Abstract
Short-term exposures at critical stages of development can lead to delayed adverse effects long after the initial stressor has been removed, a concept referred to as developmental origin of adult disease. This indicates that organisms’ phenotypes may epigenetically reflect their past exposure history as well as reflecting chemicals currently present in their environment. This concept has significant implications for environmental monitoring. However, there is as yet little or no implementation of epigenetics in environmental risk assessment. In a proof-of-principle study we exposed Daphnia magna to 5-azacytidine, a known DNA de-methylating agent. Exposures covered combinations of prenatal and postnatal exposures as well as different exposure durations and recovery stages. Growth, the transcription of genes and levels of metabolites involved in regulating DNA methylation, and methylation levels of several genes were measured. Our data shows that prenatal exposures caused significant changes in the methylome of target genes, indicating that prenatal stages of Daphnia are also susceptible to same level of change as post-natal stages of Daphnia. While the combination of pre- and postnatal exposures caused the most extreme reduction in DNA methylation compared to the control group. Furthermore, some of the changes in the methylation patterns were persistent even after the initial stressor was removed. Our results suggest that epigenetic biomarkers have the potential to be used as indicators of past chemical exposure history of organisms and provide strong support for implementing changes to the current regimes for chemical risk assessment to mimic realistic environmental scenarios.
Keywords: DNA methylation, Daphnia, Epigenetics, Prenatal exposure, Postnatal exposure, 5-azacytidine
Introduction
Although improving, ecotoxicology is still frequently focused on the relatively short-term effects of stressors on ecological or environmental health and the majority of the proposed Adverse Outcome Pathways (AOPs) do not consider long-term impacts of a stressor on the health of an individual or population (Morgan et al. 2007). There is now an increasing body of evidence to support that short-term exposures at critical stages of an individual’s development can lead to delayed adverse effects later in life, long after the initial stressor has been removed, a concept referred to as developmental origin of adult disease (Dolinoy and Jirtle 2008; Thornburg et al. 2010; Groh et al. 2015; Bhandari 2016; Bailey 2015). Furthermore, in the environment, individuals are generally exposed to relatively low levels of multiple stressors across multiple generations. Thus, the phenotypes of a population sampled from the environment may reflect its multi-generation exposure history (Groh et al. 2015; Mirbahai and Chipman 2014; Morgan et al. 2007).
As indicated by us and others, it is thought that many of these effects are partly mediated through disruption of epigenetic mechanisms (Aniagu et al. 2008; Mirbahai et al. 2011, 2013; Pegoraro et al. 2016; Rasmussen and Amdam 2015; Williams et al. 2014). Epigenetics is the study of heritable changes in gene expression that do not involve changes to the underlying DNA sequence, resulting in a change in phenotype without a change in genotype (Skinner et al. 2010; Kanwal and Gupta 2012). One of the most commonly studied epigenetic modifications is DNA methylation. The primary methyl donor for DNA methylation is S-adenosylmethionine (SAM), a metabolite generated in the cyclical cellular process called one-carbon metabolism. One-carbon metabolism is catalysed by several enzymes in the presence of dietary micronutrients, including folate, choline, betaine and other B vitamins (Locasale 2013; Lu 2000). Contaminants, such as metals (arsenic and cadmium), persistent organic pollutants or endocrine disrupting chemicals (Jaenisch and Bird 2003; Vandegehuchte and Janssen 2014) can cause changes in the methylome by interfering with the regulation of DNA methylation (e.g. one-carbon cycle) or DNA methylation machinery (e.g. DNA methyltransferases). Dysregulation of the epigenome can have significant ecological implications (Vandegehuchte and Janssen 2014; Bhandari 2016), including impacting the fitness of a range of environmentally relevant species such as Daphnia spp. (Asselman et al. 2016a, 2016b; Lyko et al. 2010; Vandegehuchte et al. 2009a, 2009b, 2010a, 2010b).
Despite such evidence, epigenetics research is limited in ecotoxicology and is not incorporated into standardised chemical risk assessment guidelines (Shaw et al. 2017). Currently, our understanding of the long-term effects of accumulative epigenetic changes on fitness and health of aquatic species is extremely limited. To improve our understanding of epigenetics in toxicology, a greatly improved understanding of the contributions of epigenetic mechanisms in regulating the responses of species to chemicals is needed. This would help to address concerns about possible adverse long-term health effects related to epigenetic changes (Marczylo et al. 2016).
Daphnia spp. are considered a keystone and indicator species in both lakes and ponds and are well-studied in terms of their ecology and response to stressors, both under laboratory conditions and in the field (Lampert 2011; OECD 2004, 2012). Furthermore, over the past few years substantial efforts have been made to not only characterise the methylome of Daphnia but to also determine its sensitivity to various environmental stressors (Asselman et al. 2016a, 2016b; Vandegehuchte et al. 2009b). Daphnia offer a variety of benefits as a model organism for epigenetic research. Daphnia’s phenotypic plasticity and eco-responsive genome coupled to a parthenogenetic life cycle allows the study of epigenetic effects in the absence of confounding genetic differences (Harris et al. 2012; Bonasio 2015). Therefore, due to their extensive use in ecotoxicology testing and their ecological importance as well as their advantages as a model organism for epigenetics research, we have used Daphnia magna as the test species in this proof-of-principle study. The standard DNA demethylating agent, 5-azacytidine was used as a stressor due to its known mechanisms of action and its potential transgenerational impact in Daphnia (Vandegehuchte et al. 2010a). 5-azacytidine is an analogue for cytosine which can be incorporated into the DNA during replication causing a cumulative effect on the DNA methylation levels at specific CpG sites (Christman 2002; Tobiasson et al. 2017). We aimed to determine whether alterations in the methylome of specific target genes involved in DNA methylation mechanisms are dependent on the life stage of exposure. In addition, we investigated whether changes in the D. magna methylome can persist after the removal of the initial stressor with DNA methylation modifying properties.
Methods
Daphnia magna culturing and experimental design
Cultures of D. magna Bham2 strain were maintained as previously described (Athanasio et al. 2016). D. magna Bham2 strain has been maintained in the laboratory conditions for over 10 years. Briefly, D. magna Bham2 strain were maintained in photoperiodic lighting (16 h of light: 8 h of dark) and temperature of 20 ± 1 °C, in high hardness COMBO medium (HH COMBO). Animals were fed every other day with Chlorella vulgaris at a concentration of ≈27,550 cells of algae per individual Daphnia. Two different exposure designs (Fig. 1) were used to investigate two hypotheses: A) exposure during different life stages will potentially induce different magnitude of DNA methylation responses (experiment setup I), and B) some DNA methylation changes can be retained after the removal of a stressor (experiment setup II). In both experimental setups (I and II) Daphnia magna Bham2 strain were maintained in either clean culture media (control group) or media containing 5-azacytidine (exposure group).
Fig. 1.
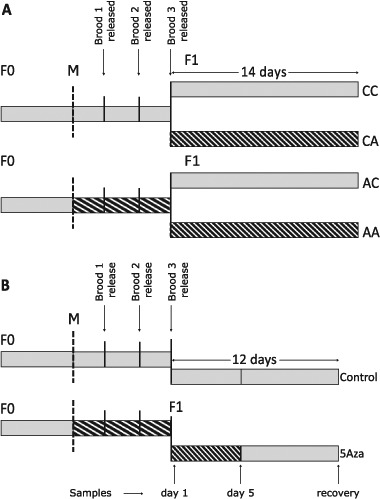
Schematic representation of Daphnia magna Bham 2 exposure to 5-azacytidine. The aim of the two exposure designs were to determine: (a) if exposure to 5-azacytidine (5-Aza) during different life stages induces different levels of DNA methylation responses, and (b) if some of the DNA methylation changes induced as a result of exposure to 5-azacytidine are retained after the removal of the chemical. F0 generation Daphnia were exposed when they reached maturation age (maturation point is shown with M and dashed lines). The first and second broods (F1 generations) were discarded. The points of release of first and second broods are shown with solid lines. The third brood (labelled F1) was transferred to either an exposure beaker (patterned bar) or to control media (solid gray bar). a The exposure continued postnatally for an additional 14 days. This exposure setup resulted in generation of four unique exposure groups: Daphnia exposed to the stressor prenatally (AC), Daphnia exposed to the stressor postnatally (CA), Daphnia exposed to the stressor pre- and postnatally (AA), and non-exposed (CC) Daphnia (C refers to control while A refers to 5-azacytidine exposure). b The exposures continued for an additional 5 days after the release of neonates from the brood chamber of the mother. Animals were sampled at day 1 and day 5 of exposure. The exposed F1 generations were then transferred to clean media and maintained for an additional 7 days (recovery period). Age matched control samples were generated following the same procedure without the addition of 5-azacytidine
Experimental set-up I
In this experiment four unique groups of Daphnia were generated (each group contained three biological replicates and each replicate contained 5 individual Daphnia) as demonstrated in Fig. 1a: 1) Daphnia exposed to the stressor prenatally (AC), 2) Daphnia exposed to the stressor postnatally (CA), 3) Daphnia exposed to the stressor pre- and postnatally (AA), and 4) non-exposed (CC) Daphnia (C refers to control while A refers to 5-azacytidine exposure). To set up the four unique treatment groups, pre- and post-natal 5-azacytidine exposed F1 populations were generated by either initiating the exposures from the time that the F1 neonates were within the ovaries of the mother (F0) (groups AA and AC) or immediately after the F1 neonates were released from the brood chamber of the mother into the media (groups CA and CC), respectively. In the prenatal exposed F1 generations, D. magna F0 generations were exposed from maturation age to 5-azacytidine (3.7mgL−1; 50% of concentration used in Vandegehuchte et al. (2010a)). First and second batches of brood releases were discarded to remove all potential first brood releases as first brood eggs will usually develop to form smaller individuals (Lampert 1993). The third brood (F1) was retained and continuously exposed for a further 14 days (group AA) or transferred and maintained in clean media for 14 days (Group AC). In the postnatal exposure group, the F1 generations were only exposed after they were released from the brood chamber (group CA) while control group (CC) was never exposed to 5-azacytidine. The Daphnia in both exposure and control groups were fed similar to the general culturing conditions. F1 samples were analysed for body length (n = 15) and changes in the methylation of selected genes using targeted bisulfite sequencing (BSP).
Experimental set-up II
D. magna F1 generations were exposed prenatally to 5-azacytidine and the exposures continued for an additional 1 day or 5 days after the release of neonates from the brood chamber of the mother (Fig. 1b). The exposed F1 generations were then transferred to clean media and maintained for an additional 7 days. Daphnia magna Bham2 has a lifespan of approximately 100 days. The experimental set up was designed to cover the development from neonates (day 1) to juveniles (day 5), and to adult daphnia (7 days of recovery = day 12). Control samples were generated following the same procedure without the addition of 5-azacytidine (Fig. 1b). The Daphnia in both exposure and control groups were fed similar to the general culturing conditions. The same sample groups were analysed for body length as well as several molecular endpoints including DNA methylation, metabolomics and gene expression. The number of Daphnia per biological replicate depends on the age and size of the Daphnia. The 1 day exposure group, 5 days exposure group and recovery group contained 50 neonates, 30 juveniles or 5 adults (age of maturation for D. magna Bham2 strain: approximately 10 days) per biological replicate, respectively. Six biological replicates were collected for each group. To prevent contamination of adult DNA samples with embryonic DNA samples, embryos were removed from the brood chamber prior to sample collection. Samples were flash frozen in liquid nitrogen and stored at −80 °C until further processing. DNA methylation was analysed using targeted bisulfite sequencing (BSP) (n = 3) while changes in the concentrations of the metabolites and expression of genes involved in one carbon pathway and regulation of DNA methylation were analysed via liquid chromatography–mass spectrometry (LC-MS) (n = 6) and reverse transcription polymerase chain reaction (RT-PCR) (n = 3), respectively.
Length measurements
Daphnia were photographed using a stereomicroscope SMZ800 (Nikon, Japan) coupled to a digital camera DS-Fi2 (Nikon, Japan). Body length was measured from top of the head to the base of the tail spine for all six groups (three time points of acute, chronic and recovery for control and 5-azacytidine groups, n = 15 individuals per group) and analysed using ImageJ software.
Sample preparation
Frozen samples from experiment I and II were homogenised in methanol and water using a microtube with ceramic beads. Samples from experiment I were then used for DNA extraction. For experiment II, after homogenising the samples, the homogenate was divided into three parts and each part was used for either RNA, DNA or metabolite extractions. DNA (n = 3 biological replicates) was extracted using MasterPure DNA purification kit (Epicentre, USA) according to the method described in our previous publication (Athanasio et al. 2016). RNA was extracted from the samples using the RNeasy Micro kit (Qiagen Ltd., UK) according to the manufacturer’s instructions. Metabolites were extracted using a bi-phasic extraction method (Taylor et al. 2009). Six biological replicates were analysed for metabolites quantification. Higher numbers of biological replicates were used for metabolomics analysis to minimize higher variability related to the method of analysis and maximize consistency.
Targeted bisulfite sequencing
We used a screening approach to identify several target genes that their methylation levels changed in response to 5-azacytidine treatment. To achieve this, first we selected 15 random genes with CpG repeats. From the initial list of genes nine genes were removed due to either cross reacting primers or no methylation being detected in the control samples. This process resulted in identified 6 genes with optimised bisulfite sequencing primers and altered methylation levels after treatment with 5-azacytidine. MethPrimer software was used to design BSP primers targeting the regions containing the CpGs (primer sequences and product size are listed in Table S2) (Li and Dahiya 2002). These genes are: host cell factor C1 (HCFC1), guanine nucleotide binding protein (G-protein), cyclin dependent kinase (cdk), galactose-1-phosphate uridylyltransferase (GALT), LIM and calponin domains-containing protein 1 (LIMCH1) and calcium-transporting ATPase type 2 C member 1 (ATP2C1) (sequences and annotation of the DMRs are presented in Table S1). These genes were used for all targeted BSP analysis. The EZ DNA Methylation-gold kit (Zymo Research Corporation, USA) was used for bisulfite conversion according to the manufacturer’s protocol. In addition, artificially methylated and unmethylated DNA samples were generated according to our previous publication (Mirbahai et al. 2011) and used to determine the efficiency of bisulfite conversion. Briefly, for each sample, 1 μg of genomic DNA was bisulfite treated and amplified using Zymo Taq DNA polymerase (Cambridge Biosciences, UK). The PCR products were confirmed via DNA gel electrophoresis followed by sequencing using an ABI3730 DNA analyzer. Analysis of BSP data was performed using the peak height for C and T bases at each CpG site obtained from the electropherogram and results are presented as percentage of methylated cytosines.
Targeted RT-PCR
Real time PCR was conducted to analyse the expression of genes involved in the one-carbon pathway as well as genes related to DNA methylation machinery (n = 3 biological replicates per condition with 3 technical replicates). Primers for genes DNA-methyltransferase 1 (DNMT1), DNA-methyltransferase 3 A (DNMT3A), DNA-methyltransferase 3B (DNMT3B), Methionine adenosyltransferase (MAT), S-adenosylhomocysteine hydrolase (SAHH), Methionine synthase reductase (MTRR), Betaine-homocysteine methyltransferase (BHMT), Methionine synthase (MS), Glycine N-methyltransferase (GNMT), Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) and Ten-eleven translocation methylcytosine dioxygenase 2 (TET2) were designed using Primer3 (Rozen and Skaletsky 2000) and synthesized by Integrated DNA technologies (Belgium). Primer sequences are presented in Table S3. Primers were validated using cDNA (80 ng) and BIOTAQ DNA polymerase (Bioline, UK) as recommended by manufacture’s guidelines. PCR products were sequenced on a capillary sequencer ABI3730 and compared to the expected sequences. RT-PCR was conducted on an AriaMx Realtime PCR System (Agilent Technologies, USA) using SensiFAST SYBR Lo-ROX kit (Bioline, UK). Three biological replicates (each with three technical replicates) were measured, containing 80 ng of cDNA with cycling parameters of 95°C for 2 min and 40 cycles of 95°C for 5 s and 60°C for 30 s. Melting curves were generated by using a final step of 65°C for 5 s and 95°C for 5 s to ensure single product amplification. Threshold cycle (CT) values were recorded in the linear phase of amplification and the data were analyzed using the delta-delta CT method of relative quantification (Livak and Schmittgen 2001). The geometrical average of beta-actin (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts was used as an internal reference for normalising the qPCR data as their expression levels did not alter across different conditions.
Targeted LC-MS/MS of one-carbon cycle metabolites
The same LC-MS/MS method that we described previously (Mirbahai et al. 2013) was used to quantify the levels of one-carbon cycle metabolites. Briefly, metabolites were extracted from the same sample used for both DNA methylation and gene expression analysis. The polar metabolites were re-suspended in 5 μL of a mixture of acetonitrile and water (1:1) and spiked with S-adenosyl-L-methionine-d3 (SAM-d3) tetra (p-toluenesulfonate) salt (final concentration of 0.125 µmol/mL; C/D/N isotopes INC) as internal standard. Samples were analyzed using a Dionex UltiMate 3000 liquid chromatography system coupled to a triple stage quadrupole (TSQ Vantage) tandem mass spectrometer equipped with Ion Max-S atmospheric pressure ionization source (Thermo Fisher Scientific). Separation was achieved using a reverse phase column with weak anion exchange properties (Acclaim Mixed-Mode WAX column, 250 × 0.3 mm internal diameter, 5 μm particle size, 120 Å pore size, Dionex, Germany) and a gradient elution of Buffer A (10 mM ammonium formate, pH 6.2), Buffer B (10 mM ammonium formate, pH 4.2, 75% acetonitrile and 25% water) and Buffer C (acetonitrile:water (1:1))(Table S4). The 10 analytes of interest, comprising methionine, choline, adenosine, betaine, sarcosine, stachydrine, glycine, dimethylglycine (DMG), S-adenosyl-L-homocysteine (SAH) and S-adenosyl-L-methionine (SAM) were measured using multiple reaction monitoring. The masses of the precursor and product ions, used for detection of the 10 metabolites and internal standard, are presented in Table S5. Quan Browser (Xcalibur 2.1 software, Thermo Fisher Scientific) was used to integrate peak areas.
Data analysis
Statistical analysis of the data was performed using SPSS (IBM, Armonk, NY, USA). Normal distribution of the data was evaluated via Shapiro-Wilk’s test and homogeneity of variance was analysed with Levenes’ test. For comparison of two or more groups with normal distribution and homogenised variance, 2-tailed independent Student’s t-test and one-way ANOVA with Tukey’s post-hoc test were used, respectively. When the requirements for normal distribution and homogeneity of variance were not met, data were analysed by applying non-parametric statistics, using a Kruskal-Wallis test (more than two independent groups) or Mann-Whitney test (two independent groups).
Results
Pre-natal exposure to 5-azacytidine can induce DNA methylation changes in the crustacean D. magna
Targeted bisulfite sequencing analysis of six selected 5-azacytidine induced differentially methylated regions (DMRs) provided an ideal controlled experiment to test the concept of life stage dependent DNA methylation response to stressors in the keystone species, D. magna. As demonstrated in Figs. 2a,b, all CpG regions in the three treatment groups (AC: prenatal exposed, CA: postnatal exposed, AA: pre- and postnatal exposed to 5-azacytidine) demonstrated lower levels of methylation compared to the control group (CC: non-exposed). This is expected as 5-azacytidine is a known demethylating agent. As demonstrated in Fig. 2, both pre- and post-natal only exposed groups (AC and CA) showed significant reduction in methylation of CpG sites. This indicates that prenatal stages of Daphnia are also susceptible to same level of change as post-natal stages of Daphnia. For example, in the AC group where exposures were restricted to the prenatal stages, DNA methylation changes were statistically significantly reduced for LIMCH1, HCFC1, ATP2C1 and GALT compared to the control group. The combination of pre- and postnatal exposures caused the most extreme reduction in DNA methylation compared to the control group. This could be linked to both longer exposure period and accumulation of changes observed during pre- and post-natal exposures. This indicates that by only limiting the exposures to the post-natal stages, the effect of stressors with DNA methylation modifying properties can be significantly underestimated compared to what it is observed in natural environments where the individual is exposed pre- and postnatally.
Fig. 2.
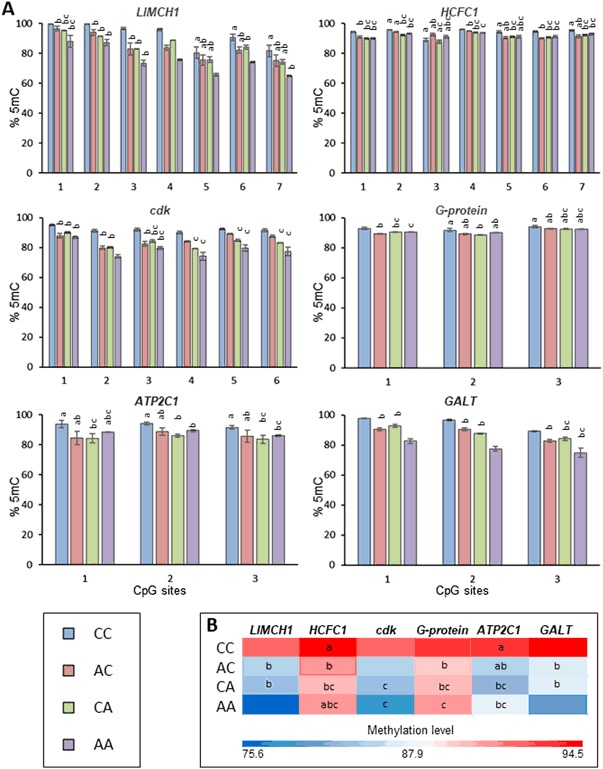
a Percentage of methylation level for CpG sites located in six different genes (LIMCH1, HCFC1, cdk, G-protein, ATP2C1, GALT) as measured via direct bisulfite sequencing. Numbers on the x-axis represents CpG sites while y-axis represents percentage of methylation level. b Heat map demonstrating averaged methylation level for each gene. Key: CC: non-exposed, AC: prenatal only exposed, CA: postnatal only exposed, AA: prenatal and postnatal exposed. Same letters indicate no difference (a: compared to control (CC), (b: compared CA to AC and AA, c: compared AC to AA; p < 0.05, t-test, n = 3 ± SD)
The body length measurements (Fig. 3) also demonstrated that prenatal exposed Daphnia (AC and AA groups) showed a significant reduction in body length from day 11 onwards compared to postnatal exposed (CA) and control (CC) groups. This indicates that pre-natal exposures are sufficient to cause same magnitude or even higher magnitude of response as post-natal exposed individuals.
Fig. 3.
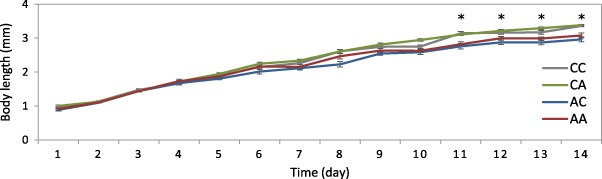
Growth rate measurements for Daphnia magna exposed to 5-azacytidine under three different exposure conditions of prenatal exposed (AC), postnatal exposed (CA), and prenatal and postnatal exposed (AA) compared to control (CC). * Significantly different in AC and AA compared to control (CC) (p < 0.05, Mann–Whitney test; n = 15 ± SEM)
Exposure to 5-azacytidine can cause enduring molecular and phenotypic effects
To further investigate if changes in DNA methylation levels caused by 5-azacytidine can persist or manifest after removal of the initial stimuli, a more extensive investigation was conducted including measuring changes in the levels of metabolites in the one-carbon pathway and expression levels of the epigenetic machinery genes.
Body length
Body length was measured after 1 day and 5 days of exposure to 5-azacytidine as well as after a seven-day period of recovery (Fig. 1b explains the exposure design). As shown in Fig. 4, no significant change in body length was observed immediately after 1 or 5 days of exposure to 5-azacytidine compared to aged matched controls. However, the group that included a recovery period demonstrated a significant reduction in body length compared to aged-matched controls. This potentially indicates an accumulative delayed effect for 5-azacytidine which only manifested itself later in life.
Fig. 4.
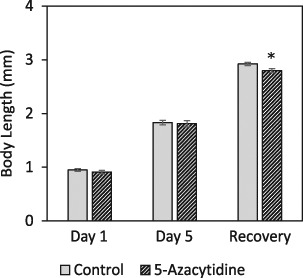
Body length of Daphnia exposed to 5-azacytidine after 1 day and 5 days of exposure and following a 7 days recovery period (See Fig. 1 for experimental design). *Significantly different from respective control (p < 0.05). Values were compared using non-parametric Mann–Whitney test between control and treatment groups (n = 15 ± SEM)
Persistent dysregulation in CpG dinucleotide methylation after removal of 5-azacytidine
As shown in Fig. 2, exposure to 5-azacytidine led to reduction in methylation levels of several CpG regions within the genome of D. magna. To test if some of the changes can persist after removal of the original stressor, methylation of the same regions was analysed after a period of recovery and compared to age-matched controls. As shown in Figs. 5, 5-azacytidine treatment caused a decrease in the methylation level of CpG sites located in HCFC1, G-protein, cdk, GALT and LIMCH1 genes. Average methylation levels for HCFC1 and cdk were reduced by 10 and 20% after 1 and 5 days of exposures to 5-azacytidine, respectively, while G-protein methylation was reduced by 7.1 and 17.6% after 1 and 5 days of exposure. For GALT and LIMCH1, the average methylation levels at day 5 were reduced by 11.6 and 9.6%, respectively. For regions in GALT and LIMCH1 genes the average methylation levels were not fully restored, showing a decrease of 14 and 8.7%. This indicated that some of the 5-azacytidine induced methylation changes are potentially persistent.
Fig. 5.
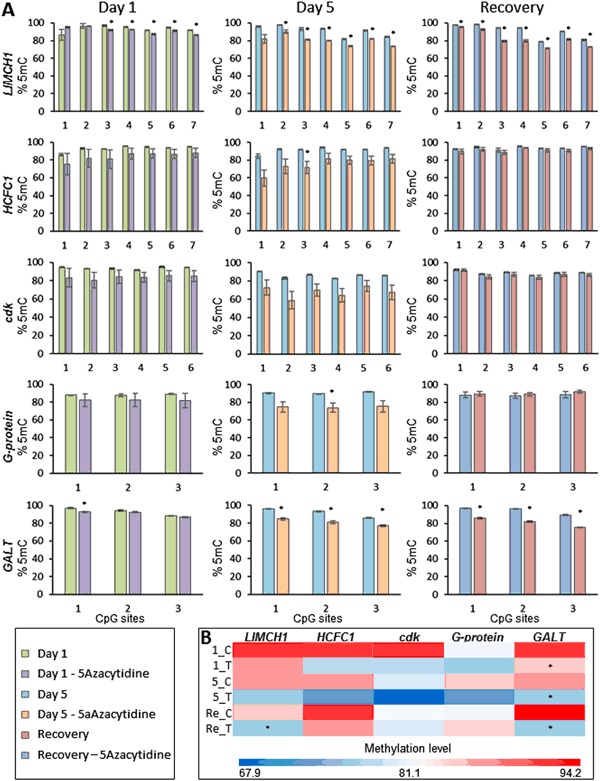
Daphnia magna F1 generations were exposed prenatally to 5-azacytidine and the exposures continued for an additional 1 day or 5 days after the release of neonates from the brood chamber of the mother. The exposed F1 generations were then transferred to clean media and maintained for an additional 7 days (recovery period). Control samples were generated following the same procedure without the addition of 5-azacytidine. a Percentage of methylation level for CpG sites located in five different genes (LIMCH1, HCFC1, cdk, G-protein, GALT) as measured via direct bisulfite sequencing. Numbers on the x-axis represents CpG sites while y-axis represents percentage of methylation level. b Heat map demonstrating averaged methylation level for each gene. * Significantly different from respective control (p < 0.05, t-test, n = 3 ± SD)
Transcriptional dysregulation of the genes involved in the one-carbon cycle and DNA methylation machinery
The expression levels of genes involved in the one-carbon cycle and DNA methylation regulation were investigated using RT-PCR and are presented as log2 fold changes between age matched control and treatment groups in Fig. 6. As expected, exposure to 5-azacytidine significantly affected the expression levels of several genes after both 1 day and 5 days of exposure. The expression level of DNMT3B was decreased after 1 day and increased after 5 days of exposure. Similar to DNMT3B, the expression level of DNMT1 was increased after 5 days of exposure. TET_1 and TET_2 genes were both downregulated after one day of exposure with expression levels of TET_1 increasing after 5 days of exposure to 5-azacytidine. In the one-carbon pathways, opposite to MTRR expression, the expression level of MAT gene was increased after one day and decreased after 5 days of exposure. Expression of the MS gene, similar to the MAT gene, was downregulated after 5 days of exposure. However, after a recovery period the expression levels of all genes with exception of GNMT were restored to normal levels. Therefore the changes observed at a transcriptional level in response to 5-azacytidine are immediate but transient and reliant on the presence of 5-azacytidine.
Fig. 6.
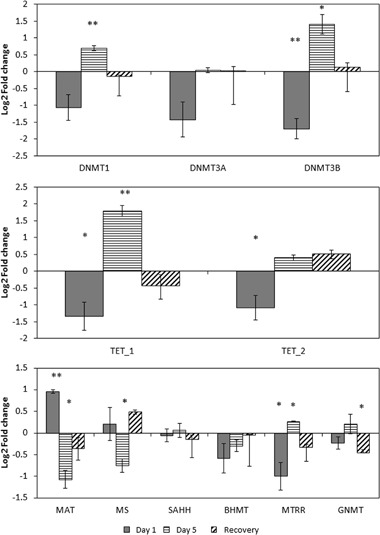
Gene expression results for the groups exposed to 5-azacytidine after 1 and 5 days of exposures and recovery period compared to age matched controls. Relative log2 fold change to control group expression (* p < 0.05, ** p < 0.01, T-test; n = 3 ± SEM). Genes involved in the one-carbon pathway and demethylation pathway: DNA-methyltransferase 1 (DNMT1), DNA-methyltransferase 3 A (DNMT3A), DNA-methyltransferase 3B (DNMT3B), Methionine adenosyltransferase (MAT), S-adenosylhomocysteine hydrolase (SAHH), Methionine synthase reductase (MTRR), Betaine-homocysteine methyltransferase (BHMT), Methionine synthase (MS), Glycine N-methyltransferase (GNMT), Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) and Ten-eleven translocation methylcytosine dioxygenase 2 (TET2)
Targeted analysis of the one-carbon cycle metabolites after exposure to 5-azacytidine
To investigate further the potential role of the one-carbon cycle in regulating the epigenetic responses to 5-azacytidine, a LC−MS/MS method was used to measure the levels of key one-carbon cycle metabolites, consisting of SAH, SAM, methionine, choline, adenosine, betaine, sarcosine, stachydrine, glycine and DMG after exposure compared to age matched controls. As shown in Fig. 7, 5-azacytidine mainly induced a statistically significant change in the levels of one-carbon cycle metabolites after 5 days of exposure, including for SAM, SAH, methionine, sarcosine and stachydrine. Most interestingly the levels of all metabolites (except betaine) recovered after the removal of the stressor.
Fig. 7.
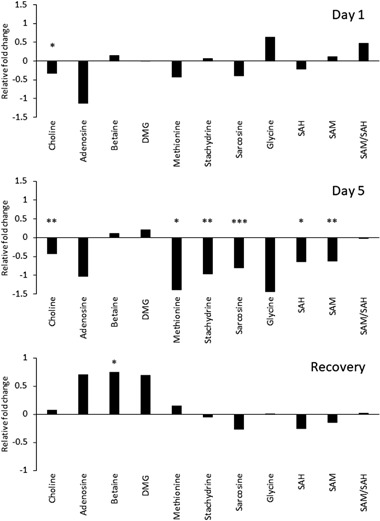
Relative changes in the levels of ten metabolites (and one metabolite ratio) involved in the one-carbon cycle in the treatment group compared to age matched control after 1 and 5 days of exposures and after recovery period. Peak area was normalised to the peak area for the internal standard (S-adenosyl-L-methionine-d3). Graph presents the relative log2 fold change of the averaged values of treatment and control group. SAH: S-adenosyl-L-homocysteine; SAM: S-adenosyl-L-methionine. Six replicates were used for each group (*p < 0.05, **p < 0.01, ***p < 0.001, Mann–Whitney test)
Discussion
It is becoming apparent through an increasing amount of epigenetics research that chemicals can induce hereto unrecognised biological effects that are not accounted for in current environmental risk assessment (Baccarelli and Bollati 2009; Shaw et al 2017). In parallel to this realisation, there is building momentum to shift away from using traditional ecotoxicity tests that do not mimic realistic environmental scenarios towards mechanistic and knowledge-based approaches in chemical risk assessment. Therefore, arguably, this is a timely opportunity to not only recognise that our current tests do not consider such concepts as epigenetic memory and developmental origin of adult disease, but to address this shortcoming. Therefore in a proof of concept study we exposed Daphnia magna to 5-azacytidine, a chemical with a known effect on DNA methylation machinery in a range of species, including Daphnia (Aniagu et al. 2008; Mirbahai and Chipman 2014; Vandegehuchte et al. 2009a, 2010a, 2010b) to investigate several concepts such as effect of life stage on exposure response and potential persistence of DNA methylation alterations. This was achieved by using two different experimental designs (see method section) and analysing the same sample for changes in the levels of specific metabolites, expression of genes and DNA methylation.
Effects on DNA methylation pathways and retention of epigenetic modifications
It is widely accepted that certain changes to the epigenome can be maintained even in the absence of the initial stressor, giving rise to the concept of epigenetic memory (Mirbahai and Chipman 2014). It is thought that these changes can have both adverse and beneficial effects for the organisms by either causing secondary, accumulative, adverse effects which can have negative impacts on the health of the organisms later on, or the changes can contribute towards phenotypic diversity and beneficial adaptation to environmental changes (Mirbahai and Chipman 2014; Vandegehuchte and Janssen 2014). Therefore, one of our aims was to investigate if epigenetic changes induced after exposure to 5-azacytidine can be maintained after removal of the stressor.
As demonstrated in Figs. 5–7, exposing the Daphnia to 5-azacytidine for 5 days induced both changes in the expression of the DNA methylation machinery and the one-carbon pathway. The one-carbon pathway is the principal biochemical pathway regulating DNA methylation, since it can alter production of the immediate methyl donor, SAM. Therefore, chronic imbalance in the concentrations of one-carbon cycle metabolites can influence DNA methylation and underlies the pathogenesis of many adverse phenotypes (Melnyk et al. 2000). However, interestingly the direction and level of transcriptional responses differed between the 1 day and 5 days of exposure to 5-azacytidine. Similarly, in a time series RNA-seq data (days 5, 9, 13, 17) obtained from human bladder cells exposed to 5-Azacytidine with 0.1uM concentration, Ding et al. (2016) demonstrated that 5-azacytidine can induce altered expression patterns of many genes on both the isoform and exon level in a time dependent manner. This potentially indicates that the expression of genes is altered as an immediate response to a stressor, in this case 5-azacytidine, whereas continuous presence of a stressor can provoke not only a different level of response but also a different type of response. However, as shown in Fig. 7, the concentrations of the one-carbon pathway metabolites were only affected after 5 days of exposure to 5-azacytidine. This is expected as in general there is a longer period between exposure and impact on concentrations of metabolites, compared with transcription. Reductions in the levels of choline, SAM and SAH indicate that chronic exposure to 5-azacytidine also depletes the production and pool of substrates required for DNA methylation which can lead to exacerbating the de-methylating effect of 5-azacytidine treatment. DNMT3B expression was firstly downregulated at day 1, however, at day 5 both DNMT1 and DNMT3B transcripts were upregulated. In this context, both SAM and SAH concentrations were decreased following 5-azacytidine exposure. According to James et al. (2002), SAH is known to act as a regulator of DNMTs expression. Often, high levels of SAH are known to repress expression of DNMTs. Here, the lower levels of SAH, caused by lower rates of cytosine methylation due to DNMTs inefficiency, could be acting as a stimulus for DNMT expression.
As shown in Fig. 5 and as expected, exposure to the de-methylating agent, 5-azacytidine, resulted in reduction in the methylation levels of CpG sites and the effect was more severe in the group exposed to 5-azacytidine for 5 days compared to 1 day. This is consistent with the mechanism of action of 5-azacytidine, where during replication the cytosine analogue is incorporated to the DNA causing a replication dependent reduction in DNA methylation levels (Lavelle et al. 2008). In addition we investigated if the cells, in a cell cycle dependent manner via de novo methylation, can restore their DNA methylation levels after removal of a demethylating chemical, such as 5-azacytidine. We observed that although most of the detected changes in both expression of the genes and concentration of the metabolites were restored after a period of recovery (transient changes), not all DNA methylation levels of CpG sites were restored to the control level. Our data is similar to what it has been observe in vitro in cell lines treated with 5-azacytdine. Several studies have shown that 5-azacytidine causes a rapid reduction at both global and gene specific methylation levels. Most interestingly, it has been demonstrated that the effects of 5-azacytidine is retained at both DNA methylation level and in some cases at gene expression levels in multiple cell lines, in some cases even after 3 months (27 cell passages) (Cosgrove and Cox 1990; Qiu et al. 2010; Kagey et al. 2010). In addition, it has been shown that the effect of 5-azacytidine at DNA methylation level can be retained for two generations in Daphnia (Vandegehuchte et al. 2010a). These findings support the idea that some of the DNA methylation changes can be much more persistent than changes at transcription and metabolite levels. Therefore, it is possible that the organism can maintain an epigenetic memory of the exposure which can later either gradually restore to the original state or lead to a permanent altered epigenetic state and long term and heritable effects on gene expression.
Phenotypic endpoints, including the measurement of body length, are usually useful to determine the physiological effects of the exposures often affecting development and growth (Lampert and Trubetskova 1996). Adverse effects on body length were only observed at a later stage, indicating a delay in visible phenotypic change, reflective of past exposure histories. This highlights an important point that phenotypic responses not always occur immediately in response to a stressor and there can be a discrepancy between observing changes at the molecular and phenotypic (e.g. reproduction and growth) levels.
Early life exposure and changes in DNA methylation levels and implications for current toxicity testing approaches
Organisms in ecosystems are usually exposed to low levels of environmental contaminants throughout their life and across multiple generations, including during critical stages of their development. Therefore, the observed phenotypes of a field population of organisms can vary dramatically compared to phenotypes measured during laboratory testing where individuals are only exposed during a specific stage of their lifespan. This exposure setup can underestimate the adverse effects of chemicals as it excludes exposures during critical stages of development. As a result there is a recent shift in the academic community towards conducting multi-generational exposures for Daphnia, covering exposures during critical stages of development (Giraudo et al. 2017; Jeong et al. 2015; Silva et al. 2017).
It has been well documented, mainly in mice and human, that environmental stressors can interfere with epigenetic programming that occur during embryogenesis and tissue differentiation (De Felici 2011; Nestor et al. 2015; Seisenberger et al. 2013). This creates an extremely sensitive window in an individual’s development where disruption in this process can lead to severe health effects (Lee Pow et al. 2017; Perera and Herbstman 2011). Therefore, the magnitude of biological response can be dependent on whether an individual encountered the stressor before or after embryogenesis (Lee Pow et al. 2017; Mersha et al. 2015). However, most importantly, this can lead to delayed adverse effects later in the life of an individual, long after the initial stressor has been removed, a concept referred to developmental origin of adult disease (Dolinoy and Jirtle 2008; Bailey 2015).
However, it is important to highlight that the degree of epigenetic reprogramming and epigenetic inheritance is dependent on sexual and asexual methods of reproduction (Verhoeven and Preite 2014; Gorelick and Carpinone 2009). Daphnia spp. can switch their reproduction mode from parthenogenesis to sexual reproduction to adapt to the external environments. Most interestingly, both apomixes and more recently, abortive meiosis (atypical automixis) have been suggested as methods of parthenogenetic reproduction for Daphnia (Hiruta et al. 2010). Although epigenetic reprogramming has not yet been investigated during either the sexual or asexual reproduction in Daphnia, it is hypothesised that if parthenogenesis occurs via a degree of meiosis, even abortive, it is less likely that they completely bypass epigenetic resetting mechanisms. It is hypothesis that depending on the level of meiosis, it is possible that the epigenetic resetting mechanisms that act during early embryonic development are unaffected, while resetting mechanisms that act during gametogenesis may or may not be affected (Verhoeven and Preite 2014). Therefore, as the genome of Daphnia is methylated and is responsive to environmental stressors (Asselman et al. 2016a, 2016b; Vandegehuchte et al. 2009b) it is highly likely that epigenetic resting events that occur during both methods of reproduction in Daphnia can be altered, although to different degrees.
Furthermore, our data (Fig. 2) demonstrates that both pre- and post-natal only exposed groups (AC and CA) have significant changes in their methylation levels compared to the control group (CC). This result indicates that both stages of Daphnia’s life are sensitive to chemical induced epigenetic changes. However, the group that was continuously exposed to 5-azacytine (AA), most reflective of exposure conditions in the natural environment, had the highest level of DNA methylation changes. This could be linked to longer duration of exposure as well as unique accumulative changes from each stage of Daphnia’s life. These data indicates that the effect of stressor in the natural environment can be dramatically underestimated if laboratory testing is only targeted to a specific stage of an animal’s life.
In conclusion, the results of this study demonstrate that a prototypical chemical that interferes with DNA methylation machinery and regulation can induce epigenetic changes that last beyond the exposure period and can be associated with phenotypic changes, such as growth, an established adverse outcome. This also highlights a challenge and a need for a method to distinguish between epigenetic changes of current versus past exposures. This may be achieved by conducting multiple detailed epigenetic profiling of chemicals with distinct mode of action in a series of comprehensive experimental designs which also include recovery stages. Furthermore, given that the largest epigenetic effects were discovered when using non-standardised exposure regimes, this strongly supports the need for working towards both a standard set of epigenetic assays for incorporation into current chemical risk assessment as well as refinement of the current OECD guidelines for chemical exposures as part of the risk assessment process. Potentially, epigenetic biomarkers that are persistent could prove effective for predicting the long-term impact of chemical exposure to the health of a population. We therefore recommend that understanding the relevance of epigenetic and multigenerational changes in response to chemical exposure should be a priority research area for risk assessment (Morgan et al. 2007).
Electronic supplementary material
Acknowledgments
Funding
This research was supported by the joint University of Birmingham and Brazil (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/BEX8572-12-7)) scholarship programme, NERC Biomolecular Analysis Facility award (NBAF863), and the University of Birmingham Fellowship Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s10646-018-1927-3) contains supplementary material, which is available to authorized users.
References
- Aniagu SO, Williams TD, Allen Y, Katsiadaki I, Chipman JK. Global genomic methylation levels in the liver and gonads of the three-spine stickleback (Gasterosteus Aculeatus) after exposure to hexabromocyclododecane and 17-β oestradiol. Environ Int. 2008;34(3):310–317. doi: 10.1016/j.envint.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Asselman J, De Coninck DI, Beert E, Janssen CR, Orsini L, Pfrender ME, Decaestecker E, De Schamphelaere KA. Bisulfite sequencing with daphnia highlights a role for epigenetics in regulating stress response to microcystis through preferential differential methylation of serine and threonine amino acids. Environ Sci Technol. 2016;51(2):acs.est.6b03870. doi: 10.1021/acs.est.6b03870. [DOI] [PubMed] [Google Scholar]
- Asselman J, De Coninck DI, Pfrender ME, De Schamphelaere KA. Gene body methylation patterns in Daphnia are associated with gene family size. Genome Biol Evol. 2016;8(4):1185–1196. doi: 10.1093/gbe/evw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasio CG, Chipman JK, Viant MR, Mirbahai L. Optimisation of DNA extraction from the Crustacean Daphnia. PeerJ. 2016;4:e2004. doi: 10.7717/peerj.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. doi: 10.1097/MOP.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA (2015) Chapter 11 - Developmental origins of adult disease: Impacts of exposure to environmental toxicants. In: Rebecca CF (ed) Systems biology in toxicology and environmental health. Academic Press, Boston, pp 239–253
- Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9):a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari RK. Medaka as a model for studying environmentally induced epigenetic transgenerational inheritance of phenotypes. Environ Epigenetics. 2016;2(1):dvv010. doi: 10.1093/eep/dvv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R. The expanding epigenetic landscape of non-model organisms. J Exp Biol. 2015;218(Pt 1):114–122. doi: 10.1242/jeb.110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-Aza-2'-Deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Cosgrove DE, Cox GS. Effects of sodium butyrate and 5-Azacytidine on DNA methylation in human tumor cell lines: variable response to drug treatment and withdrawal. Biochim Biophys Acta-Gene Struct Expr. 1990;1087(1):80–86. doi: 10.1016/0167-4781(90)90124-K. [DOI] [PubMed] [Google Scholar]
- De Felici M. Nuclear reprogramming in mouse primordial germ cells: epigenetic contribution. Stem Cells Int. 2011;2011:425863. doi: 10.4061/2011/425863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XL, Yang X, Liang G, Wang K. Isoform switching and exon skipping induced by the DNA methylation inhibitor 5-Aza-2′-Deoxycytidine. Sci Rep. 2016;6(1):24545. doi: 10.1038/srep24545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49(1):4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- Giraudo M, Cottin G, Esperanza M, Gagnon P, De Silva AO, Houde M. Transcriptional and cellular effects of benzotriazole UV stabilizers UV-234 and UV-328 in the freshwater invertebrates Chlamydomonas reinhardtii and Daphnia magna. Environ Toxicol Chem. 2017;36(12):3333–3342. doi: 10.1002/etc.3908. [DOI] [PubMed] [Google Scholar]
- Gorelick R, Carpinone J. Origin and maintenance of sex: the evolutionary joys of self sex. Biol J Linn Soc. 2009;98:707–728. doi: 10.1111/j.1095-8312.2009.01334.x. [DOI] [Google Scholar]
- Groh KJ, Carvalho RN, Chipman JK, Denslow ND, Halder M, Murphy CA, Roelofs D, Rolaki A, Schirmer K, Watanabe KH (2015) Development and application of the adverse outcome pathway framework for understanding and predicting chronic toxicity: I. challenges and research needs in ecotoxicology. Chemosphere 120:764–777 [DOI] [PubMed]
- Harris KDM, Bartlett NJ, Lloyd VK. Daphnia as an emerging epigenetic model organism. Genet Res Int. 2012;2012:147892. doi: 10.1155/2012/147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruta C, Nishida C, Tochinai S. Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res. 2010;18:833. doi: 10.1007/s10577-010-9159-2. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002;132(8):2361S–2366. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- Jeong TY, Kim HY, Kim SD. Multi-generational effects of propranolol on Daphnia magna at different environmental concentrations. Environ Pollut. 2015;206:188–194. doi: 10.1016/j.envpol.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, Reese SE, Markunas CA, Richmond RC, Xu CJ, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Kapoor-Vazirani P, McCabe MT, Powell DR, Vertino PM. Long-term stability of demethylation after transient exposure to 5-Aza-2’-Deoxycytidine correlates with sustained RNA polymerase II occupancy. Mol Cancer Res. 2010;8(7):1048–1059. doi: 10.1158/1541-7786.MCR-10-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81(4):303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Jaric I. The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes. 2017;8(3):104. doi: 10.3390/genes8030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert W. Phenotypic plasticity of the size at first reproduction in Daphnia: the importance of maternal size. Ecology. 1993;74(5):1455–1466. doi: 10.2307/1940074. [DOI] [Google Scholar]
- Lampert W (2011) Daphnia: development of a model organism in ecology and evolution, vol. 21. International Ecology Institute, Luhe
- Lampert W, Trubetskova I. Juvenile Growth Rate as a Measure of Fitness in Daphnia. Funct Ecol. 1996;10(5):631–635. doi: 10.2307/2390173. [DOI] [Google Scholar]
- Lavelle D, Saunthararajah Y, Desimone J. DNA methylation and mechanism of action of 5-azacytidine. Blood. 2008;111(4):2485. doi: 10.1182/blood-2007-10-119867. [DOI] [PubMed] [Google Scholar]
- Lee Pow CSD, Tilahun K, Creech K, Law JM, Cope WG, Kwak TJ, Rice JA, Aday DD, Kullman SW. Windows of susceptibility and consequences of early life exposures to 17β–estradiol on Medaka (Oryzias Latipes) reproductive success. Environ Sci Technol. 2017;51(9):5296–5305. doi: 10.1021/acs.est.7b01568. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods San Diego Calif. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. S-Adenosylmethionine. Int J Biochem Cell Biol. 2000;32(4):391–395. doi: 10.1016/S1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8(11):e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczylo EL, Jacobs MN, Gant TW. Environmentally induced epigenetic toxicity: potential public health concerns. Crit Rev Toxicol. 2016;46(8):676–700. doi: 10.1080/10408444.2016.1175417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular s-adenosylmethionine and s-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plasma homocysteine and pyridoxal 5’-phosphate concentrations. Clin Chem. 2000;46(2):265–272. [PubMed] [Google Scholar]
- Mersha MD, Patel BM, Patel D, Richardson BN, Dhillon HS. Effects of BPA and BPS exposure limited to early embryogenesis persist to impair non-associative learning in adults. Behav Brain Funct. 2015;11:27. doi: 10.1186/s12993-015-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbahai L, Chipman JK. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposures. Mutat Res Genet Toxicol Environ Mutagen. 2014;764–765:10–17. doi: 10.1016/j.mrgentox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Mirbahai L, Southam AD, Sommer U, Williams TD, Bignell JP, Lyons BP, Viant MR, Chipman JK. Disruption of DNA methylation via s-adenosylhomocysteine is a key process in high incidence liver carcinogenesis in fish. J Proteome Res. 2013;12:2895–2904. doi: 10.1021/pr400195u. [DOI] [PubMed] [Google Scholar]
- Mirbahai L, Williams TD, Zhan H, Gong Z, Chipman JK. Comprehensive profiling of zebrafish hepatic proximal promoter CpG island methylation and its modification during chemical carcinogenesis. BMC Genom. 2011;12:3. doi: 10.1186/1471-2164-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbahai L, Yin G, Bignell JP, Li N, Williams TD, Chipman JK. DNA methylation in liver tumorigenesis in fish from the environment. Epigenetics. 2011;6(11):1319–1333. doi: 10.4161/epi.6.11.17890. [DOI] [PubMed] [Google Scholar]
- Morgan AJ, Kille P, Stürzenbaum SR (2007) Microevolution and ecotoxicology of metals in invertebrates. Environ Sci Technol 41(4)1085–1096 [DOI] [PubMed]
- Nestor CE, Ottaviano R, Reinhardt D, Cruickshanks HA, Mjoseng HK, McPherson RC, Lentini A, Thomson JP, Dunican DS, Pennings S, et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015;16(1):11. doi: 10.1186/s13059-014-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (2004) OECD Guidelines for the Testing of Chemicals, Section 2, Test No. 202: Daphnia Sp. Acute Immobilisation Test
- OECD (2012) OECD Guidelines for the Testing of Chemicals, Section 2, Test No. 211: Daphnia Magna Reproduction Test
- Pegoraro M, Bafna A, Davies NJ, Shuker DM, Tauber E. DNA methylation changes induced by long and short photoperiods in nasonia. Genome Res. 2016;26(2):203–210. doi: 10.1101/gr.196204.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol. 2011;31(3):363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hother C, Ralfkiaer UM, Søgaard A, Lu Q, Workman CT, Liang G, Jones PA, Grønbaæk K. Equitoxic doses of 5-azacytidine and 5-aza- 2’deoxycytidine induce diverse immediate and overlapping heritable changes in the transcriptome. PLoS ONE. 2010;5(9):e12994. doi: 10.1371/journal.pone.0012994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EMK, Amdam GV. Cytosine modifications in the honey bee (Apis Mellifera) worker genome. Front Genet. 2015;5:8. doi: 10.3389/fgene.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W, Hemberger M, Dean W, Reik W, Bostick M, et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JLA, Judy JD, Kumar A, Bertsch P, Wang MB, Kirby JK. Incorporating transgenerational epigenetic inheritance into ecological risk assessment frameworks. Environ Sci Technol. 2017;51(17):9433–9445. doi: 10.1021/acs.est.7b01094. [DOI] [PubMed] [Google Scholar]
- Silva ARR, Cardoso DN, Cruz A, Pestana JLT, Mendo S, Soares AMVM, Loureiro S. Multigenerational effects of carbendazim in Daphnia magna. Environ Toxicol Chem. 2017;36(2):383–394. doi: 10.1002/etc.3541. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Weber RJM, Southam AD, Payne TG, Hrydziuszko O, Arvanitis TN, Viant MR. A new approach to toxicity testing in Daphnia magna: application of high throughput FT-ICR mass spectrometry. Metab Metab. 2009;5(1):44–58. [Google Scholar]
- Thornburg KL, Shannon J, Thuillier P, Turker MS (2010) In utero life and epigenetic predisposition for disease. NIH Public Access. In: Advances in genetics, Vol 71. Academic press, pp 57–78 [DOI] [PMC free article] [PubMed]
- Tobiasson M, Abdulkadir H, Lennartsson A, Katayama S, Marabita F, De Paepe A, Karimi M, Krjutskov K, Einarsdottir E, Grövdal M, et al. Comprehensive Mapping of the Effects of Azacitidine on DNA Methylation, Repressive/permissive Histone Marks and Gene Expression in Primary Cells from Patients with MDS and MDS-Related Disease. Oncotarget. 2017;8(17):28812–28825. doi: 10.18632/oncotarget.15807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegehuchte MB, De Coninck D, Vandenbrouck T, De Coen WM, Janssen CR. Gene Transcription Profiles, Global DNA Methylation and Potential Transgenerational Epigenetic Effects Related to Zn Exposure History in Daphnia Magna. Environ Pollut. 2010;158(10):3323–3329. doi: 10.1016/j.envpol.2010.07.023. [DOI] [PubMed] [Google Scholar]
- Vandegehuchte MB, Janssen CR (2014) Epigenetics in an ecotoxicological context. Mutat Res Genet Toxicol Environ Mutagen 764–765:36–45 [DOI] [PubMed]
- Vandegehuchte MB, Kyndt T, Vanholme B, Haegeman A, Gheysen G, Janssen CR. Occurrence of DNA methylation in Daphnia magna and influence of multigeneration Cd exposure. Environ Int. 2009;35(4):700–706. doi: 10.1016/j.envint.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Vandegehuchte MB, Lemière F, Janssen CR. Quantitative DNA-methylation in Daphnia magna and effects of multigeneration Zn exposure. Comp Biochem Physiol C Toxicol Pharmacol. 2009;150(3):343–348. doi: 10.1016/j.cbpc.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, Janssen CR. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151(3):278–285. doi: 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Preite V. Epigenetic variation in asexually reproducing organisms. Evolution. 2014;68(3):644–655. doi: 10.1111/evo.12320. [DOI] [PubMed] [Google Scholar]
- Williams TD, Mirbahai L, Chipman JK. The toxicological application of transcriptomics and epigenomics in Zebrafish and other Teleosts. Brief Funct Genom. 2014;13(2):157–171. doi: 10.1093/bfgp/elt053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


