Abstract
Species classification is challenging when taxa display limited morphological differences. In this paper, we combined morphology and DNA barcode data to investigate the complicated taxonomy of two Onychiurid Collembolan species. Thalassaphorura thalassophila and Thalassaphorura debilis are among the most common arthropod species in intertidal ecosystems and are often considered to be synonymous. Based on morphological and barcode analyses of fresh material collected in their type localities, we redescribed and compared the two species. However, their morphological distinctiveness was supported by a molecular divergence much smaller than previously reported at the interspecific level among Collembola. This divergence was even smaller than inter-population divergences recognized in the related edaphic species T. zschokkei, as well as those known between MOTUs within many Collembolan species. Our results may indicate a link between low genetic interspecific divergence and intertidal habitat, as the only biological peculiarity of the two species of interest compared to other Collembolan species analyzed to date is their strict intertidal life.
Keywords: Barcode, Genetic divergence, Intertidal ecology, Synonymy, Taxonomy, Thalassaphorura debilis, Thalassaphorura thalassophila
Introduction
The intertidal zone, a narrow littoral strip between the low and high tide marks (Mouritsen & Poulin, 2002; Raffaelli & Hawkins, 2012), is a critical interface between terrestrial and aquatic ecosystems (Raffaelli & Hawkins, 2012). It is characterized by daily cycles of submersion and exposure due to tidal movements. Environmental conditions in this ecosystem are therefore very predictable but extremely variable within a day. Many groups of marine origin, as well as some of terrestrial origin, include organisms that are well adapted to these harsh environmental conditions.
Springtails (Collembola) are the most abundant and often the most diversified hexapods in the intertidal environment (Deharveng, 2004; Joosse, 1976), where they are often found in very large numbers. This has been shown for Anurida maritima (Laboulbène, 1865) and several species of Thalassaphorura Bagnall, 1949 (Christiansen & Bellinger, 1988; Willem, 1925; Witteveen & Joosse, 1988). The genus Thalassaphorura is diverse and widely distributed. The taxonomic history of its intertidal species, traced in detail in Bellinger et al. (2015), is complex. Bagnall (1949) described the genus with Onychiurus thalassophilus Bagnall, 1937, as the type species. A few species were subsequently described in or assigned to Thalassaphorura (Fjellberg, 1998; Pomorski, 1998), and various combinations and synonyms have been proposed (Bellinger et al., 2015). More than 60 species have been assigned to the genus until now (Bellinger, Christiansen & Janssens, 1996–2018), but the taxonomic status of several species, including the intertidal species of interesthere, remains uncertain (Stach, 1954; Kaprus & Paśnik, 2017; Sun, Bedos & Deharveng, 2017). Currently, 57 valid species are recognized in the genus (Kaprus & Paśnik, 2017; Sun, Bedos & Deharveng, 2017), nine of which are halobionts or restricted to the intertidal zone (Arbea, 2017). Two of these intertidal species, namely, Thalassaphorura debilis and Thalassaphorura thalassophila, are widespread in the northern hemisphere. The intertidal ecology of these two species is well known (Moniez, 1890; Willem, 1925) compared to that of other species of the genus. Despite their unique habitat, the morphology of these species is similar to that of the non-intertidal species in the genus (Sun, Chen & Deharveng, 2010), which live in litter and soil.
Due to different placements and synonymies, the taxonomic status of the two species has been confused for a long time. T. debilis was described as Lipura debilis Moniez, 1890 and T. thalassophila as Onychiurus thalassophilus in 1937. The latter was collected from intertidal habitats in Scotland and was described as a species of the “debilis” group, differing from others by its vestigial unguiculus (Bagnall, 1937). Afterwards, it was assigned as a type species of the genus Thalassaphorura by Bagnall (1949). The generic assignation of the species was subsequently much debated. It was placed in different genera, such as Onychiurus Gervais, 1841 by Stach (1954), Spelaphorura Bagnall, 1948 by Salmon (1959), and Protaphorura Absolon, 1901 by Gisin (1960) and Hopkin (1997), and then moved back to the genus Thalassaphorura by Pomorski (1998). The old species L. debilis Moniez, 1890 was assigned to Onychiurus by Bagnall (1935), Christiansen & Bellinger (1988), Denis (1923) and Willem (1925), or to Protaphorura by Hopkin (1997) and Jordana et al. (1997). Fjellberg (1998) synonymized the two species after studying the type specimens of T. thalassophila and assuming that T. debilis is a morphologically variable species. However, re-examination of the type material and detailed studies of fresh specimens from type localities revealed consistent differences among the two species (Sun, Chen & Deharveng, 2010).
The confusing taxonomy is due to insufficient detail in the earliest descriptions of the species, unjustified synonymies, the low number of distinguishing taxonomic characters and the lack of information on intraspecific variability within the species. The characters used in the taxonomy of Thalassaphorura are as follows: the number of pseudocelli on the head, body and legs; the number of papillae of sensory organ of antennal III segment; the relative length of unguiculus; the length of anal spines; the number of chaetae in distal whorl of tibiotarsi; and the morphology and number of S-chaetae on the head and body (Sun, Bedos & Deharveng, 2017). Several of these characters are known to exhibit intra-specific polymorphism. This taxonomic uncertainty hampers meaningful studies on intertidal communities of the western Palearctic seashores, where both species are among the dominant arthropods.
In an attempt to clarify the taxonomic status of these species, we combine detailed morphological and barcode analyses of the type populations of T. debilis and T. thalassophila. In the Collembola, DNA barcoding has been used to complement morphological characters to allow species characterization in several genera, including Deutonura (Porco, Bedos & Deharveng, 2010), Heteromurus (Lukić et al., 2015), Homidia (Pan, 2015), Lepidobrya (Zhang, Greenslade & Stevens, 2017), Protaphorura (Sun et al., 2017), and Tomocerus (Zhang et al., 2014; Yu, Ding & Ma, 2017). DNA-based approaches are regarded as powerful tools for species delimitation, especially in groups of closely related species with uncertain taxonomic status (Hebert et al., 2003). Although various molecular markers have been employed at the species level, a 658-base fragment of the mitochondrial gene cytochrome c oxidase I (COI), which is widely used for barcoding animals (Hajibabaei et al., 2007), has been effective in most zoological groups, including birds (Hebert et al., 2004), fish (Ward et al., 2005), cowries (Meyer & Paulay, 2005), spiders (Barrett & Hebert, 2005), and Lepidoptera (Hajibabaei et al., 2006).
Large divergences (>5%) in DNA barcode sequences provide strong support for the taxonomic separation of two putative species (Hebert et al., 2003). However, the extent of divergence between congeneric species varies among invertebrate groups (Hebert, Ratnasingham & De Waard, 2003). Insects usually have lower interspecific divergences than non-winged arthropods. For example, average DNA barcode distances between congeneric species range from 7 to 8% in holarctic Lepidoptera (Hebert & Landry, 2010; Hausmann et al., 2011) and 9.3% in Diptera (Hebert, Ratnasingham & De Waard, 2003), to 11.5% in Hymenoptera and 13.9% in North America Ephemeroptera (Webb et al., 2012). In contrast, Collembola shows much higher divergence in COI sequences between congeneric species (Porco et al., 2012a; Yu et al., 2016), with reported values ranging from 16.35 to 24.55% (Table 1). These values are similar to divergence levels between congeneric species of other non-winged soil invertebrates, such as Scolopendromorpha (13.7–22.2% in Wesener et al., 2016) or Lithobiomorpha (13.7–24.5% in Stoev et al., 2013). Furthermore, recent molecular studies on divergences within Collembolan species have revealed divergences almost as deep as among congeneric morphological species (Cicconardi, Fanciulli & Emerson, 2013; Emerson et al., 2011; Frati et al., 2000; Katz, Giordano & Soto-Adames, 2015; Porco et al., 2012b; Soto-Adames, 2002).
Table 1. Sequence divergence at COI among Collembola for congeneric species pairs, after literature and the present work.
| Reference | Family | Genus | Number of species | Mean divergence (%) |
|---|---|---|---|---|
| This work | Onychiuridae | Thalassaphorura debilis & thalassophila | 2 | 4.3 |
| Katz, Giordano & Soto-Adames (2015) | Entomobryidae | Entomobrya | 11 | 17.83 |
| Porco et al. (2012a) | Entomobryidae | Heteromurus | 2 | 23.02 |
| Pan (2015) | Entomobryidae | Homidia | 2 | 18 |
| Porco et al. (2012a) | Hypogastruridae | Ceratophysella | 4 | 22.66 |
| Hogg & Hebert (2004) | Isotomidae | Folsomia | 4 | 17 |
| Porco et al. (2012b)* | Isotomidae | Parisotoma | 3 | 24.55 |
| Porco et al. (2012a) | Neanuridae | Bilobella | 2 | 23.19 |
| Deharveng et al. (2015) | Neanuridae | Deutonura | 4 | 18.95 |
| Porco et al. (2010) | Neanuridae | Deutonura | 5 | 20.25 |
| Porco et al. (2012a) | Neanuridae | Deutonura | 4 | 23.24 |
| Sun et al. (2017)** | Onychiuridae | Protaphorura | 13 | 16.35 |
| This work*** | Onychiuridae | Thalassaphorura | 7 | 19.4 |
| Hogg & Hebert (2004) | Sminthuridae | Sminthurides | 2 | 21 |
| Porco et al. (2012a) | Tomoceridae | Tomocerus | 3 | 19.60 |
| Yu et al. (2016) | Tomoceridae | Tomocerus | 2 | 20.4 |
| Yu, Ding & Ma (2017) | Tomoceridae | Tomocerus | 6 | 18.66 |
Notes:
Recalculated, Parisotoma notabilis excluded.
Recalculated, the MOTUs which could not be separated by morphological characters excluded.
Divergence between T. debilis and T. thalassophila excluded.
In this paper, we (i) re-describe and compare the two species T. debilis and T. thalassophila based on fresh specimens from their type localities, (ii) evaluate the congruence between DNA barcode and morphological data for the delimitation of the two species, and (iii) relate the unusually low genetic divergence with respect to clear morphological differences in the broader taxonomic and ecological context.
Material and Methods
Sampling
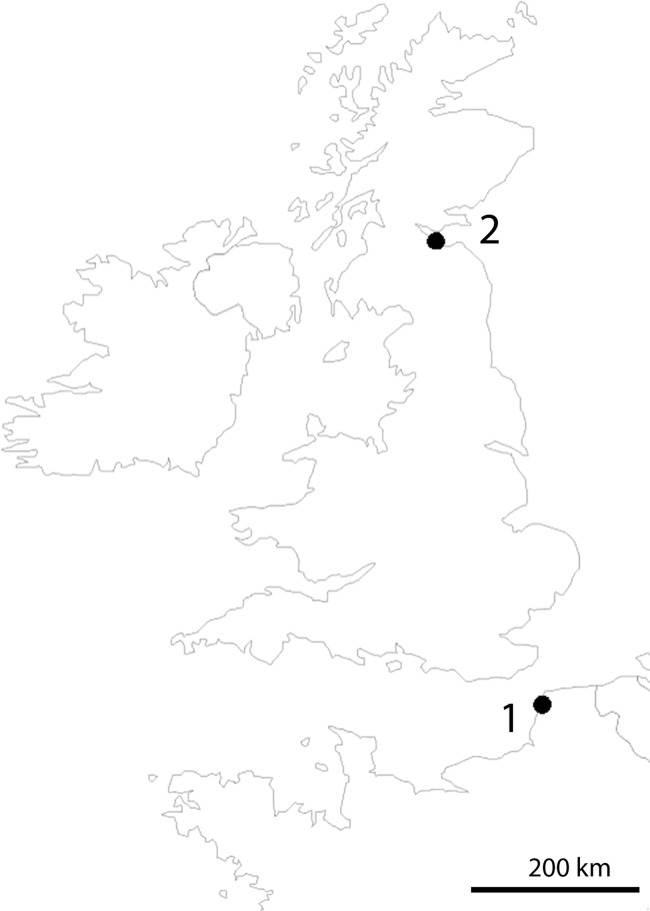
Sampling was done along the shores of Dalmeny in Scotland (type locality of T. thalassophila) and Pointe-aux-Oies in northwestern France (type locality of T. debilis) (Fig. 1). Both species were collected in the intertidal zone, where they lived in dense populations, in habitats characterized by very weak slope, rocky substrate, and abundant algae and barnacles on rocks. Specimens were picked up directly from under stones at low tide with a brush, or at the surface of the water after washing of gravels and stones in a plastic basin. Only T. thalassophila was present in the Dalmeny site, while the species co-occurred with T. debilis at Pointe-aux-Oies.
Figure 1. Location of sampling sites.
(1) Pointe-aux-Oies in France. (2) Dalmeny in Scotland.
DNA extraction and sequencing
We successfully barcoded 41 specimens, including 26 T. debilis and 15 T. thalassophila, from northwest France and Scotland, and 31 specimens belonging to five additional species (Table S1), in order to illustrate the interspecific divergence among non-marine species of the same genus. The species T. zschokkei was represented by three populations totaling 11 specimens, which were analyzed to evaluate between-populations of genetic divergence in a non-marine species living in mountain soils and mosses. Extraction and sequencing were done at the Biodiversity Institute of Ontario, University of Guelph (ON, Canada). DNA was extracted from entire specimens in 30 mL of lysis buffer and proteinase K incubated at 56 °C overnight. DNA extraction followed a standard automated protocol using 96-well glass fiber plates (Ivanova, Dewaard & Hebert, 2006). Specimens were recovered after DNA extraction for further morphological examination according to the workflow detailed in (Porco et al., 2010). The 5’ region of COI, including 658 bp used as a standard DNA barcode, was amplified using M13 tailed primers LCO1490 and HCO2198 (Folmer et al., 1994). Samples that failed to generate an amplicon were subsequently amplified with a pair of internal primers combined with full-length ones, LepF1-MLepR1 and MLepF1- LepR1 (Hajibabaei et al., 2006). A standard PCR reaction protocol was used for amplification, and products were checked on a 2% E-gel 96 Agarose (Invitrogen, Guelph, Canada). Unpurified PCR amplicons were sequenced in both directions using M13 tailed primers (Hajibabaei et al., 2005), with products subsequently purified using Agencourt CleanSEQ protocol and processed using BigDye ver. 3.1 on an ABI 3730 DNA Analyzer (Applied Biosystems, Guelph, Canada). Sequences were assembled with Sequencer 4.5 (Gene Code Corporation, Ann Arbor, MI, USA) and aligned by eye using BIOEDIT ver. 7.0.5.3 (Hall, 1999). As we observed no indels in the COI sequences, sequence alignment was unambiguous. Sequences are publicly available on BOLD (Table S1).
Data analyses
The K2P distances (Kimura, 1980) and the Neighbor-Joining tree (Saitou & Nei, 1987) were calculated in MEGA7 (Kumar, Stecher & Tamura, 2016) with 1,000 pseudo replicates and pairwise deletion and other parameters as the defaults. The frequency of K2P distances was graphed in R 3.3.2. Divergence time was estimated using *BEAST (Heled & Drummond, 2010). Specimens were assigned to species a priori by the results of above species delimitations. An uncorrelated lognormal relaxed clock was selected for each partition, the GTR+G+I for substitution mode and the Yule process for speciation priors. In the absence of available fossil calibrations in Collembola, the substitution rate (3.36% pairwise divergence per Mya) estimated by Papadopoulou, Anastasiou & Vogler (2010) was employed. An Markov Chain Monte Carlo (MCMC) chain was executed twice for 10 million generations with a sample frequency of 1,000 and the initial 5,000 generations discarded as burn-in. The effective sample size (ESS) values and convergence were checked in Tracer v1.6 (Rambaut, Suchard & Drummond, 2014).
Microscopic examination
A total of 61 specimens (30 T. debilis and 31 T. thalassophila) preserved in 95% ethanol and 25 skins retrieved following DNA extraction (16 T. debilis and nine T. thalassophila) were mounted on slides in a Marc André II solution, after clearing in lactic acid. Six type specimens (the lectotype and two paralectotypes of T. debilis and three syntypes of T. thalassophila) were examined. Photos of specimens in alcohol were taken with a Jenoptik ProgRes C10+ camera mounted on a Leica MZ16. Slides were examined with a Leica DMLB microscope with DIC. A drawing was made through a Camera lucida and improved with Photoshop Elements 9.
Terminology and abbreviations
Chaetotaxy of the labium, anal valves, and furca remnant is applied according to Fjellberg (1999), Yoshii (1996) and Weiner (1996), respectively. Tibiotarsal chaetotaxy is presented after Deharveng (1983) and is expressed as the total number of chaetae (number of chaetae in whorls A+T, B, and C, respectively). The unguiculus/unguis ratio is given according to the length of the medial line of unguiculus and the length of the inner edge of the unguis. The formulae of pseudocelli and pseudopores are presented as the number per half-tergum/sternum from head to Abd. V.
AIIIO—sensory organ of Ant. III, Abd.—abdominal segment, Ant.—antennal segment, AS—anal spine, ms—S-microchaeta, PAO—postantennal organ, pso—pseudocellus, psp—pseudopore, psx—parapseudocellus, Th.—thoracic segment, x—ventro-axial psp of Abd. IV.
Results
Family Onychiuridae Börner, 1913
Genus Thalassaphorura Bagnall, 1949
Type species: O. thalassophilus Bagnall, 1937 (Scotland)
Remarks on synonymies among halophilous species:
In his reference book on Onychiuridae, Stach (1954: 73) stated that “The synonymy of the species L. debilis Moniez, 1890 is very complicated.” Although he introduced all the forms of T. debilis that had been validly described in his key, he expressed doubt regarding the proposed synonymies and stressed that all species “should be exactly examined.” In this group with many closely related species, and in full agreement with Stach’s idea, we do not accept most synonymies that have been perpetuated in the literature, as they are not supported by explicit morphological comparisons. The only exception is the synonymy T. thalassophila = T. debilis proposed by Fjellberg (1998); however, this proposal is challenged in the present paper on combined morphological and molecular grounds. The synonymies that have to be re-assessed are the following:
-
*
Onychiurus imminutus Bagnall, 1937 is considered a synonym of Spelaphorura thalassophila by Salmon (1959: 149), based on the examination of types, but without clear justification. As the two species were collected in the same locality and are very similar, their synonymy is possible.
-
*
Onychiurus littoralis Dürkop, 1935 is considered a synonym of O. debilis by Bagnall (1937: 90, 145), without justification.
-
*
Onychiurus litoreus Folsom, 1917 is considered a synonym of O. debilis by Denis (1923: 216). This synonymy is challenged by Stach (1954: 74), and the species is listed as valid by Christiansen & Bellinger (1998: 463) under the name O. (Protaphorura) litoreus, but without discussion of its possible synonymy.
-
*
Aphorura neglecta Schaeffer, 1896 is considered a synonym of O. debilis by Denis (1931: 209), but not by Stach (1954).
Thalassaphorura debilis (Moniez, 1890)
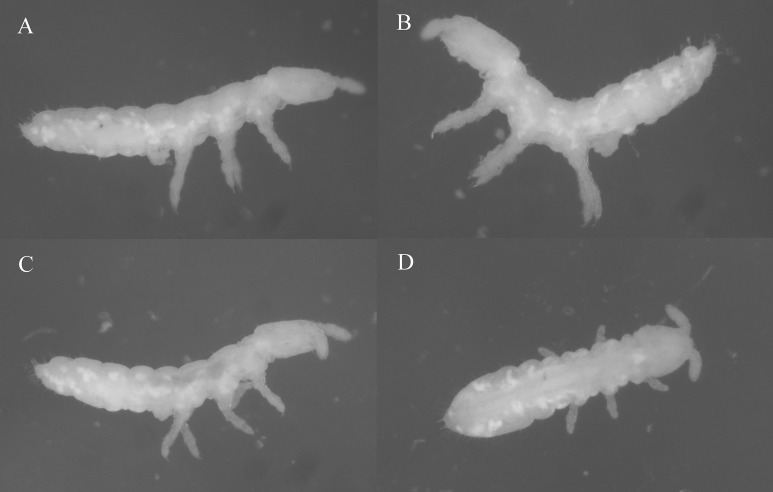
Figure 2. Species habitus in ethanol.
(A, B) Thalassaphorura debilis (Moniez, 1890). (C, D) Thalassaphorura thalassophila (Bagnall, 1937). Photos by L. Deharveng & A. Bedos.
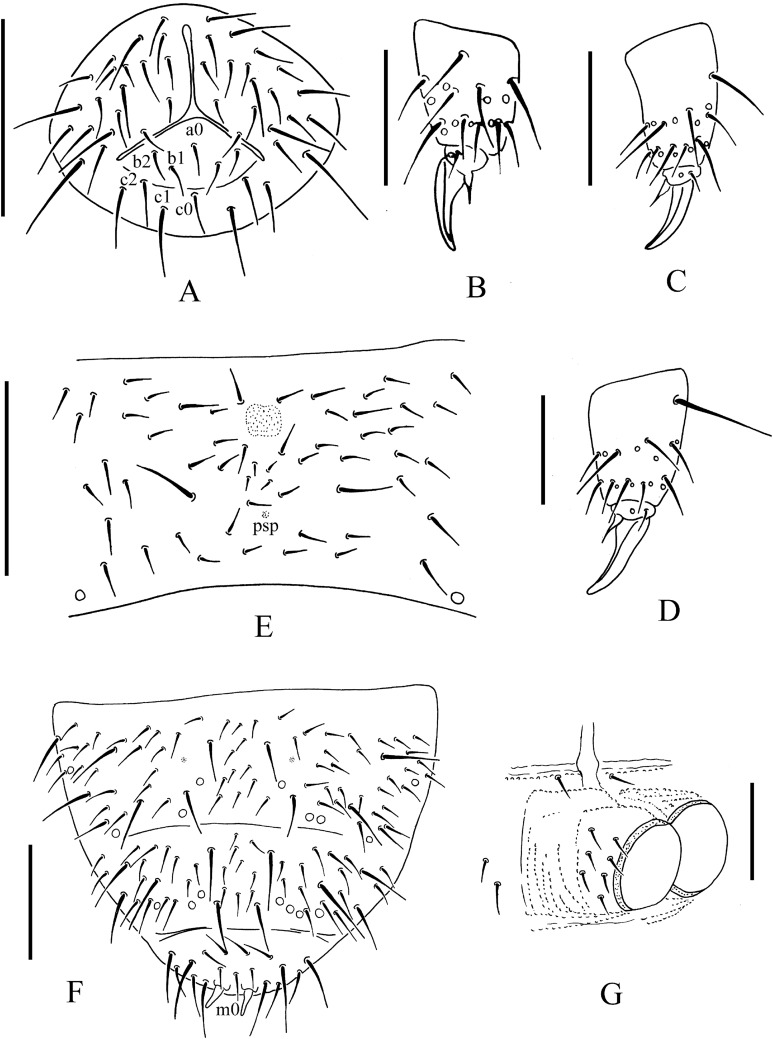
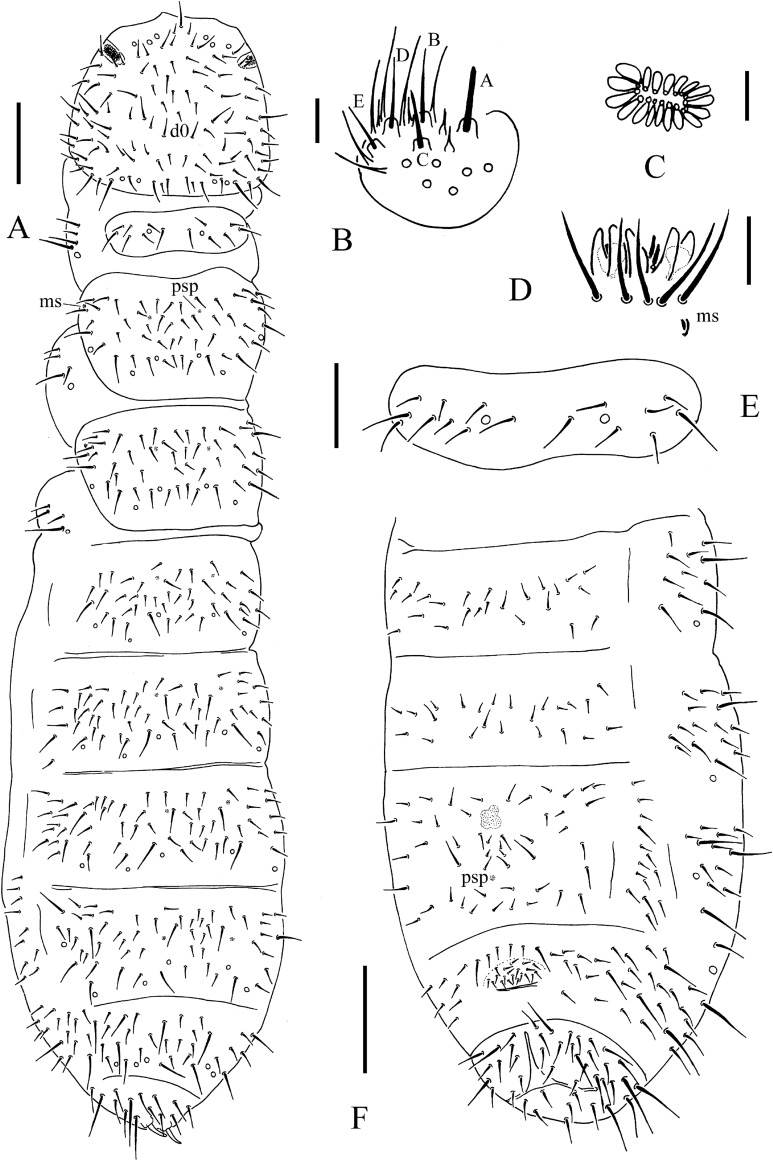
Figure 4. T. debilis.
(A) Anal valves. (B) Tibiotarsal chaetotaxy and claw of leg I. (C) Tibiotarsal chaetotaxy and claw of leg II. (D) Tibiotarsal chaetotaxy and claw of leg III. (E) Abd. IV sternum. (F) Abd. IV–VI terga. (G) Ventral tube. Scales: 0.1 mm (A, E, F), 0.05 mm (B, C, D, G).
Table 2. Comparison of the main diagnostic characters of T. debilis and T. thalassophila from different references.
| Source | Current conception | Current conception | Moniez (1890) (types) | Denis (1923) (types) | Willem (1925) | Jordana et al. (1997) | Fjellberg (1998) | Bagnall (1937) |
|---|---|---|---|---|---|---|---|---|
| Cited as | T. debilis | T. thalassophila | Lipura debilis | Onychiurus debilis | Onychiurus debilis | Protaphorura debilis | Thalassaphorura debilis | Onychiurus thalassophilus |
| Current name | T. debilis | T. thalassophila | T. debilis | T. debilis | T. debilis | T. thalassophila | T. thalassophila / debilis | T. thalassophila |
| Length (mm) | Female 1.4–2.1, male 1.4–1.65 | Female 1.32–1.93, male 1.20–1.66 | 1.1–1.2 | <1.5 | 0.95 | 1.5 | 1.4 | 1.5 |
| PAO | 13–21 | 16–23 | 23–28 | 19–20 | 17 | 18–19 | 15–20 | 16–20 |
| Dorsal pso formula | 32/1-233/3,3-4,3,4-6,3-4 | 32/133/33343 | ?2/2??/????4 | 32/1≥2≥3/≥2≥2≥2≥23 | 32/133/33354 | 32/133/33343 | 32/133/33343 | 32/133/33343 |
| Ventral pso formula | 11/000/0111(2)0 | 11/000/00000 | ? | ? | ? | 11/000/00000 | 11/000/00000 | 11/000/00000 |
| pso on subcoxae I-III | 222 | 111 | 222 | ? | 111 | 111 | 111 | 111 |
| Axial chaetae on Abd.VI | m0 | m0 (a0) | ? | ? | ? | m0 | 1 or 2 | a0 |
| Ratio of AS/clawIII | 0.47–0.77 | 0.68–1.08 | 0.5–0.6 | ≥0.5 | 0.6 | 0.7 | Variable in size | 0.62–0.86 |
| Head ventral chaetae along groove | 4+4 | 3+3 | ? | ? | ? | ? | 4–5+4–5 | 3+3 |
| Chaetae on ventral tube (anterior /distal /basal chaetae) | 1+1/7+7(8)/2+2 | 1+1/7+7/2+2 | ? | ? | ? | 1+1/7–8+7–8/2+2 | 1+1(2)/7–8+7–8/1–4+1–4 | 1+1/7+7/2+2 |
| Ratio of unguiculus/unguis | 0.27–0.47 | 0.2 | 0.3–0.4 | 0.4 | 0.4 | Short, ≤0.25 after original drawing | Variable in size, mostly 0.5 | Vestigial, reduced to a minute, stumpy process |
| Chaetae on subcoxae 1 of legs I-III | 4–5, 4–5, 4–5 | 444 | 4/4–5/? | ? | ? | 4/4/? | ? | 4(3)44 |
| Location | France: Pointe-aux-oies | France: Pointe-aux-oies; Scotland: Dalmeny | France: Pointe-aux-oies | France: Pointe-aux-oies | France: Pointe-aux-oies | Spain: Pontevedra coast | Norwegian and Danish coast | Scotland: Dalmeny |
Lipura debilis Moniez, 1890: 346
Aphorura neglecta Schaeffer, 1896: 112 after Denis (1931: 209, syn. dub.)
Onychiurus litoreus Folsom, 1917: 644 after Denis (1931: 209, syn. dub.) and Stach (1954: 74, syn. dub.)
Onychiurus debilis in Denis (1923: 216, redescription from syntypes)
Onychiurus debilis in Willem (1925: 279, redescription from specimens of the type locality)
Onychiurus littoralis Dürköp 1935: 133 after Bagnall (1937: 90, 145, syn. dub.)
Onychiurus debilis in Stach (1954: 73)
Handschiniella debilis in Salmon (1964: 162)
Onychiurus (Protaphorura) debilis in Bolger (1986: 193)
Jailolaphorura debilis in Weiner (1996: 178)
Protaphorura debilis in Skidmore (1995: 53)
Thalassaphorura debilis in Fjellberg (1998: 109)
Thalassaphorura debilis in Sun, Chen & Deharveng (2010: 24)
Material examined: Type material (examined). Denis (1923) listed eight specimens of “Onychiurus debilis” in Moniez’s collection. Only five were retrieved in the MNHN collection. Lectotype female and two paralectotype females on slides. Label, probably re-written by Denis, as «Coll. Moniez. Pointe-aux-Oies. 2.9.89». Two paralectotypes on slides (one female, one of undetermined sex). Label, probably re-written by Denis, as «Sous les Fucus. Pointe-aux-Oies. 1.9.89».
Non-type material from the type locality. France: Pas-de-Calais: Wimereux: Pointe-aux-Oies (1.361623°E, 50.463582°N), March 17, 2010, by hand and by washing of algae and sand, Sun Xin, Bedos A., Deharveng L. and Zon S. leg. (62-016, three males, three females, one juvenile on slides, including the skin of one male recovered after DNA extraction); same data (62-018, three males, one juvenile on slides, including the skins of one male and one juvenile recovered after DNA extraction). Ibid, August 5, 2010, by hand and by washing of algae and sand, Sun Xin leg. (62-044, three males, three females, two unsexed specimens, all on slides as skins of barcoded specimens recovered after DNA extraction); same data (62-045, five males, 12 females, two juveniles on slides, including the skins of one male, three females and one juvenile recovered after DNA extraction).
Redescription: Color: white. Length (without antennae): female 1.4–2.1 mm, male 1.4–1.65 mm. Body shape: cylindrical, slender, elongated, parallel-sided, with Abd. VI arched and anal spines 0.47–0.77 times as long as the inner edge of hind unguis (Figs. 2A, 2B and 3A). Granulation of body surface: regular, with more or less distinctly thinner granules on intersegment areas.
Figure 3. T. debilis.
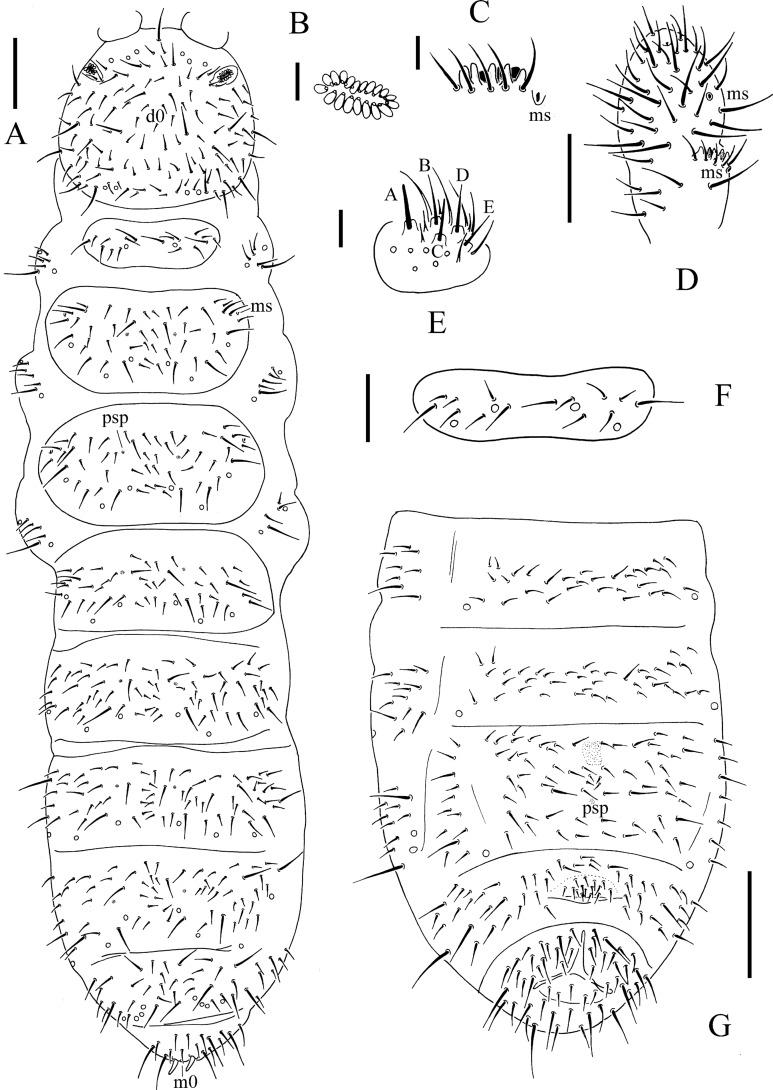
(A) Habitus, pseudopores and dorsal chaetotaxy of head and body. (B) Postantennal organ. (C) Ant. III sensory organ. (D) Antennal segments III and IV. (E) Labium. (F) Th. I tergum. (G) Abdominal II–VI sterna. Scales: 0.1 mm (A, G), 0.05 mm (D, F), 0.01 mm (B, C, E).
Pseudocelli is 32/1-233/3, 3-4, 3, 4-6, 3-4 dorsally, 11/000/0111(2)0 ventrally and 2/2/2 on subcoxae I–III (Figs. 3A, 3G and 4F). Parapseudocelli is absent. Pseudopores is 00/011/11110 dorsally, 00/111/000x0 ventrally (Figs. 3A, 3G and 4F).
S-chaetae not distinguishable from ordinary chaetae. S-microchaetae tiny and blunt, as 0/011/000000 dorsally (Fig. 3A).
The antennal basal area is not well delimited by granulation. The antennae are approximately 1.1 times as long as head. The length ratio of antennal segments I:II:III:IV is approximately 1.0:1.5:1.5:2.2. The antennal segment IV has subapical organite and basoexternal ms at approximately 1/3 length from the base (Fig. 3D). The Ant. III sensory organ is composed of five papillae, five guard chaetae, two small sensory rods and two smooth sense clubs (Fig. 3C). Ant. III has external ms just behind sensory organ (Fig. 3C). Ant. II has 13 chaetae. Ant. I has nine chaetae.
Postantennal organ is composed of 13–21 (16.0 ± 1.8 from 49 PAO) simple vesicles arranged in two rows along the axis of the organ (Fig. 3B). Dorsal cephalic chaeta d0 is present (Fig. 3A). 3+3 chaetae appear between two inner posterior pso, and p1 is anterior to others (Fig. 3A). The mandible has a strong molar plate and four apical teeth. The maxilla bears three teeth and six lamellae but is not examined in detail. The maxillary palp is simple with one basal chaeta and two sublobal hairs. The labral chaetae are 4/1, 4, 2. The labial papillae of AC type, papillae A–E are with one, four, zero, three and two guard chaetae, respectively (Fig. 3E). The labium has six proximal, four (E, F, G, and f) basomedial and six (a, b, c, d, e, e’) basolateral chaetae. Postlabial chaetae are 4+4 along the ventral groove.
The ordinary chaetae were differentiated in macro- and meso-chaetae. Th. I has 7–9+7–9 dorsal chaetae with frequent asymmetries (Figs. 3A and 3F). Th. II–III has 4–5+4–5 dorsal chaetae and Abd. I–III has 3–4+3–4 dorsal chaetae along the axial line, usually symmetrically arranged but with differences between specimens. Abd. IV–V has dorsal chaetae asymmetrically arranged along the axis; Abd. VI with m0 (Figs. 3A and 4F). Th. I–III has 1+1, 1+1 and 1+1 ventral chaetae, respectively, between the coxae.
Subcoxa 1 has 4–5, 4–5, 4–5 chaetae, and subcoxa 2 has 1, 4, 4 chaetae on legs I–III, respectively (Fig. 3A). Tibiotarsal chaetae are 18 (9, 8, 1), 18 (9, 8, 1) and 18 (9, 8, 1) chaetae on legs I–III, respectively (Figs. 4B–4D). The unguis is without teeth. The unguiculus is short, only 0.27–0.47 times as long as the inner edge the of unguis, with inner basal lamella (Figs. 4B–4D). The ventral tube has 1+1 anterior chaetae, 7+7 (rarely 7+8) distal chaetae and 2+2 basal chaetae (Fig. 4G). The furca was reduced to a finely granulated area, with four small chaetae in two rows posterior to the furcal rudiment (Figs. 3G and 4E).
The genital plate consists of 15–18 chaetae in female (Fig. 3G), 35–50 in male. The anal valves have numerous acuminate chaetae; each lateral valve with chaetae a0 and 2a1; upper valve with chaetae a0, 2b1, 2b2, c0, 2c1, 2c2 (Fig. 4A).
Habitats: On the seashore, among Fucus and barnacles or under stones in the intertidal zone.
Remarks: The type material of T. debilis was in bad condition and only a few characters could be validated, i.e., the number of pso on Th. I tergum (2) and subcoxae I–III (2, 2, 2), the ratio of unguis/unguis (0.27–0.35), and the ratio of AS/unguis (0.55–0.57).
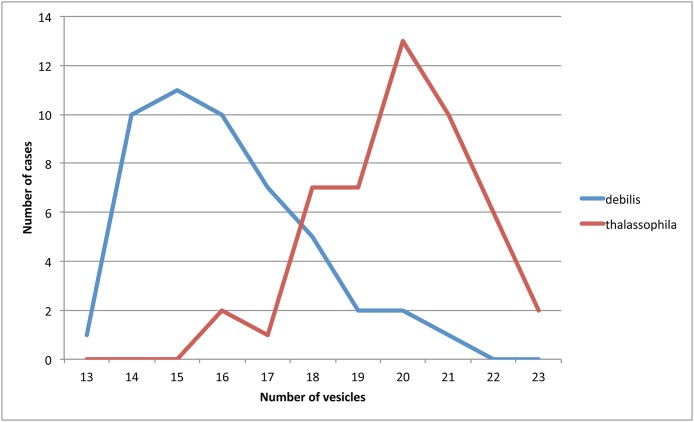
In the original description of the species by Moniez (1890), the figure of the unguiculus corresponds to T. debilis, as redefined here (approximately 1/3 of claw length), as does the number of 23–28 vesicles in the PAO given in the text. The number of PAO vesicles in the Moniez’ paratypes examined was not observable, but descriptions of the species by Denis (1923) based on eight syntypes of Moniez and by Willem (1925) and based on specimens from the type locality (Pointe-aux-Oies) state the number of vesicles as 20 and 17, respectively, which corresponds well with this redescription (Table 2). In the type locality, we found T. debilis was mixed with T. thalassophila, but in higher number. Therefore, it is possible that Moniez in 1890 included both species and described the unguiculus of a T. debilis and the PAO of a T. thalassophila.
Some characters of T. debilis are variable, especially the number of vesicles in the PAO (13–21), the number of dorsal pso (32/1-233/3, 3-4, 3, 4-6, 3-4), the number of pso on Abd. IV sternite (1 or 2), the length of unguiculus (0.27–0.47 times as long as the inner edge of unguis) and the number of chaetae on subcoxa 1 of legs (4–5). However, the length of unguiculus (short but clearly longer than that of T. thalassophila) and the presence of pseudocelli on the abdominal sterna allow separation of T. debilis from T. thalassophila. Fjellberg (1998) emphasized the former character in his work but apparently did not consider it as having a taxonomic value.
Thalassaphorura thalassophila (Bagnall, 1937)
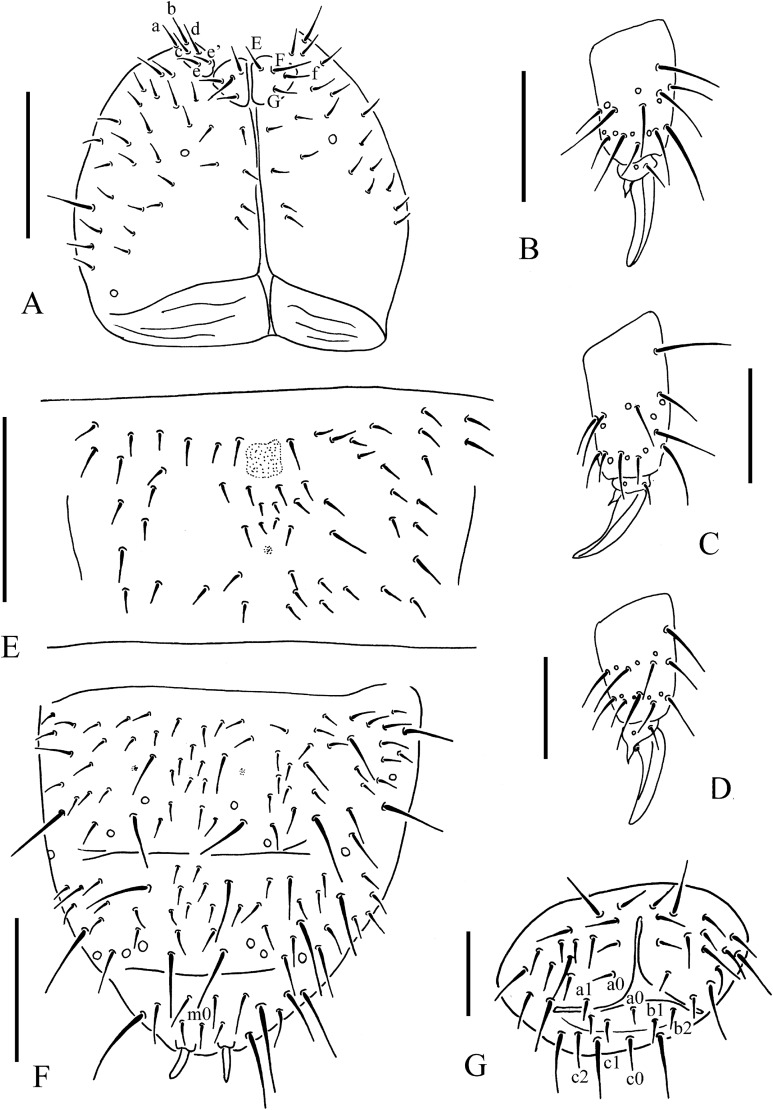
Figure 5. T. thalassophila.
(A) Habitus, pseudopores and dorsal chaetotaxy of head and body. (B) Labium. (C) Postantennal organ. (D) Ant. III sensory organ. (E) Th. I tergum. (F) Abd. II–VI sterna. Scales: 0.1 mm (A, F), 0.05 mm (E), 0.01 mm (B, C, D).
Figure 6. T. thalassophila.
(A) Ventral side of head. (B) Tibiotarsal chaetotaxy and claw of leg I. (C) Tibiotarsal chaetotaxy and claw of leg III. (D) Tibiotarsal chaetotaxy and claw of leg III (type material). (E) Abd. IV sternum. (F) Abd. IV–VI terga. (G) Anal valves. Scales: 0.1 mm (A, E, F), 0.05 mm (B, C, D, G).
Onychiurus thalassophilus Bagnall (1937: 146)
Onychiurus imminutus Bagnall, 1937: 146 after Salmon (1959: 149, syn. dub.)
Thalassaphorura thalassophila in Bagnall (1949: 504)
Onychiurus thalassophilus in Stach (1954: 44)
Spelaphorura thalassophilus (sic) in Salmon (1959: 149)
Protaphorura debilis in Jordana et al. (1997: 571)
Thalassaphorura thalassophila in Pomorski (1998: 135)
Thalassaphorura debilis in Fjellberg (1998: 109) (synonymy not accepted here)
Material examined: Type material (examined). Three female syntypes of the Bagnall type series. Great Britain, Scotland: Dalmeny Estate shore, well below the high-water mark, 12.V.35 (deposited in The Natural History Museum, London).
Non-type material examined. Great Britain, Scotland: Dalmeny Estate shore (3.310991°E, 55.983110°N), April 05, 2016, by hand and by washing of algae and sand, Sun Xin, Bedos A. and Deharveng L. (GB-011, four males, 11 females and one unsexed specimen on slides, including the skins of one male, two females and one unsexed specimen recovered after DNA extraction). France: Pas-de-Calais: Wimereux: Pointe-aux-Oies (1.361623°E, 50.463582°N), March 17, 2010, by hand and by washing of algae and sand, Sun Xin, Bedos A., Deharveng L. and Zon S. leg. (62-016, two males and one female on slides, including the skin of one female recovered after DNA extraction); same data (62-017, one male, one female and one unsexed specimen on slides, including the skins of one male and one unsexed specimen recovered after DNA extraction). Ibid, August 5, 2010, by hand and by washing of algae and sand, Sun Xin leg. (62-044, the skin on slide of one female recovered after DNA extraction); same data (62-045, three males, three females and three juveniles on slides, including the skin of one juvenile recovered after DNA extraction).
Redescription: Color: white. Length (without antennae): female 1.32–1.93 mm; male 1.20–1.66 mm. Body shape: cylindrical, slender, elongated, parallel-sided, with Abd. VI arched and anal spines 0.68–1.08 times as long as the inner edge of hind unguis (Figs. 2C, 2D and 5A). Granulation of body surface: regular, with more or less distinctly thinner granules on intersegment areas.
Pseudocelli as 32/133/33343 dorsally, 11/000/00000 ventrally and 1/1/1 on subcoxae I–III (Figs. 5A, 5F, 6A and 6F). Parapseudocelli absent. Pseudopores as 00/011/11110 dorsally, 00/111/000x0 ventrally (Figs. 5A, 5F, 6A and 6F).
The S-chaetae is not distinguishable from ordinary chaetae. The S-microchaetae is tiny and blunt, as 0/011/000000 dorsally (Fig. 5F).
The antennal basal area is not well delimited by granulation. The antennae are as long as the head. The length ratio of antennal segments I:II:III:IV is approximately 1.0:1.2:1.2:1.8. The antennal segment IV with subapical organite and basoexternal ms is at approximately 1/3 length from the base. The Ant. III sensory organ is composed of five papillae, five guard chaetae, two small rods and two smooth clubs (Fig. 5D). Antennal segment III has external ms just behind sensory organ (Fig. 5D). Ant. II has 13 chaetae. Ant. I has nine chaetae.
The PAO is composed of 16–23 (19.9 ± 1.7 from 48 PAO) simple vesicles arranged in two rows along the axis of the organ (Fig. 5C). The dorsal cephalic chaeta d0 is present (Fig. 5A). 3+3 chaetae appear between two inner posterior pso, while p1 is anterior to others (Fig. 5A). The mandible has a strong molar plate and four apical teeth. The maxilla bears three teeth and six lamellae but was not examined in detail. The maxillary palp is simple with one basal chaeta and two sublobal hairs. The labral chaetae are 4/1, 4, 2. The labial papillae are of AC type, papillae A–E with one, four, zero, three and two guard chaetae, respectively (Fig. 5B). The labium has six proximal, four (E, F, G, and F) basomedial and six (a, b, c, d, e, e’) basolateral chaetae (Fig. 6A). The postlabial chaetae are 4+4 along the ventral groove.
Ordinary chaetae were differentiated in macro- and meso-chaetae. Th. I has 6–7+6–7 dorsal chaetae (frequent asymmetries) (Figs. 5A and 5E). Th. II–Abd. III has 3–4+3–4 dorsal chaetae along the axial line, usually symmetrically arranged but with differences between specimens. Abd. IV–V has dorsal chaetae asymmetrically arranged along the axis. Abd. VI has m0 and sometimes a0 present (Figs. 5A and 6F). Th. I–III has 1+1, 1+1 and 1+1 ventral chaetae, respectively, between coxae.
Subcoxa 1 has 4, 4, 4 chaetae, and subcoxa 2 has 1, 4, 4 chaetae on legs I–III, respectively (Fig. 5A). Tibiotarsal chaetae has 18 (9, 8, 1), 18 (9, 8, 1) and 18 (9, 8, 1) on legs I–III, respectively (Figs. 6B–6D). The unguis is without teeth. The unguiculus very short, reduced to a minute, stumpy process and is 0.2 times as long as the inner edge of the unguis, with inner basal lamella (Figs. 6B–6D). The ventral tube has 1+1 anterior chaetae, 7+7 distal chaetae, and 2+2 basal chaetae. The furca is reduced to a finely granulated area, with four small chaetae in two rows posterior to the furcal rudiment (Figs. 5F and 6E).
The genital plate consists of 18–21 chaetae in females (Fig. 5F), and 40–42 in males. The anal valves have numerous acuminate chaetae; each lateral valve has chaetae a0 and 2a1; the upper valve has chaetae a0, 2b1, 2b2, c0, 2c1, 2c2 (Fig. 6G).
Habitats: Similar to T. debilis, on the seashore, among Fucus and barnacles or under stones in the intertidal zone.
Remarks: T. thalassophila is very similar to T. debilis by its habitus, non-differentiated dorsal S-chaetae, and short unguiculus. However, it can be easily distinguished by several characters (Table 2): it has shorter unguiculus, reduced to a minute and stumpy process; the papillae of AIIIO are longer and slender; there are usually more vesicles in PAO (Fig. 7) there are no pso on the abdominal sterna; there are fewer chaetae on the subcoxae; and the AS is usually longer. We did not find significant intra-specific variations in the pso formula, and the size of the unguiculus among the studied specimens of T. thalassophila is contrary to those of T. debilis. P. debilis as redescribed by Jordana et al. (1997: 571) on Spanish material is probably T. thalassophila according to the diagnostic characters, except for the number of PAO vesicles, which could correspond to another species.
Figure 7. Number of observations (ordinates) for different numbers of PAO vesicles (abscissa) in T. debilis and T. thalassophila.
Overall, T. debilis and T. thalassophila represent two species that are closely related but morphologically clearly distinct based on standards of modern Onychiuridae taxonomy (Pomorski, 1998). Therefore, the two taxa are not synonymous as proposed by Fjellberg (1998: 109) (the author described differences in unguiculus size between the two species, but did not consider them to be sufficient for separating the species).
Barcode characterization of the two species
In total, 16 (62% of barcoded specimens of T. debilis) and nine (60% of barcoded specimens of T. thalassophila) individuals were examined for morphological diagnostic characters after DNA extraction (Table S1). The remaining specimens were damaged during DNA extraction and were therefore morphologically uninformative.
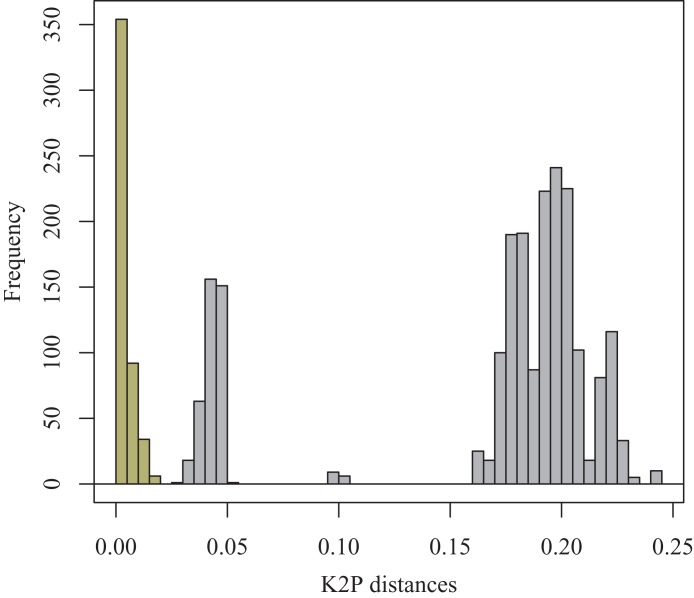
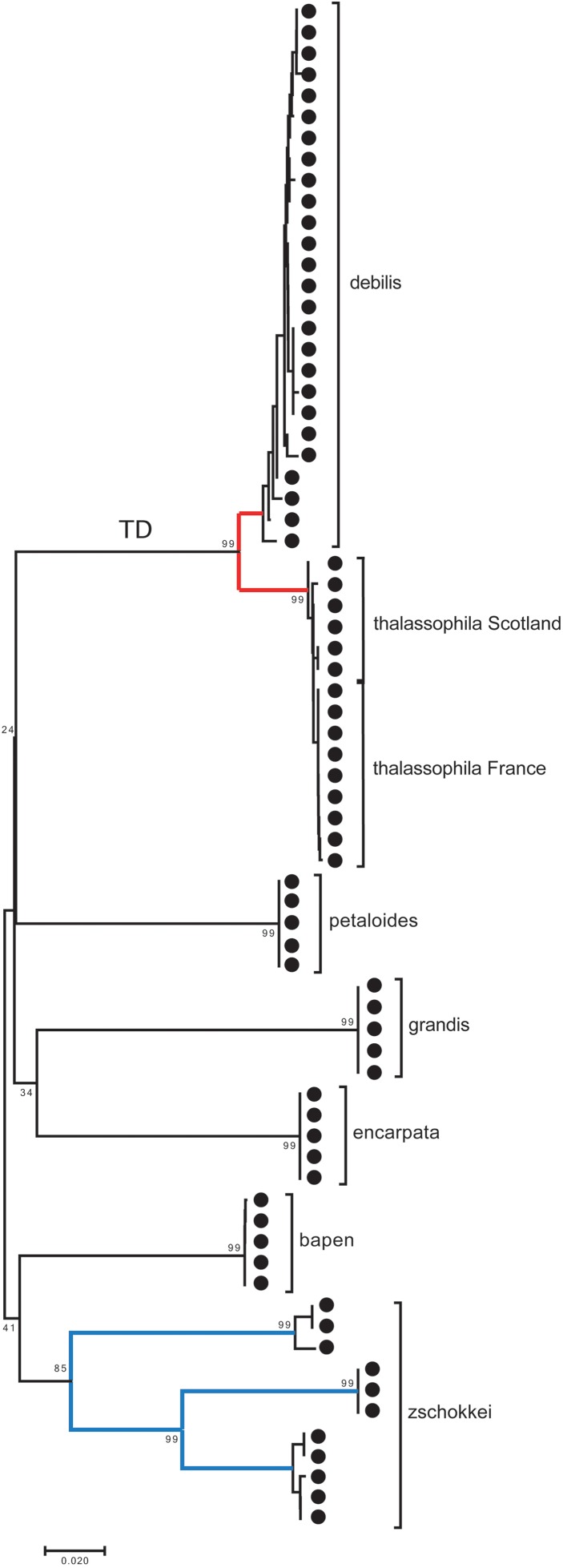
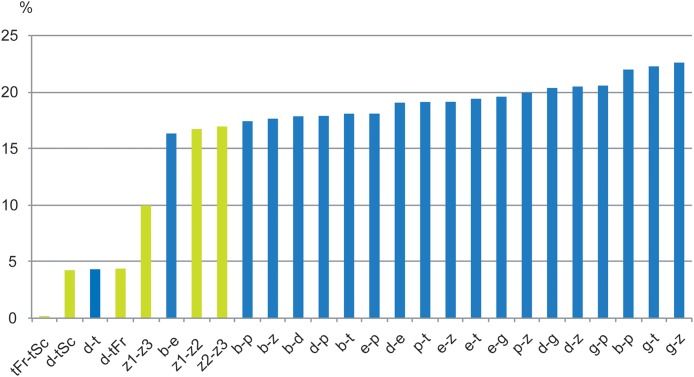
A small barcoding gap was observed at K2P distances of approximately 0.02 (Fig. 8). The two species T. debilis and T. thalassophila are clearly characterized by their barcode (Fig. 9), with a small inter-specific divergence of 4.3% and intra-specific divergence of 0.49% (0–1.9%) in T. debilis and 0.16% (0–0.3%) in T. thalassophila (Fig. S1; Tables 3 and 4). The two populations of T. thalassophila (France and Scotland) show a very low divergence (0.03%). The non-intertidal species of Thalassaphorura exhibited much higher values of inter-specific divergence (from 16.3% between T. bapen and T. encarpata to 22.6% between T. grandis and T. zschokkei), and very low intra-specific divergence, except in T. zschokkei (10.28%), which is split in well-separated MOTUs that are morphologically indistinguishable (Tables 3 and 4). Divergence time estimation indicated that the speciation event of the two species T. debilis and T. thalassophila occurred at 1.66 (0.47–3.14) Mya (Fig. S2).
Figure 8. Frequency histogram of K2P pairwise distances.
Columns of the intra-specific divergences are greenish-yellow colored.
Figure 9. Neighbor-joining tree (K2P) based on COI for the seven Thalassaphorura species, including three clusters of T. zschokkei.
The numbers at MOTU nodes are bootstrap values above 80% (1,000 replicates). TD: the branch of Thalassaphorura debilis and T. thalassophila.
Table 3. Intraspecific and intra-MOTUs divergence within the genus Thalassaphorura.
| Species | Intraspecific divergence |
|---|---|
| Thalassaphorura grandis | 0 |
| Thalassaphorura debilis | 0.004900574 |
| Thalassaphorura thalassophila | 0.001593864 |
| Thalassaphorura petaloides | 0 |
| Thalassaphorura bapen | 0 |
| Thalassaphorura zschokkei | 0.102789204 |
| Thalassaphorura encarpata | 0 |
Table 4. Molecular divergence (COI) between Thalassaphorura species (A), between populations of the T. debilis–T. thalassophila group (B), and between three populations of T. zschokkei (C).
| A | ||||||
|---|---|---|---|---|---|---|
| Species | bapen | debilis | encarpata | grandis | petaloides | thalassophila |
| debilis | 0.179 | |||||
| encarpata | 0.163 | 0.191 | ||||
| grandis | 0.220 | 0.204 | 0.196 | |||
| petaloides | 0.174 | 0.179 | 0.181 | 0.206 | ||
| thalassophila | 0.181 | 0.043 | 0.194 | 0.223 | 0.191 | |
| zschokkei | 0.176 | 0.205 | 0.191 | 0.226 | 0.199 | 0.205 |
Note:
FR, France; SC, Scotland.
Discussion
In the present study, we used specimens from the type localities of T. debilis and T. thalassophila, as the state and age of the type material on slides that precluded extraction of reliable genetic material. The combined genetic and geographic pattern of the three analyzed populations (T. debilis, T. thalassophila France and T. thalassophila Scotland) can be summarized as follows (Figs. 9 and 10; Table 3): (i) moderate but clear molecular divergence between T. debilis (France) and T. thalassophila (France and Scotland); (ii) very low molecular divergence between T. thalassophila from France and T. thalassophila from Scotland in spite of the geographic distance between them; and (iii) co-occurrence in syntopy of T. debilis and T. thalassophila in France.
Figure 10. Histograms of COI divergence in % between species, MOTUs of Thalassaphorura.
In green, between species and populations of the intertidal species debilis—thalassophila, and between three MOTUs of the edaphic species T. zschokkei; in blue, between edaphic species of the genus, and between them and the two intertidal species. Study sites: b, bapen; d, debilis; e, encarpata; g, grandis; p, petaloides, t, thalassophila; z, zschokkei (with three MOTUs: −1, −2, −3); Fr, France; Sc, Scotland.
Morphology and genetic data were congruent in support for the species status of both taxa. However, the low level of genetic divergence between T. debilis and T. thalassophila was unusual when compared to genetic differences usually observed between congeneric species of Collembola (Tables 3 and 4). Low genetic divergence associated with clear morphological differences is reported here for the first time in Collembola (Porco et al., 2012a, 2013). In Deutonura zana Deharveng, Zoughailech, Hamra-Kroua & Porco (2015), for instance, two populations geographically separated and genetically divergent at 3.7% did not reveal any morphological difference despite a thorough examination (Deharveng et al., 2015).
For other species within the genus Thalassaphorura, the interspecific divergences we measured were in line with the high values observed for other Collembola, ranging from 16.4% to 22.6% between all couples of the five non-marine species, as well as between these species and each intertidal species (Table 4). The low divergence between T. debilis and T. thalassophila was more similar to that among many winged arthropods and lower than that among three populations of closely related, morphologically indistinguishable non-marine species (Figs. 9 and 10; Table 4). This unusual pattern may be the result of our failure to detect discriminant morphological characters between populations of this last species. It also reflects different paces of morphological and molecular diversification among the Thalassaphorura species, which would potentially impact our understanding of intra- versus inter-specific variations among Collembola. Biologists using approaches for MOTU delimitations based on a barcode gap approach, e.g., ABGD (Puillandre et al., 2012), or on the use of a threshold derived from empirical data should be aware of such cases that may cause underestimation of actual diversity, as some species get overlooked.
The frequency of occurrence of the observed patterns is unknown and its origin obscure. It is probably not linked to phylogeny, as other Thalassaphorura species (Table 4) have divergences similar to other Collembolan genera. Furthermore, the estimated divergence time (0.47–3.14 Mya) between the two species is small compared to other species, suggesting that T. debilis and T. thalassophila could be two young sister species. The calibration method applied here is not optimal, as it is based, in the absence of biogeographically informative pattern, on Tenebrionidae beetles which probably have a much longer life cycle than Thalassaphorura. However, the T. zschokkei populations as well as other species of the genus analyzed here would have diverged much earlier. Therefore, the inference of a lower evolutionary pace of the T. debilis–T. thalassophila lineage cannot be ruled out. Because of these uncertainties, as well as the sympatric occurrence of the two species, the time of divergence for the two species cannot reliably be inferred.
High divergence in COI sequences between geographically distant MOTUs of the same morphological species is frequent in Collembola (Porco et al., 2013), especially among non-widespread species. This is illustrated in the dataset analyzed by Porco et al. (2012a), where populations of several species drawn from various Collembolan families were represented by MOTUs, which diverged from conspecific MOTUs by 11.33–21.47% (with less than 2% intra-population divergence), matching, in most cases, the levels of divergence observed between congeneric species of Collembola. This may indicate the presence of yet unrecognized species, especially where the different MOTUs were found in sympatry. However, in several cases, such as for Bilobella aurantiaca (Caroli, 1912), thorough morphological analysis did not reveal morphological differences between conspecific MOTUs. We observed similarly high levels of divergence without morphological differentiation between three MOTUs of the non-marine species T. zschokkei (Fig. 10; Table 4), which were from populations 40–85 km apart and spread across the Southern Alps. Conversely, the two populations of T. thalassophila studied were 660 km apart (Fig. 1), but did not show genetic divergence at COI, which is similar to divergences often observed among widely distributed species that are suspected to be dispersed by humans (Porco et al., 2013). The common assumption is that marine currents might be a powerful dispersal agent for flightless littoral arthropods (Hawes et al., 2008; Witteveen & Joosse, 1988), maintaining gene flow and explaining the very low genetic differentiation observed between populations. However, the link between wide distribution with efficient dispersal by ocean currents and low genetic divergence among populations is yet to be clearly documented for intertidal species.
The co-occurrence of two closely related species in the same microhabitat without apparent niche or trait differentiation is unusual. The two species are similar, and their minor morphological differences are probably not ecologically significant. Co-occurrences of genetically closely related and morphologically highly similar species are unknown among Collembola. When co-occurrences of morphological similar species have been reported, the taxonomic status of the species was uncertain, their microhabitat was slightly different (Rusek, 2007), or their distribution only overlapped in a narrow strip in a contact zone between parapatric forms (Deharveng, Bedos & Gisclard, 1998). Therefore, the co-existence of the morphologically similar T. debilis and T. thalassophila in the same habitats should be further investigated.
The only evident biological feature that strongly separates our two species from non-marine Thalassaphorura is their peculiar intertidal ecology, as stressed above. Whether the debilis/thalassophila case is representative of genetic patterns associated with this environment will have to be investigated in other Collembola. However, aside from the intertidal species group of Anurida maritima, very few genera or species groups are known to involve marine and non-marine species and to encompass closely related intertidal forms.
Supplemental Information
Acknowledgments
We thank Serge Zon from the Cocody University (Abidjan, Ivory Coast) for his help in field sampling; Wanda Maria Weiner from the Polish Academy of Sciences (Krakow) for helpful advice on the species taxonomy; Paul Brown from The Natural History Museum, London for the loan of the type material of T. thalassophila; David Porco from the Musée National d’Histoire Naturelle, Luxembourg; Marianne Elias and Rodolphe Rougerie from the Muséum National d’Histoire Naturelle, Paris; Feng Zhang from Nanjing Agricultural University for useful comments during the preparation of the manuscript; and Gunnar Keppel from University of South Australia, Andrew Davis from University of Goettingen for the language modification.
Funding Statement
The present study was supported by funding from the National Natural Science Foundation of China (Grant No. 41571052, 41430857), Science and Technology Development Plan Project of Jilin Province (20160520051JH), the Postdoctoral Science Foundation of China (Grant No. 2015M570281), the Excellent Young Scholars of Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences (Grant No. DLSYQ13003), the Alexander von Humboldt Foundation, Youth Innovation Promotion Association, CAS, the European project EDIT and the Parc National du Mercantour. There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Xin Sun conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Anne Bedos performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Louis Deharveng conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The sequences described here are accessible via BOLD sequence numbers which have been provided in the Supplemental Table.
Data Deposition
The following information was supplied regarding data availability:
The raw data for the main diagnostic characters of the two species are provided as a Supplemental File.
References
- Arbea (2017).Arbea I. Una nueva especie litoral de Thalassaphorura Bagnall, 1949 (Collembola: Onychiuridae) de Pontevedra, noroeste de la Península Ibérica. Arquivos Entomolóxicos. 2017;17:321–328. [Google Scholar]
- Bagnall (1935).Bagnall RS. Contributions towards a knowledge of the Scottish Onychiuridae (Collembola), I. Scottish Naturalist. 1935;214:111–117. [Google Scholar]
- Bagnall (1937).Bagnall RS. Contributions towards a knowledge of the Scottish Onychiuridae (Collembola), II. Scottish Naturalist. 1937;May-June:146–150. [Google Scholar]
- Bagnall (1949).Bagnall RS. Contributions toward a knowledge of the Onychiuridae (Collembola–Onychiuroidea). V–X. Annals and Magazine of Natural History. 1949;2(19):498–511. doi: 10.1080/00222934908654001. [DOI] [Google Scholar]
- Barrett & Hebert (2005).Barrett RDH, Hebert PDN. Identifying spiders through DNA barcodes. Canadian Journal of Zoology. 2005;83(3):481–491. doi: 10.1139/z05-024. [DOI] [Google Scholar]
- Bellinger et al. (2015).Bellinger PF, Christiansen KA, Arbea J, Janssens F. Checklist of the Collembola: Collembola species catalogue. 2015. http://www.collembola.org/ http://www.collembola.org/
- Bellinger, Christiansen & Janssens (1996–2018).Bellinger PF, Christiansen KA, Janssens F. Checklist of the Collembola of the world. 1996–2018. http://www.collembola.org http://www.collembola.org
- Bolger (1986).Bolger T. The Collembola of Ireland: a checklist and bibliography. Proceedings of the Royal Irish Academy, Section B: Biological, Geological, and Chemical Science. 1986;86(B):183–218. [Google Scholar]
- Caroli (1912).Caroli E. Contribuzioni alla conoscenza dei Collemboli italiani. I: La tribù degli Achorutini c.B. Archivio Zoologico Italiano. 1912;6(6):349–374. [Google Scholar]
- Christiansen & Bellinger (1988).Christiansen K, Bellinger P. Marine littoral collembola of North and Central America. Bulletin of Marine Science. 1988;42:215–245. [Google Scholar]
- Christiansen & Bellinger (1998).Christiansen K, Bellinger PF. Collembola of North America, north of the Rio Grande. Iowa: Grinnell College; 1998. [Google Scholar]
- Cicconardi, Fanciulli & Emerson (2013).Cicconardi F, Fanciulli PP, Emerson BC. Collembola, the biological species concept and the underestimation of global species richness. Molecular Ecology. 2013;22(21):5382–5396. doi: 10.1111/mec.12472. [DOI] [PubMed] [Google Scholar]
- Deharveng (1983).Deharveng L. Morphologie évolutive des Collemboles Neanurinae en particulier de la lignée néanurienne. Travaux du Laboratoire d’Ecobiologie des Arthropodes Edaphiques, Toulouse. 1983;4:1–63. [Google Scholar]
- Deharveng (2004).Deharveng L. Recent advances in Collembola systematics. Pedobiologia. 2004;48(5–6):415–433. doi: 10.1016/j.pedobi.2004.08.001. [DOI] [Google Scholar]
- Deharveng, Bedos & Gisclard (1998).Deharveng L, Bedos A, Gisclard C. Environmental factors, microgeographic patterns of endemism and hybrid zones in Monobella grassei (Insecta: Collembola: Neanuridae) Biological Journal of the Linnean Society. 1998;64(4):527–554. doi: 10.1111/j.1095-8312.1998.tb00348.x. [DOI] [Google Scholar]
- Deharveng et al. (2015).Deharveng L, Zoughailech A, Hamra-Kroua S, Porco D. A new species of Deutonura (Collembola: Neanuridae: Neanurinae) from north-eastern Algeria, and characterisation of two intraspecific lineages by their barcodes. Zootaxa. 2015;3920(2):281–290. doi: 10.11646/zootaxa.3920.2.4. [DOI] [PubMed] [Google Scholar]
- Denis (1923).Denis J. Notes sur les Aptérygotes. Annales de la Société Entomologique de France. 1923;14:209–246. [Google Scholar]
- Denis (1931).Denis J. Collemboles des Collections C. Schäffer et du Zoologisches Staatsinstitut und Zoologisches Museum in Hamburg. Zoologisches Staatsinstitut und Zoologisches Museum in Hamburg. 1931;44:197–242. [Google Scholar]
- Emerson et al. (2011).Emerson BC, Cicconardi F, Fanciulli PP, Shaw PJ. Phylogeny, phylogeography, phylobetadiversity and the molecular analysis of biological communities. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2011;366(1576):2391–2402. doi: 10.1098/rstb.2011.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjellberg (1998).Fjellberg A. The Collembola of Fennoscandia and Denmark: part I Poduromorpha. Fauna Entomologica Scandinavica. 1998;35:1–183. [Google Scholar]
- Fjellberg (1999).Fjellberg A. The labial palp in Collembola. Zoologischer Anzeiger. 1999;237:309–330. [Google Scholar]
- Folmer et al. (1994).Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- Frati et al. (2000).Frati F, Dell’Ampio E, Casasanta S, Carapelli A, Fanciulli PP. Large amounts of genetic divergence among Italian species of the genus Orchesella (Insecta, Collembola) and the relationships of two new species. Molecular Phylogenetics and Evolution. 2000;17(3):456–461. doi: 10.1006/mpev.2000.0854. [DOI] [PubMed] [Google Scholar]
- Gisin (1960).Gisin H. Collembolenfauna Europas. Genève: Museum d’Histoire Naturelle; 1960. [Google Scholar]
- Hajibabaei et al. (2005).Hajibabaei M, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, Mackie PM, Hebert PDN. Critical factors for assembling a high volume of DNA barcodes. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2005;360(1462):1959–1967. doi: 10.1098/rstb.2005.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajibabaei et al. (2006).Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajibabaei et al. (2007).Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics. 2007;23(4):167–172. doi: 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Hall (1999).Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hausmann et al. (2011).Hausmann A, Haszprunar G, Segerer AH, Speidel W, Behounek G, Hebert PDN. Now DNA-barcoded: the butterflies and larger moths of Germany. Spixiana. 2011;34:47–58. [Google Scholar]
- Hawes et al. (2008).Hawes T, Worland M, Bale J, Convey P. Rafting in Antarctic collembola. Journal of Zoology. 2008;274(1):44–50. doi: 10.1111/j.1469-7998.2007.00355.x. [DOI] [Google Scholar]
- Hebert et al. (2003).Hebert PDN, Cywinska A, Ball SL, De Waard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert & Landry (2010).Hebert PDN, Landry J-F. DNA barcodes for 1/1000 of the animal kingdom. Biology Letters. 2010;6(3):359–362. doi: 10.1098/rsbl.2009.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, Ratnasingham & De Waard (2003).Hebert PDN, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B: Biological Sciences. 2003;270(Suppl_1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert et al. (2004).Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLOS Biolology. 2004;2(10):e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled & Drummond (2010).Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27(3):570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg & Hebert (2004).Hogg ID, Hebert PDN. Biological identification of springtails (Hexapoda: Collembola) from the Canadian Arctic, using mitochondrial DNA barcodes. Canadian Journal of Zoology. 2004;82:749–754. doi: 10.1139/z04-041. [DOI] [Google Scholar]
- Hopkin (1997).Hopkin SP. Biology of the springtails (Insecta: Collembola) Oxford; New York, Tokyo: Oxford University Press; 1997. [Google Scholar]
- Laboulbène (1865).Laboulbène A. Recherches sur l’Anurida maritima, Insecte Thysanoure de la famille des Podurides. Annales de la Société entomologique de France. 1865;4(4):705–720. [Google Scholar]
- Ivanova, Dewaard & Hebert (2006).Ivanova NV, Dewaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Resources. 2006;6(4):998–1002. doi: 10.1111/j.1471-8286.2006.01428.x. [DOI] [Google Scholar]
- Joosse (1976).Joosse EN. Littoral apterygotes (Collembola and Thysanura) In: Cheng L, editor. Marine Insects. New York: American Elsevier; 1976. pp. 151–186. [Google Scholar]
- Jordana et al. (1997).Jordana R, Arbea JI, Simón C, Luciáñez MJ. Collembola Poduromorpha, Familia Onychiuridae, Subfamilia Onychiurinae. In: Ramos MA, et al., editors. Fauna Ibérica. Vol. 8. Madrid: Museo Nacional de Ciencias Naturales, Consejo Superior de Investigaciones Cientificas; 1997. pp. 477–641. [Google Scholar]
- Kaprus & Paśnik (2017).Kaprus I, Paśnik G. New Siberian “spineless” species of Thalassaphorura Bagnall, 1949 (Collembola, Onychiuridae), with a key to world species of the genus. Zootaxa. 2017;4362(2):225–245. doi: 10.11646/zootaxa.4362.2.3. [DOI] [PubMed] [Google Scholar]
- Katz, Giordano & Soto-Adames (2015).Katz AD, Giordano R, Soto-Adames FN. Operational criteria for cryptic species delimitation when evidence is limited, as exemplified by North American Entomobrya (Collembola: Entomobryidae) Zoological Journal of the Linnean Society. 2015;173(4):818–840. doi: 10.1111/zoj.12220. [DOI] [Google Scholar]
- Kimura (1980).Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16(2):111–120. doi: 10.1007/bf01731581. [DOI] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukić et al. (2015).Lukić M, Porco D, Bedos A, Deharveng L. The puzzling distribution of Heteromurus (Verhoeffiella) absoloni Kseneman, 1938 (Collembola: Entomobryidae: Heteromurinae) resolved: Detailed redescription of the nominal species and description of a new species from Catalonia (Spain) Zootaxa. 2015;4039(2):249–275. doi: 10.11646/zootaxa.4039.2.3. [DOI] [PubMed] [Google Scholar]
- Meyer & Paulay (2005).Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLOS Biology. 2005;3(12):e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniez (1890).Moniez R. Acariens et insectes marins des côtes du Boulonnais. Revue Biologique du Nord de la France. 1890;2:338–350. [Google Scholar]
- Mouritsen & Poulin (2002).Mouritsen KN, Poulin R. Parasitism, community structure and biodiversity in intertidal ecosystems. Parasitology. 2002;124(7):101–117. doi: 10.1017/s0031182002001476. [DOI] [PubMed] [Google Scholar]
- Pan (2015).Pan ZX. Two closely related Homidia species (Entomobryidae, Collembola) revealed by morphological and molecular evidence. Zootaxa. 2015;3918(2):285–294. doi: 10.11646/zootaxa.3918.2.9. [DOI] [PubMed] [Google Scholar]
- Papadopoulou, Anastasiou & Vogler (2010).Papadopoulou A, Anastasiou I, Vogler AP. Revisiting the insect mitochondrial molecular clock: the Mid-Aegean Trench calibration. Molecular Biology and Evolution. 2010;27(7):1659–1672. doi: 10.1093/molbev/msq051. [DOI] [PubMed] [Google Scholar]
- Pomorski (1998).Pomorski RJ. Onychiurinae of Poland (Collembola: Onychiuridae) Genus. 1998;9:1–201. [Google Scholar]
- Porco, Bedos & Deharveng (2010).Porco D, Bedos A, Deharveng L. Cuticular compounds bring new insight in the post-glacial recolonization of a Pyrenean area: Deutonura deficiens Deharveng, 1979 complex, a case study. PLOS ONE. 2010;5(12):e14405. doi: 10.1371/journal.pone.0014405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco et al. (2012a).Porco D, Bedos A, Greenslade P, Janion C, Skarżyński D, Stevens M, van Vuuren BJ, Deharveng L. Challenging species delimitation in Collembola: cryptic diversity among common springtails unveiled by DNA barcoding. Invertebrate Systematics. 2012a;26(6):470–477. doi: 10.1071/is12026. [DOI] [Google Scholar]
- Porco et al. (2013).Porco D, Decaëns T, Deharveng L, James SW, Skarżyński D, Erséus C, Butt KR, Richard B, Hebert PDN. Biological invasions in soil: DNA barcoding as a monitoring tool in a multiple taxa survey targeting European earthworms and springtails in North America. Biological Invasions. 2013;15(4):899–910. doi: 10.1007/s10530-012-0338-2. [DOI] [Google Scholar]
- Porco et al. (2012b).Porco D, Potapov M, Bedos A, Busmachiu G, Weiner WM, Hamra-Kroua S, Deharveng L. Cryptic diversity in the ubiquist species Parisotoma notabilis (Collembola, Isotomidae): a long used chimeric species? PLOS ONE. 2012b;7(9):e46056. doi: 10.1371/journal.pone.0046056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco et al. (2010).Porco D, Rougerie R, Deharveng L, Hebert PDN. Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied Arthropods in a high-throughput context: the example of Collembola. Molecular Ecology Resources. 2010;10(6):942–945. doi: 10.1111/j.1755-0998.2010.2839.x. [DOI] [PubMed] [Google Scholar]
- Puillandre et al. (2012).Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology. 2012;21(8):1864–1877. doi: 10.1111/j.1365-294x.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- Raffaelli & Hawkins (2012).Raffaelli D, Hawkins SJ. Intertidal Ecology. London: Kluwer Academic Publishers; 2012. [Google Scholar]
- Rambaut, Suchard & Drummond (2014).Rambaut A, Suchard MA, Drummond AJ. Tracer v1.6. 2014. http://tree.bio.ed.ac.uk/software/tracer/ [12 March 2018]. http://tree.bio.ed.ac.uk/software/tracer/
- Rusek (2007).Rusek J. Integration of ecological and morphological studies: Micro-distribution of. Protaphorura-species (Collembola: Onychiurinae) around a beech stem. In: Tajovsky K, Schlaghamersky J, Pizl V, editors. Contributions to Soil Zoology in Central Europe II. Proceedings of the 8th Central European Workshop on Soil Zoology. České Budějovice: ISB BC AS CR; 2007. pp. 117–120. [Google Scholar]
- Saitou & Nei (1987).Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salmon (1959).Salmon JT. Concerning the Collembola Onychiuridae. Ecological Entomology. 1959;111(6):119–156. doi: 10.1111/j.1365-2311.1959.tb02279.x. [DOI] [Google Scholar]
- Salmon (1964).Salmon JT. An index to the Collembola. Royal Society of New Zealand Bulletin. 1964;7(2):145–644. [Google Scholar]
- Skidmore (1995).Skidmore R. Checklist of Collembola (Insecta: Apterygota) of Canada and Alaska. Proceedings of the Entomological Society of Ontario. 1995;126:45–76. [Google Scholar]
- Soto-Adames (2002).Soto-Adames FN. Molecular phylogeny of the Puerto Rican Lepidocyrtus and Pseudosinella (Hexapoda: Collembola), a validation of Yoshii’s “color pattern species”. Molecular Phylogenetics and Evolution. 2002;25(1):27–42. doi: 10.1016/s1055-7903(02)00250-6. [DOI] [PubMed] [Google Scholar]
- Stach (1954).Stach J. The Apterygotan Fauna of Poland in Relation to the World-Fauna of this Group of Insects, Family: Onychiuridae. Krakow: Polska Akademia Nauk Instytut Zoologiczny; 1954. [Google Scholar]
- Stoev et al. (2013).Stoev P, Komerički MA, Akkari N, Liu MS, Zhou MX, Weigand AM, Hostens J, Hunter MCI, Edmunds SC, Porco D. Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data. Biodiversity Data Journal. 2013;1:e1013. doi: 10.3897/BDJ.1.e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Bedos & Deharveng (2017).Sun X, Bedos A, Deharveng L. Two new species of the genus Thalassaphorura Bagnall, 1949 (Collembola: Onychiuridae) from south China, with an updated key to world species of the genus. Zootaxa. 2017;4338(2):319–332. doi: 10.11646/zootaxa.4338.2.6. [DOI] [PubMed] [Google Scholar]
- Sun, Chen & Deharveng (2010).Sun X, Chen J-X, Deharveng L. Six new species of Thalassaphorura (Collembola, Onychiuridae) from southern China, with a key to world species of the genus. Zootaxa. 2010;2627:20–38. [Google Scholar]
- Sun et al. (2017).Sun X, Zhang F, Ding Y, Davies TW, Li Y, Wu D. Delimiting species of Protaphorura (Collembola: Onychiuridae): integrative evidence based on morphology, DNA sequences and geography. Scientific Reports. 2017;7(1):8261. doi: 10.1038/s41598-017-08381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward et al. (2005).Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1642):1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb et al. (2012).Webb JM, Jacobus LM, Funk DH, Zhou X, Kondratieff B, Geraci CJ, DeWalt RE, Baird DJ, Richard B, Phillips I. A DNA barcode library for North American Ephemeroptera: progress and prospects. PLOS ONE. 2012;7(5):e38063. doi: 10.1371/journal.pone.0038063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner (1996).Weiner WM. Generic revision of Onychiurinae (Collembola: Onychiuridae) with a cladistic analysis. Annales de la Société Entomologique de France. 1996;32:163–200. [Google Scholar]
- Wesener et al. (2016).Wesener T, Voigtländer K, Decker P, Oeyen JP, Spelda J. Barcoding of Central European Cryptops centipedes reveals large interspecific distances with ghost lineages and new species records from Germany and Austria (Chilopoda, Scolopendromorpha) Zookeys. 2016;564:21–46. doi: 10.3897/zookeys.564.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem (1925).Willem V. Les Collemboles marins de Wimereux. Travaux de la Station Zoologique de Wimereux. 1925;9:275–283. [Google Scholar]
- Witteveen & Joosse (1988).Witteveen J, Joosse E. The effects of inundation on marine littoral Collembola. Ecography. 1988;11(1):1–7. doi: 10.1111/j.1600-0587.1988.tb00775.x. [DOI] [Google Scholar]
- Yoshii (1996).Yoshii R. Identity of some Japanese Collembola “Deuteraphorura” Group of Onychiurus-continued. Annals of the Speleological Research Institute of Japan (Iwaizumi) 1996;14:1–15. [Google Scholar]
- Yu, Ding & Ma (2017).Yu D, Ding Y, Ma Y. Revision of Tomocerus similis Chen & Ma, with discussion of the kinoshitai complex and the distal tibiotarsal chaetae in Tomocerinae (Collembola, Tomoceridae) Zootaxa. 2017;4268(3):395–410. doi: 10.11646/zootaxa.4268.3.5. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2016).Yu D, Zhang F, Stevens MI, Yan Q, Liu M, Hu F. New insight into the systematics of Tomoceridae (Hexapoda, Collembola) by integrating molecular and morphological evidence. Zoologica Scripta. 2016;45(3):286–299. doi: 10.1111/zsc.12149. [DOI] [Google Scholar]
- Zhang, Greenslade & Stevens (2017).Zhang F, Greenslade P, Stevens MI. A revision of the genus Lepidobrya Womersley (Collembola: Entomobryidae) based on morphology and sequence data of the genotype. Zootaxa. 2017;4221(5):523–536. doi: 10.11646/zootaxa.4221.5.2. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang F, Yu D, Luo Y, Ho SYW, Wang B, Zhu C. Cryptic diversity, diversification and vicariance in the two species complexes of Tomocerus (Collembola, Tomoceridae) from China. Zoologica Scripta. 2014;43(4):393–404. doi: 10.1111/zsc.12056. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.