Abstract
Somatic mutations contribute to the heterogeneous prognosis of chronic myelomonocytic leukemia (CMML). Hypomethylating agents (HMAs) are active in CMML, but analyses of small series failed to identify mutations predicting response or survival. We analyzed a retrospective multi-center cohort of 174 CMML patients treated with a median of 7 cycles of azacitidine (n = 68) or decitabine (n = 106). Sequencing data before treatment initiation were available for all patients, from Sanger (n = 68) or next generation (n = 106) sequencing. Overall response rate (ORR) was 52%, including complete response (CR) in 28 patients (17%). In multivariate analysis, ASXL1 mutations predicted a lower ORR (Odds Ratio [OR] = 0.85, p = 0.037), whereas TET2mut/ASXL1wt genotype predicted a higher CR rate (OR = 1.18, p = 0.011) independently of clinical parameters. With a median follow-up of 36.7 months, overall survival (OS) was 23.0 months. In multivariate analysis, RUNX1mut (Hazard Ratio [HR] = 2.00, p = .011), CBLmut (HR = 1.90, p = 0.03) genotypes and higher WBC (log10(WBC) HR = 2.30, p = .005) independently predicted worse OS while the TET2mut/ASXL1wt predicted better OS (HR = 0.60, p = 0.05). CMML-specific scores CPSS and GFM had limited predictive power. Our results stress the need for robust biomarkers of HMA activity in CMML and for novel treatment strategies in patients with myeloproliferative features and RUNX1 mutations.
Keywords: Chronic myelomonocytic leukemia, Hypomethylating agents, Somatic mutations, Prognosis
Highlights
-
•
TET2mut/ASXL1wt genotype predicts higher complete response rate and prolonged survival in CMML with hypomethylating agents.
-
•
Conversely, RUNX1mut and CBLmut genotypes are associated with poorer outcome, independently of higher leukocyte count.
-
•
CPSS and GFM prognostic scores showed modest performance when calculated at initiation of hypomethylating agents.
Somatic mutations contribute to the heterogeneous prognosis of chronic myelomonocytic leukemia (CMML). Hypomethylating agents (HMAs) are active in CMML. Response and survival in MDS and AML patients treated with HMAs is difficult to predict. We explore the predictive role of recurrent somatic mutations in a large retrospective cohort of 174 HMA-treated CMMLs. Consistent with MDS studies, we report a higher response rate in TET2mut/ASXL1wt patients. We also identify a CMML-specific molecular pattern (RUNX1mut or CBLmut) associated with shorter survival. Our results can inform treatment decision in CMML, for instance by using HMAs prior to transplant in TET2mut/ASXL1wt patients.
1. Introduction
Chronic myelomonocytic leukemia (CMML) is a clonal bone marrow disorder, classified by WHO as a myelodysplastic/myeloproliferative neoplasm (MDS/MPN) [18]. It is characterized by persistent monocytosis associated with a variable degree of bone marrow blast excess, cytopenias and myeloproliferation. Its prognosis is variable but overall poor, with a median survival of 20–32 months and a risk of acute myeloid leukemia (AML) transformation of 14–29% [8,11].
Recurrent somatic mutations found in CMML affect genes encoding epigenetic regulators, signaling, splicing and transcription regulator genes [11,14,16,19]. The high frequency of TET2, ASXL1, SRSF2, and RAS pathway (NRAS, KRAS, and CBL) mutations may represent a mutational fingerprint of the disease [11,19]. Frameshift and nonsense ASXL1 mutations have invariably been shown to confer poor prognosis [8,11,19], while the poor prognostic impact of TET2 and SRSF2 mutations [11,13,21] is more controversial, and could depend on specific mutation combinations. Mutations in EZH2 [9], SETBP1 [8] and DNMT3A [22] could also be detrimental, but their impact is more difficult to assess because of their lower incidence. Recently, several prognostic scoring systems accounting for gene mutations have been developed in CMML, but most of them were developed in cohorts of untreated patients, or with heterogeneous treatments [8,11]. The prognostic value of gene mutations is highly dependent on the therapeutic context, as exemplified by the specific poor prognosis of RAS mutations in CMML in the context of allogeneic stem cell transplantation (ASCT) [30].
Retrospective studies [1,5,23] and limited prospective data [3,6,25], mostly non-randomized, have reported activity of hypomethylating agents (HMA) in CMML. In these studies, azacitidine (AZA) and decitabine (DAC) provided an overall response rate (ORR) of 40–70%, translating in median overall survivals (OS) of 12–22 months. Response to HMA is difficult to predict and is loosely correlated to survival in CMML [7]. In MDS patients treated with HMA, mutations in the epigenetic regulators TET2 [2,10,28] and ASXL1 [2] affect response rates but not overall survival (OS). In retrospective studies of CMML treated with HMA, older age, higher bone-marrow (BM) and peripheral blood (PB) blast count, higher white blood cell counts (WBC), splenomegaly and cytogenetic risk have been found to impair survival [1].
The impact of gene mutations in this setting has so far been addressed in small series, precluding the identification of mutations predicting response or survival in CMML treated with HMA [3,15,25]. Here we report the largest retrospective cohort of CMML patients treated with DAC or AZA to date with available molecular data for the most frequently mutated genes.
2. Patients and Methods
2.1. Patients
We updated clinical data from 174 patients with CMML treated with AZA or DAC between February 2007 and December 2016, in Groupe Francophone des Myélodysplasies (GFM) centers (n = 61, including Dresden), Firenze (n = 37), Mayo clinic (n = 41), Memorial Sloan Kettering (MSKCC) and Moffitt (MCC) Cancer Centers (n = 35). Patients provided written informed consent and the study was approved by each institution's IRB (GFM: PHRC MAD-06 and clinical trial EudraCT #2008-000470-21; Mayo clinic: 15-003786 and 11-005599; Firenze NCT01251627; MCC/MSKCC: 00014416). Patients with previous intensive treatment (intensive chemotherapy or ASCT) or with AML transformation prior to HMA were excluded.
CMML diagnosis and stratification was made according to WHO 2008 criteria [18]. Splenomegaly was defined as a clinical or radiological spleen enlargement. Bone marrow blasts included agranular blasts, myeloblasts and promonocytes as recommended. Cytogenetic risk was assessed according to CMML-specific cytogenetic risk classification [27]. Prognosis at initiation of treatment was evaluated according to CMML-specific prognostic scoring system (CPSS) [27] and GFM score [11]. Information on RBC-transfusion dependency was not available and was substituted by hemoglobin level (Hb < 10 g/dL) to calculate CPSS as proposed by the authors [27]. CPSS-mol [8] was assessed in patients with either available SETBP1 information, or for whom the risk was unchanged whatever the SETBP1 mutational status. Patients received HMA according to standard schedules (AZA: 75 mg/m2/d subcutaneously d1-7/28d cycles; DAC: 20 mg/m2/d intravenous d1-5/28d cycles). Responses were assessed according to MDS IWG-2006 criteria [4].
2.2. Gene Mutation Analyses
DNA extracted from peripheral blood (PB) CD14+ monocytes or bone marrow (BM) mononucleated cells (BMNCs) for GFM centers, BMNCs for Mayo Clinic and Firenze, BMNCs or PB mononucleated cells (PBMCs) for MSKCC and MCC. Analysis of somatic mutations was done by Sanger sequencing (GFM) or Next-Generation Sequencing (NGS), using custom target capture with Agilent SureSelect (Mayo Clinic), Agilent HaloPlex (Firenze), Fluidigm Access Array multiplex PCR technologies (MSKCC and MCC) followed by sequencing on Illumina platforms. The overlapping genomic regions interrogated by all platforms included exon 12 of ASXL1, and all coding exons in the SRSF2, TET2, NRAS, RUNX1, CBL, U2AF1, DNMT3A, IDH2, KRAS, SF3B1, JAK2, EZH2, IDH1 and TP53 genes. Details on mutational analysis pipelines have previously been published [11,15,20,24].
2.3. Statistical Analyses
Variables are reported as medians and interquartile ranges (IQR) and numbers and proportions for continuous and categorical variables respectively. Group comparisons for dichotomic, ordinal and continuous variables were carried by Fisher's exact tests, Kendall's correlation tests, and Mann-Whitney's tests respectively. Univariate analyses of variables influencing response rates were stratified on HMA and tested with linear regressions. All significant variables with significant impact (p < 0.05) in univariate analyses were then included in multivariate linear regressions adjusted on HMA.
OS was defined as time between initiation of HMA and date of death from any cause or date of last follow-up. AML free survival (AMLFS) was defined as the time between initiation of HMA and date of AML transformation, death or last follow-up. OS and AMLFS were obtained according to the Kaplan-Meier method and univariate analyses stratified on HMA were done with the Cox regression model. Follow-up duration was calculated with the inverse method. The prognostic impact of WBC was assessed with the log10-transformed variable (logWBC), and age, hemoglobin level, platelets count were analyzed as continuous variables. Multivariate survival analyses were performed by Cox regression followed by backward stepwise selection. The proportional hazard assumption was validated by visual inspection of Schöenfeld residuals. Interactions were studied by comparing through a likelihood ratio test Cox models including the two studied variables with or without an interaction term. The goodness-of-fit of a given model was assessed with Harrell's C concordance index (C-index), a value ranging from 0.5 (no relevance) to 1 (perfect prediction).
There was no sample size calculation prior to this retrospective study. In a post hoc power analysis using two-sided log-rank tests with an alpha risk of 0.05, a study population of 174 patients provided a power of 0.70 to detect a hazard ratio (HR) of 2.5 or higher for mutations present in only 10% of patients or to detect a milder effect (HR ≥ 1.5) for more frequent mutations (40% of patients). Mutations present in <10% of patients were thus not analyzed. Thus, only complete cases were analyzed, with imputation of missing data.
Propensity Score Matching (PSM) was performed by logistic regression using indicated variables, and patients were matched one to one by nearest neighbour according to HMA received. Quality of matching was checked by inspecting the reduction of bias for each variable and testing for differences in matched samples for each variable. All statistical analyses were stratified on HMA and two-sided, retaining p < 0.05 as statistically significant. Analyses were performed with R 3.3.2 (cran.r-project.org) or STATA 12 (Stata Corp).
3. Results
3.1. Patients Characteristics
We included 174 patients in this study, 118 men (68%) and 56 women (32%) with a median age of 72 years (Inter-quartile range [IQR] 66–78). Characteristics of patients at initiation of HMA are summarized in Table 1. Diagnosis at HMA onset was CMML-1 and CMML-2 in 64% and 36% respectively. Cytogenetic risk was low, intermediate, high risk in 70%, 13% and 17%. CPSS risk category was low in 13%, intermediate-1 in 22%, intermediate-2 in 51% and high risk in 14% of patients. CPSS-mol risk category was intermediate-1 in 1%, intermediate-2 in 30% and high-risk in 69%. Sixty-seven patients (40%) had splenomegaly at onset of HMA. Median WBC was 15.4 × 109/L [IQR 8.6–26.0]. Fifty-three patients (39%) had received a treatment before HMA (erythropoiesis stimulating agent (ESA) n = 25, G-CSF n = 3 and hydroxyurea n = 38 patients). Of note, 80% of CMML-1 patients treated with HMA in our cohort fulfilled one of the following criteria: myeloproliferation (WBC > 30 × 109/L, splenomegaly), previous ESA or hydroxyurea failure, or CPSS intermediate-2 or high.
Table 1.
Characteristics of the study population at HMA onset. Median [IQR] or N (%).
| Patients | Total (n = 174) | AZA (n = 68) | DAC (n = 106) | AZA vs DAC p=a |
|---|---|---|---|---|
| Gender, male | 118 (68%) | 45 (66%) | 73 (69%) | 0.74 |
| Age (years) | 72 [66–78] | 74 [68–79] | 71 [66–77] | 0.081 |
| Splenomegaly, yes | 67 (40%) | 24 (36%) | 43 (42%) | 0.52 |
| WBC (×109/L) | 15.4 [8.6–26.0] | 13.3 [7.7–22.8] | 17.5 [9.1–28.1] | 0.041 |
| Hb (g/dL) | 9.8 [8.7–11.8] | 9.7 [8.7–11.2] | 10.1 [8.6–11.9] | 0.52 |
| ANC (×109/L) | 7.6 [3.6–13.9] | 5.7 [3.3–12.5] | 8.7 [3.8–15.5] | 0.078 |
| Platelets (×109/L) | 84 [51–154] | 100 [56–176] | 73 [49–140] | 0.082 |
| Peripheral blasts (%) | 0 [0–1] | 0 [0–0] | 0 [0–2] | 0.074 |
| WHO 2008 | 0.66 | |||
| CMML-1 | 111 (64%) | 42 (62%) | 69 (65%) | |
| CMML-2 | 63 (36%) | 26 (38%) | 37 (35%) | |
| Cytogenetic risk (n = 172) | 0.11 | |||
| Low | 120 (70%) | 52 (78%) | 68 (65%) | |
| Intermediate | 22 (13%) | 5 (7%) | 17 (16%) | |
| High | 30 (17%) | 10 (15%) | 20 (19%) | |
| Prognostic scores | ||||
| CPSS (n = 172) | 0.86 | |||
| Low | 22 (13%) | 10 (15%) | 12 (12%) | |
| Intermediate-1 | 38 (22%) | 15 (22%) | 23 (22%) | |
| Intermediate-2 | 88 (51%) | 31 (46%) | 57 (54%) | |
| High | 24 (14%) | 11 (17%) | 13 (12%) | |
| GFM score (n = 174) | 0.21 | |||
| Low | 55 (32%) | 25 (37%) | 30 (28%) | |
| Intermediate | 60 (34%) | 23 (34%) | 37 (35%) | |
| High | 59 (34%) | 20 (29%) | 39 (37%) | |
| CPSS-mol (n = 133) | 0.007 | |||
| Low | 0 | 0 | 0 | |
| Intermediate-1 | 1 (1%) | 1 (2%) | 0 | |
| Intermediate-2 | 40 (30%) | 23 (42%) | 17 (22%) | |
| High | 61 (69%) | 31 (56%) | 61 (78%) | |
| Treatment before HMA | 53 (39%) | 25 (37%) | 28 (41%) | 0.73 |
| ESA | 25 (25%) | 10 (23%) | 15 (26%) | 0.82 |
| GCSF | 2 (2%) | 2 (5%) | 0 | 0.18 |
| Hydroxyurea | 33 (24%) | 15 (22%) | 18 (26%) | 0.69 |
| HMA treatment | ||||
| Type of HMA | 68 (39%) | 106 (61%) | ||
| Time from diagnosis to HMA (months) | 4.2 [1.1–17.5] | 1.9 [0.8–14.1] | 5.1 [1.4–20.3] | 0.07 |
| Number of HMA cycles | 7 [4–15] | 7 [4–14] | 7 [4–15] | 1.00 |
| Follow-up (months) | 36.7 [25.0–66.0] | 42.3 [16.2–66.2] | 36.4 [25.3–66.0] | 0.48 |
Mann-Whitney test, Kendall's rank correlation and Fisher exact tests for continuous, ordinal and dichotomic variables, respectively.
Sixty-eight patients (39%) received AZA and 106 patients (61%) received DAC for a median of 7 cycles for both drugs [IQR 4–15]. Median time from diagnosis to HMA onset was 4.2 months [IQR 1.1–17.5], and patients were followed for a median of 36.7 months after HMA onset. During this period, 54 patients (31%) had transformation to AML, and 17 patients (10%, median age: 58 years, range [IQR 56–65]) received ASCT, in median 5.9 months [IQR. 3.7–9.8] after HMA onset. Median OS was 23.0 months [IQR 11.6–58.0] and median AMLFS was 19.2 months [IQR 9.7–53.8].
Compared to AZA-treated patients, patients treated with DAC had comparable demographics, except for a higher WBC (DAC: 17.5 × 109/L versus AZA: 13.3 × 109/L, p = .041) and non-significant trends towards lower platelets count (DAC: 73 × 109/L versus AZA: 100 × 109/L, p = .078) and poorer cytogenetic risk (p = .11). These differences translated into a significantly poorer CPSS-mol risk (p = .007) in DAC-treated patients. A non-significant trend to poorer OS was seen in patients treated with DAC (median OS of 18.5 months for DAC versus 31.1 months for AZA, HR = 0.73, [95% CI: 0.49–1.10], p = .13). Propensity score matching on age, WBC, platelets count, peripheral blasts, cytogenetic risk, time to HMA treatment, ASXL1, CBL and IDH2 mutational statuses at HMA onset confirmed the lack of OS difference between AZA and DAC in the 161 patients (93%) that could be successfully matched (HR = 1.37 [95% CI: 0.61–3.10] for AZA compared to DAC, p = .45). All further analyses were nevertheless stratified on HMA.
3.2. Mutational Landscape
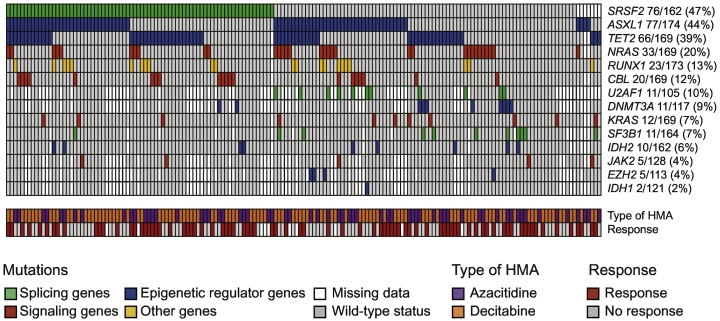
Molecular information on ASXL1, from Sanger sequencing (n = 68) or NGS (n = 106) was available for all patients, while the genotypes of other studied genes (SRSF2, TET2, NRAS, RUNX1, CBL, U2AF1, DNMT3A, IDH2, KRAS, SF3B1, JAK2, EZH2, IDH1, TP53) were available in 93–173 (53–99%) of cases (Supplementary Table 1). The most frequently mutated genes in our cohort were SRSF2 (47%), ASXL1 (44%) and TET2 (39%). Other frequently mutated genes included NRAS (20%), RUNX1 (13%), CBL (12%), U2AF1 (10%) and DNMT3A (9%), as shown in Fig. 1. There was no significant difference in the mutational spectrum of patients studied by Sanger versus NGS (Supplementary Table 1). Most mutations detected with NGS sequencing had a variant allele frequency (VAF) above 20% (Supplementary Fig. 1), a conservative threshold for the detection of somatic variants by Sanger sequencing. In particular, considering variants detected by NGS with a VAF < 20% as wild-type did not affect our main findings regarding survival analyses (not shown). DAC-treated patients had less frequent IDH2 mutations than AZA-treated patients (DAC 3% versus AZA 11%, p = .048) and a non-significant trend towards less frequent CBL mutations (DAC 9% versus AZA 17%, p = .09) and more frequent ASXL1 mutations (DAC 49% versus AZA 37%, p = .12). Their mutational spectrum was otherwise comparable (Supplementary Table 1). TET2 mutations were associated with a lower cytogenetic risk. CPSS cytogenetic risk was low in 58 (88%), intermediate in 4 (6%) and high in 4 (6%) for TET2mut patients, compared to low in 58 (57%), intermediate in 18 (18%) and high in 26 (25%) for TET2wt patients (p < .0001). Conversely, RUNX1mut patients tended to have poorer cytogenetic risk (p = .076).
Fig. 1.
Mutational landscape of the study cohort.
3.3. Impact of Clinical Variables and Somatic Mutations on Response to HMA
Response status according to IWG-2006 was available for 164 (94%) patients. Overall response rate (ORR) was 52%, including complete response (CR) in 28 (17%), partial response (PR) in 10 (6%), marrow CR (mCR) in 18 (11%), stable disease with hematologic improvement (HI) in 29 (18%), stable disease (SD) in 41 (25%) and progressive disease (PD) in 38 (23%; Supplementary Table 2). Fifty (51%) and 35 (54%) patients treated with DAC and AZA achieved response (p = .75). There was no difference in CR rate between the two HMAs (DAC 15% versus AZA 20%; p = .53).
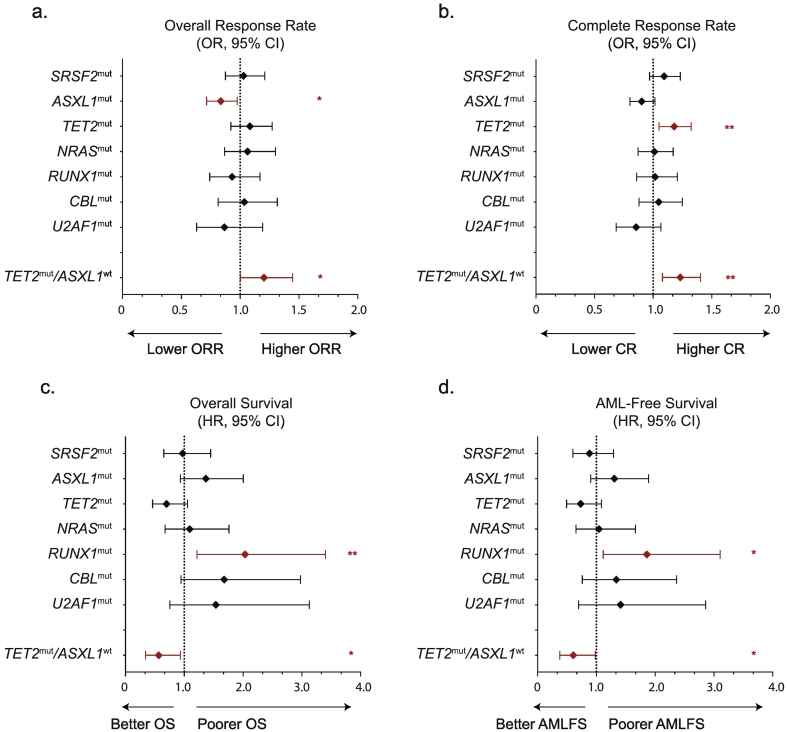
In univariate analysis stratified on HMA, mutations in ASXL1 predicted lower ORR (ASXL1mut 42% versus ASXL1wt 60%, p = .023). Twenty-six (15%) patients were ASXL1mut/TET2mut, 49 (29%) were ASXL1mut/TET2wt, 40 (24%) were ASXL1wt/TET2mut and 54 (32%) were ASXL1wt/TET2wt. TET2mut/ASXL1wt genotype predicted higher ORR (TET2mut/ASXL1wt 66% versus all other genotypes 47%, p = .048), while other mutations had no effect (Fig. 2a). TET2 mutations predicted a higher CR rate (TET2mut 26% versus TET2wt 9%, p = .005) whereas ASXL1 mutated patients tended to achieve CR less frequently (ASXL1mut 11% versus 22% for ASXL1wt, p = .088). An increased CR rate was particularly apparent in patients with the TET2mut/ASXL1wt genotype (Fig. 2b), while TET2 genotype had little influence in ASXL1mut patients (32% for TET2mut/ASXL1wt versus 11% for all other genotypes, p = .002).
Fig. 2.
Forest plots of Odds Ratios and their 95% Confidence Intervals of (a.) Overall Response and (b.) Complete Response for each genotype in a univariate linear regression adjusted on HMA. Forest plots of Hazard Ratios and their 95% Confidence Intervals for (c.) Overall Survival and (d.) AML-free Survival for each genetic or clinical variable in a univariate Cox model adjusted on HMA. A significant impact on response rate is indicated in red. *p < .05. **p < .01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Considering clinical variables (age, WBC, hemoglobin level, platelets count, all analyzed as continuous variables, WHO 2008 stratification, karyotype risk), only lower hemoglobin level (p = .001) and lower platelets count (p = .047) predicted lower ORR. Higher WBC (p = .038), lower hemoglobin level (p < .001) and higher cytogenetic risk (p = .021) predicted lower CR rates while other clinical variables had no significant impact.
In a multivariate linear regression model including type of HMA, ASXL1 and TET2 statuses, hemoglobin level and platelets count, the presence of an ASXL1 mutation predicted a lower ORR (Odds Ratio [OR] = 0.85, 95% Confidence Interval [CI]: 0.73–0.99, p = .037) independently of hemoglobin level (p = .001) and platelets count (p = .049). In a multivariate regression model including type of HMA, TET2mut/ASXL1wt genotype, WBC, hemoglobin level and cytogenetic risk, TET2mut/ASXL1wt genotype predicted higher CR rate (OR = 1.18, [95% CI: 1.04–1.34], p = .011), independently of the detrimental role of lower hemoglobin level (p < .001; Table 2).
Table 2.
Multivariate analysis for response.
| Overall response |
Complete response |
|||||
|---|---|---|---|---|---|---|
| Variables | OR | 95% CI | p= | OR | 95% CI | p= |
| AZAa | 1.00 | [0.86–1.17] | 0.96 | 1.01 | [0.91–1.13] | 0.8 |
| ASXL1mut genotype | 0.85 | [0.73–0.99] | 0.037 | – | – | – |
| TET2mut/ASXL1wtgenotype | – | – | – | 1.18 | [1.04–1.34] | 0.011 |
| Hemoglobin level | 1.07 | [1.03–1.11] | 0.001 | 1.06 | [1.03–1.09] | <0.001 |
| WBC (log10) | – | – | – | – | – | – |
| Platelets levelb | 1.01 | [1.00–1.02] | 0.049 | – | – | – |
Reference: DAC.
For every 20 × 109/L increment.
3.4. Impact of Somatic Mutations and Clinical Variables on Survival
In univariate analyses adjusted on HMA, mutations in RUNX1 (HR = 2.04, [95% CI 1.22–3.40], p = .007) and to a lesser extent CBL (HR = 1.68 [95% CI: 0.95–2.98], p = .076) and ASXL1 (HR = 1.37 [95% CI: 0.93–2.00], p = .11) mutations were associated with poorer OS (Fig. 2c). Conversely, TET2mut tended to predict prolonged OS (HR = 0.70 [95% CI: 0.46–1.05], p = .088). Mutations in SRSF2, NRAS, U2AF1, DNMT3A, IDH2, KRAS and SF3B1 had no impact on OS. RUNX1 (HR = 1.86; [95% CI: 1.12–3.10], p = .017) was the only gene to significantly predict worse AMLFS (Fig. 2d). Considering the most frequently mutated genes (SRSF2, TET2, ASXL1, RUNX1), there was no significant gene–gene interaction on overall survival. The additive effect of TET2 and ASXL1 genotypes resulted in a significantly superior outcome in TET2mut/ASXL1wt patients versus other TET2/ASXL1 genotypes (HR = 0.57, [95% CI: 0.34–0.93], p = .026). The median OS of RUNX1mut patients was 16.0 [10.3–27.6] months, compared to 24.5 [11.6–58.0] months in RUNX1wt patients (Fig. 3). The median OS of TET2mut/ASXL1wt patients was 32.5 [18.2–68.9] months, compared to 19.2 [10.3–42.2] months for other genotypes. In the 155 (89%) patients with comprehensive genotyping of 8 of the top recurrently mutated genes (SRSF2, ASXL1, TET2, NRAS, RUNX1, CBL, IDH2 and KRAS), the number of mutated genes had no significant impact on OS (p = .11) or on AMLFS (p = .27). Censoring at ASCT did not change our main findings (not shown).
Fig. 3.
Kaplan-Meier estimates of OS according to (a.) RUNX1, (b.) ASXL1, (c.) CBL, (d.) TET2, (e.) SRSF2 and (f.) TET2mut/ASXL1wt mutational status. Results of univariate analyses stratified on HMA are reported for each gene.
Clinical variables associated with poorer OS in univariate Cox models adjusted on HMA were higher WBC (logWBC: HR = 2.77, [95% CI: 1.61–4.79], p < .001), lower hemoglobin level (HR = 0.90, [95% CI: 0.82–0.99], p = .039), and to a lesser extent poorer cytogenetic risk (HR = 1.27, [95% CI: 1.00–1.60], p = .051). Time from diagnosis to HMA onset had no impact (p = .8). Higher WBC (logWBC: HR = 2.47, [95% CI: 1.46–4.17], p = .001), poorer cytogenetic risk (HR = 1.40, [95% CI: 1.12–1.75], p = .004), and to a lesser extent lower hemoglobin level (HR = 0.90, [95%CI 0.84–1.01], p = .074) and WHO-2008 stratification (HR = 1.42, [95% CI: 0.98–2.06], p = .066) predicted shorter AMLFS.
In a multivariate Cox analysis including RUNX1mut, CBLmut, TET2mut/ASXL1wt genotypes together with WBC, hemoglobin level and cytogenetic risk, RUNX1mut (p = .011), CBLmut (p = .03) and TET2mut/ASXL1wt (p = .05) retained their prognostic role, independently of higher WBC (p = .005, Table 3). In a similar model for AMLFS, only RUNX1 mutations retained poor prognostic value (p = .048) independently of higher WBC (p = .005) and high-risk cytogenetics (p = .018, Supplementary Table 3).
Table 3.
Multivariate Cox models of OS adjusted on HMA.
| Overall survival |
|||
|---|---|---|---|
| Variables | HR | 95% CI | p= |
| RUNX1mut genotype | 2.00 | [1.17–3.42] | 0.011 |
| CBLmut genotype | 1.90 | [1.06–3.40] | 0.03 |
| TET2mut/ASXL1wt genotype | 0.60 | [0.36–1.00] | 0.05 |
| WBC (log10) | 2.30 | [1.28–4.11] | 0.005 |
3.5. Relevance of CMML Prognostic Scores
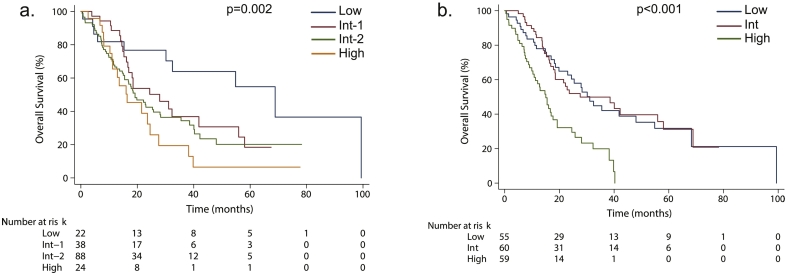
We next interrogated the prognostic relevance of CPSS and GFM scores, CMML-specific prognostic scores established in all comers, in the setting of HMA. Median OS was 68.9, 27.9, 19.2 and 16.4 months for low, intermediate-1, intermediate-2 and high CPSS risk groups, respectively (Cox model adjusted on HMA: p = .002) and 31.1, 27.6 and 15.0 months for low, intermediate and high GFM risk groups, respectively (Cox model adjusted on HMA: p < .001). Their respective C-index were 0.59 and 0.62. In particular, the GFM score, which is based on WBC, platelets, hemoglobin and ASXL1 status identified a sub-group of 59 (34%) high risk patients with a median OS of 15.0 months (Fig. 4).
Fig. 4.
Kaplan-Meier estimates of OS according to (a.) CPSS and (b.) GFM risk category. Results of univariate analyses stratified on HMA are reported for each score.
4. Discussion
In this study, we report on a large retrospective cohort of 174 patients the prognostic value of frequent gene mutations on the outcome of CMML treated with HMAs. Our results demonstrate that patients with TET2mut/ASXL1wt genotype achieve higher CR rates with a significant survival benefit, and conversely that ASXL1 mutations predict lower overall response rates. Our study also identifies the poor prognosis of RUNX1mut and CBLmut genotypes in CMML patients treated with HMA.
A number of retrospective cohorts have investigated the prognostic value of somatic gene mutations in CMML [8,11,14,19]. However, the prognostic impact of gene mutations has never been investigated so far in a large cohort of CMML patients treated by HMA. Recent studies highlight the need to re-assess the prognostic value of a given mutation in specific therapeutic settings. For instance, TP53 mutations are classically ascribed to a poor prognosis in myeloid malignancies including MDS, yet a recent study in MDS and AML reported that TP53 mutations were associated with higher rates of response to intensive schedules of decitabine [29]. Other predictive markers of response or prognosis have been suggested in CMML treated with HMA, including gene expression or methylation profiles [3,15], but these biomarkers are difficult to standardize compared to genomic analyses, were developed in smaller cohorts, and do not always predict survival.
The mutational spectrum of our cohort, dominated by frequent mutations in ASXL1, SRSF2 and TET2, is comparable to that of other published cohorts of CMML not selected for HMA treatment, highlighting both the relative homogeneity of the molecular landscape in CMML, and the broad usage of HMAs across this landscape. However, our cohort included only 39% of patients with TET2 mutations, a figure in the lower range of published series [11,19], perhaps owing to the trend towards favorable outcome of TET2 mutated patients [21].
HMA indications in our cohort reflected both the US (AZA and DAC) and European (AZA) labels of each HMA in CMML, and the inclusion criteria of two European DAC trials [3,25]. This resulted in a higher-risk CMML population, with 65% of patients having a CPSS risk intermediate-2 or high and 99% of assessable patients having CPSS-mol risk intermediate-2 or high-risk. Though the prognostic impact of gene mutations was investigated separately in both DAC trials [3,15], these analyses were hampered by their limited power. In the present study, we performed a post hoc power analysis to ensure that we could capture clinically meaningful effect (HRs > 1.5) for recurrently mutated genes (mutations occurring in >10% of patients).
Our cohort was heterogeneous in terms of biological material and technique used to assess somatic mutations. Mutations were studied on sorted monocytes or on peripheral blood or bone marrow mononucleated cells. However, previous studies have shown full clonal dominance in the bone marrow of CMML patients compared to sorted monocytes [12], and studies in MDS have shown that PBMCs are an adequate material in these entities [17]. Samples were sequenced with either Sanger sequencing or NGS with various gene panels. We checked that the mutational spectrum for frequently mutated genes were similar between the two techniques and that exclusion of mutations found by NGS with a VAF below 20%, which could have been overlooked by Sanger sequencing, did not affect our results. As only 61% of patients were assessed by NGS, we were not able to study the impact of ancestral versus secondary mutations in the context of HMA, a distinction that may hold biological relevance in CMML [12,19].
In our cohort, ASXL1 mutations predicted lower ORR together with anemia and lower platelet count. TET2 mutations did not predict ORR in multivariate analysis, but were associated with higher CR rates in ASXL1 wildtype patients, independently of the detrimental effect of anemia. These results are consistent with previous studies in MDS [2,10,28]. We assessed responses with MDS IWG 2006 criteria [4], and it is possible that the use of the novel overlap-IWG response criteria [26], which account for changes in the myeloproliferative component of CMML [7] would have captured additional impact of gene mutations on response. However, detailed data on response according to these criteria, especially spleen size and symptoms, were available in only a subset of our patients.
In univariate analyses, RUNX1 and to a lesser extent ASXL1 and CBL were the only frequently mutated genes associated with poorer outcome after adjustment on the type of HMA. Conversely, TET2mut predicted prolonged OS in patients wild-type for ASXL1. We then integrated clinical data, RUNX1mut, CBLmut and TET2mut/ASXL1wt genotypes in a multivariate analysis, and found that higher WBC, RUNX1 and CBL mutations were associated with poorer outcome, whereas TET2mut/ASXL1wt genotype predicted prolonged survival. Cytogenetic risk according to CPSS was not conserved in the multivariate analysis, perhaps because it is associated in our cohort with TET2 mutations, as previously reported [11]. These biomarkers are however different from those identified for survival after ASCT (namely RAS mutations), which remains the only curative treatment of CMML [30].
We analyzed the prognostic relevance of CPSS and GFM scores. Though both scores predicted to some extent overall survival, they failed to accurately discriminate all risk groups, further strengthening the need to carve therapy-specific prognostic scores. We could not interrogate the predictive power of the recently published CPSS-mol [8], because the mutational status of SETBP1 was only available in 32 (17%) patients and all but one evaluable patients were either intermediate-2 or high-risk by CPSS-mol.
Our analyses focused on genes mutated in >10% of CMML cases. They do not exclude the possibility that infrequently mutated genes have a strong prognostic effect in patients treated with HMA. In particular, EZH2 mutations were found in only 4% of our patients, but seemed to bare a very poor prognosis (median OS 7.03 months for EZH2mut versus 23.0 for EZH2wt, p < .001). Finally, only analysis of prospective randomized trials of HMA in CMML such as the ongoing DACOTA trial (NCT02214407) will provide information regarding the survival benefit of HMA compared to standards of care in patients whose CMML harbor the poor-risk genotypes identified in the present study.
In conclusion, our findings stress the need to design novel treatment strategies in CMML with myeloproliferative features such as high WBC or CBL mutations, and to identify druggable vulnerabilities in myeloid neoplasms with RUNX1 mutations. Nonetheless, given the limited predictive power of current prognostic scores in CMML patients treated with HMA, efforts to design robust biomarkers of HMA activity remains an important area for future research in myeloid neoplasms.
Acknowledgments
Acknowledgements
The authors would like to thank Fatiha Chermat and the clinical research associates from the GFM for their help in collecting the data. RI is supported by grants from the Gilead International Research Scholarship in Hematology/oncology and by the Laurette Fugain (ALF-2014-10) Association. ES is supported by grants from the Ligue Nationale Contre le Cancer (équipe labellisée).
Funding Sources
The GFM received funding from the French Ministry of Health (PHRC MAD-06). Janssen Inc. funded the GFM and FISM DAC trials (EudraCT #2008-000470-21; NCT01251627).
Conflicts of Interest
DS has received research funding by Celgene. CCC has received honoraria from H3 Biomedicine and Pharmacyclics. UP has received honoraria and research funding from Janssen and Celgene. VS has received honororia by Celgene, Janssen and Novartis, has received research funding by Celgene, and has served as a consultant for Abbvie and Amgen. PF has received research funding by Celgene, Janssen, Novartis, Amgen and Astex. RI has received research funding by Novartis and Janssen. The remaining authors declare no competing financial interests.
Author Contributions
MD, ES, PF and RI designed the study. MD collected the data. MD and RI performed the statistical analyses and drafted the manuscript. FY, AS, DS, TB, UP, LW, LA, VS, PF, ES and RI accrued patients and provided clinical annotations. ND, AR, OK, CCC, DS, MF, RR, EP, CP, MP and RI performed genomic analyses. All authors revised the manuscript and approved its final version. All authors had access to primary data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.04.018.
Appendix A. Supplementary data
Supplementary material
References
- 1.Adès L., Sekeres M.A., Wolfromm A., Teichman M.L., Tiu R.V., Itzykson R. Predictive factors of response and survival among chronic myelomonocytic leukemia patients treated with azacitidine. Leuk Res. 2013;37:609–613. doi: 10.1016/j.leukres.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Bejar R., Lord A., Stevenson K., Bar-Natan M., Pérez-Ladaga A., Zaneveld J. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun T., Itzykson R., Renneville A., de Renzis B., Dreyfus F., Laribi K. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: a phase 2 trial. Blood. 2011;118:3824–3831. doi: 10.1182/blood-2011-05-352039. [DOI] [PubMed] [Google Scholar]
- 4.Cheson B.D., Greenberg P.L., Bennett J.M., Lowenberg B., Wijermans P.W., Nimer S.D. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108:419–425. doi: 10.1182/blood-2005-10-4149. [DOI] [PubMed] [Google Scholar]
- 5.Costa R., Abdulhaq H., Haq B., Shadduck R.K., Latsko J., Zenati M. Activity of azacitidine in chronic myelomonocytic leukemia. Cancer. 2011;117:2690–2696. doi: 10.1002/cncr.25759. [DOI] [PubMed] [Google Scholar]
- 6.Drummond M.W., Pocock C., Boissinot M., Mills J., Brown J., Cauchy P. A multi-centre phase 2 study of azacitidine in chronic myelomonocytic leukaemia. Leukemia. 2014;28:1570–1572. doi: 10.1038/leu.2014.85. [DOI] [PubMed] [Google Scholar]
- 7.Duchmann M., Braun T., Micol J.-B., Platzbecker U., Park S., Pilorge S. Validation of response assessment according to international consortium for MDS/MPN criteria in chronic myelomonocytic leukemia treated with hypomethylating agents. Blood Cancer J. 2017;7 doi: 10.1038/bcj.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elena C., Gallì A., Such E., Meggendorfer M., Germing U., Rizzo E. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128:1408–1417. doi: 10.1182/blood-2016-05-714030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossmann V., Kohlmann A., Eder C., Haferlach C., Kern W., Cross N.C.P. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 10.Itzykson R., Kosmider O., Cluzeau T., Mansat-De Mas V., Dreyfus F., Beyne-Rauzy O. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 11.Itzykson R., Kosmider O., Renneville A., Gelsi-Boyer V., Meggendorfer M., Morabito M. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 12.Itzykson R., Kosmider O., Renneville A., Morabito M., Preudhomme C., Adès L. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–2198. doi: 10.1182/blood-2012-06-440347. [DOI] [PubMed] [Google Scholar]
- 13.Kohlmann A., Grossmann V., Klein H.U., Schindela S., Weiss T., Kazak B. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28:3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 14.Meggendorfer M., Roller A., Haferlach T., Eder C., Dicker F., Grossmann V. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–3088. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meldi K., Qin T., Buchi F., Droin N., Sotzen J., Micol J.-B. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest. 2015;125:1857–1872. doi: 10.1172/JCI78752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merlevede J., Droin N., Qin T., Meldi K., Yoshida K., Morabito M. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7 doi: 10.1038/ncomms10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohamedali A.M., Alkhatabi H., Kulasekararaj A., Shinde S., Mian S., Malik F. Utility of peripheral blood for cytogenetic and mutation analysis in myelodysplastic syndrome. Blood. 2013;122:567–570. doi: 10.1182/blood-2012-12-471847. [DOI] [PubMed] [Google Scholar]
- 18.Orazi A., Bennett J., Germing U., Brunning R., Bain B., Thiele J. Chronic myelomonocytic leukaemia. In: Swerdlow S., Campos E., Lee Harris N., Jaffe E., Pileri S., Stein H., Thiele J., Vardiman J., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press, World Health Organization; Lyon, France: 2008. pp. 76–81. [Google Scholar]
- 19.Patel B.J., Przychodzen B., Thota S., Radivoyevitch T., Visconte V., Kuzmanovic T. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31:2815–2823. doi: 10.1038/leu.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patnaik M.M., Lasho T.L., Vijayvargiya P., Finke C.M., Hanson C.A., Ketterling R.P. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6 doi: 10.1038/bcj.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patnaik M.M., Zahid M.F., Lasho T.L., Finke C., Ketterling R.L., Gangat N. Number and type of TET2 mutations in chronic myelomonocytic leukemia and their clinical relevance. Blood Cancer J. 2016;6 doi: 10.1038/bcj.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patnaik M.M., Barraco D., Lasho T.L., Finke C.M., Hanson C.A., Ketterling R.P. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. Am J Hematol. 2017;92:56–61. doi: 10.1002/ajh.24581. [DOI] [PubMed] [Google Scholar]
- 23.Pleyer L., Germing U., Sperr W.R., Linkesch W., Burgstaller S., Stauder R. Azacitidine in CMML: matched-pair analyses of daily-life patients reveal modest effects on clinical course and survival. Leuk Res. 2014;38:475–483. doi: 10.1016/j.leukres.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Sallman D.A., Komrokji R., Vaupel C., Cluzeau T., Geyer S.M., McGraw K.L. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia. 2016;30:666–673. doi: 10.1038/leu.2015.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santini V., Allione B., Zini G., Gioia D., Lunghi M., Poloni A. A phase II, multicentre trial of decitabine in higher-risk chronic myelomonocytic leukemia. Leukemia. 2018;32:413–418. doi: 10.1038/leu.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savona M.R., Malcovati L., Komrokji R., Tiu R.V., Mughal T.I., Orazi A. An international consortium proposal of uniform response criteria for myelodysplastic/myeloproliferative neoplasms (MDS/MPN) in adults. Blood. 2015;125:1857–1865. doi: 10.1182/blood-2014-10-607341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Such E., Germing U., Malcovati L., Cervera J., Kuendgen A., Della M.G. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood. 2013;121:3005–3015. doi: 10.1182/blood-2012-08-452938. [DOI] [PubMed] [Google Scholar]
- 28.Traina F., Visconte V., Elson P., Tabarroki A., Jankowska A.M., Hasrouni E. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28:78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 29.Welch J.S., Petti A.A., Miller C.A., Fronick C.C., O'Laughlin M., Fulton R.S. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375:2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshizato T., Nannya Y., Atsuta Y., Shiozawa Y., Iijima-Yamashita Y., Yoshida K. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129:2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material