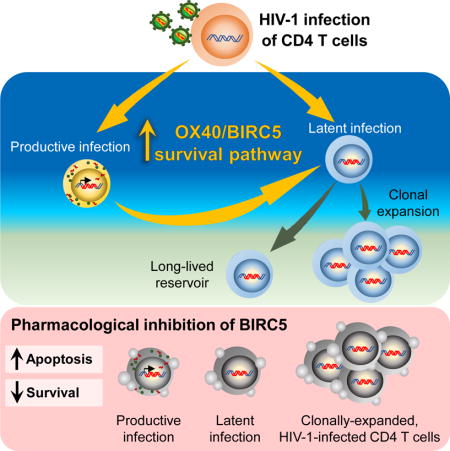
Abstract
HIV-1 infection of CD4+ T cells leads to cytopathic effects and cell demise, which is counter to the observation that certain HIV-1-infected cells possess a remarkable long-term stability and can persist lifelong in infected individuals treated with suppressive antiretroviral therapy (ART). Using quantitative mass spectrometry-based proteomics, we showed that HIV-1 infection activated cellular survival programs that were governed by BIRC5, a molecular inhibitor of cell apoptosis that is frequently overexpressed in malignant cells. BIRC5 and its upstream regulator OX40 were upregulated in productively and latently infected CD4+ T cells, and were functionally involved in maintaining their viability. Moreover, OX40-expressing CD4+ T cells from ART-treated patients were enriched for clonally-expanded HIV-1 sequences, and pharmacological inhibition of BIRC5 resulted in a selective decrease of HIV-1-infected cells in vitro. Together, these findings suggest that BIRC5 supports long-term survival of HIV-1-infected cells and may lead to clinical strategies to reduce persisting viral reservoirs.
Keywords: HIV-1, latency, viral reservoirs, BIRC5, OX40, proteomics, Survivin, apoptosis, clonal proliferation
ETOC BLURB
The factors that promote the survival and persistence of HIV-infected CD4+ T cells are not clear. Kuo et al. demonstrate that the anti-apoptotic protein BIRC5 and its upstream regulator OX40 can promote survival of HIV-1-infected reservoir CD4+ T cells, specifically during clonal proliferation. These findings point to clinical strategies that may reduce persisting viral reservoirs.
Introduction
Infection with HIV-1 causes a chronic viral illness characterized by numerically reduced and functionally defective CD4+ T cells. In part, this decline of CD4+ T cells has been attributed to viral cytopathic effects that can result from direct host-pathogen interactions, and lead to CD4+ T cell death. For instance, in resting CD4+ T cells, in particular those located in lymphoid and mucosal tissues (Munoz-Arias et al., 2015), HIV-1 reverse transcripts can be detected through the cytoplasmic microbial DNA sensor IFI16, which results in cellular apoptosis via caspase 1 activation (Monroe et al., 2014). In activated CD4+ T cells, HIV-1 induces cell death by causing DNA damage response signals that occur in response to viral integration into host chromosomes and are mediated by DNA-associated protein kinase (DNA-PK) (Cooper et al., 2013). Moreover, HIV-1 protease can induce cell death by cleaving host procaspase 8 to Casp8p41 (Cummins and Badley, 2015; Sainski et al., 2014), and HIV-1 Tat can support transcription of pro-apoptotic host cell genes through activation of the transcription factor FOXO3a (Dabrowska et al., 2008). However, the notion of HIV-1-induced CD4+ T cell death is contradictory to the clinical observation that at least some HIV-1-infected cells have a remarkable ability to survive and persist long term for decades in the presence of highly-effective suppressive antiretroviral therapy (Chun et al., 1997; Finzi et al., 1999; Siliciano et al., 2003). The ability to establish such a highly durable reservoir of virally-infected cells suggests that HIV-1 infection may induce specific host pathways that allow infected cells to resist viral cytopathic effects and support long-term survival of virally-infected cells.
BIRC5, frequently termed “Survivin”, is the smallest, but arguably the functionally most complex member of the inhibitor of apoptosis protein (IAP) family (Altieri, 2015). While it is almost ubiquitously expressed in human stem cells during embryogenesis and essential for proper fetal development, its expression in adult tissues is highly restricted (Ambrosini et al., 1997). The strongest expression in adults is observed in the cytoplasm and nucleus of cancer cells, where it is associated with chemotherapy resistance, increased metastatic activity and a higher risk for tumor recurrence; for these reasons, it represents one of the biomarkers for malignant disease prognosis most frequently used in clinical practice (Kim et al., 2010; Rodel et al., 2012; Shariat et al., 2009; Xie et al., 2016). Functionally, BIRC5 can support malignant cell growth by stabilizing the mitotic apparatus, ensuring proper segregation of chromosomes and maintaining microtubule integrity; this allows for safe and effective completion of cell divisions (Ruchaud et al., 2007). In addition, BIRC5 has a critical role for protecting cells against intrinsic and extrinsic pathways of apoptosis, most likely through indirect inhibition of caspase 9 activation, while avoiding direct biophysical interactions with apoptosis-promoting molecules (Tamm et al., 1998).
Besides its critical role as a driver of almost every cancer type in humans, increasing data suggest a vital function of BIRC5 for regulating viability and proliferation of non-malignant, physiological adult T cells, which are among the few adult human cell populations with detectable levels of BIRC5 expression (Andersson et al., 2015; Croft et al., 2009; Sharief and Semra, 2001). Within antigen-specific T cells, BIRC5 expression is controlled by OX40 (TNFRSF4, CD134), a member of the TNF receptor superfamily that is expressed on the cell surface and seems to have unique functions for protecting long-term cell viability of memory T cells, particularly at the time of clonal expansion (Song et al., 2005). For this reason, the OX40-BIRC5 pathway may have a profound influence on shaping and structuring homeostasis and pool sizes of memory T cells, and can influence efficient T cell expansion or contraction based on immunological demands.
In the present study, we employed quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics for a detailed analysis of HIV-1-infected CD4+ T cells. Our results showed that cells infected with HIV-1, both during a productive and latent infection state, upregulated BIRC5 and OX40 and were functionally dependent on BIRC5 for maintaining their viability and integrity. Moreover, cells harboring clonally-expanded HIV-1 sequences were enriched within OX40-positive CD4+ T cells from ART-treated individuals and could be targeted using pharmaceutical small-molecule antagonists of BIRC5.
Results
Proteomic profiling identifies survival programs in HIV-1-infected CD4+ T cells
Infection with HIV-1 is known to cause profound changes in cellular physiology, but molecular changes occurring in response to viral infection have been difficult to analyze at a protein expression level. Recent technical advances in liquid chromatography-tandem mass spectrometry (LC-MS/MS) enable global, quantitative profiling of changes in the proteome of CD4+ T cells at a high level of molecular resolution, and may identify activation of previously unrecognized cellular pathways and programs in HIV-1-infected cells (Greenwood et al., 2016). To profile changes in the host proteome during viral replication, in vitro activated primary CD4+ T cells from four different HIV-1-negative donors were infected with GFP-encoding R5-tropic HIV-1 in duplicate; 96 hours after infection, GFP+ and GFP− CD4+ T cells were sorted and processed for global protein expression profiling by LC-MS/MS using isobaric mass tag labeling with 10-plex tandem mass tag (TMT) reagents for quantification (see also Figure S1A). This approach identified and quantified 7761 proteins in GFP+ and GFP− CD4+ T cells, of which 552 were differentially expressed between the two cell populations based on robust statistical criteria (FDR-adjusted p<0.01) (Figure 1A, see also Table S1), and clearly separated the two cell populations in a principal component analysis (see also Figure S1B). Using established biocomputational algorithms to identify functional pathways in the proteomic signatures in GFP+ CD4+ T cells, we noted that the “cell death and survival” module accounted for a considerable number of all proteins distinguishing the GFP+ and the GFP− CD4+ T cell pools, and represented the top functional entity that differentially-expressed proteins were enriched for (Figure 1B). In contrast to previous findings emphasizing induction of cellular death signals during HIV-1 infection (Cummins and Badley, 2010), a subsequent computational analysis predicted that GFP+ HIV-1-infected CD4+ T cells activated cell survival and viability programs, while protein expression signatures of cell death, apoptosis and necrosis were mostly de-enriched in this cell population (Figure 1C). To explore key molecular factors and pathways regulating the cell survival pathways in HIV-1-infected cells, we entered all differentially-expressed proteins predicted to be involved in cell survival pathways (see also Table S2) into an in silico functional network linkage (FNL) analysis (Linghu et al., 2013), which can predict connections between functionally-related proteins and detect modules of protein-protein interactions able to drive biological processes. Proteins positioned in the center of network plots and exhibiting the most diverse range of predicted protein-protein connections included BIRC5, a cytoplasmic and nuclear protein and member of the inhibitor of apoptosis protein (IAP) family, and its upstream regulator OX40 located on the cell surface (Figure 1D/E). Moreover, BIRC5 was among the top effector molecules for the predicted upstream regulators of all differentially-expressed proteins (Figure 1F). No other members of the IAP family or alternative anti-apoptosis molecules were differentially expressed between GFP+ and GFP− CD4+ T cells (see also Figure S1C). Given the known role of BIRC5 in maintaining cancer cell survival and preserving residual reservoirs of treatment-resistant malignant cells (Altieri, 2015), and the described function of the OX40-BIRC5 pathway in the context of regulating physiological T cell survival, specifically during the vulnerable stage of clonal expansion (Song et al., 2005), we hypothesized that the OX40-BIRC5 cascade plays a key role in protecting and safeguarding the viability of HIV-1-infected CD4+ T cells.
Figure 1. HIV-1 activates survival programs in infected CD4+ T cells.
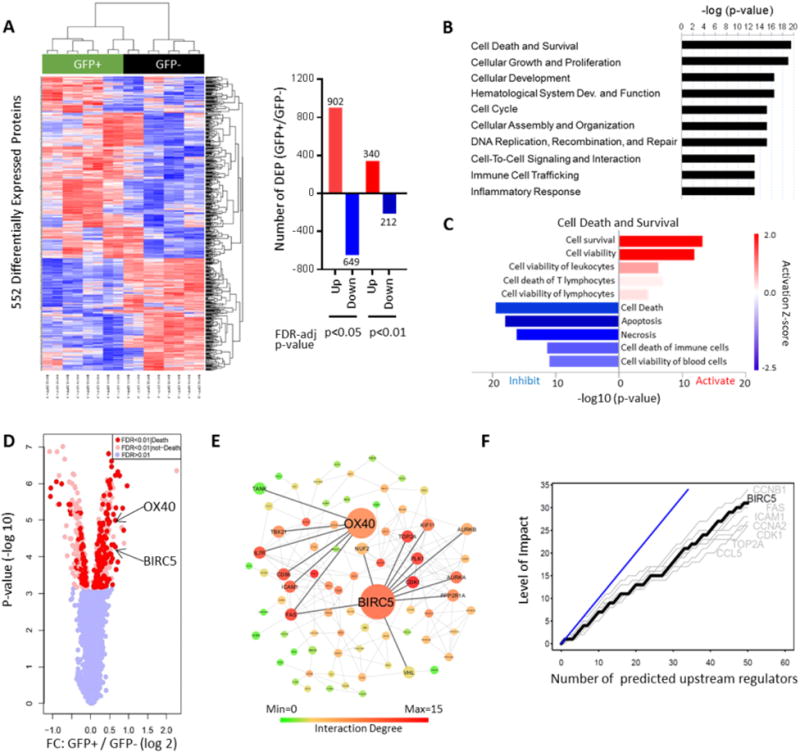
(A): Left panel: Heatmap demonstrating differentially expressed proteins between GFP+ and GFP− CD4+ T cells following in vitro infection with GFP-encoding HIV-1. Samples were run in biological duplicates; data from four patients are shown. Rows represent individual proteins detected by quantitative mass spectrometry. Right panel: Bar diagrams reflecting numbers of upregulated and downregulated proteins in GFP+ CD4+ T cells at indicated levels of FDR-adjusted significance. (B): Predicted functional annotations (“diseases and functions”) of differentially-expressed proteins (FDR-adjusted p-value <0.01), based on Ingenuity Pathway Analysis (IPA). (C): Predicted canonical pathway enrichment of differentially-expressed proteins (FDR-adjusted p-value <0.01). Z-score indicates activation vs. inhibition of indicated functions. (D): Volcano plot indicating fold-changes and corresponding FDR-adjusted p-values of all detected proteins in GFP+/GFP− cells. Proteins with predicted involvement in cell death and survival are highlighted in red. (E): Functional linkage network of differentially-expressed proteins. Positions of BIRC5 and its upstream regulator OX40 are indicated. (F): Diagram reflecting top 8 effector molecules most frequently involved in downstream signaling of predicted upstream regulators of differentially-expressed proteins. Y-axis reflects number of upstream regulators signaling via indicated effector molecules. Blue line indicates hypothetical reference molecule involved in downstream signaling of all upstream regulators. See also Figure S1, Table S1 and Table S2.
OX40 and BIRC5 maintain survival of HIV-1-infected CD4+ T cells
We subsequently conducted experiments to characterize the role of BIRC5 and OX40 in activated and resting in vitro HIV-1-infected CD4+ T cells, using flow cytometric assays. We observed that BIRC5 and OX40 expression was significantly upregulated in activated HIV-1-infected CD4+ T cells; however, the absolute difference in protein expression intensity between GFP+ and GFP− CD4+ T cells was small (Figure 2A/B), possibly due to artifacts associated with in vitro activation of CD4+ T cells. In contrast, substantial increases in BIRC5 and OX40 expression were noted in HIV-1-infected resting CD4+ T cells, relative to their GFP-negative counterparts (Figure 2C/D). An analysis of BIRC5 localization in subcellular compartments by ImageStream flow cytometry indicated that the intracellular increase of BIRC5 in resting CD4+ T cells occurred both in the nucleus and in the cytoplasm, with a tendency for a slightly more profound effect in the nucleus (Figure 2E). In addition, using a dual-reporter virus encoding for fluorescent markers allowing to distinguish CD4+ T cells with productive infection or latent HIV-1 infection (Chavez et al., 2015), we found that BIRC5 upregulation was detectable in resting CD4+ T cells with either state of infection (Figure 2F/G). Parallel evaluations of OX40 surface expression largely recapitulated the pattern observed for intracellular BIRC5 expression, with higher levels of OX40 expression being detectable on HIV-1-infected resting CD4+ T cells, both in the setting of productive and latent infection (Figure 2F/H).
Figure 2. BIRC5 and OX40 are upregulated in productively and latently HIV-1-infected CD4+ T cells.
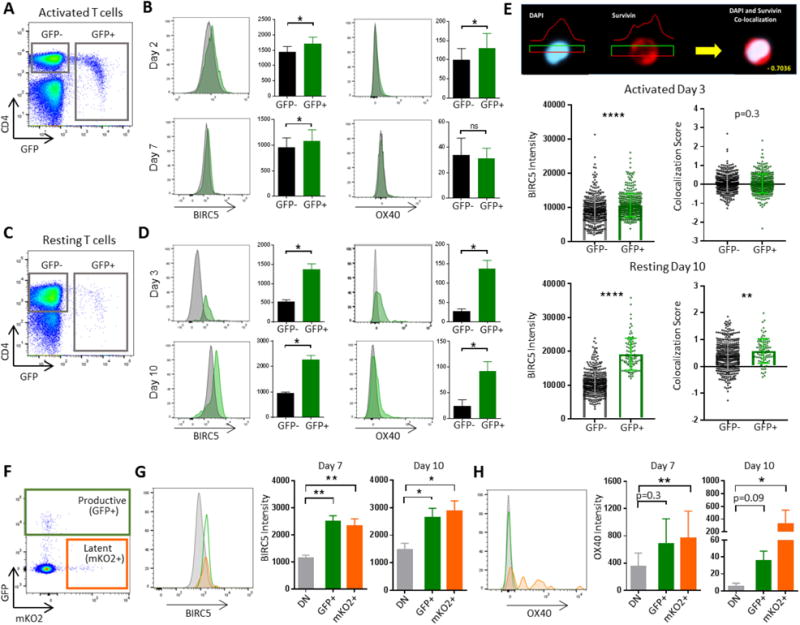
(A/C): Representative flow cytometry dot plots demonstrating gating of GFP+ and GFP− CD4+ T cells following infection of in vitro activated (A, day 7 after infection) and non-activated (C, day 10 after infection) CD4+ T cells with R5-tropic GFP-encoding HIV-1. (B/D): Histograms reflecting intracellular BIRC5 and surface OX40 expression on CD4+ T cell populations (n=6) described in (A/B) at indicated timepoints after infection. Bar diagrams summarize expression intensity of BIRC5 and OX40 on GFP+ and GFP-CD4+ T cell populations described in (A/B). *p<0.05 (Wilcoxon test). (E): Representative ImageStream analysis plots indicating subcellular location of BIRC5 in CD4+ T cells, relative to nuclear counterstain with DAPI. Left panels demonstrate intracellular BIRC5 expression on activated CD4+ T cells and non-activated CD4+ T cells at indicated timepoints after infection; right panels show co-localization scores between BIRC5 and DAPI. Each dot represents data from one individual cell. Results from one out of two experiments are shown. **p<0.01, ****p<0.0001 (Mann-Whitney U test). (F): Representative flow cytometry dot plot indicating gating of GFP+, mKO2+ and GFP−/mKO2− cells following infection of resting CD4+ T cells with dual reporter-encoding HIV-1. (G-H): Representative histograms and bar diagrams reflecting fluorescence intensity of BIRC5 (G) and OX40 (H) in cell populations described in (F). Data from n=6 study subjects are shown. (G-H): *p<0.05, **p<0.01 (Friedman test with post-hoc Dunn’s test for multiple comparison).
To assess whether BIRC5 can support the ability of HIV-1-infected CD4+ T cells to survive and resist cytopathic effects associated with HIV-1 infection, we took advantage of a previously-described in vitro experimental system in which cellular signs of HIV-1-associated cell death are particularly obvious (Cooper et al., 2013). Briefly, activated CD4+ T cells were in vitro infected with a VSV-G-pseudotyped, GFP-encoding HIV-1 construct, followed by sorting of GFP+ HIV-1-infected CD4+ T cells after 36 hours (Figure 3A). As described previously (Cooper et al., 2013), we observed that a proportion of isolated GFP+ CD4+ T cells transition to a GFP-negative phenotype during subsequent in vitro culture (Figure 3B/C); this development into a transcriptionally silent HIV-1-infection status is associated with induction of HIV-1-associated cell death in a considerable proportion of cells, while productively infected CD4+ T cells that maintain continuous GFP expression do not show noticeable signs of virally-induced cell demise (Cooper et al., 2013) (Figure 3D/E). Cells that exhibited signs of early and late apoptosis during transitioning from a GFP+ to GFP-negative phenotype, as defined based on staining with Annexin V and viability dyes, displayed the lowest intensity of BIRC5 protein expression, while maintenance of cellular integrity and viability in GFP-negative HIV-1-infected CD4+ T cells was associated with enhanced BIRC5 protein expression intensity (Figure 3F). Thus, these data suggest that BIRC5 expression can have a functional role in preserving and maintaining cell survival of HIV-1-infected cells, specifically at times when cells transition into a transcriptionally silent (latent) viral infection status.
Figure 3. BIRC5 expression is functionally associated with survival of HIV-1-infected CD4+ T cells.
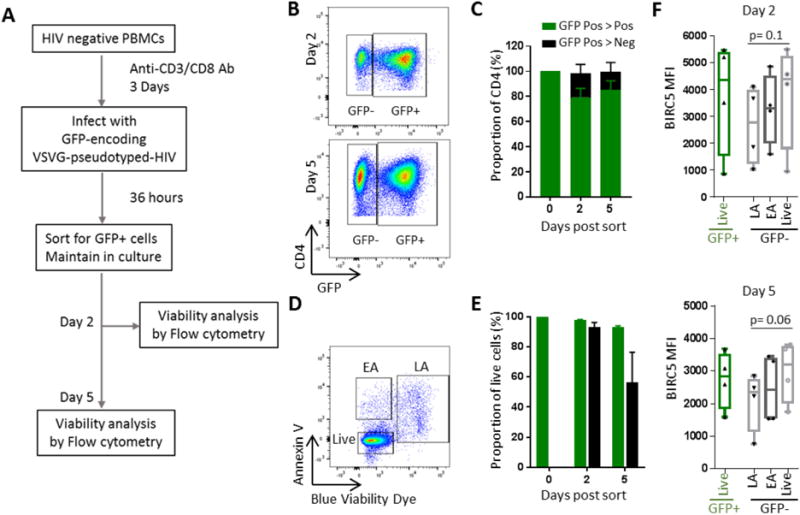
(A): Schematic overview for analyzing cell death in CD4+ T cells transitioning from productive infection to latency, as described in (Cooper et al., 2013). (B): Flow cytometry dot plots indicating gating of GFP+ and GFP− CD4+ T cells on days 2 and 5 within GFP+ CD4+ T cells sorted on day 0. (C): Bar diagram summarizing proportions of GFP+ and GFP− CD4+ T cells on indicated timepoints within GFP+ CD4+ T cells sorted on day 0. Mean and standard error from n=4 study subjects are shown. (D): Representative flow cytometry dot plot showing relative proportions of cells in early apoptosis (“EA”), late apoptosis (“LA”) or live cells (“Live”) on day 5 after sorting of GFP+ CD4+ T cells. (E): Bar diagrams demonstrating proportions of live cells within GFP+ and GFP− HIV-1-infected CD4+ T cells on indicated timepoints after sorting of GFP+ CD4+ T cells on day 0. (F): Expression intensity of BIRC5 in indicated GFP+ and GFP− CD4+ T cell populations on day 2 and day 5 after sorting of GFP+ CD4+ T cells on day 0. Differences were tested for significance using one-tailed Wilcoxon test.
OX40 enriches for CD4+ T cells encoding clonally-expanded HIV-1
To determine whether the OX40-BIRC5 pathway maintains survival of HIV-1-infected CD4+ T cells in vivo, we next focused on analyzing cells from HIV-1-infected individuals treated with suppressive antiretroviral therapy who typically maintain small reservoirs of CD4+ T cells that harbor replication-competent HIV-1 (Finzi et al., 1999; Siliciano et al., 2003). We found that BIRC5 and OX40 displayed higher protein expression intensity in CD4+ T cells expressing PD-1, TIGIT, Lag-3 and the immune activation markers HLA-DR and CD38, surface receptors previously shown to correlate with the size of HIV-1 DNA levels (Fromentin et al., 2016; Ruggiero et al., 2015) (Figure 4A–C, see also Figure S2A/B); this indicates that OX40+ CD4+ T cells represent distinct cell populations but overlap with cell types previously identified as prime locations for persistence of viral reservoirs. Using fluorescence-activated cell sorting, we isolated OX40+ and OX40− CD4+ T cells from five study individuals who had received suppressive ART for several years (Figure 4D). We subsequently subjected DNA from sorted cells to single-template amplification with primers spanning near full-length HIV-1, which allows to obtain a comprehensive view of proviral cell-associated HIV-1 DNA sequences. Consistent with prior observations (Bruner et al., 2016), we noted that viral DNA copies with gross deletions or other types of lethal mutations accounted for the vast majority of HIV-1 amplification products (Figure 4D). We noted that OX40, known to be associated with long-term survival (Rogers et al., 2001) and clonal proliferation (Song et al., 2005) of CD4+ T cells, denoted cells harboring significantly higher frequencies of total HIV-1 DNA copies (Figure 4E), and higher numbers of clonally-expanded HIV-1 DNA sequences (Figure 4F), which may be of particular importance for stabilizing the persistence of viral reservoir cells (Bui et al., 2017; Hosmane et al., 2017; Lee et al., 2017) (see also Table S3). Frequencies of intact proviral sequences were also increased in OX40-expressing CD4+ T cells in 4 of the five patients analyzed (Figure 4G–H). Together, these data show that OX40+ CD4+ T cells are enriched for clonally-expanded viral sequences, and support the hypothesis that the OX40-BIRC5 axis is functionally involved in maintaining and protecting the viability of latently-infected CD4+ T cells in ART-treated patients.
Figure 4. OX40 enriches for CD4+ T cells encoding for clonally-expanded HIV-1.
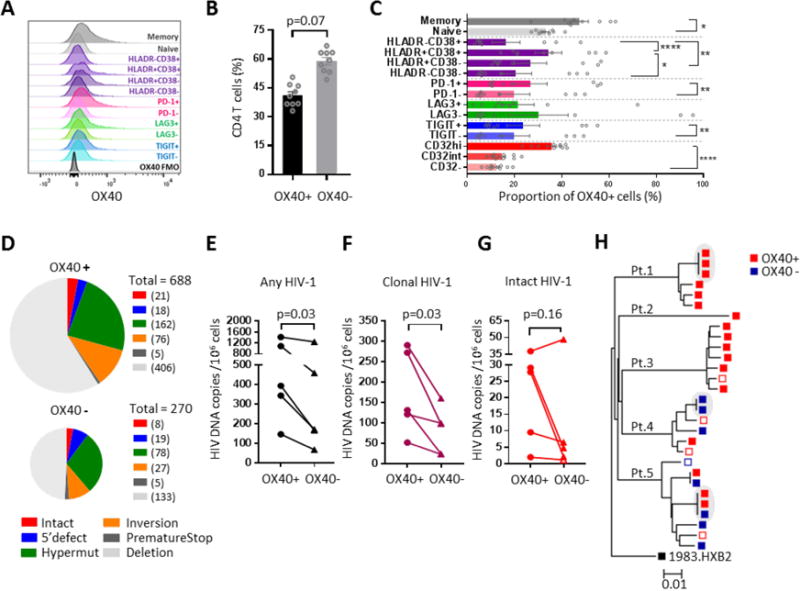
(A): Overlap histogram reflecting OX40 expression in indicated CD4+ T cell populations. Data from one representative study subject are shown. (B/C): Bar diagrams reflecting the proportions of OX40+ CD4+ T cells within total CD4+ T cells (B) and within CD4+ T cell populations with indicated phenotypic properties (C). Data from 9 ART-treated HIV-1 patients are shown. *p<0.05, **p<0.01, ****p<0.001 (Wilcoxon test or Friedman test with post-hoc Dunn’s test). (D): Pie charts reflecting HIV-1 DNA copies amplified by single-template near full-length HIV-1 PCR from OX40+ and OX40− CD4+ T cells. Color coding indicates categories of HIV-1 that are intact or show defined defects. Cumulative data from n=5 study subjects are shown, pie sizes correlate to total numbers of sequences analyzed. (E-G): Frequency of any (E), clonally-expanded (F, defined as sequences detected more than once) and intact (G) HIV-1 DNA copies in OX40+ and OX40− CD4+ T cells. P-values were calculated using a one-tailed Wilcoxon test. Open symbol reflects calculated limit of detection. (H): Phylogenetic tree of intact HIV-1 sequences retrieved from OX40+ and OX40− CD4+ T cells from the five study subjects. Shaded areas reflect identical sequences, consistent with clonal expansion. Red symbols: sequences detected in OX40+ CD4+ T cells; blue symbols: sequences detected in OX40− CD4+ T cells. Solid symbols: intact sequences, open symbols: inferred intact sequences. See also Figure S2 and Table S3.
Pharmacologic targeting of BIRC5 reduces frequency of in vitro HIV-1-infected CD4+ T cells
The availability of small molecule inhibitors of BIRC5 allowed us to assess the functional role of BIRC5 for survival of HIV-1-infected cells, and to explore the potential use of BIRC5 inhibitors for therapeutic approaches to reduce persistence of virally-infected cells. For this purpose, we focused on YM155, a pharmacological inhibitor of BIRC5 previously tested in human clinical trials in the context of malignant diseases (Clemens et al., 2015). Using resting CD4+ T cells infected in vitro with a GFP-encoding HIV-1 construct, we observed that co-culture with YM155 led to selective reductions in the frequency of virally-infected CD4+ T cells (Figure 5A–C); this was associated with pronounced increases of apoptosis and cell death markers in GFP+ CD4+ T cells, but not in GFP− cells (Figure 5D–E). In this experimental setting, reductions of virally-infected CD4+ T cells were noted when YM155 was added to CD4+ T cells prior to infection, immediately after infection or several days after infection, indicating a robust efficacy to selectively induce apoptosis in HIV-1-infected CD4+ T cells (Figure 5E). To investigate whether YM155 can also influence survival and persistence of latently-infected cells, we added YM155 to cultures of resting CD4+ T cells infected with the dual-reporter virus allowing to distinguish productively- and latently-infected CD4+ T cells (Chavez et al., 2015). We found that addition of YM155 led to substantial reductions of latently- and productively-infected CD4+ T cells over short and longer durations of co-culture (Figure 5F/G). Together, these data suggest that both latently- and productively-infected CD4+ T cells selectively depend on BIRC5 for maintaining cell survival and viability, and that pharmacological inhibition of BIRC5 is lethal for many of these cells.
Figure 5. BIRC5 inhibition causes selective reduction of in vitro HIV-1-infected CD4+ T cells.
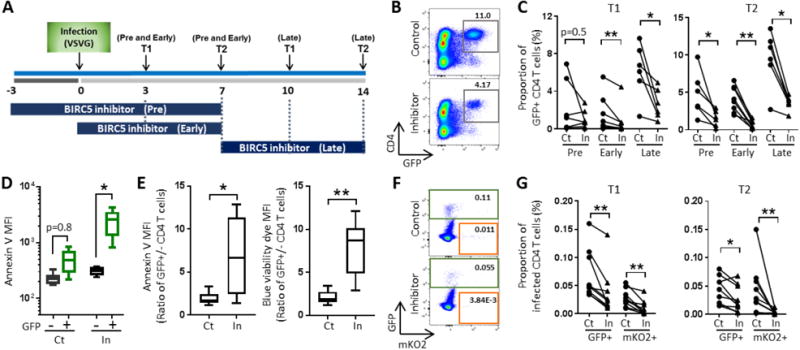
Schematic overview for experiments described in panels (B-G). Note that YM155 (pharmacological inhibitor of BIRC5) was added before (pre, n = 7), immediately after (post, n = 8) or 7 days (late, n = 6) after in vitro infection with VSV-G pseudotyped HIV-1. (B): Flow cytometry dot plots indicating proportions of resting GFP+ CD4+ T cells on day 14 after infection and treatment with YM155 (In) or DMSO as control (Ct) for 7 days. (C): Proportions of GFP+ CD4+ T cells after treatment of HIV-1-infected cells with YM155 (In) or control (Ct) at indicated timepoints. *p<0.05, **p<0.01 (Wilcoxon test). (D): Mean fluorescent intensity of Annexin V in GFP− and GFP+ CD4+ T cells after infection and co-culture with YM155 (In) or DMSO as control (Ct) for 7 days. *p<0.05 (Wilcoxon test). (E): Ratio of Annexin V and blue viability dye fluorescence intensity in GFP+ CD4+ T cells relative to GFP− counterparts 7 days after infection and treatment with YM155 or control. *p<0.05, **p<0.01 (Wilcoxon test). (F): Representative flow cytometry plots indicating proportions of GFP+ and mKO2+ CD4+ T cells 10 days after infection of resting CD4+ T cells with dual-reporter HIV-1. (G): Proportions of GFP+ and mKO2+ CD4+ T cells cultured for indicated times after infection with dual-reporter virus in the presence of YM155 or control. *p<0.05, **p<0.01 (Wilcoxon test).
Inhibition of BIRC5 decreases the number of in vivo HIV-1-infected CD4+ T cells
In our next experiments, we evaluated effects of BIRC5 inhibitors on the frequency of in vivo infected CD4+ T cells collected from ART-treated HIV-1 patients. For these experiments, patient-derived PBMCs were exposed to YM155 or control DMSO, followed by in vitro culture for defined time periods. Using a ddPCR assay to measure total HIV-1 LTR-gag DNA, we detected a significant reduction of virally-infected cells after pharmacological inhibition of BIRC5 (Figure 6A). For a more detailed analysis of the changes in viral reservoir cells after inhibition of BIRC5, we performed single-template, near full-length viral sequencing of cells after co-culture with or without YM155, using CD8 and NK cell-depleted PBMCs of eight randomly-selected ART-treated HIV-1-infected patients spanning a spectrum of different viral reservoir sizes (Figure 6B–C). These experiments showed that relative to control cells, YM155 significantly reduced the frequency of intact HIV-1 sequences (Figure 6D/G/H) and of the total number of HIV-1 sequences (Figure 6E). Viral sequences harboring hypermutations, 5-LTR defects, inversions or gross deletions were also reduced during treatment with YM155, but the degree of susceptibility to YM155 appeared to vary among CD4+ T cells carrying individual types of intact or defective HIV-1 sequences (Figure 6G). We also noted that YM155 treatment decreased the number of clonally-expanded sequences encoding for identical proviral copies (Figure 6F/G, see also Table S4). Together, these results suggest that pharmacological targeting of BIRC5 may be able to reduce in vivo infected viral reservoir cells encoding for intact HIV-1, and to decrease the frequency of clonally-expanded HIV-1 sequences.
Figure 6. Pharmacologic inhibition of BIRC5 diminishes the frequency of in vivo infected CD4+ T cells encoding for intact, near full-length HIV-1 sequences.
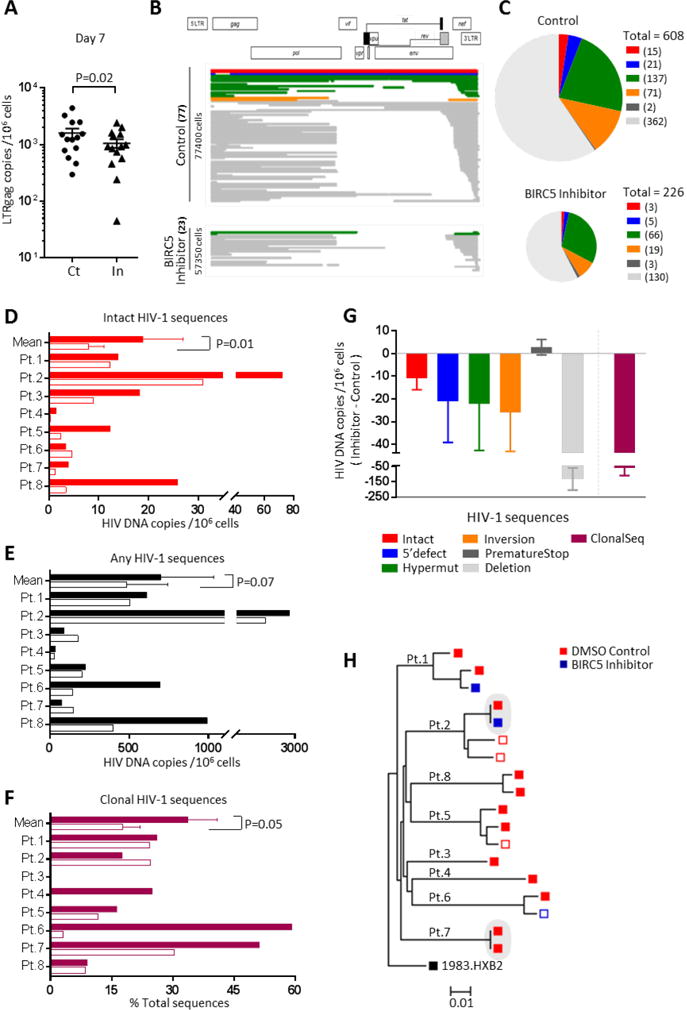
(A): HIV-1 LTR-Gag DNA copies in CD8- and NK-cell depleted PBMCs from 14 ART-treated patients co-cultured for seven days with YM155. **p<0.01 (One-tailed Wilcoxon test). (B): Diagrams highlighting viral DNA sequences generated by single-template near full-length HIV-1 PCR from CD4+ T cells isolated from an ART-treated patient (patient 8) after seven days of in vitro culture with YM 155 or control DMSO. Absolute frequencies of sequences and numbers of analyzed cells are indicated on y-axis. (C): Pie charts indicating absolute frequencies of HIV-1 DNA sequences generated by single-template near full-length HIV-1 PCR from all eight study patients, after treatment with YM155 or control. Color coding reflects intact HIV-1 DNA or sequences with defined defects. Sizes of pies correspond to total numbers of viral sequences in each condition. (D-F): Numbers of intact (D), any (E) and clonally-expanded (F) HIV-1 sequences detected in YM155-treated (open bars) and control cells (solid bars) in each of the eight study subjects. P-values were calculated using paired one-tailed Wilcoxon test. For patients 3, 4, 5, 7, 8 in panel D, no intact sequence was detected in YM155-treated conditions, respective values reflect calculated limit of detection. (G): Changes in the frequencies of indicated HIV-1 DNA sequences, or in the frequency of clusters consisting of identical viral sequences, during treatment with YM155 in all eight study patients. Data indicate mean and SEM of the difference between HIV-1 DNA sequences/million CD4+ T cells from YM155-treated and control cells. (H): Phylogenetic tree summarizing all intact HIV-1 sequences from the eight study subjects after treatment with control DMSO (red symbols) or YM155 (blue symbols). Shaded areas reflect identical sequences, consistent with clonal expansion. Solid symbols: intact sequences, open symbols: inferred intact sequences. See also Table S4.
Discussion
Owing to the tremendous progress in antiretroviral drug development, HIV-1 infection is no longer a question of life or death for the vast majority of infected individuals with access to adequate medical care (Deeks et al., 2013). However, on the level of individual viral target cells, it is clear that HIV-1 interferes profoundly with cellular death and survival pathways. Motivated by the decline of CD4+ T cells during untreated HIV-1 infection that is in some cases not fully reversed by antiretroviral therapy, significant advances have been made in understanding mechanisms by which HIV-1 induces target cell death (Doitsh and Greene, 2016). Nevertheless, it is the ability of certain HIV-1-infected CD4+ T cells to resist such death signals and to survive viral cytopathic effects that transforms HIV-1 infection into a chronic, life-long illness. In fact, if all HIV-1-infected cells would die as a result of virally-induced death programs, HIV-1 would be unable to establish extremely durable viral reservoirs that can persist for decades, despite effective antiretroviral therapy (Badley et al., 2013). Based on these considerations, we and others (Cummins et al., 2017) hypothesized that HIV-1 can initiate and activate cellular survival programs that may play a critical role for viral long-term persistence, and for the inability to cure or eradicate HIV-1 infection. In this work, we showed multiple, complementary pieces of evidence that BIRC5, an anti-apoptosis molecule known for its role in maintaining treatment-resistant reservoirs of cancer cells in the context of malignant diseases, can play a vital role in preserving long-term viability of HIV-1-infected cells: First, a global analysis of protein expression patterns using quantitative mass spectrometry-based proteomics in in vitro infected CD4+ T cells showed activation of cell survival programs that centered around BIRC5 and OX40, its upstream regulator. In addition, we observed upregulation of BIRC5 and OX40 in latently- and productively-infected resting CD4+ T cells, and higher levels of BIRC5 expression were noted in in vitro infected cells surviving HIV-1-associated death signals occurring during transition from productive to transcriptionally-silent HIV-1 infection. Moreover, in cells from ART-treated patients, we noted that OX40 surface expression denoted CD4+ T cells harboring an elevated frequency of intact viral sequences; this is consistent with a role of the OX40-BIRC5 pathway for maintaining and protecting HIV-1-infected cells in vivo, specifically during the vulnerable phase of clonal proliferation. Finally, we found that a pharmaceutical inhibitor of BIRC5 selectively reduced the number of virally-infected CD4+ T cells during in vitro culture, including those encoding for intact proviruses; this highlights the functional dependence of HIV-1-infected cells on BIRC5 and opens a perspective for reducing viral reservoir cells in clinical settings.
While BIRC5 expression seems restricted to very few cell types in adults, it is clear that expression of this marker represents a mechanism to adapt to and survive during unfavorable or vulnerable conditions (Altieri, 2010). Although most obviously detected in malignant cells, BIRC5 is also expressed under physiological conditions in CD4+ T cells, and seems to have an active role in regulating, protecting and maintaining memory CD4+ T cell responses. Our data suggest that this physiological mechanism is actively exploited by HIV-1, through an upregulation of BIRC5 in HIV-1-infected cells. Notably, given that we observed overexpression of BIRC5 in both latently- and productively-infected cells, our data suggest that signals inducing BIRC5 expression are largely independent of the transcriptional state of HIV-1. A cellular damage response, known to occur during viral chromosomal integration (Skalka and Katz, 2005), may represent a triggering signal, although other molecular signals may be required for a sustained increase of BIRC5 expression. Interestingly, a recent study raised the provocative possibility that the presence of chromosomally-integrated HIV-1, even in the absence of detectable viral gene expression, may be sufficient to increase the host cell propensity to undergo DNA damage, and promote a durable accumulation of DNA damage response signals (Piekna-Przybylska et al., 2017); the upregulation of BIRC5 and OX40 may represent a specific cell-intrinsic immune response to such events. An important question for future studies will be to determine if BIRC5 upregulation preferentially occurs in specific subsets of infected cells; given that BIRC5 is expressed in several adult human stem cells (Feng et al., 2013; Fukuda et al., 2004), it may be reasonable to hypothesize that BIRC5-mediated preservation of viral reservoir cells is most active in immature, long-lasting CD4+ T cells that sometimes can imitate stem cell behavior (Buzon et al., 2014; Jaafoura et al., 2014).
A series of recent studies have highlighted the fact that clonal proliferation of T cell encoding for intact, replication-competent virus represents a driving force for viral reservoir stabilization (Bui et al., 2017; Hosmane et al., 2017; Lee et al., 2017). Our work extends these findings by defining OX40 as a phenotypic marker for CD4+ T cells enriched for clonally-expanded HIV-1 sequences. This observation is consistent with the described functional role of the OX40-BIRC5 pathway in protecting and safeguarding T cells during the vulnerable phase of clonal proliferation (Song et al., 2005) and may help to isolate and better characterize clonally-expanded HIV-1-infected T cells in future studies. A recent study suggested that expression of CD30 (CD153, TNFRSF8), a member of the TNF surface receptor superfamily closely related to OX40, was also associated with an increased frequency of proviral HIV-1 sequences in CD4+ T cells from ART-treated patients, suggesting that the TNF receptor superfamily may be more universally involved in maintaining viral reservoir cells (Hogan et al., 2018). Based on observations in animal models, OX40 does not control initial proliferative steps but is responsible for maintaining late, long term memory T cell survival (Rogers et al., 2001; Xiao et al., 2008), and acts in conjunction with its ligand OX40L expressed on professional antigen-presenting cells (APC); how these interactions support clonal proliferation of HIV-1-infected T cells is an important area for future studies. Whether OX40 is also expressed on HIV-1-infected CD4+ T cells undergoing clonal proliferation in tissue compartments outside the peripheral blood is currently uncertain, but a recent study suggested OX40 upregulation on enteric HIV-1-infected CD4+ T cells (Yoder et al., 2017). Nevertheless, our results suggest that therapeutic targeting of OX40 may represent a possible strategy to antagonize clonal proliferation of HIV-1-infected CD4+ T cells, and to counterbalance repopulation and dissemination of virally-infected cells mediated by clonal expansion of CD4+ T cells. A recent computational analysis predicted that inhibition of HIV-1-infected CD4+ T cell proliferation could decrease the half-life of viral reservoir cells by up to 20-fold (Gerold and Hill, 2017).
Results from recent clinical studies have emphasized the high resilience of HIV-1 reservoir cells against eradication efforts. Multiple studies designed to promote viral elimination through disruption of viral latency have failed to induce meaningful reductions of viral reservoir size (Archin et al., 2012; Sogaard et al., 2015), possibly due to the ability of HIV-1-infected cells to survive during pharmacological induction of viral transcription (Shan et al., 2012) or during CD8 T cell mediated immune recognition (Huang et al., 2018). Whether BIRC5 contributes to cell survival during viral latency reversal is an interesting question that our work did not address. However, our results suggest that higher levels of BIRC5 expression during productive and latent HIV-1 infection may sensitize HIV-1-infected CD4+ T cells to BIRC5 inhibition, so that pharmacological blockade of BIRC5 may lead to a notable reduction of the viral reservoir, even when not combined with disruption of viral latency. Although the susceptibility to pharmacologic BIRC5 inhibition varied among in vivo HIV-1-infected cells and may possibly be influenced by off-target effects of the YM155 molecule, these data suggest that viral reactivation is not a prerequisite for effective targeting of latently-infected cells. Thus, pharmacological inhibition of the BIRC5-OX40 pathway may represent a promising strategy for reducing persisting reservoirs of latently infected cells in clinical settings.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Mathias Lichterfeld (mlichterfeld@partners.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patients
HIV-1-infected study participants were recruited from the Massachusetts General Hospital and the Brigham and Women’s Hospital (both in Boston, MA). PBMC samples were used according to protocols approved by the Institutional Review Board. Study subjects gave written informed consent to participate in accordance with the Declaration of Helsinki. PBMCs of 24 HIV-1 infected study participants were used for immune phenotyping study (sex distribution: 4 female and 20 male participants; ethnicity: Asian (n=1), black or African American (n=10), white (n=13); age: median of 52.5, range 25-75 years). PBMCs of 20 HIV-1 infected study participants were used for experiments with the YM155 BIRC5 inhibitor (sex distribution: 4 female and 16 male participants; ethnicity: black or African American (n=6), white (n=14); age: median of 53, range: 24- to 73 years.
METHOD DETAILS
Viral stocks
GFP-encoding R5-tropic (Ba-L) or VSV-G pseudotyped (NL4-3) HIV-1, as described previously (Unutmaz et al., 1999), were kindly provided by N. Manel and D. Littman (New York University, New York, New York, USA). A VSV-G pseudotyped dual reporter virus encoding for GFP and mKO2 was designed as described for a closely-related dual-reporter virus (Chavez et al., 2015). Viral particles were produced by transfecting 293T cells with the respective HIV-1 plasmids, using TransIT-293 (Mirus) in OptiMEM per the manufacturer’s instructions. Supernatants containing infectious retroviruses were harvested 48-72 hours after transfection, centrifuged, treated with DNase I (20 U/ml) at room temperature for 1 hour, and stored at –80°C.
Ex-vivo infection assays
For infection of activated CD4+ T cells, unselected PBMC isolated by density gradient centrifugation from HIV-1 negative donors were cultured in RPMI medium supplemented with 10% FCS and 50 IU/ml of rhIL-2, and stimulated with a CD3/CD8 bi-specific antibody as described before (Chen et al., 2011). On day 3, activated cells were infected with GFP-encoding R5-tropic (BaL) virus or VSV-G pseudotyped GFP-encoding HIV-1 (MOI = 0.5). After two hours, cells were washed twice with PBS and cultured with IL-2 supplemented medium (50 U/ml). After indicated periods of in vitro culture, cells were stained with surface antibodies and subjected to flow cytometry analysis or cell sorting. For infection of resting CD4+ T cells from HIV-1 negative donors, PBMC were infected with either the R5-tropic or VSV-G-pseudotyped GFP-encoding HIV-1 (MOI = 1) for four hours, or spinoculated (37°C, 1200g, 2 hours) with a VSV-G pseudotyped dual-reporter virus encoding for GFP and mKO2 (MOI = 2), followed by in vitro culture with IL-2 supplemented medium.
Flow cytometry and cell sorting
PBMC were stained with monoclonal antibodies to CD3 (clone OKT3), CD4+ (clone SK3 or RPA-T4), CD8 (clone SKI), CD56 (clone HCD56), CD45RO (clone UCHL1), CCR7 (clone G043H7), CD38 (clone HIT2), HLA-DR (clone L243), OX40 (clone ACT35), PD-1 (clone EH12.2H7), Lag-3 (clone 11C3C65), TIGIT (clone A15153G), CD32 (clone FUN-2) (Biolegend and Becton Dickinson), as indicated. Cells were then fixed with 2% paraformaldehyde and permeabilized using 1% Triton-X, followed by intracellular staining for BIRC5 (antibody clone EP2880Y)when indicated. Afterwards, cells were washed, and indicated cell populations were sorted in a specifically designated biosafety cabinet (Baker Hood), using a FACS Aria cell sorter (BD Biosciences) at 70 pounds per square inch. For live cell sorting, cells were stained with indicated antibodies, washed and processed to cell sorting without prior fixation. For flow cytometric analysis, cells were acquired on an LSR Fortessa flow cytometer (Becton Dickinson). Cell sorting was performed by the Ragon Institute Imaging Core Facility, and resulted in isolation of lymphocytes with the defined phenotypic characteristics of >95% purity. Data were analyzed using FlowJo software (Treestar). For imaging flow cytometry, cells were acquired using the ImageStream X MKII system (Amnis) located in the MGH Department of Pathology Flow and Image Cytometry Core. Images of the cells were analyzed with IDEAS.6 software (Amnis).
Liquid Chromatography-Mass Spectrometry
Liquid chromatography-mass spectrometry analyses were performed as previously described in detail (Mertins et al., 2013; Svinkina et al., 2015). Briefly, 1-2×106 sorted CD4+ T cells were pelleted and washed. Cellular proteins were extracted, disulfide bonds reduced and alkylated, and proteins digested with LysC followed by trypsin. The resulting peptides in each sample were subsequently labeled for quantitative analysis with one of the mass tag reagents comprising the 10-plex TMT reagent (Thermo Fisher). This approach enabled simultaneous analysis of 9 samples and a common reference sample constructed of aliquots of all 9 samples, as previously described (Mertins et al., 2016) and shown in supplemental Figure 1A. Labeled peptides from all 10 labeled samples were combined and fractionated by basic reversed-phase (bRP) chromatography to decrease sample complexity and increase the dynamic range of detection. The global proteome of each sample was measured with 24 bRP fractions on a Thermo Q Exactive Plus instrument. A normalized collision energy of 27 was used for TMT-samples. MS/MS scans were acquired at a resolution of 35,000. All MS/MS raw data were analyzed using Spectrum Mill Proteomics Workbench (Agilent Technologies) searching against a RefSeq human concatenated database. A precursor mass tolerance of 20 ppm and a product ion tolerance of 20 ppm (higher-energy collisional dissociation spectra) were allowed. Tryptic enzyme specificity was required, and up to four missed cleavages were allowed. Carbamidomethylation of cysteine and TMT-tags on lysine residues and peptide N termini were employed as fixed modifications. Lysine acetylation and methionine oxidation were set as variable modifications. Protein quantification was based on defining ratios of mass-tag reporter ions and rationing the same peptide values across samples and to the common reference as described (Mertins et al., 2016; Mertins et al., 2013). Settings in Spectrum Mill were adjusted to provide a peptide FDR of 1-2% and a protein FDR of 0-1%. Only proteins with >2 peptides and at least 2 TMT ratios in each replicate were counted as being identified and quantified. Differentially-expressed proteins between GFP+ and GFP− CD4+ T cells were analyzed using a moderated F test with the “lmFit” function in limma (Ritchie et al., 2015). Gene ontologies of differentially expressed proteins were identified using Ingenuity Pathway Analysis (IPA, Qiagen).
HIV-1 LTR-Gag DNA quantification
Sorted CD4+ T cell populations were digested with lysis buffer to extract cell lysates, or subjected to DNA extraction using the DNeasy Blood & Tissue kit (Qiagen). We amplified HIV-1 LTR-Gag DNA using digital droplet PCR (Bio-Rad), with primers and probes described previously (Buzon et al., 2014) (127bp 5’LTR-gag amplicon; HXB2 coordinates 684-810). PCR was performed using the following program: 95 for 10 min, 45 cycles of 94 for 30s and 60 for 1 min, 72 for 1 min. The droplets were subsequently read by the QX100 droplet reader and data were analyzed using QuantaSoft software (Bio-Rad) (Kiselinova et al., 2016).
Single-genome, near-full length proviral sequencing
Genomic DNA was extracted from indicated cell populations using the QIAGEN DNeasy Blood & Tissue kit (Qiagen). DNA diluted to single genome levels based on Poisson distribution statistics and ddPCR results was subjected to single-genome amplification using Invitrogen Platinum Taq and nested primers spanning near full-length HIV-1 (HXB2 coordinates 638-9632). Primers were previously published (Lee et al., 2017). PCR products were visualized by agarose gel electrophoresis. All sequences were subjected to Illumina MiSeq sequencing with a median of approximately 2500 reads per base. Resulting short reads were de novo assembled and aligned to HXB2 using MUSCLE (Edgar, 2004) to identify internal inversions, large deletions, premature/lethal stop codons, or packaging signal defects. Presence/absence of APOBEC-3G/3F-associated hypermutations was determined using the Los Alamos HIV Sequence Database Hypermut 2.0 (Rose and Korber, 2000) program. Viral sequences that lacked all mutations listed above and had <15 base pairs deletions in the sequenced 5-LTR region were classified as “intact” (Bruner et al., 2016). If a near-full length sequence showed a mapped 5’ deletion that removes the primer-binding site, but otherwise showed no lethal sequence defects, the missing 5’ sequence was inferred to be present, and this sequence was considered as an “inferred intact” HIV-1 sequence. Phylogenetic distances between sequences were examined using ClustalX-generated neighbor joining algorithms (Larkin et al., 2007).
Functional assays
To test the influence of BIRC5 inhibition on survival and persistence of HIV-1-infected CD4+ T cells, PBMCs from HIV-1-negative persons were infected with HIV-1 as described above, and incubated in R10 medium supplemented with IL-2 (25 IU/ml) in the presence of YM155 (Selleckchem) at a concentration of 200nM, or DMSO as control; at selected time points, cells were subjected to flow cytometric analysis. For in vivo infected cells derived from ART-treated HIV-1 patients, PBMC were depleted of CD8 and NK cells using immunomagnetic cell separation (autoMACS Pro, Miltenyi) and incubated in IL-2 supplemented R10 medium containing YM155 (200nM) or DMSO. Zidovudine (AZT, obtained from the NIH AIDS reagent repository #3485) was added at a concentration of 400nM to block spreading of autologous virus during in vitro culture. After indicated culture periods, cells were subjected to HIV-1 gag quantification using ddPCR, or processed to cell sorting for isolation of viable cells, determined by Blue viability dye intensity, and subsequent single-genome, near full-length viral sequencing (Lee et al., 2017).
QUANTIFICATION and STATISTICAL ANALYSIS
Data are summarized as individual data plots or pie charts. Differences were tested for statistical significance using a Mann Whitney U test, paired Wilcoxon test, or Friedman test, followed by Dunn’s test for multiple comparison where indicated.
DATA AND SOFTWARE AVAILABILITY
Mass spectrometry data were deposited in MassIVE (https://massive.ucsd.edu; accession number xx).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD3/CD8 bispecific antibody | NIH AIDS reagent repository | Cat#12277 |
| Mouse anti human CD3 monoclonal antibody (clone OKT3) | Biolegend | Cat# 317322 |
| Mouse anti human CD4 monoclonal antibody (clone SK3 or RPA-T4) | Biolegend | Cat# 344620; Cat# 300508, Cat# 353212 |
| Mouse anti human CD8 monoclonal antibody (clone SKI) | BD Biosciences | Cat# 300046 |
| Mouse anti human CD45RO monoclonal antibody (clone UCHL1) | Biolegend | Cat# 304236 |
| Mouse anti human CCR7 monoclonal antibody (clone G043H7) | Biolegend | Cat# 353204 |
| Mouse anti human CD38 monoclonal antibody (clone HIT2) | Biolegend | Cat# 303510 |
| Mouse anti human HLA-DR monoclonal antibody (clone L243) | Biolegend | Cat# 307618 |
| Mouse anti human PD-1 monoclonal antibody (clone EH12.2H7) | Biolegend | Cat# 329918 |
| Mouse anti human TIGIT monoclonal antibody (clone A15153G) | Biolegend | Cat# 372710 |
| Mouse anti human LAG-3 monoclonal antibody (clone 11C3C65) | Biolegend | Cat# 369308 |
| Mouse anti human OX40 monoclonal antibody (clone ACT35) | Biolegend | Cat# 350012, Cat# 350018 |
| Mouse anti human CD32 monoclonal antibody (clone FUN-2) | Biolegend | Cat# 303216 |
| Mouse anti human CD56 monoclonal antibody (clone HCD56) | Biolegend | Cat# 318306 |
| Rabbit anti human BIRC5 monoclonal antibody | Abcam | Cat# EP2880Y |
| Bacterial and Virus Strains | ||
| GFP-encoding R5-tropic (Ba-L) | N. Manel and D. Littman, New York University | N/A |
| VSV-G pseudotyped (NL4-3) HIV-1 | N. Manel and D. Littman, New York University | N/A |
| VSV-G pseudotyped dual reporter virus | E. Battivelli and E. Verdin, Buck Institute for Research on Aging | N/A |
| Biological Samples | ||
| Human PBMCs from HIV-negative subjects | Massachusetts General Hospital | N/A |
| HIV infected patient derived PBMCs | Massachusetts General Hospital, NIH, and Brigham and Women’s Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| rhIL-2 | NIH AIDS reagent repository | Cat#11697 |
| Annexin V | Biolegend | Cat# 550475 |
| YM155 (Sepantronium Bromide) | Selleckchem | Cat# S1130 |
| Zidovudine (AZT) | NIH AIDS reagent repository | Cat#3485 |
| Critical Commercial Assays | ||
| 10-plex TMT reagent | Thermo Fisher | Cat# 90049 |
| DNeasy Blood & Tissue kit | QIAGEN | Cat# 69504 |
| ddPCR™ Supermix for Probes | Bio-Rad | Cat#1863010 |
| Platinum Taq DNA Polymerase High Fidelity | Invitrogen | Cat# 11304102 |
| Deposited Data | ||
| Proteomic data (raw and analyzed) | This paper | |
| Oligonucleotides | ||
| Primer for HIV-1 LTR-Gag digital PCR (Forward): TCTCGACGCAGGACTCG | Buzon et al., 2014 | N/A |
| Primer for HIV-1 LTR-Gag digital PCR (Reverse): TACTGACGCTCTCGCACC | Buzon et al., 2014 | N/A |
| Probe for HIV-1 LTR-Gag digital PCR: 56-FAM/CTCTCTCCT/ZEN/TCTAGCCTC/31ABkFQ | Buzon et al., 2014 | N/A |
| Primer for RPP30 digital PCR (Forward): GATTTGGACCTGCGAGCG | Buzon et al., 2014 | N/A |
| Primer for RPP30 digital PCR (Reverse): GCGGCTGTCTCCACAAGT | Buzon et al., 2014 | N/A |
| Probe for RPP30 digital PCR: 56-FAM/CTGACCTGA/ZEN/AGGCTCT/31ABkFQ | Buzon et al., 2014 | N/A |
| Primers for near full-length HIV-1 PCR (1st Forward): AAATCTCTAGCAGTGGCGCCCGAACAG | Li et al. 2007 | N/A |
| Primers for near full-length HIV-1 PCR (1st Reverse): TGAGGGATCTCTAGTTACCAGAGTC | Li et al. 2007 | N/A |
| Primers for near full-length HIV-1 PCR (2nd Forward): GCGCCCGAACAGGGACYTGAAARCGAAAG | Li et al. 2007 | N/A |
| Primers for near full-length HIV-1 PCR (2nd Reverse): GCACTCAAGGCAAGCTTTATTGAGGCTTA | Li et al. 2007 | N/A |
| Software and Algorithms | ||
| Spectrum Mill Proteomics Workbench | Agilent Technologies | https://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter-for-life-science-research/spectrum-mill |
| Ingenuity Pathway Analysis | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/; RRID:SCR_008653 |
| IDEAS.6 | Amnis | https://www.amnis.com/index.php/ideas.html |
| Flowjo v10 | Treestar | https://www.flowjo.com/; RRID:SCR_008520 |
| Algorithms for analysis of digital PCR results output from Bio-Rad QuantaSoft software | Kiselinova et al., 2016 | N/A |
| MUSCLE | Edgar, 2004 | https://www.bioconductor.org/packages/release/bioc/html/muscle.html; RRID:SCR_011812 |
| Los Alamos HIV Sequence Database Hypermut 2.0 | Rose and Korber, 2000 | https://www.hiv.lanl.gov/content/sequence/HYPERMUT/background.html; RRID:SCR_011349 |
| Los Alamos HIV Sequence Database Gene Cutter | N/A | https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html; RRID:SCR_011349 |
| ClustalX | Larkin et al., 2007 | http://www.clustal.org |
| Prism | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
Supplementary Material
Table S1. Related to Figure 1; Differentially expressed proteins between GFP+ and GFP− CD4+ T cells after infection with GFP-encoding HIV-1.
Table S2. Related to Figure 1; Differentially expressed proteins involved in cell survival, predicted by Ingenuity Pathway Analysis (IPA).
Highlights.
HIV-1-infected CD4+ T cells activate cellular survival programs
BIRC5 and OX40 are upregulated in CD4+ T cells during latent and productive HIV-1infection
OX40-expressing CD4+ T cells from ART-treated HIV-1 patients are enriched for clonally-expanded HIV-1 sequences
Pharmacologic inhibition of BIRC5 reduces the frequency of CD4+ T cells encoding for intact HIV-1
Acknowledgments
ML is supported by NIH grants AI098487, AI106468, AI114235, AI117841, AI120008, AI124776. XGY is supported by NIH grants AI116228, AI078799, HL134539 and AI125109. This work was supported in part by the Harvard University Center for AIDS Research (CFAR), an NIH-funded program (AI060354) that is supported by the following NIH Institutes and Centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NCCAM. The authors gratefully acknowledge the support of the MGH sequencing core facility. The authors wish to thank Michael Waring and Nathalie Bonheur (Ragon Institute Imaging Core, part of the Harvard CFAR Advanced Technologies Core) for technical assistance with flow cytometry and cell sorting, and Scott Mordecai (MGH Department of Pathology Flow and Image Cytometry Core, supported in part by an NIH Shared Instrumentation Grant 1S10OD012027-01A1, FIP) for support with the Amnis ISX MKII-based ImageStream analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Study concept and design: HK, XGY, ML
Mass spectrometry-based proteomics and data analysis: RA, MS, SC
Flow cytometry and cell sorting: HK, DK
Functional cell culture assays: HK
PCR and viral sequencing: HK
Data analysis and interpretation: HK, ZO, RA, CG, HC, GQL, XGY, ML
Contribution of PBMC samples: TWC, AT, ESR
Contribution of dual-reporter virus: EB, EV
Study supervision: ML
Conflicts of Interests
The authors declare that conflicts of interests do not exist.
References
- Altieri DC. Survivin and IAP proteins in cell-death mechanisms. The Biochemical journal. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC. Survivin - The inconvenient IAP. Seminars in cell & developmental biology. 2015;39:91–96. doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature medicine. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Andersson KM, Brisslert M, Cavallini NF, Svensson MN, Welin A, Erlandsson MC, Ciesielski MJ, Katona G, Bokarewa MI. Survivin co-ordinates formation of follicular T-cells acting in synergy with Bcl-6. Oncotarget. 2015;6:20043–20057. doi: 10.18632/oncotarget.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badley AD, Sainski A, Wightman F, Lewin SR. Altering cell death pathways as an approach to cure HIV infection. Cell death & disease. 2013;4:e718. doi: 10.1038/cddis.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nature medicine. 2016;22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS pathogens. 2017;13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nature medicine. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L, Calvanese V, Verdin E. HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells. PLoS pathogens. 2015;11:e1004955. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y, et al. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. The Journal of clinical investigation. 2011;121:1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Clemens MR, Gladkov OA, Gartner E, Vladimirov V, Crown J, Steinberg J, Jie F, Keating A. Phase II, multicenter, open-label, randomized study of YM155 plus docetaxel as first-line treatment in patients with HER2-negative metastatic breast cancer. Breast cancer research and treatment. 2015;149:171–179. doi: 10.1007/s10549-014-3238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunological reviews. 2009;229:173–191. doi: 10.1111/j.1600-065X.2009.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins NW, Badley AD. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell death & disease. 2010;1:e99. doi: 10.1038/cddis.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins NW, Badley AD. Casp8p41 and HIV. Oncotarget. 2015;6:23042–23043. doi: 10.18632/oncotarget.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins NW, Sainski-Nguyen AM, Natesampillai S, Aboulnasr F, Kaufmann S, Badley AD. Maintenance of the HIV Reservoir Is Antagonized by Selective BCL2 Inhibition. Journal of virology. 2017;91 doi: 10.1128/JVI.00012-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska A, Kim N, Aldovini A. Tat-induced FOXO3a is a key mediator of apoptosis in HIV-1-infected human CD4+ T lymphocytes. Journal of immunology. 2008;181:8460–8477. doi: 10.4049/jimmunol.181.12.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382:1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Greene WC. Dissecting How CD4 T Cells Are Lost During HIV Infection. Cell host & microbe. 2016;19:280–291. doi: 10.1016/j.chom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Zhou S, Liu Y, Song D, Luan Z, Dai X, Li Y, Tang N, Wen J, Li L. Sox2 protects neural stem cells from apoptosis via up-regulating survivin expression. The Biochemical journal. 2013;450:459–468. doi: 10.1042/BJ20120924. [DOI] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature medicine. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS pathogens. 2016;12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Mantel CR, Pelus LM. Survivin regulates hematopoietic progenitor cell proliferation through p21WAF1/Cip1-dependent and -independent pathways. Blood. 2004;103:120–127. doi: 10.1182/blood-2003-05-1756. [DOI] [PubMed] [Google Scholar]
- Gerold JM, Hill AL. Estimating the contribution of proliferation to HIV-infected lymphocyte persistence. Program and abstracts of the 9th IAS Conference on HIV Science; Paris, France. 2017. [Google Scholar]
- Greenwood EJ, Matheson NJ, Wals K, van den Boomen DJ, Antrobus R, Williamson JC, Lehner PJ. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. eLife. 2016;5 doi: 10.7554/eLife.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, Thanh C, Gibson EA, Carvidi AB, Smith LCB, et al. Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30. PLoS pathogens. 2018;14:e1006856. doi: 10.1371/journal.ppat.1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, Siliciano RF. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. The Journal of experimental medicine. 2017;214:959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. The Journal of clinical investigation. 2018;128:876–889. doi: 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells. Nature communications. 2014;5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kim SM, Kim YK, Hong SP, Kim MJ, Myoung H. Evaluation of survivin as a prognostic marker in oral squamous cell carcinoma. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2010;39:368–375. doi: 10.1111/j.1600-0714.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS pathogens. 2016;12:e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. The Journal of clinical investigation. 2017;127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghu B, Franzosa EA, Xia Y. Construction of functional linkage gene networks by data integration. Methods in molecular biology. 2013;939:215–232. doi: 10.1007/978-1-62703-107-3_14. [DOI] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nature methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Arias I, Doitsh G, Yang Z, Sowinski S, Ruelas D, Greene WC. Blood-Derived CD4 T Cells Naturally Resist Pyroptosis during Abortive HIV-1 Infection. Cell host & microbe. 2015;18:463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Sharma G, Maggirwar SB, Bambara RA. Deficiency in DNA damage response, a new characteristic of cells infected with latent HIV-1. Cell cycle. 2017;16:968–978. doi: 10.1080/15384101.2017.1312225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel F, Sprenger T, Kaina B, Liersch T, Rodel C, Fulda S, Hehlgans S. Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Current medicinal chemistry. 2012;19:3679–3688. doi: 10.2174/092986712801661040. [DOI] [PubMed] [Google Scholar]
- Rogers PR, Song J, Gramaglia I, Killeen N, Croft M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity. 2001;15:445–455. doi: 10.1016/s1074-7613(01)00191-1. [DOI] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G –> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. The chromosomal passenger complex: one for all and all for one. Cell. 2007;131:230–231. doi: 10.1016/j.cell.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, De Spiegelaere W, Cozzi-Lepri A, Kiselinova M, Pollakis G, Beloukas A, Vandekerckhove L, Strain M, Richman D, Phillips A, et al. During Stably Suppressive Antiretroviral Therapy Integrated HIV-1 DNA Load in Peripheral Blood is Associated with the Frequency of CD8 Cells Expressing HLA-DR/DP/DQ. EBioMedicine. 2015;2:1153–1159. doi: 10.1016/j.ebiom.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainski AM, Dai H, Natesampillai S, Pang YP, Bren GD, Cummins NW, Correia C, Meng XW, Tarara JE, Ramirez-Alvarado M, et al. Casp8p41 generated by HIV protease kills CD4 T cells through direct Bak activation. The Journal of cell biology. 2014;206:867–876. doi: 10.1083/jcb.201405051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat SF, Karakiewicz PI, Godoy G, Karam JA, Ashfaq R, Fradet Y, Isbarn H, Montorsi F, Jeldres C, Bastian PJ, et al. Survivin as a prognostic marker for urothelial carcinoma of the bladder: a multicenter external validation study. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:7012–7019. doi: 10.1158/1078-0432.CCR-08-2554. [DOI] [PubMed] [Google Scholar]
- Sharief MK, Semra YK. Heightened expression of survivin in activated T lymphocytes from patients with multiple sclerosis. Journal of neuroimmunology. 2001;119:358–364. doi: 10.1016/s0165-5728(01)00389-7. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature medicine. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell death and differentiation. 2005;12(Suppl 1):971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS pathogens. 2015;11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, So T, Cheng M, Tang X, Croft M. Sustained survivin expression from OX40 costimulatory signals drives T cell clonal expansion. Immunity. 2005;22:621–631. doi: 10.1016/j.immuni.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, Carr SA. Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyllysine Antibodies and an Optimized Proteomic Workflow. Molecular & cellular proteomics: MCP. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer research. 1998;58:5315–5320. [PubMed] [Google Scholar]
- team., T.R.c. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2014. http://www.R-project.org/ [Google Scholar]
- Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. The Journal of experimental medicine. 1999;189:1735–1746. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Kroemer A, Gao W, Ishii N, Demirci G, Li XC. OX40/OX40L costimulation affects induction of Foxp3+ regulatory T cells in part by expanding memory T cells in vivo. Journal of immunology. 2008;181:3193–3201. doi: 10.4049/jimmunol.181.5.3193. [DOI] [PubMed] [Google Scholar]
- Xie Y, Ma X, Gu L, Li H, Chen L, Li X, Gao Y, Fan Y, Zhang Y, Yao Y, Zhang X. Prognostic and Clinicopathological Significance of Survivin Expression in Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. Scientific reports. 2016;6:29794. doi: 10.1038/srep29794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AC, Guo K, Dillon SM, Phang T, Lee EJ, Harper MS, Helm K, Kappes JC, Ochsenbauer C, McCarter MD, et al. The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria. PLoS pathogens. 2017;13:e1006226. doi: 10.1371/journal.ppat.1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 1; Differentially expressed proteins between GFP+ and GFP− CD4+ T cells after infection with GFP-encoding HIV-1.
Table S2. Related to Figure 1; Differentially expressed proteins involved in cell survival, predicted by Ingenuity Pathway Analysis (IPA).


