Abstract
U87 cells derived from human malignant gliomas and growtharrested human embryonic lung (HEL) fibroblasts were examined with respect to their response to ionizing radiation by profiling their RNAs. In the first series of experiments, cells grown in vitro were harvested and the RNAs were extracted 5 h after exposure to 1, 3, or 10 Gy. In the second series of experiments the U87 tumors were implanted in nude mice and subjected to the same doses of irradiation. The xenografts were harvested at 1, 5, or 24 h after irradiation and subjected to the same analyses. We observed and report on (i) cell-type common and cell-type specific responses, (ii) genes induced at low levels of irradiation but not at higher doses, (iii) temporal patterns of gene response in U87 xenografts that varied depending on radiation dose and temporal patterns of response that were similar at all doses tested, (iv) significantly higher up-regulation of cells in xenografts than in in vitro cultures, and (v) genes highly up-regulated by radiation. The responding genes could be grouped into nine functional clusters. The representation of the nine clusters was to some extent dependent on dose and time after irradiation. The results suggest that clinical outcome of ionizing radiation treatment may benefit significantly by taking into account both cell-type and radiation-dose specificity of cellular responses.
Keywords: cDNA arrays‖gene clusters‖time course‖radiotherapy‖transcription
Ionizing radiation (IR) has been used for nearly a century to treat human cancer (1). The objective of IR therapy is to deliver a lethal dose to cancer cells but attenuate the toxic effects of IR on adjacent normal tissue. Undesirable sequels of radiotherapy are the development of tumor resistance and damage to normal tissue (2). Various types of DNA damage, including that induced by IR, are recognized and repaired by specialized pathways first described in prokaryotes. Many of the genes involved in IR damage repair are conserved (3, 4). Transcriptional induction of DNA repair genes, immediate early genes, and a variety of cytokine and growth factor genes have been proposed as the mechanisms by which cells survive after IR in mammalian cells, tissues, and tumors (5, 6). Gene induction after the exposure of mammalian cells to IR has been reported (7–10). These reports generally describe induction of genes by IR without reference to doses used in radiotherapy or the timing of gene induction. Many in vitro studies of gene induction were performed under supraphysiologic IR doses, and therefore are of limited value in potential treatment design. Additionally, few studies of gene induction profiled RNA changes in normal tissue or in xenografted human tumors at different times after exposure to a range of IR doses. A central, unresolved issue is whether, within a range of cytoreductive doses administered in clinical practice, the gene response to IR is dose dependent. For example, conventional radiotherapy is delivered at ≈24-h intervals at 180–300 cGy/day. Recently, twice daily fractionation (110–160 cGy/day) has been studied with limited success, but experimentally based principles for rational administration of radiotherapy are as yet not available (11, 12).
Profiling of RNA on DNA arrays provides a method to study the response of thousands of genes to specific stimuli. We used this approach to profile the transcriptional response to 1, 3, or 10 Gy in primary human embryonic lung (HEL) fibroblasts and the U87 human malignant glioblastoma cell line, and in U87 tumor xenografts implanted in mice. Daily doses from 1 to 3 Gy are commonly used in radiotherapy, whereas 10 Gy is frequently used to study biochemical responses of mammalian cells to IR and in radiosurgery of some brain tumors. Here we report cell-common and cell-type specific, dose-dependent, and time-restricted patterns of gene up-regulation after IR. Analyses of the data will provide valuable information for the design of effective IR therapy and also point to adjunct therapies that could specifically target undesirable activation of specific genes by IR.
Materials and Methods
Cell Lines, Animals, and Irradiation.
The human malignant glioblastoma cell line U87 was maintained as described elsewhere (13). HEL fibroblasts were maintained as described elsewhere (14). Cells were grown to confluence, maintained in the same medium for 2 additional days, and irradiated with doses of 1, 3, or 10 Gy by using a GE Maxitron Generator operating at 250 kV, 26 mA, at a dose rate of 118 cGy/min. Samples were collected 5 h after irradiation. U87 xenografts were transplanted in athymic nude mice, and were irradiated at the same doses as described (15). Xenografts were harvested, frozen in liquid nitrogen, and stored at −80°C.
Preparation of Radiolabeled cDNA Probes and Hybridization with DNA Arrays.
Total RNA was purified as described elsewhere (16). cDNAs were prepared with reverse transcriptase of Moloney murine leukemia virus (GIBCO/BRL) in the presence of oligo(dT) and [γ-33P]ATP according to the protocol supplied by Research Genetics (Huntsville, AL).
Experimental Design, Data Acquisition, and Analysis.
The analyses described in this report are based on hybridization data from 48 GeneFilters GF211 cDNA arrays (Research Genetics). Each GeneFilters microarray consists of 5,184 distinct sequence-verified probes spotted onto a 5 × 7 cm positively charged nylon membrane, of which 4,132 spots correspond to unique human genes. The experiments led to the acquisition of ≈200,000 data points. The experiments on the response of cells grown in cell culture were done in triplicate, with purification of independent RNA samples and independent hybridizations. Quality control of hybridizations was based on internal double-spotted controls for assessment of uniformity of hybridization, estimation of reproducibility assessed by hybridization of the same sample of RNA with two different arrays (see Fig. 1 and below), and estimation of sensitivity and specificity of data acquisition by comparison with visual readings of arrays as described below. The in vivo studies were done on two independent groups of animals randomized by size of the tumor. Each dose of ionizing radiation (1, 3, or 10 Gy) and each time point (1, 5, or 24 h) was represented by two animals, and cDNA prepared from each xenograft was hybridized independently.
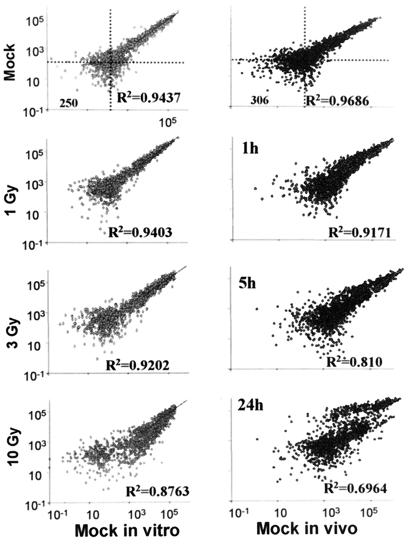
Figure 1.
Scatterplots of intensity values of independent GeneFilters GF211 arrays, hybridized with different samples of RNA. DNA arrays were hybridized and data were acquired with imagequant and normalized as described in Materials and Methods. Intensity values of one array plotted versus intensity values of the same genes on another array are shown. (Left) U87 in vitro. (Right) U87 in vivo. (Upper) Scatterplots of two arrays, hybridized with the same mock (un-irradiated) sample of RNA. Numbers at the left bottom corner of panels indicate the cutoff values for intensities (see Materials and Methods). (Lower Left) Intensity values of RNA from U87 cell cultures exposed to 1, 3, and 10 Gy, extracted from cells harvested 5 h after irradiation and plotted versus corresponding values RNA from in vitro mock-irradiated cells (see Top Left). (Lower Right) Intensity values of RNA from U87 xenografts exposed to 10 Gy, and extracted from tissues harvested at 1, 5, and 24 h after irradiation and plotted versus corresponding RNA from in vivo mock-irradiated cells (see Top Right).
The software package PATHWAYS 2.01, provided by the manufacturer for acquisition and analysis of GeneFilters GF211 data, generated many false-positives, especially for low-intensity signals. To overcome this problem, numerical signal intensity values for each hybridization spot were determined in a Storm 860 PhosphorImager with the aid of IMAGEQUANT (Molecular Dynamics).
Data Filtration.
Exported intensities of control (unirradiated) and experimental samples were further filtered, based on the following rules. (i) Negative values resulting from subtraction of background were transformed to zeros and removed from further analyses. (ii) The intensity values in each array were normalized with respect to the average intensity value of that array (17). (iii) All values less than or equal to 10% of average intensity (global mean) were transformed to zero. This cutoff value of intensity corresponds to 95.5% of specificity, calculated as true negatives/(true negatives + false positives). Estimation of numbers of true negative, true positive, and false positive data for each cutoff value was based on visual examination of array images by experienced readers. (iv) Estimation of significant levels of response was based on scatterplots of 2 independent control samples vs. each other and vs. each experimental sample (Fig. 1). For our cell culture data sets, mean ± 1 SD corresponded to 1.45 ratio and 95% confidence interval corresponded to 1.90 ratio. For in vivo data sets, the values increased to 1.54 and 2.08, respectively. We arbitrarily chose ratios of ±1.60 as cut-off values for matches of independent experiments and comparison of in vitro and in vivo data. For statistical analysis and data clustering, we used the hierarchical clustering option, provided by the JMP software (SAS Institute, Cary, NC).
Selected genes were checked by using BLAST and Human Genome Browser (http://genome.ucsc.edu/goldenPath/). Annotations of genes were based on PubMed, Online Mendelian Inheritance in Man, and other databases.
Results
Experimental Design.
We report two series of experiments. In the first series, we irradiated confluent cultures of U87 human malignant glioblastoma cells or of HEL fibroblasts with 1, 3, or 10 Gy. The cells were harvested 5 h after irradiation and the total RNA was processed and analyzed as described in Materials and Methods. The identification of genes up- or down-regulated as a consequence of irradiation was based on reproducibility of experimental results in independent experiments as described in Materials and Methods. We chose the 5 h time point because earlier studies at this time point showed that early and immediate early genes are induced by ionizing or UV light irradiation (5, 9). The key parameters of the study were as follows. Of the 4,132 genes represented in the cDNA arrays, the average number of genes whose transcripts were detected were 1,858 for mock-treated U87 cells and 1,973 for HEL fibroblasts. Of this number, those with values above 10% of the mean intensity value and which were included in these analyses were 1,470 and 1,591, respectively. The corresponding number of genes included in these analysis for 1, 3, or 10 Gy were 1,494, 1,374, and 1,318, respectively, for U87 cells and 1,495, 1,507, and 1,609, respectively, for HEL fibroblasts.
In the second series of experiments we irradiated U87 implants in the hind limb of mice as described in Materials and Methods. In this instance the mice received 1, 3, or 10 Gy, or were mock-treated, and xenografts were removed and processed for 1, 5, or 24 h. In this series of experiments we detected on the average 1,973 transcripts from mock-treated tumors. Of this number, 1,591 transcripts had intensity values above cutoff value (see Materials and Methods) and were included in these analyses. The corresponding average number of genes included in these analyses for 1, 3, or 10 Gy were 1,274, 1,303, and 1,244, respectively, for U87 cells and 1,495, 1,507 and 1,609, respectively, for HEL fibroblasts.
Analyses of Irradiation Dose-Dependent Responders in U87 and HEL Cell Cultures.
The responders were analyzed with respect to two criteria. The first compared the overall kinetics of up-regulation as a function of dose. In Table 1, the first three columns show all possible permutations of up-regulated (+) genes. The remarkable aspect of the data is the large number of genes that were up-regulated to their highest levels after 1, 3, or 10 Gy. Only a small number of genes were up-regulated to a highest level after 1 Gy and remained at the same level in cells exposed to 3 or 10 Gy. The overall impression is that more genes are transcriptionally activated as a function of dose than those whose transcripts increased after 1 Gy and then declined at higher doses.
Table 1.
Number of genes up- or down-regulated after irradiation of U87 and HEL cells in culture
| Dose, Gy
|
No. of genes
|
|||||
|---|---|---|---|---|---|---|
| 1 | 3 | 10 | U87
|
HEL
|
||
| ↑ | ↓ | ↑ | ↓ | |||
| + | − | − | 9 | 26 | 2 | 40 |
| + | + | − | 6 | 2 | 0 | 9 |
| + | − | + | 16 | 8 | 9 | 12 |
| + | + | + | 6 | 3 | 6 | 7 |
| − | + | + | 9 | 18 | 14 | 18 |
| − | + | − | 11 | 10 | 9 | 16 |
| − | − | + | 32 | 132 | 85 | 90 |
+, Responders; −, nonresponders; ↑, up-regulated; ↓, down-regulated.
The second criterion for analyses was the identification of responders common to both U87 and HEL cells. The results summarized in Table 2 show that the shared responders were 10 for 1 Gy, 11 for 3 Gy, and 48 for 10 Gy. The results indicate that the responders represented three groups, those that were U87 cell-specific, those that were HEL fibroblast-specific, and those that were common between U87- and HEL fibroblast-specific. Although the numbers were small, the bulk of the shared genes were identified in cells exposed to 10 Gy, which is consistent with the data showing that 10 Gy induced the highest number of responders.
Table 2.
Summary of responders to ionizing radiation in U87 cell grown in cell culture (U87-C) or transplanted xenografts (U87–X) or in human embryonic lung cells grown in culture (HEL-C)
| Dose, Gy | U87-C
|
HEL-C
|
U87–HEL common
|
U87–X
|
U87-C/X common
|
All cells common
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | ⇅ | ↑ | ↓ | ↑ | ↓ | ⇅ | ↑ | |
| 1 | 37 | 39 | 17 | 68 | 6 | 2 | 2 | 481 | 61 | 17 | 0 | 8 | 1 |
| 3 | 32 | 33 | 28 | 48 | 5 | 4 | 2 | 533 | 21 | 19 | 1 | 5 | 3 |
| 10 | 63 | 161 | 114 | 125 | 14 | 12 | 22 | 532 | 4 | 29 | 1 | 12 | 4 |
↑, Up regulated; ↓, down regulated; ⇅, genes up-regulated in one cell line but down-regulated in another.
Analyses of Responders to Irradiation of Implanted Xenografts of U87 Cells.
The results of the analyses carried out on responders to irradiation of xenografts of the U87 cells are shown in Table 2. There were 542 responders to 1 Gy, 554 responders to 3 Gy, and 536 responders to 10 Gy. Of this number, the 5th column of the table lists the number of responders in U87 cells grown in vitro and those in the transplanted xenografts. These numbers, 25 at 1 Gy, 25 at 3 Gy, and 42 at 10 Gy reflect the small number of responders in cultured cells in vitro. It is noteworthy, however, that the responders common to irradiated cultured cells and xenografts represent >20% of the genes up- or down-regulated after irradiation of U87 cultured cells.
Analyses of the results showed that 15 genes were up-regulated by ionizing irradiation of HEL fibroblasts and of the U87 cells cultured in vitro and in xenografts. These are identified in Table 3.
Table 3.
Common genes, responding to irradiation in U87 and HEL cell lines and U87 xenografts
| Access. no. | Group and name | U87-C
|
HEL-C
|
U87-X
|
Comments | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Gy | 3 Gy | 10 Gy | 1 Gy | 3 Gy | 10 Gy | 1 Gy | 3 Gy | 10 Gy | |||
| 1. Homeostasis/self defense | |||||||||||
| AA464246 | HLA-C | 1.28 | 1.20 | 0.51 | 0.98 | 0.91 | 2.65 | 1.71 | 1.74 | 1.53 | ↓ D |
| AA434117 | HLA-B-assot. G9a | 0.89 | 0.57 | 0.51 | 0.72 | 0.78 | 0.61 | $ | |||
| AA670408 | β2-Microglobulin | 1.02 | 2.14 | 0.53 | 1.43 | 1.82 | 5.78 | 1.80 | 1.93 | 1.24 | $ * D |
| AA778663 | 4-1BB ligand | 1.67 | 1.38 | 0.35 | 4.43 | 10.70 | 12.1 | * A | |||
| AA136271 | CD58 (LFA3) | 1.06 | 0.89 | 0.20 | 0.51 | 0.90 | 0.59 | $ | |||
| R77293 | ICAM1 | 1.22 | 0.99 | 0.42 | 2.00 | 1.47 | 2.46 | ↓ B | |||
| AA130584 | CEACAM5 | 1.77 | 1.28 | 1.88 | 3.22 | 18.63 | 3.64 | * A | |||
| N51018 | Biglycan | 1.33 | 1.22 | 1.64 | 0.47 | 0.74 | 1.69 | * E | |||
| AA399674 | SPRR2C | 0.85 | 0.89 | 0.43 | 0.82 | 0.84 | 1.79 | ↓ | |||
| T49657 | K+ channel TASK | 2.09 | 1.61 | 2.36 | 1.60 | 1.89 | 1.68 | 4.62 | 3.90 | 5.36 | $ * A |
| AA069770 | K+ channel KCNB1 | 0.56 | 1.53 | 1.59 | 10.07 | 7.12 | 6.79 | ↓ C | |||
| H14808 | Na+/K+ ATPase β2 | 1.46 | 0.93 | 2.11 | 3.07 | 2.43 | 6.45 | * A | |||
| H24316 | Aquaporin | 1.03 | 1.63 | 1.44 | 24.56 | 12.25 | 5.84 | * C | |||
| H57136 | PLM Cl− channel | 1.23 | 1.80 | 1.60 | 0.87 | 0.84 | 0.53 | 2.72 | 4.17 | 4.44 | ↓ * C |
| AA402891 | Transporter ENT2 | 1.08 | 1.19 | 1.60 | 6.07 | 36.08 | 2.61 | * C | |||
| AA191488 | Cu2+ uptake protein CTR1 | 1.72 | 1.00 | 2.46 | 2.42 | 3.20 | 4.39 | 1.05 | 1.27 | 1.80 | $ * E |
| AA490459 | Transcobalamin II | 0.76 | 0.61 | 0.55 | 0.47 | 0.83 | 0.31 | $ | |||
| H72723 | MT1B | 1.12 | 0.61 | 0.57 | 2.04 | 2.93 | 5.46 | ↓ D | |||
| H23187 | Carbonic anhydrase II | 0.84 | 1.10 | 0.59 | 4.41 | 2.50 | 2.26 | ↓B | |||
| 2. Cell–cell communications/signaling | |||||||||||
| R56211 | PDGFRβ | 1.02 | 1.12 | 1.87 | 0.92 | 1.37 | 1.85 | $ | |||
| AA486393 | IL-10 receptor β | 1.72 | 1.19 | 1.60 | 6.12 | 3.84 | 3.78 | * A | |||
| AA485226 | Vitamin D receptor | 2.31 | 0.69 | 0.64 | 6.54 | 9.35 | 3.34 | * A | |||
| H54023 | MIR-10 (LILRB2) | 1.71 | 1.00 | 1.35 | 0.48 | 0.63 | 1.98 | ↓ E | |||
| AA400973 | Lipocalin 2 | 1.16 | 0.56 | 0.49 | 0.68 | 0.62 | 0.53 | $ | |||
| AA485922 | Copine I | 1.65 | 1.06 | 1.08 | 3.84 | 3.74 | 4.24 | * A | |||
| R73545 | Flotillin 2 | 1.18 | 0.94 | 0.61 | 1.11 | 1.17 | 1.89 | ↓ | |||
| N20203 | BMP receptor II | 0.60 | 0.79 | 0.97 | 5.54 | 2.88 | 2.00 | ↓ B | |||
| AA450062 | BMP, placental | 1.45 | 1.49 | 1.60 | 1.35 | 0.82 | 1.62 | $ | |||
| AA489383 | BMP 2 | 1.71 | 1.61 | 1.80 | 0.61 | 2.14 | 1.61 | 1.77 | 1.69 | 1.06 | $ * F |
| T55558 | CSF 1 | 0.92 | 1.29 | 0.56 | 0.83 | 0.61 | 0.52 | $ | |||
| AA486072 | RANTES | 1.14 | 0.67 | 0.42 | 4.92 | 2.79 | 3.74 | ↓ A | |||
| R43320 | G-protein GNAO1 | 1.79 | 1.05 | 1.83 | 0.61 | 0.82 | 1.65 | * E | |||
| R56046 | G-protein GNAZ | 0.74 | 0.87 | 0.49 | 0.85 | 0.93 | 1.77 | 0.73 | 0.67 | 1.75 | ↓ E |
| AA458785 | Guanylate cyclase β1 | 1.74 | 1.45 | 1.86 | 3.89 | 3.75 | 4.07 | * A | |||
| R37953 | Adenylyl cyclase associated protein | 1.31 | 1.23 | 0.59 | 1.04 | 1.04 | 2.60 | ↓ | |||
| N28497 | PP2A (PPP2R1B) | 1.16 | 1.35 | 2.00 | 0.78 | 0.77 | 0.33 | 7.81 | 6.84 | 5.31 | ↓ * B |
| H15718 | Protein kinase AXL | 0.76 | 0.59 | 0.80 | 0.48 | 0.57 | 0.52 | $ | |||
| AA453789 | Protein kinase 7 | 0.94 | 1.19 | 0.60 | 0.69 | 1.04 | 0.53 | $ | |||
| R59598 | Protein kinase Syk | 0.56 | 0.64 | 0.48 | 0.54 | 0.61 | 0.76 | $ | |||
| R80779 | Protein kinase MLK-3 | 1.15 | 1.50 | 1.66 | 10.99 | 6.73 | 4.43 | * B | |||
| AA890663 | Protein kinase PAK1 | 0.64 | 1.95 | 1.80 | 0.90 | 1.06 | 0.56 | ↓ | |||
| N52958 | SLP-76 | 1.38 | 2.21 | 1.72 | 1.07 | 1.32 | 1.80 | * | |||
| H73724 | CDK6 | 1.76 | 1.53 | 2.08 | 0.47 | 0.81 | 2.01 | *E | |||
| AA464731 | Calgizzarin | 1.01 | 0.96 | 0.33 | 1.06 | 1.07 | 2.35 | ↓ | |||
| N63940 | Acetylcholinesterase | 0.83 | 0.61 | 0.87 | 0.65 | 0.62 | 0.84 | $ | |||
| 3. Cytoskeleton/motility | |||||||||||
| AA703141 | Protein 4.1 (EBP41) | 0.60 | 0.63 | 0.64 | 0.61 | 0.81 | 1.34 | $ | |||
| AA877166 | Myosin light chain 2 | 0.83 | 1.01 | 0.57 | 0.83 | 0.86 | 0.49 | $ | |||
| AA504625 | Kinesin heavy chain | 0.62 | 1.34 | 0.59 | 4.42 | 12.29 | 0.25 | * C | |||
| AA868929 | Troponin T1 | 1.01 | 0.53 | 0.27 | 0.18 | 0.12 | 1.98 | * | |||
| R44290 | β-Actin (ACTB) | 1.13 | 0.60 | 0.55 | 1.32 | 1.47 | 3.48 | 1.79 | 1.93 | 1.43 | ↓ D |
| AA634006 | Actin α-2 (ACTSA) | 1.51 | 0.80 | 0.51 | 1.01 | 1.30 | 3.16 | 1.55 | 1.55 | 1.52 | ↓ D |
| AA629189 | Keratin 4 (KRT4) | 1.08 | 1.08 | 1.62 | 1.02 | 1.32 | 0.39 | ↓ | |||
| 4. RNA synthesis/modifications | |||||||||||
| H99588 | LAF4 | 3.15 | 1.23 | 2.62 | 3.62 | 2.70 | 4.83 | 0.59 | 0.82 | 1.65 | $ * E |
| N47099 | SMAD2 | 1.12 | 1.64 | 2.04 | 0.94 | 1.90 | 0.55 | $ | |||
| AA478268 | CTBP 1 | 1.17 | 1.63 | 1.44 | 3.99 | 5.34 | 2.77 | * B | |||
| AA394127 | NF-AT3 | 1.24 | 0.96 | 0.36 | 1.15 | 0.90 | 2.19 | ↓ | |||
| AA258001 | RELB | 1.02 | 1.10 | 0.59 | 5.12 | 7.57 | 2.33 | ↓ C | |||
| AA253434 | Transcription factor HSF2 | 1.08 | 0.60 | 1.06 | 5.09 | 2.64 | 1.86 | ↓ C | |||
| AA457155 | ZNF212 | 0.49 | 0.68 | 1.32 | 2.02 | 0.87 | 2.45 | ↓ | |||
| R02346 | U1 snRNP 70 | 2.57 | 1.32 | 2.59 | 1.33 | 1.51 | 1.98 | 6.29 | 5.46 | 8.60 | $* A |
| AA496879 | RNP S1 | 1.58 | 1.70 | 2.43 | 1.18 | 1.02 | 0.47 | 7.72 | 7.05 | 4.26 | $* B |
| N26026 | Gemin 2 (SIP1) | 2.31 | 2.94 | 3.88 | 5.00 | 4.38 | 3.32 | * A | |||
| AA126911 | hnRNP A1 | 0.99 | 0.57 | 0.39 | 1.65 | 1.81 | 4.53 | ↓ | |||
| AA431440 | hnRNP-E2 | 0.81 | 0.64 | 0.61 | 0.83 | 1.09 | 2.01 | ↓ | |||
| T60163 | RNase L | 1.13 | 1.09 | 1.64 | 6.84 | 8.45 | 3.32 | * B | |||
| 5. Protein synthesis/modifications | |||||||||||
| R43973 | EF1γ | 2.23 | 1.22 | 0.54 | 1.69 | 1.30 | 3.83 | $ | |||
| R54097 | elF-2b | 0.91 | 0.63 | 0.50 | 0.55 | 0.98 | 2.15 | ↓ | |||
| AA873351 | RP L35a | 0.97 | 2.30 | 1.61 | 0.93 | 0.91 | 2.28 | 1.91 | 1.67 | 1.34 | $* D |
| T69468 | RPS4Y | 0.51 | 1.01 | 0.89 | 1.95 | 1.87 | 3.26 | ↓ | |||
| AA490011 | RPL38 | 1.43 | 0.92 | 0.22 | 0.99 | 1.40 | 2.97 | ↓ | |||
| T67270 | RPL10 | 1.97 | 1.29 | 0.61 | 1.22 | 1.31 | 2.75 | ↓ | |||
| AA464743 | RPL21 | 1.09 | 1.31 | 0.52 | 0.87 | 0.91 | 1.73 | ↓ | |||
| AA680244 | RPL11 | 1.01 | 1.18 | 0.25 | 0.84 | 1.18 | 2.94 | ↓ | |||
| W96450 | aa-tRNA synthetase FARSL | 1.10 | 1.67 | 1.46 | 1.97 | 1.84 | 1.36 | * F | |||
| AA599158 | aa-tRNA synthetase EPRS | 0.97 | 0.89 | 0.56 | 2.20 | 1.84 | 1.31 | ↓ F | |||
| AA664241 | α-NAC | 0.60 | 0.83 | 2.07 | 0.89 | 1.09 | 1.68 | $ | |||
| AA424786 | Golgin-95 | 0.90 | 0.99 | 0.57 | 10.02 | 19.14 | 2.95 | ↓ C | |||
| AA457114 | Protein B94 | 1.85 | 1.14 | 1.95 | 4.01 | 2.33 | 6.45 | * D | |||
| AA504455 | LDLC | 1.71 | 1.92 | 1.62 | 2.43 | 1.88 | 1.50 | * F | |||
| R78585 | Calumenin | 0.99 | 1.02 | 0.53 | 1.02 | 1.26 | 2.89 | 1.06 | 1.44 | 1.14 | ↓D |
| T71316 | ADP-ribosylation factor 4 | 1.25 | 0.79 | 0.48 | 0.94 | 1.05 | 3.12 | ↓ | |||
| AA455301 | Protein GPAA1 | 0.58 | 0.58 | 0.93 | 2.43 | 2.11 | 1.57 | ↓ B | |||
| N78843 | CYP-33 (PPIE) | 1.79 | 1.47 | 1.69 | 8.57 | 5.27 | 3.85 | * C | |||
| H98666 | PCOLN3 | 0.80 | 0.60 | 0.50 | 0.78 | 0.61 | 0.60 | $ | |||
| AA430524 | ACE | 1.17 | 1.70 | 1.99 | 1.78 | 2.85 | 1.66 | * A | |||
| AA410517 | Serpin PTI | 0.91 | 1.47 | 1.79 | 1.31 | 1.60 | 2.72 | $ | |||
| W61361 | Serpin CAP2 | 1.24 | 1.76 | 1.27 | 6.67 | 8.93 | 3.52 | * C | |||
| AA430512 | Serpin CAP3 | 1.22 | 1.53 | 1.69 | 6.45 | 5.37 | 4.20 | * B | |||
| AA402874 | Protective protein | 1.15 | 0.76 | 0.55 | 1.20 | 1.21 | 1.42 | ↓ D | |||
| 6 & 8. Metabolism/energy/oxidative stress | |||||||||||
| N33331 | PPARδ | 1.15 | 1.62 | 1.45 | 34.35 | 50.10 | 95.30 | * C | |||
| AA465366 | Leukotriene A4 hydrolase | 1.69 | 2.28 | 0.85 | 4.58 | 7.54 | 3.17 | * C | |||
| R55046 | MpV17 (peroxisome) | 0.75 | 0.60 | 0.57 | 1.30 | 1.69 | 2.18 | ↓ | |||
| W49667 | Fatty acid desaturase | 1.08 | 0.43 | 1.08 | 4.36 | 4.38 | 3.79 | ↓ A | |||
| W95082 | 11β-hydroxysteroid dehydrogenase | 1.07 | 0.80 | 0.45 | 0.45 | 0.82 | 0.44 | 16.25 | 5.88 | 3.48 | $ B |
| T73294 | P450 reductase | 1.78 | 1.05 | 2.01 | 0.80 | 0.71 | 2.69 | * | |||
| AA708298 | H+ ATP synthase | 1.20 | 1.02 | 0.49 | 1.11 | 1.62 | 5.38 | 1.85 | 1.46 | 1.30 | ↓D |
| H61243 | Uncoupling protein 2 | 0.93 | 0.82 | 0.53 | 1.07 | 0.93 | 1.81 | ↓ | |||
| W96179 | Glutamate-cysteine ligase | 2.67 | 2.03 | 3.18 | 6.69 | 4.97 | 4.54 | * A | |||
| AA463458 | Glutathione synthetase | 0.94 | 0.73 | 0.32 | 1.73 | 0.94 | 1.43 | ↓ | |||
| AA290738 | GSTM4 | 1.10 | 0.93 | 0.46 | 4.34 | 2.92 | 2.00 | ↓ B | |||
| R52548 | SOD-1 | 0.92 | 0.84 | 0.60 | 6.58 | 3.01 | 2.12 | ↓ B | |||
| R39463 | Aldolase C | 0.98 | 0.90 | 0.58 | 0.62 | 0.77 | 0.47 | $ | |||
| H05914 | LDHA | 0.84 | 1.27 | 0.59 | 1.15 | 1.12 | 2.68 | ↓ | |||
| AA629567 | HSP73 | 1.07 | 0.86 | 0.49 | 1.00 | 1.39 | 2.40 | ↓ | |||
| 7. DNA metabolism/chromatin structure | |||||||||||
| H15112 | Uracil-DNA glycosylase 1 | 0.45 | 0.58 | 1.64 | 0.85 | 1.36 | 1.70 | $ | |||
| N26769 | DNA glycosylase (MPG) | 0.89 | 0.94 | 0.55 | 5.88 | 3.06 | 1.70 | ↓B | |||
| AA608557 | XPE1 (DDB1) | 1.61 | 1.16 | 1.98 | 0.68 | 0.77 | 1.76 | * E | |||
| AA035095 | BCR protein 1 | 0.99 | 0.74 | 0.52 | 0.65 | 0.93 | 0.54 | $ | |||
| AA460927 | Translin | 0.53 | 1.29 | 2.49 | 0.99 | 1.51 | 2.37 | $ | |||
| AA442991 | Prothymosin α (PTMA) | 2.56 | 2.01 | 1.06 | 2.19 | 1.61 | 2.66 | $ | |||
| AA456077 | Centromere protein p27 | 0.86 | 0.72 | 0.44 | 0.62 | 0.88 | 0.41 | $ | |||
| R56871 | Chromatin assembly factor-I | 0.69 | 0.98 | 1.82 | 0.94 | 0.30 | 0.60 | ↓ | |||
| 9. Unclassified | |||||||||||
| AA683321 | PAR-5 | 1.72 | 1.27 | 1.38 | 2.26 | 2.62 | 8.38 | 0.44 | 0.72 | 1.61 | ↓ $ E |
| R06254 | Protein D54 | 1.62 | 0.92 | 1.68 | 1.26 | 1.29 | 1.87 | $ | |||
| AA406064 | BPY1 | 1.78 | 1.86 | 2.12 | 1.68 | 2.57 | 1.02 | * D | |||
| AA448289 | Protein D123 | 1.14 | 1.05 | 2.08 | 9.12 | 10.66 | 9.06 | * C | |||
| N34095 | FEZ2 | 1.14 | 1.55 | 1.69 | 2.86 | 3.20 | 3.70 | * A | |||
| R87497 | 2.19 gene | 1.85 | 2.70 | 1.71 | 4.32 | 3.50 | 5.08 | * A | |||
| AA452826 | Purkinje cell protein 4 | 0.60 | 0.80 | 0.59 | 5.26 | 3.82 | 3.32 | ↓ D | |||
Genes that responded to irradiation in both U87 and HEL cell lines in vitro or U87 grown in cell culture (U87-C) or transplanted in xenografts (U87-X) are shown. Genes distributed according cell functions. Numbers are average ratios of significant up- or down-regulation for each dose tested at 5 h after irradiation. $, consistent up- or down-regulation in both U87 and HEL in vitro;
, consistent up- or down-regulation in both U87 in vitro and U87 xenografts; ⇅, opposite response in either U87/HEL or U87-C/U87-X cell types; A–F, cluster of expression in U87 xenografts (see Fig. 1).
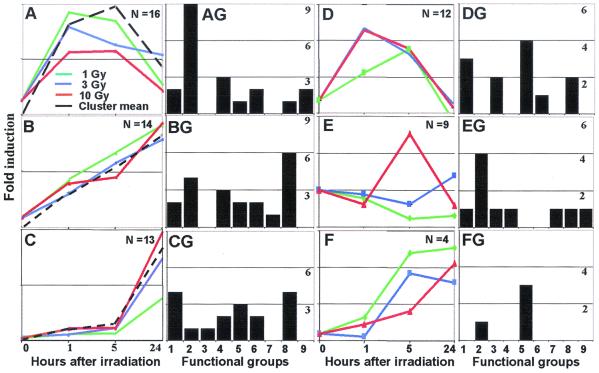
Earlier we showed that the effects of exposure to ionizing radiation between 1 and 10 Gy of cells in culture can be either dose dependent or independent. The U87 genes of cells grown in vitro or in transplanted xenografts and up-regulated by ionizing radiation were analyzed with respect to dependence on radiation dose and time of response. The results shown in Fig. 2 A–F were as follows. Fig. 2 A–D illustrates a subset of genes whose temporal response to irradiation was, for the most part, radiation dose-independent. Thus, the general pattern of response for each gene in these clusters was similar if not identical at 1, 3, and 10 Gy. Fig. 2 E and F illustrates genes whose temporal response to radiation was dose dependent. The number of genes in each cluster (N) is indicated in each panel. The genes illustrated in Fig. 2 are identified in Table 3.
Figure 2.
Representation of temporal patterns of gene expression after IR of U87 xenografts in mice. The xenografts were exposed to 1, 3, and 10 Gy and collected at 1, 5, or 24 h after irradiation. U87 genes that responded both in irradiated xenografts and in culture (Table 3) were grouped in six clusters (A–F). (AG–FG). The distribution of functional groups in each cluster. In clusters A–D, the temporal response was dose independent, although the magnitude of the response was partially dose dependent. Green, blue, and red corresponds to 1, 3, and 10 Gy, respectively. The black line shows the mean value for the entire cluster. Mean values of induction for each cluster are presented by gray lines and colored lines present examples of individual responses for glutamate-cysteine ligase (A), protein phosphatase 2A (B), and phospholemman chloride channel (C). In clusters E and F, the temporal pattern of gene response was dose dependent. Colored lines correspond to mean values of entire cluster at each dose tested. The genes are identified in Table 3. (AG–FG) A more restricted distribution based on 68 genes.
The Function of Genes Up-Regulated by Radiation.
We used a modified functional classification that was suggested by Stanton et al. (18). These groups are: (1) cell/organism defense and homeostasis; (2) cell–cell interactions and cell signaling; (3) cell cytoskeleton/motility/extracellular matrix; (4) RNA transcription processing/transport; (5) protein synthesis/modifications/transport; (6) metabolism/mitochondrion; (7) DNA metabolism/chromatin structure; (8) oxidative stress/apoptosis; and (9) unclassified. The distribution of responders by functional groups is shown in Table 3. A more restricted distribution based on a total of 68 genes is shown in Fig. 2 AG–FG. The significant data to come out of these analyses is that genes involved in cell-cell communication and signaling appear to be induced at relatively low IR levels. In contrast, genes involved in oxidative stress and apoptosis are more likely to be induced after irradiation with 3 or 10 Gy. Several groups were underrepresented, but the problem may well lie with the number of genes belonging to that group and that were included in the cDNA arrays.
Discussion
We have identified three sets of genes activated by ionizing radiation. The first set is shared by HEL fibroblasts and U87 malignant glioma cells grown in culture and harvested 5 h after irradiation with 1, 3, or 10 Gy. The second set is shared between U87 cells grown in vitro and those transplanted as xenografts in the hind limb of mice. The last and the smallest group are 15 genes induced in all irradiated cells whether grown in vitro or as mouse xenografts. Although analyses of the roles of products of the genes activated by irradiation will take considerable time and effort, the significance of certain aspects of the data are immediately apparent and are discussed below.
The response to IR consists of elements that are both cell common and cell-type specific. Because IR is administered to selectively destroy cancer cells, more detailed analyses will be necessary to determine whether U87 is representative of all malignant gliomas or is U87 specific. In the latter case, idealized treatment would require knowledge of the patterns of response of the tumor cell to irradiation.
Contrary to expectations, within the lethal range of IR administration, the response of a significant number of genes was dose dependent. As illustrated in this report in part in Table 1 and in Fig. 2 A–D, some genes were induced at low IR doses and some were induced only at high IR doses. This finding is in conflict with the prevailing notion that within certain parameters the sum total rather than individual doses predicts success of IR treatment.
Another finding of considerable interest is that in several instances the temporal pattern of gene expression was also dose dependent. The significance of this observation is difficult to assess given the small number of genes in the cDNA arrays and the fact that there were only three time points. Nevertheless, this phenomenon would not have been observed had the analyses been done at 24 h after irradiation. It is conceivable that the brief expression of certain genes may play a significant role in determining whether the cell survives or dies after irradiation.
The response of genes in xenografts was significantly higher than in cells grown in culture. The significance of this observation remains to be determined and may affect the design of future studies. It is of interest to note, however, that a fraction of genes listed in Table 3 were activated to a relatively high level. It would be of interest to determine whether the promoters of these genes are particularly sensitive to IR or whether they are induced by products of IR-inducible genes.
Current studies have identified several genes of particular interest that are inducible by IR. A few genes induced by IR in multiple systems analyzed in this study appear at first glance to be of particular interest. They are discussed below.
β2-Microglobulin is a common radiation responder (Table 3). Intracellular assembly of MHC class I heavy chains with β2-microglobulin occurs before the expression of the antigen-presenting complex on the cell surface. Treatment of human β2 microglobulin with hydroxyl radicals generated by treatment with γ radiation resulted in the disappearance of the Mr 12,000 protein and the appearance of a crosslinked complex stable under reducing conditions and in SDS (19). Augmentation of MHC class I/β2 microglobulin complexes by increasing doses of irradiation has been shown in short-term cultures established from eight human glioblastomas (29). Both MHC class I and β2-microglobulin genes were activated in the systems tested in this study. One hypothesis that could explain these results is that accelerated degradation of damaged or misfolded proteins was caused by IR.
Protein phosphatase 2A (PP2A) down-regulates the mitogen-activated protein kinase (MAPK) cascade, relays signals for cell proliferation, and appears to be linked to carcinogenesis. The PP2A holoenzyme exists in several trimeric forms consisting of a Mr 36,000 PP2A-C core catalytic subunit, Mr 65,000 structural/regulatory component, PP2A-A, and a variable regulatory subunit, PP2A-B, which confers distinct properties on the holoenzyme. Each subunit exists as multiple isoforms encoded by different genes. Consequently, the PP2A trimer exists in many configurations differing in expression pattern and specificity. The gene identified at 11q23 (20) and designated PPP2R1B encodes the structural-regulatory A subunit PP2A-A-β. This subunit is required for the interaction of the catalytic PP2A-C and variable PP2A-B subunits, and is critical for phosphatase activity. Recently it has been shown that PP2A is required for regulation of DNA-dependent protein kinase [DNA-PK (21)]. DNA-PK is a complex of DNA-PK catalytic subunit (DNA-PKcs) and the DNA end-binding Ku70/Ku80 heterodimer. DNA-PK is required for DNA double-strand break repair by the process of nonhomologous end joining. Nonhomologous end joining is a major mechanism for the repair of DNA double-strand breaks in mammalian cells. As such, DNA-PK plays essential roles in the cellular response to IR and in V(D)J recombination. In vitro, DNA-PK phosphorylation of all three protein subunits (DNA-PK cs, Ku70, and Ku80) inactivates the serine/threonine protein kinase activity of DNA-PK. Phosphorylation-induced loss of the protein kinase activity of DNA-PK was restored by the addition of the purified catalytic subunit of either protein phosphatase 1 or PP2A. Reversible protein phosphorylation is an important mechanism for the regulation of DNA-PK activity, and the protein phosphatase responsible for reactivation in vivo is a PP2A-like phosphatase.
The up-regulation by IR of several genes classified in the RNA splicing/nuclear cytoplasmic RNA transport functional group was unexpected. Two genes, the survival of motor neuron (SMN) interacting protein 1 (SIP-1 or Gemin 2) and U1 snRNP70, both belong to cluster A (Fig. 2 and Table 3). SIP-1 interacts with SMN and is involved in the assembly/metabolism of small nuclear ribonucleoproteins (snRNPs), as well as their nuclear-cytoplasmic transport (22). Also, RNPS1, in cluster C (Fig. 1 and Table 3) was a recently described general activator of pre-mRNA splicing (23). In addition, both hnRNPA1 and hnRNPE2 heterogeneous nuclear ribonucleoproteins are up-regulated after IR in both U87 and HEL cell lines (Table 3). hnRNPs mediate several RNA-related functions, including pre-mRNA splicing and mature mRNA transport to the cytoplasm. hnRNPA1 was recently isolated among 12 other hypoxia-responsive genes from cervical cancer cells, and proteomics analyses identified RNA-binding motifs containing proteins, mostly involved in RNA splicing, as major caspase-3 targets during the Fas-induced apoptosis in T cells (24). These data suggest that pathways of nuclear pre-mRNA processing and nuclear/cytoplasmic transport of RNA may be activated by IR, and may provide potential therapeutic targets.
Transcriptional activation of actin genes by IR was reported by Woloschuk et al. (25). The results reported here indicate that actin α2 and β-actin were induced in all cells subjected to IR and were co-clustered (see Fig. 1, cluster D, and Table 3). These genes are frequently classified as housekeeping genes expressed in mock-treated and stressed cells. A more likely explanation consistent with other data is that different components of the cytoskeleton may be specifically involved in the stress response and may be transcriptionally controlled through p53-dependent mechanisms (8).
The Cyp33 gene belongs to cluster C, which includes the most highly up-regulated in vivo genes. The Mr 33,000 CYP33 protein exhibits RNA-binding, peptidylprolyl cis–trans isomerase (PPIase), and protein-folding activities. CYP33 is, to our knowledge, the first example of a protein that combines RNA-binding and PPIase activities. An identical transcript was detected in a small cell lung cancer cell line (26). Recent reports indicate that Cyp33 is involved in regulation of mixed lineage leukemia (MLL)-1 (27, 28). Overexpression of the CYP33 protein in leukemia cells results in altered expression of HOX genes that are targets for regulation by MLL. These alterations are suppressed by cyclosporin and are not observed in cell lines that express a mutant MLL protein. These results suggest that binding of CYP33 to MLL modulates its effects on the expression of target genes.
Several genes associated with the endoplasmic reticulum and secretory pathway were up-regulated by IR (calumenin, golgin-95 and low-density lipoprotein receptor defect C) (see Table 3). Members of the CREC family localize to the secretory pathway of mammalian cells and include reticulocalbin, ERC-55/TCBP–49/E6BP, Cab45, calumenin, and crocalbin/CBP-50 (29). Calumenin, a calcium-binding protein, is related to the CREC family of proteins. Recent reports indicate that some CREC family members are involved in pathological activities such as malignant cell transformation, mediation of the toxic effects of snake venom toxins, and putative participation in amyloid formation. Functional significance of response of endoplasmic reticulum and Golgi-related genes to IR is so far unclear.
To properly assess the function of the responders in the environment of the irradiated cell, it will be necessary to modify or suppress the expression of the up-regulated genes. This approach may ultimately shed light on the development of resistance to IR.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA71933 and CA78766) and the United States Public Health Service.
Abbreviations
- HEL
human embryonic lung
- IR
ionizing radiation
References
- 1.Hall E J. Radiobiology for the Radiologist. 5th Ed. Philadelphia: Lippincott; 2000. pp. 5–17. [Google Scholar]
- 2.Vijayakumar S, Hellman S. Lancet. 1997;349:1–30. doi: 10.1016/s0140-6736(97)90010-6. [DOI] [PubMed] [Google Scholar]
- 3.Takanami T, Mori A, Takahashi H, Higashitani A. Nucleic Acids Res. 2000;28:4232–4236. doi: 10.1093/nar/28.21.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez B S. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallahan D E, Spriggs D R, Beckett M A, Kufe D W, Weichselbaum R R. Proc Natl Acad Sci USA. 1989;86:10104–10107. doi: 10.1073/pnas.86.24.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witte L, Fuks Z, Haimovitz-Friedman A, Vlodavsky I, Goodman D S, Eldor A. Cancer Res. 1989;49:5066–5072. [PubMed] [Google Scholar]
- 7.Komarova E A, Diatchenko L, Rokhlin O W, Hill J E, Wang Z J, Krivokrysenko V I, Feinstein E, Gudkov A V. Oncogene. 1998;17:1089–1096. doi: 10.1038/sj.onc.1202303. [DOI] [PubMed] [Google Scholar]
- 8.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman W H, Tom E, Mack D H, Levine A J. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 9.Amundson S A, Bittner M, Chen Y, Trent J, Meltzer P, Fornace A J., Jr Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 10.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. . (First Published April 17, 2001; 10.1073/pnas.091062498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulz C A, Harari P M, Mehta M P. Curr Oncol Rep. 2001;3:179–184. doi: 10.1007/s11912-001-0019-2. [DOI] [PubMed] [Google Scholar]
- 12.Crellin R P, Macmillan C H, Berridge J K, Morgan D A. Clin Oncol. 1993;5:5139–5142. doi: 10.1016/s0936-6555(05)80309-7. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka Y, Murley J S, Patel R, Grdina D J. Int J Radiat Biol. 2000;76:633–639. doi: 10.1080/095530000138295. [DOI] [PubMed] [Google Scholar]
- 14.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley J D, Kataoka Y, Advani S, Chung S M, Arani R B, Gillespie G Y, Whitley R J, Markert J M, Roizman B, Weichselbaum R R. Clin Cancer Res. 1999;5:1517–1522. [PubMed] [Google Scholar]
- 16.Khodarev N N, Advani S J, Gupta N, Roizman B, Weichselbaum R R. Proc Natl Acad Sci USA. 1999;96:12062–12067. doi: 10.1073/pnas.96.21.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman W M, Robertson D J, Vrana K E. BioTechniques. 2000;29:1042–1046. doi: 10.2144/00295rv01. [DOI] [PubMed] [Google Scholar]
- 18.Stanton L W, Garrard L J, Damm D, Garrick B L, Lam A, Kapoun A M, Zheng Q, Protter A A, Schreiner G F, White R T. Circ Res. 2000;86:939–945. doi: 10.1161/01.res.86.9.939. [DOI] [PubMed] [Google Scholar]
- 19.Capeillere-Blandin C, Delaveau T, Descamps-Latscha B. Biochem J. 1991;277:175–182. doi: 10.1042/bj2770175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S S, Esplin E D, Li J L, Huang L, Gazdar A, Minna J, Evans G A. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 21.Douglas P, Moorhead G B, Ye R, Lees-Miller S P. J Biol Chem. 2001;276:18992–18998. doi: 10.1074/jbc.M011703200. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Dreyfuss G. J Biol Chem. 2001;276:9599–9605. doi: 10.1074/jbc.M009162200. [DOI] [PubMed] [Google Scholar]
- 23.Mayeda A, Badolato J, Kobayashi R, Zhang M Q, Gardiner E M, Krainer A R. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiede B, Dimmler C, Siejak F, Rudel T. J Biol Chem. 2001;276:26044–26050. doi: 10.1074/jbc.M101062200. [DOI] [PubMed] [Google Scholar]
- 25.Woloschak G E, Chang-Liu C M. Int J Radiat Biol. 1991;59:1173–1183. doi: 10.1080/09553009114551051. [DOI] [PubMed] [Google Scholar]
- 26.Kim J O, Nau M M, Allikian K A, Makela T P, Alitalo K, Johnson B E, Kelley M J. Oncogene. 1998;17:1019–1026. doi: 10.1038/sj.onc.1202006. [DOI] [PubMed] [Google Scholar]
- 27.Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz M O. Mol Cell Biol. 2001;21:3589–3597. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honore B, Vorum H. FEBS Lett. 2000;466:11–18. doi: 10.1016/s0014-5793(99)01780-9. [DOI] [PubMed] [Google Scholar]
- 29.Klein B, Loven D, Lurie H, Rakowsky E, Nyska A, Levin I, Klein T. J Neurosurg. 1994;80:1074–1077. doi: 10.3171/jns.1994.80.6.1074. [DOI] [PubMed] [Google Scholar]