Abstract
Copper-binding metallophores, or chalkophores, play a role in microbial copper homeostasis that is analogous to that of siderophores in iron homeostasis. The best-studied chalkophores are members of the methanobactin (Mbn) family—ribosomally produced, posttranslationally modified natural products first identified as copper chelators responsible for copper uptake in methane-oxidizing bacteria. To date, Mbns have been characterized exclusively in those species, but there is genomic evidence for their production in a much wider range of bacteria. This review addresses the current state of knowledge regarding the function, biosynthesis, transport, and regulation of Mbns. While the roles of several proteins in these processes are supported by substantial genetic and biochemical evidence, key aspects of Mbn manufacture, handling, and regulation remain unclear. In addition, other natural products that have been proposed to mediate copper uptake as well as metallophores that have biologically relevant roles involving copper binding, but not copper uptake, are discussed.
Keywords: copper homeostasis, natural product, metallophore, chalkophore, methanobactin, siderophore, bioinorganic chemistry
INTRODUCTION
Metals play key roles in many biological processes. It has been estimated that up to half of all proteins contain a metal (1). While metals are purely structural elements in some proteins, the redox capabilities of transition metals are required for electron transport (2), and redox-active metals play a catalytic role in many families of enzymes (1). Metal deficiency is correspondingly a significant stressor that limits viability. However, the oxidative stress caused by surplus transition metals can also be fatal to a cell (3). All forms of life thus possess intricate metal homeostasis systems that govern both influx and efflux of metals, allowing the cell to maintain a carefully regulated pool of bioavailable metal in the correct oxidation state(s).
Efflux of transition metals has been extensively studied across all forms of life, but metal influx has received less attention. Iron acquisition in prokaryotes is the best-understood example. Despite the many biological processes that depend on iron, its low solubility under aerobic conditions and at neutral pH significantly limits its bioavailability (4). As a result, strategies have evolved to facilitate active iron uptake. The canonical microbial strategy is the use of siderophores, small iron-binding natural products that are secreted from cells and that bind extracellular iron with high affinity (5). The iron-bound siderophores are then taken back up into the cell, where the iron is liberated from the compound and incorporated into the cellular iron pool (6). These systems are particularly common in pathogenic bacteria, which compete with host cells for iron, and genomic cassettes related to siderophore biosynthesis and transport can be markers of virulence (7, 8). Similar strategies exist in fungi (9) and plants (10), but the focus of this review is bacterial metal acquisition.
Over the past 15 years, it has become increasingly apparent that this strategy is not limited to iron (11, 12). In species with a high need for other metals or in situations of metal deficiency, including host sequestration of metals from pathogenic bacteria, metallophores (biogenic ligands that facilitate metal ion uptake) can be deployed (13). Some metallophores also have secondary roles related to defense against metal toxicity. Although better known as siderophores, rhizoferrin and some desferrioxamines and pyoverdines have higher affinities for manganese than they do for iron (14–16). The broad-spectrum metallophore staphylopine binds both nickel and cobalt, and other compounds, including l-His2, l-His/2-methyl-2,4-thiazolidinedicarboxylic acid, and yet-uncharacterized metallophores, are implicated in nickel uptake as well (17–19). Staphylopine can also bind zinc, as can yersiniabactin (in a biologically relevant role), and other putative zincophores such as coelibactin have been identified (17, 20, 21). Gold toxicity can be minimized by the chelation of the metal by delftibactin (22). Even molybdenum and vanadium have possible metallophores, in the form of azotobactin, aminochelin, and protochelin (23, 24). However, the best-studied family of noniron metallophores are copper-binding natural products known as chalkophores (chalko- is derived from the Greek word for copper), and chief among them are the methanobactins (Mbns) (25).
Mbns are peptidic natural products that chelate copper via paired nitrogen-containing heterocycles and thioamide/enethiol moieties (26) (Figure 1a). They have an exceedingly high affinity for copper and bind copper from soluble or mineral sources upon secretion (27). The copper-chelated Mbn (CuMbn) is then internalized as a source of copper for cells that have a high requirement for this metal (28). Significant inroads have been made into our understanding of Mbn transport, regulation, and biosynthesis, rendering Mbns the largest and best-understood group of chalkophores. Although Mbns are the major focus of this review, other natural products with proposed roles in copper uptake and metallophores with secondary roles involving copper are addressed as well.
Figure 1.
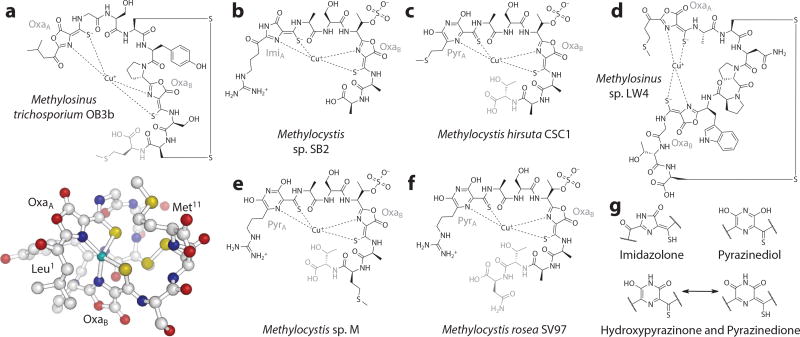
Structures of methanobactins (Mbns) characterized to date. (a) The structure of copper-chelated Mbn (CuMbn) (top) from Methylosinus (Ms.) trichosporium OB3b. The C-terminal methionine, in gray, is sometimes absent from the final compound. The crystal structure (bottom) clearly shows the distorted tetrahedral geometry of the copper binding site. (b) The structure of CuMbn from Methylocystis (Mc.) sp. SB2. Based on the precursor peptide sequences for this and other Methylocystis Mbns, additional residues are present at the C terminus of these compounds during biosynthesis but have not yet been experimentally observed. (c) The structure of Mbn from Mc. hirsuta CSC1. Determined via X-ray crystallography, this and the remaining Methylocystis Mbns have six-membered rings (assigned as a pyrazinediol) as the first heterocycle. Residues in gray are sometimes absent. (d) The structure of Ms. sp. LW4 CuMbn, which, like Ms. trichosoporium OB3b CuMbn, contains two oxazolone rings and neighboring thioamide groups. (e) The structure of Mbn from Mc. sp. M. Residues in gray are sometimes absent. (f) The structure of CuMbn from Mc. rosea SV97. Residues in gray are sometimes absent. (g) Potential identities for the “N-terminal” heterocycle in Mbns from Methylocystis species. Figure adapted with permission from References 25 and 215.
METHANOBACTINS
Copper Homeostasis in Methanotrophs
Research into prokaryotic copper homeostasis has traditionally focused on copper efflux (29). While copper enzymes play important roles across all families of life, they are comparatively few in number in prokaryotes (30), and Fenton chemistry mediated by unbound copper causes significant oxidative stress, rendering copper toxicity a significant risk (29). However, in species in which key metabolic enzymes require copper cofactors, copper deficiency is a major problem that can be addressed by the use of chalkophores. Methanotrophic bacteria, which oxidize methane as their sole carbon source, are a widespread group of bacteria with a high demand for copper (31), and Mbn was first identified in these bacteria.
In methanotrophic bacteria, two entirely unrelated families of metalloenzymes can carry out the aerobic oxidation of methane to methanol: a cytoplasmic iron enzyme, soluble methane monooxygenase (sMMO) (32), and a copper enzyme, particulate methane monooxygenase (pMMO) (33), which is found in intracytoplasmic membranes with unclear connectivity to periplasmic membranes (34) (Figure 2). Some methanotrophs possess both enzymes, and in these species, the two enzymes are reciprocally regulated by copper, with pMMO produced whenever sufficient copper is available (31, 35). pMMO is produced in massive quantities, composing nearly a fifth of the cellular protein mass (36). Since pMMO is an ~300-kDa trimeric enzyme with at least three copper sites (33), the resulting demand for copper far outstrips that of many other bacteria, and conditions that would provide more than enough copper for well-studied gram-negative Pseudomonas and Escherichia species constitute copper stress for methanotrophs. It is thus unsurprising that some of the best-understood copper acquisition (rather than copper efflux) systems have been identified in these bacteria.
Figure 2.
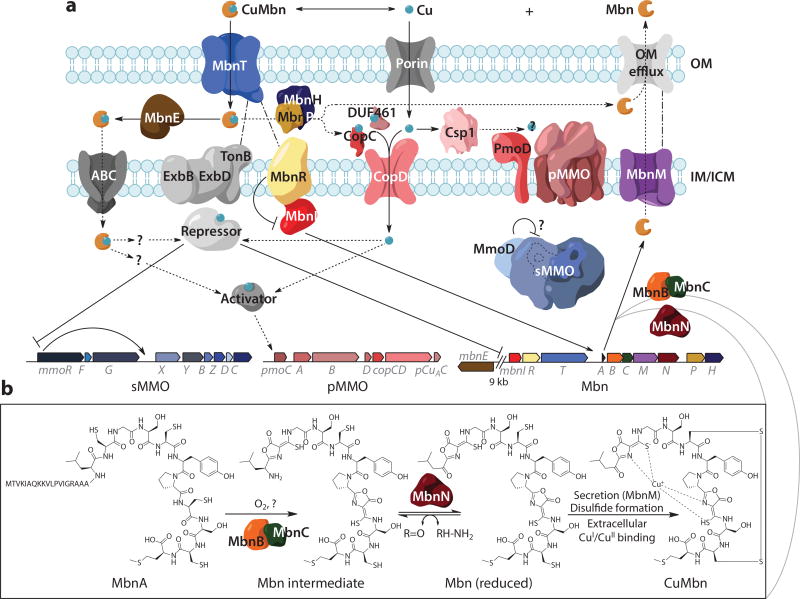
Current model for processes related to (a) copper homeostasis and methanobactin (Mbn) transport, regulation, and (b) biosynthesis in Methylosinus trichosporium OB3b. Abbreviations: ABC, ATP-binding cassette transporter; CuMbn, copper-chelated Mbn; ICM, intracytoplasmic membrane; IM, inner membrane; OM, outer membrane; pMMO, particulate methane monooxygenase; sMMO, soluble methane monooxygenase. Figure adapted with permission from References 25 and 215.
The Identification of Methanobactins in Methanotrophs
Although the importance of copper to methanotrophy had been recognized a decade earlier, the first evidence for a dedicated copper uptake system in the form of Mbns came from a set of mutant strains constructed from the methanotroph Methylosinus (Ms.) trichosporium OB3b. A set of constitutively sMMO-producing strains was developed (37), and during the extensive screening of these strains, initial evidence was obtained for the existence of small extracellular copper-binding compounds (38). Within the next five years, two groups had attempted to characterize the compound, variously described as the CBL (copper-binding ligand) and the CBC (copper-binding compound), and though the compound was not fully purified or structurally characterized, it was correctly described as having a partly peptidic nature (39, 40).
Structural and Biophysical Characteristics of Methanobactin from Methylosinus trichosporium OB3b
Structural characterization of Mbn from Ms. trichosporium OB3b was not achieved until 2004, when that compound was successfully crystallized (26) and the term chalkophore was first coined. In this structure, a single copper ion is coordinated in a distorted tetrahedral geometry by two nitrogen-containing heterocycles and two neighboring thioamide or enethiol groups (26) (Figure 1a). Initially assigned as imidazolone rings, later NMR analysis showed that the heterocycles in the Ms. trichosporium OB3b Mbn are oxazolones (OxaA and OxaB) (41). The remainder of Mbn is peptidic, although replacement of the N-terminal primary amine with a carbonyl resulted in the initial misidentification of the neighboring leucine side chain as an isopropylester group (26, 41). The C-terminal methionine group is sometimes absent in Mbn isolated from spent medium, and despite unaltered Cu(I)-binding capabilities, its reduction potential differs slightly (42). The etiology of this truncated Mbn is unclear.
The two oxazolone rings present in Mbn are responsible for two absorption features at 345 nm and 392 nm in the apo compound, and excitation at these wavelengths results in fluorescence emission in the 400–500 nm region (43). Local absorption maxima in the 330–350 nm range are common for oxazolone compounds, but extension of the heterocycle’s conjugation system results in a bathochromic shift (44), as observed for OxaA in Mbn, which is adjacent to an additional ketone group in lieu of the N-terminal amine. Copper binding by the oxazolones both quenches fluorescence emission intensity and alters their absorption maxima (43). These rings are prone to acid-catalyzed hydrolysis when not stabilized by copper binding (43, 45).
Both Cu(II) and Cu(I) can bind Mbn, but binding is reductive because, at stoichiometric levels of copper and Mbn, only EPR-silent Cu(I)Mbn is present in the final compound (46), and even after binding Cu(II), only Cu(I)Mbn is present after the first 10 minutes (47). The mechanism of Cu(II) reduction to Cu(I) has not yet been elucidated. Under certain conditions, other copper:Mbn stoichiometries are observed, at least temporarily (48, 49), and Cu2Mbn is sometimes observed when copper is added in excess.
Due to its high copper affinity, Mbn can extract copper from a wide range of otherwise biounavailable mineral sources, including borosilicate glass and humic acid species (50, 51). These observations are consistent with a major role for Mbn in liberating copper from organic-rich environments with a near-neutral pH, such as peatlands, rice paddies, lake sediments, and soils, which are some of the common habitats for methanotrophs (51, 52). Mbn can also bind metals other than copper. Some metal ions, including Ag(I), Au(III), Hg(II), Pb(II), and U(VI) appear to bind via a similar mechanism to Cu(I) and Cu(II), and for these metals, reductive binding is observed (53). Others, including Cd(II), Co(II), Fe(III), Ni(II), and Zn(II), appear to bind as bischelates and are not reduced (53). Measured binding constants for metals other than copper appear to be lower than that of Cu(I) by 10–15 orders of magnitude (53).
Early research probed not only interactions between Mbn and metals but also interactions between Mbn and the primary methanotroph copper-dependent enzyme pMMO. Mbn was initially reported to play a direct role in pMMO activity, though no evidence for a direct interaction has been found to date. There are conflicting data regarding whether or not CuMbn addition affects the activity of purified pMMO (47, 54). Apo Mbn addition has been reported to increase activity (54), but surplus copper or zinc binding can inactivate pMMO (55, 56), and it may be that apo Mbn is merely removing inhibitory metals.
Genome Mining Reveals a Wider World of Methanobactins
The mode of Mbn biosynthesis was unknown until 2010, but nonribosomal peptide synthesis (NRPS) was a common hypothesis (43, 57). This was ruled out by the sequencing of the Ms. trichosporium OB3b genome (58): An open reading frame encoding a 30-amino-acid peptide with a C-terminal sequence resembling the peptidic Mbn backbone was detected (Figure 2). In addition, a homolog of that gene (now termed mbnA), along with homologs of the surrounding genes, was identified in the genome of the nonmethanotrophic diazotrophic endophyte Azospirillum sp. B510 (45). Genome mining identified 18 putative Mbn operons in 16 species, and in subsequent years that number has increased to 74 operons in 71 species, spanning both gram-negative and gram-positive bacteria (25, 59) (Figure 3a,b; Supplemental Figure 1). Construction of a knockout strain of Ms. trichosporium OB3b in which mbnA is eliminated eradicates Mbn production (60), consistent with a role in Mbn biosynthesis.
Figure 3.
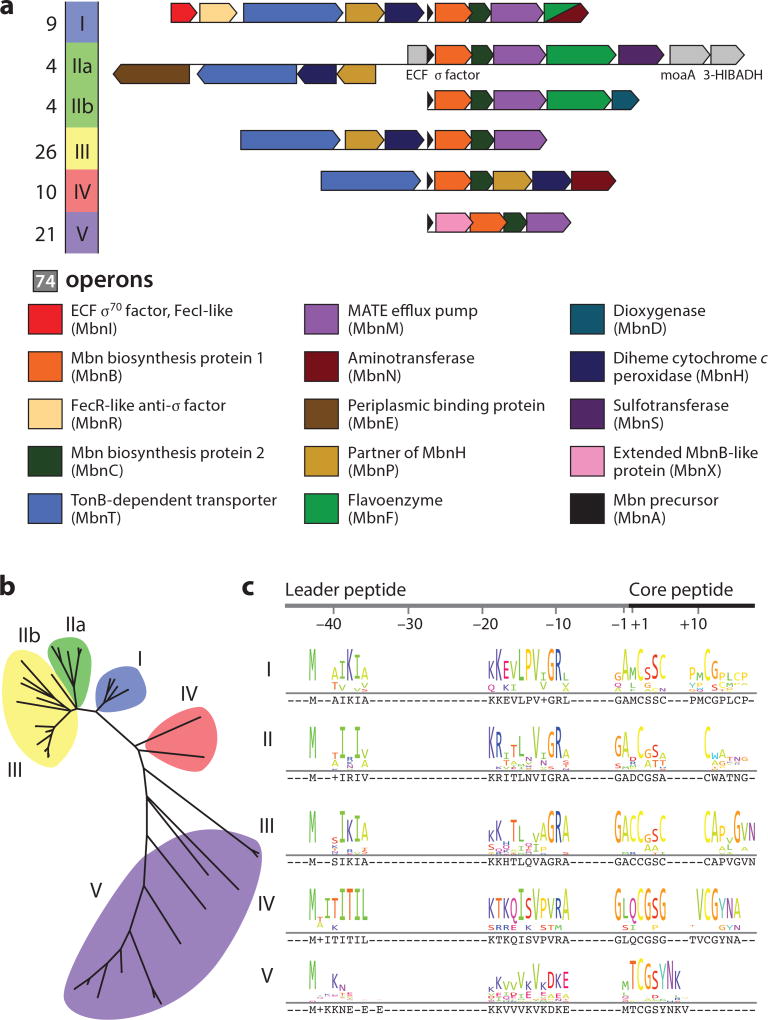
Methanobactin (Mbn) operons. (a) Operon schematics for Mbn operon groups. (b) Phylogenetic tree of MbnB protein sequences, illustrating the divisions used to define the Mbn operon groups. (c) Sequence logos for MbnA sequences from different groups. Figure adapted with permission from References 25 and 215.
The presence of mbnA, which encodes a short (22–40 amino acid) peptide in all Mbn operons (59), is consistent not with NRPS but rather with the biosynthesis of ribosomally produced, posttranslationally modified natural products (RiPPs). Mbn-related gene names, terms, and abbreviations have been accordingly been assigned following the guidelines established by the RiPP research community (61). Also present in all Mbn operons are genes encoding two proteins termed MbnB and MbnC, which are proposed to play a role in Mbn biosynthesis (59) (Figure 3a). Some operons encode additional proteins with known or putative biosynthetic roles as well as regulatory and transport proteins; the current state of knowledge regarding all these proteins is described below. Phylogenetic analysis of the three core genes (mbnABC) along with analysis of operon content supports the division of Mbn operons into five or possibly six main groups (25, 59) (Figure 3). A less granular set of subdivisions has been proposed (62), although this division is not based on phylogeny or posttranslational modifications. Groups I, IIa, and IIb are found exclusively in methanotrophs. The remaining groups comprise nonmethanotrophic bacteria, and Mbn operons in nonmethanotrophs now significantly outnumber those found in methanotrophs (25). Additional genes encoding proteins related to further posttranslational modifications, transport, and regulation are also found in many operons (59) and are discussed at greater length below.
Notably, genes encoding proteins with roles in copper homeostasis are located near Mbn operons in most nonmethanotrophic Group III–IV Mbn operons (59) (Supplemental Figure 1). Many Group III and IV operons are near genes encoding noncanonical members of the CopC family of periplasmic copper-binding proteins (63, 64), which are often adjacent to genes encoding CopD inner-membrane copper importers (65). Other periplasmic copper-binding proteins are sometimes present, including homologs of the Sco1 proteins and of the DUF461/PCuAC proteins, all implicated in assembly of the cytochrome c oxidase CuA site (66, 67). Although no specific copper need has been identified in Mbn-producing nonmethanotrophs, a P-type ATPase involved in copper import has been identified in some Pseudomonas species, consistent with active uptake of copper in at least one Mbn-producing genus of nonmethanotrophs (68).
Finally, Mbn operons are frequently associated with mobile genetic elements connected to horizontal gene transfer (59) (Supplemental Figure 1). This is unsurprising, given the broad range of species in which Mbn operons are found as well as fact that, in many genera, only a few species appear to have Mbn operons. The wide distribution of these operons, their presence in primarily nonpathogenic bacteria, and their role in trace metal acquisition suggest that they may serve as a fitness island rather than the virulence islands that are traditionally associated with siderophores (69).
Copper-Dependent Gene Regulation in Methanotrophs
Genome mining provided evidence for the conservation of genes associated with mbnA, but those assignments were based purely on bioinformatics. However, like siderophore biosynthesis, which is inhibited by excess iron (70), Mbn is produced primarily at low copper levels (40), suggesting that genes for Mbn biosynthesis should be repressed under high copper conditions. Furthermore, any regulatory patterns should be correlated with the well-attested copper-dependent reciprocal regulation of the two methane monooxygenases, referred to as the copper switch, in which sMMO is severely downregulated by copper and pMMO is highly produced under copper-abundant conditions (31) (Figure 2). Extensive time-dependent quantitative real-time PCR (qPCR) experiments in Ms. trichosporium OB3b verified these two predictions. First, copper-dependent downregulation of the Mbn operon is clearly observed: When 12.5 µM copper is added to copper-starved cells, the entire Mbn operon is downregulated, including the mbnIRTABCMNPH operon (59) as well as mbnE, a gene encoding a periplasmic binding protein that is separated from the bulk of the operon by mobile genetic elements (71). Second, the Mbn operon appears to be coregulated with the sMMO operon. These broad trends have been replicated in a whole transcriptome shotgun sequencing experiment, albeit with a simpler, time-independent copper starved/copper replete set of samples (72).
Interestingly, genes encoding regulatory proteins in both operons, including mbnI and mbnR in the mbn operon and mmoR, the recently identified mmoF (71, 73), and mmoG in the sMMO operon (Figure 2), exhibit similar patterns. All five genes are almost completely downregulated within 15 minutes of copper addition, while the transcription levels of the remaining genes in the two operons drop primarily over 5- and 24-hour time periods (71). These data suggest a model wherein MbnIR and MmoRFG activate Mbn and sMMO production, respectively, at low copper levels. Upon copper addition, the same copper-binding repressor inhibits transcription of both mbnIR and mmoRFG. As levels of the proteins encoded by those genes fall, transcription of the main operons falls as well, resulting in the characteristic copper-induced downregulation of sMMO and Mbn. This putative copper-binding repressor of mbnIR and mmoRFG has yet to be identified, although other genes encoding proteins with predicted roles in copper homeostasis were identified in this study (71).
Structural and Biochemical Characterization of Additional Methanobactins
Five Mbns other than the original compound from Ms. trichosporium OB3b have been at least partially characterized; all belong to Group I or Group IIa and are from Methylocystis (Mc.) or Methylosinus species (Figure 1b–f). The second characterized Mbn is from Mc. sp. SB2 and was investigated via NMR (45). It was reported to be smaller than the Ms. trichosporium OB3b Mbn, with a modified nine-amino-acid backbone (45) and different posttranslational modifications (Figure 1b). No intramolecular disulfide bond is present, and no N-terminal transamination occurs. Instead, an acid-insensitive heterocycle initially assigned as an imidazolone ring is the first heterocycle, and an additional posttranslational modification is present in the form of a sulfonated threonine (45).
Three additional Methylocystis Mbns have been characterized, including compounds from Mc. hirsuta CSC1 (Figure 1c), Mc. sp. M (Figure 1e), and Mc. rosea SV97 (Figure 1f), which were studied using mass spectrometry and, for the first two compounds, X-ray crystallography (74). Like Mc. sp. SB2 CuMbn, these Mbns have two nitrogen-containing heterocycles and thioamides, no intramolecular disulfide bonds, and a sulfonated threonine (45, 74). Unlike Mc. sp. SB2 CuMbn (Figure 1b), the first nitrogen-containing heterocycle in these Mbns is not a five-membered imidazolone ring but a six-membered ring depicted as a pyrazinediol group (74) (Figure 1c,e,f). Given the exceedingly high sequence similarity between Methylocystis operons, the source of the discrepancy between these pyrazinediol groups and the imidazolone of Mc. sp. SB2 was unclear. However, a reexamination of both the NMR data for Mc. sp. SB2 and the X-ray crystallography data for the remaining Methylocystis species suggests a possible answer (75). The assignment of the first Mc. sp. SB2 heterocycle rests partly on the presence of a secondary amine in that heterocycle (45). If the six-membered ring present in Methylocystis Mbn crystal structures is not a pyrazinediol but a hydroxypyrazinone or pyrazinedione tautomer (Figure 1g), a six-membered ring would be visible via crystallography and a secondary amine would be observed by NMR and would account for the long-distance interactions observed in the [13C-1H]-heteronuclear multiple-bond correlation spectroscopy (HMBC) and [15N-1H]-heteronuclear single-quantum correlation spectroscopy (HSQC) experiments performed on Mc. sp. SB2 CuMbn (45, 75). In all four characterized Methylocystis species, analysis of the mbnA genes indicates that not one but several C-terminal residues are missing from the structurally characterized Mbns (25, 45, 74). As with Ms. trichosporium OB3b, it is unclear when and how these residues are lost.
A second Methylosinus CuMbn has recently been characterized from Ms. sp. LW4 (75) (Figure 1d). This Mbn resembles the compound produced by Ms. trichosporium OB3b, despite a dissimilar peptidic backbone. Posttranslational modifications include N-terminal transamination, formation of two oxazolone/thioamide moieties from two cysteines, and intramolecular disulfide formation, and like Ms. trichosporium OB3b Mbn (Figure 1a), Mbn from this Methylosinus species binds a single copper ion (reductively in the case of Cu(II)) in what is likely to be a similar geometry (75). No C-terminal amino acid loss is observed, possibly because the C-terminal amino acid is a cysteine that stabilized by disulfide formation.
Comparing the copper binding affinities of the growing number of characterized Mbns remains somewhat challenging. To date, no equivalent of the pM scale for iron affinity in siderophores has been established for chalkophores; pM values describe the amount of unbound iron present at equilibrium under a standard set of experimental conditions (76). The only Mbn for which a similarly systematic set of measurements has been undertaken is that from Ms. trichosporium OB3b, in which proton-independent stability constants were also calculated (49). Furthermore, chalkophores present similar challenges to siderophores: Copper–chalkophore binding strength can preclude direct measurement of metal binding via common techniques. Competition experiments against compounds such as the Cu(I) chelator bathocuproine disulfonate or even other Mbns provide an alternative method that has been used for several Mbns (42, 74).
Under superstoichiometric copper-loading conditions, lower-affinity nonreductive binding of a second copper ion has been observed, for which potential binding sites include the C-terminal carboxylic group, the tyrosine phenol, the C-terminal methionine, and the two cysteines otherwise involved in a disulfide bond (49). Binding is pH sensitive, with higher affinities above neutral pH (42, 49). Despite these complexities, the copper binding properties for Mbns have been remarkably consistent: Cu(I) binding constants have converged at ~1021 M−1 (42, 74, 77). As additional structurally divergent Mbns are characterized, a wider range of binding affinities may emerge. Evidence has already been found that the presence or absence of certain posttranslational modifications such as threonine sulfonation in Methylocystis Mbns can tune affinity (74).
Current Models for Methanobactin Biosynthesis
Mbns belong to the RiPP class of natural products, but their biosynthetic machinery appears to be unique. Although heterocycle formation is common in RiPP natural products, no known RiPP biosynthetic enzymes are found in Mbn operons (61). Other features of RiPP biosynthetic machinery, including a peptide recognition element (78) often found in RiPP-related enzymes, are also absent. Instead, both new enzyme families and divergent members of well-characterized enzyme families are involved in Mbn biosynthesis.
MbnA: the precursor peptide
Like all RiPP systems, MbnA precursor peptides have multiple components, including a sequence involved in binding biosynthetic enzymes, the leader peptide, which is lost in the final natural product, and the core peptide, which forms the backbone of the final natural product (61). The positions within the MbnA sequence occupied by nitrogen-containing heterocycles in the final natural product are cysteines in the core peptide (45, 59). These cysteines targeted for modification are part of motifs; these motifs comprise the cysteine and two adjacent C-terminal residues, with the first C-terminal residue small (glycine, alanine, and sometimes serine) and the second residue frequently larger and slightly hydrophilic (most commonly serine and threonine) (59) (Figure 3c). While some RiPP systems favor specific flanking residues (79), many others do not (80, 81). In Mbns, cysteines that are not part of a modification motif remain unmodified and available to form disulfide bonds, as observed in the two structurally characterized Methylosinus Mbns (26, 75).
In a subset of operons found in certain Methylocystis species, mbnA genes are part of a longer gene encoding an extracytoplasmic function (ECF) sigma factor (59). Whether an alternate transcription start site results in the production of only the MbnA peptide or whether the full protein is produced is unclear. Precedent for the latter has been found in a subset of thiazole-oxazole modified microcins (TOMMs) produced from larger genes encoding a nitrile hydrolase-like sequence in lieu of a standard leader peptide (82). This extended leader peptide sequence is biochemically relevant in those TOMM families (83) but has not yet been investigated for Mbns.
MbnB and MbnC: the core biosynthesis machinery
Two genes encoding proteins known as MbnB and MbnC are present in all Mbn operons. MbnB is a member of the uncharacterized DUF692 family, while MbnC is a smaller protein of unknown function that belongs to no previously known family. Their conservation (Figure 3a) despite the lack of a known function led to suggestions that these two proteins play a role in Mbn biosynthesis (59). While originally annotated as two open reading frames in the Ms. trichosporium OB3b operon (45), mbnB is a single gene in other Mbn operons, and the pseudogene observed in the published Ms. trichosporium OB3b genome is not present in all lineages of that strain (71). Although RiPPs containing amino acid–derived heterocycles such as thiazol(in)es and (methyl)oxazol(in)es are common, particularly in the TOMM family (84, 85), no precedents for enzymatic oxazolone biosynthesis have been found. The sole characterized oxazolone-containing natural product family, jadomycins, receives these modifications as part of the nonenzymatic incorporation of amino acids (86). Thioamides are slightly less uncommon in natural products (87), with thioviridamide (88) and closthioamide (89) of particular interest.
However, none of the biosynthetic machinery found in any of these enzyme families is related to MbnB or MbnC. Azol(in)e formation in TOMM systems involves the recently characterized ATP-binding YcaO family (90, 91), but neither MbnB nor MbnC bears any similarity to those enzymes, and both lack nucleotide-binding motifs. Thioamide biosynthesis is less well understood, but a YcaO family enzyme has been proposed to be responsible for thioamide biosynthesis in thioviridamide as well (88). Additionally, unlike many enzymes involved in RiPP biosynthesis, MbnB and MbnC lack RiPP recognition elements (RREs), which are involved in precursor peptide binding (78).
Instead, recent work indicates that MbnB and MbnC form an entirely novel heterodimeric iron-containing enzyme (MbnBC) that carries out a four-electron oxidation on the MbnA substrate, forming an oxazolone and thioamide moiety from a cysteine in a single concerted rearrangement (92). The iron active site is located in the MbnB subunit, and some ambiguity remains regarding the composition and oxidation state of the active cofactor, with coupled di- and trinuclear iron centers as possibilities. Importantly, this mechanism of heterocycle formation appears to apply to MbnA/MbnBC pairs from all five operon groups, although Group V is sufficiently divergent so that its enzyme and substrate are not compatible with the other four groups. An alternative mechanism involving a catalytic thiol and several biosynthetically unusual steps has been suggested (62), but even apart from the lack of conserved cysteines in MbnB and MbnC, this model does not fit the biochemical data, which support an oxygen- and iron-dependent mechanism (92) (Figure 2b).
MbnS, MbnF, MbnD, and MbnX enact additional posttranslational modifications
Along with MbnA, MbnB, and MbnC, some Mbn operons encode other proteins with predicted roles in Mbn biosynthesis (Figure 3a). Like MbnB and MbnC, these enzymes also lack RREs (78). MbnS, a 3′-phosphoadenosyl-5′-phosphosulfate-dependent sulfotransferase, is likely responsible for threonine sulfonation in Group IIa Mbns (59, 93, 94) (Figure 1b–f). MbnF, a flavin-dependent oxidoreductase, is predicted to play a role in (hydroxy)pyrazin(edi)one biosynthesis in Group I and Group II Mbns (Figure 1c–f), catalyzing the transformation from oxazolone to (hydroxy)pyrazin(edi)one (59). The closest characterized flavin-dependent monooxygenase is involved in rebeccamycin biosynthesis (95), but flavin enzymes catalyze a wide range of biosynthetic reactions (96). MbnD is a predicted dioxygenase that is believed to play a role in the biosynthesis of Group IIb Mbns, though the reaction that it catalyzes has not been identified and no Mbns from that subfamily have been structurally characterized (59). Group V Mbn operons also contain mbnX, a gene encoding a DUF692 enzyme that is only distantly related to MbnB (59). Both Group IIb and Group V MbnAs lack a second cysteine modification moiety (and in the case of Group V, another cysteine at all), meaning that they cannot bind copper via a canonical Mbn ligand set; one possibility might be modification of another amino acid to provide replacement metal-binding ligand(s). Finally, both Group I and Group IV operons contain genes encoding pyridoxal-5′-phosphate-dependent aminotransferases, MbnNs, believed to perform N-terminal transamination reactions (59).
A role in oxazolone biosynthesis has also been proposed for Ms. trichosporium OB3b MbnN owing to the absence of the first Mbn oxazolone ring in an Mbn intermediate secreted by an ΔmbnN strain (97). Given that no neighboring transamination reaction is necessary for the completion of the second oxazolone in any Mbn, why the first oxazolone would require it is unclear, particularly when aminotransferases are only present in a few Mbn operons (25). However, the observed mass is consistent with acid-catalyzed oxazolone hydrolysis (45), which in combination with the predicted lack of an N-terminal transamination reaction, provides a potential explanation for the missing oxazolone ring. Indeed, addition of copper during purification stabilizes the Mbn intermediate and prevents oxazolone degradation (92); the role of MbnN is thus predicted to be catalyzing a simple transamination reaction on this intermediate. A biosynthetic role has also been proposed for the diheme cytochrome c peroxidase MbnH and its partner protein MbnP (62); however, mbnPH genes are commonly associated with genes related to CuMbn uptake (25), and while the Ms. sp. LW4 Mbn operon lacks mbnPH, its Mbn contains oxazolone/thioamide moieties (75). On the basis of in vitro experiments with MbnBC, it does not appear that there is a role for either MbnN or MbnPH in oxazolone/thioamide biosynthesis (92).
Finally, all of the proposed biosynthetic enzymes are cytoplasmic. There are RiPP systems in which leader peptide cleavage or additional posttranslational modifications occur in the periplasm (98), but this is not likely to be the case in Mbn systems owing to the cytoplasmic nature of MbnN and MbnF, which require cytoplasmic leader peptide cleavage to access the N terminus of the core peptide. One gene conspicuously absent in all Mbn operons is a protease responsible for leader peptide cleavage. Whether MbnB or MbnC is capable of catalyzing MbnA leader peptide cleavage is unclear, although this proteolytic step has not been observed in vitro (92), or whether an unknown process is responsible.
Efflux and Influx of Methanobactins
Transport is vital for metallophores. Mature metallophores are secreted into the environment, where they can liberate otherwise biounavailable metal. The metal-bound compounds must then be taken back up into the cell, where the metal is released. These processes have been studied extensively in siderophores, and in particular, many active importers have been identified. Mbn trafficking appears to follow a similar model.
Methanobactin secretion via MbnM
The secretion of Mbn is thought to be mediated primarily by a group of inner membrane efflux pumps designated MbnMs (59). Genes encoding MbnMs are even present in the divergent gram-positive Group V operons (Figures 2 and 3a; Supplemental Figure 1). Group IV Mbn operons lack mbnM genes, and whether these operons are no longer functional or whether a different exporter is involved is unclear (25). MbnM belongs to the multidrug and toxic compound extrusion (MATE) family (99). MATE proteins function as proton/sodium antiporters (100), and previously identified members of this family are primarily involved in the efflux of cationic xenobiotic compounds (101). Confirmed export of a native natural product like Mbn by MbnM would significantly broaden the range of MATE-transported compounds.
Internalization of copper-chelated methanobactin via MbnT
Evidence for active transport of CuMbn predated both the classification of Mbn operons via genome mining (59) and the confirmation that the Ms. trichosporium OB3b operon is regulated by copper (71). CuMbn uptake is inhibited via proton pump uncouplers such as carbonyl cyanide-m-chlorophenylhydrazone, indicating an energy-dependent uptake process, while soluble copper is taken up by a passive process that is disrupted by porin inhibitors such as spermine (102). Furthermore, apo Mbn inhibits copper uptake via CuMbn (102), a phenomenon that is observed in siderophore uptake as well (103) and is consistent with the presence of a specific transporter.
Genome mining identified a candidate CuMbn importer, a TonB-dependent transporter (TBDT) found in Groups I–IV Mbn operons (59) and initially named MbnT (Figures 2 and 3a). TBDTs are responsible for the energy-dependent import of various metabolites, including siderophores (104) as well as heme, thiamine, cobalamin, colicins, sugars, and small soluble nickel and cobalt chelates (105). Additional bioinformatic analysis revealed that MbnTs fall into several classes. One group, the MbnT1s, are associated with Group I Mbns and are paired with ECF sigma factors (MbnIs) and anti–sigma factors (MbnRs) (59). These MbnT1s possess an ~100-amino-acid periplasmic N-terminal extension that mediates interactions between the outer membrane MbnT and the inner membrane MbnR, which then interacts with the cytoplasmic MbnI, allowing for cell surface-to-cytoplasm signaling upon the binding of Mbn (106) (Figure 2). Similar systems exist for enterobactin (107), pyoverdine (108), and other siderophores.
However, this signaling pathway is not present in other Mbn operon groups. The genes encoding MbnT2s, a second set of TBDTs found in Group II Mbn operons, do not display significant sequence similarity to mbnT1s, lack neighboring mbnIR genes, and lack the N-terminal extension necessary for trans-periplasmic signal transduction (28). A third group of TBDTs, the MbnT3s, are found in the nonmethanotrophic Groups III–IV. These importers resemble heme-importing TBDTs and are not closely related to MbnT2 or MbnT1s (28). No mbnT genes are found in Group V operons or elsewhere in the genomes of these species.
Siderophore piracy is common among organisms with a high need for iron (109), and chalkophore piracy can occur among methanotrophs. Mc. hirsuta CSC1 cells take up Ms. trichosporium OB3b CuMbn and vice versa, and Ms. trichosporium OB3b also take up CuMbn from Mc. sp. SB2. In both cases, the Mbn-mediated copper influx can trigger the copper switch (74, 110). Mbn piracy is consistent with the presence of multiple mbnT1 and mbnT2 (and mbnIRT) genes in the genomes of many species lacking Mbn operons, including other methanotrophs as well as ammonia oxidizers (which produce an ammonia-oxidizing homolog of the copper enzyme pMMO) (28). Many methanotrophs in fact have multiple mbnT homologs, complicating functional studies of this transporter family. In Ms. trichosporium OB3b, two additional mbnT1 homologs are part of non-operon copper-regulated mbnIRTPH cassettes, but their regulatory patterns differ from those of the in-operon mbnT1 (28). These proteins possibly function in the uptake of non-native Mbns.
Disruption of the in-operon mbnT1 gene of Ms. trichosporium OB3b significantly decreases uptake of copper provided as CuMbn (but not soluble copper), despite intact biosynthesis and secretion of the chalkophore (111). The continued uptake of soluble copper is consistent with earlier reports of a second passive method for soluble copper uptake in this species (102). In a similar mutant strain, isotopically labeled 65CuMbn import by the wild-type but not ΔmbnT1 strain alters the copper isotopic ratio of the wild-type cells (28). These results can be replicated in Escherichia coli cells heterologously expressing MbnT1 from Ms. trichosporium OB3b (28), providing strong evidence for the correct identification of this transporter.
Binding of CuMbn to MbnT transporters is observed in vitro. Surface plasmon resonance, a method not yet commonly used for siderophore/receptor studies, provides evidence for binding of Ms. trichosporium OB3b CuMbn to both its cognate MbnT1 (Kd value of 6 µM) and the in-operon MbnT2 from Mc. rosea SV97 (Kd value of 18 µM) (28). Although TBDTs can be quite specific, their promiscuity depends on what portion of the substrate is recognized. The peptidic backbones of Mbns can differ significantly, but the geometries of their copper binding sites are similar (26, 74). FpvA from Pseudomonas aeruginosa ATCC 15692 recognizes pyoverdine metal binding sites, permitting some binding to noncognate pyoverdines (112), and Mbn binding to MbnT could conceivably be similar. How these binding affinities correlate to internalization rates of noncognate Mbns has not yet been elucidated.
A new role for a periplasmic binding protein, MbnE
In many siderophore families, internalized compounds are transferred to chaperone proteins after import into the periplasm. Such a system had previously been proposed for Mbns (113), and Group IIa Mbn operons do in fact encode a gene for a periplasmic binding protein (PBP), MbnE (28) (Figures 2 and 3a). A similar gene adjacent to the Mbn operon in Ms. trichosporium OB3b is copper regulated, while other Ms. trichosporium OB3b PBPs belonging to the same protein family (solute binding protein family 5) are not (28, 71). Bioinformatic analyses indicate that MbnEs are related to the microcin C7 transporter YejA and the oligopeptide transporter OppA/AppA (28, 114, 115), both of which also bind substrates with peptidic backbones. Moreover, a 1.9-Å resolution crystal structure of the apo MbnE from Mc. parvus OBBP suggests both significant homology with AppA and the presence of a binding cavity sufficiently large to accommodate Mbn (28). Notably, MbnEs from three species (Ms. trichosporium OB3b, Mc. hirsuta CSC1, and Mc. parvus OBBP) bind cognate Mbns with Kd values of 6–12 µM but do not interact with noncognate Mbns (28). However, in the absence of an operon-adjacent ABC importer, the fate of CuMbns after MbnE binding has not yet been ascertained. What happens to CuMbns after internalization in species that lack MbnE homologs is also not clear.
Copper release from methanobactin
The mechanism of copper release from CuMbn has not been explored experimentally. Some siderophores are degraded upon uptake (116), but degradation of a complex compound like Mbn is potentially wasteful. Recycling of Mbn after copper release is another possibility, similar to recycling of pyoverdine (117), carboxymycobactin (118), and ferrichrome (119). One way to release copper without chalkophore degradation could involve copper oxidation, analogous to iron reduction for release from some siderophores (120) and consistent with the lower binding affinity of Mbn for Cu(II) than for Cu(I) (42). Given the ability of Mbn to reduce Cu(II), additional proteins are possibly required to facilitate copper release.
Two candidates for such partner proteins are MbnH and MbnP, encoded by operons in Groups I–IV (Figure 3a). MbnH is MauG-like diheme cytochrome c peroxidase, and genes encoding MbnH homologs are almost always found adjacent to the gene encoding MbnP, a hypothetical protein with several highly conserved tryptophan residues (59). These proteins are reminiscent of MopE/CorA, copper-binding proteins secreted by some methanotrophs. MopE/CorA proteins bind copper using a posttranslationally modified tryptophan that is likely generated by a diheme cytochrome c peroxidase encoded by a neighboring gene (121, 122). However, there is no significant sequence similarity between MopE/CorA and MbnP, and though MbnP and MbnH are predicted to be periplasmic, there is no indication that they are secreted. Alternatively, MbnH might itself oxidize or even modify CuMbn, directly effecting copper release.
The location of copper release is also unclear. CuMbns interact with MbnEs (28), which may reflect ultimate internalization to the cytoplasm and release there. However, copper release mediated by MbnPH would be periplasmic. In Ms. trichosporium OB3b, many periplasmic proteins implicated in copper-related processes are present, including CopC, CopD, DUF461/PCuAC, and Sco1 proteins (71) as well as the so-called copper sponge Csp1 (123) and the uncharacterized protein PmoD (71). Finally, pMMO itself houses a copper active site in the periplasmic portion of the PmoB subunit (55, 124), which is believed to be the final destination of most copper trafficked into methanotroph cells. For siderophores, demetalation can occur in the cytoplasm (e.g., pyochelin; 125) or the periplasm (e.g., pyoverdine; 117), and more work is needed to determine which is the better model for CuMbn copper release.
Secondary Roles for Methanobactin
Historically, metallophores were considered primarily in the context of their role in metal uptake and metal homeostasis. However, many metallophores, including methanobactin, appear to have a broad range of secondary roles, ranging from regulatory functions to protection against toxicity caused by metal or reactive oxygen species to biomedically relevant antibiotic or therapeutic functions (12).
Signaling factor
Self-regulation of production and uptake machinery, often via anti–sigma factor/sigma factor pairs that interact with the TBDT, is well documented for a range of siderophores, including enterobactin and pyoverdine (126). As described above, mbnIRT cassettes may play a similar role in the import of Mbn in both Mbn producers and in species that are engaging in chalkophore piracy (28) (Figure 2). Other regulatory schemes are observed in some siderophore systems, such as the cytoplasmic uptake of pyochelin, which is required for activation of the AraC-family regulator PchR (127), but it is not known if similar systems are present in Mbn operon groups without mbnIRT cassettes.
A direct role has been proposed for Mbn in the methanotroph copper switch. Disruption of the mbnA gene dampens the effect of copper addition on both the sMMO and pMMO operons (60). Knockout of the sMMO operon, including a gene encoding a protein of unknown function termed MmoD, also perturbs the copper switch, as does knockout of mmoD alone (128). It was thus proposed that MmoD and Mbn work together to effect this regulatory switchover (60). However, neither Mbn nor sMMO operons are necessary for fully triggering the copper switch. Species lacking Mbn operons can still switch between sMMO and pMMO production (35, 73), indicating that Mbn is not primarily responsible for copper-dependent repression of the sMMO operon. Moreover, species lacking either the sMMO operon (and thus MmoD) or both the sMMO and Mbn operons still exhibit copper-responsive phenotypes, including decreased Mbn production and intracytoplasmic membrane formation (77, 129, 130).
MmoD involvement in the copper switch is particularly problematic. MmoD has no observed copper binding capacity and has an unusual set of conserved residues for a putative copper-binding protein. It also does not bind heparin, commonly used as a proxy for DNA binding (71). Despite reports to the contrary (62), mmoD is downregulated in response to copper in at least two species (71, 73), which would be unexpected for a copper-binding repressor. Finally, in the obligate sMMO-producing Methylocella silvestris BL2, which has MmoD but not the pMMO or Mbn operons, copper addition does not repress sMMO (131). Biochemical evidence for interactions between MmoD and the sMMO enzyme complex has been obtained, although the role of those interactions in vivo is uncertain; sMMO is active in vitro without MmoD (132, 133). These considerations suggest that Mbn and/or MmoD are not the primary regulatory elements governing the copper switch.
A second proposed model includes Mbn, MbnI, and MmoR, a transcription factor encoded by the sMMO operon (62). However, most methanotrophs lack Mbn operons, and it seems correspondingly unlikely that the variable genomic environments of multiple σ70-regulated pMMO operons (134) have been retrofitted to support a σECF regulatory binding site for MbnI or an Mbn-binding riboswitch. This is also true of sMMO operons, for which σN but not σECF binding sites have been reported (135). There is no indication that MmoR might bind Mbn, particularly as similar regulators exist for distant sMMO homologs with no role in methane oxidation and no known copper switch (136, 137). Taken together, any direct role for Mbn in signaling beyond MbnIRT-mediated regulation of the mbn operon is speculative. In a more conservative model (Figure 2), most regulatory changes associated with Mbn production and CuMbn uptake result from the increase in the pool of bioavailable copper, with direct Mbn-mediated signaling limited to the MbnI regulon, likely consisting of just the mbn operon and possibly the non-operon mbnIRTPH clusters (28, 71).
Protection from metal toxicity
As mentioned above, Mbn binds some metals other than copper reductively (53). For example, Mbn binds Au(III), mediating the production of gold nanoparticles by reducing the metal to Au(0) (138). Although the measured affinity for copper is higher, optical spectroscopic data indicate that gold may be able to compete with copper to some extent, inhibiting copper uptake and the ensuing copper switch (139). The biological relevance of gold binding is not clear: Both gold toxicity (140) and mitigation of gold toxicity via biomineralization (141) are observed in other proteobacteria, such as Cupriavidus metallidurans CH34, but unlike methanotrophs, these species are known to tolerate extreme levels of heavy metals (142, 143).
Mercury binding to Mbns from Ms. trichosporium OB3b and Mc. sp. SB2 has also been investigated in some detail. Mbn can bind Hg(II) in the absence of and, to a lesser extent, the presence of copper, although the binding constant is 15 orders of magnitude lower than that of copper; because copper does not easily displace bound mercury, a role in protection against mercury toxicity has been suggested (144, 145). The affinity and specificity are somewhat compound specific, with Mc. sp. SB2 Mbn exhibiting less selectivity for copper (144, 145). A role for Mbn in methylmercury (CH3Hg+) degradation has also been suggested, although Mbn does not demethylate mercury on its own (146). It may be that the ability of Mbn to reduce methylmercury makes the substrate available to the demethylating enzyme (53, 147). While some methanotroph species have been found in environments replete in heavy metals (148), methanotrophs are present in a wide range of environments (52, 149), from both freshwater and marine sediments (150) to peat bogs (151), arctic tundra (152), and volcanic soil (153), and it is unclear if interactions between Mbn and mercury are relevant to the behavior of methanotrophs under common biological conditions.
Superoxide dismutase activity
The ability of Mbn not just to bind Cu(I) but to reductively bind Cu(II) (46), producing Cu(I)Mbn, hinted that CuMbn might be capable of more redox chemistry. CuMbn has been reported to exhibit oxidase, superoxide dismutase (SOD), and hydrogen peroxide reductase activities (54). Biological superoxide production and secretion by heterotrophic bacteria may be a source for oxidative stress in environments frequented by methanotrophs, such as lake sediments and terrestrial soils (154). Thus, SOD activity of secreted Mbns may be functionally relevant.
Antibiotic activities
Mbn from Ms. trichosporium OB3b has been reported to exhibit antibiotic activity against gram-positive bacteria (155). Any mechanism remains hypothetical, but the antibiotic activity of redox-active metal-binding natural products such as bleomycins is well documented (156), and uptake of surplus copper can be cytotoxic in its own right (29). Given that Mbn operons from Groups I–IV contain TBDTs predicted to enable CuMbn uptake along with additional genes with predicted roles in periplasmic copper homeostasis (59), a primary antibiotic role seems less likely in these groups.
By contrast, Mbns from Group V may well function as antibiotics or quorum-sensing compounds. All Group V species lack operon-associated and non-operon mbnT genes. While Group V compounds have not yet been characterized, the presence of only a single modifiable cysteine in their MbnA sequences (Figure 3c) suggests a notably different copper coordination environment, if they indeed bind copper (25). Several Group V Mbn operons are found in Streptomyces species, which are prolific antibiotic producers. Some Streptomyces species play a symbiotic role, producing antibiotics that protect their hosts from other bacteria, and a similar phenomenon is observed in several Group V bacteria, including entomopathogenic Photorhabdus and Xenorhabdus species (157, 158). The Vibrio species that together form the third-largest group of Group V Mbn producers also possess a range of poorly characterized biosynthetic machinery for secondary metabolites (159). Further research is necessary to determine the role(s) of the Mbn-like compounds in these species.
Therapeutics
Mbns have also attracted interest as drug candidates. Similar to the treatment of hemochromatosis with the siderophore desferrioxamine (160, 161), Mbn is a potential therapeutic for Wilson disease, a human genetic disorder of copper overload (162). Current treatments have significant side effects, are incapable of mobilizing copper in both oxidation states, and/or chelate other physiologically relevant transition metals (163, 164). In a rat model of Wilson disease, Mbn can reverse acute liver failure (165, 166). Copper overload may also play a role in a range of neurological disorders (167), and if that role is verified, Mbns might be widely applicable as copper-chelating drugs. Finally, the modularity of Mbn biosynthesis and its peptidic nature could enable biological generation of diverse Mbn-like drug candidates, potentially with additional features such as the ability to utilize tissue-directed peptide targeting.
CHALKOPHORES BEYOND THE METHANOBACTIN FAMILY
Copper Uptake Mechanisms in Non-Methanobactin-Producing Methanotrophs
The production of Mbns by two species lacking Mbn operons (59), Methylococcus capsulatus (Bath) and Methylomicrobium album BG8 (168), has been reported. If these species do produce a chalkophore, it is not one directly related to the Mbn family. These compounds have significantly lower calculated copper affinities than other Mbns, bind iron, and contain non-negligible amounts of Cu(II) (168). As such, they are possibly metallophores that target some other metal but are capable of binding copper. The secreted protein MopE may instead substitute for high-affinity chalkophores in these species. In MopE, a tryptophan-to-kynurenine posttranslational modification likely carried out by a diheme cytochrome c peroxidase generates a high-affinity copper binding site (121, 169, 170).
Coproporphyrin III
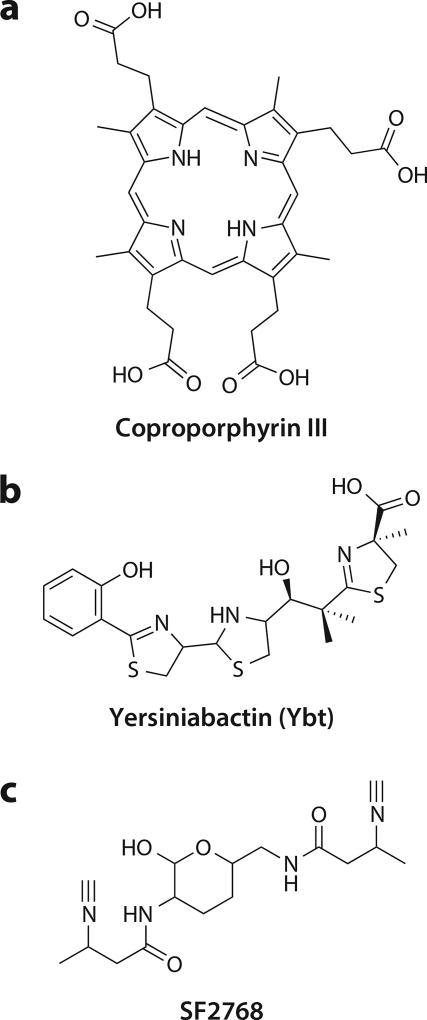
Methanotrophs are not the only bacteria that rely on copper enzymes for key metabolic processes. Under anaerobic conditions or in the presence of nitrate, Paracoccus denitrificans uses copper enzymes for both respiration and denitrification (171, 172). Under copper-deficient conditions, less cell yield is obtained and a red compound is secreted (173). The addition of Cu-coproporphyrin III to the medium allows recovery of growth and limits secretion of the chromophore, identified as coproporphyrin III (Figure 4a), which turns red upon binding zinc in copper-free medium (173). This putative copper acquisition role is not universal, however. Coproporphyrin III is secreted at higher copper levels in a strain of Rubrivivax gelatinosus that lacks the copper efflux pump CopA, which is inconsistent with a role in copper influx (174).
Figure 4.
Structures of putative chalkophores. (a) Coproporphyrin III is a porphyrin. Although not traditionally considered siderophores, porphyrin (heme) uptake is a source of iron for many species. (b) Yersiniabactin (Ybt) metal binding is partly mediated by heterocycles, which are not among the traditional catecholate, hydroxymate, and phenolate siderophore ligands. (c) SF2768 has unusual diisonitrile groups that are likely involved in copper binding.
Yersiniabactin
As an iron-binding natural product produced by Yersinia pestis, the pathogen responsible for the bubonic plague, yersiniabactin (Ybt) (Figure 4b) was discovered nearly 20 years ago and was identified as a siderophore and a key component of the Yersinia high-pathogenicity island (175, 176). However, like a small number of other siderophores, including pyochelin and anguibactin, several of the iron-coordinating ligands are not the classical siderophore hydroxymate and phenolate-catecholate groups but rather are thiazoline and thiazolidine groups (175, 177). This divergent ligand set proved to be biochemically relevant when it was shown that Ybt produced by uropathogenic E. coli binds copper during infection (178). Under these conditions, Ybt binds Cu(II) competitively with Fe(III), protecting the cell from copper toxicity. Furthermore, this protective effect is not simply due to copper sequestration: Catecholate siderophores can bind Cu(II) and reduce it to the more cytotoxic Cu(I) form, and the higher copper affinity of Ybt eliminates that possibility (178). Copper release in cytokine-stimulated macrophages and copper overload in bacteria internalized by macrophages suggest that high external copper levels are a significant risk for pathogenic bacteria. Thus, a secondary protective role for less-specific metallophores is plausible (179).
Like CuMbn, copper-bound Ybt (CuYbt) exhibits SOD activity, potentially providing protection against phagocytic killing (180). Similar to staphylopine, Ybt’s role as a broad-spectrum metallophore means that it is capable of interacting with more than just copper. Ybt has also been reported to play a role in Zn(II) acquisition in Y. pestis, although conflicting reports exist regarding its affinity for zinc, particularly in the presence of iron, and a range of metal-Ybt complexes are imported in a TonB-dependent manner (20, 181). Cu(II)Ybt does not competitively inhibit Fe(III)Ybt uptake, suggesting that if the compound acts as a true chalkophore to mediate copper acquisition, it does not do so under common growth conditions (181). However, under copper starvation conditions, uropathogenic E. coli does take up Cu(II)Ybt, followed by dissociation of the copper within the cell (and incorporation into the cellular copper pool) and finally recycling of this bifunctional siderophore/chalkophore (182). Notably, this process involves Cu(II)Ybt, unlike many of the other chalkophores that prefer Cu(I); this allows the chalkophore to coopt the siderophore reduction machinery, the YbtFQ enzyme complex, to liberate the metal after internalization, rather than requiring a second set of enzymes for copper uptake.
SF2768 and Other Diisonitrile Natural Products
A diisonitrile natural product that binds copper, SF2768 is produced via NRPS in the gram-positive bacterium Streptomyces thioluteus (183) (Figure 4c). Like siderophore and chalkophore operons, the SF2768 operon contains exporters [members of the major facilitator superfamily (MFS)] as well as importers (an ABC transporter annotated as an iron chelate importer). SF2768 binds Cu(I) and Cu(II), and like Mbn, it appears to be able to bind Cu(II) reductively, followed by reinternalization after copper binding (184). Similar operons were identified in other gram-positive species, and characterization of the resulting compounds in Mycobacterium species indicated that they form diisonitrile lipopeptides with a possible role in zinc homeostasis (185). The broader structural and metal-binding diversity of these diisonitrile metallophores has yet to be explored, but it is clear that Mbns are not the only potential chalkophores found in gram-positive bacteria.
COPPER-BINDING SIDEROPHORES AND METALLOPHORES
Nicotianamine, Staphylopine, and Pseudopaline: Multi-Metal Metallophores
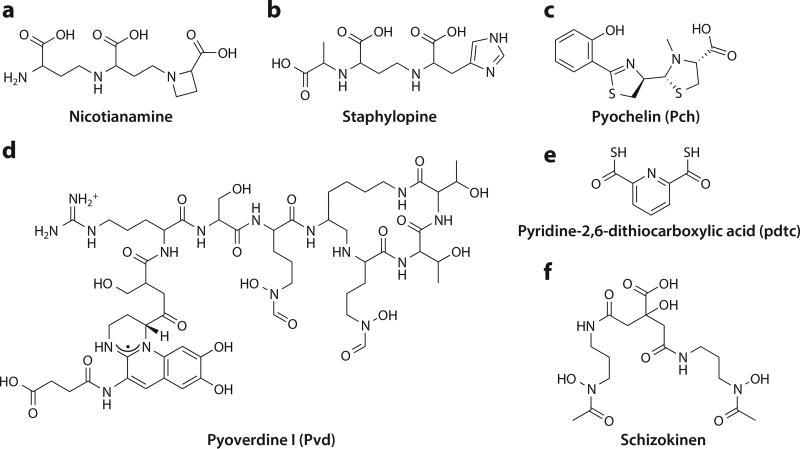
Plants also secrete metallophores, and as in bacteria, the best-known metallophores are iron-binding compounds known as phytosiderophores. However, some phytosiderophores are involved in the acquisition of other metals. Nicotianamine (Figure 5a) has been implicated in iron, copper, nickel, and zinc homeostasis (186). In some species, such as Brassica carinata, nicotianamine plays a role in copper acquisition during periods of copper starvation (187). Nicotianamine and related phytosiderophores are found primarily in eukaryotic and archaeal species, but nicotianamine synthase (NAS), a key S-adenosylmethionine (SAM)–dependent enzyme in nicotianamine biosynthesis, has prokaryotic homologs, including one in the genome of Staphylococcus aureus (17).
Figure 5.
Structures of select copper-binding siderophores. (a) Nicotianamine and (b) staphylopine share structural characteristics and biosynthesis genes, despite differing origins in eukaryotic and bacterial species. (c) Pyochelin (Pch) has a metal-chelating hydroxyphenol-thiazolyl-thiazolidinyl backbone similar to that of Yersiniabactin (Ybt). (d) Pyoverdine I (Pvd) is a member of the large family of primarily peptidic pyoverdine siderophores. (e) Pyridine-2,6-dithiocarboxylic acid (pdtc) is a small metal-binding secondary metabolite. (f) Schizokinen was originally identified in gram-positive Bacillus and cyanobacterial Anabaena species.
In S. aureus, the gene for the NAS homolog (cntL) is accompanied by an additional two biosynthetic genes: a gene encoding an epimerase (cntK) and a gene encoding an NAD(P)H-dependent dehydrogenase (cntM) (17). Also found in the operon are genes encoding an ABC family transporter thought to be involved in nickel import (cntABCDEF) and a MFS exporter (cntE) (17). Incubation of heterologously expressed biosynthetic components of the operon (CntK, CntL, and CntM) in the presence of l-histidine, SAM, pyruvate, and NADPH results in production of a natural product termed staphylopine, which exhibits significant structural similarity to nicotianamine (Figure 5b). Staphylopine binds divalent metal ions with similar Kd values and affinity order (Cu2+ > Ni2+ > Co2+ > Zn2+ > Fe2+) to nicotianamine (17). Strains with genetic disruptions of the key biosynthesis or importer genes exhibit reduced levels of Zn, Ni, Cu, and Co and decreased metal uptake when exposed to soluble extracellular metal, although results for copper appear to be affected by swift efflux (17). It is not yet clear what the primary role of this metallophore is in S. aureus or how a broad-spectrum metallophore is regulated in different environments. Staphylopine-like operons are found in other pathogenic species, including P. aeruginosa, in which the products of those genes are needed for growth within the host environment (188). Pseudopaline, the opine metallophore produced by these staphylopine-like operons, has recently been identified, and evidence that it plays a role in zinc and nickel import is available (189–191).
Pyochelin and Pyoverdine
The siderophores pyochelin (Pch) and pyoverdine (Pvd) are also produced by P. aeruginosa. Members of the partially peptidic Pvd family have a higher affinity for iron than Pch (192) (Figure 5c,d). Pch, like Ybt, contains thiazoline and thiazolidine groups, which facilitate binding of other divalent metal ions, including Cu(II) and Zn(II). While Pch binds Fe(III) in monochelate and bischelate modes, Cu(II)Pch and Zn(II)Pch complexes are exclusively bischelate. Pch is only weakly selective for Fe(III) over Cu(II) and Zn(II) (193). Similar to Ybt, Pch may not be a true chalkophore under standard physiological conditions. Binding of Cu(II) to Pch and binding of Cu(II)Pch to the Pch importer are significant, but little uptake of Cu(II)Pch is observed in comparison with other divalent metal ions, including Ni(II) and Co(II) (193).
Pvds are also capable of binding a range of divalent metal ions, although the ligands have not been identified (Figure 5d). However, Cu(II)Pvd cannot inhibit uptake of Fe(III)Pvd and is taken up into the cell at a rate 37-fold lower than that of Fe(III)Pvd (194). As with Ybt, Pvd and Pch may sequester copper outside of the cell, playing a protective role (195). In support of this notion, not only does copper stress upregulate Pvd (and not Pch) biosynthesis (196), but also import of CuPvd upregulates Pvd biosynthetic machinery and not the Pvd importer, the TBDT FpvA (195). While both Pch and Pvd can bind copper and play some protective role against copper toxicity, Pvd appears to be more biologically relevant in this process. Operons for both siderophores are downregulated in conditions of copper starvation, suggesting that neither siderophore is primarily involved in copper uptake (197). Finally, Pseudomonas species are among the nonmethanotrophic species in which Mbn homologs have been detected, and the presence of both MbnT3 TBDTs and copper homeostasis machinery suggests that Mbns in these species are the primary chalkophores (59).
Pyridine-2,6-Dithiocarboxylic Acid
Pyridine-2,6-dithiocarboxylic acid (pdtc) (Figure 5e) is also secreted by Pseudomonas species. Like Pch and Pvd, it binds a range of metals in varying arrangements and stoichiometries, and like CuYbt, copper-bound pdtc (Cu-pdtc) is capable of redox cycling (198). Interestingly, like Mbn, copper binding is mediated by both nitrogen and sulfur ligands (198), and unsurprisingly, pdtc can also bind other soft and easily polarizable metals. Stabilization of harder transition metals like nickel and iron requires bischelate formation, and despite demonstrated iron binding (199), pdtc’s role in iron uptake may rely partly on its ability to reduce iron (198, 200). Nevertheless, uptake of Fe(III)-pdtc and of Zn-pdtc (201, 202) has been the major biological focus, and whether pdtc is a true chalkophore is unclear. Cu-pdtc appears to have other unusual properties, including the ability to dechlorinate the carcinogen carbon tetrachloride (203).
Schizokinen and Marine Cyanobacterial Metallophores
Schizokinen is a citrate-containing hydroxamate siderophore originally identified in gram-positive Bacillus and cyanobacterial Anabaena species (204) (Figure 5f). The copper-binding abilities of siderophores produced by Anabaena species were noted as early as 1980 (205). While copper binding was clearly a secondary role for this siderophore, schizokinen’s copper-binding abilities were proposed as an explanation for the resistance of cyanobacteria to copper sulfate treatments during algal blooms (206). There is no evidence that the TBDT that imports Fe(III)-schizokinen can import copper-bound schizokinen (207), but alternate copper uptake methods exist, including the TBDT IacT, which appears to be involved in both iron and copper uptake (208).
The marine environment contains other copper-complexing compounds, including classes of uncharacterized compounds with higher affinity (L1) and lower affinity (L2) (209). Other specifically cyanobacterial metallophores appear to have roles related to copper, often for protection against copper toxicity (210). Under copper stress, Synechococcus species secrete an uncharacterized copper chelator with a binding affinity similar to that of L1 (211, 212). These compounds appear to be distinct from the schizokinen-like marine siderophores known as synechobactins (213), but whether they are primarily siderophores or their major role is in prevention of copper toxicity is unclear. The identification of marine metallophores with metal preferences other than iron remains an area of active research (214).
Supplementary Material
SUMMARY POINTS.
Metallophores other than siderophores play major roles in metal homeostasis. The best-studied nonsiderophores are the Mbn copper-binding chalkophores.
Mbns are RiPPs. The copper ligands comprise nitrogen-containing heterocycles and neighboring thioamide groups generated by core posttranslational modifications.
Mbn operons are found in a wide range of nonmethanotrophic bacteria, including gram-positive bacteria and human pathogens.
Increasing genetic and biochemical evidence has been found for the roles and mechanisms of the proteins involved in Mbn biosynthesis, regulation, and transport.
Other compounds with proposed chalkophore roles exist, indicating that chalkophore production is not restricted to methanotrophs or Mbns alone.
In some cases, metallophores that prefer other metals can bind copper with relatively high affinity, and copper binding that does not result in copper uptake may be a biologically relevant function of several siderophores.
FUTURE ISSUES.
Characterization of new Mbns from additional species is necessary to reveal the full structural and biochemical diversity of the family.
Specific aspects of Mbn biochemistry remain to be elucidated, including mechanisms of leader peptide cleavage as well as mechanisms of copper release.
The roles of Mbns in nonmethanotrophs are unexplored. Such studies will provide new insight into the roles and mechanisms of copper influx in a wide range of bacteria.
Further work is required to uncover how Mbn production is regulated, and for methanotrophs in particular, regulatory processes of interest include any role in the copper switch.
The roles of additional compounds currently classified as siderophores in the homeostasis of metals other than iron should be explored. For copper in particular, siderophores with nitrogen-containing heterocycles merit investigation.
Chalkophores and copper-binding siderophores should be pursued as drug candidates for diseases in which copper overload or mismetalation occurs.
Acknowledgments
Research is supported by National Institutes of Health grant GM118035 and Department of Energy grant DE-SC0016284 (A.C.R.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Thomson A, Gray H. Bio-inorganic chemistry. Curr. Opin. Chem. Biol. 1998;2:155–58. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- 2.Winkler JR, Gray HB. Electron flow through metalloproteins. Chem. Rev. 2014;114:3369–80. doi: 10.1021/cr4004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valko M, Morris H, Cronin MTD. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 4.Raymond KN, Carrano CJ. Coordination chemistry and microbial iron transport. Acc. Chem. Res. 1979;12:183–90. doi: 10.1021/acs.accounts.5b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lankford CE. Siderophore, a low-molecular weight carrier of iron. Crit. Rev. Microbiol. 1973;2:273–331. [Google Scholar]
- 6.Raymond KN, Allred BE, Sia AK. Coordination chemistry of microbial iron transport. Acc. Chem. Res. 2015;48:2496–505. doi: 10.1021/acs.accounts.5b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker KW, Skaar EP. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014;38:1235–49. doi: 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer LD, Skaar EP. Transition metals and virulence in bacteria. Annu. Rev. Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 2008;46:149–87. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- 10.Buděšínský M, Budzikiewicz H, Procházka Ž. Nicotianamine, a possible phytosiderophore of general occurrence. Phytochemistry. 1980;19:2295–97. [Google Scholar]
- 11.Springer SD, Butler A. Microbial ligand coordination: consideration of biological significance. Coord. Chem. Rev. 2016;306:628–35. [Google Scholar]
- 12.Johnstone TC, Nolan EM. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44:6320–39. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer SM, Duckworth OW, Harrington JM, Schenkeveld WDC. Metallophores and trace metal biogeochemistry. Aquat. Geochem. 2014;21:159–95. [Google Scholar]
- 14.Harrington JM, Parker DL, Bargar JR, Jarzecki AA, Tebo BM, et al. Structural dependence of Mn complexation by siderophores: donor group dependence on complex stability and reactivity. Geochim. Cosmochim. Acta. 2012;88:106–19. [Google Scholar]
- 15.Parker DL, Sposito G, Tebo BM. Manganese(III) binding to a pyoverdine siderophore produced by a manganese(II)-oxidizing bacterium. Geochim. Cosmochim. Acta. 2004;68:4809–20. [Google Scholar]
- 16.Parker DL, Lee S-W, Geszvain K, Davis RE, Gruffaz C, et al. Pyoverdine synthesis by the Mn(II)-oxidizing bacterium Pseudomonas putida GB-1. Front. Microbiol. 2014;5:202. doi: 10.3389/fmicb.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science. 2016;352:1105–9. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 18.Chivers PT, Benanti EL, Heil-Chapdelaine V, Iwig JS, Rowe JL. Identification of Ni-(L-His)2 as a substrate for NikABCDE-dependent nickel uptake in Escherichia coli. Metallomics. 2012;4:1043–50. doi: 10.1039/c2mt20139a. [DOI] [PubMed] [Google Scholar]
- 19.Lebrette H, Borezee-Durant E, Martin L, Richaud P, Boeri Erba E, Cavazza C. Novel insights into nickel import in Staphylococcus aureus: the positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator. Metallomics. 2015;7:613–21. doi: 10.1039/c4mt00295d. [DOI] [PubMed] [Google Scholar]
- 20.Bobrov AG, Kirillina O, Fetherston JD, Miller MC, Burlison JA, Perry RD. The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol. 2014;93:759–75. doi: 10.1111/mmi.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesketh A, Kock H, Mootien S, Bibb M. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3(2) Mol. Microbiol. 2009;74:1427–44. doi: 10.1111/j.1365-2958.2009.06941.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, et al. Gold biomineralization by a metallophore from a gold-associated microbe. Nat. Chem. Biol. 2013;9:241–43. doi: 10.1038/nchembio.1179. [DOI] [PubMed] [Google Scholar]
- 23.Wichard T, Mishra B, Myneni SCB, Bellenger J-P, Kraepiel AML. Storage and bioavailability of molybdenum in soils increased by organic matter complexation. Nat. Geosci. 2009;2:625–29. [Google Scholar]
- 24.Liermann LJ, Guynn RL, Anbar A, Brantley SL. Production of a molybdophore during metal-targeted dissolution of silicates by soil bacteria. Chem. Geol. 2005;220:285–302. [Google Scholar]
- 25.Dassama LMK, Kenney GE, Rosenzweig AC. Methanobactins: from genome to function. Metallomics. 2016;9:7–20. doi: 10.1039/c6mt00208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–15. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 27.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. PNAS. 2007;104:12040–45. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dassama LMK, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. Methanobactin transport machinery. PNAS. 2016;113:13027–32. doi: 10.1073/pnas.1603578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osman D, Cavet JS. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008;65:217–47. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- 30.Festa RA, Thiele DJ. Copper: an essential metal in biology. Curr. Biol. 2011;21:R877–83. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 32.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–43. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–82. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 34.Stanley SH, Prior SD, Leak DJ, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol. Lett. 1983;5:487–92. [Google Scholar]
- 35.Nielsen AK, Gerdes K, Murrell JC. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 36.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, et al. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 2003;185:5755–64. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phelps PA, Agarwal SK, Speitel GE, Georgiou G. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl. Environ. Microbiol. 1992;58:3701–8. doi: 10.1128/aem.58.11.3701-3708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitch MW, Graham DW, Arnold RG, Agarwal SK, Phelps PA, et al. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 1993;59:2771–76. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Téllez CM, Gaus KP, Graham DW, Arnold RG, Guzman RZ. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl. Environ. Microbiol. 1998;64:1115–22. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, et al. Copper-binding compounds from Methylosinus trichosporium OB3b. J. Bacteriol. 1998;180:3606–13. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behling LA, Hartsel SC, Lewis DE, DiSpirito AA, Choi DW, et al. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J. Am. Chem. Soc. 2008;130:12604–5. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Ghazouani A, Baslé A, Firbank SJ, Knapp CW, Gray J, et al. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg. Chem. 2011;50:1378–91. doi: 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 43.Kim HJ, Galeva N, Larive CK, Alterman M, Graham DW. Purification and physical-chemical properties of methanobactin: a chalkophore from Methylosinus trichosporium OB3b. Biochemistry. 2005;44:5140–48. doi: 10.1021/bi047367r. [DOI] [PubMed] [Google Scholar]
- 44.Blanco-Lomas M, Funes-Ardoiz I, Campos PJ, Sampedro D. Oxazolone-based photoswitches: synthesis and properties. Eur. J. Org. Chem. 2013;2013:6611–18. [Google Scholar]
- 45.Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow NL, et al. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry. 2010;49:10117–30. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hakemian AS, Tinberg CE, Kondapalli KC, Telser J, Hoffman BM, et al. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I) J. Am. Chem. Soc. 2005;127:17142–43. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi DW, Antholine WE, Do YS, Semrau JD, Kisting CJ, et al. Effect of methanobactin on the activity and electron paramagnetic resonance spectra of the membrane-associated methane monooxygenase in Methylococcus capsulatus Bath. Microbiology. 2005;151:3417–26. doi: 10.1099/mic.0.28169-0. [DOI] [PubMed] [Google Scholar]
- 48.Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, et al. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45:1442–53. doi: 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 49.Pesch M-L, Christl I, Hoffmann M, Kraemer SM, Kretzschmar R. Copper complexation of methanobactin isolated from Methylosinus trichosporium OB3b: pH-dependent speciation and modeling. J. Inorg. Biochem. 2012;116:55–62. doi: 10.1016/j.jinorgbio.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 50.Kulczycki E, Fowle DA, Knapp CW, Graham DW, Roberts JA. Methanobactin-promoted dissolution of Cu-substituted borosilicate glass. Geobiology. 2007;5:251–63. [Google Scholar]
- 51.Pesch M-L, Hoffmann M, Christl I, Kraemer SM, Kretzschmar R. Competitive ligand exchange between Cu–humic acid complexes and methanobactin. Geobiology. 2012;11:44–54. doi: 10.1111/gbi.12010. [DOI] [PubMed] [Google Scholar]
- 52.Ho A, Kerckhof F-M, Lüke C, Reim A, Krause S, et al. Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ. Microbiol. Rep. 2012;5:335–45. doi: 10.1111/j.1758-2229.2012.00370.x. [DOI] [PubMed] [Google Scholar]
- 53.Choi DW, Do YS, Zea CJ, McEllistrem MT, Lee S-W, et al. Spectral and thermodynamic properties of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV), and Zn(II) binding by methanobactin from Methylosinus trichosporium OB3b. J. Inorg. Biochem. 2006;100:2150–61. doi: 10.1016/j.jinorgbio.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 54.Choi DW, Semrau JD, Antholine WE, Hartsel SC, Anderson RC, et al. Oxidase, superoxide dismutase, and hydrogen peroxide reductase activities of methanobactin from types I and II methanotrophs. J. Inorg. Biochem. 2008;102:1571–80. doi: 10.1016/j.jinorgbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–19. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sirajuddin S, Barupala D, Helling S, Marcus K, Stemmler TL, Rosenzweig AC. Effects of zinc on particulate methane monooxygenase activity and structure. J. Biol. Chem. 2014;289:21782–94. doi: 10.1074/jbc.M114.581363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balasubramanian R, Rosenzweig AC. Copper methanobactin: a molecule whose time has come. Curr. Opin. Chem. Biol. 2008;12:245–49. doi: 10.1016/j.cbpa.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein LY, Yoon S, Semrau JD, DiSpirito AA, Crombie A, et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J. Bacteriol. 2010;192:6497–98. doi: 10.1128/JB.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenney GE, Rosenzweig AC. Genome mining for methanobactins. BMC Biol. 2013;11:17. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, et al. Methanobactin and MmoD work in concert to act as the “copper-switch” in methanotrophs. Environ. Microbiol. 2013;15:3077–86. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 61.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–60. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DiSpirito AA, Semrau JD, Murrell JC, Gallagher WH, Dennison C, Vuilleumier S. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev. 2016;80:387–409. doi: 10.1128/MMBR.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Koay M, Maher MJ, Xiao Z, Wedd AG. Intermolecular transfer of copper ions from the CopC protein of Pseudomonas syringae Crystal structures of fully loaded CuICuII forms. J. Am. Chem. Soc. 2006;128:5834–50. doi: 10.1021/ja058528x. [DOI] [PubMed] [Google Scholar]
- 64.Lawton TJ, Kenney GE, Hurley JD, Rosenzweig AC. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry. 2016;55:2278–90. doi: 10.1021/acs.biochem.6b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cha JS, Cooksey DA. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl. Environ. Microbiol. 1993;59:1671–74. doi: 10.1128/aem.59.5.1671-1674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dash BP, Alles M, Bundschuh FA, Richter O. Protein chaperones mediating copper insertion into the CuA site of the aa3-type cytochrome c oxidase of Paracoccus denitrificans. Biochim. Biophys. Acta. 2015;1847:202–11. doi: 10.1016/j.bbabio.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, et al. Mechanism of CuA assembly. Nat. Chem. Biol. 2008;4:599–601. doi: 10.1038/nchembio.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewinson O, Lee AT, Rees DC. A P-type ATPase importer that discriminates between essential and toxic transition metals. PNAS. 2009;106:4677–82. doi: 10.1073/pnas.0900666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004;2:414–24. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 70.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1999;181:6223–29. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kenney GE, Sadek M, Rosenzweig AC. Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics. 2016;8:931–40. doi: 10.1039/c5mt00289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu W, Semrau JD. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl. Microbiol. Biotechnol. 2017;101:8499–516. doi: 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 73.Larsen Ø, Karlsen OA. Transcriptomic profiling of Methylococcus capsulatus (Bath) during growth with two different methane monooxygenases. MicrobiologyOpen. 2015;5:254–67. doi: 10.1002/mbo3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.El Ghazouani A, Baslé A, Gray J, Graham DW, Firbank SJ, Dennison C. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. PNAS. 2012;109:8400–4. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kenney GE, Goering AW, Ross MO, DeHart CJ, Thomas PM, et al. Characterization of methanobactin from Methylosinus sp. LW4. J. Am. Chem. Soc. 2016;138:11124–27. doi: 10.1021/jacs.6b06821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris WR, Carrano CJ, Cooper SR, Sofen SR, Avdeef AE, et al. Coordination chemistry of microbial iron transport compounds. 19. Stability constants and electrochemical behavior of ferric enterobactin and model complexes. J. Am. Chem. Soc. 1979;101:6097–104. [Google Scholar]
- 77.Bandow NL, Gilles VS, Freesmeier B, Semrau JD, Krentz B, et al. Spectral and copper binding properties of methanobactin from the facultative methanotroph Methylocystis strain SB2. J. Inorg. Biochem. 2012;110:72–82. doi: 10.1016/j.jinorgbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 2015;11:564–70. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J. Am. Chem. Soc. 2012;134:5309–16. doi: 10.1021/ja211675n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins—ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010;86:1213–25. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Molohon KJ, Melby JO, Lee J, Evans BS, Dunbar KL, et al. Structure determination and interception of biosynthetic intermediates for the plantazolicin class of highly discriminating antibiotics. ACS Chem. Biol. 2011;6:1307–13. doi: 10.1021/cb200339d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010;8:70. doi: 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fuchs SW, Lackner G, Morinaka BI, Morishita Y, Asai T, et al. A lanthipeptide-like N-terminal leader region guides peptide epimerization by radical SAM epimerases: implications for RiPP evolution. Angew. Chem. Int. Ed. Engl. 2016;128:12330–33. doi: 10.1002/anie.201602863. [DOI] [PubMed] [Google Scholar]
- 84.Ortega MA, van der Donk WA. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 2016;23:31–44. doi: 10.1016/j.chembiol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox CL, Doroghazi JR, Mitchell DA. The genomic landscape of ribosomal peptides containing thiazole and oxazole heterocycles. BMC Genom. 2015;16:778. doi: 10.1186/s12864-015-2008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rix U, Zheng J, Remsing Rix LL, Greenwell L, Yang K, Rohr J. The dynamic structure of jadomycin B and the amino acid incorporation step of its biosynthesis. J. Am. Chem. Soc. 2004;126:4496–97. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 87.Banala S, Süssmuth RD. Thioamides in nature: in search of secondary metabolites in anaerobic microorganisms. ChemBioChem. 2010;11:1335–37. doi: 10.1002/cbic.201000266. [DOI] [PubMed] [Google Scholar]
- 88.Izawa M, Kawasaki T, Hayakawa Y. Cloning and heterologous expression of the thioviridamide biosynthesis gene cluster from Streptomyces olivoviridis. Appl. Environ. Microbiol. 2013;79:7110–13. doi: 10.1128/AEM.01978-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lincke T, Behnken S, Ishida K, Roth M, Hertweck C. Closthioamide: an unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed. Engl. 2010;122:2055–57. doi: 10.1002/anie.200906114. [DOI] [PubMed] [Google Scholar]
- 90.Dunbar KL, Chekan JR, Cox CL, Burkhart BJ, Nair SK, Mitchell DA. Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. Nat. Chem. Biol. 2014;10:823–29. doi: 10.1038/nchembio.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dunbar KL, Melby JO, Mitchell DA. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat. Chem. Biol. 2012;8:569–75. doi: 10.1038/nchembio.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kenney GE, Dassama LMK, Pandelia M-E, Gizzi AS, Martinie RJ, et al. The biosynthesis of methanobactin. Science. 2018;359:1411–16. doi: 10.1126/science.aap9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medzihradszky KF, Darula Z, Perlson E, Fainzilber M, Chalkley RJ, et al. O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol. Cell. Proteom. 2004;3:429–40. doi: 10.1074/mcp.M300140-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.Wang T, Cook I, Falany CN, Leyh TS. Paradigms of sulfotransferase catalysis: the mechanism of SULT2A1. J. Biol. Chem. 2014;289:26474–80. doi: 10.1074/jbc.M114.573501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howard-Jones AR, Walsh CT. Staurosporine and rebeccamycin aglycones are assembled by the oxidative action of StaP, StaC, and RebC on chromopyrrolic acid. J. Am. Chem. Soc. 2006;128:12289–98. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 96.Walsh CT, Wencewicz TA. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 2012;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gu W, Baral BS, DiSpirito AA, Semrau JD. An aminotransferase is responsible for the deamination of the N-terminal leucine and required for formation of oxazolone ring A in methanobactin of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2017;83:e02619–16. doi: 10.1128/AEM.02619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lagedroste M, Smits SHJ, Schmitt L. Substrate specificity of the secreted nisin leader peptidase NisP. Biochemistry. 2017;56:4005–14. doi: 10.1021/acs.biochem.7b00524. [DOI] [PubMed] [Google Scholar]
- 99.Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006;27:587–93. doi: 10.1016/j.tips.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 100.He X, Szewczyk P, Karyakin A, Evin M, Hong W-X, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–94. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta. 2009;1794:763–68. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 102.Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 2011;286:37313–19. doi: 10.1074/jbc.M111.284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schalk IJ, Yue WW, Buchanan SK. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 2004;54:14–22. doi: 10.1111/j.1365-2958.2004.04241.x. [DOI] [PubMed] [Google Scholar]
- 104.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: Do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 2008;33:330–38. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 106.Koebnik R. TonB-dependent trans-envelope signalling: The exception or the rule? Trends Microbiol. 2005;13:343–47. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 107.Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 2003;6:173–80. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 108.Rédly GA, Poole K. FpvIR control of fpvA ferric pyoverdine receptor gene expression in Pseudomonas aeruginosa: demonstration of an interaction between FpvI and FpvR and identification of mutations in each compromising this interaction. J. Bacteriol. 2005;187:5648–57. doi: 10.1128/JB.187.16.5648-5657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol. Microbiol. 2012;86:628–44. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ul-Haque MF, Kalidass B, Vorobev AV, Baral BS, DiSpirito AA, Semrau JD. Methanobactin from Methylocystis strain SB2 affects gene expression and methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2015;81:2466–73. doi: 10.1128/AEM.03981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gu W, Farhan Ul Haque M, Baral BS, Turpin EA, Bandow NL, et al. A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2016;82:1917–23. doi: 10.1128/AEM.03884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Greenwald J, Nader M, Celia H, Gruffaz C, Geoffroy V, et al. FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol. Microbiol. 2009;72:1246–59. doi: 10.1111/j.1365-2958.2009.06721.x. [DOI] [PubMed] [Google Scholar]
- 113.Kenney GE, Rosenzweig AC. Chemistry and biology of the copper chelator methanobactin. ACS Chem. Biol. 2012;7:260–68. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, et al. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 2007;189:8361–65. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sleigh SH, Tame JR, Dodson EJ, Wilkinson AJ. Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine. Biochemistry. 1997;36:9747–58. doi: 10.1021/bi970457u. [DOI] [PubMed] [Google Scholar]
- 116.Schalk IJ, Guillon L. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids. 2013;44:1267–77. doi: 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 117.Imperi F, Tiburzi F, Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. PNAS. 2009;106:20440–45. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jones CM, Wells RM, Madduri AVR, Renfrow MB, Ratledge C, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. PNAS. 2014;111:1945–50. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hannauer M, Barda Y, Mislin GLA, Shanzer A, Schalk IJ. The ferrichrome uptake pathway in Pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J. Bacteriol. 2010;192:1212–20. doi: 10.1128/JB.01539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miethke M, Hou J, Marahiel MA. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry. 2011;50:10951–64. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- 121.Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J. Biol. Chem. 2008;283:13897–904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- 122.Johnson KA, Ve T, Larsen Ø, Pedersen R-B, Lillehaug JR, et al. CorA is a copper repressible surface-associated copper(I)-binding protein produced in Methylomicrobium album BG8. PLOS ONE. 2014;9:e87750. doi: 10.1371/journal.pone.0087750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vita N, Platsaki S, Baslé A, Allen SJ, Paterson NG, et al. A four-helix bundle stores copper for methane oxidation. Nature. 2015;525:140–43. doi: 10.1038/nature14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ross MO, Rosenzweig AC. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 2016;22:307–19. doi: 10.1007/s00775-016-1419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reimmann C. Inner-membrane transporters for the siderophores pyochelin in Pseudomonas aeruginosa and enantio-pyochelin in Pseudomonas fluorescens display different enantioselectivities. Microbiology. 2012;158:1317–24. doi: 10.1099/mic.0.057430-0. [DOI] [PubMed] [Google Scholar]
- 126.Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, et al. Signal transduction pathway of TonB-dependent transporters. PNAS. 2007;104:513–18. doi: 10.1073/pnas.0609887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Youard ZA, Reimmann C. Stereospecific recognition of pyochelin and enantio-pyochelin by the PchR proteins in fluorescent pseudomonads. Microbiology. 2010;156:1722–82. doi: 10.1099/mic.0.037796-0. [DOI] [PubMed] [Google Scholar]
- 128.Yan X, Chu F, Puri AW, Fu Y, Lidstrom ME. Electroporation-based genetic manipulation in Type I methanotrophs. Appl. Environ. Microbiol. 2016;82:2062–69. doi: 10.1128/AEM.03724-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Collins ML, Buchholz LA, Remsen CC. Effect of copper on Methylomonas albus BG8. Appl. Environ. Microbiol. 1991;57:1261–64. doi: 10.1128/aem.57.4.1261-1264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brantner CA, Buchholz LA, McSwain CL, Newcomb LL, Remsen CC, Collins MLP. Intracytoplasmic membrane formation in Methylomicrobium album BG8 is stimulated by copper in the growth medium. Can. J. Microbiol. 1997;43:672–76. [Google Scholar]
- 131.Theisen AR, Ali MH, Radajewski S, Dumont MG, Dunfield PF, et al. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 2005;58:682–92. doi: 10.1111/j.1365-2958.2005.04861.x. [DOI] [PubMed] [Google Scholar]
- 132.Merkx M, Lippard SJ. Why OrfY? Characterization of MMOD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath) J. Biol. Chem. 2002;277:5858–65. doi: 10.1074/jbc.M107712200. [DOI] [PubMed] [Google Scholar]
- 133.Lipscomb JD. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 1994;48:371–99. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 134.Ali MH, Murrell JC. Development and validation of promoter-probe vectors for the study of methane monooxygenase gene expression in Methylococcus capsulatus Bath. Microbiology. 2009;155:761–71. doi: 10.1099/mic.0.021816-0. [DOI] [PubMed] [Google Scholar]
- 135.Stafford GP, Scanlan J, McDonald IR, Murrell JC. rpoN, mmoR and mmoG genes involved in regulating the expression of soluble methane monooxygenase in Methylosinus trichosporium OB3b. Microbiology. 2003;149:1771–84. doi: 10.1099/mic.0.26060-0. [DOI] [PubMed] [Google Scholar]
- 136.Kurth EG, Doughty DM, Bottomley PJ, Arp DJ, Sayavedra-Soto LA. Involvement of BmoR and BmoG in n-alkane metabolism in “Pseudomonas butanovora”. Microbiology. 2008;154:139–47. doi: 10.1099/mic.0.2007/012724-0. [DOI] [PubMed] [Google Scholar]
- 137.Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, et al. Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ. Microbiol. Rep. 2010;3:297–307. doi: 10.1111/j.1758-2229.2010.00225.x. [DOI] [PubMed] [Google Scholar]
- 138.Xin JY, Lin K, Wang Y, Xia C-G. Methanobactin-mediated synthesis of gold nanoparticles supported over Al2O3 toward an efficient catalyst for glucose oxidation. Int. J. Mol. Sci. 2014;15:21603–20. doi: 10.3390/ijms151221603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kalidass B, Ul-Haque MF, Baral BS, DiSpirito AA, Semrau JD. Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of copper. Appl. Environ. Microbiol. 2014;81:1024–31. doi: 10.1128/AEM.03151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zammit CM, Weiland F, Brugger J, Wade B, Winderbaum LJ, et al. Proteomic responses to gold(III)-toxicity in the bacterium Cupriavidus metallidurans CH34. Metallomics. 2016;8:1204–16. doi: 10.1039/c6mt00142d. [DOI] [PubMed] [Google Scholar]
- 141.Reith F, Etschmann B, Grosse C, Moors H, Benotmane MA, et al. Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. PNAS. 2009;106:17757–62. doi: 10.1073/pnas.0904583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLOS ONE. 2010;5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.von Rozycki T, Nies DH. Cupriavidus metallidurans: evolution of a metal-resistant bacterium. A. Van Leeuw. J. Microbiol. 2009;96:115–39. doi: 10.1007/s10482-008-9284-5. [DOI] [PubMed] [Google Scholar]
- 144.Vorobev AV, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, et al. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2013;79:5918–26. doi: 10.1128/AEM.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baral BS, Bandow NL, Vorobev AV, Freemeier BC, Bergman BH, et al. Mercury binding by methanobactin from Methylocystis strain SB2. J. Inorg. Biochem. 2014;141C:161–69. doi: 10.1016/j.jinorgbio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 146.Lu X, Gu W, Zhao L, Farhan Ul Haque M, DiSpirito AA, et al. Methylmercury uptake and degradation by methanotrophs. Sci. Adv. 2017;3:e1700041. doi: 10.1126/sciadv.1700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Boden R, Murrell JC. Response to mercury (II) ions in Methylococcus capsulatus (Bath) FEMS Microbiol. Lett. 2011;324:106–10. doi: 10.1111/j.1574-6968.2011.02395.x. [DOI] [PubMed] [Google Scholar]
- 148.Baesman SM, Miller LG, Wei JH, Cho Y, Matys ED, et al. Methane oxidation and molecular characterization of methanotrophs from a former mercury mine impoundment. Microorganisms. 2015;3:290–309. doi: 10.3390/microorganisms3020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol. Rev. 1996;60:439–71. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Costello AM, Lidstrom ME. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 1999;65:5066–74. doi: 10.1128/aem.65.11.5066-5074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dedysh SN, Belova SE, Bodelier PLE, Smirnova KV, Khmelenina VN, et al. Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 2007;57:472–79. doi: 10.1099/ijs.0.64623-0. [DOI] [PubMed] [Google Scholar]
- 152.Wartiainen I, Hestnes AG, McDonald IR, Svenning MM. Methylocystis rosea sp. nov., a novel methanotrophic bacterium from Arctic wetland soil, Svalbard, Norway (78 degrees N) Int. J. Syst. Evol. Microbiol. 2006;56:541–47. doi: 10.1099/ijs.0.63912-0. [DOI] [PubMed] [Google Scholar]
- 153.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. Methanotrophy below pH1 by a new Verrucomicrobia species. Nature. 2007;450:874–78. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 154.Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T. Widespread production of extracellular superoxide by heterotrophic bacteria. Science. 2013;340:1223–26. doi: 10.1126/science.1237331. [DOI] [PubMed] [Google Scholar]
- 155.Johnson CL. PhD Thesis. Iowa State Univ; Ames, IA: 2006. Methanobactin: a potential novel biopreservative for use against the foodborne pathogen Listeria monocytogenes. [Google Scholar]
- 156.Hecht SM. Bleomycin: new perspectives on the mechanism of action. J. Nat. Prod. 2000;63:158–68. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 157.Seipke RF, Kaltenpoth M, Hutchings MI. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev. 2012;36:862–76. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 158.Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009;13:224–30. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 159.Mansson M, Gram L, Larsen TO. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs. 2011;9:1440–68. doi: 10.3390/md9091440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wöhler F. The treatment of haemochromatosis with desferrioxamine. Acta. Haematol. 1963;30:65–87. doi: 10.1159/000208110. [DOI] [PubMed] [Google Scholar]
- 161.Moeschlin S, Schnider U. Treatment of primary and secondary hemochromatosis and acute iron poisoning with a new, potent iron-eliminating agent (desferrioxamine-B) N. Engl. J. Med. 1963;269:57–66. [PubMed] [Google Scholar]
- 162.Ala A, Walker AP, Ashkan K, Dooley JS, Schilsky ML. Wilson’s disease. Lancet. 2007;369:397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- 163.Medici V, Rossaro L, Sturniolo GC. Wilson disease—a practical approach to diagnosis, treatment and follow-up. Digest. Liver Dis. 2007;39:601–9. doi: 10.1016/j.dld.2006.12.095. [DOI] [PubMed] [Google Scholar]
- 164.Ding X, Xie H, Kang YJ. The significance of copper chelators in clinical and experimental application. J. Nutr. Biochem. 2011;22:301–10. doi: 10.1016/j.jnutbio.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 165.Summer KH, Lichtmannegger J, Bandow NL, Choi DW, DiSpirito AA, Michalke B. The biogenic methanobactin is an effective chelator for copper in a rat model for Wilson disease. J. Trace Elem. Med. Biol. 2011;25:36–41. doi: 10.1016/j.jtemb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 166.Lichtmannegger J, Leitzinger C, Wimmer R, Schmitt S, Schulz S, et al. Methanobactin reverses acute liver failure in a rat model of Wilson disease. J. Clin. Invest. 2016;126:2721–35. doi: 10.1172/JCI85226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Desai V, Kaler SG. Role of copper in human neurological disorders. Am. J. Clin. Nutr. 2008;88:855S–8S. doi: 10.1093/ajcn/88.3.855S. [DOI] [PubMed] [Google Scholar]
- 168.Choi DW, Bandow NL, McEllistrem MT, Semrau JD, Antholine WE, et al. Spectral and thermodynamic properties of methanobactin from γ-proteobacterial methane oxidizing bacteria: a case for copper competition on a molecular level. J. Inorg. Biochem. 2010;104:1240–47. doi: 10.1016/j.jinorgbio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 169.Karlsen OA, Berven FS, Stafford GP, Larsen Ø, Murrell JC, et al. The surface-associated and secreted MopE protein of Methylococcus capsulatus (Bath) responds to changes in the concentration of copper in the growth medium. Appl. Environ. Microbiol. 2003;69:2386–88. doi: 10.1128/AEM.69.4.2386-2388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, et al. The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper. PLOS ONE. 2012;7:e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Koepke J, Olkhova E, Angerer H, Müller H. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: new insights into the active site and the proton transfer pathways. Biochim. Biophys. Acta. 2009;1787:635–45. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 172.Haltia T, Brown K, Tegoni M, Cambillau C, Saraste M, et al. Crystal structure of nitrous oxide reductase from Paracoccus denitrificans at 1.6 Å resolution. Biochem. J. 2003;369:77–88. doi: 10.1042/BJ20020782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Anttila J, Heinonen P, Nenonen T, Pino A, Iwaï H, et al. Is coproporphyrin III a copper-acquisition compound in Paracoccus denitrificans? Biochim. Biophys. Acta. 2011;1807:311–18. doi: 10.1016/j.bbabio.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 174.Azzouzi A, Steunou A-S, Durand A, Khalfaoui-Hassani B, Bourbon M-L, et al. Coproporphyrin III excretion identifies the anaerobic coproporphyrinogen III oxidase HemN as a copper target in the Cu+-ATPase mutant copA− of Rubrivivax gelatinosus. Mol. Microbiol. 2013;88:339–51. doi: 10.1111/mmi.12188. [DOI] [PubMed] [Google Scholar]
- 175.Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145:1181–90. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 176.Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microb. Infect. 2001;3:561–69. doi: 10.1016/s1286-4579(01)01412-5. [DOI] [PubMed] [Google Scholar]
- 177.Payne SM. Detection, isolation, and characterization of siderophores. Method. Enzymol. 1994;235:329–44. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- 178.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 2012;8:731–36. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stafford SL, Bokil NJ, Achard MES, Kapetanovic R, Schembri MA, et al. Metal ions in macrophage antimicrobial pathways: emerging roles for zinc and copper. Biosci. Rep. 2013;33:e00049. doi: 10.1042/BSR20130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chaturvedi KS, Hung CS, Giblin DE, Urushidani S, Austin AM, et al. Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol. 2013;9:551–61. doi: 10.1021/cb400658k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koh E-I, Hung CS, Parker KS, Crowley JR, Giblin DE, Henderson JP. Metal selectivity by the virulence-associated yersiniabactin metallophore system. Metallomics. 2015;7:1011–22. doi: 10.1039/c4mt00341a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Koh E-I, Robinson AE, Bandara N, Rogers BE, Henderson JP. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat. Chem. Biol. 2017;13:1016–21. doi: 10.1038/nchembio.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Amano S-I, Sakurai T, Endo K, Takano H, Beppu T, et al. A cryptic antibiotic triggered by monensin. J. Antibiot. 2011;64:703. doi: 10.1038/ja.2011.69. [DOI] [PubMed] [Google Scholar]
- 184.Wang L, Zhu M, Zhang Q, Zhang X, Yang P, et al. Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS Chem. Biol. 2017;12:3067–75. doi: 10.1021/acschembio.7b00897. [DOI] [PubMed] [Google Scholar]
- 185.Harris NC, Sato M, Herman NA, Twigg F, Cai W, et al. Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria. PNAS. 2017;114:7025–30. doi: 10.1073/pnas.1705016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Stephan UW, Schmidke I, Stephan VW, Scholz G. The nicotianamine molecule is made-to-measure for complexation of metal micronutrients in plants. Biometals. 1996;9:84–90. [Google Scholar]
- 187.Irtelli B, Petrucci WA, Navari-Izzo F. Nicotianamine and histidine/proline are, respectively, the most important copper chelators in xylem sap of Brassica carinata under conditions of copper deficiency and excess. J. Exp. Bot. 2009;60:269–77. doi: 10.1093/jxb/ern286. [DOI] [PubMed] [Google Scholar]
- 188.Gi M, Lee K-M, Kim SC, Yoon J-H, Yoon SS, Choi JY. A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci. Rep. 2015;5:14644–59. doi: 10.1038/srep14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McFarlane JS, Lamb AL. Biosynthesis of an opine metallophore by Pseudomonas aeruginosa. Biochemistry. 2017;56:5967–71. doi: 10.1021/acs.biochem.7b00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mastropasqua MC, D’Orazio M, Cerasi M, Pacello F, Gismondi A, et al. Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol. Microbiol. 2017;106:543–61. doi: 10.1111/mmi.13834. [DOI] [PubMed] [Google Scholar]
- 191.Lhospice S, Gomez NO, Ouerdane L, Brutesco C, Ghssein G, et al. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 2017;7:17132. doi: 10.1038/s41598-017-16765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schalk IJ, Guillon L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ. Microbiol. 2013;15:1661–73. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 193.Brandel J, Humbert N, Elhabiri M, Schalk IJ, Mislin GLA, Albrecht-Gary A-M. Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans. 2012;41:2820–34. doi: 10.1039/c1dt11804h. [DOI] [PubMed] [Google Scholar]
- 194.Braud A, Hoegy F, Jezequel K, Lebeau T, Schalk IJ. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009;11:1079–91. doi: 10.1111/j.1462-2920.2008.01838.x. [DOI] [PubMed] [Google Scholar]
- 195.Braud A, Geoffroy V, Hoegy F, Mislin GLA, Schalk IJ. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ. Microbiol. Rep. 2010;2:419–25. doi: 10.1111/j.1758-2229.2009.00126.x. [DOI] [PubMed] [Google Scholar]
- 196.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7242–56. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Frangipani E, Slaveykova VI, Reimmann C, Haas D. Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J. Bacteriol. 2008;190:6706–17. doi: 10.1128/JB.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cortese MS, Paszczynski A, Lewis TA, Sebat JL, Borek V, Crawford RL. Metal chelating properties of pyridine-2,6-bis(thiocarboxylic acid) produced by Pseudomonas sp. and the biological activities of the formed complexes. Biometals. 2002;15:103–20. doi: 10.1023/a:1015241925322. [DOI] [PubMed] [Google Scholar]
- 199.Stolworthy JC, Paszczynski A, Korus R, Crawford RL. Metal binding by pyridine-2,6-bis(monothiocarboxylic acid), a biochelator produced by Pseudomonas stutzeri and Pseudomonas putida. Biodegradation. 2001;12:411–18. doi: 10.1023/a:1015067729660. [DOI] [PubMed] [Google Scholar]
- 200.Ockels W, Römer A, Budzikiewicz H, Korth H, Pulverer G. An Fe (II) complex of pyridine-2,6-di-(monothiocarboxylic acid)-a novel bacterial metabolic product. Tetrahedron Lett. 1978;19:3341–42. [Google Scholar]
- 201.Leach LH. Identification and characterization of Pseudomonas membrane transporters necessary for utilization of the siderophore pyridine-2,6-bis(thiocarboxylic acid) (PDTC) Microbiology. 2006;152:3157–66. doi: 10.1099/mic.0.29116-0. [DOI] [PubMed] [Google Scholar]
- 202.Leach LH, Morris JC, Lewis TA. The role of the siderophore pyridine-2,6-bis (thiocarboxylic acid) (PDTC) in zinc utilization by Pseudomonas putida DSM 3601. Biometals. 2006;20:717–26. doi: 10.1007/s10534-006-9035-x. [DOI] [PubMed] [Google Scholar]
- 203.Lewis TA, Paszczynski A, Gordon-Wylie SW, Jeedigunta S, Lee CH, Crawford RL. Carbon tetrachloride dechlorination by the bacterial transition metal chelator pyridine-2,6-bis(thiocarboxylic acid) Environ. Sci. Technol. 2001;35:552–59. doi: 10.1021/es001419s. [DOI] [PubMed] [Google Scholar]
- 204.Plowman JE, Loehr TM, Goldman SJ, Sanders-Loehr J. Structure and siderophore activity of ferric schizokinen. J. Inorg. Biochem. 1984;20:183–97. [Google Scholar]
- 205.McKnight DM, Morel FMM. Copper complexation by siderophores from filamentous blue-green algae. Limnol. Oceanogr. 1980;25:62–71. [Google Scholar]
- 206.Clarke SE, Stuart J, Sanders-Loehr J. Induction of siderophore activity in Anabaena spp. and its moderation of copper toxicity. Appl. Environ. Microbiol. 1987;53:917–22. doi: 10.1128/aem.53.5.917-922.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nicolaisen K, Moslavac S, Samborski A, Valdebenito M, Hantke K, et al. Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. J. Bacteriol. 2008;190:7500–7. doi: 10.1128/JB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nicolaisen K, Hahn A, Valdebenito M, Moslavac S, Samborski A, et al. The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta. 2010;1798:2131–40. doi: 10.1016/j.bbamem.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 209.Moffett JW. Temporal and spatial variability of copper complexation by strong chelators in the Sargasso Sea. Deep-Sea Res. Part I. 1995;42:1273–95. [Google Scholar]
- 210.Mann EL, Ahlgren N, Moffett JW, Chisholm SW. Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 2002;47:976–88. [Google Scholar]
- 211.Moffett JW, Brand LE. Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol. Oceanogr. 1996;41:388–95. [Google Scholar]
- 212.Wiramanaden CIE, Cullen JT, Ross ARS, Orians KJ. Cyanobacterial copper-binding ligands isolated from artificial seawater cultures. Mar. Chem. 2008;110:28–41. [Google Scholar]
- 213.Ito Y, Butler A. Structure of synechobactins, new siderophores of the marine cyanobacterium Synechococcus sp. PCC 7002. Limnol. Oceanogr. 2005;50:1918–23. [Google Scholar]
- 214.Boiteau RM, Till CP, Ruacho A, Bundy RM, Hawco NJ, et al. Structural characterization of natural nickel and copper binding ligands along the US GEOTRACES Eastern Pacific Zonal Transect. Front. Mar. Sci. 2016;3:243. [Google Scholar]
- 215.Kenney GE, Rosenzweig AC. Methanobactins: maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 2018 doi: 10.1074/jbc.TM117.000185. In press. https://doi.org/10.1074/jbc.TM117.000185. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.