Abstract
Rationale
Organs of the body require vascular networks to supply oxygen and nutrients and maintain physiological function. The blood vessels of different organs are structurally and functionally heterogeneous in nature. To more precisely dissect their distinct in vivo function in individual organs, without potential interference from off-site targets, it is necessary to genetically target them in an organ-specific manner.
Objective
To generate a genetic system that targets vascular endothelial cells in an organ- or tissue-specific manner and to exemplify the potential application of intersectional genetics for precise, target-specific gene manipulation in vivo.
Methods and Results
We took advantage of two orthogonal recombination systems, Dre-rox and Cre-loxP, to create a genetic targeting system based on intersectional genetics. Using this approach, Cre activity was only detectable in cells that had expressed both Dre and Cre. Applying this new system, we generated a coronary endothelial cell specific Cre (CoEC-Cre) and a brain endothelial cell specific Cre (BEC-Cre). Through lineage tracing, gene knockout and over-expression experiments, we demonstrated that CoEC-Cre and BEC-Cre efficiently and specifically target blood vessels in the heart and brain, respectively. By deletion of VEGFR2 using BEC-Cre, we showed that VEGF signaling regulates angiogenesis in the central nervous system and also controls the integrity of the blood-brain barrier.
Conclusions
We provide two examples to illustrate the use of intersectional genetics for more precise gene targeting in vivo, namely manipulation of genes in blood vessels of the heart and brain. More broadly, this system provides a valuable strategy for tissue-specific gene manipulation that can be widely applied to other fields of biomedical research.
Keywords: Dual recombinases, gene manipulation, Cre-loxP, Dre-rox, blood vessels, animal model, angiogenesis, vessel, genetic technology
Subject Terms: Angiogenesis, Vascular Biology
Introduction
Blood vessels are essential to the proper function of organs and tissues in development, homeostasis, repair and regeneration. While the endothelial cells lining the innermost layer of blood vessels are conduits for passively delivering blood, they actively modulate tissue function and control organ growth during development and regeneration.1–3 In response to tissue injury, endothelial cells orchestrate self-renewal and differentiation of tissue-specific resident stem cells for repair.4 The paracrine factors and chemokines secreted by endothelial cells induce regenerative lung alveolarization and promote liver regeneration, without provoking fibrosis.3, 5 The response of endothelial cells to injury varies considerably between tissues and they express a combination of transcription factors, chemokines and growth factors, that is unique to each organ.6 Moreover, diversity in endothelial structure confers organ-specific function. For instance, endothelial cells of the kidney and liver are fenestrated to facilitate the filtration of the blood and metabolic substances, respectively; in contrast, brain endothelial cells possess tight junctions that restrict the free passage of substances between the circulating blood and brain tissues.7, 8 Therefore, it is increasingly apparent that endothelial cells display marked heterogeneity in structure, phenotype and function between different tissues.9, 10
The ability to precisely delineate the function of organotypic vasculature in vivo requires a novel strategy to genetically target endothelial cells in a tissue-specific manner. Conventional genetic tools, such as Tie2 or VE-Cadherin (Cdh5) promoter-driven Cre recombinase, target endothelial cells almost ubiquitously in most organs of the body.11, 12 The development of tissue-specific vascular Cre tools to enhance precision of genetic targeting would advance our understanding of endothelial cell function in organ formation and disease. Recent transcriptomic profiling as well as other technologies have provided tremendous insight into the unique profiles of angiocrine factors, adhesion molecules, chemokines, transcription factors and metabolic regulators of endothelial cells in different tissues.6 However, none of these genes, individually, define endothelial cells of a particular organ; rather, a combination of several genes is required to constitute a unique molecular signature. Thus, in practice, the targeting of a single endothelial cell type of an organ requires a new strategy that utilizes a combination of different markers to achieve the requisite specificity.
To date, genetic targeting tools have been built on the most widely used Cre-loxP homologous recombination system.13 Thousands of Cre and floxed mouse alleles (loxP flanked genes) have been generated to address diverse questions regarding cell lineage and gene function. The precision of this system largely depends on the specificity of Cre expression, which is usually driven by a single gene promoter.14 Although very powerful, this conventional approach has inherent limitations, such as incomplete specificity, which has led to some discrepancies, and even controversies, in data interpretation.15 Recently, we and others sought a more precise targeting approach, utilizing different site-specific recombinases, under the control of multiple promoters, for genetic lineage tracing to study endogenous cell fates.16–20 However, it remained unclear whether this approach could be utilized for genetic manipulation in vivo, e.g. knockout of conventional floxed alleles. Here, we report a gene targeting system that exploits two recombination systems, Dre-rox and Cre-loxP, for their sequential intersectional genetics, whereby Dre-rox controls Cre expression such that Cre activity is restricted within, and defined by, the intersection of two gene expression domains. As proof-of-principle, we first validated the intersectional genetic system by generation of new mouse lines. These were then applied towards genetic targeting, achieving both gene knockout and over-expression, selectively in endothelial cells of distinct organ systems, namely the heart and brain. This valuable technology has broad applicability as a means to target organ-specific vasculatures and can also be extended to target further cellular compartments in other key organs to facilitate better understanding of gene functions.
Methods
The authors will make all data, analytic methods and study materials available to other researchers. The authors declare that all data that support the findings of this study are available within the article and its Online Data Supplement.
Mouse generation, breeding and genotyping
All animal studies were carried out in accordance with the guidelines in the Institutional Animal Care and Use Committee (IACUC) of the Institute for Nutritional Sciences and the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Science. Nrg1-CrexER, Nrg1-CreER, Wt1-CrexER, Tie2-Dre, Mfsd2a-CrexER mouse lines were generated by homologous recombination using CRISPR/Cas9 technology. For the Nrg1-CrexER line, CrexER was generated by insertion of two rox sequences flanking ERT2: Cre-rox-ER-rox. cDNA encoding CrexER was inserted into the translational start codon ATG of the Nrg1 gene, and followed by a polyadenylation sequence. For the Nrg1-CreER line, CreERT2 cDNA, followed by a polyadenylation sequence, was inserted into the last coding exon of Nrg1 gene, and a 2A peptide sequence was used to link the Nrg1 coding region and CreERT2 cDNA to allow expression of both Nrg1 and CreERT2. For the Wt1-CrexER mouse line, the cDNA encoding CrexER was inserted into the translational start site of the Wt1 gene and followed by a polyadenylation sequence. For the Tie2-Dre mouse line, cDNA encoding Dre recombinase, followed by a polyadenylation sequence, was inserted into the translation start site of the Tie2 gene. For the Mfsd2a-CrexER mouse line, the cDNA encoding CrexER was inserted into the translation start site of the Mfsd2a gene. These mouse lines were generated by Shanghai Biomodel Organism Co., Ltd., China. R26-LacZ, R26-rox-LacZ, R26-tdTomato, R26-rox-tdTomato, ACTB-Cre, CAG-Dre, Kdr-flox, R26-iDTR, and Mfsd2a-CreER mice were described previously.21–27 A list of genomic PCR primer for these mice was included in Online Table I. All experimental mice were maintained on 129, C57BL6 and ICR mixed backgrounds. Tamoxifen (sigma, T5648) was dissolved in corn oil and administered to mice at the indicated time. Adult animals received 0.1 mg tamoxifen per gram mouse body weight by oral gavage.
In situ hybridization
In situ hybridization was performed according to a protocol described previously.18 Briefly, dissected embryos were fixed overnight in fresh 4% paraformaldehyde (PFA) and embedded in OCT (Sakura) after dehydration in 30% sucrose. Cryosections of 8-10 μm thickness were treated with hybridization buffer containing 1 μg/ml digoxigenin (DIG)-labeled probes overnight at 65°C. Slides were washed for 10 minutes in MABT buffer at room temperature and slides were washed in SSC buffer for 1 hour at 65°C. After incubating with blocking buffer (10% sheep serum and 2% blocking reagent in MABT buffer) at room temperature for 1 hour, slides were stained with anti-DIG antibody (Roche, 11093274910) diluted in blocking buffer at 4°C overnight. Then the slides were washed in MABT buffer and equilibrated in NTMT buffer, signals were developed with NBT and BCIP (Promega, S3771) in the dark. Images were obtained on an Olympus microscope (BX53). The following primers were used to construct Nrg1 probes: forward, GACTAGTGCTGTCTGCTTTTCCTCCCTTAC, reverse, ATAAGAATGCGGCCGCCCTCATCCTCCACTATCCTCAATG.
X-gal staining
Embryos were dissected in cold PBS and fixed in LacZ fix solution (0.2% glutaraldehyde, 5mM EGTA (pH 7.3), and 0.1M MgCl2 in PBS) for 30 min on ice with gentle shaking. After washing with LacZ wash buffer (0.01% sodium deoxycholate, 0.02% Nonidet-P40, and 2mM MgCl2 in 100mM sodium phosphate buffer) three times for 30 min, embryos were stained with LacZ staining buffer containing 1 mg/ml X-gal at 37 °C overnight to the desired extent. During the staining process, embryos were protected from light. Then embryos were washed with LacZ wash buffer three times and whole mount images were acquired using a Zeiss stereomicroscope (AXIO Zoom. V16).
Immunostaining
Immunostaining was performed as previously described.28 In detail, dissected tissues were fixed in 4% PFA (Sigma) at 4°C for 20-60 minutes, depending on the tissue size. Afterwards, tissues were washed in PBS for 3 times, dehydrated in 30% sucrose overnight at 4°C then embedded in OCT (Sakura) for cryosectioning. Cryosections (8-10 μm) were obtained and air-dried at room temperature. For staining, sections were washed in PBS for 5 min, incubated in blocking buffer containing 5% normal donkey serum (Jackson Immunoresearch), 0.1% Triton X-100 in PBS for 30 minutes at room temperature. Sections were then stained with the primary antibodies overnight at 4°C. Signals were developed with Alexa fluorescence antibodies (Invitrogen). HRP-conjugated secondary antibodies with tyramide signal amplification kit (PerkinElmer) were used to amplify weak signals. Nuclei were counterstained with 4'6-diamidino-2-phenylindole (DAPI, Vector lab). The following antibodies were used: RFP/tdTomato (Rockland, 600-401-379, 1:1000), Estrogen receptor (ESR, Abcam, ab27595, 1:1), NeuN (Millipore, MAB377X, 1:400), VE-cadherin (R & D, AF1002, 1:100), Flk-1/VEGFR2 (BD Biosciences, 550549, 1:100), CD31/PECAM (BD Biosciences, 553370, 1:500), cardiac Troponin I (TNNI3, Abcam, ab56357, 1:100), cytokeratin 19 (Developmental Studies Hybridoma Bank, TROMA-III, 1:500), HNF4a (Santa Cruz, sc-6556, 1:100), E-cadherin (Cell signaling, 3195, 1:100), HB-EGF (R & D, AF-259-NA, 1:100), Ter-119 (eBioscience, 14-5921-85, 1:400), Glut1 (Merk/Millipore, 07-1401, 1:200), PLVAP (BD pharmingen, 553849, 1:500), CTNNB1 (Santa Cruz, sc-59737, 1:100), Ki67 (Thermo Fisher Scientific, RM-9106-S0, 1:200). Immunostaining images were acquired using an Olympus fluorescence microscope (BX53), a Zeiss stereomicroscope (AXIO Zoom. V16), a Zeiss confocal laser scanning microscope (LSM510) or an Olympus confocal microscope (FV1200).
FACS analysis of brain endothelial cells
Brains of neonatal mice were dissected to remove pial membrane and minced into 1-2mm fragments in cold PBS. Then tissues were dissociated in Type IV collagenase (400U/ml, Worthington) and DNase I (32U/ml, Worthington) in PBS with Ca2+ and Mg2+ (Hyclone) at 37 °C on a rotator for 30 min. After disaggregation, cells were washed in cold PBS and passed through a 40-μm filter followed by centrifugation for 5min at 500g. Supernatant was discarded and cell pellet was resuspended in 20%BSA and centrifuged at 1000g for 15min to remove the myelin. The red blood cells were lysed with RBC lysis buffer. Afterwards, the cells were washed in PBS and stained with the Violet Dye (Invitrogen, L34955) to exclude dead cells according to the manufacture’s instruction. After washing in PBS, the cells were blocked with CD16/32 (eBioscience, 14-0161) and stained with CD31-APC (eBioscience, 17-0311) to label endothelial cells at 4°C for 30 min. The stained cells were analyzed using a FACS Aria II Flow Cytometer (BD Bioscience).
Hypoxia analysis
Hypoxia levels were assessed by Hypoxyprobe-1 Omni Kit (Hypoxyprobe Inc), according to the manufacturer’s protocols.29 Mice were injected intraperitoneally with 60~100 mg/kg pimonidazole HCl. One hour later, brains were collected and fixed in 4% PFA, then washed in PBS, dehydrated in 30% sucrose and frozen in OCT. 10-μm sections were stained with anti-pimonidazole. Signals were developed with 488-Alexa Fluor conjugated donkey anti-rabbit antibody (Invitrogen). Immunostaining images were acquired with a Zeiss confocal laser scanning microscope (LSM510) or an Olympus confocal microscope (FV1200).
Examination of integrity of the blood-brain barrier (BBB)
This method was performed as described previously.30 P0 mice were anesthetized with isoflurane. The beating heart was injected via the left ventricle with 10μl of 10-kDa dextran tetramethylrhodamine (4 mg/ml, Invitrogen, D3312). After 5 min of circulation, brains were collected and fixed in 4% PFA at 4°C overnight, then washed in PBS, dehydrated in 30% sucrose and embedded in OCT (Sakura) for cryosectioning. 10-μm sections were post-fixed in 4%PFA for 15 min, followed by washing in PBS, incubated in blocking buffer containing 5% normal donkey serum (Jackson Immunoresearch) and 0.1% Triton X-100 in PBS for 30 minutes at room temperature. Sections were then stained with fluorescein labeled Griffonia (Bandeiraea) Simplicifolia Lectin I (GSL I or BSL I, Vector Lab, FL-1101-5, 1:500) overnight at 4 °C. Immunostained tissue images were acquired using a Zeiss confocal laser scanning microscope (LSM510) or an Olympus confocal microscope (FV1200).
Cell ablation experiments
For specific cell ablation experiments, adult Wt1-CrexER;Tie2-Dre;R26-iDTR mice were intraperitoneally injected with diphtheria toxin (DT) dissolved in PBS at a dose of 20 ng/g bodyweight for two consecutive days. Control mice received intraperitoneal injection of PBS. Hearts were collected two days after the last diphtheria toxin injection.
TUNEL assay
Apoptosis was detected using the In Situ Cell Death Detection Kit, Fluorescein (Roche, 11684795910) according to the manufacture’s instruction. Briefly, sections were fixed in 4% PFA for 10 minutes at room temperature, followed by 3 washes in PBS. Afterwards, sections were incubated with blocking buffer containing 5% normal donkey serum (Jackson Immunoresearch) and 0.1% Triton X-100 in PBS for 5 minutes at 4 °C. Sections were then incubated with TUNEL reaction mixture in a humidified chamber for 1 hour at 37 °C. After washing in PBS, sections were stained with antibodies referring to the procedure described in Immunostaining. Images were acquired using a Zeiss confocal laser scanning microscope (LSM510) or an Olympus confocal microscope (FV1200).
Statistical analysis
All data were acquired from five independent samples and presented as mean values ± standard error of the mean (SEM). All mice were randomly assigned to different experimental groups and investigators were blinded to genotype. Sample sizes were designed with adequate power according to the literature and our previous studies. Statistical comparisons between data sets were made with analysis of normality and variance, followed by a two-sided unpaired Student's t-test for comparing differences between two groups. ANOVA test was used for comparison between 3 or more groups. Significance was accepted when P < 0.05.
Results
Design of a new genetic targeting system by combining Dre-rox and Cre-loxP
To achieve more precise, organ-selective gene manipulation, we took advantage of two heterospecific recombination systems: Cre-loxP and Dre-rox. Dre recombinase is similar to Cre, but catalyzes recombination between different sequences, named rox sites.31 To first verify that these two systems achieve orthogonal homologous recombination events, we crossed constitutive Cre or Dre (ACTB-Cre or CAG-Dre) with R26-tdTomato and Rosa26-rox-tdTomato reporter lines.22–24 By detection of tdTomato expression, we found that Cre-loxP and Dre-rox were heterospecific in homologous recombination (Figure 1A), in that they showed no tdTomato by either Cre-rox or Dre-loxP recombination (Figure 1A). Similar results were obtained using R26-LacZ and R26-rox-LacZ reporter lines.21, 22 We then combined these two orthogonal recombination systems, utilizing two different promoters to drive Dre and Cre for an intersectional genetics strategy (Figure 1B). One key consideration in the design of this system was that the ultimate effector should be a singular Cre recombinase by intersectional genetics, such that the existing extensive mouse resource of available loxP-based alleles could be utilized for functional studies. To achieve this, we designed a strategy that permitted sequential Dre-mediated Cre expression in targeted cells that have expressed both Dre and Cre recombinases (Figure 1C). We generated a switchable CreER (CrexER), by insertion of two rox sites to flank the estrogen receptor (ER or ESR) of the original CreER cDNA (Figure 1C). By default, the Cre-ER fusion protein was restricted to the cytoplasm.32 We predicted that Dre-rox recombination would remove ER to release Cre activity, upon cytoplasmic-nuclear translocation. Thus, this system incorporated two basic elements: A-Dre and B-CrexER, in which Dre recombinase was driven by gene A and CrexER was driven by gene B. The ultimate readout of this system was Cre, and its activity was only present in cells that expressed genes A and B. Due to the sequential recombination (Dre-rox first, Figure 1C) in the intersection of the expression domains of gene A and B (Figure 1B), we refer to this experimental strategy as sequential intersectional genetics.
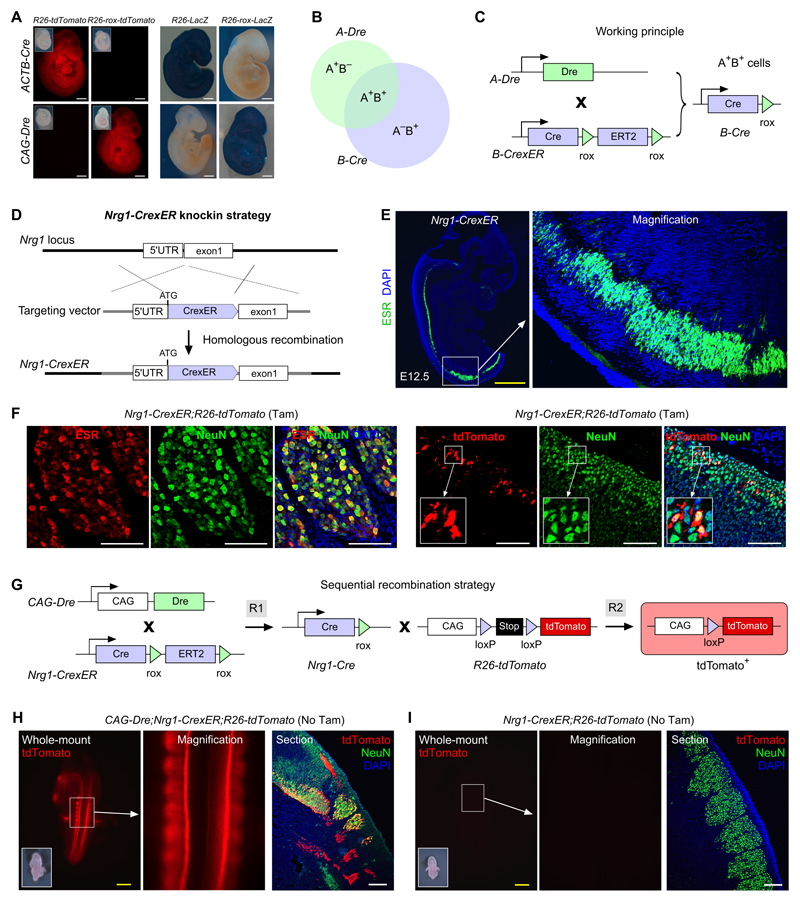
Figure 1. Proof-of-principle for a sequential recombination strategy for genetic targeting.
A, Whole-mount view of E9.5 embryos by tdTomato or LacZ reporter. Inserts indicate the bright-field images. B, Schematic showing intersectional genetic targeting of A+B+ cells by A-Dre and B-Cre recombinases. C, Working principle showing how A-Dre mediated recombination (R1) results in removal of rox flanked ERT2, yielding Cre expression in cells that have expressed Dre. D, Schematic showing strategy for Nrg1-CrexER mouse generation. E, Immunostaining for ESR on E12.5 Nrg1-CrexER sections. F, Immunostaining for ESR, tdTomato and NeuN on E12.5 Nrg1-CrexER;R26-tdTomato embryonic sections. Tamoxifen (Tam) was administered at E9.5. G, Sequential recombination strategy for genetic lineage tracing by crossing CAG-Dre, Nrg1-CrexER and R26-tdTomato mice lines. R1 and R2 denotes Dre-rox and Cre-loxP recombinations respectively. H,I, Whole-mount view and staining on sections showed tdTomato+ signals in E12.5 CAG-Dre;Nrg1-CrexER;R26-tdTomato embryos (H), but not in the littermate Nrg1-CrexER;R26-tdTomato embryos (I). Inserts indicate bright-field images of embryos. Each figure is representative of 5 individual samples. Scale bars, yellow 1 mm; white, 100 µm.
Proof-of-principle study for sequential intersectional genetics
As proof-of-principle for this genetic strategy and use of CrexER in an animal model, we first needed to test if Cre-rox protein is effective after rox-ER-rox recombination by Dre. We selected a gene with a clearly defined recombination pattern, which should not be germline or sporadically activated by constitutive Cre-rox recombinase. Neither Wt1 nor Mfsd2a genes, that would be used in our subsequent experiments, were initially selected for this proof-of-principle test, because of previously reported sporadic expression during early embryonic development.33 Instead, we used the Neuregulin 1 (Nrg1) gene, implicated in neural development,34 and generated a Nrg1-CrexER line, in which CrexER cDNA was knocked into the endogenous Nrg1 locus (Figure 1D). Immunostaining for estrogen receptor (ESR), as a surrogate for endogenous Nrg1, showed a neural crest expression pattern (Figure 1E), consistent with Nrg1 in situ hybridization data (Online Figure IA) and previous studies.34, 35 In addition, Nrg1-CrexER labeled a subset of neuronal cells in the dorsal root ganglion of the neural crest after tamoxifen induction (Figure 1F), which was similar to the Nrg1-CreER labeling pattern (Online Figure IB,C). To test if Cre could be released from CrexER and perform subsequent recombination, we crossed Nrg1-CrexER with CAG-Dre to permit Dre-mediated excision of rox-flanked ER, and assessed subsequent Cre-loxP recombination (Figure 1G). Without tamoxifen induction, we could detect tdTomato in the neural crest in the CAG-Dre;Nrg1-CrexER;R26-tdTomato mice (Figure 1H), demonstrating Cre release from CrexER specifically and efficiently in vivo. There was no detectable signal in the littermate Nrg1-CrexER;R26-tdTomato control mouse without tamoxifen treatment (Figure 1I), indicating no leakiness of CrexER in the absence of Dre-mediated ER excision. These data validated the feasibility of our strategy for Dre-mediated Cre expression in transgenic mouse lines and confirmed the utility of this novel CrexER module for sequential intersectional genetic targeting in vivo.
Generation of coronary endothelial cell specific Cre (CoEC-Cre)
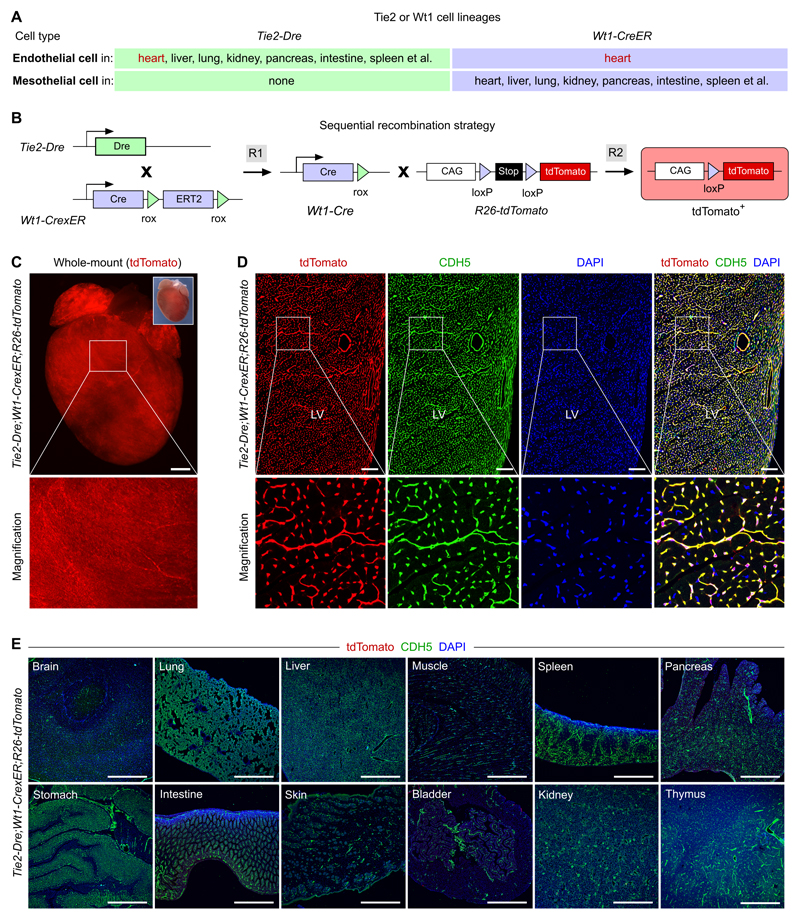
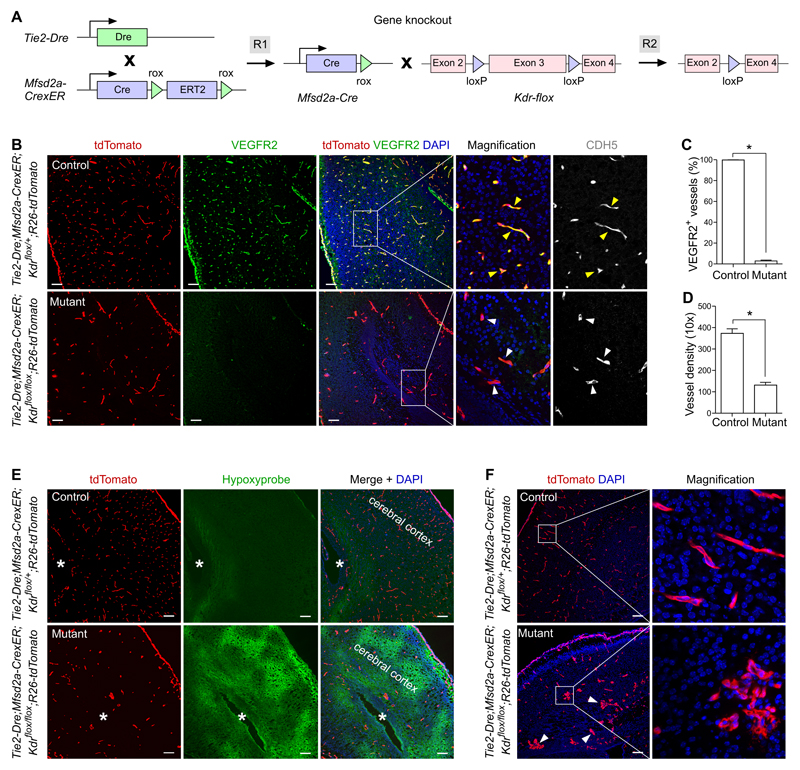
We chose the blood vessels of heart and brain as two cases in point to test our new system. Application of sequential intersectional genetics to the coronary endothelium requires two markers which intersect only in vascular endothelial cells of the heart. The mesothelial cell marker, Wt1, was recently reported to express in developing coronary blood vessels, but not in other vessels.36 In addition to epicardial cells, Wt1-CreER labels coronary endothelial cells in the embryonic heart.33, 37 A second marker, Tie2, is pan-endothelial cell marker, and expressed in most vessels as well as certain populations of myeloid cells.38 With knowledge of these gene expression profiles, we aimed to test whether the intersection of Wt1 and Tie2 could direct the precise targeting of coronary vessels (Figure 2A). We first generated a Wt1-CrexER mouse line by insertion of CrexER in frame with the translational start codon (ATG) of the Wt1 gene (Online Figure IIA). Immunostaining for ESR showed that CrexER was restricted to the proepicardium and epicardial cells in the developing heart at E9.5 and E10.5 (Online Figure IIB,C). For the second marker, Tie2, we generated a Tie2-Dre mouse allele by knocking in a Dre cassette in frame with the ATG of the endogenous Tie2 gene (Online Figure IIIA) and characterized this mouse by crossing with a rox reporter line R26-rox-tdTomato. (Online Figure IIIB). Whole-mount images and immunostaining on tissue sections confirmed that Tie2-Dre labeled endothelial cells in multiple organs (Online Figure IIIC-E). Based on the Tie2-Dre and Wt1-CreER expression domains (Figure 2A), we generated Tie2-Dre;Wt1-CrexER mice to determine if Cre activity is specific to coronary vascular endothelial cells (Figure 2B). Whole-mount fluorescence and immunostaining of heart sections showed both specific and efficient labeling of coronary vessels (Figure 2C,D). Quantification of the percentage of tdTomato+ coronary vascular endothelial cells showed labeling of 97.15 ± 0.61% of endothelial cells in the compact myocardium. Immunostaining for tdTomato and VE-cad (CDH5) on other tissue sections showed only rare tdTomato+ cells (Figure 2E and Online Figure IV for magnification). As internal controls, we did not detect any tdTomato+ cells in the tissues of Tie2-Dre;R26-tdTomato, thus ruling out Dre-loxP recombination (Online Figure VA-C). Nor did we observe any tdTomato+ cells in tissues of Wt1-CrexER;T26-tdTomato mice without tamoxifen treatment, confirming no leakiness of Cre-loxP recombination by Wt1-CrexER (Online Figure VD-F). These data demonstrate that Cre-loxP recombination occurred only in the presence of both Tie2-Dre and Wt1-CrexER. Taken together, Tie2-Dre;Wt1-CrexER (hereafter named CoEC-Cre) genetically targeted blood vessels in the heart but not in other organs.
Figure 2. Genetic targeting of coronary vessels by Tie2-Dre;Wt1-CrexER line.
A, Cell lineages targeted by Tie2-Dre and Wt1-CreER. B, Schematic showing the working principle for sequential Dre and Cre recombination. The final readout is Cre-loxP recombination. C, Whole-mount fluorescent images of Tie2-Dre;Wt1-CrexER;R26-tdTomato heart from 6 week old mice. Insert indicates bright-field image of the same heart. D, Immunostaining for tdTomato and CDH5 on heart section shows CDH5+tdTomato+ coronary vessels. LV, left ventricle. E, Immunostaining for tdTomato and CDH5 on sections from organs of 6 week old Tie2-Dre;Wt1-CrexER;R26-tdTomato mice. Scale bars, 1 mm in C,E; 100 µm in D. Each image is representative of 5 individual samples.
Using CoEC-Cre for gene manipulation in coronary vascular endothelial cells
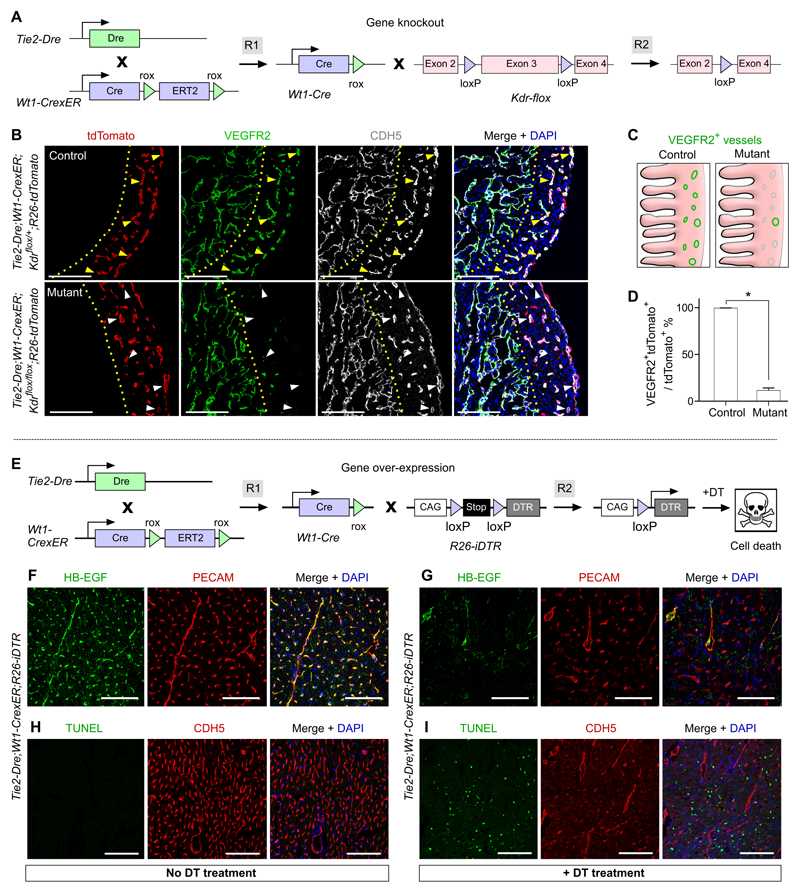
To examine whether this new genetic system could be utilized for gene deletion using conventional floxed mouse alleles, we selected VEGFR2 (Kdr) as an exemplar. Kdr is universally expressed in endothelial cells of most organs,39 but its selective targeting in coronary arteries has not previously been possible. We therefore crossed CoEC-Cre (Tie2-Dre;Wt1-CrexER) with the Kdr flox line25 to test whether Kdr could be specifically deleted in the coronary vessels but not in vessels of other organs (Figure 3A). To allow simultaneous tracing of targeted cells, we generated Tie2-Dre;Wt1-CrexER;Kdrflox/flox;R26-tdTomato (mutant) and compared it with littermate Tie2-Dre;Wt1-CrexER;Kdrflox/+;R26-tdTomato (control) mice. Immunostaining for tdTomato, VEGFR2 and CDH5 on E14.5 heart sections revealed a substantial loss of VEGFR2 in tdTomato+CDH5+ coronary endothelial cells within the compact myocardium of mutant hearts, compared with controls (Figure 3B,C). However, VEGFR2 expression was unaffected in endocardial cells of trabecular myocardium (Figure 3B), demonstrating that the CoEC-Cre specifically targeted Wt1-expressing coronary endothelial cells, but not endocardial endothelial cells, which do not express Wt1 in the embryonic heart. Quantification of the percentage of VEGFR2+tdTomato+ vascular endothelial cells in tdTomato+ vessels showed a significant reduction in the mutant compared with controls (11.82 ± 2.68 vs. 99.68 ± 0.16% in mutant and control, respectively, Figure 3D), indicating efficient deletion of Kdr in coronary vascular endothelial cells. The deletion efficiency of Kdr did not reach the same efficiency as that of cell labeling, which was likely due to a difference in Cre recombination efficiency at individual loxP allels and also the number of recombination events required in that of Kdrflox/flox needs both alleles to be recombined, while R26-tdTomato requires only one. Furthermore, we found that, in the mutant heart, VEGFR2+tdTomato+ cells displayed a higher degree of proliferation, compared with VEGFR2–tdTomato+ cells (49.92 ± 4.66% vs. 7.86 ± 2.08% respectively, Online Figure VI), suggesting an enhanced proliferation in endothelial cells that evade VEGFR2 deletion. By examining VEGFR2 expression in other organs of the same mice, we could not detect any noticeable reduction in the proportion of VEGFR2+ vessels in mutant compared with control mice (data now shown).
Figure 3. Gene knockout and over-expression in coronary endothelial cells.
A, Schematic figure showing the working principle for Kdr gene deletion in coronary endothelial cells. B, Immunostaining for tdTomato, VEGFR2 and CDH5 on E14.5 heart sections of Tie2-Dre;Wt1-CrexER;Kdrflox/+;R26-tdTomato (control) or Tie2-Dre;Wt1-CrexER;Kdrflox/flox;R26-tdTomato (mutant) littermate embryos. Yellow arrowheads indicate VEGFR2+CDH5+ vascular endothelial cells in the control heart; white arrowheads indicate VEGFR2–CDH5+ vascular endothelial cells in the mutant heart. Dotted yellow lines indicate border between trabecular myocardium (left) and compact myocardium (right). C, Schematic figure showing VEGFR2+ vessels (green) in the compact myocardium. D, Quantification of the percentage of VEGFR2+tdTomato+ vascular endothelial cells in tdTomato+ vessels in the control and mutant groups. Data are mean ± SEM.; *P < 0.05; n = 5. E, Schematic figure showing the working principle for over-expression of DTR in coronary endothelial cells. F,G, Immunostaining for HB-EGF and PECAM on heart sections of adult Tie2-Dre;Wt1-CrexER;R26-iDTR mouse treated without DT (F) and with DT (G). H,I, Immunostaining for TUNEL and CDH5 on heart sections of same mice. Scale bars, 100 µm. Each figure is representative of 5 individual samples.
To test if our Cre tool could be utilized to drive over-expression of genes in coronary endothelial cells, but not in other types of vessels, we selected to over-express human diphtheria toxin receptor (DTR), which is not endogenously expressed in mouse.40 We crossed CoEC-Cre with the R26-iDTR mouse,26 in which DTR expression is controlled by Cre-loxP recombination (Figure 3E). Diphtheria toxin (DT) binds with DTR and the A subunit of DT enters into the cytoplasm and catalyzes the inactivation of elongation factor to terminate protein synthesis, inducing apoptotic death of target cells.41 We sought to express DTR specifically in coronary endothelial cells and assess whether administration of DT could ablate DTR-expressing endothelial cells. Since DTR was identified as a membrane-anchored heparin-binding EGF-like growth factor (HB-EGF precursor),42 we used HB-EGF antibody to detect DTR expression in mouse tissues. Immunostaining for HB-EGF and PECAM on heart sections showed expression of DTR exclusively in coronary endothelial cells (Figure 3F). We did not detect HB-EGF+ endothelial cells in other tissues or organs (Online Figure VII). We next treated mice with DT for two days and collected heart samples two days after the last DT treatment for analysis. Immunostaining for HB-EGF and PECAM showed severe disruption of endothelial cell morphology, suggestive of cell death (Figure 3G). Apoptosis was confirmed by an increased number of TUNEL+ cells in the heart sections from DT-treated Tie2-Dre;Wt1-CrexER;R26-iDTR mice, compared with No DT treatment (Figure 3H,I). Taken together, these gene knockout and over-expression data demonstrated that our new genetic system permits gene manipulation specifically in coronary endothelial cells, without impacting other organ systems.
Generation of brain endothelial cell specific Cre (BEC-Cre)
Unraveling the signaling pathways that control the integrity and function of brain endothelial cells has important implications for diseases of the central nervous system, such as stroke, epilepsy and Alzheimer’s disease.43 Generation of a Cre tool that specifically and efficiently targets brain endothelial cells would aid functional studies of blood-brain barrier (BBB) regulation. Previous work showed that Mfsd2a was expressed in periportal hepatocytes and also in brain endothelial cells of the central nervous system.27, 30 To independently map Mfsd2a expression, we took advantage of the recently generated Mfsd2a-CreER line27 and stained tissue sections with ESR, as a surrogate for endogenous Mfsd2a. Immunostaining data showed that Mfsd2a was expressed in brain endothelial cells, and also in neurons, hepatocytes and epithelial cells of the intestinal villus (Online Figure VIIIA-D). Mfsd2a expression was further confirmed by collecting tissues from Mfsd2a-CreER;R26-tdTomato mice at 24-48 hours after tamoxifen treatment (Online Figure VIIIE-H). These data convincingly demonstrated expression of Mfsd2a in brain endothelial cells but also in other cell types.
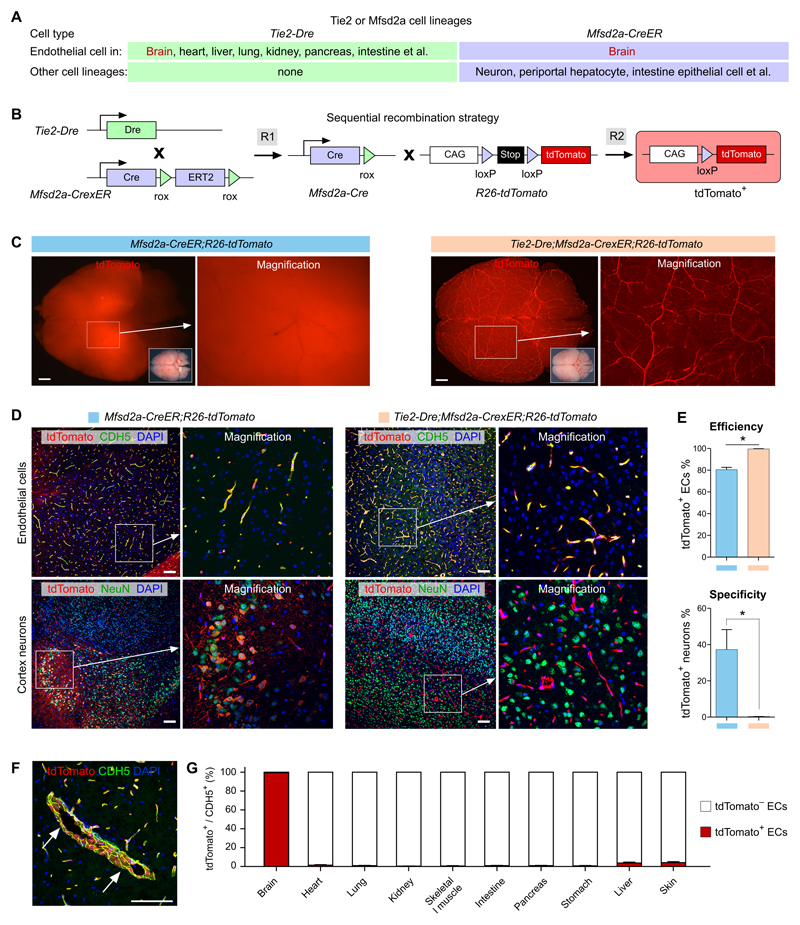
Based on Tie2-Dre and Mfsd2a-CreER targeting domains, we predicted that their intersection would target brain endothelial cells but not neurons or other cell lineages (Figure 4A). We therefore generated a Mfsd2a-CrexER mouse allele, in which cDNA encoding CrexER was inserted into the ATG of the first exon of the Mfsd2a gene (Online Figure IXA). Immunostaining for ESR, as a surrogate for CrexER, on Mfsd2a-CrexER mouse tissue sections showed ESR expression in periportal hepatocytes, brain endothelial cells and also a subset of neuronal cells (Online Figure IXB-C). There was no leakiness in Cre activity without tamoxifen induction (Online Figure IXD-E). These data confirmed the successful generation of the Mfsd2a-CrexER mouse line, which we then crossed to generate the compound Tie2-Dre;Mfsd2a-CrexER line (hereafter named BEC-Cre) for a sequential intersectional genetics study (Figure 4B). Compared with Mfsd2a-CreER that targeted both brain vessels and neurons, BEC-Cre targeted brain blood vessels efficiently and specifically, without labeling any neurons (Figure 4C,D). Quantification of the percentage of tdTomato+ endothelial cells in the brain showed a significant increase in BEC-Cre compared with Mfsd2a-CreER (99.56 ± 0.24% vs. 80.44 ± 2.18%, Figure 4E), while the percentage of tdTomato+ neurons was negligible in BEC-Cre compared with Mfsd2a-CreER (0.13 ± 0.14% vs. 37.25 ± 11.01%, Figure 4E). It should be noted that, in addition to the specificity, BEC-Cre targeted endothelial cells in the brain more efficiently than conventional Mfsd2a-CreER (Figure 4E). Flow cytometric analysis confirmed the efficient labeling of brain endothelial cells, showing 97.02 ± 0.31% of endothelial cells were tdTomato+ labeled by BEC-Cre (Online Figure X). We also observed that large vessels were also robustly labeled by BEC-Cre (Figure 4F). Large vessels do not themselves express Mfsd2a,27, 30 but they are tdTomato+ by virtue of their formation from Mfsd2a+ preliminary vascular plexus that was previously labeled by BEC-Cre. Additionally, BEC-Cre did not target any periportal hepatocytes, nor significant number of blood vessels in other organs or tissues (Online Figure XI). Quantification of the percentage of tdTomato+ endothelial cells in brain and other organs or tissues confirmed the efficiency and specificity of the BEC-Cre mouse line (Figure 4G).
Figure 4. Specific and efficient brain endothelial cell targeting.
A, Cell lineages targeted by Tie2-Dre and Mfsd2a-CreER. B, Schematic showing the working principle for sequential Dre and Cre recombinations. The final readout is Cre-loxP recombination. C, Whole-mount fluorescence view of brains from adult Mfsd2a-CreER;R26-tdTomato or Tie2-Dre;Mfsd2a-CrexER;R26-tdTomato mice. For Mfsd2a-CreER, Tamoxifen was induced at 6 weeks and samples were collected 2 days later. Inserts are bright-field images of brains. D, Immunostaining for tdTomato and CDH5 or NeuN on brain sections of Mfsd2a-CreER;R26-tdTomato or Tie2-Dre;Mfsd2a-CrexER;R26-tdTomato mice. E, Quantification of the percentage of tdTomato+ endothelial cells (ECs) or tdTomato+ neurons. Data are mean ± SEM.; *P < 0.05; n = 5. F, Immunostaining for tdTomato and CDH5 shows large brain vessels (arrow) are also tdTomato+. G, Quantification of the percentage of tdTomato+ vascular endothelial cells in different organs, collected from adult Tie2-Dre;Mfsd2a-CrexER;R26-tdTomato mice. Scale bars, 1 mm in C; 100 µm in D,F.
VEGFA-VEGFR2 signaling is required for angiogenesis and integrity of the BBB
To test if BEC-Cre could be utilized for gene deletion, we again used VEGFR2 as an example, due to its ubiquitous endothelial cell expression that allows us to assess the relative specificity for deletion in vessels of the brain. For specific knockout of Kdr in brain endothelial cells, we generated Tie2-Dre;Mfsd2a-CrexER;Kdrflox/flox;R26-tdTomato (mutant) and compared it with littermate Tie2-Dre;Mfsd2a-CrexER;Kdrflox/+;R26-tdTomato (control) mice (Figure 5A). The mutant was lethal at an early neonatal stage, as no mutants were alive by postnatal day 10 (P10), out of 11 litters (>120 mice genotyped). We therefore collected brain samples at late embryonic (E15.5-E18.5) and early neonatal stages (P0-P1). In the neonatal brain, we could hardly detect any VEGFR2+ blood vessels in the mutant brain, compared with controls (Figure 5B). Quantification data showed a significant reduction of the percentage of VEGFR2+ vessels in the mutant compared with control mice (2.91 ± 0.82% vs. 99.79 ± 0.12% in mutant and control, respectively, Figure 5C), indicating efficient deletion of Kdr in brain endothelial cells. In contrast, we did not detect any loss of VEGFR2 expression in vessels in other organs or tissues of mutant mice (Online Figure XII), demonstrating the specificity of Kdr deletion in brain vessels. We also observed a significant reduction in vessel density in the brain of mutants compared with the controls (131.33 ± 13.19 vs. 373.32 ± 21.25 vessels per 10x field in mutant and control respectively, Figure 5D), while there was no significant change in vessel density in other organs or tissues (Online Figure XII). Mutant brain also showed an enlarged avascular zone, and hypoxyprobe staining revealed a significantly increased hypoxia in the mutant brain, compared with the control (Figure 5E), confirming the impact of reduced brain blood vessel density on tissue oxygenation. In addition to reduced angiogenesis, the morphology of vessels in mutant brain was abnormal, as many mutant vessels were dilated and twisted in a cluster (Figure 5F), similar to glomeruloid-like vascular bodies. Taken together, we utilized brain vessel specific knockout to circumvent early lethality by conventional whole-body Kdr knockout,39 and demonstrated that Kdr was required for angiogenesis in the central nervous system.
Figure 5. Specific knockout of VEGFR2 in brain endothelial cells.
A, Schematic figure showing the working principle for sequential Dre and Cre recombinations that ultimately delete the flox flanked Kdr gene in brain endothelial cells. B, Immunostaining for tdTomato, VEGFR2 and CDH5 on P0 brain sections of Tie2-Dre;Mfsd2a-CrexER;Kdrflox/+;R26-tdTomato (control) or Tie2-Dre;Mfsd2a-CrexER;Kdrflox/flox;R26-tdTomato (mutant) mice. Yellow arrowheads indicate VEGFR2+CDH5+ endothelial cells in the control brain; white arrowheads indicate VEGFR2–CDH5+ endothelial cells in the mutant brain. C, Quantification of VEGFR2+ endothelial cells in the control and mutant groups. Data are mean ± SEM.; *P < 0.05; n =5. D, Quantification of the vessel density in a 10x field. Data are mean ± SEM.; *P < 0.05; n = 5. E, Immunostaining for tdTomato and Hypoxyprobe on the control and mutant tissue sections. Asterisks indicate lateral ventricle in cerebral cortex. F, Immunostaining for tdTomato on brain sections shows abnormal vessels (arrowheads) in mutant compared with control. Scale bars, 100 µm. Each figure is representative of 5 individual samples.
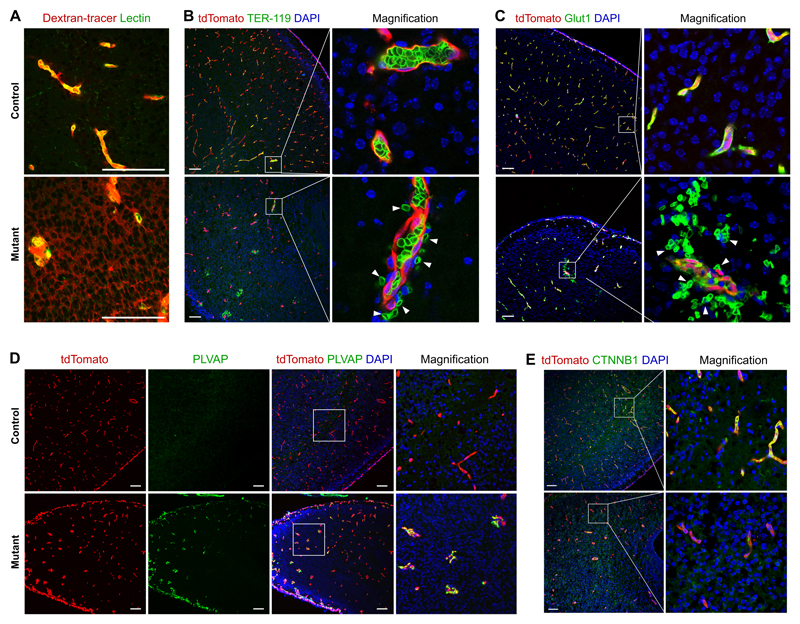
Finally, we examined whether Kdr was required for the integrity of the BBB. By injection of 10-kDa dextran tracer into the circulation, we found that dextran tracer leaked out of capillaries and was taken up by non-vascular brain parenchymal cells of the cortex (Figure 6A). Immunostaining for tdTomato and TER-119, an erythrocyte marker,44 revealed the presence of TER-119+ blood cells outside the tdTomato+ blood vessels (Figure 6B). Consistent with this, when we stained tissue sections with glucose transporter 1 (Glut1), a BBB marker also expressed in erythrocytes,45 we found a significant leakage of Glut1+ blood cells into the extra-vascular regions of the mutant brain (Figure 6C), further confirming that loss of Kdr impairs vascular integrity. The mutant brains also showed changes in endothelial cell protein composition, which was consistent with a loss of barrier function. In particular, plasmalemma vesicle-associated protein (PLVAP), an important component of endothelial fenestrations,46 which was normally absent from BBB endothelial cells (control, Figure 6D), was significantly induced in the mutant brain (Figure 6D). Canonical WNT signaling is required for proper assembly and differentiation of blood vessels in brain.47, 48 In our Kdr mutants, we found that expression of beta-catenin (CTNNB1) was significantly reduced, compared with littermate controls (Figure 6E). Taken together, these data indicate that the VEGF signaling pathway regulates essential genes related to BBB endothelial cell integrity.
Figure 6. Vegfa-Vegfr2 signaling regulates BBB integrity.
A, Images of cerebral sections of 10-kDa Dextran-tracer injected mice after capillary labelling with lectin. B,C, Immunostaining for tdTomato and TER-119 (B) or Glut1 (C) on brain sections shows a significant leakiness of blood cells (arrowheads). D, Immunostaining for tdTomato and PLVAP (a component of endothelial fenestration) shows increased PLVAP expression in the mutant E, Immunostaining for tdTomato and CTNNB1 on brain sections shows reduced CTNNB1 expression in the mutant. Scale bars, 100 µm. Each image is representative of 5 individual samples.
Discussion
Conventional genetic targeting takes advantage of Cre recombinase driven by one specific gene promoter that is active in, but not necessarily unique to, a particular cell population. Current biological studies require more refined gene manipulation. However, it is technically challenging to define a subpopulation of cells by a single marker. In many cases, a combination of two or more markers is required, as commonly accepted in the immunological studies. Our new system exploited the concept of intersectional genetics to distinguish a subpopulation of cells from other cell lineages based on the expression of two genes. Indeed, this approach has previously been used to increase precision of genetic lineage tracing.18–20 While the dual-recombinase strategy is useful for lineage tracing, it cannot be directly applied to the study of gene function, as the loxP flanked allele is only responsive to Cre but not simultaneously to Dre. For that reason, we generated this novel genetic targeting system, in which Dre-rox recombination first releases Cre from CreER, to then allow Cre targeting of conventional loxP alleles. This genetic strategy incorporates the idea of intersectional genetics and sequential recombination. Defining Cre activity by two distinct gene promoters, rather than one, confers a substantially greater level of precision in genetic targeting than could be achieved previously with Cre controlled by a single gene promoter. It is important to note that the effector of this system remains Cre recombinase, such that all previously generated conventional loxP alleles can be usefully incorporated. While this strategy aims principally at enhanced specificity, it is also highly efficient in gene deletion and over-expression in the experimental examples we studied. This study provided two examples of relevance to the cardiovascular field, but the technology reported here could be utilized more broadly in other fields when a more precise genetic targeting is required. On the basis that two, but not one, marker could specifically define a subpopulation of cells, this system can be employed for functional interrogation of the subpopulation of cells in vivo, in addition to their lineage tracing.
While this system is specific and efficient, there are limitations, caveats and scope for future improvements. Firstly, this system depends on two distinct markers to define a cell population. However, sometimes two markers are insufficient to precisely define a population. Some, such as immune cells or specific subtypes of neurons, are usually defined by multiple cell surface markers that typically exceed two. One possible solution is to utilize additional orthogonal recombinases, such as Flpe49 or the recently reported Nigri recombinase,50 to generate an additional intersection that reflects three or more genes. However, crossing of three or more lines (Flpe, Dre, Cre or more) to loxP alleles require vast resources and time, which will limit widespread application, and, moreover, is incompatible with the ethical principle of reducing animal breeding numbers. We believe that, with the advent of powerful single cell sequencing, more specific markers will emerge that will distinguish subpopulations of cells more precisely with fewer markers. Secondly, this system does not distinguish between cells that expressed the two genes driving recombination results either successively or concomitantly. It is therefore important that recombination results are interpreted and informed by careful expression maps of the genes that drive recombinases. Thirdly, this system needs to be modified for temporal control of gene expression. The current system, based on Dre-controlled Cre expression, is not manually inducible, as Dre is constitutively active. To impose temporal control, we could use DreER, which works under tamoxifen induction22 and on a different strategy, such as rox-stop-rox-Cre. This would require intersectional genes to be concurrently active genes in the target cells rather than on relying on developmentally active genes, as reported here. Alternatively, establishment of an efficient Split Cre51 approach, using genes that define specific cell populations, could also be tested for inducible Cre release. These modifications merit further investigation and refinement in future studies. Fourthly, knock-in of Cre or Dre cDNA into the genomic locus at the translational start codon replaces the endogenous gene. We therefore recommend insertion of recombinase cDNA into the 3’ UTR of the gene to avoid potential haploinsufficiency phenotypes. While genetic approaches facilitate the study of gene function, increasingly, research requires enhanced precision in both spatial and temporal control of gene expression, which, in turn, calls for improved, more innovative methodologies for gene manipulation in the future.
Supplementary Material
Novelty and Significance.
What Is Known?
Cre-loxP mediated genomic engineering depends on a single gene promoter that drives Cre.
Two intersectional gene promoters could define one cell population more precisely.
Endothelial cells are heterogenous in gene expression, structure and function among different organs.
Precise genetic targeting of blood vessels in organ or tissue specific manner remains challenging.
What New Information Does This Article Contribute?
Combination of Dre-rox and Cre-loxP recombination systems enhances the precision of genetic targeting.
Sequential intersection genetics permits more precise gene manipulation in specific cell population.
Coronary vessel specific Cre (CoEC-Cre) or brain vessel specific Cre (BEC-Cre) are generated and validated for gene manipulation.
VEGF signaling regulates angiogenesis and the integrity of the blood-brain barrier.
More precise genetic targeting is important to better understand the gene function in organ development, diseases and tissue regeneration. In this study, a sequential intersectional genetics mediated by dual recombination systems Dre-rox and Cre-loxP permits more precise gene manipulation. This dual recombinases-mediated gene targeting system could be used for generation of coronary or brain blood vessels specific Cre tools. By these new Cre lines, gene deletion or over-expression could be achieved in coronary vessels or brain vessels efficiently and specifically. While blood vessels are heterogenous in function among different organs, this new genetic system would help us to delineate the molecular mechanism regulating their distinct functions in organ-specific manner.
Acknowledgments
We thank Shanghai Model Organisms Center, Inc. (SMOC) for mouse generation and research platforms at the Institute of Biochemistry and Cell Biology, Institutes for Nutritional Sciences, National Center for Protein Science Shanghai.
Sources of Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS, XDB19000000, XDA16020204), National key Research & Development Program of China (2016YFC1300600, 2017YFC1001303, SQ2018YFA010021, SQ2018YFA010113), National Science Foundation of China (31730112, 91639302, 31625019, 81761138040, 31571503, 31501172, 31601168, 31701292, 91749122), Youth Innovation Promotion Association of CAS (2015218, 2060299), Key Project of Frontier Sciences of CAS (QYZDB-SSW-SMC003), International Cooperation Fund of CAS, Shanghai Science and Technology Commission (17ZR1449600, 17ZR1449800), Shanghai Yangfan Project (16YF1413400, 18YF1427600) and Rising-Star Program (15QA1404300), National Postdoctoral Program for Innovative Talents (2016M600337, 2017M611634, 2017M621552, 2016LH0042), China Postdoctoral Innovative Talent Support Program (BX201700267), China Young Talents Lift Engineering (YESS20160050, 2017QNRC001), The Pearl River Talent recruitment Program of Guangdong Province, Astrazeneca, Boehringer Ingelheim, Sanofi-SIBS Fellowship, British Heart Foundation Fellowship (FS/13/4/30045) and Royal Society-Newton Advanced Fellowship (NA170109).
Nonstandard Abbreviations and Acronyms
- CoEC-Cre
Coronary endothelia cell specific Cre
- BEC-Cre
Brain endothelia cell specific Cre
- BBB
Blood-brain barrier
- DTR
Diphtheria toxin receptor
- DT
Diphtheria toxin
- HB-EGF
Heparin-binding EGF-like growth factor
- ESR
Estrogen receptor
- Glut1
Glucose transporter 1
- PLVAP
Plasmalemma vesicle-associated protein
Footnotes
Disclosures
None.
In April 2018, the average time from submission to first decision for all original research papers submitted to Circulation Research was 13.36 days.
References
- 1.Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 3.Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, Sato TN, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding BS, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, Worgall S, Shido K, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding BS, Schachterle W, Liu Y, Rosenwaks Z, Butler JM, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013;19:1584–1596. doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreone BJ, Chow BW, Tata A, Lacoste B, Ben-Zvi A, Bullock K, Deik AA, Ginty DD, Clish CB, Gu C. Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron. 2017;94:581–594.e5. doi: 10.1016/j.neuron.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 10.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science. 2017;357 doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 11.Forde A, Constien R, Grone HJ, Hammerling G, Arnold B. Temporal Cre-mediated recombination exclusively in endothelial cells using Tie2 regulatory elements. Genesis. 2002;33:191–197. doi: 10.1002/gene.10117. [DOI] [PubMed] [Google Scholar]
- 12.Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 14.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Tian X, Pu WT, Zhou B. Cellular Origin and Developmental Program of Coronary Angiogenesis. Circ Res. 2015;116:515–530. doi: 10.1161/CIRCRESAHA.116.305097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lao Z, Raju GP, Bai CB, Joyner AL. MASTR: a technique for mosaic mutant analysis with spatial and temporal control of recombination using conditional floxed alleles in mice. Cell Rep. 2012;2:386–396. doi: 10.1016/j.celrep.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann M, Stillhard P, Wildner H, Seruggia D, Kapp V, Sanchez-Iranzo H, Mercader N, Montoliu L, Zeilhofer HU, Pelczar P. Binary recombinase systems for high-resolution conditional mutagenesis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Pu W, Tian X, Huang X, He L, Liu Q, Li Y, Zhang L, He L, Liu K, Gillich A, et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet. 2016;48:537–543. doi: 10.1038/ng.3536. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, Zhou B. Endocardium Minimally Contributes to Coronary Endothelium in the Embryonic Ventricular Free Walls. Circ Res. 2016;118:1880–1893. doi: 10.1161/CIRCRESAHA.116.308749. [DOI] [PubMed] [Google Scholar]
- 20.He L, Li Y, Li Y, Pu W, Huang X, Tian X, Wang Y, Zhang H, Liu Q, Zhang L, Zhao H, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 22.Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 23.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Pu W, Liu Q, He L, Huang X, Tian X, Zhang L, Nie Y, Hu S, Lui KO, Zhou B. Endocardium Contributes to Cardiac Fat. Circ Res. 2016;118:254–265. doi: 10.1161/CIRCRESAHA.115.307202. [DOI] [PubMed] [Google Scholar]
- 25.Haigh JJ, Morelli PI, Gerhardt H, Haigh K, Tsien J, Damert A, Miquerol L, Muhlner U, Klein R, Ferrara N, Wagner EF, et al. Cortical and retinal defects caused by dosage-dependent reductions in VEGF-A paracrine signaling. Dev Biol. 2003;262:225–241. doi: 10.1016/s0012-1606(03)00356-7. [DOI] [PubMed] [Google Scholar]
- 26.Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 27.Pu W, Zhang H, Huang X, Tian X, He L, Wang Y, Zhang L, Liu Q, Li Y, Li Y, Zhao H, et al. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat Commun. 2016;7:13369. doi: 10.1038/ncomms13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Hu T, He L, Huang X, Tian X, Zhang H, He L, Pu W, Zhang L, Sun H, Fang J, et al. Genetic targeting of sprouting angiogenesis using Apln-CreER. Nat Commun. 2015;6:6020. doi: 10.1038/ncomms7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J, Zhang H, He L, Huang X, Li Y, Pu W, Yu W, Zhang L, Cai D, Lui KO, Zhou B. Genetic Fate Mapping Defines the Vascular Potential of Endocardial Cells in the Adult Heart. Circ Res. 2018;122:984–993. doi: 10.1161/CIRCRESAHA.117.312354. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, Gu C. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauer B, McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 33.Rudat C, Kispert A. Wt1 and Epicardial Fate Mapping. Circ Res. 2012;111:165–169. doi: 10.1161/CIRCRESAHA.112.273946. [DOI] [PubMed] [Google Scholar]
- 34.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 35.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 36.Duim SN, Kurakula K, Goumans MJ, Kruithof BP. Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Zhou B, Pu WT. Genetic Cre-loxP Assessment of Epicardial Cell Fate Using Wt1-Driven Cre Alleles. Circ Res. 2012;111:e276–e280. doi: 10.1161/CIRCRESAHA.112.275784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Zhang Z, Zhong TP, et al. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013;23:1075–1090. doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 40.Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 41.Yamaizumi M, Mekada E, Uchida T, Okada Y. One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 42.Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, Pamer EG, et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran KA, Zhang X, Predescu D, Huang X, Machado RF, Göthert JR, Malik AB, Valyi-Nagy T, Zhao YY. Endothelial β-Catenin Signaling Is Required for Maintaining Adult Blood-Brain Barrier Integrity and Central Nervous System Homeostasis. Circulation. 2016;133:177–186. doi: 10.1161/CIRCULATIONAHA.115.015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, Katsura Y. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- 45.Montel-Hagen A, Blanc L, Boyer-Clavel M, Jacquet C, Vidal M, Sitbon M, Taylor N. The Glut1 and Glut4 glucose transporters are differentially expressed during perinatal and postnatal erythropoiesis. Blood. 2008;112:4729–4738. doi: 10.1182/blood-2008-05-159269. [DOI] [PubMed] [Google Scholar]
- 46.Stan RV, Kubitza M, Palade GE. PV-1 is a component of the fenestral and stomatal diaphragms in fenestrated endothelia. Proc Natl Acad Sci U S A. 1999;96:13203–13207. doi: 10.1073/pnas.96.23.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, McMahon AP. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322:1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, Wang Y, Tischfield M, Williams J, Smallwood PM, Rattner A, Taketo MM, Nathans J. Canonical WNT signaling components in vascular development and barrier formation. J Clin Invest. 2014;124:3825–3846. doi: 10.1172/JCI76431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 50.Karimova M, Splith V, Karpinski J, Pisabarro MT, Buchholz F. Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering. Sci Rep. 2016;6:30130. doi: 10.1038/srep30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, Kirchhoff F. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One. 2009;4:e4286. doi: 10.1371/journal.pone.0004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.