Abstract
Accumulation of aggregated α-synuclein into Lewy bodies is thought to contribute to the onset and progression of dopaminergic neuron degeneration in Parkinson's disease (PD) and related disorders. Although protein aggregation is associated with perturbation of proteostasis, how α-synuclein aggregation affects the brain proteome and signaling remains uncertain. In a mouse model of α-synuclein aggregation, 6% of 6215 proteins and 1.6% of 8183 phosphopeptides changed in abundance, indicating conservation of proteostasis and phosphorylation signaling. The proteomic analysis confirmed changes in abundance of proteins that regulate dopamine synthesis and transport, synaptic activity and integrity, and unearthed changes in mRNA binding, processing and protein translation. Phosphorylation signaling changes centered on axonal and synaptic cytoskeletal organization and structural integrity. Proteostatic responses included a significant increase in the levels of Lmp7, a component of the immunoproteasome. Increased Lmp7 levels and activity were also quantified in postmortem human brains with PD and dementia with Lewy bodies. Functionally, the immunoproteasome degrades α-synuclein aggregates and generates potentially antigenic peptides. Expression and activity of the immunoproteasome may represent testable targets to induce adaptive responses that maintain proteome integrity and modulate immune responses in protein aggregation disorders.
Keywords: Neurodegeneration, Parkinson's disease, Dopaminergic neurons, Immunoproteasome, Proteostasis
Highlights
-
•
Relative quantification of 6215 proteins and 8183 phosphopeptides in a mouse model of endogenous aggregation of α-synuclein
-
•
The immunoproteasome component Lmp7, is increased in the mouse model and postmortem human brains with Parkinson's and DLB
-
•
Lmp7 may represent an adaptive response to protein aggregation through aggregate elimination and innate immunity activation
Aggregated a-synuclein is a component of Lewy bodies a hallmark of Parkinson's disease. Changes in the brain proteome and phosphorylated peptides were quantified upon aggregation of α-synuclein in a mouse model. The levels and activity of LMP7, a component of the immunoproteasome, which is linked to proteostasis were increased indicating a potential adaptive response to α-synuclein aggregation.
1. Introduction
Cells have developed protein homeostasis networks to maintain proper cellular function and combat potentially toxic protein aggregation [[1], [2], [3]]. Failure to sustain proteostasis upon protein aggregation may contribute to the pathogenesis of several neurodegenerative diseases of aging such as Parkinson's disease, Alzheimer's disease and Amyotrophic lateral sclerosis [[1], [2], [3]]. These diseases are characterized by progressive misfolding and aggregation of proteins and ultimately neuron death.
However, it remains unclear how intracellular aggregation of proteins leads to neuron dysfunction and death. Studies in cellular model systems have shed some light on the pathological mechanisms of endogenous protein aggregation. Dynamic changes in the proteome of cultured cells following intracellular aggregation of artificial synthetic proteins indicated that the formation of amyloid-like aggregates attracted several interacting proteins, which were functionally linked to protein synthesis and quality control [4]. These findings were recently expanded to show that not only artificial synthetic proteins enriched in β-sheet structure but also fragments of mutant huntingtin and TAR DNA-binding protein 43 (TDP-43) peptides, which aggregate in human diseases, cause disturbances in the proteome by interfering with nuclear-cytoplasmic protein and RNA transport [5]. Collectively, these studies highlight the impact of protein aggregation on the proteome and proteostasis, and provide the intriguing hypothesis that the re-allocation of cellular resources to combat changes in the proteome upon endogenous protein aggregation leads to dysfunction and ultimately neuron death.
To explore, for the first time, protein aggregation induced changes in the proteome and signaling through phosphorylation in vivo we capitalized on the development of a mouse model of α-synuclein aggregation and the use of quantitative mass spectrometry (MS)-based proteomic technologies. α-Synuclein is a 140 amino acid protein that is predominantly localized to vesicles in pre-synaptic terminals [[6], [7], [8]] participating in the regulation of neurotransmitter release and synaptic plasticity [[9], [10], [11]]. Several point mutations, as well as multiplications, of the α-synuclein gene are associated with familial Parkinson's disease (PD) [[12], [13], [14]]. Moreover, highly organized amyloid-like fibrils and non-amyloid amorphous aggregates of non-mutant α-synuclein are deposited into Lewy bodies, cytoplasmic inclusions that serve as histopathological hallmarks of sporadic PD and other related neurodegenerative disorders [[15], [16], [17]].
To investigate the pathological mechanisms of α-synuclein aggregation, Luk et al. developed a mouse model of α-synuclein aggregation [18]. In this model, unilateral injection of preformed fibrils (PFFs) of recombinant wild type mouse α-synuclein into the striatum of non-transgenic mice induces progressive aggregation of endogenous α-synuclein, first in regions proximal to the injection site 30 days post injection (dpi) with further involvement of distally interconnected regions by 90 and 180 dpi. Injected mice developed significant dopaminergic neuron degeneration and impaired balance and motor coordination at 180 dpi [18,19]. Importantly, degeneration and α-synuclein inclusions within the nigrostriatal dopaminergic system are confined to the injected side and absent in the non-injected contralateral side of the brain at 180 dpi. Additionally, injection of PFFs into α-synuclein null (Snca−/−) mice fails to induce these effects, indicating that endogenous α-synuclein is required for aggregation and dopamine neuron degeneration [18]. This PFF injection model has been reproduced in mice, rats and non-human primates [[20], [21], [22], [23], [24]]. Therefore, this model (both wild type and Snca −/− injected mice) provides an opportunity for in vivo study of quantitative changes in the proteome upon aggregation of α-synuclein using MS-based proteomics and phosphoproteomics when used in combination with Stable Isotope Labeling in Mammals (SILAM) [25].
2. Methods
2.1. Animals
Wild type female, 2–3 month old, C57BL6/C3H mice were obtained from the Jackson Laboratories (Bar Harbor, ME). Snca−/− mice were maintained on a C57BL6 background. 13C-Stable Isotope Labeling in Mammals (SILAM) mouse brain tissue (C57BL6 female, l-Lysine-13C6, 97%) was purchased from Cambridge Isotope Laboratories, Inc. All housing, breeding, and procedures were performed according to the NIH Guide for the Care and Use of Experimental Animals and approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
2.2. Stereotaxic Injection of PFFs
For stereotaxic injections, the PFFs were diluted in sterile PBS and fragmented using a Bioruptor bath sonicator (Diagenode, Denville, NJ). Sonication was performed at high power for 10 cycles (30s on, 30s off, at 10 °C). Mice were anesthetized with ketamine hydrochloride (100 mg/kg, i.p.) and xylazine (10 mg/kg, i.p.). For each animal, PFFs were stereotaxically targeted into the ventral striatum (AP: +0.2 mm Bregma, lateral: 2.0 mm from midline, depth: 3.6 mm beneath the dura), dorsal striatum (AP: +0.2 mm, lateral: 2.0 mm, depth: 2.6 mm), and overlaying cortex (AP: +0.2 mm, lateral: 2.0 mm, depth: 0.8 mm). Injections were made through a single needle tract using 10 μL syringes (Hamilton, NV) at a rate of 0.1 μL per min (2.5 μL total per site) with the needle in place for ≥5 min at each target. Animals were monitored regularly following recovery from surgery. Mice were sacrificed at 90 days post injection by overdose with ketamine/xylazine. For biochemical studies, dorsal striatum and ventral midbrain from ipsilateral and contralateral sides were dissected and stored at −80 °C until used. For histological studies, the brain and spinal cord were removed after transcardial perfusion with PBS and underwent overnight post-fixation in either neutral buffered formalin (Fisher Scientific) or 70% ethanol (in 150 mM NaCl, pH 7.4), before being processed and embedded in paraffin.
2.3. Immunohistochemistry and Neuron Counting for Mouse Brain
Immunohistochemistry for α-synuclein phosphorylated at Ser-129 and tyrosine hydroxylase (TH) were performed on 6 μm thick coronal sections as previously described [18]. Digitized images of stained sections were acquired using a Perkin Elmer Lamina scanner at 20× magnification. Midbrain dopaminergic neurons belonging to the substantia nigra pars compacta and the ventral tegmental area were quantified from TH-immunostained coronal sections spanning the entire extent of the midbrain (every 9th section). Only intact neurons with visible nuclei and TH positive staining were included in the counting based on established criteria [26]. Statistical analysis between groups was compared using unpaired t-test.
2.4. Sample Preparation and LC-MS/MS Analysis
For each mouse injected, the midbrain and striatum of the injected and non-injected sides were individual dissected and kept separate. Two midbrain and striatum regions of the injected hemisphere were combined to generate one biological sample for the proteomic analysis. The same approach was employed for the non-injected side. Four biological samples for wild type and three for Snca−/− for each injected and non-injected side were analyzed through the proteomic workflow. Homogenates were prepared as described previously [27]. Briefly, brains were homogenized with a tissue grinder in cold urea buffer: 8 M urea, 75 mM NaCl, 50 mM Tris HCl pH 8.0, 1 mM EDTA, 2 μg/mL aprotinin (Sigma, A6103), 10 μg/mL leupeptin (Roche, 11017101001), 1 mM PMSF (Sigma, 78830), 10 mM NaF, 5 mM sodium butyrate, 5 mM iodoacetamide (Sigma, A3221), Phosphatase Inhibitor Cocktail 2 (1:100, Sigma, P5726), and Phosphatase Inhibitor Cocktail 3 (1:100, Sigma, P0044). Following 10 min centrifugation at 20,000g, protein concentration was determined by a BCA assay (Thermo, 23235). The supernatant was then combined with 13C-labeled brain lysates in a 1:1 ratio (5 μg). Samples were reduced for 45 min with 5 mM dithiothreitol followed by alkylation with 20 mM iodoacetamide for 45 min. Samples were then diluted 1:4 with 50 mM Tris HCl pH 8.0 (to reduce urea concentration to 2 M), then digested overnight with trypsin (Promega, V5111) at 37 °C overnight. 1% formic acid was added to the digests to remove urea by pelleting. The tryptic peptides were desalted by ultraMicro-Spin Vydac C18 column (Nestgroup, Inc., SUMSS18V). After peptide separation by high-pH reverse phase chromatography, 95% of peptides were combined in a concatenated pattern into 12 fractions for phosphoproteomic analysis. Lyophilized phosphopeptides fractions were re-suspended in 50% acetonitrile/0.1% trifluoroacetic acid (TFA) and then diluted 1:1 with 100% acetonitrile/0.1% TFA. These samples were then enriched for phosphorylation by incubation with 10 μL immobilized metal affinity chromatography (IMAC) for 30 min. Enriched IMAC beads were the loaded onto C18 silica-packed stage tips washed twice with 50 μL of 80% acetonitrile/0.1% TFA and 100 μL of 1% formic acid. Phosphopeptides were then eluted from IMAC beads with three washes of 70 μL 500 mM dibasic sodium phosphate, pH 7.0, (Sigma, S9763) and 2 washes of 100 μL of 1% formic acid. Elution from stage tips was then performed with 60 μL of 50% acetonitrile/0.1% formic acid. Washes were performed on a tabletop centrifuge at a maximum speed of 3500g. The peptides were analyzed by mass spectrometry (MS) and the data was analyzed with MaxQuant (described below). The SILAM ratio of light/heavy generated from the MaxQuant was converted to log2 scale and the median of the SILAM ratios therefore was calculated. If the SILAM ratio was close to 1:1, a larger scale sample prep was performed similarly as described above. Protein (2 mg heavy: 2 mg light) was digested with trypsin/Lys-C mix (Promega, V5073) at 1:25 enzyme: protein ratio. This protease mix has been reported to enhance mass spectrometry-based proteomics analysis by reducing the missed cleavages at lysine residue of a given peptide [44]. The peptide fragments were desalted on tC18 SepPak cartridge (Waters, WAT036815) and the peptides were lyophilized and stored in −80 °C. For reverse phase-HPLC, the peptides were reconstituted in 20 mM ammonium formate, pH 10.0. Peptide concentration was determined by UV280 before they were separated by high-pH reverse phage chromatography (Acquity UPLC H-Class instrument, Waters) to 72 fractions. Solvent A (2% acetonitrile, 5 mM ammonium formate, pH 10) and solvent B (90% acetonitrile, 5 mM ammonium formate, pH 10) were used to separate peptides with a ZPRBAX 300Extend-C18 column (4.6 mm x 250 mm, 5 Micron, Agilent). The gradient for separation was 1 mL/min flow rate as at 9 min, 100% A; 13 min, 94% A; 63 min, 71.5%; 68.5 min, 66% A; 81.5 min, 40% A; 83 min; 0% A; at 88–120 min with 1.2 mL/min with 100%A. Five percent of the samples were removed and recombined in a concatenated pattern into 24 fractions for proteomics analysis.
Peptide digests were analyzed on a hybrid LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled with a NanoLC Ultra (Eksigent Technologies). Mobile phase A consisted of 1% methanol/0.1% formic acid and mobile phase B consisted of 1% methanol/0.1% formic acid/79% acetonitrile. Peptides were eluted into the MS at 200 nL/min with each RP-LC run comprising a 15 min sample load at 3% B and a 90 min linear gradient from 5 to 45% B. The mass spectrometer repetitively scanned m/z from 300 to 1800 (R = 240,000 for LTQ-Orbitrap). FTMS full scan maximum fill time was set to 500 ms, while ion trap MSn fill time was 50 ms; microscans were set at one. FT preview mode, charge state screening, and monoisotopic precursor selection were all enabled with rejection of unassigned and 1+ charge states.
2.5. Database Searching, Construction of the Mouse Brain Reference Proteome and Proteomic Data Analysis
Protein identification was performed with MaxQuant (1.5.1.2) using a mouse UniProt database. Carbamidomethyl was defined as a fixed modification. The False Discovery Rate for peptides was set at 1%. Fragment ion tolerance was set to 0.5 Da. The MS/MS tolerance was set at 20 ppm. The minimum peptide length was set at 7 amino acids. The re-quantification option was left unchecked and the match-between-runs was turned on. For a protein to be quantified, the peptide must be identified at least once in light and once in heavy. To construct a unified set of literature and experimental proteins, UniProt accessions were cross-referenced by gene or protein name. For literature proteins, we selected several studies that used MS-based proteomics to identify proteins in the unperturbed mouse brain [[28], [29], [30]]. A reference proteome was generated combining the literature proteome with proteins identified in our experimental runs (Data file S4).
The Light-to-Heavy (L/H) ratio in the non-injected side was divided by the L/H ratio in the injected side to compute the injected/non-injected ratio of ratios. Resulting lists of proteins were analyzed using the Perseus software (http://www.coxdocs.org/doku.php) [31]. The H/L ratios reported by MaxQuant were inverted and log2 transformed. The mean, standard deviation, coefficient of variation, and principle component analysis (PCA) were calculated to examine the precision and repeatability of experiments. We used Z-score to scale the data by subtracting the mean of each column from the values and dividing by the standard deviation of the column.
Student's t-test and group averages were calculated and visualized in volcano plots within Perseus. Lists of statistically significant proteins with p-value <0.05 were selected and DAVID Bioinformatics Resources V6.8 was employed for Gene Ontology (GO) analysis. GO data was visualized using GO plot package in R [32]. The gene network analysis was performed using GeneMANIA (http://genemania.org/) prediction server [33]. Motif-x software tool was used for motif analysis of phosphoproteome using IPI Mouse Proteome as background.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009647.
2.6. Western Blot Analysis of Mouse Tissue
Mouse brain samples were extracted using the same method described above for the MS analysis. For each analysis, 5–20 μg of sample was added per lane, separated on 12% Bis-Tris Pre-Cast gels (Thermo Fisher) and transferred to PVDF membranes using 7.5% BSA in TBS for blocking. All primary antibodies were used overnight at 1:1000. Antibodies were used against TH (tyrosine hydroxylase; EMD Millipore, 657012 RRID:AB_696697), DAT (dopamine transporter; EMD Millipore, MAB369 RRID:AB_2190413), Ddc (aromatic-L-amino-acid decarboxylase; Abcam, ab3905 RRID:AB_304145), NSE (neuron specific enolase; Abcam, ab53025 RRID:AB_881756), PKC-β2 (protein kinase C beta-2; Abcam, ab32026 RRID:AB_779042), Akt (protein kinase B; Cell Signaling, 9272 RRID:AB_329827), Lmp7 (proteasome subunit beta type-8; Abcam, ab3329 RRID:AB_303708), and α-synuclein clone D37A6 (Cell Signaling Technology, #4179 RRID:AB_1904156) or clone Syn211 (Sigma-Aldrich Cat# S5566, RRID:AB_261518). Antigen-antibody complexes were detected using an Odyssey LC scanner (LiCor) after incubation with appropriate secondary antibodies. Densitometry was used to quantify intensity of protein bands.
2.7. Western Blot Analysis and Proteasome Activity of Human Tissue
Brain samples (amygdala, cortex) from dementia with Lewy bodies and non-disease brains that have been described previously [34,35] were extracted using high salt buffer containing 1% Triton-X100 (150 mM NaCl, 50 mM Tris, pH 7.6). Protease inhibitors were added to buffer prior to use. To collect triton-soluble fractions, samples were homogenized, sonicated and centrifuged at 15,000 rpm for 30 min. 500 μL of buffer was added to each sample. Protein concentrations were determined by BCA assay (Thermo Fisher). Samples were separated in a 12% Bis-Tris Pre-Cast gel (Thermo Fisher) and transferred to a PVDF membrane and blocked using 7.5% BSA in TBS. For each analysis 40 μg of sample was added per lane. All primary antibodies were used overnight at 1:1000. Antibodies were used against Lmp7 (proteasome subunit beta type-8; Abcam, ab3329 RRID:AB_303708), β5 (proteasome subunit beta type-5; Invitrogen, PA1-977 RRID:AB_2172052) and NSE (neuron specific enolase; Abcam, ab53025 RRID:AB_881756). Antigen-antibody complexes were detected using an Odyssey LC scanner (LiCor) after incubation with appropriate secondary antibodies. Densitometry was used to quantify intensity of protein bands. For proteasome activity, the brain samples were homogenized as described above in the absence of protease inhibitors and freshly prepared samples were analyzed using the proteasome activity fluorometric assay (BioVision K245). Assays were performed in a 96-well plate at 37 °C using 5 μg of brain lysate per experimental condition. Each condition was analyzed with or without the proteasome inhibitor lactacystin (Enzo Life Science PI104). After 60 min, the fluorescence with inhibitor was subtracted from the fluorescence without inhibitor in each sample. Three biological replicates were quantified for each experimental condition. Statistical analysis between groups was compared using unpaired t-test.
2.8. Immunohistochemistry for Human Brain
Human brain sections were obtained from the Brain Bank of the Honolulu Asian Aging Study [36]. The study is approved by the Kuakini Medical Center Institutional Review Board and participants signed informed consents. Five cases of PD and five healthy controls were examined. DAB and fluorescent Immunohistochemistry for Lmp7, IBA1, and GFAP was performed on 10 μm thick tissue sections. For colorimetric DAB staining, formalin fixed paraffin embedded sections were rehydrated in ethanol and diH2O and submerged in H2O2 for 30 min to quench endogenous peroxidase activity. Antigen retrieval was accomplished by microwaving the slides in a pressure cooker with R Buffer U (Electron Microscopy Sciences, 62,706-13). The tissue sections were then blocked at room temperature in 2% normal horse serum and incubated with Lmp7 antibody (Abcam, Ab3329, 1:5000) at 4 °C for 18 h. Slides were rinsed in 0.1 M Tris and incubated with a biotinylated secondary antibody, ABC, and DAB solution according to Vectastain instructions (Vector Labs, PK-6200 and SK-4100). Images were acquired using a Nikon Eclipse E800 microscope. For fluorescent triple labeling, tissue sections were also rehydrated in ethanol and diH2O and subjected to the same antigen retrieval and blocking steps as described previously. Slides were then incubated with the same Lmp7 antibody at 1:2500 concentration, along with antibodies against IBA1 (ionized calcium binding adaptor molecule 1; Abcam, ab5076 RRID:AB_2224402, 1:1000) and GFAP (glial fibrillary acidic protein; Millipore, NE1015 RRID:AB_2043416, 1:2000) antibodies at 4 °C for 18 h. Slides were then rinsed in 0.1 M Tris and incubated with corresponding Alexa Fluor-conjugated secondary antibodies (Invitrogen, A10042 RRID:AB_2534017; A21202 RRID:AB_141607; A21447 RRID:AB_2535864) for 1 h at room temperature. Slides were rinsed again in 0.1 M Tris and incubated with Hoechst 33,342 (Life Technologies, H3570), mounted with Fluormount G (Southern Biotech, 0100–01) and cover-slipped. I mages were acquired using a Nikon A1RSI laser scanning confocal microscope.
2.9. In Vitro Degradation Assay
Purified human WT α-synuclein was aggregated at 5 mg/mL for 7 days at 1400 rpm at 37 °C. The fibrils generated from the aggregation of α-synuclein were used as substrate for the in vitro degradation assays. Myelin Basic Protein (Sigma M1891) was used at 25 μM and α-synuclein was used at 1 mg/mL. Human Immunoproteasome 20S (Enzo BML-PW8720) and Human Proteasome 20S (Enzo BML-PW9645) were used at a ratio of 0.11:1 proteasome:α-synuclein. Human Proteasome Activator 11S complex (BML-PW9420) was added at a final concentration of 500 nM. Reactions were incubated at 37 °C agitating at 600 rpm, and samples were removed at indicated time points. Samples were separated in a 12% Bis-Tris Pre-Cast gel (Thermo Fisher) and stained with colloidal blue. Densitometry was used to quantify protein degradation of monomer bands of α-synuclein. Initial time point was considered 100% for each experimental condition. Three biological replicates were quantified for each experimental condition. For mass spectrometry analysis of the immunoproteasome peptides, the samples were removed following the indicated incubation time and frozen prior to processing. For the processing, a modified enhanced filter aided sample preparation (eFASP) was used. Briefly, 10 kDa cutoff microcon filters (Millipore) were first passivated in a solution of 5% Tween 20 overnight to reduce peptide binding, followed by extensive washes with dH2O. The samples were then transferred to the filters and centrifuged at 14,000 ×g for 10 min, retaining the flow through containing the peptides. Sample volume was then reduced to roughly 50 μL by SpeedVac. Contaminants in the sample were removed using C18 Stage Tips (Thermo Electron) and the samples were transferred to autosampler vials. LC-MS/MS conditions and instrument settings proceeded as above with the exception that the samples were not fractionated prior to analysis. The raw data was searched against a database constructed with the human sequence of α-synuclein, proteasome and immunoproteasome subunits using MaxQuant (1.5.1.2) with peptide cleavage set to nonspecific and a FDR of 1%.
2.10. Cell Treatments
U-251 cells were grown in high glucose DMEM supplemented with 10% FBS, 1% penicillin and streptomycin, 1% glutamine and 1% HEPES. Cells were plated at low density and allowed to adhere for 24 h, after which cells were pretreated with 0.1 μg/mL human interferon gamma (Pepro Tech Inc.) alone or in conjunction with the Lmp7 inhibitor ONX 0914 (Cayman Chemicals) at a final concentration of 0.2 μM. Following incubation overnight, 5 μg/mL α-synuclein fibrils were added to the media and incubated for 4d. The cells were washed 3× with PBS and lysed in PBS + 1% Trition-X100. 12 μg of protein was loaded into each lane and the western proceeded as described above. Primary antibodies to α-synuclein (clone Syn211, SigmaAldrich, S5566 RRID:AB_261518), Lmp7 (proteasome subunit beta type-8; Abcam, ab3329), and Actin (Sigma Aldrich, A2066 RRID:AB_476693) were incubated with the membrane at a concentration of 1:1000 overnight at 4 °C (Syn211) or for 1 h at room temp (Lmp7, Actin). IR Dye conjugated anti-mouse or anti-rabbit secondary was incubated at 1:5000 for 1 h and the blots were visualized using an Odyssey LC scanner (LiCor).
3. Results
3.1. Mouse Model of α-Synuclein Aggregation
To identify proteomic and signaling perturbations induced by the aggregation of α-synuclein, a modified version of the recently developed and independently verified mouse model was employed [18,20,21]. α-Synuclein aggregation in non-transgenic mice was induced by unilateral injection of PFFs into three locations, the motor cortex, dorsal and ventral-striatum. PFFs were generated by aggregating purified recombinant mouse monomers of α-synuclein to insoluble amyloid-like fibrils exhibiting typical β-sheet secondary structure, then sonicating them to produce smaller, β-sheet PFFs (Fig. S1). The changes in the proteome and phosphoproteome were quantified 90 dpi, a time point that is characterized by aggregation of α-synuclein, but prior to significant dopaminergic neuron loss [18].
Although this model has been extensively validated, dopaminergic neurons belonging to the substantia nigra pars compacta were stereologically quantified from tyrosine hydroxylase (TH) stained coronal sections spanning the entire extent of the midbrain at 90 dpi [18]. A non-significant 19 ± 6% loss of substantia nigra dopaminergic neurons was quantified in the injected side of wild type mice, (n = 3, t-test, p-value = .0507) as compared to the non-injected side (Fig. S2C). In contrast, no changes in TH staining was observed in the injected side of Snca−/− mice (Fig. S2C), consistent with the previous results [18]. An antibody that recognizes phosphorylated α-synuclein at Ser-129 (pS129), a marker for α-synuclein inclusions in human disorders was used to stain brain tissue obtained at 90 dpi [[37], [38], [39]]. Consistent with the previous report [18], Lewy body-like inclusions immunoreactive for pS129 were abundant in the substantia nigra (Fig. S2A) and striatum (Fig. S2B) ipsilateral to the injection site by 90 dpi. In contrast, no staining was detected in the same areas on the non-injected side at this time point. Total levels of mouse α-synuclein are unchanged in the injected and non-injected side (Fig. S2E). Moreover, there were no detectable pS129 α-synuclein inclusions in the brains of Snca−/− mice injected with PFFs in the same manner (Fig. S2). Collectively, these data confirmed the presence of PFF-triggered α-synuclein aggregation in the ipsilateral injected side of wild type mice, which is associated with mild TH-positive neuron loss, consistent with previous findings in this model [18].
3.2. Acquisition and Overview of the Proteome and Phosphoproteome
A quantitative proteomic workflow depicted in Fig. 1A was implemented to quantify changes in the relative abundance of proteins in both wild type and Snca−/− mice. The method incorporated the use of [13C6-lysine]-SILAM mouse brain as an internal “heavy” standard. Detailed examination of the [13C6-lysine]-containing internal reference peptides showed excellent reproducibility (average Pearson correlation coefficient = 0.87, 80% overlap) (Fig. S3). Although the [13C6-lysine] internal standard was derived from the entire brain, whereas the “light” samples were derived from combining the midbrain and striatum regions only, we report high correlations and tighter spreads of the corresponding heavy-to-light peptide pairs (average Pearson correlation = 0.96) than the heavy-to-heavy peptide pairs (Fig. S3). This improvement likely arises from the coincident sample processing workflow of the SILAM experiment as opposed to the parallel sample processing shown by the heavy-to-heavy comparisons. This analysis validated the use of [13C6-lysine]-SILAM mouse brain as an appropriate standard for accurate proteome quantification.
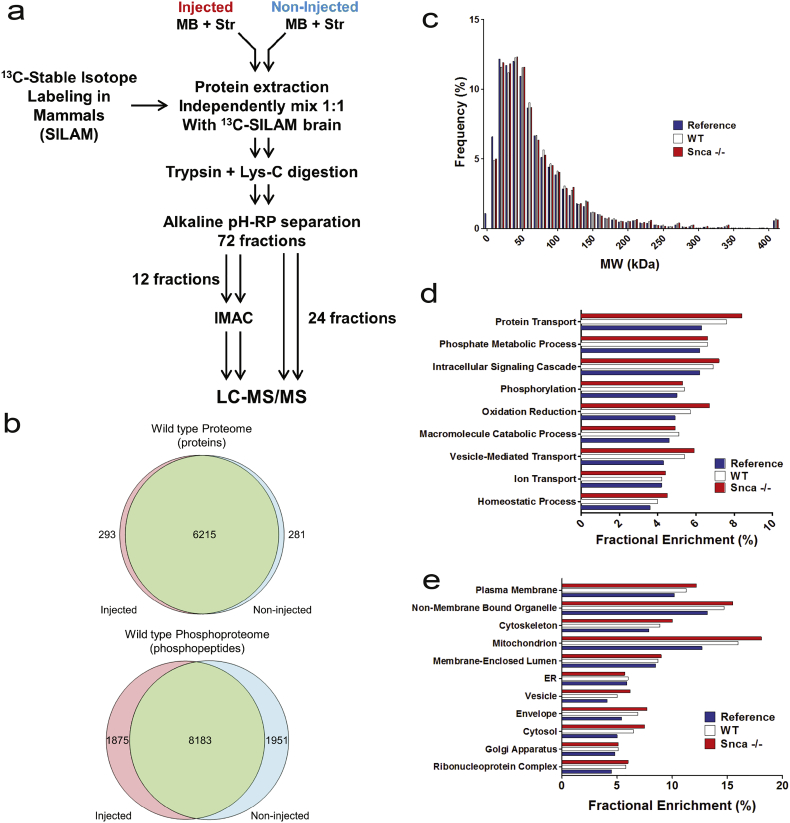
Fig. 1.
Workflow for the acquisition and overall description of proteomes. (a) Overview of proteomic workflow. The midbrain and striatum from the brains of two mice were dissected and combined to generate one sample replicate. The ipsilateral injected and the contralateral non-injected sides were kept separate and processed throughout the workflow independently. Immobilized metal affinity chromatography (IMAC) was used to enrich for phosphorylated peptides. (b) Venn diagrams depicting the number of proteins and phosphopeptides quantified in 4 independent sample replicates in the injected side and non-injected side in wild type mice. The acquired proteomes in wild type and Snca−/− showed typical distribution in terms of molecular weight, cellular location and major biological processes when compared to a whole mouse brain proteome: (b) Distribution of the proteins quantified in wild type and Snca−/− mice based on protein molecular weight. (d) Gene ontology enrichment analysis using cellular components (e) Gene enrichment using biological process and KEGG pathways.
The quantitative proteomic analysis was restricted to brain regions of pathological interest, which include the midbrain and the striatum. These two regions, dissected from two mice, were combined to generate sufficient protein for the proteomic analysis. Using this approach the proteomic analysis included four replicates derived from the injected and non-injected sides of wild type and three replicates obtained from each side of the similarly injected Snca−/− mice. Combined analysis of the replicates in wild type mice applying a false discovery rate of 1% and removal of contaminants and reversed sequences resulted in the identification of 6508 proteins with a light-to-heavy (L/H) ratio in the injected side, 6496 in the non-injected side, with 6215 proteins identified in both sides (Fig. 1B). Analysis in the Snca−/− mice resulted in the identification of 4661 proteins with L/H ratio in the injected side, 4994 in the non-injected side, with 4460 proteins identified in both sides (Fig. S4A). Immobilized metal affinity chromatography (IMAC) enrichment for phosphopeptides resulted in the identification of 10,058 phosphopeptides in the injected and 10,134 in the non-injected side of the wild type mice; 8183 phosphopeptides were common between the two sides. Similarly, 10,285 and 9775 phosphopeptides were identified in the Snca−/− mice; 8031 were shared between the two sides (Data file S1 and Fig. S4B).
In order to determine the extent by which the proteins quantified in this study are reflective of the proteins expressed in the mouse brain, and to protect against potential biases during the acquisition of the proteomic data, we constructed a global mouse brain proteome. For the mouse brain proteome we combined our non-injected wild type proteome with proteins identified by mass spectrometry in unperturbed mouse brain from several existing studies [[28], [29], [30],40]. The resulting proteome comprised of 11,055 unique proteins was curated through UniProt (http://www.uniprot.org). The distribution of the proteins quantified in our study was evaluated based on their molecular weight, cellular localization and biological function against the reference proteome. The proteins quantified were representative of the whole mouse brain proteome covering the entire range of molecular weights >10 kDa to >400 kDa (Fig. 1C) all major cellular compartments (Fig. 1D) and major biological processes (Fig. 1E). Together, these analyses indicated that the number of proteins quantified through this workflow in the midbrain and striatum are compatible with recent studies [40,41] and are representative of the proteins expressed in the mouse brain.
3.3. Analysis of the Proteome and Phosphoproteome Reveals Selective Disease-Related Changes
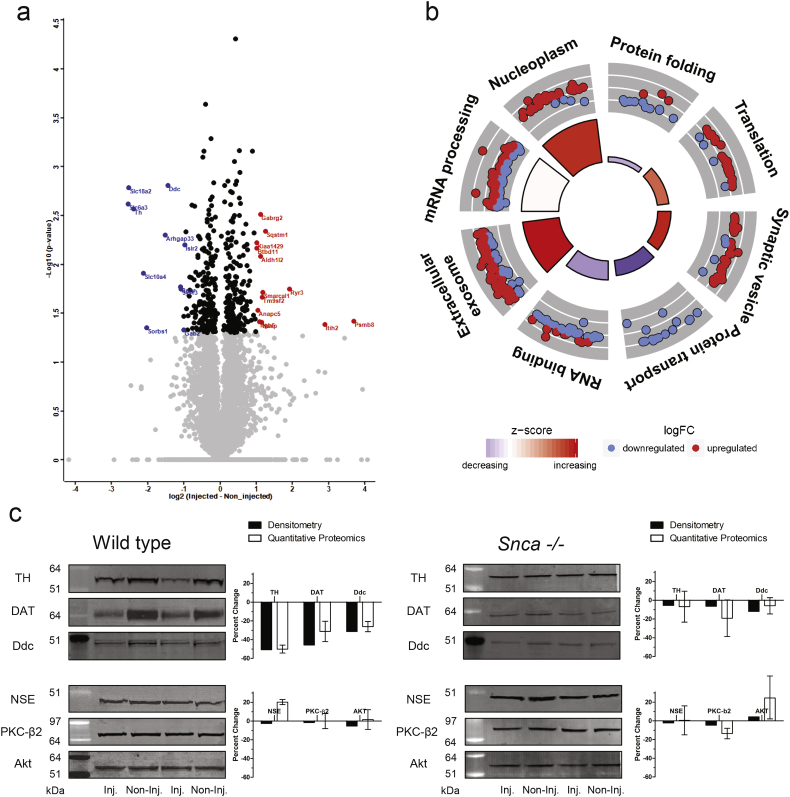
Changes in the relative abundance of proteins were quantified by a ratiometric method, dividing the L/H ratio of the non-injected side by the L/H ratio in the injected side. The data was Log2 transformed, the coefficient of variance for each experiment determined, and the data scaled using the standard z-score. Statistically significant changes in proteins (t-test, p < 0.05) between the injected and non-injected sides were identified (Data file S2) and typical volcano plots integrating the p-value and the magnitude change were generated. The relative abundance of 377 of 6215 in wild type and 112 of 4460 proteins in the Snca−/− mice changed significantly in the injected side (Fig. 2A; Fig. S5). Similarly, 132 of 8183 phosphopeptides in wild type and 109 of 8031 in the Snca−/− mice showed significant change (Fig. 3A; Fig. S5). There was a minimal overlap between the proteins that showed changes in the injected side of the wild type and Snca−/− mice (7 proteins), indicating that the changes in the wild type were driven primarily by the aggregation of α-synuclein. This analysis indicated that aggregation of α-synuclein does not induce large remodeling of the proteome or phosphorylation-dependent signaling. Ontological and functional analysis of the 377 proteins is depicted in the circular plot (Fig. 2B) in which the outer circle is a scatter plot for each biological term of the logFC (fold change) of the enriched proteins and the size of the inner trapezoids correspond to the adjusted p-value and the colour indicates the z-score. Some of these functional protein clusters (extracellular exosome, synaptic vesicle) have been previously linked to α-synuclein and may represent adaptive response to maintain proteostasis and synaptic function.
Fig. 2.
Analysis of Quantified Proteins. (a) Volcano plot showing the 377 proteins (black circles) that changed significantly (p < 0.05) in relative abundance in the injected side as compared to the contralateral non-injected side of wild type mice. The red dots indicate the proteins with >2-fold increase and the blue dots the proteins with >2-fold decrease in relative abundance. (b) Ontological analysis of the 377 proteins, which showed significant changes in relative abundance between the ipsilateral injected side and the contralateral non-injected side in the wild type mice. The circular plot depicts the enriched functional networks of proteins. The outer circle is a scatter plot for each biological term of the logFC of the enriched proteins. Within each network, a protein is depicted as a dot, red for upregulated and blue for downregulated. The size of the inner trapezoids correspond to the adjusted p-value and the colour indicates the z-score as calculated by GOplot algorithm. (c) Representative immunoblots from two independent biological replicates for the significantly changed proteins TH, DAT, and Ddc paired with NSE, PKC-β2 and Akt as unchanged protein controls. The graphs indicate the mean ± sem for protein levels quantified by the SILAM-MS based method and the average of two independent western blot analysis. The relative abundance of NSE, PKC-β2 and Akt did not change in the injected wild type or Snca−/− mice.
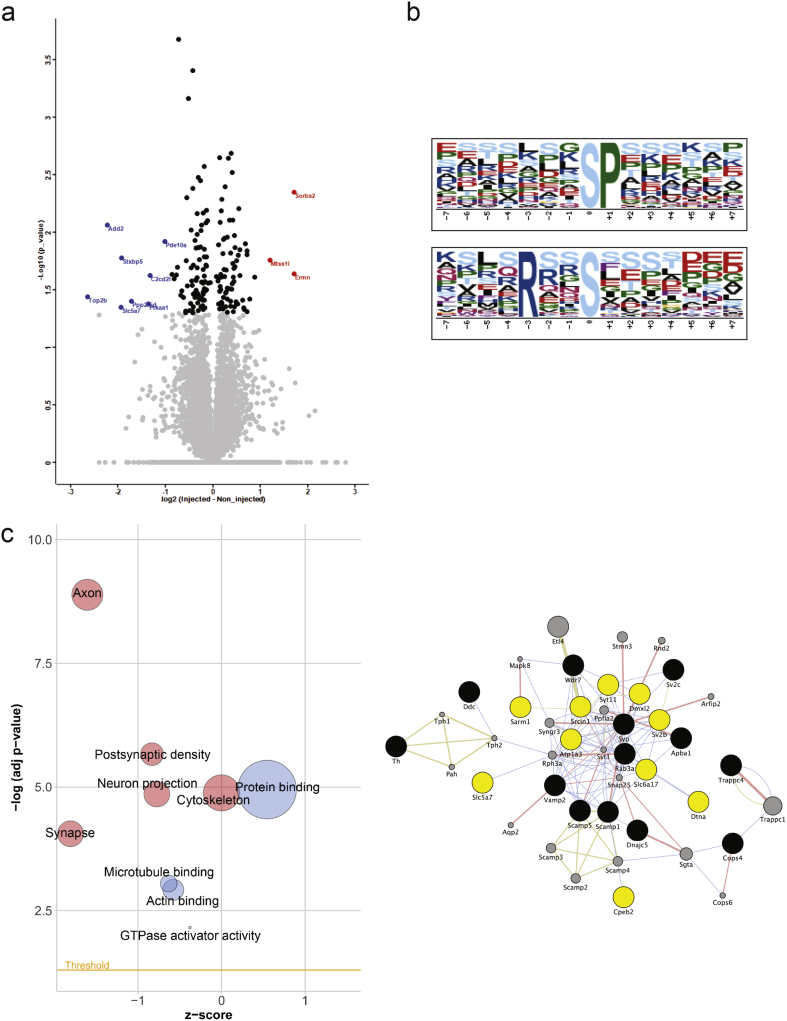
Fig. 3.
Analysis of Quantified Phosphopeptides. (a) Volcano plot showing the 132 phosphorylated proteins (black circles) that changed significantly (p < 0.05) in relative abundance in the injected side as compared to the contralateral non-injected side of wild type mice. The red dots indicate the phosphoproteins with >2-fold increase and the blue dots the phosphoproteins with >2-fold decrease in relative abundance. (b) Motif analysis of the 132 phosphopeptides, which showed a significant change in relative abundance. Enrichment for two motifs was discovered; the XXSP motif, a known target for phosphorylation by MAP kinase and RXXS indicating substrates for CaMK-II kinase. (c) Bubble-plot indicating the cellular components (red) and molecular functions (blue) that were significantly enriched among the 132 phosphopeptides. The area of the displayed circles is proportional to the number of proteins. (d) Construction of a synaptic network from the 377 proteins and 132 phosphopeptides. Black circles indicate proteins that are functionally assigned to synaptic vesicles and showed significant changes in relative abundance. The yellow circles indicate phosphoproteins assigned to synapse and showed significant changes in relative abundance. The gray circles are proteins imported based on known interactions. Physical interactions are depicted as pink edges; associations based on co-localization (blue) and shared protein domains (gold).
The proteomic analysis identified five proteins selectively expressed in dopaminergic neurons among the proteins that decreased in abundance in the injected side of wild type mice. The analysis showed a significant decline in the levels of TH 50 ± 3%, aromatic-L-amino-acid decarboxylase (Ddc) 26 ± 4%, dopamine transporter (DAT; Slc6a3) 31 ± 9%, synaptic vesicular monoamine transporter-2 (VMAT2; Slc18a2) 42 ± 8% and Slc10a4 (specific function is unclear but is specifically expressed in dopaminergic neurons) [42,43] 44 ± 3%. In contrast, the relative abundance of monoamine oxidase A (MAO-A) and catechol-O-methyl transferase (COMT) that metabolize dopamine and are expressed in both glia cells and dopaminergic neurons, did not change significantly (increased by 3 ± 4% and 8 ± 4%, respectively). This finding indicates that, despite possible dilution of the proteome by multiple cell types, our quantitative proteomic approach was capable of identifying specific changes in dopaminergic neurons. Moreover, the decline in the abundance of proteins critical for dopamine synthesis, transport, and recycling exceeded the loss of dopamine neurons (Fig. 2S) suggesting ongoing dopamine neuron dysfunction consistent with the progressive injury in the setting of α-synuclein aggregation. As a reference, the abundance of TH, Ddc and DAT did not change in the injected side of Snca−/− mice when compared to the contralateral, non-injected side.
Finally, the changes in the levels of TH, DAT and Ddc quantified by SILAM-MS-based methodology were further confirmed by western blot analysis. The western blot analysis matched closely the MS-based quantification showing similar decline in the levels of TH, DAT and Ddc in the injected versus the non-injected side (Fig. 2C). The levels of TH, DAT and Ddc were unchanged in the injected side of Snca−/− mice when quantified by western blot (Fig. 2C). As controls for the western blots, we probed for unchanged proteins as determined by the MS analysis. NSE, PKC-β2 and Akt, showed no change in the levels between injected and non-injected side in wild type and Snca−/− mice (Fig. 2C) by both MS-based quantification and western.
Although a relatively small number of phosphopeptides showed significant change in the injected side of the wild type mice (132 phosphopeptides identified in 125 proteins, Fig. 3A), the data was mined for kinases and functional pathways impacted by α-synuclein aggregation. First, we performed a motif analysis to identify possible sequences targeted by kinase families. This analysis revealed the enrichment for two motifs (Fig. 3B). The first motif, X(any amino acid)-X-Serine-Proline is a known minimal sequence motif for phosphorylation by mitogen-activated protein kinases (MAPK), which have been implicated in α-synuclein aggregation related disorders [[44], [45], [46], [47]]. The second motif, Arginine-X-X-Serine-X indicates the minimal motif for phosphorylation by Ca2+/calmodulin-dependent protein kinase II (CaMK-II), a serine/threonine kinase enriched at synaptic sites [[48], [49], [50]]. To further explore phosphorylation signal networks, bubble plots of the enriched cellular components and molecular functions were constructed using the z-score and the negative logarithm of the adjusted p-value (Fig. 3C). The changes in phosphorylation signaling localize primarily in synapses and axons, and functionally relate to proteins that regulate axonal and synaptic cytoskeletal organization and structural integrity (Fig. 3C, Data file S3). Moreover, when the changes in synapse associated phosphopeptides were combined with changes in synaptic vesicle proteins (Fig. 2B), the integrated network highlighted numerous physical interactions indicative of functional changes to synaptic architecture and secretory processes in response to α-synuclein aggregation occurring before significant neuron loss (Fig. 3D).
3.4. Upregulation of the Immunoproteasome in α-Synuclein Aggregation Disorders
Among the significantly changed proteins a prominent increase in the relative levels of the proteasome subunit beta type-8 (Psmb8; common name, low molecular mass protein 7; Lmp7), was quantified in the injected side of wild type mice (Fig. 2A). The increase in relative abundance of Lmp7 by MS in the injected side of wild type mice was validated by western blot analysis (Figs. S6 and S7). Lmp7 is one of the three subunits of the catalytic core of the immunoproteasome [[51], [52], [53], [54]], an inducible form of the proteasome assembled in response to inflammatory stimuli, primarily IFNγ [[55], [56], [57], [58]]. The immunoproteasome is defined by the replacement of catalytic subunits β1, β2 and β5 of the proteasome with proteasome subunit beta type-9 (low molecular mass protein 2; Lmp2), proteasome subunit beta type-10 (multicatalytic endopeptidase complex subunit-1; MECL-1) and Lmp7, respectively [[51], [52], [53], [54]].
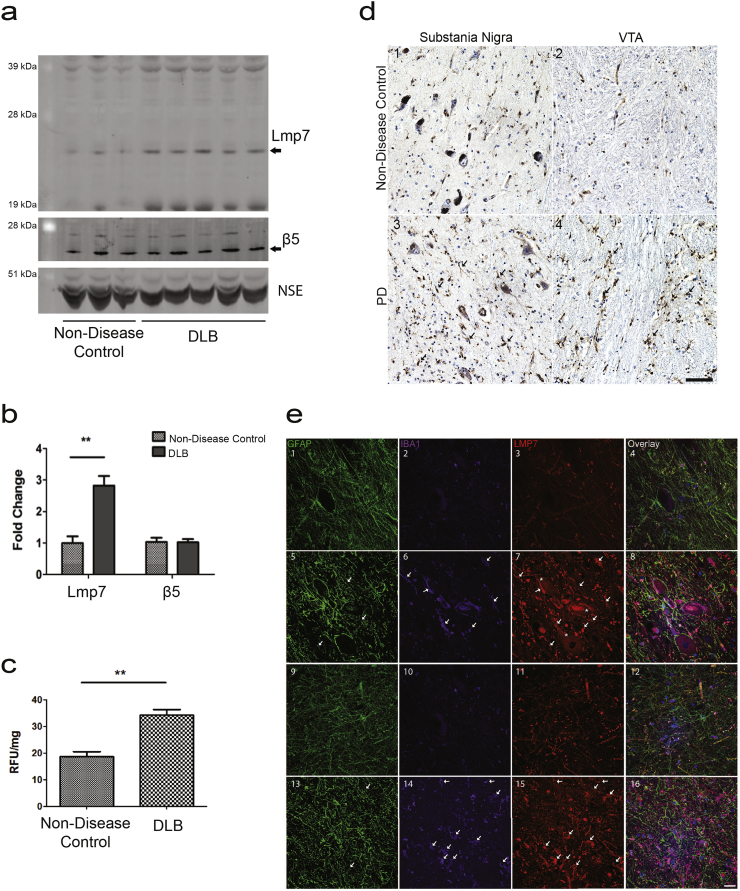
Since there are no previous studies linking α-synuclein aggregation to immunoproteasome induction and its potential role in proteostasis, we examined whether a similar increase of the immunoproteasome levels and activity occurs in human disease. We compared the levels of Lmp7 in the brains of dementia with Lewy bodies (DLB), which contain aggregated α-synuclein [16,17,35], and non-disease control subjects. The analysis revealed a 3-fold increase in the levels of Lmp7 in DLB as compared to controls but no change in the levels of constitutive proteasome catalytic subunit β5 (Fig. 4A and B). Chymotrypsin-like activity, which is selectively elevated in the immunoproteasome [52,53], was quantified to evaluate if the increase in protein levels correlates with activity. An 80% increase in the lactacystin-sensitive, chymotrypsin-like activity was quantified in the DLB brain homogenates as compared to controls (Fig. 4C).
Fig. 4.
The Immunoproteasome in Human Disease Driven by α-Synuclein Aggregation. (a) Representative western blot analysis in human DLB and non-disease control brains for Lmp7 and the proteasome protein β5. (b) Densitometric analysis of western blots normalized to the levels of NSE protein indicate a significant increase (**p < 0.01, t-test) in Lmp7 levels in DLB brains (n = 5) compared with controls, (n = 3). (c) Concomitant with the increase in Lmp7 protein levels the chymotrypsin-like activity, a proxy for immunoproteasome activity, is increased by 189% in DLB brain compared with control brain (**p < 0.01, n = 3, t-test). (d) Immunohistochemical staining revealed increased immunoreactivity for Lmp7 in PD substantia nigra and VTA. (d1) Representative staining for Lmp7b in control substantia nigra and (d2) VTA. Increased Lmp7 staining in both neurons (asterisks) and glia (arrows) in PD substantia nigra (d3) and VTA (d4). Nuclei were counterstained with hematoxylin (blue). Scale bar = 50 μm. (e) Co-localization of Lmp7 with glial markers. Representative fluorescent staining for the astrocyte marker, GFAP; microglial marker, IBA1; and Lmp7. Overlay of GFAP, IBA1, LMP7 and Hoechst nuclear stain is displayed in the right column. Staining in non-disease control substantia nigra (e1–4) and in VTA (e5–8). Increased staining for Lmp7 in the PD substantia nigra (e9–12) and in the VTA (e12–16). Lmp7 co-localized with both astrocytes and microglia. Staining increased in glial processes and soma (arrows) and neurons (asterisks) in PD substantia nigra relative to normal controls. Lmp7 staining increased in astrocytes and microglia (arrows) of PD VTA relative to non-disease controls. Scale bar = 20 μm.
Immunohistochemical analysis of PD (n = 5) and non-disease control tissue (n = 5) showed increased staining for Lmp7 in both the substantia nigra and ventral tegmental area (VTA) in PD tissue (Fig. 4D). Increased staining for Lmp7 was observed in both neurons and glia in the substantia nigra of PD cases but not in control cases (Fig. 4D and E). Neuronal staining occurred in pigmented and non-pigmented neurons in PD substantia nigra. In PD ventral tegmental area, Lmp7 staining occurred primarily in glia, while neuronal Lmp7 was not detected in PD or control VTA. In glia and neurons, Lmp7 staining was detected throughout both cell bodies and processes. Lmp7 localization was further characterized by fluorescent triple labeling of astrocytes, microglia and Lmp7 (Fig. 4E). In the VTA of PD cases, 47 ± 10% of IBA-1 positive cells, a marker for microglia, and 40 ± 4% of GFAP, an astrocytic marker, co-localized with Lmp7 (n = 4). Overall, the biochemical and histological data demonstrate activation of the immunoproteasome in human diseases characterized by α-synuclein aggregation.
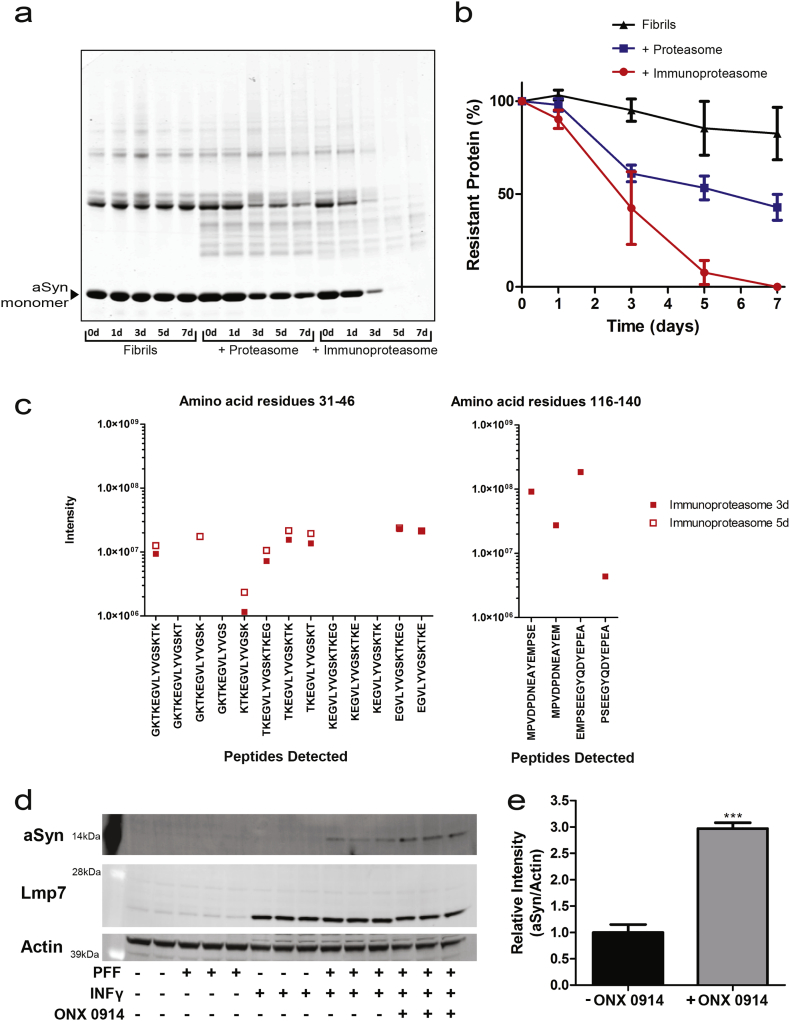
To investigate the potential significance of the immunoproteasome we explored its ability to degrade α-synuclein fibrils in vitro. For these experiments, we used the 20S immunoproteasome and the 20S proteasome, which degrade proteins without the need for ubiquitin tagging or the presence of the 19S regulatory particle [59]. First, we secured that the purified immunoproteasome degraded myelin basic protein (MBP), a selective substrate [60]. After 22 h, 96% of MBP was degraded by the immunoproteasome and 69% by the proteasome (Fig. S8). Next, α-synuclein fibrils generated from purified human monomeric α-synuclein with typical β-sheet secondary structure were used as substrate. The immunoproteasome was capable of degrading α-synuclein fibrils in a time dependent manner achieving >50% degradation by 72 h and nearly complete degradation after 5 days of incubation (Fig. 5A and B). We further explored the possible consequences of immunoproteasome-mediated α-synuclein fibril degradation. Since one of the main functions of the immunoproteasome is to produce peptides for antigen presentation, we tested if the degradation products exhibit peptide sequences that have been found to elicit MHC I and MHC II responses [61]. Mass spectrometry detected 1111 unique peptide sequences covering the entire 140 α-synuclein amino acid residues (Data file S5). Further refinement of the detected peptides indicated that the immunoproteasome produced multiple peptides within the antigenic regions of α-synuclein [61], specifically between residues 31–46 and 116–140 (Fig. 5C). Finally, to demonstrate that the immunoproteasome degrades α-synuclein fibrils in intact cells, we used U-251 cells, a human glioblastoma cell line that does not express endogenous α-synuclein and minimally detectable basal levels of Lmp7. The only source of α-synuclein in this cell model is the exogenous α-synuclein PFFs added to the culture media. In these cells, α-synuclein PFF uptake appears to be dependent on inflammatory activation as evidence by the lack of uptake of exogenous α-synuclein in PFF only treated cells (Fig. 5D). The link between inflammatory activation and α-synuclein uptake has been previously documented [62]. Concurrent treatment with INFγ induces expression of Lmp7 and intracellular accumulation of exogenous α-synuclein. INFγ only treated cells show the expected increase in Lmp7, but there is no α-synuclein available to detect in these cells. Intracellular accumulation of the exogenous α-synuclein PFFs was quantified in the absence or presence of the Lmp7 specific inhibitor ONX-0914 [63]. Inhibition of the immunoproteasome results in greater intracellular accumulation of α-synuclein (Fig. 5D and E) without altering Lmp7 levels. Taken together, the data suggest a possible neuroprotective role for the induction of the immunoproteasome to maintain proteostatic networks in response to α-synuclein aggregation.
Fig. 5.
Degradation of α-Synuclein preformed fibrils by the immunoprotesome.
(a) Representative immunoblot for the SDS-soluble fraction of α-synuclein fibrils in the absence or presence of purified 20S proteasome or immunoproteasome for the indicated amount of time. Incubation of pre-formed α-synuclein fibrils with purified 20S proteasome or immunoproteasome resulted in a time-dependent decrease of oligomeric and monomeric α-synuclein. (b) Quantification of the ~14 kDa α-synuclein band corresponding to monomeric protein in the SDS-soluble fraction. (n = 3). (c) Analysis by mass spectrometry of the α-synuclein peptides generated after the degradation of α-synuclein fibrils by the purified immunoproteasome. The relative intensity of the peptides generated within the presumed antigenic regions, residues 31–46 and residues 116–140 is shown after 3 and 5 days of incubation. (d) Immunoblot analysis of α-synuclein and Lmp7 in U-251 cells. Cell were treated with 0.1 μg/mL INFγ for 24 h prior to exposure to α-synuclein preformed fibrils. To ascertain the contribution of immunoproteasome in the degradation of internalized α-synuclein fibrils cells were treated with the Lmp7 specific inhibitor ONX-0914. (e) Quantification of the normalized intensity α-synuclein in U-251 cells after induction of Lmp7 and α-synuclein preformed fibril uptake in the absence or presence of the Lmp7 specific inhibitor ONX-0914, (***p < 0.001, n = 3, t-test).
4. Discussion
Here, we quantified changes in the proteome and phosphorylation signaling in a validated, non-transgenic mouse model of α-synuclein aggregation. The proteomic approach achieved quantification of >6000 proteins and 8000 phosphoproteins, with typical representation of proteins based on size, localization, and biological function consistent with similar MS-based proteomic studies using the entire mouse brain [30] or specific regions of mouse brain [41] indicating a reasonable coverage and representation of the proteome. The proteomic workflow enabled the quantification of 46 proteins that have been linked to Parkinson's disease or associated with Lewy body formation and 220 of 226 proteins that were recently linked to α-synuclein [64]. Contrary to expectations, the changes in the proteome and phosphorylation signaling pathways induced by aggregation of α-synuclein before significant dopaminergic neuron death are minimal and for the most part proteostasis is preserved. Despite the absence of widespread remodeling of the proteome, the data provided unique insights for the responses to protein aggregation, revealing selective changes in protein clusters that participate in dopamine neurotransmission, maintenance of neuronal and synaptic architecture and immune responses. These selective changes were absent in PFF-treated Snca−/− mice.
Ontological analysis of the changes in the proteome and phosphoproteome revealed both expected and novel findings. Notably, and consistent with the anticipated pathology, the data revealed selective and significant decrease in the relative abundance of proteins that regulate dopamine biosynthesis (TH, Ddc), transport and storage (VMAT-2) and recycling (DAT). The decline in the levels of TH, Ddc, VMAT-2 and DAT was greater than the magnitude of neuron loss supporting previous observations that alteration in synaptic proteins and function precedes dopaminergic neuron death [[65], [66], [67], [68]]. This observation is further reinforced by the quantification of significant changes in proteins that regulate synaptic vesicles and signaling through phosphorylation for proteins participating in the organization synaptic architecture and function. These results support previous reports [67,69,70] including observations in humans [69,71] that perturbations of synaptic protein function and cytoskeletal integrity precede neurodegeneration in α-synuclein aggregation disorders.
Additional pathways impacted by the aggregation of α-synuclein included changes in mRNA processing, binding, protein transport and nucleoplasmic function. These changes are consistent with the alterations in nuclear-cytoplasmic protein and RNA transport observed in models of mutant huntingtin and TDP-43 neurotoxicity [5]. Significant changes in relative abundance were also quantified for 112 proteins that are annotated as extracellular and proteins associated with exosomes. Augmentation of extracellular protein release, including exosomes, in response to α-synuclein has been reported [[72], [73], [74]] and may represent neuronal attempt to remove potential toxic species of α-synuclein, which can be engulfed and degraded by surrounding glia. Although this hypothesis implies a neuroprotective strategy to accommodate proteostasis, it may also propagate α-synuclein toxicity and activate inflammatory and immune glia responses.
The analysis unearthed an increase in the levels of Lmp7, one of the three cardinal subunits of the immunoproteasome, which may represent a response to protein aggregation and activation of innate immunity, since the immunoproteasome is primarily induced by inflammatory stimuli, namely IFNγ [75,76]. Increased Lmp7 levels in the mouse model of α-synuclein aggregation and, critically, in DLB and PD brains has not been previously documented. However, induction of the immunoproteasome is not specific for α-synuclein aggregation. Levels and activity of the immunoproteasome are elevated in postmortem brains of patients with Huntington's and Alzheimer's disease [[77], [78], [79], [80]]. In Huntington's disease, Lmp2-positive staining was predominantly within neurons and overlapped with approximately 5% of cortical huntingtin protein aggregates [78]. In the Alzheimer's disease brains increased labeling for the immunoproteasome was associated with reactive glia that surrounded amyloid-β plaques [79,80]. Taken in conjunction with our findings that Lmp7 staining was increased in both neurons and glia in the substantia nigra and co-localized with microglia in the ventral tegmental area in cases with PD, these data indicate that neurons and glia attempt to counter protein aggregation by inducing the formation of the immunoproteasome.
The immunoproteasome serves at least two known biological functions. The major function relates to the generation of peptides presented by MHC class I molecules on the cell surface for recognition by CD8+ T cells [[51], [52], [53],81]. The immunoproteasome also removes ubiquitin-tagged oxidatively-modified misfolded proteins [[55], [56], [57], [58]]. Both of these functions may be critical for protein aggregation diseases and particularly for PD, DLB and related disorders characterized by aggregation of α-synuclein. Our data indicates that the immunoproteasome degrades α-synuclein PFFs in vitro and in cells, implicating the induction of the immunoproteasome as a potential pathway for elimination of α-synuclein aggregates. Moreover, the degradation of α-synuclein PFFs resulted in the generation of potentially antigenic peptides that can induce both MHC I and MHC II responses.
Expression of MHC I in dopaminergic neurons in control (non-disease) and PD postmortem tissues has been reported [82,83]. Moreover, the selective expression of MHC I in dopaminergic neurons renders them susceptible to CD8+ T cell mediated injury and death [83]. A recent study has extended these observations to show that peptides derived from α-synuclein elicit both MHC I and MHC II responses to drive cytokine secretion from CD4+ T cells and CD8+ cytotoxic T cells in peripheral mononuclear cells from patients with PD [61]. Therefore, the induction of the immunoproteasome in PD may relate to the degradation of α-synuclein and the generation of antigenic peptides for T cell presentation. This function may be viewed as detrimental for dopaminergic neurons as activation of T cells elicits cytotoxic effects in neurons.
However, the immunoproteasome may play an important protective role. Primates and mice with longer lifespans have elevated levels of the immunoproteasome [84]. This may derive from the role of the immunoproteasome in degrading oxidatively modified, ubiquitin-tagged, aggregated proteins [[55], [56], [57], [58]]. The capacity of the immunoproteasome to degrade proteins modified by oxidants may also be a vital response to neurodegeneration since an association between oxidized proteins and increased immunoproteasome has been reported [85]. Oxidative stress and oxidized proteins have been long considered in the pathological cascade of neurodegenerative diseases including PD and related α-synuclein aggregation disorders [16,86,87]. Although several forms of autophagy and the lysosomal pathway may participate in clearing α-synuclein and related intermediates such as α-synuclein oligomers [88], the immunoproteasome may be a viable strategy to influence neuroinflammation as well as clearance of potentially toxic α-synuclein species. Given these findings, the timely and appropriate induction of the immunoproteasome by small molecules or other activators may limit the progression of pathology in neurodegenerative disorders. A recent study provided a proof-of-concept for this suggestion by identifying small molecules known to inhibit the p38 MAP kinase arm of MAPK signaling as potent activators of the proteasome [89]. Interestingly, the activation of the proteasome by the p38 MAPK inhibitors was shown to increase removal of overexpressed α-synuclein in cells providing a foundation for future studies that target the immunoproteasome [89].
Data uncovered a significant induction of Lmp7, one of the three principal subunits of the immunoproteasome. The induction of immunoproteasome has been associated with removal of misfolded proteins and preservation of proteostasis. Increased Lmp7 levels were also documented in postmortem human brain with Parkinson's disease and increased levels and proteolytic activity were documented in postmortem brains with DLB disease, which is characterized by α-synuclein aggregation.
Acknowledgments
Acknowledgements
We thank Dr. Benoit Giasson, (Center for Translational Research in Neurodegenerative Disease, University of Florida) for providing the human DLB and control postmortem brains.
Funding Sources
The work was supported by National Institute of Health grants NINDS NS088322 (KCL); U54 HD 086984 (SS and HI) and NIA-AG13966 (HI). The information contained in the manuscript is the sole responsibility of the authors, no funding source was involved in the collection, analysis, or interpretation of the data nor the manuscript preparation.
Conflicts of Interest
The authors declare that they have no conflicts of interest with the contents of the manuscript.
Author Contributions
SU participated in all aspects of data collection, generated the samples for MS-based analysis, performed western blot analyses, evaluated the immunoproteasome activity in human brains and helped with the writing of the manuscript. MD and NG generated and analyzed data, generated figures and helped with editing. HF and AKY performed statistical and bioinformatic analyses of the data; HF generated figures. KCL and EL performed the stereotactic injections, evaluated α-synuclein aggregation and neuron viability in the mouse model. HD, CMcK and SS performed the proteomic analysis and supported MS-based data analysis. DM screened and evaluated antibodies used for immunohistochemistry. PE supported the construction of the mouse brain reference proteome and provided bioinformatics support. DB and JED performed and analyzed the immunohistological studies in human tissues. HI planned, organized and participated in the analysis and discussion of all the data and writing of the manuscript. HI is the Gisela and Dennis Alter research professor at the Children's Hospital of Philadelphia Research Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.05.007.
Appendix A. Supplementary data
Supplementary Figures
Supplementary data file 1-Protein and phosphopeptides identified in wildtype and α-synuclein null brains
Supplementary data file 2-Significantly changed proteins and phosphopeptides
Supplementary data file 3-Pathway enrichment; GO term analysis
Supplementary data file 4-Reference mouse brain proteome
Supplementary data file 5-Peptides generated from the in vitro degradation of α-synuclein aggregates by the immunoproteasome
References
- 1.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Douglas P.M., Dillin A. Protein homeostasis and aging in neurodegeneration. J. Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 4.Olzscha H., Schermann S.M., Woerner A.C., Pinker S., Hecht M.H., Tartaglia G.G. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 5.Woerner A.C., Frottin F., Hornburg D., Feng L.R., Meissner F., Patra M. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 6.Maroteaux L., Campanelli J.T., Scheller R.H. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George J.M., Jin H., Woods W.S., Clayton D.F. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron. 1995;15:361–372. doi: 10.1016/0896-6273(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 8.Davidson W.S., Jonas A., Clayton D.F., George J.M. Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 9.Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W.H., Castillo P.E. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 10.Chandra S., Fornai F., Kwon H.-B., Yazdani U., Atasoy D., Liu X. Double-knockout mice for alpha- and beta-synucleins: effect on synaptic functions. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14966–14971. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemani V.M., Lu W., Berge V., Nakamura K., Onoa B., Lee M.K. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 13.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 14.Farrer M., Kachergus J., Forno L., Lincoln S., Wang D.S., Hulihan M. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 15.Spillantini M.G., Schmidt M.L., Lee V.M.-Y., Trojanowski J.Q., Jakes R., Goedert M. α-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 16.Giasson B.I., Duda J.E., Murray I.V., Chen Q., Souza J.M., Hurtig H.I. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 17.Duda J.E., Giasson B.I., Mabon M.E., Lee V.M.-Y., Trojanowski J.Q. Novel antibodies to synuclein show abundant striatal pathology in Lewy body diseases. Ann. Neurol. 2002;52:205–210. doi: 10.1002/ana.10279. [DOI] [PubMed] [Google Scholar]
- 18.Luk K.C., Kehm V., Carroll J., Zhang B., O'Brien P., Trojanowski J.Q. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk K.C., Covell D.J., Kehm V.M., Zhang B., Song I.Y., Byrne M.D. Molecular and biological compatibility with host alpha-Synuclein influences fibril pathogenicity. Cell Rep. 2016;16:3373–3387. doi: 10.1016/j.celrep.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacino A.N., Brooks M., Thomas M.A., McKinney A.B., Lee S., Regenhardt R.W. Intramuscular injection of α-synuclein induces CNS α-synuclein pathology and a rapid-onset motor phenotype in transgenic mice. Proc. Natl. Acad. Sci. 2014;111:10732–10737. doi: 10.1073/pnas.1321785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osterberg V.R., Spinelli K.J., Weston L.J., Luk K.C., Woltjer R.L., Unni V.K. Progressive aggregation of alpha-synuclein and selective degeneration of Lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Rep. 2015;10:1252–1260. doi: 10.1016/j.celrep.2015.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paumier K.L., Luk K.C., Manfredsson F.P., Kanaan N.M., Lipton J.W., Collier T.J. Intrastriatal injection of pre-formed mouse α-synuclein fibrils into rats triggers α-synuclein pathology and bilateral nigrostriatal degeneration. Neurobiol. Dis. 2015;82:185–199. doi: 10.1016/j.nbd.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peelaerts W., Bousset L., Van der Perren A., Moskalyuk A., Pulizzi R., Giugliano M. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature. 2015;522:340–344. doi: 10.1038/nature14547. [DOI] [PubMed] [Google Scholar]
- 24.Shimozawa A., Ono M., Takahara D., Tarutani A., Imura S., Masuda-Suzukake M. Propagation of pathological α-synuclein in marmoset brain. Acta Neuropathol. Commun. 2017;5:12–26. doi: 10.1186/s40478-017-0413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C.C., MacCoss M.J., Howell K.E., Matthews D.E., Yates J.R. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y., Yuan Y., Halliday G., Rusznák Z., Watson C., Paxinos G. A cytoarchitectonic and chemoarchitectonic analysis of the dopamine cell groups in the substantia nigra, ventral tegmental area, and retrorubral field in the mouse. Brain Struct. Funct. 2012;217:591–612. doi: 10.1007/s00429-011-0349-2. [DOI] [PubMed] [Google Scholar]
- 27.Mertins P., Quao J.W., Patel J., Udeshi N.D., Clauser K.R., Mani D.R. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Qian W.J., Chin M.H., Petyuk V.A., Barry R.C., Liu T. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price J.C., Guan S., Burlingame A., Prusiner S.B., Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walther D.M., Mann M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 32.Walter W., Sanchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 33.Montojo J., Zuberi K., Rodriguez H., Bader G.D., Morris Q. GeneMANIA: fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutherford N.J., Lewis J., Clippinger A.K., Thomas M.A., Adamson J., Cruz P.E. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res. 2013;1524:62–73. doi: 10.1016/j.brainres.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waxman E.A., Duda J.E., Giasson B.I. Characterization of antibodies that selectively detect alpha-synuclein in pathological inclusions. Acta Neuropathol. 2008;116:37–46. doi: 10.1007/s00401-008-0375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White L., Petrovitch H., Ross G.W., Masaki K.H., Abbott R.D., Teng E.L. Prevalence of dementia in older Japanese-American men in Hawaii: the Honolulu-Asia aging study. JAMA. 1996;276:955–960. [PubMed] [Google Scholar]
- 37.Anderson J.P., Walker D.E., Goldstein J.M., de Laat R., Banducci K., Caccavello R.J. Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 38.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 39.Colom-Cadena M., Pegueroles J., Herrmann A.G., Henstridge C.M., Muñoz L., Querol-Vilaseca M. Synaptic phosphorylated α-synuclein in dementia with Lewy bodies. Brain. 2017;140:3204–3214. doi: 10.1093/brain/awx275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung S.Y., Choi J.M., Rousseaux M.W., Malovannaya A., Kim J.J., Kutzera J. An anatomically resolved mouse brain proteome reveals Parkinson disease-relevant pathways. Mol. Cell. Proteomics. 2017;16:581–593. doi: 10.1074/mcp.M116.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larhammar M., Patra K., Blunder M., Emilsson L., Peuckert C., Arvidsson E. SLC10A4 is a vesicular amine-associated transporter modulating dopamine homeostasis. Biol. Psychiatry. 2015;77:526–536. doi: 10.1016/j.biopsych.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Patra K., Lyons D.J., Bauer P., Hilscher M.M., Sharma S. A role for solute carrier family 10 member 4, or vesicular aminergic-associated transporter, in structural remodelling and transmitter release at the mouse neuromuscular junction. Eur. J. Neurosci. 2015;41:316–327. doi: 10.1111/ejn.12790. [DOI] [PubMed] [Google Scholar]
- 44.Ferrer I., Blanco R., Carmona M., Puig B., Barrachina M., Gómez C. Active, phosphorylation-dependent mitogen-activated protein kinase (MAPK/ERK), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), and p38 kinase expression in Parkinson's disease and dementia with Lewy bodies. J. Neural. Transm (Vienna). 2001;108:1383–1396. doi: 10.1007/s007020100015. [DOI] [PubMed] [Google Scholar]
- 45.Iwata A., Maruyama M., Kanazawa I., Nukina N. Alpha-Synuclein affects the MAPK pathway and accelerates cell death. J. Biol. Chem. 2001;276:45320–45329. doi: 10.1074/jbc.M103736200. [DOI] [PubMed] [Google Scholar]
- 46.Wilms H., Rosenstiel P., Romero-Ramos M., Arlt A., Schäfer H., Seegert D. Suppression of MAP kinases inhibits microglial activation and attenuates neuronal cell death induced by alpha-synuclein protofibrils. Int. J. Immunopathol. Pharmacol. 2009;22:897–909. doi: 10.1177/039463200902200405. [DOI] [PubMed] [Google Scholar]
- 47.Zhu J.H., Guo F., Shelburne J., Watkins S., Chu C.T. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;4:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller S.G., Kennedy M.B. Regulation of brain type II Ca2+/calmodulin-dependent protein kinase by autophosphorylation: a Ca2+−triggered molecular switch. Cell. 1986;44:861–870. doi: 10.1016/0092-8674(86)90008-5. [DOI] [PubMed] [Google Scholar]
- 49.Picconi B., Gardoni F., Centonze D., Mauceri D., Cenci M.A., Bernardi G. Abnormal Ca2+−calmodulin-dependent protein kinase II function mediates synaptic and motor deficits in experimental parkinsonism. J. Neurosci. 2004;24:5283–5291. doi: 10.1523/JNEUROSCI.1224-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriguchi S., Yabuki Y., Reduced Fukunaga K. Calcium/calmodulin-dependent protein kinase II activity in the hippocampus is associated with impaired cognitive function in MPTP-treated mice. J. Neurochem. 2012;2012(120):541–551. doi: 10.1111/j.1471-4159.2011.07608.x. [DOI] [PubMed] [Google Scholar]
- 51.Driscoll J., Brown M.G., Finley D., Monaco J.J. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 52.Gaczynska M., Rock K.L., Spies T., Goldberg A.L. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber E.M., Basler M., Schwab R., Heinemeyer W., Kirk C.J., Groettrup M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 54.Cascio P., Hilton C., Kisselev A.F., Rock K.L., Goldberg A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seifert U., Bialy L.P., Ebstein F., Bech-Otschir D., Voigt A., Schröter F. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 56.Nathan J.A., Spinnenhirn V., Schmidtke G., Basler M., Groettrup M. Immuno- and constitutive proteasomes do not differ in their abilities to degrade ubiquitinated proteins. Cell. 2013;152:1184–1194. doi: 10.1016/j.cell.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebstein F., Voigt A., Lange N., Warnatsch A., Schröter F., Prozorovski T. Immunoproteasomes are important for proteostasis in immune responses. Cell. 2013;152:935–937. doi: 10.1016/j.cell.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Yun Y.S., Kim K.H., Tschida B., Sachs Z., Noble-Orcutt K.E., Moriarity B.S. mTORC1 coordinates protein synthesis and immunoproteasome formation via PRAS40 to prevent accumulation of protein stress. Mol. Cell. 2016;61:625–639. doi: 10.1016/j.molcel.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moscovitz O., Ben-Nissan G., Fainer I., Pollack D., Mizrachi L., Sharon M. The Parkinson's-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015;6(6609):1–13. doi: 10.1038/ncomms7609. [DOI] [PubMed] [Google Scholar]
- 60.Raule M., Cerruti F., Cascio P. Enhanced rate of degradation of basic proteins by 26S immunoproteasomes. Biochim. Biophys. Acta. 2014;1843:1942–1947. doi: 10.1016/j.bbamcr.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Sulzer D., Alcalay R.N., Garretti F., Cote L., Kanter E., Agin-Liebes J. T cells from patients with Parkinson's disease recognize α-synuclein peptides. Nature. 2017;546:656–661. doi: 10.1038/nature22815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee H., Suk J., Patrick C., Bae E., Chio J., Rho S. Direct transfer of alpha-synucelin from neuron to astroglia causes inflammatory responses in synucleinopathies. J. Biol. Chem. 2010;285:9262–9272. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muchamuel T., Basler M., Aujay M.A., Suzuki E., Kalim K.W., Lauer C. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 64.Chung C.Y., Khurana V., Yi S., Sahni N., Loh K.H., Auluck P.K. In situ peroxidase labeling and mass-spectrometry connects alpha-Synuclein directly to endocytic trafficking and mRNA metabolism in neurons. Cell Syst. 2017;4:242–250.e4. doi: 10.1016/j.cels.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson-Lewis V., Jakowec M., Burke R.E., Przedborski S. Time course and morphology of dopaminergic neuronal death caused by the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Neurodegeneration. 1995;4:257–269. doi: 10.1016/1055-8330(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 66.Ara J., Przedborski S., Naini A.B., Jackson-Lewis V., Trifiletti R.R., Horwitz J. Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Proc. Natl. Acad. Sci. U. S. A. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung C.Y., Koprich J.B., Siddiqi H., Isacson O. Dynamic changes in presynaptic and axonal transport proteins combined with striatal neuroinflammation precede dopaminergic neuronal loss in a rat model of AAV alpha-synucleinopathy. J. Neurosci. 2009;29:3365–3373. doi: 10.1523/JNEUROSCI.5427-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia-Reitböck P., Anichtchik O., Bellucci A., Iovino M., Ballini C., Fineberg E. SNARE protein redistribution and synaptic failure in a transgenic mouse model of Parkinson's disease. Brain. 2010;133:2032–2044. doi: 10.1093/brain/awq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milber J.M., Noorigian J.V., Morley J.F., Petrovitch H., White L.R., Ross W.G. Lewy pathology is not the first sign of degeneration in vulnerable neurons in PD. Neurology. 2012;79:2307–2314. doi: 10.1212/WNL.0b013e318278fe32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rockenstein E., Nuber S., Overk C.R., Ubhi K., Mante M., Patrick C. Accumulation of oligomer-prone α-synuclein exacerbates synaptic and neuronal degeneration in vivo. Brain. 2014;137:1496–1513. doi: 10.1093/brain/awu057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott D.A., Tabarean I., Tang Y., Cartier A., Masliah E., Roy S. A pathologic cascade leading to synaptic dysfunction in alpha-synuclein-induced neurodegeneration. J. Neurosci. 2010;30:8083–8095. doi: 10.1523/JNEUROSCI.1091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S.D., Ntzouni M., Margaritis L.H. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010;30:6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danzer K.M., Kranich L.R., Ruf W.P., Cagsal-Getkin O., Winslow A.R., Zhu L. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minakaki G., Menges S., Kittel A., Emmanouilidou E., Schaeffner I., Barkovits K. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy. 2018;14:98–119. doi: 10.1080/15548627.2017.1395992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T. Interferon gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- 76.Heink S., Ludwig D., Kloetzel P., Kru E. IFN-y-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aso E., Lomoio S., López-González I., Joda L., Carmona M., Fernández-Yagüe N. Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer's disease. Brain Pathol. 2012;22:636–653. doi: 10.1111/j.1750-3639.2011.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dıaz-Hernandez M., Hernández F., Martín-Aparicio E., Gómez-Ramos P., Morán M.A., Castaño J.G. Neuronal induction of the immunoproteasome in Huntington's disease. J. Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mishto M., Bellavista E., Santoro A., Stolzing A., Ligorio C., Nacmias B. Immunoproteasome and LMP2 polymorphism in aged and Alzheimer's disease brains. Neurobiol. Aging. 2006;27:54–66. doi: 10.1016/j.neurobiolaging.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 80.Orre M., Kamphuis W., Dooves S., Kooijman L., Chan E.T., Kirk C.J. Reactive glia show increased immunoproteasome activity in Alzheimer's disease. Brain. 2013;136:1415–1431. doi: 10.1093/brain/awt083. [DOI] [PubMed] [Google Scholar]
- 81.Kincaid E.Z., Che J.W., York I., Escobar H., Reyes-Vargas E., Delgado J.C. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2011;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lindå H., Hammarberg H., Piehl F., Khademi M., Olsson T. Expression of MHC class I heavy chain and beta2-microglobulin in rat brainstem motoneurons and nigral dopaminergic neurons. J. Neuroimmunol. 1999;101:76–86. doi: 10.1016/s0165-5728(99)00135-6. [DOI] [PubMed] [Google Scholar]
- 83.Cebrián C., Zucca F.A., Mauri P., Steinbeck J.A., Studer L., Scherzer C.R. MHC-I expression renders catecholaminergic neurons susceptible to T-cell-mediated degeneration. Nat. Commun. 2014;5:3633. doi: 10.1038/ncomms4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pickering A.M., Lehr M., Miller R.A. Lifespan of mice and primates correlates with immunoproteasome expression. J. Clin. Invest. 2015;125:2059–2068. doi: 10.1172/JCI80514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teoh C.Y., Davies K.J.A. Potential roles of protein oxidation and the immunoproteasome in MHC class I antigen presentation: the ‘PrOxI’ hypothesis. Arch. Biochem. Biophys. 2004;423:88–96. doi: 10.1016/j.abb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 86.Ischiropoulos H., Beckman J.S. Oxidative stress and nitration in neurodegeneration: cause, effect or association? J. Clin. Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Souza J.M., Giasson B.I., Chen Q., Lee V.M.-Y., Ischiropoulos H. Dityrosine cross-linking promotes formation of stable alpha-synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J. Biol. Chem. 2000;275:18344–18349. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 88.Ebrahimi-Fakhari D., Cantuti-Castelvetri I., Fan Z., Rockenstein E., Masliah E., Hyman B.T. Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of α-synuclein. J. Neurosci. 2011;31:14508–14520. doi: 10.1523/JNEUROSCI.1560-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leestemaker Y., de Jong A., Witting K.F., Penning R., Schuurman K., Rodenko B. Proteasome activation by small molecules. Cell Chem. Biol. 2017;24:725–736. doi: 10.1016/j.chembiol.2017.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary data file 1-Protein and phosphopeptides identified in wildtype and α-synuclein null brains
Supplementary data file 2-Significantly changed proteins and phosphopeptides
Supplementary data file 3-Pathway enrichment; GO term analysis
Supplementary data file 4-Reference mouse brain proteome
Supplementary data file 5-Peptides generated from the in vitro degradation of α-synuclein aggregates by the immunoproteasome