ABSTRACT
The genus Cryptococcus includes several species pathogenic for humans. Until recently, the two major pathogenic species were recognized to be Cryptococcus neoformans and Cryptococcus gattii. We compared the interaction of murine macrophages with three C. gattii species complex strains (WM179, R265, and WM161, representing molecular types VGI, VGIIa, and VGIII, respectively) and one C. neoformans species complex strain (H99, molecular type VNI) to ascertain similarities and differences in the yeast intracellular pathogenic strategy. The parameters analyzed included nonlytic exocytosis frequency, phagolysosomal pH, intracellular capsular growth, phagolysosomal membrane permeabilization, and macrophage transcriptional response, assessed using time-lapse microscopy, fluorescence microscopy, flow cytometry, and gene expression microarray analysis. The most striking result was that the intracellular pathogenic strategies of C. neoformans and C. gattii species complex strains were qualitatively similar, despite the species having separated an estimated 100 million years ago. Macrophages exhibited a leaky phagolysosomal membrane phenotype and nonlytic exocytosis when infected with either C. gattii or C. neoformans. Conservation of the intracellular strategy among species that separated long ago suggests that it is ancient and possibly maintained by similar selection pressures through eons.
KEYWORDS: Cryptococcus neoformans, macrophages, virulence
INTRODUCTION
The genus Cryptococcus includes several species pathogenic for humans. Until recently, the two major pathogenic species were recognized to be Cryptococcus neoformans and Cryptococcus gattii. However, accumulating genomic evidence indicates that both of these denominations include more diversity than was previously recognized, such that the splitting of these species into several distinct species may be necessary. However, given taxonomic uncertainties, it has been proposed that the term “species complex” be utilized until these issues are resolved and agreed upon by the field (1). Hence, in this paper we employ the species complex nomenclature and refer to the two major groupings as C. neoformans species complex and C. gattii species complex. Despite the genetic diversity within and between these two groups, they cause strikingly similar disease.
Based on the coincidence in dates between the separation of the two cryptococcal species complexes and the breakup of the supercontinent Gondwanaland, we recently proposed that the initial speciation trigger was continental drift, which helps explain the global distribution of the various genotypes (2, 3). In addition to causing very similar disease in humans and animals, the cryptococcal complexes share very similar virulence factors, despite separation of the various lineages in ancient geologic time. Given that the morphology, biology, and virulence characteristics of the two species complexes are very similar and that these differ from those of other related species, it is possible that they descended from a common Pangean ancestor that also possessed these traits (2, 4). Consequently, the comparative study of the pathogenic strategies employed by the two species complexes could provide important insight into distant common ancestors and the mechanisms responsible for the emergence and maintenance of virulence.
The C. gattii species complex comprises a diverse set of organisms that include strains responsible for an outbreak of cryptococcosis in immunocompetent patients in British Columbia and the Pacific Northwest of the United States (5–7). Cases of cryptococcosis caused by both species complexes number about 223,000, with approximately 181,000 deaths (8). The majority of these cases were caused by C. neoformans species complex strains. However, the sister species complex of C. neoformans, C. gattii, is remarkable in that strains of this complex often cause disease in patients with no known immunological defects (9, 10). The C. gattii species complex has been categorized into serotypes B and C, each with four molecular subtypes: VGI, VGII, VGIII, and VGIV (10). C. gattii species complex strains were originally thought to be restricted to the tropics and subtropics, but as the outbreaks in British Columbia and the Pacific Northwest of the United States demonstrate, this microbe has adapted to more temperate climates (9). Thus, the outbreaks in the Vancouver Islands and the Pacific Northwest of the United States, which had higher incidence rates than areas of endemicity in Australia, have stimulated interest in C. gattii biology (10, 11).
Alveolar macrophages (Mac), dendritic cells (DC), and neutrophils are the early responders to Cryptococcus infection, while CD4+ T helper cells are the main effector cell of the adaptive cellular immune response to Cryptococcus (12). There have been inconsistent reports about which cryptococcal species complex is more virulent, but C. gattii Vancouver Island outbreak strains (mainly VGIIa strains) elicit weaker immune responses with lower levels of interleukin 6 (IL-6), tumor necrosis factor alpha, monocyte chemotactic protein 1, Th1, and Th17 cytokines than strains not associated with the outbreak (13–15). The VGII strains further inhibit neutrophil and CD4+ T helper cell migration to the lung (13, 16, 17). A possible mechanism for this reduced protective immunity is inhibition of DC maturation, which may be related to C. gattii polysaccharide capsule effects (18).
Although there are differences in the degree of the immune response, the sets of virulence factors employed by the two species complexes are remarkably similar. The cryptococcal capsule, one of the major virulence factors of Cryptococcus, is mainly composed of glucuronoxylomannan (GXM), which has several immunomodulatory properties (9, 19). However, it has been reported that the GXM capsule structure of C. gattii strains differs from that of C. neoformans strains and may be responsible for the different degrees of host activation (20) associated with the two species complexes. Urease is another major virulence factor shared by both species complexes and one that is proposed to have several roles, though these remain under debate (21). Mice infected with urease-deficient C. neoformans mutants lived longer and had less dissemination than mice infected with wild-type strains (21). Additionally, C. gattii species complex strains have also been shown to possess several unique virulence factors differing from those of C. neoformans, such as a deubiquitinase, an increased rate of intracellular proliferation, and the ability to induce host mitochondrial morphology changes (22).
C. gattii Vancouver Island outbreak strains have a significantly higher intracellular proliferation rate than nonoutbreak C. gattii strains, correlating with a greater fungal burden in the lungs (23, 24). The intracellular proliferation rate and host mitochondrial morphology of the C. gattii strains are correlated with increased virulence (24). This suggests that the intracellular pathogenic strategy in macrophages has some role in the increased virulence. In this study, we examined and compared the intracellular pathogenic strategy of strains belonging to the two species complexes.
RESULTS
Phagocytosis of Cryptococcus gattii and Cryptococcus neoformans species complex strains.
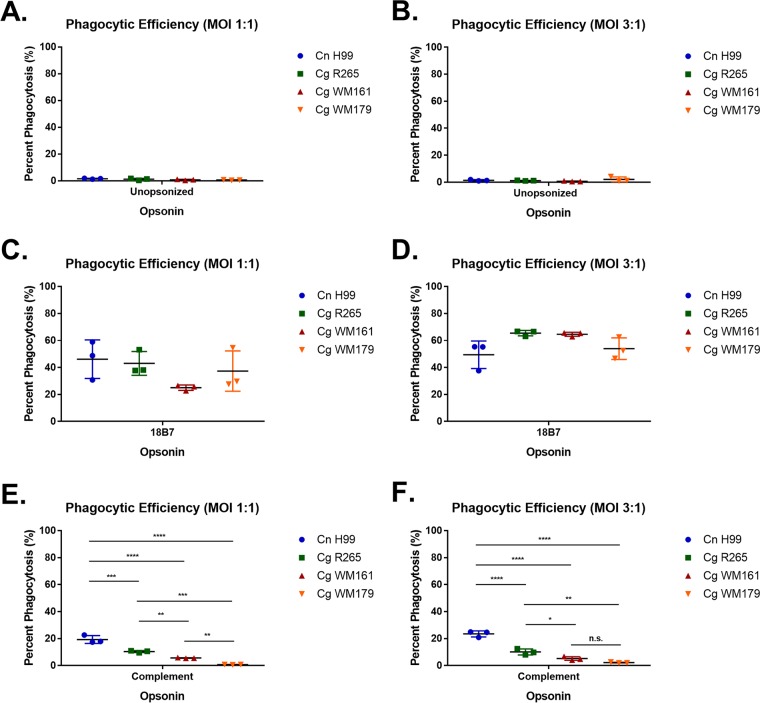
First we compared the efficiency of phagocytosis of C. neoformans and C. gattii species complex strains, as the capsule structure has been shown to be different enough to potentially affect the binding of antibody and influence the activation of complement or its deposition (19). The efficiency of phagocytosis was greatest for all strains when opsonized with monoclonal antibody (MAb) 18B7, a monoclonal antibody against the GXM of C. neoformans (24), although complement could also trigger significant phagocytosis. We noted no major differences in strain phagocytosis when opsonized with the 18B7 antibody, but we measured significant differences when opsonized with complement, such that C. neoformans H99 was ingested at a higher rate than any of the C. gattii strains (Fig. 1).
FIG 1.
Phagocytic efficiency of BMDM when ingesting Cryptococcus strains. BMDM were infected with various cryptococcal strains at an MOI of 1:1 or 3:1 without opsonin (A and B), MAb 18B7 (C and D), or guinea pig complement (E and F) for 1.5 h. Shown is the mean with standard deviation for triplicates under each condition. Cn, C. neoformans; Cg, C. gattii. Statistics were calculated using one-way ANOVA, which compared every column with every other column. ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; n.s., not significant (P > 0.05).
Phagolysosomal acidification.
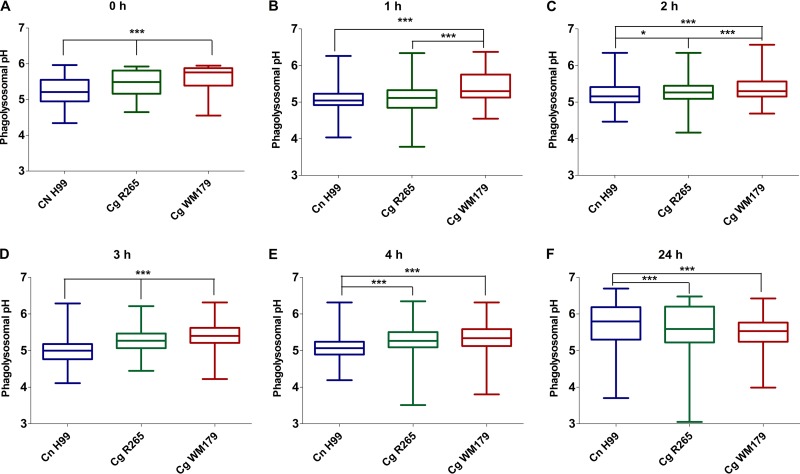
The phagolysosomal pH in bone marrow-derived macrophages (BMDM) was measured at 0, 1, 2, 3, 4, and 24 h postinfection (Fig. 2A to F). At about 10 min after phagocytosis (time zero), C. neoformans H99-infected BMDM had a lower mean phagolysosomal pH (5.2 ± 0.4) than BMDM infected with either of the C. gattii strains (Fig. 2A) (P < 0.001). At both 1 and 2 h postinfection, C. gattii WM179-infected BMDM had a higher phagolysosomal pH than BMDM infected with any of the other tested strains (Fig. 2B and C). At 3 h postinfection, there was a significant difference in the pH between BMDM infected with the three Cryptococcus strains (P < 0.001), and C. gattii WM179-infected BMDM had the most basic phagolysosomal pH of 5.4 ± 0.4 (Fig. 2D). At 4 h postinfection, both C. gattii R265- and C. gattii WM179-infected BMDM had a similar phagolysosomal pH of 5.3 ± 0.4, while the C. neoformans H99-infected BMDM pH was more acidic at 5.1 ± 0.4 (Fig. 2E). At 24 h postinfection, the phagolysosomal pH of C. neoformans H99-infected BMDM increased to 5.7 ± 0.6, which was less acidic than both C. gattii R265- and C. gattii WM179-infected BMDM (Fig. 2F). Overall, C. neoformans H99 had a more acidic phagosome than the C. gattii strains tested up to the 24-h time point, when the phagosomal pH became less acidic than the phagosomal pH for the C. gattii strains.
FIG 2.
Phagolysosomal pH in infected BMDM. (A to F) The phagolysosomal pH in BMDM infected with cryptococcal strains at 0 to 24 h postinfection (p.i.) was determined using dual-ratio fluorescence microscopy. The pH remained essentially the same over the time intervals within and among strains, as well as between experiments. Three biological replicates were carried out for each strain and time point, with at least cells 5 cells/field and 7 to 10 fields being imaged in each replicate. Shown are data pooled from three independent experiments. Data are graphed as means and upper and lower quartiles with minimum and maximum value tails. P values were determined using one-way ANOVA and Tukey's test for multiple comparisons. ***, P < 0.001; *, P < 0.05.
Apoptosis in C. neoformans- and C. gattii-infected bone marrow-derived macrophages.
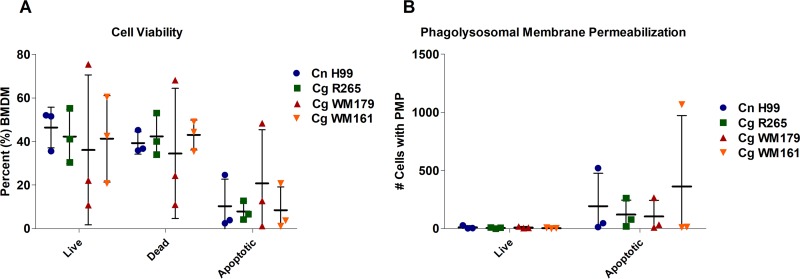
Lysosomal membrane damage is associated with C. neoformans virulence within the macrophage (25). We first determined the percentage of infected live, dead, and apoptotic macrophage populations over a 24-h period (Fig. 3). No statistically significant differences in the capacity to cause death or apoptosis were found among all Cryptococcus strains tested, based on SYTOX viability dye exclusion and membrane potential, as measured by use of the ratiometric dye F2N12S. We next sought to determine whether there were differences in phagolysosomal membrane permeabilization (PMP) between C. neoformans- and C. gattii-infected macrophages. The dye LysoTracker Deep Red accumulates in phagolysosomes, but permeabilized phagosomes cannot accumulate this dye (Fig. 3B). All strains displayed the capacity to cause PMP in host macrophages, and we could not measure differences in their capacity to cause PMP.
FIG 3.
Cell viability and PMP in C. neoformans- and C. gattii-infected BMDM. (A) The percentages of populations of live, dead, and apoptotic infected BMDM were determined. (B) Number of cells that have phagolysosomal membrane permeabilization, determined by selecting the LysoTracker Deep Red-negative population of infected cells. Three independent biological replicates were performed for each experiment (shown). No statistically significant difference was determined (two-way ANOVA).
Cryptococcal capsule size after infection.
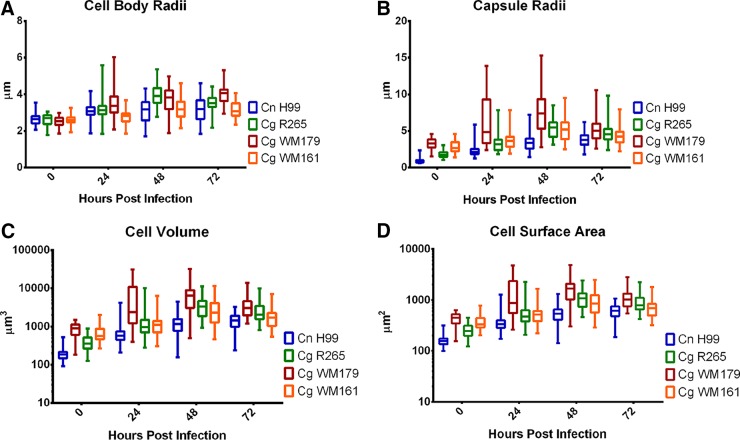
Next, we examined whether the capsule size of the Cryptococcus strains differed after infection, since we have noted that a larger capsule size may lead to more membrane damage (unpublished). We measured the cell body, capsule radius, cell volume, and cell surface area of the cryptococcal strains at 0, 24, 48, and 72 h postinfection (Fig. 4). Across each time interval, no statistically significant difference in the total surface area of the cells was observed between strains (Fig. 4A). However, at both 24 and 72 h, C. gattii WM179 had the largest average cell body radius of 3.6 μm and 4.0 μm, respectively (P < 0.0001) (P values are described in Table S1 in the supplemental material). The average capsule radius was consistently larger for C. gattii WM179 than for the other strains at each time point at 7.4 μm (Fig. 4B). These observations were consistent with those of previous work that showed that VGI molecular type strains have larger capsules (26).
FIG 4.
Changes in cell body and capsule radii, volume, and surface area of cryptococcal strains after infection. C. neoformans H99, C. gattii R265, C. gattii WM179, and C. gattii WM161 cells were analyzed via bright-field microscopy at 0, 24, 48, and 72 h postinfection. Cells were visualized by bright-field microscopy of India ink-stained slides. The capsule was identified as the area of exclusion of India ink. Measurements were taken of cryptococcal cell body radii (A) and capsule radii (B), from which cell volumes (C) and cell surface areas (D) were calculated. Data were pooled from three independent experiments, performed in triplicate. Data are graphed as means and upper and lower quartiles with minimum and maximum value tails. See Tables S1 to S4 in the supplemental material for the results of two-way ANOVA and the number of experiments for each group.
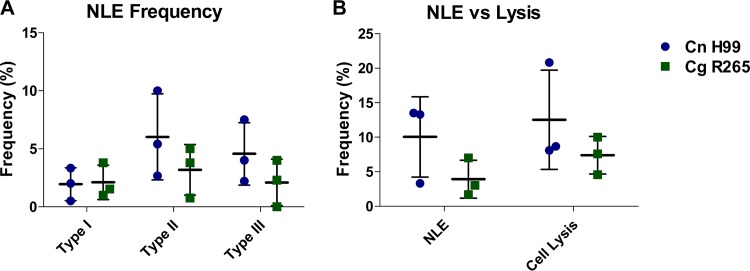
NLE in C. neoformans- and C. gattii-infected macrophages.
Nonlytic exocytosis (NLE) events have been reported in C. neoformans-infected macrophages and have been hypothesized to assist in dissemination to the central nervous system (CNS) via a Trojan horse mechanism (27). C. gattii has been shown to be less neurotropic than C. neoformans strains (16). Thus, we hypothesized that there would be a lower frequency of NLE events associated with C. gattii infections of murine macrophages. We found no statistically significant differences between strains (Fig. 5).
FIG 5.
Nonlytic exocytosis (NLE) frequency in C. neoformans- and C. gattii-infected BMDM over a 24-h period. (A) Type I (complete exocytosis), type II (partial exocytosis), and type III (cell-to-cell transfer) frequencies in BMDM infected with C. neoformans H99 and C. gattii R265. (B) Total frequency of NLE compared to cell lysis frequency in BMDM infected with C. neoformans H99 and C. gattii R265. Three biological replicates (movies) for each strain were performed. Bars represent the standard deviation. No statistically significant difference was determined (two-way ANOVA).
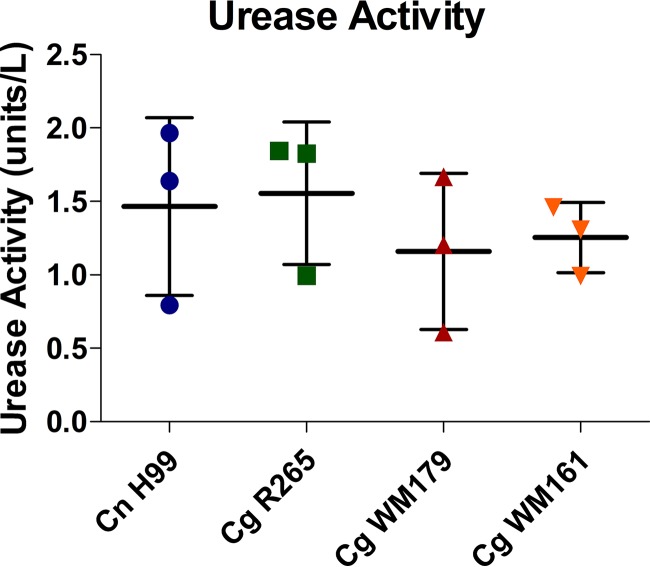
C. neoformans and C. gattii urease activity.
Urease is considered an important virulence factor in Cryptococcus infection that has the potential to affect the pH of phagolysosomes (21). In particular, urease-deficient strains have reduced intracellular proliferation within macrophages, suggesting an important role for this enzyme (21). Urease activity ex vivo was found to have no statistically significant difference among the strains (Fig. 6).
FIG 6.
Urease activity ex vivo following 2 h of incubation with urea. Activity was determined using the urease activity assay (Sigma-Aldrich). Three independent experiments with four replicate wells per strain were performed. Shown are the average and standard deviation from all three experiments. No significant difference was found (Tukey's test).
Macrophage transcriptional response.
Using gene expression microarrays, we compared the transcriptional responses of C. neoformans H99-infected, C. gattii R265-infected, and uninfected BMDM. We selected 10 genes for which the responses were validated by quantitative PCR (qPCR) (Table S2), and the validation data supported the results from the microarray analyses. Genes with significant differences in the levels of expression in the uninfected controls between 24 h and 2 h, referred to as time-only genes (1,435 genes), were removed from the filtered gene lists prior to comparison between infection groups and time points. These time-only genes constituted about 50% of the total gene response for infected versus uninfected samples (Fig. S1 and Table S1). This significant genetic response derived from the collection/culture times for uninfected macrophages is well documented (28).
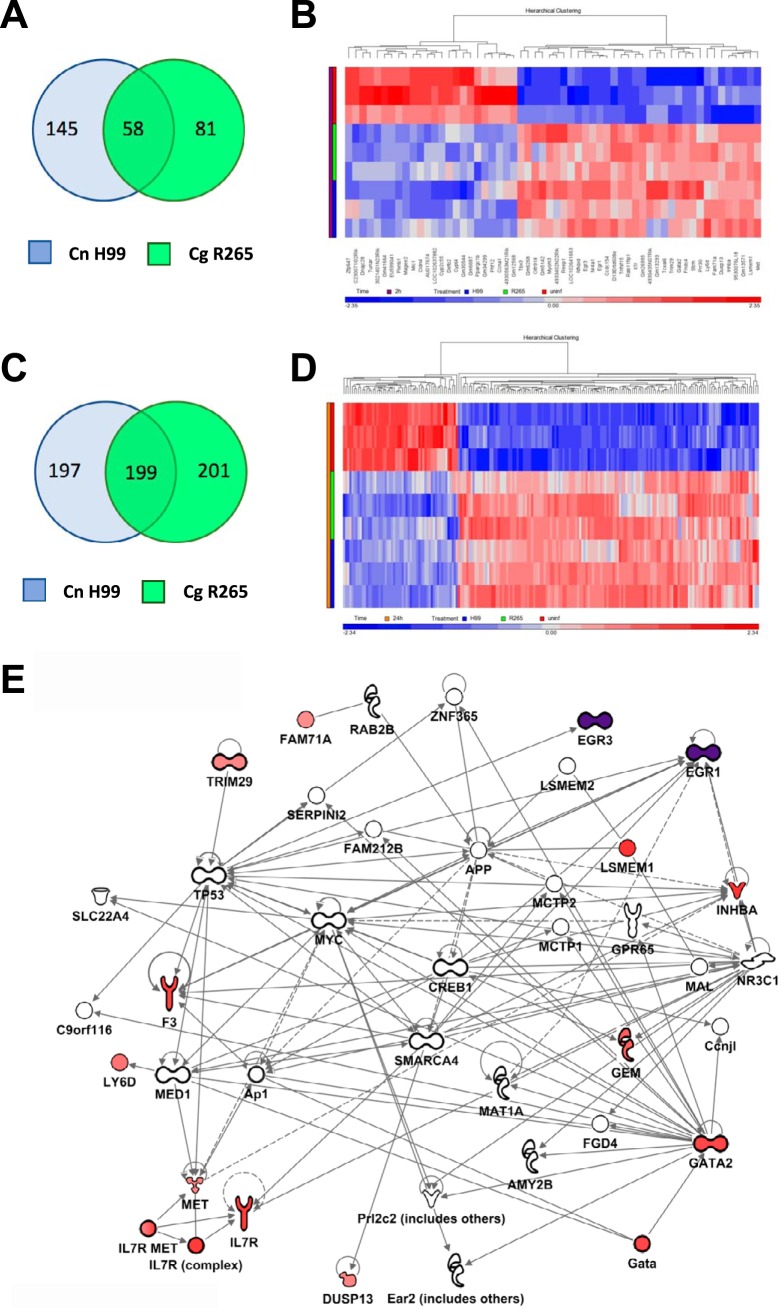
Analysis of genes regulated differently from their regulation in the control at 2 h of infection showed that C. neoformans H99- and C. gattii R265-infected cells respond with transcriptional changes in 203 and 139 genes, respectively, of which 58 genes were shared (Fig. 7). At 24 h, C. neoformans H99- and C. gattii R265-infected cells responded with transcriptional changes in 396 and 400 genes, respectively, of which 199 genes were shared, leading us to initially posit that 30 to 50% of the genetic response of macrophages to infection by the two cryptococcal strains is similar. However, when we analyzed the genes uniquely regulated by each of the two species representatives, we found no instance of genes being regulated in opposite directions for each strain. Analysis of the clustering maps (Fig. S1) showed a stronger clustering by infection time than by strain, suggesting that the host response caused by infection with the two strains is indeed remarkably similar, which implies that the macrophage responses are very similar. This conclusion would also imply that investigators attempting to detect genetic response differences in hosts would need a very robust and sensitive method, as the differences in the responses caused by infection with the different strains in this model are probably quite small.
FIG 7.
Genes differentially regulated in BMDM upon C. neoformans H99 and C. gattii R265 infection. (A) Venn diagram showing genes differentially regulated at 2 h compared with the regulation of genes from uninfected samples. (B) Heatmap of genes in common between both strains at 2 h. (C) Venn diagram showing genes differentially regulated at 24 h compared with the regulation of genes from uninfected samples. (D) Heatmap of genes in common between both strains at 24 h. The color scale in the heatmaps corresponds to Z-scores of standardized intensity values for each sample. (E) Network analysis of the genes upregulated under all 4 conditions with the addition of Egr1 and Egr3, which were the top upregulated genes at 2 h. Red shading denotes the intensity of upregulation at 24 h. Purple shading denotes the intensity of upregulation at 2 h. The experiment was performed in BMDM derived from three mice across all conditions.
For both C. neoformans H99- and C. gattii R265-infected cells, the transcriptional profile was different at 2 and 24 h, with only 13 genes being affected under all 4 conditions (strain and time) (Table 1). Using Ingenuity Pathways Analysis software, we built a network comprising these 13 genes together with the most strongly upregulated genes in 2 h and 24 h to decipher some of the pathways responsive to cryptococcal infection (Fig. 7E). Some of these genes are transcription factors (Gata2, Egr1, Egr3, Inhba) and have been associated with monocyte/macrophage growth and differentiation (29–31). The IL-7 receptor (IL-7R) network was also affected. The role of IL-7R in the monocyte/macrophage response is not well defined, but its importance for the development of T cells (as well as B cells for mice but not for humans) is underscored by the fact that the genetic deficiency causes SCID-type syndromes (32). Trim29 is associated with alterations in susceptibility to pneumonia of viral and bacterial etiologies (33). The Met gene has been associated with Stat3 activation, but the functions of Met, Dusp13, Lsmem, and Fam71a are not clear at this time and will be the subject of future work (34).
TABLE 1.
Genes upregulated at both time points (2 h and 24 h) in both C. neoformans H99 and C. gattii R265
| Gene name | Gene or protein encoded by gene | Fold change in expression ata: |
|||
|---|---|---|---|---|---|
| 2 h |
24 h |
||||
| C. neoformans H99 | C. gattii R265 | C. neoformans H99 | C. gattii R265 | ||
| Gm26995 | Predicted gene, 26995 | 2.155 | 2.385 | 3.150 | 2.883 |
| 9530076L18 | Uncharacterized protein 9530076L18 | 3.433 | 3.383 | 18.279 | 9.576 |
| Il7r | Interleukin 7 receptor | 3.446 | 3.550 | 3.684 | 3.413 |
| Lsmem1 | Leucine-rich single-pass membrane protein 1 | 3.643 | 3.718 | 9.998 | 9.391 |
| Inhba | Inhibin beta-A | 3.543 | 3.334 | 16.095 | 9.907 |
| Ly6d | Lymphocyte antigen 6 complex, locus D | 2.497 | 2.216 | 2.337 | 3.402 |
| D13Ertd608e | DNA segment, chromosome 13, ERATO Doi 608, expressed | 3.393 | 4.042 | 3.474 | 2.614 |
| Gata2 | GATA binding protein 2 | 3.238 | 2.754 | 4.082 | 3.757 |
| Dusp13 | Dual-specificity phosphatase 13 | 2.208 | 2.060 | 6.100 | 5.908 |
| Trim29 | Tripartite motif-containing 29 | 2.161 | 2.675 | 2.310 | 2.819 |
| Fam71a | Family with sequence similarity 71, member A | 2.047 | 2.127 | 9.913 | 2.927 |
| Met | Met proto-oncogene | 2.159 | 2.111 | 2.985 | 2.835 |
| Gm13571 | Predicted gene, 13571 | 2.853 | 2.423 | 4.772 | 4.480 |
The fold changes in expression by infected versus uninfected cells at 2 h and 24 h are shown.
DISCUSSION
The C. neoformans and C. gattii species diverged approximately 80 million to 100 million years ago, roughly correlating with the time of separation of Africa and South America, where these species originated, respectively (2, 35–38). Despite these species' long period of separation and geographical distribution, they are remarkably similar, such that they were considered the same species for over a century. For example, they share core virulence factors, such as a capsule, melanin, urease, and the ability to grow at 37°C (9, 10). These characteristics are not shared by other closely related cryptococcal species, which suggests conservation from a Pangean ancestor (2, 39). Despite causing similar disease in humans and animals, there appear to be quantitative differences between the two species complexes with regard to the immune response that they elicit. In general, C. gattii species complex strains induce a less protective immune response in immunocompetent hosts that is associated with structural differences in the capsular polysaccharide and that reduces DC activation (18, 20).
Given the conservation of virulence factors, the clinical similarities in the cryptococcal diseases caused by strains of the two species complexes, and the fact that Cryptococcus is an intracellular pathogen of alveolar macrophages, we were interested in examining the intracellular survival strategy of C. gattii within macrophages (40). Both C. gattii and C. neoformans required opsonization by either complement or antibodies, as the capsule of both cryptococcal species complexes is inherently antiphagocytic (12). Despite significant structural differences in the capsular polysaccharide, we observed very little difference in the efficiency of phagocytosis after antibody opsonization but significant differences in complement-dependent opsonization (18, 20). IgG is the preferred opsonin because it so efficient, with the caveat that its high efficiency may mask differences in the pathogen surface molecule. Complement manifests great interstrain variability in opsonizing efficiency, depending on where C3 is deposited (19), and we hypothesize that this is related to antigenic differences in the capsule of each of these strains. A detailed study of the surface molecules exposed by C. gattii versus C. neoformans and of the phagocytic efficiency could shed some light on this conundrum.
After macrophage ingestion, cells of both C. neoformans and C. gattii species complexes reside in mature phagolysosomes of macrophages (40, 41). Measurement of the phagolysosomal pH after the ingestion of cells of both species complexes revealed that all phagosomes acidified, with the pH remaining relatively constant over 4 h. At 24 h postinfection, the phagolysosomal pH was higher in phagolysosomes containing C. neoformans H99 cells than in those containing cells of either strain of C. gattii (25), but we observed no measurable differences in the PMP of infected macrophages or host macrophage death. C. gattii WM179, a VGI molecular type, has a larger capsule than all other strains before infection and after 24, 48, and 72 h postinfection. Therefore, it seems that in the first 24 h of interaction with murine macrophages, C. neoformans and C. gattii do not display striking differences in the capacity to damage host cells.
C. neoformans-infected macrophages can expunge fungal cells, leaving both the macrophage and the fungus intact (42). This mechanism is believed to assist in dissemination of the organism to the central nervous system (CNS) in vivo (43). When comparing the frequency of nonlytic exocytosis for C. gattii R265-infected macrophages compared with that for C. neoformans H99-infected macrophages, we were unable to demonstrate a statistically significant difference. C. gattii species complex VGIIa strains establish disease primarily in the lungs and rarely disseminate. C. neoformans species complex strains predominately result in severe CNS disease (10, 16). In vitro studies have shown that C. neoformans species complex strains transmigrate at greater frequencies than C. gattii species complex strains (27).
Urease is important for hydrolyzing urea to produce ammonia and CO2 and is found in many organisms, including bacteria and plants (21). The presence of urease activity within microorganisms is associated with virulence and increased pathogenesis (21). Urease is a major virulence factor of C. neoformans, is thought to play a role in phagolysosome survival, and has been shown to have a role in aiding transmigration through the blood-brain barrier (BBB) (21, 44). Because urease is important for migration across the BBB and C. gattii strains are less prone to cause meningoencephalitis, we reasoned that urease activity may contribute to differential transmigration. We found similar urease activity among all strains examined. We note that the assay used in our study to measure urease activity is extremely sensitive, such that differences in culture age or incubation period produced vastly different results. Overall, factors other than urease activity may explain the preferential infection site of C. gattii.
Transcriptional responses had a high degree of overlap for macrophages infected with C. neoformans H99 or C. gattii R265. Our results suggest that some genes could be novel regulators in defense against Cryptococcus spp. Gata2 is a major immune regulator whose absence is associated with severe immunodeficiency, including susceptibility to fungal infections (45). Egr1 and Egr3, coding for hematopoietic cell growth factors, were also implicated in the response to fungal disease (46). Stat1 and Ifng are major regulators of the monocyte/macrophage response to Cryptococcus, but we did not find Stat1 or Ifng expression to be modulated in our model (47–50). However, Gata2 and Egr1 are predicted to interact with or are found to be coexpressed with Stat1 and Ifng by data-mining algorithms (String, v10.5). Future work will dissect and validate the importance of these regulators in fungal disease. Critically, we did not find major differences in host genetic responses to either strain of Cryptococcus spp., implicating that C. neoformans and C. gattii are largely undistinguishable to the host macrophage and perhaps that macrophage susceptibility to both pathogens is similar. Consistent with the notion that the intracellular pathogenic strategy is globally conserved, though differing in details, is the recent report that both C. neoformans and C. gattii damage DNA in human peripheral monocytes, a process that was associated with nitric oxide (NO) in C. gattii but not C. neoformans (51). In mouse models, C. gattii R265 has a preference for causing lung infections over brain infections, mimicking the human disease pattern (16). However, comparison between these two strains does not allow definitive conclusions regarding species differences. Further studies involving multiple strains of each species are needed to extend these observations from the strain to the species level. To this point, we do not know how the C. gattii disease prevalence and impact would compare with the C. neoformans disease prevalence and impact if both species had the same geographical distribution. As this work was performed in murine macrophages, further work should seek to establish if the results are applicable to other mouse models or primary human macrophages. However, it is noteworthy that most facets of cryptococcal infection in murine macrophages that have been investigated in human cells are conserved across both species (52).
Both cryptococcal species complexes have been isolated from soil and trees, where the microbial communities also include amoebae (53–56). On the basis of similarities between the intracellular pathogenic strategy of C. neoformans in mammalian macrophages and amoebae, we proposed that the virulence originated by selection pressures in the environment independent of the vertebrate host (57, 58). The similarities between C. neoformans and C. gattii macrophage intracellular strategies suggest that both experienced parallel selection pressures after their divergence into separate species. Given that the fungal and amoeba lineages predate the emergence of metazoans, that phagocytosis is an ancient mechanism of food acquisition, and that amoebae and cryptococcal yeasts are likely to have interacted since distant geological epochs by virtue of occupying the same ecological niches, the similarities in C. neoformans and C. gattii intracellular strategies suggest that these interactions occurred as early as in the common Pangean ancestor and thus represent an ancient solution to the challenge of survival from amoeboid host predation (59, 60). Hence, the finding that C. neoformans and C. gattii intracellular pathogenic strategies are similar is consistent with and supportive of the theory that cryptococcal virulence emerged from survival strategies selected by amoeboid predators and accidentally proved suitable for survival in metazoan hosts (61).
Our results are also relevant to the current taxonomy controversy involving the cryptococcal field (1). After decades of nomenclature stability, genome sequencing of many strains has led to the proposal that each of the C. neoformans and C. gattii species complexes be split into several distinct species (36). In response, some have argued that the overwhelming similarities in clinical presentation and biology override the genetic difference and instead suggest the species complex nomenclature used in this paper (1). Our results provide fodder for both camps. On one hand, the conservation of the intracellular pathogenic strategy supports the wider species complex view, encompassing C. neoformans and C. gattii as causative agents of the same disease. On the other hand, outbreaks such as the Vancouver Island outbreak may be caused by a particularly virulent isolate of one species or a series of serendipitous events of an unusual high exposure dose.
In summary, our study reveals that despite ancient separation, the interaction of cells from both cryptococcal species complexes with murine macrophages was qualitatively similar. Hence, the overall intracellular pathogenic strategy of both cryptococcal species complexes appears to be conserved. Conservation of intracellular strategies among species with such temporally distant separation implies that these evolved strategies are ancient and possibly maintained by common continuous and extant selection pressures.
MATERIALS AND METHODS
Fungal strains.
Cryptococcus gattii species complex strains R265 (VGIIa), WM179 (VGI), and WM161 (VGIII) were obtained from ATCC (Manassas, VA), and Cryptococcus neoformans species complex strain H99 (VNI) was originally obtained from John Perfect (Durham, NC). The strains were stored at −80°C. Frozen stocks were streaked onto Sabouraud dextrose agar (SDA) and incubated at 30°C. Liquid suspensions of cryptococcal cultures were grown in SDA overnight at 30°C.
Mice.
All experiments used C57BL/6 female mice 8 to 16 weeks of age obtained from the Jackson Laboratory. All animal procedures were performed with prior approval from Johns Hopkins University (JHU) Animal Care and Use Committee (IACUC), under approved protocol number M015H134. Mice were handled and euthanized with CO2 in an appropriate chamber, followed by thoracotomy as a secondary means of death in accordance with guidelines on Euthanasia of the American Veterinary Medical Association. JHU is accredited by AAALAC International, in compliance with Animal Welfare Act regulations and Public Health Service (PHS) Policy, and has a PHS Approved Animal Welfare Assurance with the NIH Office of Laboratory Animal Welfare. JHU Animal Welfare Assurance Number is D16-00173 (A3272-01). JHU utilizes the United States Government laws and policies for the utilization and care of vertebrate animals used in testing and research and follows training guidelines for appropriate animal use in a research and teaching setting.
Macrophages.
Primary macrophages were obtained via isolation of bone marrow progenitors from C57BL/6 mice. Mice were euthanized, the skin of each leg was peeled back, and muscle was cut away to reveal the bone. An L929 cell supernatant was obtained by collecting supernatants from L929 cells at confluence. Once the skin and muscle were removed, the tibia and femur were removed and placed in Dulbecco modified Eagle medium (DMEM) feeding medium (20% L929 cell supernatant, 10% fetal bovine serum [FBS], 0.1% β-mercaptoethanol, 1% penicillin-streptomycin [Pen-Strep], 1% minimal essential medium nonessential amino acids [MEM-NEA], 1% GlutaMAX, 1% HEPES buffer). The marrow was then flushed out with feeding medium, filtered through 100-μm-pore-size filters to remove aggregates or bone debris, and centrifuged at 350 × g for 5 min at 4°C. The supernatant was then discarded and the pellet was suspended in 120 ml of feeding medium. The resuspended solution was seeded onto 100-mm tissue culture-treated petri dishes (Corning) and incubated at 37°C for 6 days.
J774.16 (J774) murine macrophage-like cells from frozen stocks were plated onto 6-well plates containing DMEM complete medium (10% FBS, 10% medium NCTC-109 (Gibco, Gaithersburg, MD), 1% Pen-Strep, 1% MEM-NEA).
Phagocytic efficiency.
BMDM were seeded onto the wells of a 96-well plate containing BMDM feeding medium at a density of 1 × 105/ml. The BMDM were incubated overnight at 37°C with gamma interferon (IFN-γ; Bioscience) and lipopolysaccharide (LPS; Bioscience) at 100 U/ml and 0.5 μg/ml, respectively. Cryptococcal cultures for infection were opsonized with either MAb 18B7, which binds capsular glucuronoxylomannan, at a concentration of 10 μg/ml (24) or 20% guinea pig complement (MP Biomedical). BMDM were infected at a multiplicity of infection (MOI) of 1:1 or 3:1 (cryptococcal cells/BMDM). BMDM were incubated for 1.5 h at 37°C before imaging.
Measurement of phagolysosomal pH.
Approximately 1.25 × 105 BMDM were seeded onto the wells of a 24-well tissue culture plate containing 12-mm glass coverslips and DMEM feeding medium. Macrophages were incubated at 37°C and activated overnight with LPS and IFN-γ at 0.5 μg/ml and 100 U/ml, respectively. Macrophages were infected at an MOI of 3:1 (about 3.75 × 105 fungal cells/well) with cryptococcal strains opsonized with MAb 18B7 conjugated to Oregon Green (Molecular Probes) at a concentration of 10 μg/ml (62). The plate was immediately spun down at 350 × g for 1 min to facilitate contact with the monolayer. Infected macrophages were then incubated at 37°C for 10 min to initialize phagocytosis. For each strain of the two Cryptococcus species complexes, a standard curve was generated to determine the pH at various time points after phagocytosis was initiated. Calibration buffers were made for pH 3.0 to pH 7.0 at increments of 0.5 pH unit. Buffers were composed of 140 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM glucose. For solutions with a pH of less than 5.0, acetic acid was added to the mixture to adjust the pH. For solutions with a pH of between 5.5 and 6.5, 2-(N-morpholino)ethanesulfonic acid (MES) was used. For solutions with a pH of greater than 7.0, HEPES was used as the buffer. The pH was then adjusted to the desired values using either 1 M KOH or 1 M HCl.
For the standard curve, wells were incubated and washed with the desired pH calibration buffer for 5 min at 37°C. Nigericin (stock concentration at 5 mg/ml) was added at a final concentration of 5 μg/ml to allow the intracellular pH to be dictated by the extracellular pH. The 12-mm coverslips were then removed and placed culture side down on a 35-mm culture dish (MatTek), and images were obtained on the fluorescein isothiocyanate channel (440 nm and 488 nm) using a QImaging Retiga 1300 camera attached to an Olympus AX70 microscope. For the experimental samples, coverslips were placed in Hanks buffered saline solution (HBSS) medium and imaged on the Olympus AX70 microscope (Olympus). Imaging was done through the use of MetaFluor fluorescence ratio imaging software (Molecular Devices) for Olympus. Analysis of the images was also performed on MetaFluor software, and the pH was determined according to the fluorescence ratio of 440 nm/488 nm and comparisons with the pH standard curve.
Measurement of phagolysosomal membrane permeabilization.
BMDM or J774 cells were seeded into a 6-well plate at approximately 1 × 106 cells per 2 ml of feeding medium and activated overnight with LPS and IFN-γ (0.5 μg/ml and 100 U/ml, respectively). On the following day, cells were infected with Cryptococcus stained with Uvitex 2B stain (which binds to the cell wall of fungi) at an MOI of 1:2 for a period of 24 h. Cultures were opsonized with MAb 18B7 at a concentration of 10 μg/ml. One hour before the incubation period, LysoTracker Deep Red was added to each sample well and the single-stain control. Once the 24 h of the incubation period was finished, supernatant was collected and transferred to a corresponding 15-ml Falcon tube. Each well was washed with Hanks buffered saline solution (HBSS; Gibco), and the cells were collected and transferred to the corresponding Falcon tube. After the wash, 1 ml of Cellstripper solution (Corning) was added to each well and the plate was incubated at 37°C for 5 min. Cells were then collected and centrifuged for 5 min at 350 × g. After centrifugation, the supernatant was discarded and the pellet was suspended in HBSS, stained with CD11b-phycoerythrin (PE; clone M1/70; eBioscience), and incubated for 5 min, followed by centrifugation for 5 min at 350 × g. The supernatant was discarded, and the pellet was suspended in HBSS and placed on ice. F2N12S and SYTOX-PE-Cy5.5 AADvanced dead cell stain (Thermo Fisher Scientific) was added 5 min before analysis with a Becton Dickinson LSRII flow cytometer (BD Biosciences). The data were further analyzed with FlowJo software (FlowJo Inc., a subsidiary of BD Biosciences).
Phagosomal capsule growth measurement.
BMDM were seeded in 24-well plates at a density of 2.5 × 104 cells per well. BMDM were activated for 16 h with IFN-γ and LPS at concentrations of 100 U/ml and 0.5 μg/ml, respectively. After activation, the BMDM were infected for 1 h with cultures of Cryptococcus at an MOI of 1 and opsonized with 10 μg/ml MAb 18B7. After infection, the wells were washed three times with BMDM feeding medium. At each time point, extracellular Cryptococcus was removed by washing each well three times with BMDM feeding medium. The BMDM were then lysed by replacing the medium with 200 μl distilled H2O, incubated for 10 min, and then pipetted to ensure lysis. The released intracellular Cryptococcus cells were then pelleted by centrifugation at 15,600 × g for 5 min. The pellet was resuspended in 15 μl phosphate-buffered saline (PBS), 10 μl of which was stained with India ink and imaged at a ×100 magnification under a bright-field microscope. Cryptococcus capsule and cell body measurements were manually assessed using ImageJ and Fiji software according to India ink exclusion (63).
Measurement of nonlytic exocytosis.
BMDM were seeded at approximately 5 × 104 cells onto 35-mm MatTek culture dishes (Corning) containing BMDM feeding medium and incubated at 37°C for 2 h to induce adherence to the plate. Following the 2 h of incubation, macrophages were then activated overnight using LPS and IFN-γ at a concentration of 0.5 μg/ml and 100 U/ml, respectively. A Cryptococcus suspension was prepared, and MAb 18B7 was added to the suspension at a concentration of 10 μg/ml to enhance phagocytosis. The Cryptococcus strains were added at an MOI of 3:1 (approximately 1.5 × 105 cells). The infected macrophages were then incubated at 37°C for 2 h and then washed to remove extracellular cryptococcal cells. Time-lapse images were taken every 4 min for a period of 24 h at 37°C with 10% CO2 at a ×10 magnification on the Axiovert 200M microscope (Zeiss). Time-lapse movies were analyzed using Axio Vision software (Zeiss), and the number of cells exhibiting nonlytic exocytosis was recorded by operator visual analysis of the movie.
Urease activity assay.
Cryptococcus cultures were grown in liquid SDA for 17 h. Cultures were washed, and the cells were counted and diluted to a concentration of approximately 1.11 × 107 cells/ml. Approximately 1 × 106 cells were added onto a 96-well plate. To measure urease activity, a urease activity assay (catalog number MAK120; Sigma-Aldrich) was used and the manufacturer's protocol was followed. Briefly, an ammonium standard that was at a concentration of 500 μM was prepared and serially diluted. Then, 90 μl of the sample or the ammonium standard was added to the wells of a 96-well plate. Next, 10 μl of urea was added and the plate was incubated for 2 h at 30°C. Reagent A (from urease activity assay kit, catalog no. MAK120; Sigma-Aldrich) was added to stop the urease reaction. Then, reagent B (from urease activity assay kit, catalog no. MAK120; Sigma-Aldrich) was added to develop the plate and the plate was incubated at room temperature for 30 min. The plate was then read on an EMax Plus microplate reader (Molecular Devices) at a wavelength of 650 nm. Urease activity was determined by the following equation: urease activity = [(Asample − Ablank) · n]/(t · s), where Asample and Ablank are the absorbance of the sample and the blank, respectively; n is the dilution factor; t is the incubation time (in minutes); and s is the slope of the standard curve.
Statistics.
All statistical measurements for the above-described methods were made using the statistical software in Prism software. Unless otherwise noted, a one-way analysis of variance (ANOVA) and Tukey's multiple-comparison test were used to compare all groups.
Macrophage mRNA gene expression array.
BMDM were washed five times with 1× PBS, removed with a cell scraper, and centrifuged at 350 × g for 5 min. The cell pellet was then suspended in TRIzol reagent and gently broken up, and the debris was removed by centrifugation at 8,000 × g. The supernatant was then flash frozen and stored at −80°C. Total RNA was extracted using a PureLink RNA minikit (Ambion/Life Technologies) according to the manufacturer's protocol, including on-column DNase treatment. Following elution of purified RNA from the PureLink columns with nuclease-free water, quantitation was performed using a NanoDrop spectrophotometer, and quality assessment was determined by RNA LabChip analysis on an Agilent Bioanalyzer 2100 system or with RNA Screen tape on an Agilent TapeStation 2200 system. One hundred nanograms of total RNA was processed for hybridization to Agilent SurePrint G3 mouse (v2) 8x60K gene expression arrays according to Agilent's one-color microarray-based analysis (low-input QuickAmp labeling) protocol, including cDNA synthesis, cRNA synthesis with Cy3 labeling and purification, fragmentation, hybridization, and posthybridization washing. Spike-In controls were utilized and processed according to Agilent's one-color RNA Spike-In kit protocol. The arrays were scanned in an Agilent G2600D SureScan microarray scanner using scan protocol AgilentG3_GX_1color. Agilent's Feature Extraction software was used to assign grids, provide raw image files per array, and generate quality control (QC) metric reports from the microarray scan data. The QC metric reports were used for quality assessment of all hybridizations and scans. Txt files from the Feature Extraction software were imported into the Partek Genomics Suite (v6.6; Partek) for detailed analyses of gene expression. Within Partek, the gene expression work flow was followed for Agilent Feature Extraction files, including import of gProcessedSignal, quantile normalization, and log transformation base 2.0. For genes for which multiple probes were present, a summarization of probes was performed (merge probes by gene symbol with the mean of the intensity values). The batch effect removal tool (analysis of variance [ANOVA]) was used to correct for effects of culture batch and array (slide). A two-way ANOVA with linear contrasts for treatment (infection/strain) and time (2 h or 24 h) versus the control (uninfected cells) was performed with outputs of P value, fold change, and mean ratio. A linear contrast of uninfected controls (uninfected cells at 24 h versus uninfected cells at 2 h) was also performed to generate a time-only gene list. The cutoff criteria for filtering gene lists were significant P values (P < 0.01) with fold changes of greater than 2 or less than −2. Heatmaps were generated from the filtered gene lists, as well as lists from Venn diagram comparisons. The data derived from Partek analysis were imported to Ingenuity Pathways Analysis software (Qiagen) to identify molecular networks of relevance and the pathways and biological processes that were most significantly related in the data set.
Gene expression validation by qPCR.
Ten genes from the filtered gene lists were selected for validation by qPCR. Selection criteria included the greatest differential expression in infection groups (infection/strain and time) versus the controls, genes with functions relevant to phagosomal damage, and genes similar to those in previous data sets (64). Four housekeeping genes, those for ribosomal protein L13 (rpl13a), thyroid hormone receptor-interactor 6 (Trip6), β-actin, and glyceraldehyde-3-phosphate dehydrogenase (Gapdh), were run for normalization and comparison. Selection criteria for housekeeping genes included comparison of means and assessment of standard deviations across all ANOVA linear contrast groups, as well as transcript abundance. A SuperScript III First-Strand cDNA kit (Invitrogen/Thermo Fisher) was used to synthesize cDNA from 500 ng total RNA according to the manufacturer's recommended protocol. Assays on Demand TaqMan probe assays were run for all selected genes (ABI/Thermo Fisher) in 20-μl-volume reaction mixtures with 1 μl cDNA and the TaqMan Gene Expression master mix in a StepOne Plus instrument, utilizing standard protocols. StepOne Plus software (ABI/Thermo Fisher) was used to determine the threshold cycle (CT). Detailed analysis was performed in Partek Genomics Suite (v6.6). Gapdh was selected for normalization based on comparison of CT means and standard deviations for each housekeeping gene across groups. ANOVA, with linear contrasts, was used as described above to determine P values and fold changes in expression.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge Nina Grossman for her careful editing of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00946-17.
REFERENCES
- 1.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castañeda E, Chang YC, Chen J, Cogliati M, Dromer F, Ellis D, Filler SG, Fisher MC, Harrison TS, Holland SM, Kohno S, Kronstad JW, Lazera M, Levitz SM, Lionakis MS, May RC, Ngamskulrongroj P, Pappas PG, Perfect JR, Rickerts V, Sorrell TC, Walsh TJ, Williamson PR, Xu J, Zelazny AM, Casadevall A. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2:e00357-. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Freij JB, Hann-Soden C, Taylor J. 2017. Continental drift and speciation of the Cryptococcus neoformans and Cryptococcus gattii species complexes. mSphere 2:e00103-. doi: 10.1128/mSphere.00103-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelthaler DM, Meyer W. 2017. Furthering the continental drift speciation hypothesis in the pathogenic Cryptococcus species complexes. mSphere 2:e00241-. doi: 10.1128/mSphere.00241-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca A, Vilgalys R, Heitman J. 2009. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot Cell 8:353–361. doi: 10.1128/EC.00373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, MacDougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, Kronstad JW, Bartlett KH. 2007. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol 73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrnes EJ, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chatuverdi S, Chaturvedi V, Chatuverdi V, Heitman J. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog 7:e1002205. doi: 10.1371/journal.ppat.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 3099:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, Idnurm A, Bahn YS. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med 4:a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen SCA, Meyer W, Sorrell TC. 2014. Cryptococcus gattii infections. Clin Microbiol Rev 27:980–1024. doi: 10.1128/CMR.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA. 2009. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis 15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson JF, Johnston SA. 2015. Immunity to Cryptococcus neoformans and C. gattii during cryptococcosis. Fungal Genet Biol 78:76–86. doi: 10.1016/j.fgb.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng PY, Sham A, Kronstad JW. 2009. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun 77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leongson K, Cousineau-Côté V, Goupil M, Aumont F, Sénéchal S, Gaboury L, Jolicoeur P, Kronstad JW, de Repentigny L. 2013. Altered immune response differentially enhances susceptibility to Cryptococcus neoformans and Cryptococcus gattii infection in mice expressing the HIV-1 transgene. Infect Immun 81:1100–1113. doi: 10.1128/IAI.01339-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4:e5862. doi: 10.1371/journal.pone.0005862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ. 2012. The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3:e00103-. doi: 10.1128/mBio.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angkasekwinai P, Sringkarin N, Supasorn O, Fungkrajai M, Wang YH, Chayakulkeeree M, Ngamskulrungroj P, Angkasekwinai N, Pattanapanyasat K. 2014. Cryptococcus gattii infection dampens Th1 and Th17 responses by attenuating dendritic cell function and pulmonary chemokine expression in the immunocompetent hosts. Infect Immun 82:3880–3890. doi: 10.1128/IAI.01773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huston SM, Ngamskulrungroj P, Xiang RF, Ogbomo H, Stack D, Li SS, Timm-McCann M, Kyei SK, Oykhman P, Kwon-Chung KJ, Mody CH. 2016. Cryptococcus gattii capsule blocks surface recognition required for dendritic cell maturation independent of internalization and antigen processing. J Immunol 196:1259–1271. doi: 10.4049/jimmunol.1501089. [DOI] [PubMed] [Google Scholar]
- 19.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urai M, Kaneko Y, Ueno K, Okubo Y, Aizawa T, Fukazawa H, Sugita T, Ohno H, Shibuya K, Kinjo Y, Miyazaki Y. 2016. Evasion of innate immune responses by the highly virulent Cryptococcus gattii by altering capsule glucuronoxylomannan structure. Front Cell Infect Microbiol 5:101. doi: 10.3389/fcimb.2015.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feder V, Kmetzsch L, Staats CC, Vidal-Figueiredo N, Ligabue-Braun R, Carlini CR, Vainstein MH. 2015. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J 282:1406–1418. doi: 10.1111/febs.13229. [DOI] [PubMed] [Google Scholar]
- 22.Bielska E, May RC. 2016. What makes Cryptococcus gattii a pathogen? FEMS Yeast Res 16:fov106. doi: 10.1093/femsyr/fov106. [DOI] [PubMed] [Google Scholar]
- 23.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A 106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrer RA, Voelz K, Henk DA, Johnston SA, Fisher MC, May RC, Cuomo CA. 2016. Microevolutionary traits and comparative population genomics of the emerging pathogenic fungus Cryptococcus gattii. Philos Trans R Soc Lond B Biol Sci 371:20160021. doi: 10.1098/rstb.2016.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis MJ, Eastman AJ, Qiu Y, Gregorka B, Kozel TR, Osterholzer JJ, Curtis JL, Swanson JA, Olszewski MA. 2015. Cryptococcus neoformans-induced macrophage lysosome damage crucially contributes to fungal virulence. J Immunol 194:2219–2231. doi: 10.4049/jimmunol.1402376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandes KE, Dwyer C, Campbell LT, Carter DA. 2016. Species in the Cryptococcus gattii complex differ in capsule and cell size following growth under capsule-inducing conditions. mSphere 1:e00350-. doi: 10.1128/mSphere.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorrell TC, Juillard P-G, Djordjevic JT, Kaufman-Francis K, Dietmann A, Milonig A, Combes V, Grau GER. 2016. Cryptococcal transmigration across a model brain blood-barrier: evidence of the Trojan horse mechanism and differences between Cryptococcus neoformans var. grubii strain H99 and Cryptococcus gattii strain R265. Microbes Infect 18:57–67. doi: 10.1016/j.micinf.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Andreu N, Phelan J, de Sessions PF, Cliff JM, Clark TG, Hibberd ML. 2017. Primary macrophages and J774 cells respond differently to infection with Mycobacterium tuberculosis. Sci Rep 7:42225. doi: 10.1038/srep42225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migliaccio AR, Bieker JJ. 2011. GATA2 finds its macrophage niche. Blood 118:2647–2649. doi: 10.1182/blood-2011-06-362772. [DOI] [PubMed] [Google Scholar]
- 30.Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado JDD, Popovich PG, Partida-Sanchez S, Guerau-De-Arellano M. 2015. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izquierdo E, Cuevas VD, Fernández-Arroyo S, Riera-Borrull M, Orta-Zavalza E, Joven J, Rial E, Corbi AL, Escribese MM. 2015. Reshaping of human macrophage polarization through modulation of glucose catabolic pathways. J Immunol 195:2442–2451. doi: 10.4049/jimmunol.1403045. [DOI] [PubMed] [Google Scholar]
- 32.Cramer SD, Aplan PD, Durum SK. 2016. Therapeutic targeting of IL-7Rα signaling pathways in ALL treatment. Blood 128:473–478. doi: 10.1182/blood-2016-03-679209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin R, Lu H, Li XC, Liu Y-J, Zhang Z. 2016. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol 17:1373–1380. doi: 10.1038/ni.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannoni P, Pietra G, Travaini G, Quarto R, Shyti G, Benelli R, Ottaggio L, Mingari MC, Zupo S, Cutrona G, Pierri I, Balleari E, Pattarozzi A, Calvaruso M, Tripodo C, Ferrarini M, de Totero D. 2014. Chronic lymphocytic leukemia nurse-like cells express hepatocyte growth factor receptor (c-MET) and indoleamine 2,3-dioxygenase and display features of immunosuppressive type 2 skewed macrophages. Haematologica 99:1078–1087. doi: 10.3324/haematol.2013.091405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. 2008. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol 9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Souto ACP, Bonfietti LX, Ferreira-Paim K, Trilles L, Martins M, Ribeiro-Alves M, Pham CD, Martins L, Dos Santos W, Chang M, Brito-Santos F, Santos DCS, Fortes S, Lockhart SR, Wanke B, Melhem MSC, Lazéra MS, Meyer W. 2016. Population genetic analysis reveals a high genetic diversity in the Brazilian Cryptococcus gattii VGII population and shifts the global origin from the Amazon Rainforest to the semi-arid desert in the northeast of Brazil. PLoS Negl Trop Dis 10:e0004885. doi: 10.1371/journal.pntd.0004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litvintseva AP, Mitchell TG. 2012. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathog 8:e1002495. doi: 10.1371/journal.ppat.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petter R, Kang BS, Boekhout T, Davis BJ, Kwon-Chung KJ. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 147:2029–2036. doi: 10.1099/00221287-147-8-2029. [DOI] [PubMed] [Google Scholar]
- 40.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levitz SM, Nong SH, Seetoo KF, Harrison TS, Speizer RA, Simons ER. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun 67:885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 43.Nicola AM, Robertson EJ, Albuquerque P, Derengowski LDS, Casadevall A. 2011. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. mBio 2:e00167-11. doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J Clin Invest 120:1683–1693. doi: 10.1172/JCI41963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu AP, McReynolds LJ, Holland SM. 2015. GATA2 deficiency. Curr Opin Allergy Clin Immunol 15:104–109. doi: 10.1097/ACI.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenblatt MB, Aliprantis A, Hu B, Glimcher LH. 2010. Calcineurin regulates innate antifungal immunity in neutrophils. J Exp Med 207:923–931. doi: 10.1084/jem.20092531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leopold Wager CM, Hole CR, Wozniak KL, Wormley FL. 2016. Cryptococcus and phagocytes: complex interactions that influence disease outcome. Front Microbiol 7:105. doi: 10.3389/fmicb.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan RR, Casadevall A, Oh J, Scharff MD. 1997. T cells cooperate with passive antibody to modify Cryptococcus neoformans infection in mice. Proc Natl Acad Sci U S A 94:2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoag KA, Lipscomb MF, Izzo AA, Street NE. 1997. IL-12 and IFN-gamma are required for initiating the protective Th1 response to pulmonary cryptococcosis in resistant C.B-17 mice. Am J Respir Cell Mol Biol 17:733–739. doi: 10.1165/ajrcmb.17.6.2879. [DOI] [PubMed] [Google Scholar]
- 50.Leopold Wager CM, Hole CR, Wozniak KL, Olszewski MA, Wormley FL. 2014. STAT1 signaling is essential for protection against Cryptococcus neoformans infection in mice. J Immunol 193:4060–4071. doi: 10.4049/jimmunol.1400318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrade JCBN, Gatto M, Rodrigues DR, Soares ÂMVDC, Calvi SA. 2018. Cryptococcus neoformans and gattii promote DNA damage in human peripheral blood mononuclear cells. Med Mycol 56:344–349. doi: 10.1093/mmy/myx046. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez M, Burn T, Luo Y, Pirofski L, Casadevall A. 2009. The outcome of Cryptococcus neoformans intracellular pathogenesis in human monocytes. BMC Microbiol 9:51. doi: 10.1186/1471-2180-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Randhawa HS, Kowshik T, Chowdhary A, Prakash A, Khan ZU, Xu J. 2011. Seasonal variations in the prevalence of Cryptococcus neoformans var. grubii and Cryptococcus gattii in decayed wood inside trunk hollows of diverse tree species in north-western India: a retrospective study. Med Mycol 49:320–323. doi: 10.3109/13693786.2010.516457. [DOI] [PubMed] [Google Scholar]
- 54.Randhawa HS, Kowshik T, Chowdhary A, Preeti Sinha K, Khan ZU, Sun S, Xu J. 2008. The expanding host tree species spectrum of Cryptococcus gattii and Cryptococcus neoformans and their isolations from surrounding soil in India. Med Mycol 46:823–833. doi: 10.1080/13693780802124026. [DOI] [PubMed] [Google Scholar]
- 55.Anderson OR. 2014. Microbial communities associated with tree bark foliose lichens: a perspective on their microecology. J Eukaryot Microbiol 61:364–370. doi: 10.1111/jeu.12116. [DOI] [PubMed] [Google Scholar]
- 56.Ruiz A, Neilson JB, Bulmer GS. 1982. Control of Cryptococcus neoformans in nature by biotic factors. Sabouraudia 20:21–29. doi: 10.1080/00362178285380051. [DOI] [PubMed] [Google Scholar]
- 57.Casadevall A. 2012. Amoeba provide insight into the origin of virulence in pathogenic fungi. Adv Exp Med Biol 710:1–10. doi: 10.1007/978-1-4419-5638-5_1. [DOI] [PubMed] [Google Scholar]
- 58.Steenbergen JN, Shuman HA, Casadevall A. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. 2008. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol 25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 60.Broderick NA. 2015. A common origin for immunity and digestion. Front Immunol 6:72. doi: 10.3389/fimmu.2015.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casadevall A, Pirofski L-A. 2007. Antibody-mediated protection through cross-reactivity introduces a fungal heresy into immunological dogma. Infect Immun 75:5074–5078. doi: 10.1128/IAI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canton J, Grinstein S. 2017. Measuring phagosomal pH by fluorescence microscopy. Methods Mol Biol 1519:185–199. doi: 10.1007/978-1-4939-6581-6_12. [DOI] [PubMed] [Google Scholar]
- 63.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coelho C, Souza ACO, Derengowski LDS, de Leon-Rodriguez C, Wang B, Leon-Rivera R, Bocca AL, Gonçalves T, Casadevall A. 2015. Macrophage mitochondrial and stress response to ingestion of Cryptococcus neoformans. J Immunol 194:2345–2357. doi: 10.4049/jimmunol.1402350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.