27) MRL/lpr) was analyzed in technical duplicates.
MMP12 mRNA analyses
Relative expression levels of MMP12 mRNA was measured in PMA-matured THP-1 cells treated with 20 ng/mL IFN-γ, 30 ng/mL IL-4, or PBS for 24 h (n = 3, N = 2 for each condition). After isolating RNA using the RNeasy Mini system (Qiagen), total RNA (1 μg) was amplified (Ambion MessageAmp II amplification kit, Thermo Scientific) and labeled by ULS aRNA fluorescent labeling (Kreatech Biotechnology, Amsterdam, The Netherlands). ULS-Cy3 labeled RNA was mixed 1:1 with ULS-Cy5 labeled control RNA (equimix of all amplified RNA samples) and hybridized to the CLIP-CHIPTM human protease and inhibitor targeted microarray63 for 18 h, 42 °C in duplicate with three biological replicates (i.e., six data points/gene). The microarray slides were scanned (428 ArrayScanner, MWG Biotech, High Point, NC, USA) and analyzed using ImaGene 6.1 (BioDiscovery, El Segundo, CA, USA). Data were normalized using the Bioconductor-based CARMAWeb software package44 and statistically analyzed and visualized with the MulitExperiment viewer 4.8.1 from TIGR (TM4.org) using the significance analysis of microarrays algorithm as described45, 63.
RNA expression relationships in human systemic lupus erythematosus
We searched three major databases (19/07/2017) GEO (26,622 human array or sequencing-based RNA datasets)64, ArrayExpress (70,672 datasets of 2,242,940 assays)65, and Gemma66 (4118 datasets)) data series relating to human lupus transcript analyses and individually examined more than 239 datasets. Although there are 152 data series for human lupus in GEO and 130 in ArrayExpress, only five in total had longitudinal data, which were downloaded from Gemma. We compared RNA expression levels of MMP12 and IFNG genes in untreated SLE patients vs. healthy subjects in the GSE11909 dataset; and of untreated healthy reference subjects vs. treated SLE patients in the GSE37356 dataset. For each gene, microarray probes with the lowest p-value were selected and the multiple testing FDR-corrected q-values calculated by a linear modeling approach and taken directly from Gemma. All plots and analyses were done in R.
Human lupus nephritis analyses and grade scores
Five kidney biopsies from non-SLE patients with mild to moderate arteriosclerosis and healthy, non-lupus, non-inflammatory kidney disease, and kidney samples from SLE patients were classified independently by one blinded pathologist and one nephrologist: Class I, minimal mesangial lupus nephritis; Class II, mesangial proliferative lupus nephritis; Class III, focal lupus nephritis; Class IV, diffuse segmental (IV-S) or global (IV-G) lupus nephritis; Class V, membranous lupus nephritis; Class VI, advanced sclerosing lupus nephritis. Healthy (n = 5) and active (A) (n = 5) SLE Grade III-(A), IV-Global-(A), and IV-Segmental-(A) kidney biopsies were selected for immunochemical analysis.
Glomeruli were examined and quantified for each antibody from all human biopsies of the healthy kidneys (n = 43 glomeruli) and SLE patient kidneys (n = 46 glomeruli) and scored according to these classes: Class I, normal glomeruli by light microscopy; Class II, mesangial hypercellularity or mesangial matrix expansion by light microscopy with immune deposits; Class III, active or inactive focal, segmental or global endocapillary or extracapillary glomerulonephritis involving <50% of all glomeruli; Class IV, active or inactive focal, segmental or global endocapillary or extracapillary glomerulonephritis involving >50% of all glomeruli; Class V, global or segmental subepithelial immune deposits or their morphologic sequelae by light microscopy; and Class VI, >90% of glomeruli sclerosed.
Statistical analyses
A normal distribution of the data were shown in all cases and hence only parametric tests were required. Standard deviation and analysis of variance were used to analyze data variability. The Student’s t-test determined statistical significance between two treatments. A p-value <0.05 was considered statistically significant.
Human ethics approvals
The UBC/Providence Health Care British Columbia Research Ethics Board approved the patient consenting procedures and research protocols for studies using human samples (approval #H15-00953), informed consent was obtained from all patients and the research was conducted in accordance with the Declaration of Helsinki.
Data availability
All data are available from the authors upon reasonable request.
Electronic supplementary material
Acknowledgements
We thank Dr. Georgina Butler for editorial help and Dr. Gabriela Cohen-Freue, Department of Statistics UBC for statistical evaluation and advice. We thank Drs. Steven Shapiro, Harvard School of Medicine, Boston, MA and V. Wee Yong, University of Calgary, Calgary, AB for Mmp12–/– mice. C.M.O. holds a Canada Research Chair in Protease Proteomics and Systems Biology. Access to imaging facilities was provided by Dr. Edward Conway, Center for Blood Research and the UBC Life Sciences Institute Imaging Facility. This work was supported by the Canadian Institutes of Health Research grants (MOP-37937 to C.M.O.) and by the British Columbia Proteomics Network (C.M.O.).
Author contributions
A.D. conceived, performed, and planned most experiments, analyzed the data and wrote the paper. N.F. performed data mining analysis of SLE patients in Fig. 1. P.P. designed and supervised the data mining analysis of SLE patients in Fig. 1. U.E. performed the in silico structure analysis in Fig. 2, and data analysis of the MALDI-TOF experiments. N.S. and T.K. contributed to the MALDI-TOF experiments. N.S. performed the top down MS analyses locating the MMP12 cleavage sites. N.S. is a Canadian Michael Smith Foundation of Health Research trainee and an Australian CJ Martin Early Career Fellow. J.M. contributed to Figs. 3 and 4. C.L.B. crossed the Mmp12+/+ and Mmp12–/– mice on the B10.RIII and MRL/lpr backgrounds and performed the CFA-induced arthritis experiments in Fig. 5. R.K. managed mouse colonies, performed mRNA measurements, contributed with the arthritis models in Fig. 5 and provided laboratory management for the entire project. P.J. contributed to the SLE patient histology analysis. J.R. interpreted and provided advice on the clinical results. V.D. designed and synthesized Rxp470.1. S.J.B. selected the SLE patients and supervised both the mouse and human kidneys histological analyses in Figs. 6 and 7. C.M.O. conceived the project, the in vivo and in vitro experiments, wrote and edited the paper, analyzed data, supervised, and enthusiastically provided support for the project.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-04717-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroder K, Hetzog JP, Ravasi T, Hume AD. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 4.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchant DJ, et al. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat. Med. 2014;20:493–502. doi: 10.1038/nm.3508. [DOI] [PubMed] [Google Scholar]
- 7.Pham CTN. Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 8.Dufour A, Overall CM. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol. Sci. 2013;34:233–242. doi: 10.1016/j.tips.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K, et al. HIV-induced metalloproteinase processing of the chemokine stromal cell derived factor-1 causes neurodegeneration. Nat. Neurosci. 2003;6:1064–1071. doi: 10.1038/nn1127. [DOI] [PubMed] [Google Scholar]
- 10.Berahovich RD, et al. Proteolytic activation of alternative CCR1 ligands in inflammation. J. Immunol. 2005;174:7341–7351. doi: 10.4049/jimmunol.174.11.7341. [DOI] [PubMed] [Google Scholar]
- 11.Fortelny N, et al. Network analyses reveal pervasive functional regulation between proteases in the human protease web. PLoS Biol. 2014;12:e1001869. doi: 10.1371/journal.pbio.1001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McQuibban GA, et al. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 13.McQuibban GA, et al. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood. 2002;100:1160–1167. [PubMed] [Google Scholar]
- 14.Vergote D, et al. Proteolytic processing of SDF-1alpha reveals a change in receptor specificity mediating HIV-associated neurodegeneration. Proc. Natl Acad. Sci. USA. 2006;103:19182–19187. doi: 10.1073/pnas.0604678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korman BD, et al. Inflammatory expression profiles in monocyte-to-macrophage differentiation in patients with systemic lupus erythematosus and relationship with atherosclerosis. Arthritis Res. Ther. 2014;16:R147. doi: 10.1186/ar4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller CHT, Maher SG, Young HA. Clinical Use of Interferon-gamma. Ann. N. Y. Acad. Sci. 2009;1182:69–79. doi: 10.1111/j.1749-6632.2009.05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skurkovich S, Skurkovich B, Kelly J. Anticytokine therapy, particularly anti-IFN-gamma, in Th1-mediated autoimmune diseases. Expert Rev. Clin. Immunol. 2005;1:11–25. doi: 10.1586/1744666X.1.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Martinez J, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Goncalves DaSilva A, Yong VW. Matrix metalloproteinase-12 deficiency worsens relapsing-remitting experimental autoimmune encephalomyelitis in association with cytokine and chemokine dysregulation. Am. J. Pathol. 2009;174:898–909. doi: 10.2353/ajpath.2009.080952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver A, et al. An elevated matrix metalloproteinase (MMP) in an animal model of multiple sclerosis is protective by affecting Th1/Th2 polarization. FASEB J. 2005;19:1668–1670. doi: 10.1096/fj.04-2030fje. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro SD, Kobayashi DK, Ley TJ. Cloning and characterization of a unique elastolytic metalloproteinase produced by human alveolar macrophages. J. Biol. Chem. 1993;268:23824–23829. [PubMed] [Google Scholar]
- 22.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc. Natl Acad. Sci. USA. 1996;93:3942–3946. doi: 10.1073/pnas.93.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellac CL, et al. Macrophage matrix metalloproteinase-12 dampens inflammation and neutrophil influx in arthritis. Cell Rep. 2014;9:618–632. doi: 10.1016/j.celrep.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Gronski TJ, et al. Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J. Biol. Chem. 1997;272:12189–12194. doi: 10.1074/jbc.272.18.12189. [DOI] [PubMed] [Google Scholar]
- 25.Madala SK, et al. Matrix metalloproteinase 12-deficiency augments extracellular matrix degrading metalloproteinases and attenuates IL-13-dependent fibrosis. J. Immunol. 2010;184:3955–3963. doi: 10.4049/jimmunol.0903008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houghton AMA, Hartzell WOW, Robbins CSC, Gomis-Rüth FXF, Shapiro SDS. Macrophage elastase kills bacteria within murine macrophages. Nature. 2009;460:637–641. doi: 10.1038/nature08181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dean RA, et al. Macrophage-specific metalloelastase (MMP-12) truncates and inactivates ELR+ CXC chemokines and generates CCL2, -7, -8, and -13 antagonists: potential role of the macrophage in terminating polymorphonuclear leukocyte influx. Blood. 2008;112:3455–3464. doi: 10.1182/blood-2007-12-129080. [DOI] [PubMed] [Google Scholar]
- 28.Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JL, et al. A selective matrix metalloproteinase-12 inhibitor retards atherosclerotic plaque development in apolipoprotein E-knockout mice. Arterioscler. Thromb. Vasc. Biol. 2011;31:528–535. doi: 10.1161/ATVBAHA.110.219147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devel L, et al. Insights from selective non-phosphinic inhibitors of MMP-12 tailored to fit with an S1’ loop canonical conformation. J. Biol. Chem. 2010;285:35900–35909. doi: 10.1074/jbc.M110.139634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthys P, Vermeire K, Heremans H, Billiau A. The protective effect of IFN-gamma in experimental autoimmune diseases: a central role of mycobacterial adjuvant-induced myelopoiesis. J. Leukoc. Biol. 2000;68:447–454. [PubMed] [Google Scholar]
- 32.Thiel DJ, et al. Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex. Structure. 2000;8:927–936. doi: 10.1016/S0969-2126(00)00184-2. [DOI] [PubMed] [Google Scholar]
- 33.Randal M, Kossiakoff AA. The structure and activity of a monomeric interferon-gamma:alpha-chain receptor signaling complex. Structure. 2001;9:155–163. doi: 10.1016/S0969-2126(01)00567-6. [DOI] [PubMed] [Google Scholar]
- 34.Lundell D, et al. The carboxyl-terminal region of human interferon gamma is important for biological activity: mutagenic and nmR analysis. Protein Eng. 1991;4:335–341. doi: 10.1093/protein/4.3.335. [DOI] [PubMed] [Google Scholar]
- 35.Leinikki PO, Calderon J, Luquette MH, Schreiber RD. Reduced receptor binding by a human interferon-gamma fragment lacking 11 carboxyl-terminal amino acids. J. Immunol. 1987;139:3360–3366. [PubMed] [Google Scholar]
- 36.Karonitsch T, et al. Activation of the interferon-gamma signaling pathway in systemic lupus erythematosus peripheral blood mononuclear cells. Arthritis Rheum. 2009;60:1463–1471. doi: 10.1002/art.24449. [DOI] [PubMed] [Google Scholar]
- 37.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol. Res. 2011;50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat. Protoc. 2007;2:1269–1275. doi: 10.1038/nprot.2007.173. [DOI] [PubMed] [Google Scholar]
- 39.Balomenos D, Rumold R, Theofilopoulos AN. Interferon-gamma is required for lupus-like disease and lymphoaccumulation in MRL-lpr mice. J. Clin. Invest. 1998;101:364–371. doi: 10.1172/JCI750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billiau A, Matthys P. Modes of action of Freund’s adjuvants in experimental models of autoimmune diseases. J. Leukoc. Biol. 2001;70:849–860. [PubMed] [Google Scholar]
- 41.Ratkay LG, Tait B, Tonzetich J, Waterfield JD. Lpr and MRL background gene involvement in the control of adjuvant enhanced arthritis in MRL-lpr mice. J. Autoimmun. 1994;7:561–573. doi: 10.1006/jaut.1994.1041. [DOI] [PubMed] [Google Scholar]
- 42.Kim K, et al. Interferon-gamma gene polymorphisms associated with susceptibility to systemic lupus erythematosus. Ann. Rheum. Dis. 2010;69:1247–1250. doi: 10.1136/ard.2009.117572. [DOI] [PubMed] [Google Scholar]
- 43.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front. Neuroendocrinol. 2014;35:347–369. doi: 10.1016/j.yfrne.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard KM, et al. Definition of IFN-γ-related pathways critical for chemically-induced systemic autoimmunity. J. Autoimmun. 2012;39:323–331. doi: 10.1016/j.jaut.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vira H, et al. Role of MMP-7 in the pathogenesis of systemic lupus erythematosus (SLE) Lupus. 2017;26:937–943. doi: 10.1177/0961203316682855. [DOI] [PubMed] [Google Scholar]
- 48.Keyszer G, et al. Circulating levels of matrix metalloproteinases MMP-3 and MMP-1, tissue inhibitor of metalloproteinases 1 (TIMP-1), and MMP-1/TIMP-1 complex in rheumatic disease. Correlation with clinical activity of rheumatoid arthritis versus other surrogate markers. J. Rheumatol. 1999;26:251–258. [PubMed] [Google Scholar]
- 49.Butler GS, Tam EM, Overall CM. The canonical methionine 392 of matrix metalloproteinase 2 (gelatinase A) is not required for catalytic efficiency or structural integrity: probing the role of the methionine-turn in the metzincin metalloprotease superfamily. J. Biol. Chem. 2004;279:15615–15620. doi: 10.1074/jbc.M312727200. [DOI] [PubMed] [Google Scholar]
- 50.Starr AE, Overall CM. Chapter 13. Characterizing proteolytic processing of chemokines by mass spectrometry, biochemistry, neo-epitope antibodies and functional assays. Methods Enzymol. 2009;461:281–307. doi: 10.1016/S0076-6879(09)05413-5. [DOI] [PubMed] [Google Scholar]
- 51.Ealick SE, et al. Three-dimensional structure of recombinant human interferon-gamma. Science. 1991;252:698–702. doi: 10.1126/science.1902591. [DOI] [PubMed] [Google Scholar]
- 52.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 53.Dunbrack RL. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002;12:431–440. doi: 10.1016/S0959-440X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 54.Walter MR, et al. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 55.London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server—high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39:W249–W253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformat. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.DeLano WL. The case for open-source software in drug discovery. Drug Discov. Today. 2005;10:213–217. doi: 10.1016/S1359-6446(04)03363-X. [DOI] [PubMed] [Google Scholar]
- 59.Shiogama K, et al. Visualization of neutrophil extracellular traps and fibrin meshwork in human fibrinopurulent inflammatory lesions: I. Light Microscopic Study. Acta Histochem. Cytochem. 2016;49:109–116. doi: 10.1267/ahc.16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narasaraju T, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011;179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wernick RM, et al. Reliability of histologic scoring for lupus nephritis: a community-based evaluation. Ann. Intern. Med. 1993;119:805–811. doi: 10.7326/0003-4819-119-8-199310150-00006. [DOI] [PubMed] [Google Scholar]
- 62.Renner K, et al. IL-3 contributes to development of lupus nephritis in MRL/lpr mice. Kidney Int. 2015;88:1088–1098. doi: 10.1038/ki.2015.196. [DOI] [PubMed] [Google Scholar]
- 63.Kappelhoff R, et al. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the CLIP-CHIP™ DNA microarray. Biochim. Biophys. Acta. 2017;1864:2210–2219. doi: 10.1016/j.bbamcr.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 64.Barrett T, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolesnikov N, et al. ArrayExpress update—simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoubarev A, et al. Gemma: a resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics. 2012;28:2272–2273. doi: 10.1093/bioinformatics/bts430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the authors upon reasonable request.