Abstract
The roles of immunodeficiency and combined antiretroviral therapy (cART) in shaping the gut microbiota in HIV-1-infected subjects (HISs) have not been described thoroughly by time-series investigations. In this study, 36 antiretroviral-naïve HISs were enrolled to prospectively assess alterations in the fecal microbiota and plasma markers of microbial translocation and inflammation with cART. At baseline, the species α-diversity of the fecal microbiota was significantly lower in HISs with a CD4+ T cell count <300/mm3 than in HISs with a CD4+ T cell count >300/mm3 (Shannon index: Median 2.557 vs. 2.981, P = 0.006; Simpson index: Median 0.168 vs. 0.096, P = 0.004). Additionally, the baseline α-diversity indices correlated with CD4+ T cell counts (Shannon index: r = 0.474, P = 0.004; Simpson index: r = −0.467, P = 0.004) and the specific plasma biomarkers for microbial translocation and inflammation. After cART introduction, the species α-diversity of fecal microbiota in HISs with CD4+ T cell counts <300/mm3 was significantly restored (Shannon index: Median 2.557 vs. 2.791, P = 0.007; Simpson index: Median 0.168 vs. 0.112, P = 0.004), while the variances were insignificant among HISs with CD4+ T cell counts >300/mm3 (Shannon index: Median 2.981 vs. 2.934, P = 0.179; Simpson index: Median 0.096 vs. 0.119, P = 0.082). Meanwhile, with cART introduction, alterations in the gut microbial composition were more significant in the subgroup with CD4+ T cell counts >300/mm3, corresponding to increases in the specific plasma inflammatory markers. These findings implicated the interactive roles of immunodeficiency and cART for affecting gut microbiota in HIV-1-infected individuals, providing new insights into intestinal microbiome dysbiosis related to HIV-1 infection.
Introduction
There are an estimated 10–100 trillion microorganisms inhabiting the human gastrointestinal tract1. The symbiotic relationship that is established between the intestinal microbiota and the host plays a crucial role in maintaining host health2. Human immunodeficiency virus type 1 (HIV-1) preferentially infects and destroys CD4+ T lymphocytes3. Even during early HIV-1 infection, the T helper cells residing in gut-associated lymphoid tissue are depleted, which induces disintegration and dysfunction of the intestinal mucosal barrier4. Consequently, with the loss of protection by the intestinal mucosal barrier, microbial products (such as lipopolysaccharides) can intrude into the lamina propria of the gut and the systemic circulation in a process known as microbial translocation5,6. In addition, the composition of the intestinal microbiota shifts in HIV-1-infected individuals compared with that in non-HIV-infected individuals7, which is an indicator of host immune status8.
Combined antiretroviral therapy (cART) effectively inhibits HIV-1 replication in the host. However, even with sustained viral suppression, a chronic immune activation and hyper-inflammatory state manifests in HIV-1-infected individuals9, and this state may be associated with the increased incidence of specific diseases such as cancer, metabolic disorders and cardiovascular disease10,11. Given that the gut microbiota is closely linked to the host’s immune profile, the microbial dysbiosis in HIV-1-infected individuals likely significantly contributes to the excessive risk of inflammation-related diseases12. Considering that the findings from previous cross-sectional studies may be biased by confounding factors that impact the gut microbiome, such as dietary patterns and male homosexual behaviors13,14, the roles of the host’s immune status, and cART on the intestinal microbiota in HIV-1-infected subjects have not been clearly determined15–18. Hence, we present this study with the aim of prospectively observing the impact of short-term cART on the intestinal microbiota of HIV-1-infected subjects with varying immune statuses.
Materials and methods
Study settings and design
This prospective cohort study was performed at the Shanghai Public Clinical Health Center (SPHCC) affiliated with Fudan University from December 2015 to June 2017. Thirty-six HIV-1-infected subjects (HISs) were consecutively enrolled. At the time of enrollment, all HISs were older than 18 years and had not initiated cART. The exclusion criteria included inflammatory bowel disease, infectious gastroenteritis, antibiotic/probiotic administration within 4 weeks prior to sample collection and HIV-related complications.
At the time of recruitment, both fecal and blood samples were collected from the HISs. Thereafter, all HISs were given cART (tenofovir, lamivudine, and efavirenz) according to the current recommendations in China and underwent regular follow-up. Approximately 12 months later, the HISs were recalled to collect fecal and blood samples. The study was approved by the SPHCC Institutional Review Board, and informed consent was obtained from all participants.
Laboratory assays for T cell populations, HIV viral, load and plasma biomarkers of microbial translocation and inflammation
Immediately after sample collection, plasma and peripheral blood mononuclear cells (PBMCs) were separated from whole blood and stored at −80 °C for batch assays. T cell population assessments and plasma HIV-1 RNA load assays were performed by using flow cytometry and Cobas Amplicor (Roche, USA) at the clinical diagnostic laboratory of the SPHCC. Customized kits (MILLIPLEX® MAP; Merck Millipore, Germany) were used to quantify 38 cytokines in the plasma samples (Supplementary Table 1). In addition, enzyme-linked immunosorbent assays (ELISAs) were performed to quantify plasma markers for microbial translocation, including endotoxin core immunoglobulin M (EndoCAb IgM, Hycult Biotech, The Netherlands), lipopolysaccharide binding protein (LBP, Hycult Biotech, The Netherlands), soluble CD14 (sCD14, R&D Systems, USA), soluble CD163 (sCD163, R&D Systems, USA), and intestinal fatty acid-binding protein (I-FABP, R&D Systems, USA). All assays were performed following the manufacturer’s instructions. Measured values below the limit of detection were considered zero. For values beyond the detection range, the value of the upper limit was assigned.
DNA extraction, amplification, and 16S rRNA gene sequencing
Fecal samples were stored at -80 degrees C after collection until DNA extraction. The samples were processed, and DNA was extracted using an EZNA® Stool DNA Kit (Omega Bio-Tek, USA) according to the manufacturer’s instructions. 16S rRNA gene libraries were established according to the 16S Metagenomic Sequencing Library Preparation Guide (Illumina, USA). The targeted V3-V4 region of the 16S rRNA gene from extracted DNA was PCR-amplified with the universal primers 341F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 805R (5′-GACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′). The PCR products were purified by AMPure XP® beads (Beckman Coulter, UK), and each PCR amplicon was quantified using Qubit 2.0 (Life Technologies, USA) and pooled at equal concentrations for sequencing. 16S rRNA gene sequencing was performed on an Illumina MiSeq platform (Illumina, USA) at Sangon Biotech in Shanghai, China.
Sequence analyses
All reads were demultiplexed, preprocessed, and subsequently analyzed with the Quantitative Insights into Microbial Ecology (QIIME) software package ver. 1.8.0. Sequence numbers were randomly subsampled to 10,000 per sample to ensure an even amount of information for analysis. Operational taxonomic units (OTUs) were generated using USEARCH software version 7.0 (http://www.drive5.com/uparse/) at a similarity threshold of 97%. The taxonomic classification was constructed using the Bayesian classifier with Silva database 128 (http://www.arb-silva.de) as a reference.
Statistical and bioinformatic analysis
For cross-sectional analyses of characteristics and diversity indices between groups, differences were estimated by Student’s t-test, the Mann–Whitney U test or the chi-square test, as appropriate. For comparisons between baseline and follow-up samples, a paired t-test or Wilcoxon signed-rank test was performed depending on the distribution of the variable. Each correlation analysis was assessed using nonparametric Spearman’s rank tests and corrected by the Benjamini and Hochberg false discovery rate (BH FDR) test. Multivariate linear regression models were constructed to examine the factors associated with α-diversity indices. Two-tailed significance was set at P < 0.05.
The species richness and α-diversity of the fecal microbiome were indicated by the number of observed species (SOB) and Shannon and Simpson indices. β-diversity was assessed by principal coordinate analysis (PCoA). Weighted UniFrac distances were utilized to estimate the sample distributions in each group, and an Adonis significance analysis was performed for each pairwise comparison.
To identify the differentially abundant bacterial taxa between groups, linear discriminant analyses (LDA) coupled with effect size measurements (LefSe) were performed. The primary bacterial taxa for differentiating between groups were identified as those taxa with relative abundances >0.1% (in any group) showing an LDA score > 4, which were further tested using nonparametric paired Wilcoxon signed-rank tests (P < 0.05).
Statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, USA), SPSS version 22 (IBM, USA), and the R programming environment (http://www.r-project.org/).
Results
Clinical and demographic characteristics of the participants
The demographic and clinical characteristics of the 36 HISs in this cohort are described in Table 1. At baseline in this study, all HISs were cART-naïve with detectable plasma viral loads (median 4.68lg10 copies/ml, IQR 4.18–5.12lg10 copies/ml), and most patients exhibited moderate immunodeficiency (median CD4+ T cell count: 331 cells/mm3, IQR: 247–429 cells/mm3). After their enrollment, the median follow-up duration was 15 months (IQR: 14–16 months). At the last visit, 32 patients (88.9%) had undetectable viral loads, and the remaining 4 patients had extremely low levels (median: 41 copies/ml, range: 20–96 copies/ml). The median recovery of CD4+ T cells from baseline was 138 cells/mm3 (IQR: 87–248 cells/mm3). When all enrolled subjects were divided by baseline CD4+ T cell count (300/mm3), there were no significant differences in age (P = 0.791), gender (P = 0.715), sexual preference (P = 0.760), baseline HIV viral load (P = 0.234), and cART duration (P = 0.968) between the two subgroups (Table 1).
Table 1.
Demographics and clinical characteristics of participants
| Overall N = 36 | CD4+ T cells <300/mm3 N = 14 | CD4+ T cells >300/mm3 N = 22 | P valuea | |
|---|---|---|---|---|
| Age, years, median (IQR) | 33 (29–40) | 32 (29–38) | 33 (29–44) | 0.791b |
| Male gender, N (%) | 34 (94.4%) | 13 (92.9%) | 21 (95.5%) | 0.715c |
| Man who have sex with man, N (%) | 30 (83.3%) | 12 (85.7%) | 18 (81.8%) | 0.760c |
| Baseline CD4+ T cell/mm3, median (IQR) | 331 (247–429) | 227 (150–263) | 414 (345–484) | <0.001b |
| Last visit CD4+ T cell/mm3, median (IQR) | 467 (342–619) | 370 (272–454) | 581 (400–792) | 0.001d |
| Baseline CD4+/CD8+ ratio, %, median (IQR) | 39.57 (25.33–39.57) | 27.22 (14.48–42.49) | 52.87 (32.12–67.93) | 0.014d |
| Last visit CD4+/CD8+ ratio, %, median (IQR) | 63.12 (48.64–93.49) | 55.30 (39.83–62.98) | 89.45 (53.90–100.07) | 0.005d |
| Baseline HIV RNA lg10 copies/ml, median (IQR) | 4.68 (4.18–5.12) | 4.70 (4.25–5.41) | 4.66 (3.62–5.08) | 0.234d |
| cART duration, months, median (IQR) | 15 (14–16) | 15 (14–16) | 15 (14–17) | 0.968b |
aCompared between subjects with CD4+ T cells <300/mm3 and >300/mm3
bAnalyzed by Mann–Whitney U test
cAnalyzed by chi-square test
dAnalyzed by t-test
Baseline species α-diversity of fecal microbiota correlated with CD4+ T cell counts and plasma biomarkers for microbial translocation and inflammation in HISs
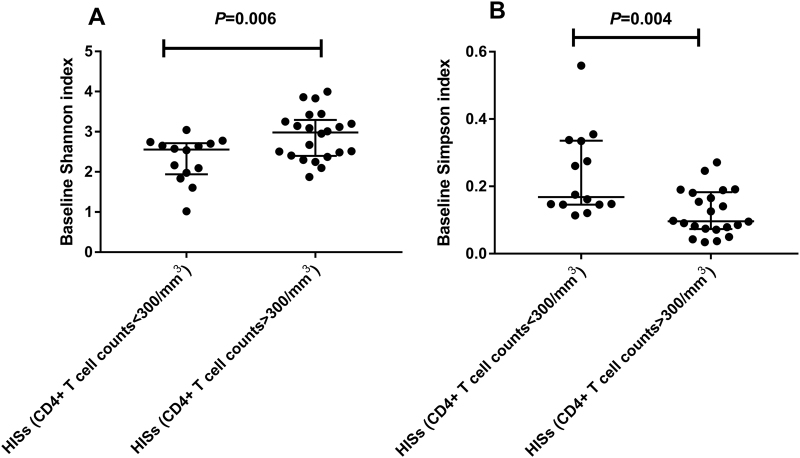
At baseline, the species α-diversity of fecal microbiota was significantly lower in HISs with a CD4+ T cell count <300/mm3 than in HISs with a CD4+ T cell count >300/mm3 (Fig. 1), with a decrease in the Shannon index (Median 2.557 vs. 2.981, P = 0.006) as well as an increase in the Simpson index (Median 0.168 vs. 0.096, P = 0.004). However, the species richness index (SOB) did not significantly differ between the two subgroups at baseline (Median 102 vs. 120, P = 0.256).
Fig. 1. Differences in species α-diversity of fecal microbiota between HISs subgrouped by CD4+ T cell counts at 300/mm3.
a Shannon index: t test; b Simpson index: Mann–Whitney test
The bivariate correlation analysis revealed that the Shannon index was positively correlated with the CD4+ T cell count (r = 0.474, P = 0.004) and negatively correlated with plasma markers for microbial translocation and inflammation at baseline, including lipopolysaccharide binding protein (LBP; r = −0.389, P = 0.028), fibroblast growth factor (FGF)-2 (r = −0.391, P = 0.022), interferon (IFN)-α2 (r = −0.418, P = 0.014), interleukin (IL)-1β (r = −0.342, P = 0.048), IL-3 (r = −0.345, P = 0.046), IL-4 (r = −0.382, P = 0.026), IL-7 (r = −0.379, P = 0.027), IL-9 (r = −0.368, P = 0.032), IL-15 (r = −0.402, P = 0.019), and macrophage inflammatory protein (MIP)-1β (r = −0.390, P = 0.023). Conversely, the baseline Simpson index was negatively correlated with the CD4+ T cell count (r = −0.467, P = 0.004) and positively correlated with LBP (r = 0.425, P = 0.015), FGF2 (r = 0.357, P = 0.038), IFN-α2 (r = 0.398, P = 0.020), IL-4 (r = 0.443, P = 0.009), IL-6 (r = 0.353, P = 0.041), IL-10 (r = 0.352, P = 0.041), IL-15 (r = 0.411, P = 0.016), and MIP-1β (r = 0.460, P = 0.006). After the BH FDR correction with a threshold at q < 0.2, the CD4+ T cell count (q = 0.176), LBP (q = 0.152), IL-9 (q = 0.158), IL-7 (q = 0.171), IL-4 (q = 0.190), and IL-1β (q = 0.192) were significantly correlated with the Shannon index, and the CD4+ T cell count (q = 0.176), IL-4 (q = 0.127), MIP-1β (q = 0.136), IL-15 (q = 0.139), IFN-α2 (q = 0.145), and LBP (q = 0.169) were significantly correlated with the Simpson index.
During multivariate linear regression analysis, the baseline CD4+ T cell count (standardized β-coefficient = 0.499, P < 0.001), IL-7 (standardized β-coefficient = −0.524, P < 0.001), and EndoCab IgM (standardized β-coefficient = −0.273, P = 0.038) were identified as factors that independently correlated with the Shannon index. Additionally, the baseline CD4+ T cell count (standardized β-coefficient = −0.590, P < 0.001) and IL-7 (standardized β-coefficient = 0.494, P < 0.001) were independently correlated with the Simpson index.
cART introduction affected gut microbiome diversity and plasma markers for inflammation and microbial translocation in HISs
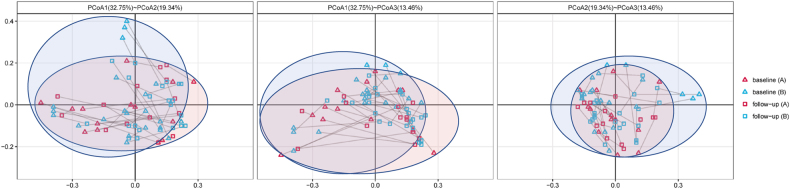
Overall, cART introduction did not impact the species richness (SOB: Median 114 vs. 110 P = 0.952) and α-diversity (Shannon index: Median 2.645 vs. 2.890, P = 0.589; Simpson index: Median 0.146 vs. 0.121, P = 0.560) of the gut microbiota in HISs in this cohort. For β-diversity (Fig. 2), a statistically significant difference was observed in the Adonis analysis comparing the microbial community profiles of the paired baseline and cART-treated samples (Adonis analysis of weighted UniFrac distance metric, P = 0.001). Additionally, microbial translocation markers, including sCD163 (median 893.70 vs. 596.70 ng/ml, P < 0.001) and I-FABP (median 20.14 vs. 9.20 ng/ml, P < 0.001), significantly decreased with alterations in specific inflammatory biomarkers (Supplementary Table 2) after cART.
Fig. 2.
Characterization of β-diversity of intestinal microbiota in HISs at baseline and follow-up by PCoA (the paired baseline and follow-up samples were connected with lines. a HISs with baseline CD4+ T cell counts <300/mm3; b HISs with baseline CD4+ T cell counts >300/mm3
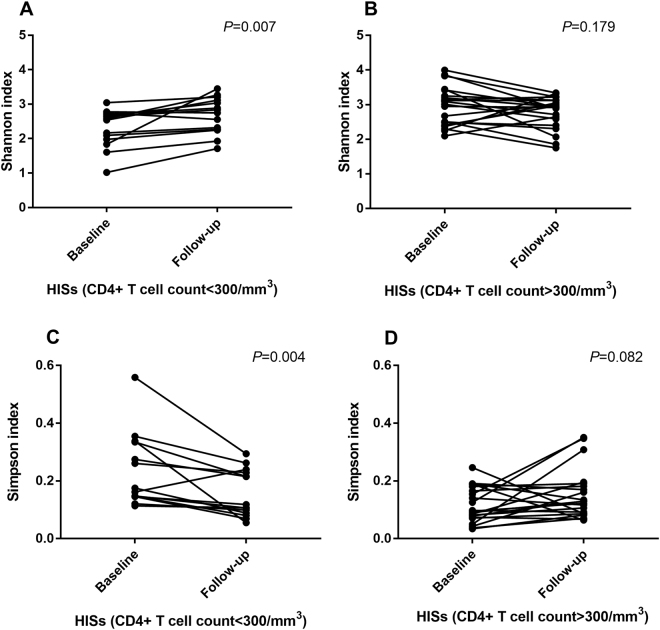
In the subgroup analysis of HISs with CD4+ T cell counts <300/mm3 at baseline, the species α-diversity of fecal microbiota in HISs with CD4+ T cell counts <300/mm3 was significantly restored (Shannon index: Median 2.557 vs. 2.791, P = 0.007; Simpson index: Median 0.168 vs. 0.112, P = 0.004) after the introduction of cART, as shown in Fig. 3a, c. On the other hand, among HISs with CD4+ T cell counts >300/mm3, the α-diversity of the gut microbiota decreased slightly after the introduction of cART (Fig. 3b, d), while the variances were insignificant (Shannon index: Median 2.981 vs. 2.934, P = 0.179; Simpson index: Median 0.096 vs. 0.119, P = 0.082).
Fig. 3.
Paired comparisons (pairedt test) of α-diversity indices in HISs before and after cART treatment. a Analysis of the Shannon index in HISs with baseline CD4+ T cell counts <300/mm3; b Analysis of the Shannon index in HISs with baseline CD4+ T cell counts >300/mm3; c Analysis of the Simpson index in HISs with baseline CD4+ T cell counts <300/mm3; d Analysis of the Simpson index in HISs with baseline CD4+ T cell counts >300/mm3)
Similar to the results described for the α-diversity indices, different alteration patterns in plasma inflammatory biomarkers were observed after cART introduction in the subgroup analysis (Supplementary Table 2). Among HISs with CD4+ T cell counts <300/mm3, the major variances in inflammatory biomarkers after cART primarily manifested as decreased levels of interferon gamma-induced protein (IP)-10 (P < 0.001), transforming growth factor (TGF)-α (P = 0.019), monocyte chemoattractant protein (MCP)-1 (P = 0.002), and IL-4 (P = 0.028), despite the increase in IL-5 (P = 0.011). For HISs with CD4+ T cell counts >300/mm3, in addition to the decreased IP-10 (P = 0.001) and TGF-α (P = 0.046) levels consistent with those in the subgroup with lower CD4+ T cell counts, increased levels of MIP-1β (P = 0.001), macrophage-derived chemokine (MDC, P = 0.004), fractalkine (P = 0.006), epidermal growth factor (EGF, P = 0.014), granulocyte colony-stimulating factor (G-CSF, P = 0.033), granulocyte-macrophage colony-stimulating factor (GM-CSF, P = 0.039), IL-1β (P = 0.042), IL-1RA (P = 0.049), fibroblast growth factor-2 (FGF-2, P = 0.044), and growth-regulated protein (GRO, P = 0.046) were observed.
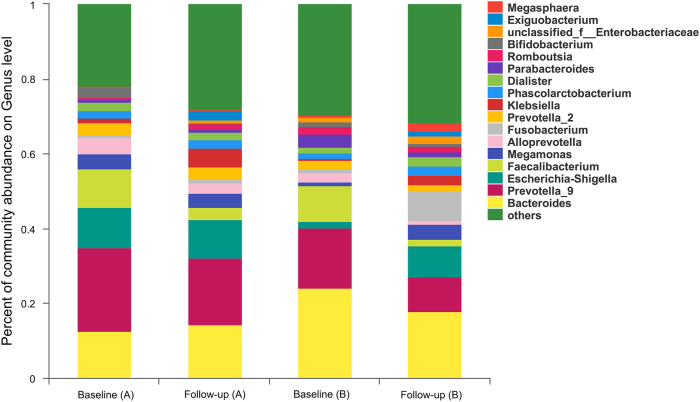
Gut microbial composition was altered after cART introduction
With the introduction of cART, the landscape of the gut microbial composition in HISs was distinguishable from that of baseline (Fig. 4). As described for the overall analysis compared with baseline (Table 2), the Proteobacteria (LDA = 4.66, P = 0.002) and Fusobacteria (LDA = 4.27, P = 0.021) phyla with their sub-taxonomies, including the classes Gammaproteobacteria (LDA = 4.67, P = 0.03) and Fusobacteria (LDA = 4.27, P = 0.021), the orders Enterobacteriales (LDA = 4.60, P = 0.008) and Fusobacteriales (LDA = 4.27, P = 0.021), the families Enterobacteriaceae (LDA = 4.60, P = 0.008) and Fusobacteriaceae (LDA = 4.27, P = 0.020), and the genera Klebsiella (LDA = 4.12, P < 0.001), and Fusobacterium (LDA = 4.27, P = 0.017), were relatively increased after cART, while the Bacteroidetes phylum (LDA = 4.78, P = 0.027) with its subtaxonomies, including the class Bacteroidia (LDA = 4.78, P = 0.027) and the order Bacteroidales (LDA = 4.78, P = 0.027), were depleted. Additionally, the relative richness of the Ruminococcaceae family (LDA = 4.63, P = 0.006) and the Faecalibacterium genus (LDA = 4.55, P = 0.001) belonging to the Firmicutes phylum decreased after cART. In the subgroup with baseline CD4+ T cell counts >300/mm3, variations in the gut microbiota composition after cART were similar to those in the bulk analysis, and differences in the order Fusobacteriales, family Fusobacteriaceae and genus Fusobacterium became insignificant (Table 2). For HISs with CD4+ T cell counts 300/mm3 at baseline, only relative enrichment of the Bacillales order (LDA = 4.06, P = 0.001) with its sub-taxonomies Family_XII_o_Bacillales (LDA = 4.05, P = 0.001) and genus Exiguobacterium (LDA = 4.05, P = 0.001) were observed after cART administration (Table 2).
Fig. 4. The grouped microbial composition at the genus level at baseline and follow-up.
a HISs with baseline CD4+ T cell counts <300/mm3; b HISs with baseline CD4+ T cell counts >300/mm3
Table 2.
Alterations of gut microbiota composition in HIV positive participants with cART introduction
| Overall (N=36) | CD4+ T cell count <300/mm3 (N=14) | CD4+ T cell count >300/mm3 (N=22) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Change | Category | Taxonomy | LDAa | P b | Category | Taxonomy | LDAa | P b | Category | Taxonomy | LDAa | P b |
| Enrichment | Phylum | Fusobacteria | 4.27 | 0.021 | Order | Bacillales | 4.06 | 0.001 | Phylum | Fusobacteria | 4.52 | 0.021 |
| Phylum | Proteobacteria | 4.66 | 0.002 | Family | Family_XII_o_Bacillales | 4.05 | <0.001 | Phylum | Proteobacteria | 4.77 | 0.005 | |
| Class | Fusobacteriia | 4.27 | 0.021 | Genus | Exiguobacterium | 4.05 | <0.001 | Class | Fusobacteriia | 4.52 | 0.021 | |
| Class | Gammaproteobacteria | 4.67 | 0.003 | Class | Gammaproteobacteria | 4.78 | 0.004 | |||||
| Order | Fusobacteriales | 4.27 | 0.021 | Order | Enterobacteriales | 4.75 | 0.004 | |||||
| Order | Enterobacteriales | 4.60 | 0.008 | Family | Enterobacteriaceae | 4.75 | 0.004 | |||||
| Family | Fusobacteriaceae | 4.27 | 0.020 | Genus | Klebsiella | 4.09 | 0.001 | |||||
| Family | Enterobacteriaceae | 4.60 | 0.008 | |||||||||
| Genus | Fusobacterium | 4.27 | 0.017 | |||||||||
| Genus | Klebsiella | 4.12 | <0.001 | |||||||||
| Depletion | Phylum | Bacteroidetes | 4.78 | 0.027 | Phylum | Bacteroidetes | 4.97 | 0.010 | ||||
| Class | Bacteroidia | 4.78 | 0.027 | Class | Bacteroidia | 4.97 | 0.010 | |||||
| Order | Bacteroidales | 4.78 | 0.027 | Order | Bacteroidales | 4.97 | 0.010 | |||||
| Family | Ruminococcaceae | 4.63 | 0.006 | Family | Ruminococcaceae | 4.73 | 0.005 | |||||
| Genus | Faecalibacterium | 4.55 | <0.001 | Genus | Faecalibacterium | 4.59 | <0.001 | |||||
aAnalyzed by linear discriminant analysis effect size method
bAnalyzed by paired Wilcoxon’s signed-rank test
Discussion
Recent studies have described the occurrence of intestinal microbial translocations and microbial dysbiosis in HIV-1-infected individuals19–23; these processes are regarded as important pathways for systemic immune activation involving HIV disease progression. The causal relationship between altered intestinal microbiota composition, HIV-1 infection-induced host immune dysfunction and cART remains an open question in HIV research15. In this study, we observed that host immune status and cART interactively influenced the alterations of gut microbiota in HISs with viral suppression, which is discussed below.
α-Diversity has been widely measured during evaluations of gut microbial diversity in HISs. In previous studies, compared with non-HIV-1-infected individuals, lower α-diversities of intestinal microbiota were observed in HISs8,21, while such discrepancies did not reach significance in other studies15,17, and an earlier study reached a contradictory conclusion23. In this study, we found that baseline CD4+ T cell counts were closely associated with α-diversity indices of the intestinal microbiota in HISs. In addition, in subgroup comparisons, significantly lower α-diversity indices were observed in subjects with CD4+ T cell counts <300/mm3. Hence, our findings implied that the conflicting results in previous studies investigating gut microbial diversity in HISs are attributable to the immune-heterogeneous subjects enrolled in different studies.
Currently, findings regarding the role of cART in restoring gut microbiota diversity in HISs are conflicting, as summarized in recent literature reviews15,24. In our study, alterations in α-diversity indices after a median of 15 months of cART were not significant in an overall analysis, which corresponded to observations from a previous prospective study8. However, in the subgroup with baseline CD4+ T cell counts <300/mm3, the increase in α-diversity indices was significant after cART. Conversely, in subjects with CD4+ T cell counts >300/mm3 at baseline, the α-diversity of the gut microbiota slightly decreased. Because of the established biological link between gut barrier function, microbiome diversity, and systemic inflammation25–27, the findings described above were echoed by alterations in specific plasma inflammation biomarkers in this study. In a subgroup of HISs with CD4+ T cell counts <300/mm3, the intensity of systemic inflammation was alleviated after cART, which was indicated by the significant decreases in IP-10, TGF-α, MCP-1, and IL-4. By contrast, in HISs with CD4+ T cell counts >300/mm3, we found several plasma markers, including MIP-1β, MDP, IL-1β, FGF-2, EGF, GRO, G-CSF, and GM-CSF, that were elevated after cART, which closely correlated with host inflammation status28–34. In summarizing previous studies, the impact of cART on gut microbiota diversity could be contributed by reconstituting the host’s mucosal immune system as well as direct effects on specific bacterial populations4,35. Therefore, in this study, we proposed that intestinal microbial α-diversity in cART-naive subjects with severe immunosuppression was restored via the positive impact of antiviral therapy on immune reconstitution of the host, which was indicated by the alleviation of systemic inflammation, as observed in previous studies27,36. In subjects with greater immune competence prior to cART, lower restoration due to immune recovery may be counteracted by the direct influences of cART drugs on gut microbial diversity, which is further discussed below.
HIV-1 infection is associated with host intestinal microbial dysbiosis6, while the influence of cART introduction in altering gut microbial composition in HISs has not been fully estimated19–23. In our cohort, the alteration patterns in terms of gut microbial composition in HISs with cART introduction were differentially correlated with immune status prior to treatment, which was more drastic in subjects with CD4+ T cell counts >300/mm3. In light of the findings described above, cART and host immune status appear to interactively contribute to alterations in the gut microbiota composition. The mechanisms by which cART influences the gut microbiota are not fully understood. In a cross-sectional study including 45 HIV-infected patients on three different cART regimens, different cART regimens were associated with diverse profiles in terms of gut microbiota composition, while integrase strand transfer inhibitor-based cART was associated with comparable systemic inflammation and microbial diversity to that of uninfected controls37. In a recent study assessing the in vitro antibacterial activity of antiretroviral drugs, efavirenz and zidovudine showed potential inhibition of Bacillus subtilis and Escherichia coli separately35. Given that the regimen containing efavirenz was administered in our study, potential disruption of the intestinal microbiota by efavirenz itself and other cART drugs deserves further consideration and investigation.
We acknowledge certain limitations to this study. First, because of the relatively short follow-up duration, we were unable to observe the long-term influences of cART on the gut microbiome. Additionally, although fecal samples, which were used in our study, are most commonly employed for analysis of the intestinal microbiota, integration analysis based on both fecal and intestinal biopsy samples would be optimal38.
Conclusions
Collectively, the results of our study showed that immune status and cART were the key factors interactively affecting the gut microbiome in HIV-1-infected individuals. All of the above findings provide new insights into interpreting previous studies and the design of future investigations on the intestinal microbiota in HISs.
Electronic supplementary material
Acknowledgements
This study was sponsored by the National Natural Science Foundation of China (NSFC no. 81571977), the Shanghai Municipal Commission of Health and Family Planning (no. 20164Y0015) and the Medical Science Support Program of the Shanghai Science and Technology Committee (no. 16411960400).
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Jun Chen, Email: qtchenjun@163.com.
Hongzhou Lu, Email: luhongzhou@fudan.edu.cn.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0117-y).
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragic T, Litwin V, Allaway GP. HIV-1 entry into CD4 plus cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 4.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 2012;10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 5.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 6.Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS. 2016;11:182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, et al. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg. Microbes Infect. 2016;5:e31. doi: 10.1038/emi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowak P, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, et al. Anti-retroviral therapy decreases but does not normalize indoleamine 2,3-dioxygenase activity in hiv-infected patients. Plos ONE. 2014;9:e100446. doi: 10.1371/journal.pone.0100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paiardini M, Muller-Trutwin M. HIV-associated chronic immune activation. Immunol. Rev. 2013;254:78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nix LM, Tien PC. Metabolic syndrome, diabetes, and cardiovascular risk in HIV. Curr. HIV/AIDS Rep. 2014;11:271–278. doi: 10.1007/s11904-014-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat. Rev. Nephrol. 2016;12:169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- 13.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noguera-Julian M, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SX, et al. Complexities of gut microbiome dysbiosis in the context of HIV infection and antiretroviral therapy. Clin. Pharmacol. Ther. 2016;99:600–611. doi: 10.1002/cpt.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two way street. AIDS. 2016;30:2737–2751. doi: 10.1097/QAD.0000000000001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tincati C, Douek DC, Marchetti G. Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res Ther. 2016;13:19. doi: 10.1186/s12981-016-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubourg G, Surenaud M, Lévy Y, Hüe S, Raoult D. Microbiome of HIV-infected people. Microb. Pathog. 2017;106:85–93. doi: 10.1016/j.micpath.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Dillon S, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHardy IH, et al. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutlu EA, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. plos Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013;5:193ra91–ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubourg G. Impact of HIV on the human gut microbiota: challenges and perspectives. Human Micro. J. 2016;2:3–9. doi: 10.1016/j.humic.2016.10.001. [DOI] [Google Scholar]
- 25.Bäckhed F. Programming of host metabolism by the gut microbiota. Ann. Nutr. Metab. 2011;58(Suppl. 2):44–52. doi: 10.1159/000328042. [DOI] [PubMed] [Google Scholar]
- 26.Scher JU, et al. Decreased bacterial diversity characterizes an altered gut microbiota in psoriatic arthritis and resembles dysbiosis of inflammatory bowel disease. Arthritis Rheumatol. (Hoboken, NJ) 2015;67:128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenabian MA, et al. Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J. Infect. Dis. 2015;212:355–366. doi: 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- 28.Sherry BTOP, et al. Resolution of the two components of macrophage inflammatory protein 1, and cloning and characterization of one of those components, macrophage inflammatory protein 1 beta. J. Exp. Med. 1988;168:2251–2259. doi: 10.1084/jem.168.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantovani A, Gray PA, Van Damme J, Sozzani S. Macrophage-derived chemokine (MDC) J. Leukoc. Biol. 2000;68:400–404. [PubMed] [Google Scholar]
- 30.Gaidt MM, Hornung V. Alternative inflammasome activation enables IL-1beta release from living cells. Curr. Opin. Immunol. 2017;44:7–13. doi: 10.1016/j.coi.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zittermann SI, Issekutz AC. Basic fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte recruitment to inflammation by enhancing endothelial adhesion molecule expression. Am. J. Pathol. 2006;168:835–846. doi: 10.2353/ajpath.2006.050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasza A. IL-1 and EGF regulate expression of genes important in inflammation and cancer. Cytokine. 2013;62:22–33. doi: 10.1016/j.cyto.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Alam R. Chemokines in allergic inflammation. J. Allergy Clin. Immunol. 1997;99:273–277. doi: 10.1016/S0091-6749(97)70042-3. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8:533–545. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 35.Shilaih M, et al. Antibacterial effects of antiretrovirals, potential implications for microbiome studies in HIV. Antivir. Ther. 2017;23:91–94. doi: 10.3851/IMP3173. [DOI] [PubMed] [Google Scholar]
- 36.Ahmad A, et al. Combination antiretroviral therapy and indoleamine 2,3-dioxygenase in HIV infections: challenges and new opportunities. AIDS. 2016;30:1839–1841. doi: 10.1097/QAD.0000000000001168. [DOI] [PubMed] [Google Scholar]
- 37.Villanueva-Millan MJ, Perez-Matute P, Recio-Fernandez E, Lezana Rosales JM, Oteo JA. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J. Int AIDS Soc. 2017;20:1–13. doi: 10.7448/IAS.20.1.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyler AD, Smith MI, Silverberg MS. Analyzing the human microbiome: a “how to” guide for physicians. Am. J. Gastroenterol. 2014;109:983–993. doi: 10.1038/ajg.2014.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.