Abstract
CD1d-restricted natural killer (NK) T cells reactive with the glycolipid α-galactosylceramide (α-GalCer) are a distinct lymphocyte sublineage. They express an invariant Vα14-Jα18 T cell receptor (TcR), but the role of the β chain has been controversial. Here, we have used CD1d tetramers to identify and isolate NK T cells based on their antigen specificity. In mice lacking germline Vβ8, most of the α-GalCer-reactive T cells express either Vβ2 or Vβ7, strong Vβ selection being revealed by the lack of an increase in other Vβ regions. By contrast to the selection for complementarity determining region (CDR) 3β sequences in some anti-peptide responses, α-GalCer-reactive T cells have polyclonal CDR3β sequences. There is little CDR3β sequence redundancy between organs or individual mice, and, surprisingly, there also is no evidence for organ-specific CDR3β sequence motifs. These data argue against a T cell receptor-mediated self-reactivity for tissue-specific CD1d-bound ligands. Each NKT clone is represented by only 5–10 cells. This clone size is similar to naive conventional T cells, and much lower than that reported for memory T cells, although NK T cells have an activated/memory phenotype.
Keywords: lipid antigen‖T lymphocyte‖T cell antigen receptor‖tetramer‖antigen specificity
Natural killer (NK) T cells are a distinct subset of mature lymphocytes that coexpress NK receptors and T cell receptors (TcRs; refs. 1–6). Two major subsets of NK T cells have been identified. CD1d-independent NK T cells can develop in the absence of CD1d (7), a nonclassical class I antigen-presenting molecule that presents lipid antigens. In the mouse, these CD1d-independent NK T cells are mainly double negative (DN) and CD8+, they have a diverse TcR repertoire (5, 7, 8), and, although some of them express a naive phenotype, they also include some conventional MHC class I-restricted memory CD8+ cells (9). By contrast, most CD1d-dependent NK T cells in the mouse express a semiinvariant TcR composed of a Vα14-Jα18 rearrangement that preferentially associates with either Vβ8, Vβ7, or Vβ2 (1, 10–13). These cells are almost uniformly reactive to the marine sponge-derived glycolipid α-galactosylceramide (α-GalCer; refs. 14 and 15). CD1d-restricted NK T cells differ from CD1d-independent NK T cells in that they express a memory or activated phenotype (CD62L−CD69+CD44+; refs. 5 and 7) and they are either CD4+ or CD4/CD8 DN cells. Additionally, there are some CD1d-dependent cells with diverse TcRs that lack α-GalCer reactivity, and some of these T lymphocytes also are NK1.1+ (16–19). To distinguish between these two populations of CD1d-dependent cells, we refer in this paper to α-GalCer-reactive cells as NK T cells, a population that is synonymous, or nearly so, with the Vα14-Jα18 NK T cells.
Until recently, the lack of specific markers capable of readily discriminating between the different NK T cell populations greatly complicated their study. To circumvent this problem, we and others have recently produced tetramers of CD1d molecules loaded with α-GalCer (20, 21). Analysis of lymphocyte populations with these tetramers confirms that more than 80% of the NK T cells in the thymus and liver are α-GalCer reactive and Vα14-Jα18 expressing T cells. By contrast, in the spleen and bone marrow, only ≈50% of the NK T cells are α-GalCer reactive (20, 21).
Relatively little is known about the structural interactions between the TcR and the α-GalCer/CD1d complex. The murine NK T TcR requires the α-anomeric linkage of the sugar to the hydrophobic lipid tails, the equatorial orientation of the 2′-OH group of the sugar, and the presence of a 3′-OH on the sphingosine base (14, 22). Therefore, like the TcR of conventional T cells, the TcRs of CD1d-restricted NK T cells show a high degree of specificity for antigen.
The development of α-GalCer/CD1d tetramers enables us to provide a complete and quantitative clonal description of the major NK T cell population based on antigen specificity. Therefore, this approach offers a unique opportunity to characterize the diversity of TcRs capable of recognizing a defined glycolipid antigen. Our goals in this report were to use these newly developed CD1d tetramers to address three issues. First, we wished to investigate the functional role of the β chain in the recognition of α-GalCer. Second, because previous results suggested that Vα14+ CD1d-restricted NK T cells exhibit an organ-specific pattern of reactivity for a presumed autologous self-ligand (23, 24), we wished to examine whether these T lymphocytes exhibited organ-specific patterns of complementarity determining region (CDR) 3β sequences. Third, because NK T cells have an activated/memory phenotype, we wished to determine whether particular clones, identified by signature CDR3β sequences, had undergone extensive clonal expansion. In each case, the data reveal novel properties of NK T cells that distinguish them from conventional T lymphocytes.
Materials and Methods
Mice.
C57BL/6, C57L/J, and C57BR/J mice were purchased from The Jackson Laboratory. All mice were maintained at the La Jolla Institute for Allergy and Immunology vivarium. Mice of both sexes were used between 7 to 11 weeks of age. NK T transgenic mice, expressing a rearranged Vα14 TcR with the canonical sequence and the Vβ8.2 TcR from the H-Y peptide (Smcy-encoded)/Db-specific TcR were characterized previously (25). For experiments with primed animals, mice were injected i.v. with 4 μg of α-GalCer or an equivalent volume of vehicle and analyzed at the times indicated.
Flow Cytometry.
The following mAb conjugates were used in this study: FITC, phycoerythrin (PE)-, or Tricolor-labeled anti-TcRβ clone H57-597, FITC-labeled anti-Vβ8.1/8.2 clone MR5-2, FITC-labeled anti-Vβ7 clone TR310, FITC-labeled anti-Vβ2 clone B20.6, FITC-labeled anti-Vβ12 clone MR11-1, Tricolor-labeled anti-CD4 clone GK1.5, allophycocyanin (APC)-labeled anti-CD8α clone 53-6.7, FITC-labeled rat anti-mouse IL-4, and FITC-labeled Rat anti-mouse IFN-γ. All mAbs were purchased from PharMingen. PE-labeled α-GalCer/CD1d tetramer was prepared as previously described (20). Intracellular stainings were performed by using the Cytofix/Cytoperm Plus kit (PharMingen) following the manufacturer's protocol.
FACS Sorting.
Before FACS sorting, splenocytes and thymocytes were enriched for tetramer+ cells by using anti-PE microbeads (Miltenyi Biotec, Auburn, CA) as previously described (20). MACS-enriched α-GalCer/CD1d-reactive T cells were then stained with PE-labeled α-GalCer/CD1d tetramer and FITC-labeled anti-TcRβ mAbs and purified by cell sorting with a FACStar flow cytometer (Becton Dickinson). Purity of the sorted populations was >98%. The number of lymphocytes sorted ranged from between 1.25 and 2.5 × 105 cells. After sorting, the cells were suspended in Trizol reagent (Life Technologies, Grand Island, NY), and total RNA was extracted following the manufacturer's protocol.
Vβ Repertoire Analysis.
The CDR3 spectratyping method has been described elsewhere (26), and the oligonucleotides used to analyze the Vβ and Vα repertoires have been described previously (27, 28). Each V-C PCR product was used as a template for extension or run-off reactions with oligonucleotides labeled with fluorescent tag. The fluorescent run-off products generated varied in size depending on CDR3 length. Run-off products were subjected to capillary electrophoresis in an automated DNA sequencer (Applied Biosystems), and CDR3 size distribution and signal intensities were analyzed with genescan software (Perkin-Elmer).
Sequencing.
The cloning and sequencing methods have been described elsewhere (29). Briefly, Vβ-Cβ PCR products were purified by gel electrophoresis and were directly cloned in pCR4 TOPO TA vector (Invitrogen). Sequencing reactions were carried out directly on PCR product generated from LacZ− colonies by using M13(–20) primer and with the ABI PRISM Big Dye Terminator Cycle Sequencing ready Reaction Kit (Applied Biosystems). CDR3 region sequences were extracted and analyzed by using software designed for this purpose.
Results
CD1d-Restricted NK T Cells Are Present in Mice That Lack the Vβ8 Gene Segment.
We compared the frequencies of α-GalCer/CD1d tetramer-reactive T cells in the liver, the spleen, and the thymus of C57BL/6, C57L/J, and C57BR/J mice. The proportion of α-GalCer/CD1d-reactive T cells in the different organs of C57BL/6 mice was similar to that reported previously (20). Vβa haplotype C57L/J and C57BR/J mice have a deletion in the central portion of the TcRβ locus that includes the germ line Vβ8.2 segment (30), which is the one most commonly used by NK T cells (1, 2, 4). These mice retain tetramer-binding T cells, however, in thymus, liver, and spleen (data not shown). Vβ6, Vβ10, and Vβ14 are not highly represented among the α-GalCer-reactive T cells in C57BL/6 mice. These Vβ segments are not deleted in Vβa haplotype mice, but their frequency did not increase markedly among the tetramer-binding lymphocytes from C57BR/J mice (Table 1). In fact, Vβ7 and Vβ2 together comprise ≈95% of the tetramer+ cells in the thymus and liver of these mice. Therefore, in the absence of Vβ8, the use of Vβ segments other than Vβ7 and Vβ2 by α-GalCer-reactive NK T cells does not increase.
Table 1.
Relative usage of Vβ2, Vβ6, Vβ7, Vβ8.1/8.2, Vβ10, and Vβ14 gene segments among the tetramer+ cells in various organs
| Mouse | Organ | % usage in tetramer+ cells*
|
|||||
|---|---|---|---|---|---|---|---|
| Vβ2 | Vβ6 | Vβ7 | Vβ8.1/8.2 | Vβ10 | Vβ14 | ||
| C57BL/6 (n = 3) | Thymus | 15 | 0.8 | 16.5 | 47.7 | 1.6 | 1.8 |
| Liver | 12 | 1.1 | 16.0 | 55.0 | 0.7 | 1.7 | |
| Spleen | ND | 1.1 | 10.7 | 45.0 | 1.5 | 1.0 | |
| C57BR/J (n = 3) | Thymus | 48 | 0.4 | 46.4 | 0.6 | 2.0 | 1.0 |
| Liver | 44 | 0.3 | 51.0 | 0.7 | 2.0 | 0.4 | |
| Spleen | ND | 1.0 | 32.7 | 0.5 | 3.1 | 0.9 | |
ND, not done.
SE is less than 20% of the mean value for TcRVβ determination.
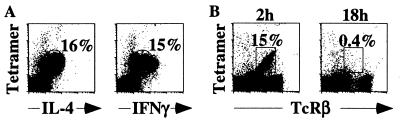
C57BR/J mice were injected with α-GalCer and analyzed 2 h later. Mononuclear liver cells were then surface stained with α-GalCer/CD1d tetramers, and intracellularly with anti-IL-4 or anti-IFN-γ antibodies. Two hours after injection, all tetramer+ liver cells derived from Vβa mice responded to α-GalCer by producing IL-4 and IFN-γ (Fig. 1A). Eighteen hours after the injection, no α-GalCer/CD1d-reactive T cells could be detected in the liver of C57BR/J mice (Fig. 1B), as we previously reported for C57BL/6 mice (20). These results demonstrate that the response to α-GalCer injection, namely rapid cytokine production followed by disappearance of the cells, is not altered by the absence of Vβ8+ NK T cells.
Figure 1.
Normal α-GalCer responsiveness in mice lacking Vβ8. (A) Intracellular cytokine staining with α-IL-4 and α-IFN-γ mAbs of lymphocytes isolated from the liver of a C57BR/J mouse 2 h after i.v. injection of α-GalCer. (B) Comparison of α-GalCer/CD1d tetramer staining of lymphocytes from the liver of C57BR/J mice at 2 h and 18 h after i.v. injection of α-GalCer. These data are representative of three animals tested in each condition.
CD1d-Restricted NK T Cells Use Polyclonal Vβ Rearrangements.
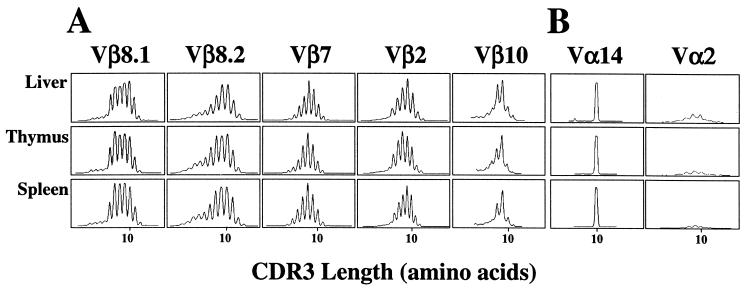
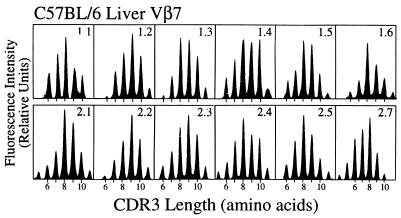
We used fragment length analysis of the CDR3β regions to examine in greater detail the Vβ repertoire of α-GalCer/CD1d-reactive T cells. These lymphocytes were purified based on CD1d tetramer reactivity, and PCR was carried out on cDNA from these cells to amplify the rearranged Vβ8.1, Vβ8.2, Vβ7, Vβ2, and Vβ10 gene segments. Amplifications were also carried out with a Cα primer and either a Vα14 or a control Vα2 primer. The single 10-aa peak with Vα14, combined with the weak Vα2 signal, confirmed the strong enrichment obtained for cells with the canonical Vα14 rearrangement by the tetramer-based enrichment (Fig. 2B). TcR β chain diversity was assessed by run-off reactions with an oligonucleotide that hybridizes with the Cβ gene. The profile for each Vβ-Cβ combination yielded a typical bell-shaped distribution of the CDR3 lengths resolved in six to eight distinct peaks (Fig. 2A), similar to the pattern reported for polyclonal, naive T splenocytes (26, 31). Identical polyclonal patterns were obtained when each of the 12 Jβ-specific primers was used instead of Cβ for the run off reactions (Fig. 3), demonstrating the usage of all Jβ segments, as well as the absence of any bias in CDR3 length.
Figure 2.
Analysis of TcR diversity of α-GalCer reactive T cells. α-GalCer/CD1d tetramer+ cells were sorted by flow cytometry from the liver, thymus, and spleen of C57BL/6 mice. Lymphocytes from the organs of two mice were pooled. (A) The CDR3β profiles are depicted for selected Vβ-Cβ PCR amplifications with the indicated Vβ primers. The intensity of fluorescence is represented in arbitrary units as a function of CDR3 length, in amino acids. (B) The stringency of the sort for tetramer+ cells was confirmed by analysis of the Vα chain usage. Vα-Cα PCR amplifications were performed with primers for Vα14 and primers for Vα2, a negative control for Vα regions typically not expressed by α-GalCer reactive T cells. Two independent sorting experiments were performed, and each gave similar results.
Figure 3.
Diverse Vβ-Jβ rearrangements of α-GalCer/CD1d-reactive T cells. Fragment length analysis profiles of Vβ7-Jβ products are depicted for tetramer-binding cells from the liver. The intensity of fluorescence is represented in arbitrary units as a function of CDR3 length in amino acids. The results from this analysis are representative of the polyclonality of all Vβ-Jβ rearrangements in tetramer-reactive cells observed in the liver, spleen, and thymus for Vβ8.1, Vβ8.2, Vβ7, and Vβ2. Two independent experiments gave similar results.
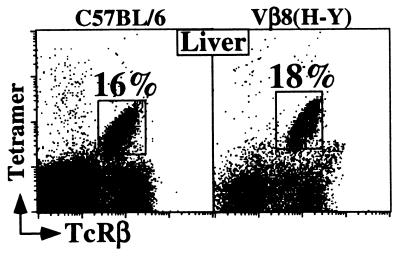
Altogether, these results suggest that the TcR Vβ repertoire of α-GalCer/CD1d-reactive T cells is restricted primarily by the Vβ segment used (Vβ8/7/2). The data further suggest that many rearrangements derived from these Vβs can encode a TcR specific for α-GalCer presented by CD1d, as long as the invariant Vα14 rearrangement is present. To test this hypothesis further, we used the α-GalCer/CD1d tetramers to analyze mice transgenic for a rearranged Vβ8.2 gene that was not derived from an α-GalCer-reactive T cell. We chose the well-studied Vβ8.2-Dβ2-Jβ2.3 rearrangement, which when associated with a Vα3 rearrangement, recognizes an Smcy-encoded male-derived peptide (H-Y) presented by Db (25). Strikingly, we identified a substantial population of α-GalCer/CD1d-reactive T cells in the Vβ8.2 transgenic animals (Fig. 4 and data not shown), emphasizing the lack of stringent selection for CDR3β sequences in the TcRs of the NK T cells.
Figure 4.
α-GalCer/CD1d tetramer+ populations are detected in HY Vβ8.2 TcR transgenic mice. The FACS dot plots display the α-GalCer/CD1d tetramer vs. TcRβ profiles of cell suspensions obtained from the liver of C57BL/6 and Vβ8.2 (anti-H-Y) transgenic mice. The expression of the Vβ8.2 transgene by tetramer+ T cells in the liver of transgenic mice was ensured by the absence of staining with another endogenous Vβ (anti-Vβ7) Abs (data not shown). The percentage of tetramer+ cells from one representative experiment is shown; three mice from each strain were analyzed.
α-GalCer-Reactive T Cells Have a Small Clone Size.
Nucleotide sequence analysis of the CDR3β regions of tetramer binding T lymphocytes was carried out to determine whether α-GalCer-reactive T cells exhibit selection for particular CDR3 sequence motifs, and whether identical clones are found in different organs. To do this, PCR was carried out on cDNA from sorted tetramer+ cells by using primers specific for Vβ7 and Jβ1.2. These segments were selected because they are less frequently used than many others, with Vβ7 representing ≈10–17% of the total for NK T cell Vβs (Table 1) and Jβ1.2 estimated to be used ≈6% of the time (32). By selecting a V-J combination that accounts for only 1–2% of the total pool of α-GalCer-reactive T cells, we intended to reduce the number of sequences that would have to be carried out to obtain a representative picture of the total repertoire. Sequencing of this rearrangement was performed on samples from two individual mice and revealed the expected polyclonal population (Table 2 and data not shown). For example, of the 97 sequences done on tetramer+ cells extracted from the thymus of mouse 1, 66 of them were unique clones of varying CDR3 length found only once in the pool. The remaining 31 sequences were redundant, usually having been found either two or three times. This pool of 31 redundant sequences contains 14 distinct sequences. Similar results were reproducibly obtained when the liver and spleen of two individuals mice were analyzed (Table 2).
Table 2.
Sequence analysis of tetramer+ cells utilizing a Vβ7Jβ1.2 rearrangement in individual C57BL/6 mice
| Organ | CDR3β nucleotide sequences
|
Consensus§ | Clone size | ||||
|---|---|---|---|---|---|---|---|
| Total sequences | Unique* | Redundant† | MLE‡ | ||||
| Thymus | Mouse 1 | 97 | 66 | 31 (14) | 241 | SLGGANSDY | 5 |
| Mouse 2 | 164 | 85 | 79 (33) | 233 | SLGGANSDY | 7 | |
| Liver | Mouse 1 | 85 | 73 | 12 (5) | 481 | SLGGANSDY | 4 |
| Mouse 2 | 154 | 66 | 88 (31) | 152 | SLGGANSDY | 4 | |
| Spleen | Mouse 1 | 144 | 64 | 80 (29) | 151 | SLGGANSDY | 8 |
| Mouse 2 | 208 | 103 | 105 (39) | 253 | SLGGGNSDY | 7 | |
Number of unique sequences identified once in the organ.
Number of sequences identified more than once in the organ. Number in parentheses indicates the number of unique sequences within this redundant subset.
MLE (maximum likelihood estimate) of the number of distinct sequences, calculated with a 95% confidence interval.
Sequence of 9-aa CDR3 that is generated by selecting the most highly represented (>20%) amino acid for each position of the CDR3.
Based on the extent of redundancy in the sequences found, a likely number of distinct sequences present in the sample can be estimated. This calculation is defined as the maximum likelihood estimate (MLE; refs. 29 and 33). The MLE shown in Table 2 represents the likely number of distinct Vβ7-Jβ1.2 sequences in the samples, with a 95% confidence. For example, it is estimated that ≈480 distinct Vβ7-Jβ1.2 sequences would have been found in the liver of mouse 1, had the sequencing been done exhaustively. One can estimate the size of the entire Vβ repertoire for all tetramer+ cells in any given sample as follows: (the MLE for Vβ7-Jβ1.2)/(the frequency of Vβ7 in tetramer+ cells × the frequency of Jβ1.2 rearranged with Vβ7). For instance, in the spleen of mouse 1, if we divide 151 by (0.11 × 0.06), we determine that there were ≈2.4 × 104 distinct Vβ-Jβ rearrangements in the sample. The number of sorted cells for this sample was 2 × 105, so the size of the average clone would be ≈8 cells. For the thymus and liver, the calculated clone sizes were similar and reproducible (Table 2).
α-GalCer-Reactive T Cells Do Not Exhibit Organ-Specific CD3Rβ Sequence Motifs.
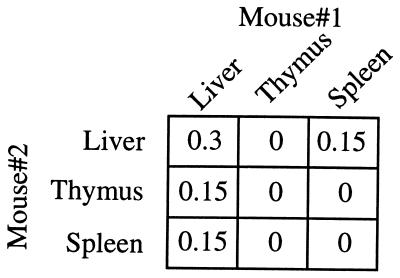
There was little overlap in CDR3β sequences when tetramer+ cells from different organs were compared. Although at least 85 Vβ7-Jβ1.2 sequences were obtained in each case, there were no common sequences found in the thymus and liver, only 4 common sequences between the liver and spleen, and 6 between the thymus and spleen of mouse 1. Similar results were obtained when the sequences derived from another individual mouse were analyzed (data not shown). Likewise, the Vβ7-Jβ1.2 sequences derived from the NK T cells found in the different organs of distinct animals were mostly nonoverlapping (Fig. 5).
Figure 5.
NK T cell repertoires of distinct animals are mostly nonoverlapping. α-GalCer/CD1d tetramer+ cells were sorted by flow cytometry from the liver, thymus, and spleen of two individual C57BL/6 mice. The percentages, less than 1% in every case, of recurrent sequences of the Vβ7-Jβ1.2 CDR3 region of NK T cells between the different organs of two individual mice are presented.
Despite this relative lack of overlap of common sequences between organs and mice, it was not possible to identify selection for particular sequences in any of the organs analyzed. For example, considering the Vβ7-Jβ1.2 CDR3β sequences that are 9 aa in length, the same consensus sequence was identified in the thymus (SLGGANSDY), liver, and spleen in both individual mice analyzed (Table 2). The criterion for selecting the consensus was >20% representation, because it was not possible to assign consensus amino acids for most positions with more stringent criteria.
Discussion
α-GalCer-reactive T cells constitute the majority of NK T cells in the mouse (20, 21), and there is increasing evidence suggesting that these lymphocytes have important immune regulatory functions (34–41). It is clear from previous studies that nearly all of these cells have an invariant Vα14-Jα18 TcRα chain, although their β chains are somewhat more diverse (1, 10–12). We purified these cells based on their binding to the newly developed α-GalCer/CD1d tetramers. The TcRβ gene rearrangements contained in these cells provide a marker for individual clones among the tetramer binding lymphocytes. By analyzing these rearrangements in cells analyzed ex vivo without any further manipulation, we have gained important insights into the diversity, specificity, and distribution of NK T cell clones with a well defined specificity. Furthermore, because the tetramers detect the majority of the NK T cells in the mouse, it is reasonable to infer that the rules governing the selection of TcRs responsive to this marine sponge-derived lipid also may apply to the natural ligand driving the expansion of these cells.
In agreement with previous reports that used the NK1.1 marker (12, 35), we find evidence for strong selection for the use of a few Vβ segments in α-GalCer-reactive T cells. In the absence of a germ line Vβ8 segment, most of the tetramer binding cells are either Vβ2+ or Vβ7+, and there is no evidence for the proportional increase in other Vβ segments. In the absence of pairing constraints between specific Vβ chains and the canonical Vα14 rearrangement (35), the selection for Vβ8, -7, and -2 is likely due to interactions involving the germ line-encoded CDR1 and CDR2 regions within these Vβ segments, providing an increased probability of specificity for CD1d and particular lipids such as α-GalCer. With regard to α-GalCer responsiveness, however, we detect no difference in cells that do not express Vβ8. By contrast to this strong Vβ segment selection, the results from CDR3 length analysis and nucleotide sequencing clearly indicate that there is little or no selection for particular CDR3β regions. Consistent with this, a rearranged Vβ8.2 transgene derived from a conventional CD8+ peptide-reactive T cell clone could drive the differentiation of a normal number of α-GalCer-reactive T cells. Additionally, whereas N regions are prevalent in the TcRβ rearrangements of NK T cells, perhaps reflecting their origin relatively late in ontogeny (3, 42–44), a normal α-GalCer-reactive T lymphocyte population could be generated in mice lacking TdT (data not shown).
The role of the CDR3β in NK T cell specificity had previously been controversial. The conclusions reached here do not agree with those in two earlier analyses of α-GalCer-reactive T cells, which reported the selection for charged and acidic CDR3β amino acids (45, 46). This observation of selection for amino acids could have resulted from the sampling of relatively small lymphocyte populations in the earlier reports and to other selective events due to the cell culture or cell fusion required to obtain the T cells that were previously analyzed. An analysis of short-term cultured Vα24/Vβ11 DN clones from the blood of healthy human donors found repeated CDR3β sequences (47). Therefore, it remains possible that α-GalCer-reactive NK T cells in humans represent relatively few clones that have undergone significant expansion. On the other hand, the results here are in agreement with those from recent TcR sequence analyses (48, 49). In these reports, however, either DN or CD4+ cells selected for NK1.1+ and TcRβ expression were analyzed. Some α-GalCer-reactive T cells do not express NK1.1, however, and the correlation between NK1.1, TcRβ expression, and α-GalCer reactivity varies significantly in different organs. Therefore, it was not possible to correlate directly the diverse CDR3β sequences with α-GalCer/CD1d specificity in these earlier studies.
A number of peptide-specific T cell responses by conventional T cells show strong selection for CDR3β amino acids (26, 50–53). The strong selection for CDR3α sequences in the α-GalCer-reactive T cells, combined with the lack of selection for CDR3β, suggests that the TcR of NK T cells has a distinct mode of interaction with the complex of antigen bound to CD1 on the antigen-presenting molecule. This mode may not be entirely unique to this sublineage, however, because the crystal structure of the 2C TcR shows a strong involvement of CDR3α in peptide/MHC class I recognition, with little involvement for CDR3β amino acids (54, 55). Interestingly, the 2C TcR also contains a Vβ8.2 segment.
The results from sequencing the TcRβ regions demonstrate very little overlap in the α-GalCer-reactive T cell clones found in the thymus, spleen, and liver, but at the same time, no evidence for any organ-specific selection for Vβ8.2, Vβ7, or Vβ2, or for particular CDR3β sequence motifs. Whereas sequence analysis has been carried out for Vβ7-Jβ1.2 rearrangements only, the results from the Jβ run off reactions, demonstrating Gaussian distributions and more than 70–80 visible peaks with the different Jβ primers, strongly suggest that this conclusion can be generalized to other Vβ-Jβ combinations. Previously, it had been suggested that Vα14+ NK T cells in different organs might be preferentially reactive with autologous CD1d-bound ligands found in those organs, such that splenic NK T cells would respond preferentially to splenic APCs, and thymic NK T cells would prefer APC from the thymus (23, 24). This conclusion was based on the analysis of Vα14+ NK T cell hybridomas, some of which show autoreactivity for CD1d+ APCs in the absence of α-GalCer. Because the only differences in the TcRs of these cells are in CDR3β, we consider it unlikely that these preferential responses reflect TcR specificity. More likely, they result from the differential expression of accessory molecules on either the T cells and/or the antigen-presenting cells. Consistent with this possibility, we have found that the CD1d autoreactivity of different cultures or subclones of a Vβ8.2/Vα14+ hybridoma can vary greatly despite maintenance of equivalent TcR levels (unpublished observations), providing evidence that molecules besides the TcR might contribute to CD1d autoreactivity of NK T hybridomas.
The estimated clone size for the α-GalCer-reactive T cells is ≈5–10 cells. This size is comparable to what has been observed for conventional, naive T cells (29, 56), but much smaller than that for memory T cells (57). Work done in peptide antigen responses has led to the estimate that a given naive clone consisting of 30 cells will divide ≈10 times on responding to antigen, expanding to 3 × 104 lymphocytes (58). An estimated 5% will survive and become long-term memory cells, leading to a clonal population of 1.5 × 103 cells (59, 60). The small clone size of the α-GalCer-reactive T cells is surprising, given that they closely resemble memory/activated T lymphocytes with regard to both their cell surface phenotype and their ability to rapidly secrete cytokines. It remains possible, however, that α-GalCer-reactive NK T cells are not truly antigen experienced, despite their phenotype. Alternatively, in mice, cells with different CDR3β regions may be undergoing equivalent stimulation, with their clonal expansion limited by factors that define and limit the NK T cell niche. The immune response of conventional T cells to peptide antigens is usually composed of very few clones that expand greatly to complete their effector functions (58, 59, 61). In sharp contrast, and perhaps because of a requirement for an end-stage effector function (62, 63), the α-GalCer/CD1d-reactive T cell population is maintained at stable levels by the presence of a large number of different clones with the same specificity, capable of responding quickly without significant expansion.
Acknowledgments
We thank Chris Lena for his help with the cell sorting at the La Jolla Institute for Allergy and Immunology facility. We also thank Drs. Frits Koning and Antoine Attinger for critical review of this manuscript. This project was funded by National Institutes of Health Grants RO1 CA52511 and AI45053 (to M.K.), a grant from the Human Frontiers of Science Program, and a fellowship from the Cancer Research Institute (to L.G.). The work conducted in the Laboratory of Molecular Biology of the Gene was supported by grants from the European Economic Community (Biotechnologies CT 94-2005), Ligue Nationale Contre le Cancer, and Association pour la Recherche Contre le Cancer. This is manuscript number 415 from the La Jolla Institute for Allergy and Immunology.
Abbreviations
- DN
double negative
- NK
natural killer
- CDR
complementarity determining region
- α-GalCer
α-galactosyl ceramide
- TcR
T cell receptor
- PE
phycoerythrin
- APC
allophycocyanin
- MLE
maximum likelihood estimate
References
- 1.Lantz O, Bendelac A. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohteki T, MacDonald H R. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald H R. J Exp Med. 1995;182:633–638. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendelac A, Rivera M N, Park S H, Roark J H. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 5.Hammond K J, Pelikan S B, Crowe N Y, Randle-Barrett E, Nakayama T, Taniguchi M, Smyth M J, van Driel I R, Scollay R, Baxter A G, Godfrey D I. Eur J Immunol. 1999;29:3768–3781. doi: 10.1002/(SICI)1521-4141(199911)29:11<3768::AID-IMMU3768>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Godfrey D I, Hammond K J, Poulton L D, Smyth M J, Baxter A G. Immunol Today. 2000;21:573–583. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 7.Eberl G, Lees R, Smiley S T, Taniguchi M, Grusby M J, MacDonald H R. J Immunol. 1999;162:6410–6419. [PubMed] [Google Scholar]
- 8.Emoto M, Zerrahn J, Miyamoto M, Perarnau B, Kaufmann S H. Eur J Immunol. 2000;30:2300–2311. doi: 10.1002/1521-4141(2000)30:8<2300::AID-IMMU2300>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Coles M C, McMahon C W, Takizawa H, Raulet D H. Eur J Immunol. 2000;30:236–244. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 10.Arase H, Arase N, Ogasawara K, Good R A, Onoe K. Proc Natl Acad Sci USA. 1992;89:6506–6510. doi: 10.1073/pnas.89.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Int Immunol. 1995;7:1157–1161. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 12.Ohteki T, MacDonald H R. J Exp Med. 1996;183:1277–1282. doi: 10.1084/jem.183.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimamura M, Ohteki T, Beutner U, MacDonald H R. Eur J Immunol. 1997;27:1576–1579. doi: 10.1002/eji.1830270638. [DOI] [PubMed] [Google Scholar]
- 14.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 15.Burdin N, Brossay L, Koezuka Y, Smiley S T, Grusby M J, Gui M, Taniguchi M, Hayakawa K, Kronenberg M. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 16.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behar S M, Podrebarac T A, Roy C J, Wang C R, Brenner M B. J Immunol. 1999;162:161–167. [PubMed] [Google Scholar]
- 18.Zeng D, Dick M, Cheng L, Amano M, Dejbakhsh-Jones S, Huie P, Sibley R, Strober S. J Exp Med. 1998;187:525–536. doi: 10.1084/jem.187.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behar S M, Cardell S. Semin Immunol. 2000;12:551–560. doi: 10.1006/smim.2000.0273. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda J L, Naidenko O V, Gapin L, Nakayama T, Taniguchi M, Wang C R, Koezuka Y, Kronenberg M. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. J Immunol. 1998;161:5124–5128. [PubMed] [Google Scholar]
- 23.Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. J Immunol. 1998;160:3681–3688. [PubMed] [Google Scholar]
- 24.Park S H, Roark J H, Bendelac A. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- 25.Bluthmann H, Kisielow P, Uematsu Y, Malissen M, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Nature (London) 1988;334:156–159. doi: 10.1038/334156a0. [DOI] [PubMed] [Google Scholar]
- 26.Pannetier C, Even J, Kourilsky P. Immunol Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 27.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casanova J L, Romero P, Widmann C, Kourilsky P, Maryanski J L. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. J Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 30.Behlke M A, Chou H S, Huppi K, Loh D Y. Proc Natl Acad Sci USA. 1986;83:767–771. doi: 10.1073/pnas.83.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochet M, Pannetier C, Regnault A, Darche S, Leclerc C, Kourilsky P. Eur J Immunol. 1992;22:2639–2647. doi: 10.1002/eji.1830221025. [DOI] [PubMed] [Google Scholar]
- 32.Kato T, Suzuki S, Sasakawa H, Masuko K, Ikeda Y, Nishioka K, Yamamoto K. Eur J Immunol. 1994;24:2410–2414. doi: 10.1002/eji.1830241022. [DOI] [PubMed] [Google Scholar]
- 33.Arstila T P, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 34.Sumida T, Sakamoto A, Murata H, Makino Y, Takahashi H, Yoshida S, Nishioka K, Iwamoto I, Taniguchi M. J Exp Med. 1995;182:1163–1168. doi: 10.1084/jem.182.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendelac A, Hunziker R D, Lantz O. J Exp Med. 1996;184:1285–1293. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gombert J M, Herbelin A, Tancrede-Bohin E, Dy M, Carnaud C, Bach J F. Eur J Immunol. 1996;26:2989–2998. doi: 10.1002/eji.1830261226. [DOI] [PubMed] [Google Scholar]
- 37.Hammond K J L, Poulton L D, Palmisano L J, Silveira P A, Godfrey D I, Baxter A G. J Exp Med. 1998;187:1047–1056. doi: 10.1084/jem.187.7.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson S B, Kent S C, Patton K T, Orban T, Jackson R A, Exley M, Porcelli S, Schatz D A, Atkinson M A, Balk S P, Strominger J L, Hafler D A. Nature (London) 1998;391:177–181. doi: 10.1038/34419. [DOI] [PubMed] [Google Scholar]
- 39.Zeng D, Lewis D, Dejbakhsh-Jones S, Lan F, Garcia-Ojeda M, Sibley R, Strober S. J Exp Med. 1999;189:1073–1081. doi: 10.1084/jem.189.7.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falcone M, Yeung B, Tucker L, Rodriguez E, Sarvetnick N. J Exp Med. 1999;190:963–972. doi: 10.1084/jem.190.7.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamshiev A, Donda A, Prigozy T I, Mori L, Chigorno V, Benedict C A, Kappos L, Sonnino S, Kronenberg M, De Libero G. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 42.Takahama Y, Kosugi A, Singer A. J Immunol. 1991;146:1134–1141. [PubMed] [Google Scholar]
- 43.Bendelac A. Curr Opin Immunol. 1995;7:367–374. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 44.Hammond K, Cain W, van Driel I, Godfrey D. Int Immunol. 1998;10:1491–1499. doi: 10.1093/intimm/10.10.1491. [DOI] [PubMed] [Google Scholar]
- 45.Kawano T, Tanaka Y, Shimizu E, Kaneko Y, Kamata N, Sato H, Osada H, Sekiya S, Nakayama T, Taniguchi M. Int Immunol. 1999;11:881–887. doi: 10.1093/intimm/11.6.881. [DOI] [PubMed] [Google Scholar]
- 46.Burdin N, Brossay L, Degano M, Iijima H, Gui M, Wilson I A, Kronenberg M. Proc Natl Acad Sci USA. 2000;97:10156–10161. doi: 10.1073/pnas.97.18.10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Apostolou I, Cumano A, Gachelin G, Kourilsky P. J Immunol. 2000;165:2481–2490. doi: 10.4049/jimmunol.165.5.2481. [DOI] [PubMed] [Google Scholar]
- 49.Ronet C, Mempel M, Thieblemont N, Lehuen A, Kourilsky P, Gachelin G. J Immunol. 2001;166:1755–1762. doi: 10.4049/jimmunol.166.3.1755. [DOI] [PubMed] [Google Scholar]
- 50.Cibotti R, Cabaniols J P, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos J M, Kourilsky P. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman J D, Moss P A, Goulder P J, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 52.Gapin L, Bravo de Alba Y, Casrouge A, Cabaniols J P, Kourilsky P, Kanellopoulos J. J Immunol. 1998;160:1555–1564. [PubMed] [Google Scholar]
- 53.Wang F, Ono T, Kalergis A M, Zhang W, DiLorenzo T P, Lim K, Nathenson S G. Proc Natl Acad Sci USA. 1998;95:5217–5222. doi: 10.1073/pnas.95.9.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 55.Garcia K C, Degano M, Pease L R, Huang M, Peterson P A, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 56.Bousso P, Casrouge A, Altman J D, Haury M, Kanellopoulos J, Abastado J P, Kourilsky P. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 57.Bousso P, Kourilsky P. Semin Immunol. 1999;11:423–431. doi: 10.1006/smim.1999.0200. [DOI] [PubMed] [Google Scholar]
- 58.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 59.Sourdive D J, Murali-Krishna K, Altman J D, Zajac A J, Whitmire J K, Pannetier C, Kourilsky P, Evavold B, Sette A, Ahmed R. J Exp Med. 1998;188:71–82. doi: 10.1084/jem.188.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blattman J N, Sourdive D J, Murali-Krishna K, Ahmed R, Altman J D. J Immunol. 2000;165:6081–6090. doi: 10.4049/jimmunol.165.11.6081. [DOI] [PubMed] [Google Scholar]
- 61.Maryanski J L, Jongeneel C V, Bucher P, Casanova J L, Walker P R. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 62.Benlagha K, Bendelac A. Semin Immunol. 2000;12:537–542. doi: 10.1006/smim.2000.0276. [DOI] [PubMed] [Google Scholar]
- 63.Matsuda J L, Kronenberg M. Curr Opin Immunol. 2001;13:19–25. doi: 10.1016/s0952-7915(00)00176-x. [DOI] [PubMed] [Google Scholar]