Abstract
Since their discovery in the 1960s, the family of Fe(II)/2-oxoglutarate-dependent oxygenases has undergone a tremendous expansion to include enzymes catalyzing a vast diversity of biologically important reactions. Recent examples highlight roles in controlling chromatin modification, transcription, mRNA demethylation, and mRNA splicing. Others generate modifications in tRNA, translation factors, ribosomes, and other proteins. Thus, oxygenases affect all components of molecular biology’s central dogma in which information flows from DNA to RNA to proteins. These enzymes also function in biosynthesis and catabolism of cellular metabolites, including antibiotics and signaling molecules. Due to their critical importance, ongoing efforts have targeted family members for the development of specific therapeutics. This review provides a general overview of recently characterized oxygenase reactions and their key biological roles.
Keywords: non-heme iron oxygenase, chromatin modification, transcription, translation, biosynthesis, biodegradation
Chemistry and Diversity of Fe(II)/2-Oxoglutarate Oxygenases
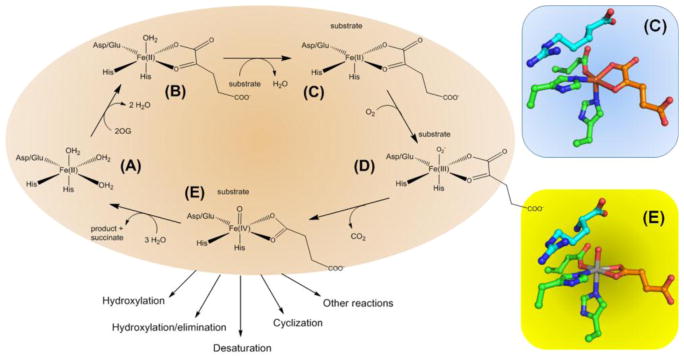
The Fe(II)/2-oxoglutarate (2OG, see Glossary)-dependent oxygenases catalyze a wide array of biochemical reactions [1–4] using a common double-stranded β-helix fold [5], and bioinformatics studies reveal a significant untapped potential for identifying additional family members in bacteria, metazoa, and plants [6–8]. The mechanisms of these enzymes have been intensively investigated [9], although questions remain to be answered about the chemistry of specific transformations. A simplified and generalized reaction cycle (Figure 1, Key Figure) illustrates several key features of the oxygenase active sites. (A) Fe(II) typically is bound to a 2-His-1-carboxylate motif with three water molecules coordinated to one face of the metal. (B) 2OG displaces two water molecules as it chelates the Fe(II) with its keto group opposite the carboxylate metal ligand. (C) The primary substrate binds near the metallocenter, displaces the remaining water molecule, and triggers the subsequent oxygen-related chemistry. (D) The binding of dioxygen generates an Fe(III)-superoxo species. (E) Oxidative decarboxylation of 2OG leads to release of CO2 and formation of an Fe(IV)-oxo (ferryl) species with bound succinate. Potentially all subsequent reactions utilize this common intermediate, leading to product formation and rebinding of water to restore the original species. We depict active site views of two states of VioC, a representative enzyme that hydroxylates the C3 atom of L-Arg. The substrate- and 2OG-bound state of VioC (C) illustrates both the open coordination site for binding dioxygen and the flexibility in the position of the 2OG C1 carboxylate. The substrate- and succinate-bound form of VioC containing vanadyl ion (E) provides a stable structural mimic of the unstable ferryl intermediate state [10]. The spectroscopic properties of catalytic intermediates are best understood for TauD, a sulfonate-degrading oxygenase [11]. Below, we focus on the biochemical roles for members of this diverse set of enzymes.
Figure 1.
Generalized Mechanism of 2OG-Dependent Oxygenases. (A) Active site metallocenter with Fe(II) coordinated by a 2-His-1-carboxylase motif. (B) 2OG-bound protein. (C) Creation of an O2-binding site in the substrate- and 2OG-bound state. (D) Fe(III)-superoxo species. (E) Ferryl intermediate that is common to all further chemistry. Also depicted are two views of the VioC active site: (C) Fe(II)-containing protein with bound L-Arg and 2OG (PDB access code 6ALM) and (E) vanadyl-containing protein (as a mimic of the ferryl species) with bound L-Arg and succinate (PDB access code 6ALR).
Relationship of 2OG-Dependent Oxygenases to the Central Dogma
Several members of the 2OG-dependent oxygenase family have regulatory or structural roles related to information flow from DNA to RNA to proteins. The enzymatic functions vary from removing chromatin modifications to creating hypermodification sites in tRNA to hydroxylating transcriptional or translational regulatory factors [12]. In this section, we provide a general overview that highlights several key areas where these proteins have significant impacts on processes related to the central dogma of molecular biology [12, 13].
2OG-Dependent Oxygenases Involved in Chromatin Protein Modification
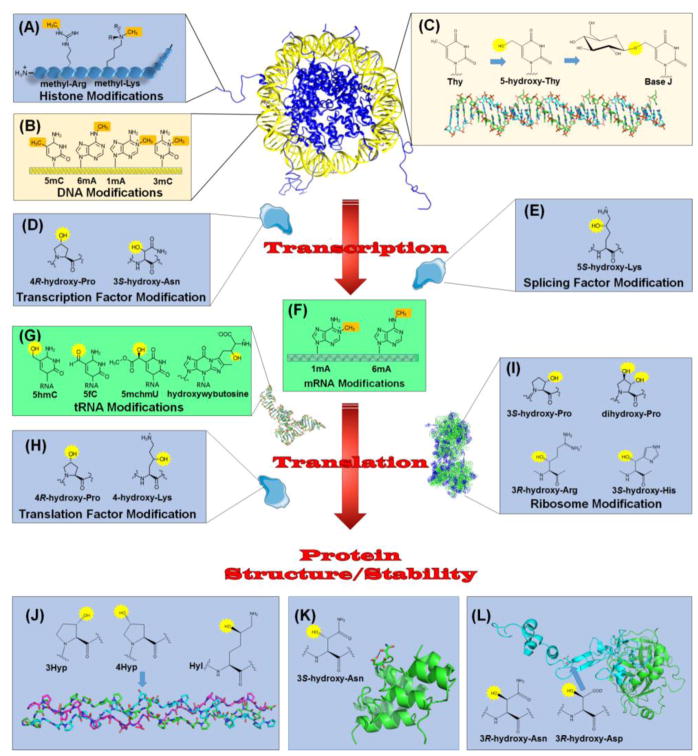
Eukaryotes package DNA into chromatin by interaction with histone proteins (H2A, H2B, H3, and H4) possessing N-terminal regions that undergo modifications including methylations of Lys and Arg residues (Figure 2A), with consequent transcriptional regulatory effects. A large family of 2OG-dependent oxygenases reverses these methylated sites with exquisite sequence and methylation status specificity. These enzymes contain a Jumonji C (JmjC) catalytic domain [14], comprising a distinct structural sub-family [5], and include both N-methyl-lysine demethylases (i.e. KDMs) [15] that target mono-, di-, and tri-methylated Lys along with N-methyl-arginine demethyases (i.e. RDMs) [16] that may target mono- and di-methyl Arg. The biological relevance of RDMs is not established and other investigators have proposed an alternative role for a subset of JmjC proteins in recognizing methylated arginine residues leading to histone endoproteolysis, thus resulting in tailless histones [17]. Hydroxylation of the methyl groups yield hemiaminal intermediates that decompose to release formaldehyde. These enzymes are associated with human cancers and genetic diseases, leading to great interest in identifying specific drugs to control these reactions [18–20] (Box 1).
Figure 2.
Relationship of 2OG-Dependent Oxygenases to the Central Dogma. Chromatin regulation involves methylation of (A) specific Lys and Arg residues in the N-terminal tails of histones and (B) several types of bases in DNA. Demethylases and other 2OG-dependent oxygenases function to reverse these modifications. (C) Base J is a DNA modification found in certain kinetoplasts. Selected thymidine bases (Thy) are modified by a 2OG-dependent hydroxylase to form 5-hydroxy-Thy, then glycosylated by a separate enzyme. (D) Regulated RNA synthesis involves transcription factors, such as HIF, modified by 2OG-dependent hydroxylases. (E) An oxygenase also modifies a splicing factor as part of the maturation of precursor RNA. (F) 2OG-dependent enzymes remove methylation marks in mRNA. (G) Representatives of these enzymes demethylate, hydroxylate, and hypermodify bases in tRNA. (H) Translation factors undergo hydroxylations catalyzed by 2OG-dependent oxygenase. (I) Several hydroxylations are introduced into ribosomal proteins by these enzymes. In addition, other proteins are hydroxylated related to structure/stability. (J) Collagen contains 4R-hydroxy-Pro (4Hyp), 3S-hydroxy-Pro (3Hyp), and 5R-hydroxy-Lys (Hyl) residues synthesized by 2OG-dependent hydroxylases. The structure is shown for a collagen model peptide (PDB access code 3ABN) that is rich in Pro and Gly residues and contains 4Hyp (arrow); three peptides (depicted in green, cyan, and magenta) form a triple-helical structure. (K) Some ankyrin repeat domain (ARD) proteins undergo hydroxylation at the 3S position of Asn residues, leading to protein stabilization. The structure shown (PDB access code 2ZGD) is from a synthetic consensus sequence hydroxylated by FIH. (L) Activated factor IX (PDB access code 1PFX) has a large catalytic chain (green) and a small chain (cyan) containing an EGF domain with a 3R-hydroxy-Asp residue (arrow). Also shown is the structure of 3R-hydroxy-Asn. DNA bases are shown in yellow, RNA components are in green, and protein sidechains are in blue. The methylation sites targeted for hydroxylation (A, B, and F) and the introduced hydroxylations (C, D, E, G, H, I, J, K, and L) are highlighted in brown and yellow, respectively.
Box 1. 2OG-Dependent Oxygenases as Potential Therapeutic Targets.
The potential for regulating specific oxygenases by use of selective inhibitors has garnered a great deal of interest as a means to control the associated biological activities. An early review of oxygenase inhibitors summarized the kinetic and structural properties of many small molecules that interact with this family of enzymes [103]; typical issues noted were lack of potency and specificity. More recently, many groups have attempted to identify more potent and specific inhibitors of histone demethylases associated with epigenetic modifications, the hypoxic response-related enzymes for promoting erythropoeisis, and other specific enzymes linked to dysfunctions ranging from ischemia, inflammation, obesity, to cancer. Several inhibitors focusing on JmjC proteins are therapeutic targets for various cancers [19]. Other inhibitors of HIF prolyl hydroxylases with known modes of action are now in clinical trials for anemia [28]. A ligand-based NMR screening method was developed and applied to γ-butyrobetaine hydroxylase [104] to replace the cardioprotective effects of meldonium (3-(2,2,2-trimethylhydrazinium)propionate), a currently available inhibitor that is mis-used as an athletic performance enhancer. To illustrate one direction for future development, a notable recent study investigated the interactions of a suite of ligands with 41 human oxygenases by use of a mass spectrometry-based ligand competition approach [20]. Despite these advances, the discovery of specific drugs with desirable characteristics continues to be a challenge.
2OG-Dependent Oxygenases Involved in Chromatin DNA Modification
Many, but not all, eukaryotes regulate DNA packaging and transcription by controlling the methylation status of cytosine bases within CpG islands of their DNA. Methyltransferases form 5-methylcytosine (5mC, Figure 2B), and three isozymes of the ten-eleven translocation (TET) hydroxylase family participate in reversing this DNA methylation [21]. The TET enzymes catalyze sequential conversion of 5mC to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine, with the demethylation process completed by action of a thymine DNA glycosylase and base excision repair. The demethylation activities exhibit important distinctions in different biological contexts (e.g., paternal zygote, primordial germ cells, embryonic stem cells, or neurons) and genetic dysfunctions of these enzymes are associated with malignancy [21].
A second form of methylation-dependent regulation of DNA has been found in mammalian embryonic stem cells [22]. In this case, the modification is N6-methyladenine (6mA, Figure 2B), with methylation leading to transcriptional (epigenetic) silencing. ALKBH1 reverses this methylation by forming an unstable hemiaminal and eliminating formaldehyde, similar to the demethylation reactions of the KDM and RDM proteins (vide supra).
In addition to the chromatin-specific oxygenases with regulatory roles, selected family members act to repair lesions caused by alkylation damage to DNA. For example, Escherichia coli AlkB demethylates the predominant lesions 1-methyladenine and 3-methylcytosine (1mA and 3mC, Figure 2B) [23]. Analogous enzymes are present in other bacteria, some viruses, and most eukaryotic organisms. Although humans and other mammals synthesize nine AlkB homologs (ALKBH1-ALKBH8 and the fat mass and obesity-associated gene product, FTO), most of these enzymes exhibit distinct functions. ALKBH2 serves as the primary mammalian DNA repair enzyme according to mouse knockout studies [24]. We mention several ALKBH orthologs elsewhere in this review, such as ALKBH1 described in the prior paragraph.
A final type of chromatin modification by a 2OG-dependent oxygenase involves a modified base occuring in the DNA of kinetoplastids such as the pathogenic unicellular eukaryotes Trypanosoma brucei, T. cruzi, and Leishmania species [25]. A hydroxylase first acts on a subset of thymidine bases, and the resulting 5-hydroxy-Thy is modified by a glycosylase to form base J (Figure 2C). This base is prevalent in repetitive DNA elements such as telomeric repeats. Surprisingly, the function of this modification remains unclear.
Roles of 2OG-Dependent Oxygenases in Regulation/Control of Transcription
The archetype example illustrating oxygenase control of transcription centers on the hypoxia-inducible factor (HIF), a critical oxygen-dependent transcription regulatory protein [26, 27]. During anoxic or low oxygen conditions, HIF is stable and induces the expression of more than 100 genes such as those regulating formation of blood vessels and red blood cell production (erythropoiesis) to combat ischemia. In the presence of sufficient oxygen, three isoenzymes containing the prolyl hydroxylase domain (PHD) modify two conserved prolyl residues in the α-subunit of HIF. These 4R-hydroxy-Pro sites (Figure 2D) enhance the binding affinity for the von Hipple-Lindau ubiquitin ligase, resulting in HIF ubiquitination, targeting to the proteasome, and destruction of the transcription factor. Inhibitors of these enzymes are of great interest for treating anemia (Box 1), and enzyme-bound structural studies have been reported for several inhibitors in clinical trials [28]. Another oxygenase acting on this transcription regulatory protein is factor inhibiting HIF (FIH). This protein is active at lower oxygen concentrations than the PHDs [29], and when O2 is present, FIH modifies a specific Asn residue in the C-terminal region of the transcriptional activation domain. The resulting 3δ-hydroxy-Asn (Figure 2D) hinders binding by the p300/CBP transcriptional activators, further reducing HIF activity.
RNA Splicing Control by 2OG Oxygenases
Splicing of precursor RNAs occurs in many organisms, and at least one known oxygenase regulates this process. The JmjC-domain protein JMJD6 mediates the hydroxylation of specific lysyl residues (forming 5S-hydroxy-Lys, Figure 2E) in splicing factor U2AF65, which in part controls the range of alternative splicing events that are observed [30]. JMJD6 also has been implicated, along with bromodomain-containing protein 4 (Brd4), in a transcriptional pause release function acting via enhancers [31]; however, it is unknown if the oxygenase activity is required for this process.
2OG-Dependent Oxygenases Acting on mRNA
Messenger RNA is a target of post-transcriptional regulation by methylation of particular bases, and specific oxygenases function to remove these modifications prior to translation. For example, 1mA (Figure 2F) occurs in thousands of eukaryotic gene transcripts in more structured regions around canonical and alternative translation initiation sites [32]. ALKBH3 demethylates 1mA and several other modified bases in RNA, and the presence of ALKBH3 positively correlates with translation efficiency of the expressed protein. Similarly, 6mA is a common modification in RNA (Figure 2F), and both ALKBH5 and FTO reverse this type of methylation [33]. Furthermore, the 5′ cap of mRNA typically contains 7-methylguanosine followed by N6,2′-O-dimethyladenosine, a modification that stabilizes mRNA in eukaryotic cells; FTO demethylates this base preferentially over 6mA [34]. Efforts to obtain selective inhibitors of these various enzymes have been challenging due to the similarities in their active sites [20].
Another target for hydroxylation in RNA is 5mC. All three TET proteins were shown to form 5-hydroxymethylcytosine in vitro [35]. This activity has a regulatory role in promoting myelopoiesis during pathogen infection in mammalian cells [36]. Furthermore, in pluripotent stem cells TET2 is recruited to chromatin by an RNA-binding protein with bound RNA; the oxygenase then hydroxylates 5mC in the RNA leading to its degradation [37].
2OG-Dependent Oxygenases Required for Translation
Translation requires tRNA molecules for which oxygenases play important roles in demethylation, hydroxylation, and hypermodification. For example, the frequency of 1mA increases in mitochondrial tRNA for ALKBH1 knockout cells, consistent with demethylation of this lesion by ALKBH1 [38]. This enzyme also participates in the synthesis of 5-hydroxymethyl-2′-O-methylcytidine and 5-formyl-2′-O-methylcytidine (5hmC and 5fC, Figure 2G) in the anticodon of mitochondrial tRNAMet [38], and it forms the latter species in the anticodon of cytoplasmic tRNALeu [39]. ALKBH8 exhibits two activities involving the tRNA anticodon loop; its methylase domain converts 5-carboxymethyluridine to 5-methoxycarbonylmethyluridine, then its oxygenase domain hydroxylates this species [40, 41] to form 5S-methyoxycarbonylhydroxymethyluridine (mchm5U, Figure 2G). A second type of tRNA hypermodification involves the TYW5-catalyzed hydroxylation of the tricyclic base 7-(α-amino-α-carboxypropyl)wyosine, or wybutosine, to form hydroxywybutosine (Figure 2G) [42]. This modification, found in eukaryotes and archaea, likely affects translational fidelity by ensuring use of the proper reading frame during translation.
Translation factors also are hydroxylated by these enzymes [43]. Elongation factor-Tu (EF-Tu) and elongation factor-1A form a complex that binds to the aminoacyl-tRNA delivering an amino acid to the A site within the mature ribosome. In some Gram-negative bacteria EF-Tu is hydroxylated to form a 4R-hydroxy-prolyl residue (Figure 2H), e.g. by the Pseudomonas prolyl hydroxylase domain containing protein (PPHD) enzyme [44], although the direct biological role still remains unclear [45]. A second example is the eukaryotic elongation release factor-1 (ERF-1) which is modified at C4 of a lysyl residue (Figure 2H) by JMJD4 [46]. This modification occurs in the domain that interacts with the stop codon, specifically with the uridine nucleotide, by forming a hydrogen bond that increases the ability to terminate translation.
Oxygenases also act directly on the ribosome [43, 47], and the different types of hydroxylations on different ribosomal subunits can alter the rate and efficiency of translation [45]. In humans, the structurally characterized protein OGFOD1 [48] forms a 3 S-hydroxy-prolyl residue (Figure 2I) in ribosomal protein S23 (RPS23/uS12) [49]. Inactivation of the gene encoding this mammalian oxygenase leads to formation of stress granules, arrest of translation, and impairment of growth. The same ribosomal protein is dihydroxylated (Figure 2I) by Tpa1p in Saccharomyces cerevisiae, and by related proteins in Schizosaccharomyces pombe and Ostreococcus tauri [50]. RPS23 hydroxylation in yeast determines viability due to suppression of the nonsense codon. A similar situation exists for Drosophila which possess the homolog, Sudestada1. RNAi-mediated inhibition of the fly enzyme led to a decrease in cell size, fewer cell numbers, and a reduction in translational efficiency [51].
In contrast to these prolyl modifications, the E. coli enzyme YcfD generates a 3R-hydroxy-arginyl residue (Figure 2I) in RPL16/uL16 [47]. The thermophilic bacterium Rhodothermus marinus also contains a YcfD homolog with this same activity, but it exhibits oxygen limitation at high temperatures [52]. Furthermore, one of the human orthologs of this enzyme, NO66, catalyzes the 3S-hydroxylation of histidyl residues (Figure 2I) in RPL8/uL2, and another human ortholog, MINA53, may do the same for RPL27a/eL27[47]. A high percentage (90–95%) of these proteins are modified in various cancer cell lines [47], normal cell tissue, and different tissue cell lines, leading to the proposal that histidyl hydroxylation promotes protein binding to helix 93 of the 28S subunit which causes conformational changes in the peptidyl transferase center of the mature ribosome [53].
Roles of 2OG-Dependent Oxygenases in Protein Structure/Stability
In this section, we briefly describe three examples in which 2OG-dependent oxygenases function after protein synthesis to assist in the structure or stability.
Collagen, the most abundant protein in mammals, contains extensively modified subunits that form triple helical filaments providing structural strength to connective tissues [54]. The crystal structure of a collagen model peptide [(Pro-Pro-Gly)4-4Hyp-Asp-Gly-(Pro-Pro-Gly)4] (Figure 2J) includes the most common collagen modification 4R-hydroxy-Pro (4Hyp), highlights the predominance of Pro and Gly residues, and illustrates a typical sequence repeat found in the protein [55]. Prolyl 4R-, prolyl 3S-, and lysyl 5R-hydroxylases generate 4Hyp, 3S-hydroxy-Pro (3Hyp), and 5R-hydroxy-Lys (Hyl) in collagen and selected other proteins. These modifications enhance the structural integrity of the final product through stereoelectronic and other effects. The crystal structure of a collagen lysyl hydroxylase was recently reported for a giant virus that infects the human lung [56], revealing features that are conserved in the human enzymes. Mutations in human genes for these oxygenases can lead to Ehlers-Danlos syndrome [57].
The ankyrin repeat domain (ARD) forms a hairpin/α-helix/loop/α-helix motif of ~33 residues and occurs in more than 250 human proteins. The example shown (Figure 2K) includes a highly conserved asparaginyl residue that is hydroxylated by FIH, mentioned earlier as being involved in hypoxic signaling. Formation of the 3S-hydroxy-Asn residue enhances the protein stability due to the formation of a hydrogen bond with an aspartyl residue located two residues earlier in the sequence [58]. Similar stabilization by side chain hydroxylation may be associated with other ARD-containing proteins [59, 60].
Secreted proteins and the extracellular domains of membrane-bound proteins in animals often contain an epidermal growth factor (EGF)-like domain of 30–40 residues. These EGF domains typically contain multiple disulfide bonds, glycosylation, and calcium-binding sites. For coagulation factors VII, IX, and X, complement factors, thrombomodulin, thrombospondin, proteins C, S, Z, and other examples, the EGF domain additionally possesses a 3R-hydroxy-Asp or 3R-hydroxy-Asn residue. The same enzyme, aspartyl (asparaginyl) β-hydroxylase, carries out both of these modifications. The structure of activated factor IX [61] depicts 3R-hydroxy-Asp within an EGF domain of its light chain (Figure 2L). In the absence of the hydroxylase, mice exhibit developmental defects and have an increased incidence of neoplasia, thus highlighting the importance of these modifications [62].
Roles of 2OG-Dependent Oxygenases in Biosynthesis
Biosynthetic pathways involving 2OG-dependent oxygenases are used to generate a tremendous diversity of metabolites [1, 2]. We present here only a few examples, all published later than 2014, illustrating the multiple types of chemistry catalyzed by these enzymes.
Hydroxylation
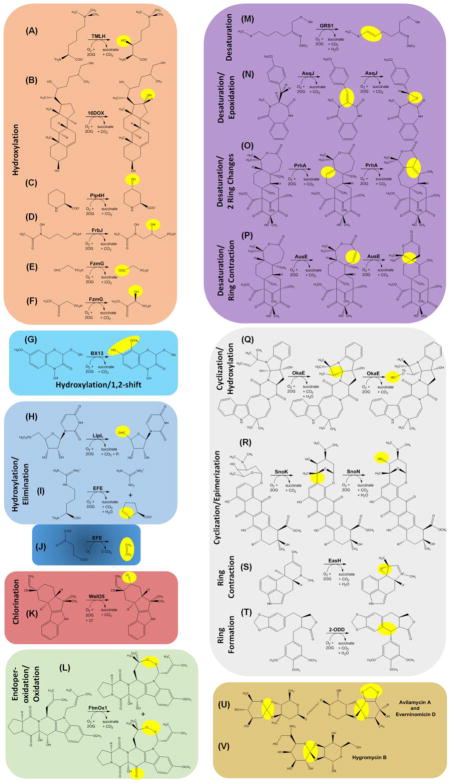
Among the enzymes that catalyze a prototypical hydroxylation reaction is trimethyllysine hydroxylase (TMLH); e.g., the human enzyme converts (2S)-Nε-trimethyllysine to (2S,3S)-3-hydroxy-Nε-trimethyllysine (Figure 3A) [63]. Subsequent aldolase, dehydrogenase, and hydroxylase reactions convert this product to carnitine which is used to transport fatty acids across membranes in animals or as an osmoprotectant in some bacteria [64]. Another recently described oxygenase involved in lipid metabolism is the potato steroid 16α-hydroxylase (16DOX) that acts on 22,26-dihydroxycholesterol (Figure 3B) [65]. This enzyme, broadly found in Solanaceae, participates in pathways that generate steroidal glycoalkaloids such as α-solanine and α-chaconine and is a candidate for gene silencing to reduce the toxicity from these poisons. Other hydroxylases are responsible for generating various hydroxypipecolic acids that are widespread in nature and incorporated into several antibiotics and alkaloids. A recent example is the L-pipecolic acid trans-4-hydroxylase (Pip4H) (Figure 3C) from Fusarium oxysporum and other fungi [66]. The synthesis of selected phosphonate-containing antibiotics also involves hydroxylases. For example, the structurally characterized FrbJ from Streptomyces rubellomurinus [67] catalyzes a reaction (Figure 3D) yielding the anti-malarial compound FR-33289. Similarly, FzmG from Streptomyces lavendofoliae catalyzes two hydroxylation reactions (Figure 3E and 3F) during the synthesis of fosfazinomycin, an anti-fungal compound [68].
Figure 3.
Representative Biosynthetic Reactions of 2OG-Dependent Oxygenases. (A–F) Selected hydroxylation reactions. (G) A hydroxylation reaction coupled with a 1,2-shift of a methoxy group. (H–I) Hydroxylation reactions followed by spontaneous elimination reactions. The enzyme catalyzing reaction (I) also catalyzes reaction (J) (see Box 2). (K) A chlorination reaction. (L) An endoperoxide-forming reaction, along with an oxidation reaction. (M–P) Desaturation reactions, in most cases accompanied by ring formation or ring rearrangement reactions. (Q–T) Additional ring transformation reactions. (U–V) Examples of orthoether linkages and methylenedioxy groups synthesized by 2OG-dependent oxygenases. See text for details.
Hydroxylation with a Secondary Reaction
A maize oxygenase used for synthesis of a toxic 8-O-methylated benzoxazinoid illustrates an intriguing twist on the simple hydroxylation reaction [69]. This enzyme, BX13, hydroxylates the C7 position of DIMBOA-Glc (2-(2,4,-dihydroxy-7-methoxy-1,4,-bezoxazin-3-one)-β-D-glucopyranose) while affecting a 1,2-shift of a methoxy group (Figure 3G). Two other alternatives to the simple hydroxylation reaction involve bacterial oxygenases for which elimination follows hydroxylation. In one case, LipL from Streptomyces sp. SANK 60405 (or Cpr19 from Amycolatopsis sp. SANK 60206) hydroxylates the 5′-methylene group of UMP (Figure 3H) to generate an unstable intermediate that eliminates phosphate and forms uridine-5′-aldehyde [70]. Further metabolism incorporates this compound into caprazamycin, cupuramycin, liposidomycin, and other antibiotics. The second example is the especially intriguing ethylene-forming enzyme (EFE, see Box 2) from Pseudomonas syringae pv. phaseolicola PK2 for which the structure was recently elucidated [71, 72]. This enzyme catalyzes two reactions: hydroxylation of L-Arg at C5 resulting in release of guanidine and formation of L-Δ--1-pyrroline-5-carboxylate (Figure 3I) and the L-Arg dependent synthesis of ethylene from 2OG (Figure 3J). The first (minor) reaction has no known physiological role, but we speculate it may have been the ancestral activity of the enzyme and used for L-Arg degradation. The second (major) reaction produces a plant hormone that may stimulate release of nutrients into the environment for use by the microorganism.
Box 2. Multiple Reactions Catalyzed by a Single Enzyme.
Several representatives of the 2OG-dependent oxygenases catalyze more than one reaction. For example, we have already discussed FzmG (Figure 3E and 3F), AsqJ (Figure 3N), PhnA (Figure 3O), AusE (Figure 3P), and OkaE (Figure 3Q). As a case study, we highlight the ethylene-forming enzyme (EFE). Ethylene is of interest both as a precursor for the synthesis of plastics, fibers, and other materials and for its potential use as a fuel source. Commercial production of ethylene typically involves the thermal cracking of natural gas and petroleum; however, this energy-intensive process releases greenhouse gases so a biological source is desirable. Plants make ethylene, but do so using the specialized metabolite 1-aminocyclopropane-1-carboxylate. In contrast, bacteria and fungi containing EFE produce ethylene directly from 2OG while also catalyzing (at about half the flux) the 2OG-dependent hydroxylation of L-Arg to form guanidine and L-Δ-1-pyrroline-5-carboxylate (Figure 3I and 3J). Recent structural and biochemical studies have partially uncovered the basis for this dual reactivity [71, 72]. EFE with bound 2OG and L-Arg has an apparent dioxygen binding site in an off-line configuration, i.e. pointing away from the L-Arg C5 atom undergoing hydroxylation. Also of note, EFE exhibits an unusual twisted peptide bond involving the metal-ligating Asp residue, this side chain has an atypical geometry of binding to the metal, and the environment surrounding the 2OG is hydrophobic. A requirement for a ferryl flip and a blocking Phe residue disfavor L-Arg hydroxylation, while the other features may facilitate the decarboxylative fragmentation of 2OG to generate ethylene. EFE variants that reduce ethylene formation while allowing for the hydroxylation of L-Arg were identified, but the more desirable variants that hinder L-Arg metabolism while allowing for ethylene formation have not yet been reported.
Halogenation
In contrast to the hydroxylation chemistry described thus far, some oxygenases catalyze halogenase reactions [73]. A recent example is the structurally characterized WelO5 from the cyanobacterium Hapalosiphon welwitschii UTEX B1830 [74]. This enzyme coordinates Fe(II) using two His residues, with the carboxylate position replaced by Ala and with a halide bound to the metal. During catalysis, a cis-haloferryl intermediate abstracts a hydride from the substrate with transfer of a halide radical forming the product; i.e., 12-epi-fisherindole U converts to 12-epi-fisherindole G by chloride addition at C13 (Figure 3K). Similar halogenation reactions are carried out by WelO5* from the IC-52-3 strain of this species [75] and by AmbO5 from Fischerella ambigua [76] involved in welwitindolelinone and ambiguine alkaloid syntheses, respectively.
Endoperoxidation
Another type of oxygenase chemistry is endoperoxidation (Figure 3L) as catalyzed by the structurally characterized enzyme FtmOx1 from Aspergillus fumigatus [77]. This enzyme converts fumitremorgin B to verruculogen, a mycotoxin, and a second product formed by oxidation of a particular alcohol to a keto group.
Desaturation
Several recent studies have described biosynthetic desaturation reactions catalyzed by oxygenases. One example is radish GRS1 that forms the alkene-containing molecule glucoraphasatin (Figure 3M). This compound and related glucosinolates are used as defense molecules in Brassicaceae [78]. In several other cases, subsequent reactions accompany the initial desaturation step. AsqJ, a structurally characterized enzyme from Aspergillus nidulans [79], catalyzes sequential desaturation and epoxidation of 4′-methoxycyclopeptin (Figure 3N), with the epoxide later undergoing elimination and non-enzymatic rearrangement to yield 4′-methoxyviridicatin. This molecule incorporates into a variety of quinolone alkaloids with anti-bacterial, anti-viral, anti-malarial, and other activities. Mechanistic and computational studies of AsqJ have confirmed the key role of the ferryl intermediate in both enzymatic reactions [80–83]. Fungal monoterpenoids such as paraherquonin and acetoxydehydroaustin also exhibit a variety of biological activities. The biosynthetic pathways for these compounds utilize the PrhA and AusE oxygenases, obtained from strains of Penicillium brasilianum, which act on the same substrate, preaustinoid A1, but with distinct outcomes [84]. PrhA desaturates a six-member ring then catalyzes a remarkable rearrangement involving expansion of that ring and contraction of an adjacent seven-membered ring (Figure 3O). In contrast, the sequence-related AusE desaturates the seven-membered ring then contracts it to the spiro-ring species (Figure 3P). Structures of AusE and PrhA were recently elucidated and revealed the importance of three key residues in controlling the specificity [85].
Ring Transformation
Continuing the theme of oxygenases that catalyze ring transformations, we describe enzymes in the okaramine, nogalmycin, cycloclavine, and orthosomycin biosynthetic pathways. OkaE from Penicillium simplicissimum strain ATCC 90288 generates a four-membered azetidine ring in okaramine A and then catalyzes a hydroxylation reaction (Figure 3Q) during the synthesis of indole alkaloids; these compounds exhibit potent insecticidal activities due to the selective inhibition of glutamate-gated chloride channels [86]. SnoK catalyzes a cyclization reaction between an aromatic ring and a sugar unit and SnoN catalyzes an epimerization (Figure 3R) during synthesis of anthracycline antibiotics in Streptomcyes nogalater [87]. The Aspergillus japonicas pathway for synthesis of the ergot alkaloid cycloclavine was reproduced in Saccharomyces cerevisiae [88]. Of interest in this recombinant system is the EasH-catalyzed ring contraction reaction with formation of a cyclopropyl group (Figure 3S). Finally, mayapple 2-ODD is a ring-forming oxygenase (Figure 3T) in the pathway for synthesis of an etoposide aglycone podophyllotoxin [89]. Although the corresponding biosynthetic reactions have not been established, oxidative cyclization reactions are used for synthesis of the orthoether linkages and methylenedioxy bridges of avilamycin A and everninomycin D (Figure 3U), and the orthoester feature of hygromycin B (Figure 3V). Crystal structures were elucidated for AviO1 of Streptomyces viridochromogenes, EvdO1 and EvdO2 of Micromonospora carbonacea, and HygX of S. hydroscopicus that are responsible for these reactions [90].
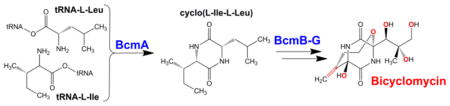
A final oxygenase-dependent biosynthetic pathway of interest makes the antibiotic bicyclomycin (Box 3). Synthesis of this 2,5-diketopiperazine alkaloid in Streptomyces sapporonensis ATCC 21532 features five distinct members of this enzyme family that catalyze a fascinating series of hydroxylation, desaturation, and epoxidation reactions [91, 92].
Box 3. Applications of 2OG-Dependent Oxygenases.
The multitude of substrates utilized by 2OG-dependent oxygenases has attracted the interest of industrial chemists who wish to functionalize non-activated C-H bonds during the synthesis of desired products. Brute force screening of genes that encode putative members of this enzyme family against a series of potential substrates succeeded in identifying five new activities among 131 open reading frames tested [105]. An alternative approach focuses effort on a single broad-specificity enzyme to expand its known substrate range. For example, the structurally-characterized leucine 5-hydroxylase GriE from Streptomyces strain DSM 40835 [106] was shown to hydroxylate 13 different substrates and has been used for chemoenzymatic synthesis of manzacidin C [107]. These approaches are certain to expand to include designed mutations and evolved strains to enhance desirable activities for other family members.
Roles of 2OG-Dependent Oxygenases in Biodegradation
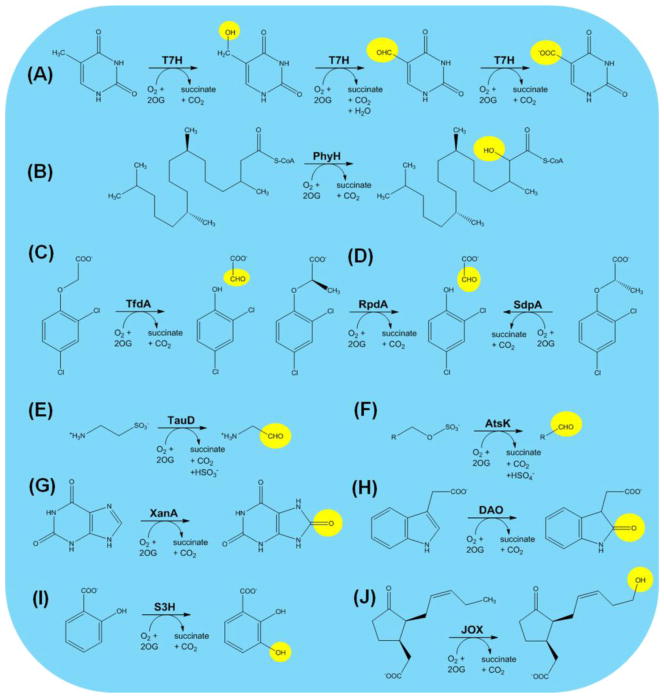
In contrast to the biosynthetic enzymes described above, many oxygenases function to decompose their substrates for metabolite recycling, nutrient provision, or regulatory purposes. An example of metabolite recycling uses thymine 7-hydroxylase (T7H) to catalyze three sequential hydroxylation reactions that transform thymine into 5-hydroxymethyluracil, 5-formyluracil, and 5-carboxyuracil (Figure 4A). An iso-orotate decarboxylase converts 5-carboxyuracil into uracil that is recycled into pyrimidine nucleotide biosynthesis. The hydroxylation reactions are highly reminiscent of those catalyzed by the TET proteins acting on 5-methylcytosine in DNA, as described earlier. Several structures and biochemical properties of T7H from Neurospora crassa have been characterized [93].
Figure 4.
Representative Biodegradative Reactions of 2OG-Dependent Oxygenases. (A) Thymine 7-hydroxylase (T7H). (B) Phytanoyl-CoA hydroxylase (PhyH). (C) 2,4-Dichlorophenoxyacetic acid hydroxylase (TfdA). (D) (R)- and (S)-specific dichlorophenoxypropionic acid hydroxylases (RdpA and SdpA). (E) Taurine hydroxylase (TauD). (F) Alkylsulfate hydroxylase (AtsK). (G) Xanthine hydroxylase (XanA). (H) Auxin dioxygenase (DAO). (I) Salicyclic acid 3-hydroxylase (S3H). (J) Jasmonic acid oxygenase (JOX).
Selected examples illustrate carbon utilization by hydroxylases. Phytanic acid (3,7,11,15-tetramethylhexadecanoic acid) resembles a fatty acid, but this polyisoprenoid cannot be degraded by β-oxidation because of its 3-methyl group. To circumvent this problem, mammalian cells create the coenzyme A (CoA) adduct, hydroxylate C2 of phytanoyl-CoA using PhyH (Figure 4B), cleave the C1–C2 linkage to generate formyl-CoA and pristanal, and oxidize the aldehyde to pristanic acid that is a substrate for β-oxidation [94]. Substitutions in PhyH can cause Refsum disease with its associated retinitis pigmentosa, polyneuropathy, ataxia, and other dysfunctions, and structural and biochemical studies of PhyH reveal the basis of the multiple disease mutations [94]. In certain bacteria aryloxyalkanoate herbicides are metabolized by a pathway involving a hydroxylase. For example, Cupriavidis necator TfdA hydroxylates 2,4-dichlorophenoxyacetate (Figure 4C) to produce the corresponding phenol and acetaldehyde [95]. Similarly, Sphingomonas herbicidovorans MG uses RdpA and SdpA to degrade specifically the (R) and (S) enantiomers of 2-phenoxypropionic acid (Figure 4D). Homology modeling and mutagenesis studies provide insights into the basis for specificity by these enzymes [96]. Of agricultural interest, the bacterial genes have been used to develop herbicide-tolerant transgenic crops plants [97].
Certain biodegradative oxygenases provide the cell with nutritional sources of sulfur or nitrogen. The best studied example is TauD, an E. coli enzyme that hydroxylates C1 of taurine (2-aminoethanesulfonate) to release sulfite and aminoacetaldehyde (Figure 4E) [11]. Sulfite satisfies the cellular sulfur needs. Other bacteria such as Pseudomonas putida S-313 catalyze a similar reaction to decompose alkylsulfates (Figure 4F) using the sequence-related and structurally-elucidated enzyme AtsK [98]. In this case, sulfate is the released sulfur source. Aspergillus nidulans and other fungi use xanthine as a nitrogen source by action of XanA [99]. This enzyme generates uric acid, like the molybdopterin cofactor-containing xanthine dehydrogenase, but does so using hydroxylase chemistry (Figure 4G).
We conclude this section by describing three plant enzymes that use hydroxylases to inactivate signaling molecules. Auxin (indole-3-acetic acid) is an essential plant hormone that functions in growth and development. The first step in its degradation involves hydroxylation by DAO to produce 2-oxoindole-3-acetic acid (Figure 4H) [100]. Salicyclic acid (2-hydroxybenzoic acid) is a phytohormone with roles in defense, flowering, leaf senescence, and thermogenesis. Its decomposition begins with salicylic acid 3-hydroxylase (S3H) (Figure 4I) [101]. Finally, the lipid-derived plant hormone jasmonic acid accumulates during wounding or attack by pathogens and functions in plant defense. When this compound is no longer required by the plant, it is inactivated by hydroxylation (Figure 4J) using jasmonic acid oxygenase (JOX) [102].
Concluding Remarks
The reactions catalyzed by the Fe(II)/2OG oxygenases are not limited to the selected recent examples discussed in this review. As supported by bioinformatic studies that reveal the existence of numerous undefined orthologs, new members of this enzyme family are certain to be discovered and characterized. In addition, there is great potential in using these enzymes for industrial applications (Box 4). Although tremendous strides have been achieved in deciphering the enzyme structures, mechanisms, and biological roles for this group of enzymes, many questions remain to be elucidated (see Outstanding Questions). We hope this review has sparked interest in this remarkable family of enzymes.
Box 4. Multiple 2OG-Dependent Oxygenases in a Single Pathway.
An interesting case study illustrating how multiple oxygenases act within a single pathway is that for the biosynthesis of bicyclomycin. Several strains of Streptomyces produce this broad-spectrum antibiotic that acts by inhibiting Rho, the prokaryotic transcription termination factor. Bicyclomycin has a unique diketopiperazine unit that contains primary, secondary, and two tertiary hydroxyl groups along with an exo-methylene moiety (see figure). The nonribosomal biosynthesis of this compound was recently associated with genes in the bcmABCDEFG cluster and selected intermediate steps were uncovered [91, 92]. The process begins with a cyclodipeptide synthase (BcmA) acting on leucine- and isoleucine-charged tRNA molecules to produce cyclo(L-Ile-L-Leu). Five 2OG-dependent oxygenases (BcmB, C, E, F, and G) and one cytochrome P450 (BcmD) complete the oxidative functionalization of eight unactivated C-H bonds: BcmE hydroxylates the terminal carbon atom from Ile, BchC hydroxylates the tertiary carbon from Leu, BcmG hydroxylates a primary carbon atom from Leu, BcmB probably desaturates and forms an epoxide in the side chain from Leu, a spontaneous heterocyclization likely occurs next, BcmD hydroxylates a tertiary carbon atom of the ring, and BcmF desaturates this species to form the exo-methylene unit. Further understanding of these complex reactions will provide insight into the development of antibiotic analogues and may have other applications in synthetic biology.
Outstanding Questions.
Given the large number of genes that encode uncharacterized family members, what other roles do these enzymes play in biology?
How can one develop inhibitors that are specific to single enzymes (or to a targeted subset of enzymes) given the common protein fold and metallocenter architecture?
Which of these enzymes are most amenable for use in desired industrial applications and how can we enhance their reactivity for a non-native substrate?
Trends.
Structural studies of 2OG-dependent oxygenases provide keen insights into the mechanisms of these enzymes, including recent visualization of the key ferryl intermediate by use of a stable structural mimic.
2OG-dependent oxygenases are increasingly associated with genetic disorders and cancers, stimulating intense efforts to develop specific therapeutic agents directed at these enzymes.
2OG-dependent hydroxylases participate at multiple steps in transcriptional and translational processes.
The list of oxygenases involved in cellular biosynthesis and catabolism continues to expand rapidly, especially in pathways for anti-microbial compounds.
The types of chemical reactions catalyzed by these oxygenases continues to diversify, and industrial applications of these activities are growing.
Acknowledgments
A grant from the National Institutes of Health (GM063584) partially supported work in the Hausinger laboratory related to this topic.
Glossary
- Alkaloid
nitrogenous organic compound found in plants.
- Chromatin
a complex of DNA and histones in eukaryotes.
- Demethylase
an enzyme that removes a methyl group from a substrate.
- Desaturation
the removal of two hydrogen atoms to form a double bond.
- Endoperoxidation
formation of a heterocyclic molecule containing a peroxide O-O unit in the ring.
- Epoxidation
formation of a cyclic ether with a three-atom ring.
- Ferryl
iron (oxidation state of 4)-oxo species, a potent oxidant.
- Halogenase
an enzyme that incorporates a halogen atom into a substrate.
- Hemiaminal
functional group with a hydroxyl and amino group attached to the same carbon atom.
- Hydroxylase
an enzyme that incorporates a hydroxyl (OH) group into a substrate.
- Orthologs
genes in different species that evolved from a common ancestor.
- 2-Oxoglutarate
also known as α-ketoglutarate, a central metabolic intermediate found in the citric acid cycle.
- Oxygenase
enzyme that incorporates molecular oxygen into the substrate(s).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hausinger RP. Biochemical diversity of 2-oxoglutarate-dependent oxygenases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 1–58. [Google Scholar]
- 2.Wu LF, et al. Ferrous iron and α-ketoglutarate dependent dioxygenases in the biosynthesis of microbial natural products. Biochim Biophys Acta. 2016;1864:453–470. doi: 10.1016/j.bbapap.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Islam MS, et al. 2-Oxoglutarate-dependent oxygenases. Annu Rev Biochem. 2018 doi: 10.1146/annurev-biochem-061516-044724. in press. [DOI] [PubMed] [Google Scholar]
- 4.Hagel JM, Facchini PJ. Expanding the roles for 2-oxoglutarate-dependent-oxygenases in plant metabolism. Natural Prod Rep. 2018 doi: 10.1039/c7np00060j. in press. [DOI] [PubMed] [Google Scholar]
- 5.Aik WS, et al. Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. [Google Scholar]
- 6.Jia B, et al. Integrative view of 2-oxoglutarate/Fe(II)-dependent oxygenase diversity and functions in bacteria. Biochim Biophys Acta. 2017;1861:323–334. doi: 10.1016/j.bbagen.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Jia B, et al. Large-scale examination of functional and sequence diversity of 2-oxoglutarate/Fe(II)-dependent oxygenases in Metazoa. Biochim Biophys Acta. 2017;1861:2922–2933. doi: 10.1016/j.bbagen.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Kawai Y, et al. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014;78:328–343. doi: 10.1111/tpj.12479. [DOI] [PubMed] [Google Scholar]
- 9.Martinez S, Hausinger RP. Catalytic mechanisms of Fe(II)- and 2-oxoglutarate-dependent oxygenases. J Biol Chem. 2015;290:20702–20711. doi: 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell AJ, et al. Visualizing the reaction cycle in an iron(II)- and 2-(oxo)-glutarate-dependent hydroxylase. J Am Chem Soc. 2017;139:13830–13836. doi: 10.1021/jacs.7b07374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proshlyakov DA, et al. Spectroscopic analyses of 2-oxoglutarate-dependent oxygenases: TauD as a case study. J Biol Inorg Chem. 2017;22:367–379. doi: 10.1007/s00775-016-1406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ploumakis A, Coleman ML. OH, the places you’ll go! Hydroxylation, gene expression, and cancer. Molec Cell. 2015;58:729–741. doi: 10.1016/j.molcel.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Markolovic S, et al. Protein hydroxylation catalyzed by 2-oxoglutarte-dependent oxygenases. J Biol Chem. 2015;290:20712–20722. doi: 10.1074/jbc.R115.662627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markolovic S, et al. Structure-function relationships of human JmjC oxygenases -- demethylases versus hydroxylases. Curr Opin Struct Biol. 2016;41:62–72. doi: 10.1016/j.sbi.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X, Trievel RC. JmjC lysine demethylases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 210–245. [Google Scholar]
- 16.Walport LJ, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun. 2016;7:11974. doi: 10.1038/ncomms11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, et al. Specific recognition of arginine methylated histone tails by JMJD5 and JMJD7. Sci Rep. 2018;8:3275. doi: 10.1038/s41598-018-21432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak R, et al. Advances and challenges in understanding histone demethylase biology. Curr Opin Chem Biol. 2016;33:151–159. doi: 10.1016/j.cbpa.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 19.Bonnici J, et al. Inhibitors of both N-methyl lysyl- and arginyl-demethylase activities of the JmjC oxygenases. Phil Trans R Soc B. 2017 doi: 10.1098/rstb.2017.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joberty G, et al. Interrogating the druggability of the 2-oxoglutarate-dependent dioxygenase target class by chemical properties. 2016;11:2002–2020. doi: 10.1021/acschembio.6b00080. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Zhang Y. TET-mediated active DNA demethylation: Mechanism, function, and beyond. Nat Rev Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 22.Wu TP, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trewick SC, et al. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 24.Ringvoll J, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds D, et al. 2-Oxoglutarate-dependent hydroxylase involved in DNA base J (b-D-glucopyranosyloxymethyluracil) synthesis. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. [Google Scholar]
- 26.Wilkins SE, et al. The role of 2-oxoglutarate-dependent oxygenases in hypoxia sensing. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. [Google Scholar]
- 27.Pugh CW, Ratcliffe PJ. New horizons in hypoxia signaling pathways. Exp Cell Res. 2017;356:116–121. doi: 10.1016/j.yexcr.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh TL, et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem Sci. 2017;8:7651–7668. doi: 10.1039/c7sc02103h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarhonskaya H, et al. Kinetic investigations of the role of factor inhibiting hypoxia-inducible factor (FIH) as an oxygen sensor. J Biol Chem. 2015;290:19726–19742. doi: 10.1074/jbc.M115.653014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi J, et al. JMJD6 and U2AF65 co-regulate alternative splicing in both JMJD6 enzymatic activity dependent and independent manner. Nucleic Acids Res. 2017;45:3503–3518. doi: 10.1093/nar/gkw1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominissini D, et al. The dynamic N1-methyladenosine methylome in eukaryote messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou S, et al. N6-Methyladenosine: A conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep. 2016;6:25677. doi: 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauer J, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;(541) doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu L, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Q, et al. Tet2 promotes pathogen infection - induced myelopoiesis through mRNA oxidation. Nature. 2018;554:123–127. doi: 10.1038/nature25434. [DOI] [PubMed] [Google Scholar]
- 37.Guallar D, et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Genet. 2018 doi: 10.1038/s41588-018-0060-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawarada L, et al. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401–7415. doi: 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haag S, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–2119. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Born E, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 41.Fu Y, et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M, et al. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 2010;39:1576–1585. doi: 10.1093/nar/gkq919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang Q, et al. Modifying the maker: Oxygenases target ribosome biology. Translation. 2015;3:e1009331. doi: 10.1080/21690731.2015.1009331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scotti JS, et al. Human oxygen sensing may have its origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc Natl Acad Sci USA. 2014;111:13331–13336. doi: 10.1073/pnas.1409916111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz MJ, et al. Hydroxylation and translational adaptation to stress: Some answers lie beyond the STOP codon. Cell Molec Life Sci. 2016;73:1881–1893. doi: 10.1007/s00018-016-2160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng T, et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Molec Cell. 2014;53:1–10. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8:960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horita S, et al. Structure of the ribosomal oxygenase OGFOD1 provides insights into the regio- and stereoselectivity of prolyl hydroxylases. Structure. 2015;23:1–14. doi: 10.1016/j.str.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singleton RS, et al. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci USA. 2014;111:4031–4036. doi: 10.1073/pnas.1314482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loenarz C, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA. 2014;111:4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katz MJ, et al. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc Natl Acad Sci USA. 2014;111:4025–4030. doi: 10.1073/pnas.1314485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekirnik R, et al. XcfDRM is athermophilic oxygen-dependent ribosomal protein uL16 oxygenase. Extremophiles. 2018 doi: 10.1007/s00792-018-1016-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanshina DD, et al. Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. FEBS J. 2015;282:1554–1566. doi: 10.1111/febs.13241. [DOI] [PubMed] [Google Scholar]
- 54.Myllyharju J. Collagen hydroxylases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 149–167. [Google Scholar]
- 55.Okuyama K, et al. Crystal structure of the collagen model peptide (Pro-Pro-Gly)4-Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.0 Å resolution. Biopolymers. 2013;99:436–447. doi: 10.1002/bip.22198. [DOI] [PubMed] [Google Scholar]
- 56.Guo HF, et al. Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding. Nat Commun. 2018;9:512. doi: 10.1038/s41467-018-02859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylase. Adv Enzymol Rel Areas Molec Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 58.Kelly L, et al. Asparagine β-hydroxylation stabilizes the ankyrin repeat domain fold. Mol Biosyst. 2009;5:52–58. doi: 10.1039/b815271c. [DOI] [PubMed] [Google Scholar]
- 59.Yang M, et al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor inhibiting hypoxia-inducible factor (FIH) J Biol Chem. 2011;286:7648–7660. doi: 10.1074/jbc.M110.193540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, et al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the posttranslational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 2011;278:1086–1097. doi: 10.1111/j.1742-4658.2011.08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandstetter H, et al. X-ray structure of clotting factor IXa: Active site and module structure related to Xase activity and hemophilia B. Proc Natl Acad Sci USA. 1995;92:9796–9800. doi: 10.1073/pnas.92.21.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinchuk JE, et al. Absence of post-translational aspartyl β-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. Proc Natl Acad Sci USA. 2002;277:12970–12977. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 63.Reddy YV, et al. Evidence that trimethyllysine hydroxylase catalyzes the formation of (2S,3S)-3-hydroxy-Nε-trimethyllysine. Org Lett. 2017;19:400–403. doi: 10.1021/acs.orglett.6b03608. [DOI] [PubMed] [Google Scholar]
- 64.Vaz FM, van Vlies N. Dioxygenases of carnitine biosynthesis: 6-N-trimethyllysine and g-butyrobetaine hydroxylases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 324–337. [Google Scholar]
- 65.Nakayasu M, et al. A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol. 2017;175:120–133. doi: 10.1104/pp.17.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hibi M, et al. Novel enzyme family found in filamentous fungi catalyzing trans-4-hydroxylation of L-pipecolic acid. Appl Environ Microbiol. 2016;82:2070–2077. doi: 10.1128/AEM.03764-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li C, et al. Structural analysis of a phosphonate hydroxylase with an access tunnel at the back of the active site. Acta Crystallogr. 2016;F32:362–368. doi: 10.1107/S2053230X16004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z, et al. Biosynthesis of fosfazinomycin is a convergent process. Chem Sci. 2015;6:1282–1287. doi: 10.1039/c4sc03095h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Handrick V, et al. Biosynthesis of 8-O-methylated benzoxazinoid defense compounds in maize. The Plant Cell. 2016;28:1682–1700. doi: 10.1105/tpc.16.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goswami A, et al. Evidence that oxidative dephosphorylation by the nonheme Fe(II), α-ketoglutarate:UMP oxygenase occurs by stereospecific hydroxylation. FEBS Lett. 2017;591:468–478. doi: 10.1002/1873-3468.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z, et al. Structural and steroelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate. Proc Natl Acad Sci USA. 2017;114:4667–4672. doi: 10.1073/pnas.1617760114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez S, et al. Structures and mechanisms of the non-heme Fe(II)- and 2-oxoglutarate-dependent ethylene-forming enzyme: Substrate binding creates a twist. J Am Chem Soc. 2017;139:11980–11988. doi: 10.1021/jacs.7b06186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith JL, Khare D. Recent advances in the structural and mechanistic biology of non-haem Fe(II), 2-oxoglutarate and O2-dependent halogenases. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. pp. 401–413. [Google Scholar]
- 74.Mitchell AJ, et al. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat Chem Biol. 2016;12:636–640. doi: 10.1038/nchembio.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu Q, Liu X. Characterization of non-heme iron aliphatic halogenase Wel05* from Hapalosiphon welwitschii IC-52–3: Identification of a minimal protein sequence motif that confers enzymatic chlorination specificity in the biosynthesis of welwitindolelinones. Beilstein J Org Chem. 2017;13:1168–1173. doi: 10.3762/bjoc.13.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hillwig ML, et al. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: Implications for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew Chem Int Ed. 2016;55:5780–5784. doi: 10.1002/anie.201601447. [DOI] [PubMed] [Google Scholar]
- 77.Yan W, et al. Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme. Nature. 2015;527:539–543. doi: 10.1038/nature15519. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Kakizaki T, et al. A 2-oxoglutarate-dependent dioxygenase mediates the biosynthesis of glucoraphasatin in radish. Plant Physiol. 2017;173:1583–1593. doi: 10.1104/pp.16.01814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bräuer A, et al. Structure of the dioxygenase AsqJ: Mechanistic insights into a one-pot multistep quinolone antibiotic biosynthesis. Angew Chem Int Ed. 2016;55:422–426. doi: 10.1002/anie.201507835. [DOI] [PubMed] [Google Scholar]
- 80.Chang W-c, et al. Mechanistic investigation of a non-heme iron enzyme catalyzed epoxidation in (−)-4′-methoxycyclopenin biosynthesis. J Am Chem Soc. 2016;138:10390–10393. doi: 10.1021/jacs.6b05400. [DOI] [PubMed] [Google Scholar]
- 81.Su H, et al. Mechanistic insights into the decoupled desaturation and epoxidation catalyzed by dioxygenase AsqJ involved in the biosynthesis of quinolone alkaloids. ACS Catalysis. 2017;7:5534–5543. [Google Scholar]
- 82.Song X, et al. Mechanistic insights into dioxygen activation, oxygen atom exchange and substrate epoxidation by AsqJ dioxygenase from quantum mechanical/molecular mechanical calculations. Phys Chem Chem Phys. 2017;19:20188–20197. doi: 10.1039/c7cp02687k. [DOI] [PubMed] [Google Scholar]
- 83.Liao H-J, et al. Insights into desaturation of cyclopeptin and its C3-epimer catalyzed by a non-heme iron enzyme: Structural characterization and mechanism elucidation. Angew Chem Int Ed. 2018 doi: 10.1002/anie.201710567. in press. [DOI] [PubMed] [Google Scholar]
- 84.Matsuda Y, et al. Discovery of key dioxygenases that diverged in the paraherquonin and acetoxydehydroaustin pathways in Penicillium brasilianum. J Am Chem Soc. 2016;138:12671–12677. doi: 10.1021/jacs.6b08424. [DOI] [PubMed] [Google Scholar]
- 85.Nakashima Y, et al. Structure function and engineering of multifunctional non-heme iron dependent oxygenases in fungal meroterpenoid biosynthesis. Nat Commun. 2018;9:104. doi: 10.1038/s41467-017-02371-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lai CY, et al. Biosynthesis of complex indole alkaloids: Elucidation of the concise pathway of okaramines. Angew Chem Int Ed. 2017;56:9478–9482. doi: 10.1002/anie.201705501. [DOI] [PubMed] [Google Scholar]
- 87.Siitonen V, et al. Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway. Proc Natl Acad Sci USA. 2016;113:5251–5256. doi: 10.1073/pnas.1525034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jakubczyk D, et al. Discovery and reconstitution of the cycloclavine biosynthetic pathway--enzymatic formation of a cyclopropyl group. Angew Chem Int Ed. 2015;54:5117–5121. doi: 10.1002/anie.201410002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:12224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCulloch KM, et al. Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase family. Proc Natl Acad Sci USA. 2015;112:11547–11552. doi: 10.1073/pnas.1500964112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meng S, et al. A six-oxidase cascade for tandem C-H bond activation revealed by reconstitution of bicyclomycin biosynthesis. Angew Chem Int Ed. 2018;57:719–723. doi: 10.1002/anie.201710529. [DOI] [PubMed] [Google Scholar]
- 92.Patteson JB, et al. Identification of the biosynthetic pathway for the antibiotic bicyclomycin. Biochemistry. 2018;57:61–65. doi: 10.1021/acs.biochem.7b00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W, et al. Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi. Nucleic Acids Res. 2015;43:10026–10038. doi: 10.1093/nar/gkv979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wanders RJA, et al. Phytanoyl-CoA hydroxylase: A 2-oxoglutarate-dependent dioxygenase crucial for fatty acid alpha-oxidation in humans. In: Schofield CJ, Hausinger RP, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society for Chemistry; 2015. pp. 338–349. [Google Scholar]
- 95.Fukumori F, Hausinger RP. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 96.Müller TA, et al. Structural basis for the enantiospecificities of R- and S-phenoxypropionate/α-ketoglutarate dioxygenases. Protein Sci. 2006;15:1356–1368. doi: 10.1110/ps.052059406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Griffin SL, et al. Characterization of aryloxyalkanoate dioxygenase-12, a nonheme Fe(II)/α-ketoglutarate-dependent dioxygenase, expressed in transgenic soybean and Pseudomonas fluorescens. J Agric Food Chem. 2013;61:6589–6596. doi: 10.1021/jf4003076. [DOI] [PubMed] [Google Scholar]
- 98.Müller I, et al. Crystal structure of the alkylsulfatase AtsK: Insights into the catalytic mechanism of the Fe(II) α-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43:3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- 99.Montero-Moràn GM, et al. Purification and characterization of the FeII- and α-ketoglutarate-dependent xanthine hydroxylase from Aspergillus nidulans. Biochemistry. 2007;46:5293–5304. doi: 10.1021/bi700065h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao Z, et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27:113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 101.Zhang K, et al. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA. 2013;110:14807–14812. doi: 10.1073/pnas.1302702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caarls L, et al. Arabidopsis JASMONATE-INDUCED OXYGENASES downregulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc Natl Acad Sci USA. 2017;114:6388–6393. doi: 10.1073/pnas.1701101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rose NR, et al. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40:4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 104.Khan A, et al. Development and application of ligand-based NMR screening assays for γ-butyrobetaine hydroxylase. MedChemComm. 2016;7:873–880. [Google Scholar]
- 105.Baud D, et al. Synthesis of mono- and dihydroxylated amino acids with new α-ketoglutarate-dependent dioxygenases: Biocatalytic oxidation of C-H bonds. ChemCatChem. 2014;6:3012–3017. [Google Scholar]
- 106.Lukat P, et al. Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity. Chem Sci. 2017;8:7521–7527. doi: 10.1039/c7sc02622f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zwick CR, III, Renata H. Remote C-H hydroxylation by an a-ketoglutarate-dependent dioxygenase enables efficient chemoenzymatic synthesis of manzacidin C and proline analogs. J Am Chem Soc. 2018;140:1165–1169. doi: 10.1021/jacs.7b12918. [DOI] [PubMed] [Google Scholar]