Abstract
IMPORTANCE
MicroRNAs (miRNAs) are promising multiple sclerosis (MS) biomarkers. Establishing the association between miRNAs and magnetic resonance imaging (MRI) measures of disease severity will help define their significance and potential impact.
OBJECTIVE
To correlate circulating miRNAs in the serum of patients with MS to brain and spinal MRI.
DESIGN, SETTING, AND PARTICIPANTS
A cross-sectional study comparing serum miRNA samples with MRI metrics was conducted at a tertiary MS referral center. Two independent cohorts (41 and 79 patients) were retrospectively identified from the Comprehensive Longitudinal Investigation of Multiple Sclerosis at the Brigham and Women’s Hospital. Expression of miRNA was determined by locked nucleic acid–based quantitative real-time polymerase chain reaction. Spearman correlation coefficients were used to test the association between miRNA and brain lesions (T2 hyperintense lesion volume [T2LV]), the ratio of T1 hypointense lesion volume [T1LV] to T2LV [T1:T2]), brain atrophy (whole brain and gray matter), and cervical spinal cord lesions (T2LV) and atrophy. The study was conducted from December 2013 to April 2016.
MAIN OUTCOMES AND MEASURES
miRNA expression.
RESULTS
Of the 120 patients included in the study, cohort 1 included 41 participants (7 [17.1%] men), with mean (SD) age of 47.7 (9.5) years; cohort 2 had 79 participants (26 [32.9%] men) with a mean (SD) age of 43.0 (7.5) years. Associations between miRNAs and MRIs were both protective and pathogenic. Regarding miRNA signatures, a topographic specificity differed for the brain vs the spinal cord, and the signature differed between T2LV and atrophy/destructive measures. Four miRNAs showed similar significant protective correlations with T1:T2 in both cohorts, with the highest for hsa.miR.143.3p (cohort 1: Spearman correlation coefficient rs = −0.452, P = .003; cohort 2: rs = −0.225, P = .046); the others included hsa.miR.142.5p (cohort 1: rs = −0.424, P = .006; cohort 2: rs = −0.226, P = .045), hsa.miR.181c.3p (cohort 1: rs = −0.383, P = .01; cohort 2: rs = −0.222, P = .049), and hsa.miR.181c.5p (cohort 1: rs = −0.433, P = .005; cohort 2: rs = −0.231, P = .04). In the 2 cohorts, hsa.miR.486.5p (cohort 1: rs = 0.348, P = .03; cohort 2: rs = 0.254, P = .02) and hsa.miR.92a.3p (cohort 1: rs = 0.392, P = .01; cohort 2: rs = 0.222, P = .049) showed similar significant pathogenic correlations with T1:T2; hsa.miR.375 (cohort 1: rs = −0.345, P = .03; cohort 2: rs = −0.257, P = .022) and hsa.miR.629.5p (cohort 1: rs = −0.350, P = .03; cohort 2: rs = −0.269, P = .02) showed significant pathogenic correlations with brain atrophy. Although we found several miRNAs associated with MRI outcomes, none of these associations remained significant when correcting for multiple comparisons, suggesting that further validation of our findings is needed.
CONCLUSIONS AND RELEVANCE
Serum miRNAs may serve as MS biomarkers for monitoring disease progression and act as surrogate markers to identify underlying disease processes.
Multiple sclerosis (MS) involves autoimmune-related inflammatory demyelination of the central nervous system white matter and gray matter (GM).1 The disease may transect axons and cause brain and spinal cord neuronal loss and atrophy.2 Magnetic resonance imaging (MRI) is a standard tool for the diagnosis and therapeutic monitoring of MS.3–5 Recent MRI technology allows visualization of tissue atrophy6,7 and GM involvement7,8 beyond the traditional focal white matter lesion assessments.9
Among lesion measures, T2 hyperintense areas are histologically nonspecific but can estimate the burden of overt demyelinating foci.9 Persisting T1 hypointense lesions (black holes) specifically indicate irreversible tissue destruction.10 The total burden of such lesions and the proportion of T2 lesions showing T1 hypointensity11 provide a better indication of physical disability,12 quality of life,13 and unemployment.14
Quantification of atrophy with MRI can estimate axonal loss, neuronal loss, and neurodegenerative changes, which complement lesion measurements.15,16 Brain atrophy, particularly of GM, links to physical disability and cognitive impairment after adjusting for the effect of lesions.17,18 Spinal cord involvement is associated with disability19 and provides unique information vs brain MRI.20,21
MicroRNAs (miRNAs)are small, noncoding RNAs that regulate gene expression by binding to complementary sequences in untranslated messenger RNA (mRNA) regions, resulting in translational repression or mRNA degradation.22–24 Dysregulated miRNA expression links to a range of immunologic and other diseases.25–27 Due to their detectability and stability in plasma, serum, and cerebrospinal fluid25,27,28 as well as the development of sensitive methods for their detection and quantification,29 miRNAs are attractive biomarkers.
Previous studies showed changes in miRNA expression in brain tissue, serum and plasma, and immune cells from patients with MS and in experimental autoimmune encephalomyelitis.30–33 Previous studies have also shown associations between MS disability or disease progression and the expression of miRNAs.34,35
Magnetic resonance imaging measures of lesions and atrophy aid in defining the pathobiology reflected by a candidate biomarker.36 Since miRNAs regulate several cellular pathways, we hypothesized that different miRNA expression patterns would show associations with specific MRI measures of cerebral and spinal cord involvement.
Methods
Participants
As part of the Comprehensive Longitudinal Investigation of Multiple Sclerosis cohort study of clinical, MRI, and blood phenotyping in more than 2000 patients at Brigham and Women’s Hospital,37 we retrospectively identified patients with (1) age 18 to 55 years, (2) MS diagnosis,38 (3) absence of other major medical disorders, and (4) no more than 30 days between serum and brain MRI acquisition (cohort 1) or both spine and brain MRI acquisition (cohort 2). Patients were evaluated by an MS specialist at the time of MRI for neurologic disability by the Expanded Disability Status Scale (EDSS) score, an ordinal scale ranging from low (0) to high (10) disability.39 Two independent cohorts (eTable 1 in the Supplement) were tested and analyzed for miRNA-MRI associations. Cohort 1 had 41 participants (7 [17.1%] men), with a mean (SD) age of 47.7 (9.5) years; cohort 2 had 79 participants (26 [32.9%] men), with a mean (SD) age of 43.0 (7.5) years. The study was conducted from December 2013 to April 2016.
The study was approved by the Partners Health Care Human Research Committee. The participants provided written informed consent; there was no financial compensation.
Samples
Serum samples were collected in red-top Vacutainer tubes without additives, centrifuged at 2000 rpm for 10 minutes to separate serum, and stored at −70°C until RNA extraction. Isolation of RNA was then performed (miRcury kit; Exiqon) and converted to complementary DNA using a synthesis kit (Exiqon). Prepared complementary DNA samples were stored at −20°C. Locked nucleic acid, green-based, real-time quantitative polymerase chain reaction Human Panel I and II (LNA-SYBR; Exiqon) containing 652 miRNAs and 752 miR-NAs were used for profiling the first set and second sets, respectively. Normalization was performed using the mean miRNA expression of 10 miRNAS (hsa.let.7d.3p, hsa.miR.103a.3p, hsa.miR.106a.5p, hsa.miR.126.3p, hsa.miR.15b.5p, hsa.miR.19a.3, hsa.miR.20a.5p, hsa.miR.30b.5p, hsa.miR.425.5p, and hsa.miR.92a.3p), with the best stability index determined by Norm Finder software. Normalized cycle quantification (Cq) was calculated as mean Cq – assay Cq.
MRI Acquisition
Patients were scanned on one of two 1.5T MRI units (Signa; GE Healthcare). For brain imaging, the sequences covered the whole head; were free of intersection gaps; and included (1) axial, T2-weighted, conventional spin-echo dual-echo (repetition time (TR)/echo time 1 (TE1)/TE2, 3000/30/80 milliseconds; voxel size, 0.9375 × 0.9375 × 3 mm); (2) axial T1-weighted spinecho (TR/TE, 467–733/20 milliseconds; voxel size, 0.9375 × 0.9375 × 3 mm); (3) (cohort 2 only) sagittal, 3-dimensional, magnetization-prepared rapid gradient-echo (TR/TE, 8.6/3.8 milliseconds; voxel size, 0.94 × 0.94 × 1.2 mm); and (4) axial T1-weighted spin-echo imaging, which was repeated 5 to 7 minutes after a 0.1-mmol/kg intravenous infusion of gadolinium. For the cervical spinal cord imaging (cohort 2), patients were scanned on the same MRI unit, including axial T2-weighted fast spin-echo covering the whole cervical spinal cord, with a voxel size of 0.35 × 0.35 × 3mm. The mean (SD) TR was 3561 (431) milliseconds, and the TE was 103 (1.3) milliseconds. Thus, cohort 2 had a 3-dimensional, high-resolution brain acquisition and a spinal cord image set in addition to conventional brain imaging that was obtained in both cohorts.
MRI Analysis
Lesion Measures
Whole-brain T1 hypointense (T1LV) and T2 hyperintense (T2LV) lesion volume (both cohorts) and whole cervical (C1 to C7) spinal cord T2LV (cohort 2) were expert traced using an edge-finding tool in Jim, version 7 software (Xinapse Systems Ltd, http://www.xinapse.com/). Brain T2 lesions required hyperintensity on both the early- and late-echo T2 images. Brain T1 hypointense lesions (black holes) were defined as containing some degree of visible hyperintensity on the T2 images but were free of gadolinium enhancement (to reduce the detection of benign and transient lesions).40 As an index of the destructive potential of the lesions, we calculated the intrapatient ratio of T1LV to T2LV (T1:T2) based on its unique and valuable role in previous studies.11,41,42
Brain Atrophy
For cohort 2, given the availability of a high-resolution image set, we used a fully automated algorithm to derive normalized whole-brain parenchymal (BPV) and GM(GMV) volumes(SIENAX, version 5.0; Analysis Group,http://fsl.fmrib.ox.ac.uk).14,43 Given the ineffective contouring of the deep central and posterior foss a GM from the SIENAX output, we performed manual masking of the GMV maps to remove those regions to calculate the cortical GMV (cGMV). For cohort 1 (Figure 1), we relied on the dual-echo images to estimate brain atrophy using a fully automated pipeline (SPM, version 12; Wellcome Trust Centre for Neuroimaging, www.fil.ion.ucl.ac.uk/spm/). This derived brain parenchymal fraction (BPF) (a validated estimation of whole-brain atrophy6) normalized as the ratio of brain parenchymal tissue (GM plus white matter) volume divided by the intracranial volume.44 We also calculated the global cerebral GM fraction (GMF) in an analogous manner.
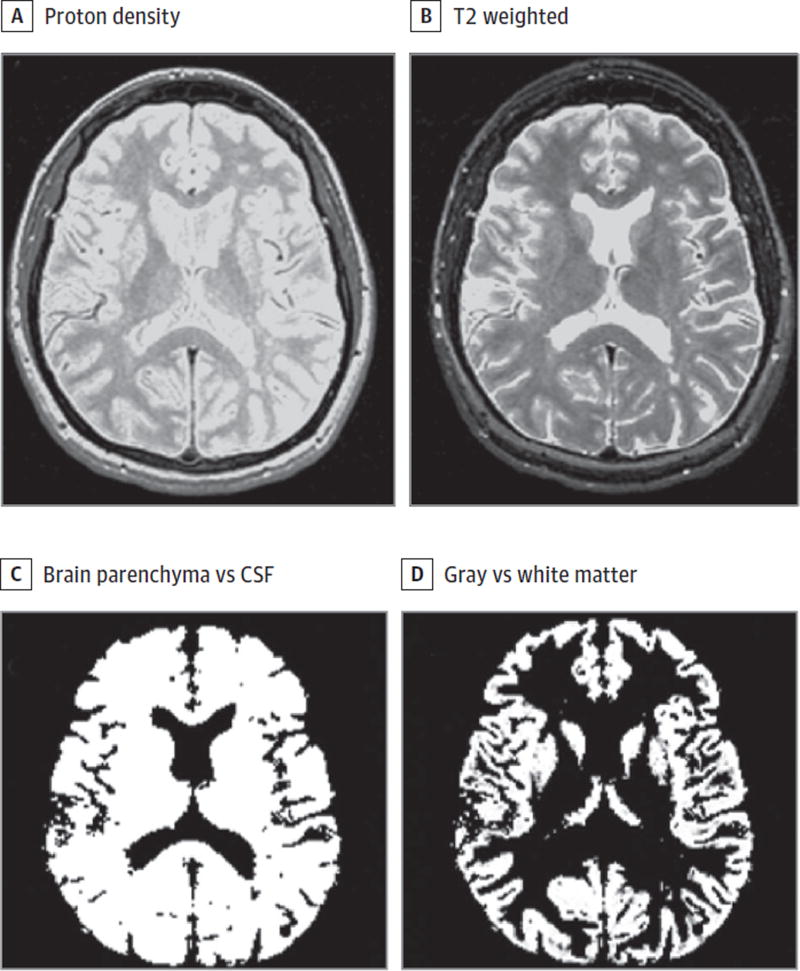
Figure 1. Magnetic Resonance Imaging (MRI) Segmentation Procedure to Derive Brain Volume (Atrophy) Measures in Cohort 1.
Native axial dual-echo proton density (A) and T2-weighted (B) images were applied to a fully automated statistical parametric mapping pipeline (version 12), resulting in an intracranial volume segmented into brain vs cerebrospinal fluid (CSF) compartments used to derive brain parenchymal fraction (C) and gray vs white matter maps (D) used to derive global cerebral gray matter fraction. A patient with relapsing-remitting multiple sclerosis from cohort 1 is shown.
Spinal Cord Atrophy
In cohort 2, axial T2-weighted images of the cervical cord (C1-C5) were applied to a validated active surface tool45 using the Jim, version 7 software package. Manual adjustments were made as necessary to capture the spinal cord contour. Spinal cord volume from C1 to C5 was normalized by dividing by the number of axial sections46 to generate the upper cervical spinal cord area (UCCA). Our group has previously shown that cord volumes derived from T2-weighted images are similar to those obtained from T1-weighted images.47
Reliability and Blinding
Analysis of MR images was performed by operators (F.K., R.C., S. Tauhid, S. Tummala, S.L.D., and G.K.) blinded to clinical and biomarker information. Five metrics required operator input (brain T2LV, brain T1LV, cGMV, UCCA, and spinal cord T2LV). Regarding brain T2LV, brain T1LV, and UCCA, our procedures and their high reliability have been well established in previously published studies.47,48 In the present data set, intrarater reliability was high as assessed from 5 randomly chosen cases. For spinal cord T2LV, the mean coefficient of variation was 3.53%; for cGMV, the mean coefficient of variation was 0.44%.
Statistical Analysis
Spearman correlation coefficients (rs) estimated the association between miRNA expression (normalized Cq) and MRI and disability (EDSS score). Spearman correlations were used so that participants with miRNA levels below detection limits (missing or undetected values) could contribute to the analysis. Undetected expression values were assigned a value lower than the smallest observed value from all participants. P Values were adjusted for multiple comparisons using the false discovery rate. The miRNA-MRI associations were observed as either pathogenic, shown by high miRNA expression associated with high MRI severity, or protective, shown by association with lower MRI-indicated severity. In the interpretation of the correlation coefficients, MRI-indicated severity was reflected in 2 possible directions depending on the metric: (1) for lesion severity (T2LV and T1:T2), a higher number represented increased disease severity, and (2) for atrophy measures (BPF, GMF, BPV, cGMV, and UCCA), a higher number represented lower disease severity (less atrophy). Demographic characteristics were compared using 2-sample, 2-tailed t tests for continuous variables and χ2 tests for dichotomous variables. Data analysis was performed from April 2016 to June 2016. Statistical analysis was completed in R, version 3.0.2 (http://www.r-project.org) and Stata/IC, version 14 (http://www.stata.com).
Results
Correlation Between MRI and Disability
The MRI measures of destructive pathology (T1:T2 ratio and atrophy), but not brain and spinal cord T2LV, showed significant associations with EDSS (eTable 2 in the Supplement). The strongest correlation was for atrophy of the brain (GMF: rs = −0.507; P < .001; BPV: rs = −0.470; P < .001) and spinal cord (UCCA: rs = −0.389; P < .001).
Correlation Between miRNAs and MRI
The 10 miRNAs showing the strongest MRI association in each cohort are provided in Tables 1 and 2 (eTable 3 and eTable 4 in the Supplement present all correlations). In cohort 1, the miRNAs that demonstrated strong correlations were similar for BPF and GMF. However, the miRNAs associated with lesions (T2LV and T1:T2 ratio) were mostly different from those associated with atrophy, suggesting different pathologic processes underlying focal lesions (inflammatory demyelination) vs neurodegeneration (axonal and neuronal loss). Similar to those in cohort 1, miRNAs in cohort 2 that showed a strong correlation with whole-brain atrophy also indicated a strong correlation with cerebral GM atrophy. However, a different set of miRNAs correlated with spinal cord vs brain atrophy. In addition, only a small overlap was found between miRNAs associated with brain vs spinal cord T2LV. Thus, there was a topographic specificity in brain- vs spinal cord–related miRNAs. Although we found several miRNAs associated with MRI outcomes, none of these remained significant after false discovery rate correction.
Table 1.
Cohort 1: Highest Correlations Between miRNAs and Clinical/MRI Data
| miRNA | Spearman Correlation Coefficienta |
Protective/ Pathogenic miRNA |
P Value | |
|---|---|---|---|---|
| Uncorrected | Correctedb | |||
| Brain T2LV | ||||
| hsa.miR.628.3p | 0.506 | Pathogenic | .001 | .32 |
| hsa.miR.195.5p | 0.464 | Pathogenic | .002 | .48 |
| hsa.miR.92b.3p | 0.412 | Pathogenic | .007 | .80 |
| hsa.miR.145.5p | 0.407 | Pathogenic | .009 | .80 |
| hsa.miR.32.3p | −0.378 | Protective | .02 | .80 |
| hsa.miR.296.5p | −0.369 | Protective | .02 | .80 |
| hsa.miR.511 | 0.367 | Pathogenic | .02 | .80 |
| hsa.miR.501.5p | −0.361 | Protective | .02 | .80 |
| hsa.miR.627 | 0.359 | Pathogenic | .02 | .80 |
| hsa.miR.139.3p | 0.356 | Pathogenic | .02 | .80 |
| Brain T1:T2 Ratio | ||||
| hsa.miR.331.3p | −0.490 | Protective | .001 | .27 |
| hsa.miR.338.3p | −0.476 | Protective | .002 | .27 |
| hsa.miR.433 | −0.471 | Protective | .002 | .27 |
| hsa.miR.143.3p | −0.452 | Protective | .003 | .32 |
| hsa.miR.181c.5p | −0.433 | Protective | .005 | .34 |
| hsa.miR.183.5p | 0.432 | Pathogenic | .005 | .34 |
| hsa.miR.142.5p | −0.424 | Protective | .006 | .36 |
| hsa.miR.365a.3p | −0.416 | Protective | .007 | .36 |
| hsa.miR.92a.3p | 0.392 | Pathogenic | .01 | .50 |
| hsa.miR.337.3p | −0.390 | Protective | .01 | .50 |
| BPF | ||||
| hsa.miR.101.3p | 0.493 | Protective | .001 | .52 |
| hsa.miR.324.3p | 0.411 | Protective | .008 | .63 |
| hsa.miR.19a.3p | 0.389 | Protective | .01 | .63 |
| hsa.miR.21.5p | 0.388 | Protective | .01 | .63 |
| hsa.miR.548l | −0.384 | Pathogenic | .01 | .63 |
| hsa.miR.211.5p | −0.378 | Pathogenic | .02 | .63 |
| hsa.miR.590.5p | 0.377 | Protective | .02 | .63 |
| hsa.miR.99b.3p | −0.374 | Pathogenic | .02 | .63 |
| hsa.miR.32.5p | 0.374 | Protective | .02 | .63 |
| hsa.miR.660.5p | 0.371 | Protective | .02 | .63 |
| Whole-brain GMF | ||||
| hsa.miR.101.3p | 0.547 | Protective | <.001 | .11 |
| hsa.miR.324.3p | 0.494 | Protective | .001 | .20 |
| hsa.miR.379.3p | −0.480 | Pathogenic | .001 | .20 |
| hsa.miR.340.3p | −0.469 | Pathogenic | .002 | .20 |
| hsa.let.7a.5p | −0.466 | Pathogenic | .002 | .20 |
| hsa.miR.19a.3p | 0.430 | Protective | .005 | .32 |
| hsa.miR.20a.5p | 0.423 | Protective | .006 | .32 |
| hsa.let.7e.5p | −0.422 | Pathogenic | .006 | .32 |
| hsa.miR.15b.3p | 0.420 | Protective | .007 | .32 |
| hsa.miR.106b.5p | 0.410 | Protective | .008 | .35 |
| EDSS | ||||
| hsa.miR.19a.3p | −0.627 | Protective | <.001 | .005 |
| hsa.miR.101.3p | −0.608 | Protective | <.001 | .005 |
| hsa.miR.30e.5p | −0.569 | Protective | <.001 | .01 |
| hsa.miR.19b.3p | −0.562 | Protective | <.001 | .01 |
| hsa.miR.29c.3p | −0.553 | Protective | <.001 | .02 |
| hsa.miR.32.5p | −0.520 | Protective | <.001 | .04 |
| hsa.miR.195.5p | 0.503 | Pathogenic | .001 | .049 |
| hsa.miR.142.5p | −0.450 | Protective | .003 | .14 |
| hsa.miR.27a.3p | −0.443 | Protective | .004 | .14 |
| hsa.miR.136.3p | −0.440 | Protective | .004 | .14 |
Abbreviations: BPF, brain parenchymal fraction; EDSS, Expanded Disability Status Scale; GMF, gray matter fraction; miRNA, microRNA; MRI, magnetic resonance imaging; T1:T2, ratio of T1 hypointense to T2 hyperintense lesion volume; T2LV, T2 hyperintense lesion volume.
Estimated Spearman correlation coefficients are shown: the sign of the Spearman correlation indicates the direction of association between the miRNA expression level and the MRI measure of disease activity; its value reflects the strength of the association. In the interpretation of the meaning of the correlation coefficients, MRI-indicated severity was reflected in 2 possible directions: (1) for lesion severity (T2LV, T1:T2 ratio), a higher number represented increased disease severity, and (2) for atrophy measures (BPF, GMF), a higher number represented lower disease severity (less atrophy). The miRNA-MRI associations were observed in either direction: (1) a pathogenic relationship shown by a higher miRNA expression was associated with greater severity of MRI-indicated involvement, and (2) a protective relationship shown by a higher miRNA expression was associated with lower severity of MRI-indicated involvement.
Corrected for multiple comparisons using false discovery rate.
Table 2.
Cohort 2: Highest Correlations Between miRNAs and Clinical/MRI Data
| miRNA | Spearman Correlation Coefficienta |
Protective/Pathogenic miRNA |
P Value | |
|---|---|---|---|---|
| Uncorrected | Correctedb | |||
| Brain T2LV | ||||
| hsa.miR.210.3p | 0.359 | Pathogenic | .001 | .50 |
| hsa.miR.362.5p | −0.333 | Protective | .003 | .50 |
| hsa.miR.92a.1.5p | 0.327 | Pathogenic | .003 | .50 |
| hsa.miR.1914.3p | 0.324 | Pathogenic | .004 | .50 |
| hsa.miR.330.5p | −0.305 | Protective | .006 | .66 |
| hsa.miR.577 | 0.296 | Pathogenic | .008 | .66 |
| hsa.miR.572 | −0.292 | Protective | .009 | .66 |
| hsa.miR.154.3p | 0.281 | Pathogenic | .01 | .66 |
| hsa.miR.30b.5p | −0.279 | Protective | .01 | .66 |
| hsa.miR.671.5p | −0.274 | Protective | .02 | .66 |
| Brain T1:T2 Ratio | ||||
| hsa.miR.548a.3p | −0.365 | Protective | <.001 | .18 |
| hsa.miR.515.5p | −0.348 | Protective | .002 | .18 |
| hsa.miR.27b.3p | −0.348 | Protective | .002 | .18 |
| hsa.miR.223.3p | −0.345 | Protective | .002 | .18 |
| hsa.miR.574.3p | −0.344 | Protective | .002 | .18 |
| hsa.miR.23b.3p | −0.342 | Protective | .002 | .18 |
| hsa.miR.23a.3p | −0.337 | Protective | .002 | .18 |
| hsa.miR.197.3p | −0.332 | Protective | .003 | .18 |
| hsa.miR.25.5p | 0.323 | Pathogenic | .003 | .18 |
| hsa.miR.1271.5p | −0.317 | Protective | .005 | .25 |
| BPV | ||||
| hsa.miR.484 | −0.392 | Pathogenic | <.001 | .19 |
| hsa.miR.627.5p | 0.335 | Protective | .003 | .41 |
| hsa.miR.671.5p | 0.330 | Protective | .003 | .41 |
| hsa.miR.320b | −0.323 | Pathogenic | .004 | .41 |
| hsa.miR.362.5p | 0.322 | Protective | .004 | .41 |
| hsa.miR.30a.3p | −0.317 | Pathogenic | .004 | .40 |
| hsa.miR.548d.5p | 0.300 | Protective | .007 | .48 |
| hsa.miR.486.5p | −0.288 | Pathogenic | .01 | .48 |
| hsa.miR.132.5p | 0.287 | Protective | .01 | .48 |
| hsa.miR.135a.5p | 0.281 | Protective | .01 | .48 |
| cGMV | ||||
| hsa.miR.484 | −0.386 | Pathogenic | <.001 | .24 |
| hsa.miR.610 | −0.329 | Pathogenic | .003 | .58 |
| hsa.miR.627.5p | 0.312 | Protective | .005 | .58 |
| hsa.miR.340.5p | 0.306 | Protective | .006 | .58 |
| hsa.miR.188.5p | −0.302 | Pathogenic | .007 | .58 |
| hsa.miR.7.5p | −0.301 | Pathogenic | .007 | .58 |
| hsa.miR.934 | −0.285 | Pathogenic | .01 | .58 |
| hsa.miR.556.5p | −0.279 | Pathogenic | .01 | .58 |
| hsa.miR.615.3p | 0.277 | Protective | .01 | .58 |
| hsa.miR.25.3p | −0.277 | Pathogenic | .01 | .58 |
| Spinal T2LV | ||||
| hsa.miR.132.5p | −0.349 | Protective | .002 | .88 |
| hsa.miR.548j.5p | 0.325 | Pathogenic | .003 | .88 |
| hsa.miR.937.3p | 0.305 | Pathogenic | .006 | .88 |
| hsa.miR.342.5p | 0.285 | Pathogenic | .01 | .88 |
| hsa.miR.433.3p | 0.273 | Pathogenic | .02 | .88 |
| hsa.miR.150.5p | 0.263 | Pathogenic | .02 | .88 |
| hsa.miR.155.5p | 0.263 | Pathogenic | .02 | .88 |
| hsa.miR.10a.5p | 0.261 | Pathogenic | .02 | .88 |
| hsa.miR.202.5p | −0.254 | Protective | .02 | .88 |
| hsa.miR.943 | 0.244 | Pathogenic | .03 | .88 |
| Normalized UCCA | ||||
| hsa.miR.130b.3p | −0.350 | Pathogenic | .002 | .56 |
| hsa.miR.135a.5p | 0.337 | Protective | .002 | .56 |
| hsa.miR.148b.5p | 0.329 | Protective | .003 | .56 |
| hsa.miR.374a.5p | 0.272 | Protective | .02 | .86 |
| hsa.miR.101.3p | 0.261 | Protective | .02 | .86 |
| hsa.miR.1538 | −0.249 | Pathogenic | .03 | .86 |
| hsa.miR.1468.5p | −0.247 | Pathogenic | .03 | .86 |
| hsa.miR.1247.5p | 0.245 | Protective | .03 | .86 |
| hsa.miR.190a.5p | 0.245 | Protective | .03 | .86 |
| hsa.miR.30a.3p | −0.242 | Pathogenic | .03 | .86 |
| EDSS | ||||
| hsa.miR.199a.5p | −0.380 | Protective | <.001 | .23 |
| hsa.miR.25.5p | 0.350 | Pathogenic | .002 | .23 |
| hsa.miR.551b.3p | −0.350 | Protective | .002 | .23 |
| hsa.miR.496 | −0.349 | Protective | .002 | .23 |
| hsa.miR.301a.3p | −0.323 | Protective | .004 | .37 |
| hsa.miR.181c.3p | −0.320 | Protective | .004 | .37 |
| hsa.miR.301b | −0.308 | Protective | .006 | .37 |
| hsa.miR.136.3p | −0.305 | Protective | .006 | .37 |
| hsa.miR.15a.5p | 0.298 | Pathogenic | .008 | .37 |
| hsa.let.7b.5p | 0.294 | Pathogenic | .009 | .37 |
Abbreviations: BPV, whole-brain parenchymal volume; cGMV, cortical gray matter volume; EDSS, Expanded Disability Status Scale; miRNA, microRNAs; MRI, magnetic resonance imaging; T1:T2, ratio of T1 hypointense to T2 hyperintense lesion volume; T2LV, T2 hyperintense lesion volume; UCCA, upper cervical spinal cord area.
Estimated Spearman correlation coefficients are shown: the sign of the Spearman correlation indicates the direction of association between the miRNA expression level and the MRI measure of disease activity; its value reflects the strength of the association. In the interpretation of the meaning of the correlation coefficients, MRI-indicated severity was reflected in 2 possible directions: (1) for lesion severity (T2LV, T1:T2 ratio), a higher number represented increased disease severity, and (2) for atrophy measures (BPV, cGMV, and UCCA), ahigher number represented lower disease severity (less atrophy). The miRNA-MRI associations were observed in either direction: (1) a pathogenic relationship shown by a higher miRNA expression was associated with greater severity of MRI-indicated involvement, and (2) a protective relationship shown by a higher miRNA expression was associated with lower severity of MRI-indicated involvement.
Corrected for multiple comparisons using false discovery rate.
Correlations Common Between miRNAs and MRI in Both Cohorts
Four miRNAs showed similar significant protective correlations with T1:T2 in both cohorts, with the highest for hsa.miR.143.3p (cohort 1: Spearman correlation coefficient rs = −0.452, P = .003; cohort 2: rs = −0.225, P = .046); the others included hsa.miR.142.5p (cohort 1: rs = −0.424, P = .006; cohort 2: rs = −0.226, P = .045), hsa.miR.181c.3p (cohort 1: rs = −0.383, P = .01; cohort 2: rs = −0.222, P = .049), and hsa.miR.181c.5p (cohort 1: rs = −0.433, P = .005; cohort 2: rs = −0.231, P = .04). In the 2 cohorts, hsa.miR.486.5p (cohort 1: rs = 0.348, P = .03; cohort 2: rs = 0.254, P = .02) and hsa.miR.92a.3p (cohort 1: rs = 0.392, P = .01; cohort 2: rs = 0.222, P = .049) showed similar significant pathogenic correlations with T1:T2; hsa.miR.375 (cohort 1: rs = −0.345, P = .03; cohort 2: rs = −0.257, P = .022) and hsa.miR.629.5p (cohort 1: rs = −0.350, P = .03; cohort 2: rs = −0.269, P = .02) showed significant pathogenic correlations with brain atrophy. Thus, a common set of miRNAs was as sociated with measures of destructive pathology/neurodegeneration in both cohorts. The miRNAs that were significantly correlated in the same direction with MRI are summarized in Table 3 and Figure 2. Four miRNAs (hsa.miR.142.5p, hsa.miR.143.3p, hsa.miR.181c.3p, and hsa.miR.181c.5p) were significantly correlated (protective) with T1:T2 ratio in both cohorts, with hsa.miR.143.3p correlating the highest (cohort 1: rs = −0.452, P = .003; cohort 2: rs = −0.225, P = .046). Two miRNAs (hsa.miR.486.5p and hsa.miR.92a.3p) significantly correlated (pathogenic) with T1:T2 ratio in both cohorts. Two miRNAs (hsa.miR.375 and hsa.miR.629.5p) significantly correlated (pathogenic) with brain atrophy in both cohorts. Thus, a common set of miRNAs was associated with measures of destructive pathology/neurodegeneration in both cohorts.
Table 3.
miRNAs Showing Significant Correlations With MRI Measures in Both Cohort1 and Cohort2
| miRNA | Description | Relevant Pathways Involveda |
Cell Types/Tissues Expressing |
Relevant Findings in Previous Publications |
|---|---|---|---|---|
| hsa.miR.142.5p | Negative correlation with T1:T2 ratio (protective) | Axonal guidance signaling, IL-6 signaling, B-cell activating factor signaling | PBMCs, neurons | Upregulated in both PBMCs and brain white matter lesions from patients with MS and mouse model; normalizes with BMT and GA treatment; in patient’s serum, negative correlation with EDSS49 |
| hsa.miR.143.3p | Negative correlation with T1:T2 ratio (protective) | NF-κB signaling pathway, T-cell receptor signaling pathway, leukocyte transendothelial migration | CD4 T cells, macrophages, dendritic cells, neurons | Markedly decreased in serum of patients with AD vs age-matched controls50 |
| hsa.miR.181c.3p | Negative correlation with T1:T2 ratio (protective) | Axonal guidance signaling, TGF-β signaling | B cells, retinal cells, and in brain | Upregulated in MS CSF vs other neurologic diseases, upregulated in RR MS vs SP MS28 |
| hsa.miR.181c.5p | Negative correlation with T1:T2 ratio (protective) | TGF-β signaling pathway, neurotrophin signaling pathway, T-cell receptor signaling pathway | Brain | Released from the brain following ischemia and associated with neurogenesis51 |
| hsa.miR.375 | Negative correlation with BPF (pathogenic) | No known targets | Brain, spinal cord | CSF levels lower in patients with AD vs age- and sex-matched HCs52 |
| hsa.miR.486.5p | Positive correlation with T1:T2 ratio (pathogenic) | Integrin signaling, leukocyte extravasation signaling | Muscle | Correlates with EDSS; upregulated in patients with MS vs HCs, patients with progressive MS, those with other neurologic diseases, and those with other autoimmune diseases53 |
| hsa.miR.629.5p | Negative correlation with BPF (pathogenic) | IL-10 signaling, axonal guidance signaling | Whole blood | Lower expression found in blood samples from patients with MS treated with natalizumab and developed PML vs patients who did not develop PML54 |
| hsa.miR.92a.3p | Positive correlation with T1:T2 ratio (pathologic) | B-cell receptor signaling, chemokine signaling, neuregulin signaling | Brain, B cells | Its cluster, miR17-92, is overexpressed in B cells and plasma of patients with MS vs controls34,55 |
Abbreviations: AD, Alzheimer disease; BMT, bone marrow transplantation; BPF, brain parenchymal fraction; CSF, cerebrospinal fluid; EDSS, Expanded Disability Status Scale; GA, glatiramer acetate; HC, healthy controls; IL, interleukin; miRNA, microRNA; MRI, magnetic resonance imaging; MS, multiple sclerosis; NF-κB, nuclear factor κB; PBMCs, peripheral blood mononuclear cells; PML, progressive multifocal leukoencephalopathy; RR, relapsing-remitting; SP, secondary progressive; T1:T2, ratio of T1 hypointense to T2 hyperintense lesion volume; TGF-β, transforming growth factor β.
Ingenuity pathway analysis.
Figure 2. MicroRNA (miRNA) Correlation With Magnetic Resonance Imaging (MRI) Measures of Disease Severity.
miRNAs showed significant correlations with MRI measures of disease severity in both cohort 1 and cohort 2. BPF indicates whole-brain parenchymal fraction; BPV, whole-brain parenchymal volume; and T1:T2, ratio of T1 hypointense to T2 hyperintense lesion volume.
Correlations Between miRNAs and Disability
Several miRNAs showed significant associations with EDSS score in both cohorts (Tables 1 and 2). Seven of these associations remained significant after false discovery rate correction (Table 1).
Discussion
We evaluated the association between serum miRNA and MRI measures of disease severity in MS, including lesions and atrophy, in the brain and spinal cord. Several key findings emerged: both protective and pathogenic associations were found between miRNAs and MRI; a topographic specificity, different between the brain and spinal cord, was identified; a pathobiologic specificity was found with T2 lesions showing a different miRNA signature vs atrophy and destructive measures; and T2 lesions demonstrated the weakest link to both miRNA and disability.
MicroRNAs have been proposed as biomarkers relevant to diagnosis, stage of disease, and response to treatment in MS.33–35,53 Magnetic resonance imaging techniques have long contributed to the diagnosis and monitoring of the disease.3 Imaging advances have allowed measurement of heterogeneous pathologic processes, allowing clustering of patients based on these differences.15,56 To further understand the correlation between pathogenic and protective miRNAs and the relevance of their association with MRI in the present study, we performed an Ingenuity Pathway Analysis (https://targetexplorer.ingenuity.com/) to identify the most relevant experimentally observed gene targets of the miRNAs.
Among the identified miRNAs, reduced expression of hsa.miR.143.3p correlated with increased T1:T2 ratio as a protective association. Previous studies indicate that hsa.miR.143.3p is inversely associated with brain tissue damage, with decreased levels in the serum in persons with Alzheimer disease, minimal cognitive impairment, and vascular dementia vs the levels in healthy individuals.50 Deregulated expression of hsa.miR.143 is present in synaptoneurosomes isolated from prion-infected mice at both asymptomatic and symptomatic stages of the disease.57 The expression of hsa.miR.143 in neuronal stem cells is increased following the administration of insulinlike growth factor-158 (known to support the proliferation and enhanced survival of neuronal stem cells).59 An analysis of the downstream cascade of hsa.miR.143 has exposed many potential targets, such as platelet-derived growth factor receptor-α, which play a significant role in the differentiation of oligodendrocytes, as trocytes, and neuroprogenitors.60 Another target, mitogen-activated protein kinase, regulates differentiation of neuronal stem cells.61 Taken together, the above results suggest the potential role of hsa.miR.143.3p in neuroprotection and repair, consistent with the present findings.
Similarly, hsa.miR.142.5p, which also showed a protective correlation with T1:T2 ratio in our study, is known to be downregulated in cerebrospinal fluid and plasma from patients with Alzheimer disease.62 As predicted by pathway analysis, hsa.miR.142.5p targets nuclear factor erythroid-like 2 and plays a key role in regulating genes involved in responses to free radical production upon injury and inflammation.63
The pathogenic miRNA, hsa.miR.92a.3p, showed a pathogenic relationship with T1:T2 ratio in our study. This miRNA belongs to the miR17–92 cluster, which is overexpressed in B cells55 and plasma34 in patients with MS vs healthy individuals serving as controls. In mice, miR17–92 favors proinflammatory Th17 polarization.64 Furthermore, CD4 T-cell–specific ablation of the miR17–92 cluster mitigates experimental autoimmune encephalomyelitis by inhibiting the effector function of Th17 cells.
Another pathogenic miRNA in our study with regard to T1:T2 ratio (hsa.miR.486.5p) primarily targets phosphatase and tensin homologue (PTEN) and forkhead Box O1 (FOXO1).65,66 Both PTEN and FOXO1 are involved in the PI3K/Akt signaling pathway. T-cell receptor and cytokine signaling activate this pathway, which leads to Akt phosphorylation and inactivation of FOXO1, which is a factor critical for T-cell homeostasis, homing, and Treg induction. PTEN is a suppressor of the phosphatidylinositol 3-kinase/protein kinase B (P13K/Akt) pathway67,68; therefore, enhanced expression of hsa.miR.486.5p may lead to downregulation of PTEN and FOXO1, followed by activation of the P13K/Akt pathway and T-cell dysfunction. The other pathogenic miRNA with regard to T1:T2 ratio in our study, hsa.miR.92a.3p, also targets PTEN69,70 and other proteins, including IKAROS family zinc finger 1 (IKZF1),71 which is a transcription factor that has been shown to regulate hematopoietic cell development. Studies indicate that IKZF1 represses the expression of Tbet transcription factor, a regulator of Th1 cells. In addition, IKZF1 represses the B-cell production of IgG2c, an IgG whose levels are upregulated in autoimmune disorders.72,73 Therefore, one can speculate that hsa.miR.92a.3p plays a pathogenic role by repressing gene targets known to regulate immune cell activity and prevent inflammation.
miRNA hsa.miR.181c.5p showed a protective correlation with T1:T2 ratio in our study. This miRNA targets TRA, CD69, and AICDA—all known to mediate proinflammatory pathogenic effects by modulating T-cell activation or autoantibody production by B cells.74–79 Therefore, a reduction in protective miRNA levels may enhance inflammatory responses by increased expression of its targets. Another target of hsa.miR.181c.5p includes matrix metallopeptidase 14 (MMP14),80 which is implicated in cell migration and infiltration. Loss of MMP14 in mice was shown to impair monocyte migration, transendothelial invasion, cytokine release, and infiltration of T cells into sites of inflammation.81 These findings suggest that the presence of MMP14 due to low levels of this protective miRNA may correlate with disease progression by allowing entry of immune cells into the central nervous system.
An important observation in this study was the topographic specificity of the relationships that we detected between miRNAs and MRI. Different sets of miRNAs were associated with spinal cord MRI measures of disease severity (lesions or atrophy) vs brain measures. This divergence is in line with previous laboratory and clinical studies showing that specific genetic susceptibility is associated with a predilection for spinal cord lesions82 and that different effector T-cell subsets induce predominantly spinal cord (Th1) or brain (Th17) parenchymal infiltration and inflammation.83 In further support of this concept, a study found no correlation between brain and spinal cord MRI involvement in MS.21 Other studies have shown the complementary information obtained by combining brain and spinal cord MRI to characterize disease severity.20,84
We observed a limited overlap between miRNAs associated with cerebral lesions (T2LV) vs those associated with tissue destruction (ie, atrophy [BPF, BPV, GMF, and cGMV] or severe lesions [T1:T2]). Such findings are consistent with the weak association that has been detected between T2LV and brain atrophy.15,56 These observations suggest a decoupling between focal inflammatory demyelination in white matter and diffuse cerebral neurodegeneration.15 A range of factors other than T2 lesions may be more important determinants of tissue loss, such as remyelination and repair capacity,85 meningeal inflammation,86 microglial proliferation,87 and toxins or oxidative stress.88
Regarding lesion measures, T2LV showed the weakest association with either disability or miRNA expression. This observation is consistent with the notion that T2 lesions are nonspecific for the nature and severity of tissue injury, but the development of lesional T1 hypointensity9 has a higher clinical relevance. This would explain why a T1:T2 lesion index shows more utility than T2LV in tracking therapeutic effects and providing clinical relevance in MS.11,89
Strengths and Limitations
To our knowledge, this is the first study that includes 2 large, independent patient cohorts; a highly specific, locked nucleic acid–based, quantitative polymerase chain reaction platform for miRNA expression29; and quantitative MRI analysis that includes the brain and spinal cord. One of the limitations is the lack of traditional discovery and validation phases, that is, the patients in cohort 2 were different from those in cohort 1 in their treatment history and the miRNA panel. However, this difference might indicate better generalizability of our findings given their consistency despite these variations. Among the miRNAs showing a significant correlation with disability, several correlations remained significant after correcting for multiple comparisons. In contrast, none of the correlations with MRI remained significant following this correction. These limitations temper our conclusions and suggest the need for further validation of our findings in future studies. Some of the miRNAs that significantly correlated with disease severity measures in the present study have also been related to MS or experimental autoimmune encephalomyelitis (Table 3). However, these correlations do not necessarily represent causation, although the evidence described above suggests such a relationship. Circulating miRNAs could be a byproduct of routine microvesicle secretion and cell death or be actively secreted to contribute to intracellular communication.90
Conclusions
Further studies in larger MS cohorts, other neurodegenerative diseases, and healthy individuals serving as controls may indicate whether the miRNAs identified in this study are exclusive to MS or generalized. A study with longitudinal design, with serum samples drawn at disease onset, would allow the evaluation of the ability of miRNA expression to determine subsequent disease evolution.
Key Points.
Question
Are serum microRNAs associated with brain and spinal cord magnetic resonance imaging involvement in multiple sclerosis?
Findings
In this cross-sectional cohort study of 120 patients with multiple sclerosis, microRNA magnetic resonance imaging associations were both pathogenic and protective. A topographic specificity differed for the brain vs the spinal cord, and the microRNA signature differed between magnetic resonance imaging T2 lesions and atrophy measures.
Meaning
Serum microRNAs may serve as multiple sclerosis biomarkers for monitoring disease progression and act as surrogate markers to identify distinct underlying disease processes, such as inflammation vs tissue destruction.
Acknowledgments
Conflict of Interest Disclosures: Dr Healy has received grant support from Genzyme, Merck-Serono, Novartis, and Verily and consulting fees from Biogen. Dr Diaz-Cruz has received research support from EMD Serono and Verily. Dr Kivisakk has received research support from Biogen, EMD Serono, Sanofi, and Verily. Dr Chitnis has served as a paid consultant for Biogen, Teva, Novartis, Roche, and Sanofi and received research support from Biogen, Merck-Serono, Novartis, and Verily. Dr Weiner has served as a paid consultant for Gerson Lehrman, Genentech, and Tiziana and received research support from EMD Serono, Miragen, Sanofi, Teva, and Verily. Dr Gandhi has served as a paid consultant for Biogen and received research support from Biogen, EMD Serono, Genzyme, and Novartis. Dr Bakshi has served as a paid consultant for AbbVie, EMD Serono, Genentech, and Novartis and received research support from Biogen, EMD Serono, Novartis, and Sanofi Genzyme. No other disclosures were reported.
Funding/Support: This study was funded by a research grant from Biogen, Inc, who conducted a medical accuracy review of this manuscript, and by Watercove Charitable Foundation.
Footnotes
Author Contributions: Drs Gandhi and Bakshi contributed equally to the project, had full access to all the data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Weiner, Gandhi, Bakshi.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Regev, Healy, Gandhi, Bakshi.
Critical revision of the manuscript for important intellectual content: Khalid, Paul, Chu, Tauhid, Tummala, Diaz-Cruz, Raheja, Mazzola, von Glehn, Kivisakk, Dupuy, Kim, Chitnis, Weiner, Gandhi, Bakshi.
Statistical analysis:, Healy.
Obtained funding: Weiner, Gandhi, Bakshi.
Administrative, technical, or material support: All authors.
Study supervision: Weiner, Gandhi, Bakshi.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation or approval of the manuscript; and decision to submit the manuscript for publication.
Meeting Presentations: This work was presented in preliminary form at the 2015 annual meeting of the European Committee on Treatment and Research in Multiple Sclerosis; October 9, 2015; Barcelona, Spain; and at the 2016 annual meeting of the American Academy of Neurology; April 19, 2016; Vancouver, British Columbia, Canada.
References
- 1.Kutzelnigg A, Lassmann H. Pathology of multiple sclerosis and related inflammatory demyelinating diseases. Handb Clin Neurol. 2014;122:15–58. doi: 10.1016/B978-0-444-52001-2.00002-9. [DOI] [PubMed] [Google Scholar]
- 2.Geurts JJ, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. Lancet Neurol. 2012;11(12):1082–1092. doi: 10.1016/S1474-4422(12)70230-2. [DOI] [PubMed] [Google Scholar]
- 3.Filippi M, Rocca MA, Arnold DL, et al. EFNS guidelines on the use of neuroimaging in the management of multiple sclerosis. Eur J Neurol. 2006;13(4):313–325. doi: 10.1111/j.1468-1331.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- 4.Filippi M, Wolinsky JS, Comi G CORAL Study Group. Effects of oral glatiramer acetate on clinical and MRI-monitored disease activity in patients with relapsing multiple sclerosis: a multicentre, double-blind, randomised, placebo-controlled study. Lancet Neurol. 2006;5(3):213–220. doi: 10.1016/S1474-4422(06)70327-1. [DOI] [PubMed] [Google Scholar]
- 5.Bakshi R, Thompson AJ, Rocca MA, et al. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol. 2008;7(7):615–625. doi: 10.1016/S1474-4422(08)70137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakshi R, Dandamudi VS, Neema M, De C, Bermel RA. Measurement of brain and spinal cord atrophy by magnetic resonance imaging as a tool to monitor multiple sclerosis. J Neuroimaging. 2005;15(4 suppl):30S–45S. doi: 10.1177/1051228405283901. [DOI] [PubMed] [Google Scholar]
- 7.Filippi M. MRI measures of neurodegeneration in multiple sclerosis: implications for disability, disease monitoring, and treatment. J Neurol. 2015;262(1):1–6. doi: 10.1007/s00415-014-7340-9. [DOI] [PubMed] [Google Scholar]
- 8.Pirko I, Lucchinetti CF, Sriram S, Bakshi R. Gray matter involvement in multiple sclerosis. Neurology. 2007;68(9):634–642. doi: 10.1212/01.wnl.0000250267.85698.7a. [DOI] [PubMed] [Google Scholar]
- 9.Zivadinov R, Bakshi R. Role of MRI in multiple sclerosis; I: inflammation and lesions. Front Biosci. 2004;9:665–683. doi: 10.2741/1251. [DOI] [PubMed] [Google Scholar]
- 10.van Walderveen MA, Kamphorst W, Scheltens P, et al. Histopathologic correlate of hypointense lesions on T1-weighted spin-echo MRI in multiple sclerosis. Neurology. 1998;50(5):1282–1288. doi: 10.1212/wnl.50.5.1282. [DOI] [PubMed] [Google Scholar]
- 11.Kim G, Tauhid S, Dupuy SL, et al. An MRI-defined measure of cerebral lesion severity to assess therapeutic effects in multiple sclerosis. J Neurol. 2016;263(3):531–538. doi: 10.1007/s00415-015-8009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truyen L, van Waesberghe JH, van Walderveen MA, et al. Accumulation of hypointense lesions (“black holes”) on T1 spin-echo MRI correlates with disease progression in multiple sclerosis. Neurology. 1996;47(6):1469–1476. doi: 10.1212/wnl.47.6.1469. [DOI] [PubMed] [Google Scholar]
- 13.Janardhan V, Bakshi R. Quality of life and its relationship to brain lesions and atrophy on magnetic resonance images in 60 patients with multiple sclerosis. Arch Neurol. 2000;57(10):1485–1491. doi: 10.1001/archneur.57.10.1485. [DOI] [PubMed] [Google Scholar]
- 14.Tauhid S, Chu R, Sasane R, et al. Brain MRI lesions and atrophy are associated with employment status in patients with multiple sclerosis. J Neurol. 2015;262(11):2425–2432. doi: 10.1007/s00415-015-7853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauhid S, Neema M, Healy BC, Weiner HL, Bakshi R. MRI phenotypes based on cerebral lesions and atrophy in patients with multiple sclerosis. J Neurol Sci. 2014;346(1–2):250–254. doi: 10.1016/j.jns.2014.08.047. [DOI] [PubMed] [Google Scholar]
- 16.Sormani MP, Arnold DL, De Stefano N. Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann Neurol. 2014;75(1):43–49. doi: 10.1002/ana.24018. [DOI] [PubMed] [Google Scholar]
- 17.Sanfilipo MP, Benedict RH, Sharma J, Weinstock-Guttman B, Bakshi R. The relationship between whole brain volume and disability in multiple sclerosis: a comparison of normalized gray vs. white matter with misclassification correction. Neuroimage. 2005;26(4):1068–1077. doi: 10.1016/j.neuroimage.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Sanfilipo MP, Benedict RH, Weinstock-Guttman B, Bakshi R. Gray and white matter brain atrophy and neuropsychological impairment in multiple sclerosis. Neurology. 2006;66(5):685–692. doi: 10.1212/01.wnl.0000201238.93586.d9. [DOI] [PubMed] [Google Scholar]
- 19.Oh J, Seigo M, Saidha S, et al. Spinal cord normalization in multiple sclerosis. J Neuroimaging. 2014;24(6):577–584. doi: 10.1111/jon.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bot JC, Barkhof F, Polman CH, et al. Spinal cord abnormalities in recently diagnosed MS patients: added value of spinal MRI examination. Neurology. 2004;62(2):226–233. doi: 10.1212/wnl.62.2.226. [DOI] [PubMed] [Google Scholar]
- 21.Cohen AB, Neema M, Arora A, et al. The relationships among MRI-defined spinal cord involvement, brain involvement, and disability in multiple sclerosis. J Neuroimaging. 2012;22(2):122–128. doi: 10.1111/j.1552-6569.2011.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTRofribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica. 2009;137(2):159–164. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 25.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haghikia A, Haghikia A, Hellwig K, et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79(22):2166–2170. doi: 10.1212/WNL.0b013e3182759621. [DOI] [PubMed] [Google Scholar]
- 29.Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 30.Guerau-de-Arellano M, Smith KM, Godlewski J, et al. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain. 2011;134(pt 12):3578–3589. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisele S, Krumbholz M, Fischer MT, et al. Prospects of transcript profiling for mRNAs and microRNAs using formalin-fixed and paraffin-embedded dissected autoptic multiple sclerosis lesions. Brain Pathol. 2012;22(5):607–618. doi: 10.1111/j.1750-3639.2012.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thamilarasan M, Koczan D, Hecker M, Paap B, Zettl UK. MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun Rev. 2012;11(3):174–179. doi: 10.1016/j.autrev.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Gandhi R. miRNA in multiple sclerosis: search for novel biomarkers. Mult Scler. 2015;21(9):1095–1103. doi: 10.1177/1352458515578771. [DOI] [PubMed] [Google Scholar]
- 34.Gandhi R, Healy B, Gholipour T, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73(6):729–740. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 35.Huang Q, Xiao B, Ma X, et al. MicroRNAs associated with the pathogenesis of multiple sclerosis. J Neuroimmunol. 2016;295–296:148–161. doi: 10.1016/j.jneuroim.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Eikelenboom MJ, Petzold A, Lazeron RH, et al. Multiple sclerosis: neurofilament light chain antibodies are correlated to cerebral atrophy. Neurology. 2003;60(2):219–223. doi: 10.1212/01.wnl.0000041496.58127.e3. [DOI] [PubMed] [Google Scholar]
- 37.Gauthier SA, Glanz BI, Mandel M, Weiner HL. A model for the comprehensive investigation of a chronic autoimmune disease: the multiple sclerosis CLIMB study. Autoimmun Rev. 2006;5(8):532–536. doi: 10.1016/j.autrev.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtzke JF. On the origin of EDSS. Mult Scler Relat Disord. 2015;4(2):95–103. doi: 10.1016/j.msard.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Oommen VV, Tauhid S, Healy BC, et al. The effect of fingolimod on conversion of acute gadolinium-enhancing lesions to chronic T1 hypointensities in multiple sclerosis. J Neuroimaging. 2016;26(2):184–187. doi: 10.1111/jon.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakshi R, Neema M, Healy BC, et al. Predicting clinical progression in multiple sclerosis with the Magnetic Resonance Disease Severity Scale. Arch Neurol. 2008;65(11):1449–1453. doi: 10.1001/archneur.65.11.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moodie J, Healy BC, Buckle GJ, et al. Magnetic Resonance Disease Severity Scale (MRDSS) for patients with multiple sclerosis: a longitudinal study. J Neurol Sci. 2012;315(1–2):49–54. doi: 10.1016/j.jns.2011.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu R, Tauhid S, Glanz BI, et al. Whole brain volume measured from 1.5T versus 3T MRI in healthy subjects and patients with multiple sclerosis. J Neuroimaging. 2016;26(1):62–67. doi: 10.1111/jon.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma J, Sanfilipo MP, Benedict RH, Weinstock-Guttman B, Munschauer FE, III, Bakshi R. Whole-brain atrophy in multiple sclerosis measured by automated versus semiautomated MR imaging segmentation. A JNR Am J Neuroradiol. 2004;25(6):985–996. [PMC free article] [PubMed] [Google Scholar]
- 45.Horsfield MA, Sala S, Neema M, et al. Rapid semi-automatic segmentation of the spinal cord from magnetic resonance images: application in multiple sclerosis. Neuroimage. 2010;50(2):446–455. doi: 10.1016/j.neuroimage.2009.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Healy BC, Arora A, Hayden DL, et al. Approaches to normalization of spinal cord volume: application to multiple sclerosis. J Neuroimaging. 2012;22(3):e12–e19. doi: 10.1111/j.1552-6569.2011.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim G, Khalid F, Oommen VV, et al. T1-vs. T2-based MRI measures of spinal cord volume in healthy subjects and patients with multiple sclerosis. BMC Neurol. 2015;15:124. doi: 10.1186/s12883-015-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bermel RA, Sharma J, Tjoa CW, Puli SR, Bakshi R. Asemiautomated measure of whole-brain atrophy in multiple sclerosis. J Neurol Sci. 2003;208(1–2):57–65. doi: 10.1016/s0022-510x(02)00425-2. [DOI] [PubMed] [Google Scholar]
- 49.Ma X, Zhou J, Zhong Y, et al. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci. 2014;11(8):810–818. doi: 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong H, Li J, Huang L, et al. Serum MicroRNA profiles serve as novel biomarkers for the diagnosis of Alzheimer’s disease. Dis Markers. 2015;2015:625659. doi: 10.1155/2015/625659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun M, Yamashita T, Shang J, et al. Time-dependent profiles of micro RNA expression induced by ischemic preconditioning in the gerbil hippocampus. Cell Transplant. 2015;24(3):367–376. doi: 10.3727/096368915X686869. [DOI] [PubMed] [Google Scholar]
- 52.Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer’s disease. PLoS One. 2015;10(5):e0126423. doi: 10.1371/journal.pone.0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regev K, Paul A, Healy B, et al. Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3(5):e267. doi: 10.1212/NXI.0000000000000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muñoz-Culla M, Irizar H, Castillo-Triviño T, et al. Blood miRNA expression pattern is a possible risk marker for natalizumab-associated progressive multifocal leukoencephalopathy in multiple sclerosis patients. Mult Scler. 2014;20(14):1851–1859. doi: 10.1177/1352458514534513. [DOI] [PubMed] [Google Scholar]
- 55.Sievers C, Meira M, Hoffmann F, Fontoura P, Kappos L, Lindberg RL. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol. 2012;144(1):70–79. doi: 10.1016/j.clim.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Bielekova B, Kadom N, Fisher E, et al. MRI as a marker for disease heterogeneity in multiple sclerosis. Neurology. 2005;65(7):1071–1076. doi: 10.1212/01.wnl.0000178984.30534.f9. [DOI] [PubMed] [Google Scholar]
- 57.Boese AS, Saba R, Campbell K, et al. MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol Cell Neurosci. 2016;71:13–24. doi: 10.1016/j.mcn.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Pati S, Supeno NE, Muthuraju S, et al. MicroRNA profiling reveals unique miRNA signatures in IGF-1 treated embryonic striatal stem cell fate decisions in striatal neurogenesis in vitro. Biomed Res Int. 2014;2014:503162. doi: 10.1155/2014/503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Supeno NE, Pati S, Hadi RA, et al. IGF-1 acts as controlling switch for long-term proliferation and maintenance of EGF/FGF-responsive striatal neural stem cells. Int J Med Sci. 2013;10(5):522–531. doi: 10.7150/ijms.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song J, Cho KJ, Cheon SY, et al. Apoptosis signal-regulating kinase 1 (ASK1) is linked to neural stem cell differentiation after ischemic brain injury. Exp Mol Med. 2013;45:e69. doi: 10.1038/emm.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sørensen SS, Nygaard AB, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia—an exploratory study. Transl Neurodegener. 2016;5:6. doi: 10.1186/s40035-016-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang S, Deng C, Lv J, et al. Nrf2 weaves an elaborate network of neuroprotection against stroke [published online February 5, 2016] Mol Neurobiol. doi: 10.1007/s12035-016-9707-7. [DOI] [PubMed] [Google Scholar]
- 64.Liu S-Q, Jiang S, Li C, Zhang B, Li Q-J. miR-17-92 cluster targets phosphatase and tensin homology and ikaros family zinc finger 4 to promote TH17-mediated inflammation. J Biol Chem. 2014;289(18):12446–12456. doi: 10.1074/jbc.M114.550723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang LS, Li L, Li L, et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015;125(8):1302–1313. doi: 10.1182/blood-2014-06-581926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander MS, Casar JC, Motohashi N, et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest. 2014;124(6):2651–2667. doi: 10.1172/JCI73579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton RH, Turka LA. Regulation of T cell homeostasis and responses by pten. Front Immunol. 2012;3:151. doi: 10.3389/fimmu.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 69.Zhang H, Cao H, Xu D, Zhu K. MicroRNA-92a promotes metastasis of nasopharyngeal carcinoma by targeting the PTEN/AKT pathway. Onco Targets Ther. 2016;9:3579–3588. doi: 10.2147/OTT.S105470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ke T-W, Wei P-L, Yeh K-T, Chen WT-L, Cheng Y-W. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol. 2015;22(8):2649–2655. doi: 10.1245/s10434-014-4305-2. [DOI] [PubMed] [Google Scholar]
- 71.Mavrakis KJ, Van Der Meulen J, Wolfe AL, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43(7):673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu S, Ou X, Huo J, et al. Mir-17-92 regulates bone marrow homing of plasma cells and production of immunoglobulin G2c. Nat Commun. 2015;6:6764. doi: 10.1038/ncomms7764. [DOI] [PubMed] [Google Scholar]
- 73.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48(9–10):1272–1278. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 74.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression inordered stages of cellular development. Genes Dev. 2007;21(5):578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Podshivalova K, Salomon DR. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit Rev Immunol. 2013;33(5):435–476. doi: 10.1615/critrevimmunol.2013006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saki N, Abroun S, Soleimani M, et al. Involvement of microRNA in T-cell differentiation and malignancy. Int J Hematol Oncol Stem Cell Res. 2015;9(1):33–49. [PMC free article] [PubMed] [Google Scholar]
- 77.González-Amaro R, Cortés JR, Sánchez-Madrid F, Martín P. Is CD69 an effective brake to control inflammatory diseases? Trends Mol Med. 2013;19(10):625–632. doi: 10.1016/j.molmed.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Podojil JR, Miller SD. Molecular mechanisms of T-cell receptor and costimulatory molecule ligation/blockade in autoimmune disease therapy. Immunol Rev. 2009;229(1):337–355. doi: 10.1111/j.1600-065X.2009.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsu HC, Yang P, Wu Q, et al. Inhibition of the catalytic function of activation-induced cytidine deaminase promotes apoptosis of germinal center B cells in BXD2 mice. Arthritis Rheum. 2011;63(7):2038–2048. doi: 10.1002/art.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Kuscu C, Banach A, et al. miR-181a-5p Inhibits cancer cell migration and angiogenesis via downregulation of matrix metalloproteinase-14. Cancer Res. 2015;75(13):2674–2685. doi: 10.1158/0008-5472.CAN-14-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klose A, Zigrino P, Mauch C. Monocyte/macrophage MMP-14 modulates cell infiltration and T-cell attraction in contact dermatitis but not in murine wound healing. Am J Pathol. 2013;182(3):755–764. doi: 10.1016/j.ajpath.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 82.Sombekke MH, Lukas C, Crusius JB, et al. HLA-DRB1*1501 and spinal cord magnetic resonance imaging lesions in multiple sclerosis. Arch Neurol. 2009;66(12):1531–1536. doi: 10.1001/archneurol.2009.278. [DOI] [PubMed] [Google Scholar]
- 83.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM. Differential regulation of central nervous system autoimmunity by TH1 and TH17 cells. Nat Med. 2008;14(3):337–342. doi: 10.1038/nm1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bakshi R, Neema M, Tauhid S, et al. An expanded composite scale of MRI-defined disease severity in multiple sclerosis: MRDSS2. Neuroreport. 2014;25(14):1156–1161. doi: 10.1097/WNR.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olsen JA, Akirav EM. Remyelination in multiple sclerosis: cellular mechanisms and novel therapeutic approaches. J Neurosci Res. 2015;93(5):687–696. doi: 10.1002/jnr.23493. [DOI] [PubMed] [Google Scholar]
- 86.Zurawski J, Lassmann H, Bakshi R. Use of magnetic resonance imaging to visualize leptomeningeal inflammation in patients with multiple sclerosis: a review [published online November 28, 2016] JAMA Neurol. doi: 10.1001/jamaneurol.2016.4237. [DOI] [PubMed] [Google Scholar]
- 87.Giannetti P, Politis M, Su P, et al. Microglia activation in multiple sclerosis black holes predicts outcome in progressive patients: an in vivo [11C](R)-PK11195-PET pilot study. Neurobiol Dis. 2014;65:203–210. doi: 10.1016/j.nbd.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 88.Haider L, Simeonidou C, Steinberger G, et al. Multiple sclerosis deep grey matter: the relation between demyelination, neurodegeneration, inflammation and iron. J Neurol Neurosurg Psychiatry. 2014;85(12):1386–1395. doi: 10.1136/jnnp-2014-307712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beer A, Biberacher V, Schmidt P, et al. Tissue damage within normal appearing white matter in early multiple sclerosis: assessment by the ratio of T1- and T2-weighted MR image intensity. J Neurol. 2016;263(8):1495–1502. doi: 10.1007/s00415-016-8156-6. [DOI] [PubMed] [Google Scholar]
- 90.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]