Abstract
Chemical use in society is growing rapidly and is one of the five major pressures on biodiversity worldwide. Since empirical toxicity studies of pollutants generally focus on a handful of model organisms, reliable approaches are needed to assess sensitivity to chemicals across the wide variety of species in the environment. Phylogenetic comparative methods (PCM) offer a promising approach for toxicity extrapolation incorporating known evolutionary relationships among species. If phylogenetic signal in toxicity data is high – i.e., closely related species are more similarly sensitive as compared to distantly related species – PCM could ultimately help predict species sensitivity when toxicity data are lacking. Here, we present the largest ever test of phylogenetic signal in toxicity data by combining phylogenetic data from fish with acute mortality data for 42 chemicals spanning 10 different chemical classes. Phylogenetic signal is high for some chemicals, particularly organophosphate pesticides, but not necessarily for many chemicals in other classes (e.g., metals, organochlorines). These results demonstrate that PCM may be useful for toxicity extrapolation in untested species for those chemicals with clear phylogenetic signal. This study provides a framework for using PCM to understand the patterns and causes of variation in species sensitivity to pollutants.
Keywords: Ecotoxicology, Ecological Risk Assessment, Fish, Phylogenetic Comparative Methods, Phylogeny, Organophosphate, Organochlorine, Evolutionary Toxicology
INTRODUCTION
The number of chemicals manufactured and released into the environment has grown rapidly, with an estimated 85,000 and 100,000 chemicals registered commercially in the United States (U.S. EPA, 2017a) and Europe (Schwarzenbach et al., 2006), respectively. Moreover, global chemical production is projected to quadruple by 2050 from 2000 levels (Wilson and Schwarzman, 2009). Because of their pervasive use in society, many types of chemicals, including pesticides, industrial chemicals, pharmaceuticals, and household cleaning products can be found across all environmental media (Pool and Rusch, 2014), leading to widespread exposure of many species and ecosystems. Chemical pollutants have therefore been indicated as one of the five major pressures on biodiversity worldwide (Secretariat of the Convention on Biological Diversity, 2010), whose loss threatens the health of ecosystems and the services they provide (Hooper et al., 2012; Köhler and Triebskorn, 2013; Millennium Ecosystem Assessment, 2005; Vörösmarty et al., 2010). Surprisingly, only a few studies have investigated chemical impacts on biodiversity, and have found that pollutants can lead to dramatic regional species losses (Beketov et al., 2013) and even pose continental-scale risks to species diversity (Malaj et al., 2014).
On of the challenges to understanding the range of chemical impacts across species and ecosystems is the lack of empirical toxicity data for most chemicals and species (Strempel et al., 2012). Bold initiatives such as the European Union’s (EU) Registration, Evaluation, Authorization, and Restriction of Chemicals (REACH) legislation (European Commission, 2006), which requires that chemical manufacturers and importers provide basic hazard data for all high-volume chemicals, are helping to reduce this knowledge gap. Yet, regulatory toxicity tests focus on a small number of model organisms to represent potential chemical impacts on the wide range of species in the environment (Backhaus et al., 2012; OECD, 2017; U.S. EPA, 2017b). Reliance on these surrogate species can be attributed, in part, to the significant resources required for toxicity testing and the small number of laboratory test species available. However, a key yet unanswered question is to what extent model organisms’ responses to toxic chemicals are representative of the highly diverse range of species found in nature. To address this major information gap, we need tools that allow us to understand and ultimately predict variation in sensitivity across a wide range of species.
Phylogenetic comparative methods (PCM) (Felsenstein, 1985; Harvey and Pagel, 1991) provide a promising framework for examining cross-species patterns in chemical sensitivity using fundamental principles from evolutionary biology. Central to PCM is the assumption that species exhibit similarity in phenotypic characteristics in direct proportion to their degree of phylogenetic relatedness (i.e., their shared evolutionary history) (Felsenstein, 1985; Harvey and Pagel, 1991). “Phylogenetic signal” is the degree to which trait variation among species corresponds to their common evolution, and it is detected in many types of traits (Freckleton et al., 2002). Most importantly, phylogenetic signal is particularly strong for body size and physiological traits, the latter of which are integral to how organisms respond to chemical exposures. Thus, we expect substantial phylogenetic signal in species sensitivity to pollutants. Should this be the case, PCM could ultimately help predict species sensitivity using known phylogenetic relationships between tested and untested species.
A major issue when working with cross-species data is that phylogenetic pseudoreplication may be caused by the presence of phylogenetic signal in the data (i.e., the degree of similarity in trait values between species due to their common ancestry). As a result, cross-species data cannot be treated as independent, violating a fundamental statistical assumption (Felsenstein, 1985; Harvey and Pagel, 1991; Freckleton et al 2002, Pagel 1999). PCM accounts for phylogenetic pseudoreplication by explicitly incorporating phylogeny of the studied species into statistical models (Felsenstein, 1985; Harvey and Pagel, 1991). Thus, PCM can help explain both patterns of species phenotypic values given their phylogenetic relationships and incorporate phylogenetic relationships into statistical models when investigating the effect of different explanatory factors (e.g., ecology, life history, morphology) on these same phenotypic values (Felsenstein, 1985; Harvey and Pagel, 1991, Freckleton et al 2002, Pagel 1999). For example, Chiari et al. (2015) show that there is strong phylogenetic signal in species sensitivity to copper sulfate in amphibians, but also that temperature explains more of the variation in acute mortality once phylogeny has been accounted for using PCM.
Crucially, PCM provides an important theoretical framework for understanding the biological basis for variation in species sensitivity to chemicals. It is now well known that adverse effects from chemicals are the result of many interactions across multiple levels of biological organization, from molecular initiating events all the way up to apical outcomes (e.g., survival, growth, reproduction), and even population effects (Ankley et al., 2010). To some degree, variation in adverse outcomes among species is due to structural differences in target receptor proteins that define a chemical’s mechanism of action (LaLone et al., 2016). Like most protein sequences, these sequences are generally conserved among closely related species. Many other factors can influence sensitivity in a given species, including general morpho-physiological characteristics, such as body size and metabolic rate, the rate of chemical uptake (Buchwalter and Luoma, 2005), the ability to biotransform or eliminate a given compound (Groh et al., 2015; van den Hurk et al., 2017), behavioral responses to chemical exposure (Sandahl et al., 2005; Scholz et al., 2000), or life history strategies (Stark et al., 2004). Thus, variation in chemical sensitivity cannot be fully explained by examining effects at any single level of biological organization and many of the characteristics underpinning sensitivity are likely to exhibit some level of phylogenetic signal. PCM integrates all these levels by placing the phenotype (e.g., measures of chemical sensitivity) into a phylogenetic framework in which closely related species are expected to respond more similarly at all of these levels.
The few previous studies applying PCM or other phylogenetic approaches to toxicity data indicate that there is a strong correspondence between evolutionary relationships and species sensitivity (Buchwalter et al., 2008; Carew et al., 2011; Chiari et al., 2015; Guénard et al., 2011, 2014; Hammond et al., 2012; Poteat and Buchwalter, 2014). While these studies have revealed important patterns, they have primarily looked at either a single chemical or a small group of related chemicals: so drawing general conclusions for the many diverse chemicals currently released in the environment is difficult. Here, we greatly expand the scope of previous studies by examining acute toxicity of 42 different chemicals spanning multiple classes (e.g., pesticides, metals) across a wide range of fish species. Fish are the most highly tested organisms in regulatory toxicology (Knöbel et al., 2012) as they provide essential information on sensitivity to toxicants in two different environments (saltwater and freshwater), are important for human consumption, and represent organisms at higher trophic levels in aquatic ecosystem function. Therefore, fish are an ideal focal group for examining phylogenetic patterns in species sensitivity to chemicals and its utility for assessing ecological risk.
MATERIALS AND METHODS
Species sensitivity data for fish, measured as LC50 (median lethal concentration, 50%), were acquired from the United States Environmental Protection Agency’s (U.S. EPA) Web-based Interspecies Correlation Estimation (Web-ICE) Database Version 3.3 (Raimondo et al., 2015). The Web-ICE database represents a curated and highly standardized database of acute toxicity values. Experimental temperature data were also compiled from Web-ICE because it affects species sensitivity to a multitude of chemicals (Chiari et al., 2015). In order to maximize uniformity among the dataset, LC50 and experimental temperature values were only used if they were generated by studies conducted with juvenile fish over a 96-hour chemical exposure duration. Only chemicals with LC50 data for 10 or more unique fish species were used to ensure sufficient statistical power, which in comparative studies is the number of species. If more than one LC50 value was available for a given species-chemical pair, the geometric mean was calculated. If toxicity studies for that species were conducted at different experimental temperatures, the geometric mean of those temperatures was used. Mode of action (MOA) – the general physiological interaction through which a chemical causes toxicity in an organism – was assigned to each chemical using EPA’s MOAtox classification scheme (Barron et al., 2015).
Phylogenetic signal was estimated from log10-transformed LC50 data using the phylogenetic generalized least squares model (PGLS) implemented in the package caper version 0.5.2 (Orme, 2013) for the R computing environment (R Core Team, 2016). We used a published phylogenetic tree (Rabosky et al., 2013) generated from a matrix of 13 genes in all PGLS analyses. The degree of phylogenetic signal in the data is quantified through the parameter λ, which can vary between 0 and 1 (Freckleton et al., 2002; Pagel, 1997). A λ value of 0 indicates no phylogenetic signal (independent evolution) and species can be considered statistically independent. A value of 1 indicates very strong phylogenetic signal conforming to a Brownian motion model of evolution (i.e., the degree of similarity between species due to common ancestry is directly proportional to the time of shared evolution). Because λ can take any value between these two extremes, caper generates tests evaluating to what extent the estimated maximum likelihood λ value for the data differs from the extremes using a standard likelihood ratio test (Freckleton et al., 2002; Pagel, 1997). In a PGLS linear model with independent explanatory variables, λ is estimated on the model’s residuals (Freckleton et al., 2002; Pagel, 1999).
We estimated λ values for individual chemicals. For all chemicals, λ was first estimated for a PGLS model with LC50 as the dependent variable and experimental temperature as the independent variable. For chemicals in which there was no significant relationship between LC50 and temperature, λ was then estimated for LC50 alone. Visual inspection of the diagnostic plots generated in caper indicated that all the assumptions of linear models with predictor variables were met, and we did not identify any outliers, defined as any species whose residual was over three standard deviations from the mean of residuals. We then used a χ2 statistical test in R to evaluate the impact of sample size on detecting a λ significantly greater than 0 versus not significantly different from 0 for chemicals with 15 or more species versus less than 15 species.
We also tested for the potential confounding effect of species adaptation to salinity levels on LC50, as it is highly debated whether freshwater and saltwater fish species differ in their sensitivity to chemicals due to differences in underlying physiology or due to the physical properties of chemicals in freshwater versus saltwater (Wheeler et al., 2002). To this end, we coded species adaptation to salinity as a binary variable (freshwater/saltwater). We added salinity as a second predictor (with temperature, if significant) in PGLS models for the four chemicals (carbaryl, chlorpyrifos, copper, and zinc) with at least five freshwater and five saltwater species. The threshold for significance for predictors in all analyses was α=0.05.
RESULTS AND DISCUSSION
Our dataset includes 42 chemicals with LC50 data for 10 or more fish species in the phylogenetic tree (Table 1). Most of these chemicals are pesticides, predominantly insecticides, including 12 organophosphates (OP), 9 organochlorines, 5 carbamates, and 3 pyrethroids. Other chemicals such as metals or inorganics have a wide range of industrial or agricultural uses. Temperature is a significant predictor of LC50 for 12 of the 42 chemicals analyzed, but the effect of temperature on toxicity varies among chemicals within each class (Table 1). When significant, temperature is positively associated with LC50 (i.e., higher temperature corresponds to lower sensitivity) for 9 chemicals, but negatively associated for EPN, parathion and aminocarb (Table 1). Overall, regardless of whether we also include experimental temperature (depending on whether it is a significant predictor of LC50 or not), the phylogenetic signal is significantly greater than 0 for 10 of the 42 chemicals examined. Only for one chemical, parathion, are temperature and phylogenetic signal both significant. The λ values are intermediate to high (λ>0.5) for all of the OPs except fenitrothion (λ=0.223) and coumaphos (λ=0), and significantly greater than 0 in 7 out of the 12 OPs (p<0.05; Table 1). Conversely, the phylogenetic signal for organochlorines – the next most represented chemical class in the dataset – is weak, with only 3 of 9 chemicals with λ>0.5, and only one, Lindane, with a λ value significantly greater than 0 (λ=0.574). Of chemicals among other classes, only 2 show phylogenetic signal significantly higher than 0: bioethanomethrin (λ=0.942), a pyrethroid insecticide, and captan (λ=0.912), a phthalamide fungicide. There is no obvious pattern in λ values based on chemical MOA (Table 1). Salinity (t12=-4.0, df=12, p<0.05), but not temperature (t12=0.92, p=0.37), is a significant predictor of LC50 only for a single chemical, chlorpyrifos.
Table 1.
Phylogenetic signal in fish LC50 values by chemical class
| Chemical | Class | Mode of Action* | N | Temp t-Value |
Temp p-Value |
λ Value |
|---|---|---|---|---|---|---|
| Azinphos-methyl | Organophosphate | AChE | 18 | 0.79 | 0.44 | 0.995a |
| Chlorpyrifos | Organophosphate | AChE | 15 | 0.67 | 0.51 | 0.584b |
| Coumaphos | Organophosphate | AChE | 10 | 0.11 | 0.91 | 0.0b |
| Diazinon | Organophosphate | AChE | 13 | 0.43 | 0.67 | 1.0a |
| Dichlorvos | Organophosphate | AChE | 10 | 0.12 | 0.91 | 1.0a |
| EPN | Organophosphate | AChE | 12 | −2.3 | 0.043 | 0.981 |
| Fenitrothion | Organophosphate | AChE | 14 | 1.4 | 0.18 | 0.223b |
| Malathion | Organophosphate | AChE | 25 | 1.7 | 0.095 | 0.979a |
| Methylparathion | Organophosphate | AChE | 14 | 1.7 | 0.12 | 0.723a |
| Parathion | Organophosphate | AChE | 11 | −2.6 | 0.027 | 1.0a |
| Phorate | Organophosphate | AChE | 10 | −1.1 | 0.30 | 0.845 |
| Trichlorfon | Organophosphate | AChE | 15 | 0.37 | 0.72 | 0.891a,b |
| Aminocarb | Carbamate | AChE | 11 | −2.4 | 0.039 | 0.97 |
| Carbaryl | Carbamate | AChE | 33 | 2.6 | 0.013 | 0.0b |
| Carbofuran | Carbamate | AChE | 10 | 0.51 | 0.62 | 1.0 |
| Methomyl | Carbamate | AChE | 11 | −2.0 | 0.075 | 0.0b |
| Mexacarbate | Carbamate | AChE | 17 | 0.76 | 0.46 | 0.0b |
| Chlordane | Organochlorine | Neurotoxicity | 14 | 7.7 | <0.001 | 0.0 |
| DDT | Organochlorine | Neurotoxicity | 25 | 0.97 | 0.34 | 0.246b |
| Endosulfan | Organochlorine | Neurotoxicity | 14 | −0.33 | 0.74 | 0.0 |
| Endrin | Organochlorine | Neurotoxicity | 20 | 1.7 | 0.11 | 0.0b |
| Heptochlor | Organochlorine | Neurotoxicity | 10 | 0.16 | 0.88 | 1.0 |
| Lindane | Organochlorine | Neurotoxicity | 15 | 1.6 | 0.16 | 0.574a |
| Methoxychlor | Organochlorine | Neurotoxicity | 18 | 3.3 | 0.004 | 0.0b |
| Pentochlorophenol | Organochlorine | Neurotoxicity | 20 | −0.82 | 0.42 | 0.833b |
| Toxaphene | Organochlorine | Neurotoxicity | 15 | 1.9 | 0.08 | 0.0b |
| Cadmium | Metal | MIOI | 13 | 2.9 | 0.014 | 0.0b |
| Chromium (VI) | Metal | MIOI | 15 | 1.1 | 0.28 | 0.0b |
| Copper | Metal | MIOI | 46 | 2.6 | 0.012 | 0.601b |
| Zinc | Metal | MIOI | 28 | 2.9 | 0.001 | 0.0b |
| Bioethanomethrin | Pyrethroid | Neurotoxicity | 11 | −1.2 | 0.27 | 0.942a |
| Permethrin | Pyrethroid | Neurotoxicity | 16 | 1.4 | 0.18 | 0.0b |
| S-Bioallethrin | Pyrethroid | Neurotoxicity | 10 | 3.0 | 0.017 | 0.817 |
| 3-Chloro-3-nitrosalicylanilide | Nitroaromatic | Non-polar Narcosis | 10 | 0.86 | 0.41 | 0.0b |
| Clonitralid | Nitroaromatic | ETI | 10 | −0.30 | 0.77 | 0.0 |
| Dinitramine | Nitroaromatic | ETI | 10 | –c | –c | 0.322b |
| Ammonia | Inorganic | ETI | 26 | 1.1 | 0.28 | 0.0 |
| Potassium Permanganate | Inorganic | Reactivity | 11 | 0.20 | 0.85 | 0.0 |
| 4-Nonylphenol | Phenol | Polar Narcosis | 10 | 1.1 | 0.31 | 0.0b |
| Phenol | Phenol | Polar Narcosis | 10 | 2.8 | 0.022 | 0.69 |
| Antimycin A | Antibiotic | ETI | 14 | 2.9 | 0.013 | 0.514b |
| Captan | Phthalamide | Non-polar Narcosis | 10 | −0.16 | 0.87 | 0.912a |
Mode of action classified according to Barron et al. (2015). AChE: acetylcholinesterase inhibition; Neurotoxicity: MIOI: Metallic iono/osmoregulatory impairment; ETI: electron transport inhibition.
λ significantly different from 0
λ significantly different from 1
All species tested at 12°C
As for any statistical model, the power to estimate λ and differentiate between the estimated value and its extremes, 0 or 1, decreases with small sample sizes (Freckleton et al., 2002). However, using a χ2 test, we find no significant difference in the proportion of chemicals with λ values significantly greater than 0 (χ2 value=1.08; p=0.11) between chemicals with less than 15 species versus those with 15 or more species. Still, since OPs and carbamates possess the same MOA (cholinesterase inhibition), it is still possible that lack of consistency in phylogenetic signal between these classes is due to sample size.
Visual display of the LC50 values for OPs shows that sensitivity levels cluster by fish taxonomic family for most chemicals (Figure 1a), which explains the high level of phylogenetic signal found for this chemical class. Conversely, toxicity values do not cluster at the family level for organochlorines (Figure 1b). Inspection of LC50 data overlaid onto the fish phylogenetic tree (Figure 2) shows that individual fish clades exhibit consistent levels of sensitivity across several OPs. Perciform (e.g., Lepomis macrochirus) and salmoniform (e.g., Oncorhynchus mykiss) fish species are typically the most sensitive (lowest LC50 values), while cyprinodontiform fish (e.g., Poecilia reticulata) are moderately sensitive, and cypriniform (e.g., Cyprinus carpio) and siluriform (e.g., Ictalurus punctatus) fish were generally the least sensitive.
Figure 1.
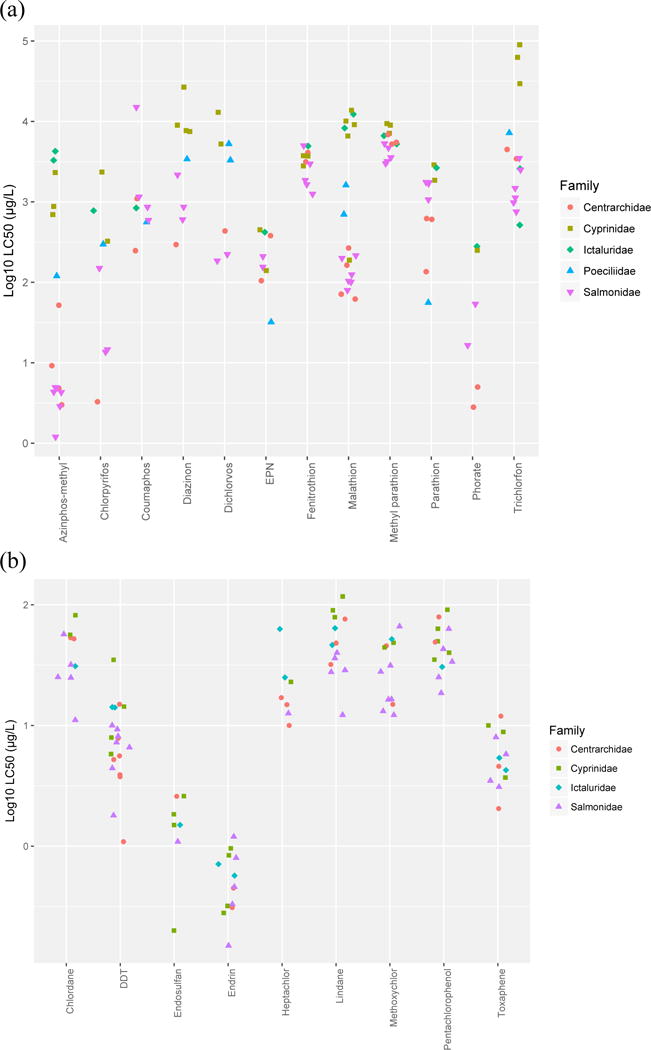
Fish sensitivity to (a) organophosphate insecticides and (b) organochlorine compounds organized by taxonomic family. Only families with 10 or more species in the dataset are represented.
Figure 2.
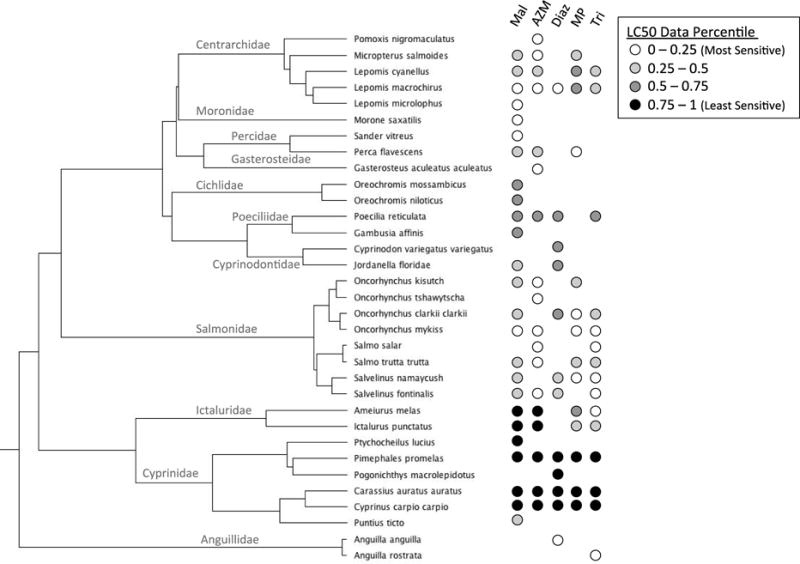
Phylogeny of fish with toxicity data overlaid from five organophosphates (malathion (Mal), azinphos-methyl (AZM), diazinon (Diaz), methyl parathion (MP), and trichlorfon (Tri)) with strong phylogenetic signal (i.e., λ > 0.5 and significantly different from 0). Toxicity data percentiles represent scaling of the LC50 for each species-chemical pair as a proportion between the lowest and highest log-LC50 value across all species for a given chemical. Fish family names are included in the tree next to corresponding taxon branches.
These results have important implications for developing reliable predictions of toxicity for species that cannot be easily tested, as well as better understanding the biological basis of toxicity variation across species and chemical classes. Our study uses the largest toxicity dataset to date to test whether sensitivity differences among species can be explained by their evolutionary relationships. Surprisingly, we find that this holds true for some, but not necessarily all, classes of chemicals. In contrast, most earlier studies applying PCM to chemical response data – although few in number – report an association with phylogenic relationships (Buchwalter et al., 2008; Carew et al., 2011; Chiari et al., 2015; Hammond et al., 2012). Our finding of mixed phylogenetic signal among chemicals suggests there are potential data limitations or biological factors that could influence the relationship between phylogeny and species sensitivity.
Small species sample sizes, heterogeneous chemical representation, or laboratory experimental effects are all potential data limitations that could explain the lack of consistent phylogenetic signal across chemicals. Although our study is by far the largest to date to measure phylogenetic signal in toxicity data, there remain few chemicals with appreciable species data, most of which are pesticides. This is because the bulk of chemical testing in fish, for example, is still conducted on common model organisms such as rainbow trout (Oncorhynchus mykiss), fathead minnow (Pimephales promelas), and bluegill (Lepomis macrochirus) (OECD, 2017; U.S. EPA, 2017b). Moreover, there are historically stricter regulatory data requirements for pesticides versus industrial chemicals in the U.S. Although we do not find that λ values differ significantly for chemicals above or below a sample size threshold of 15 fish species, much larger species-chemical datasets would likely generate more robust λ estimates (Freckleton et al., 2002). Although toxicity data used in our study are consistent across exposure duration and life stage, other experimental variables such as exposure method (e.g., static versus flow-through), stability of chemical over the exposure duration, water quality, use of solvents, animal source and husbandry, and researcher technique could all produce variation in results. Future datasets with larger species sample sizes would help assess and limit the impact of such experimental factors.
There are also biological factors that could explain the mixed phylogenetic signal across chemicals and chemical classes, and could help guide future studies. Interspecific variation in proteins that are critical to a chemical’s mechanism of action can change the conformation of ligand-binding domains and influence binding affinities of various xenobiotic agents (LaLone et al., 2016). Future work could investigate protein variation among fish species for OPs, which inhibit acetylcholinesterase (Fukuto, 1990), versus organochlorines, which either inhibit sodium channels or GABA-gated chloride channels (Coats, 1990). It is possible that the protein variation in OP targets is more strongly associated with phylogeny and thus responsible for the more consistent phylogenetic signal observed in this group of chemicals. Species sensitivity variation can also arise from differences in chemical uptake and bioaccumulation (Buchwalter et al., 2008) or the ability to biotransform or eliminate a compound. The latter was recently demonstrated in several groups of fish with varying sensitivities corresponding to different induction rates of Cytochrome P450 detoxification enzymes (van den Hurk et al., 2017). Thus, follow-up studies could examine interspecific variation in xenobiotic molecular targets, and adsorption, distribution, metabolism, and elimination of chemicals with strong versus absent phylogenetic signal.
General morpho-physiological characteristics of species such as body size, metabolic rate, and energy use can also influence species responses to chemicals (Muller et al., 2010). Such factors may not only influence acute toxicity, as examined in our study, but also the physiological and resource challenges posed by sublethal or chronic chemical exposures. It is important to note that such broad characteristics or organisms, can – and usually do – exhibit strong phylogenetic signal themselves (Freckleton et al., 2002; Garland et al., 2005). Thus, application of PCM to toxicity data is not only important for understanding how species evolutionary history has shaped patterns in species sensitivity, but also to statistically account for the influence of phylogeny on other explanatory factors related to physiology or ecology.
Given the pervasive use of chemicals in society, which is expected to increase over the coming decades, it is imperative to develop approaches to understand and assess risk to the wide breadth of species in the environment using limited resources. Evolutionary biology provides a useful framework with which to form expectations about how diverse sets of species will respond to chemical exposure based on their degree of shared evolutionary history. Our study serves as an important step in grasping the power and usefulness of PCM for understanding the patterns and causes of variation in species sensitivity to pollutants. It suggests that the relationship between phylogeny and acute toxicity data is not consistent among chemicals or chemical classes. Future work should focus on disentangling the experimental versus biological factors that contribute to this pattern of mixed phylogenetic signal. Such further examination would benefit from increased representation of diverse fish species in toxicity tests beyond pesticides and the use of a wider array of fish species in toxicity tests outside of the major regulatory aquatic toxicity test organisms.
Acknowledgments
The authors would like to thank David Buchwalter for insightful comments on the manuscript and Sandy Raimondo and Crystal Lilavois of the U.S. EPA Gulf Ecology Division for access to the Web-ICE data used for this study. This paper has been reviewed according to EPA’s journal clearance guidelines, but do not necessarily reflect the views or policies of the Agency.
References
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29:730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Backhaus T, Snape J, Lazorchak J. The impact of chemical pollution on biodiversity and ecosystem services: the need for an improved understanding. Integr Environ Assess Manag. 2012;8:575–576. doi: 10.1002/ieam.1353. [DOI] [PubMed] [Google Scholar]
- Barron MG, Lilavois CR, Martin TM. MOAtox: A comprehensive mode of action and acute aquatic toxicity database for predictive model development. Aquat Toxicol. 2015;161:102–107. doi: 10.1016/j.aquatox.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Beketov MA, Kefford BJ, Schäfer RB, Liess M. Pesticides reduce regional biodiversity of stream invertebrates. Proc Natl Acad Sci. 2013;110:11039–11043. doi: 10.1073/pnas.1305618110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter DB, Luoma SN. Differences in Dissolved Cadmium and Zinc Uptake among Stream Insects: Mechanistic Explanations. Environ Sci Technol. 2005;39:498–504. doi: 10.1021/es0404421. [DOI] [PubMed] [Google Scholar]
- Buchwalter DB, Cain DJ, Martin CA, Xie L, Luoma SN, Garland T. Aquatic insect ecophysiological traits reveal phylogenetically based differences in dissolved cadmium susceptibility. Proc Natl Acad Sci. 2008;105:8321–8326. doi: 10.1073/pnas.0801686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew ME, Miller AD, Hoffmann AA. Phylogenetic signals and ecotoxicological responses: potential implications for aquatic biomonitoring. Ecotoxicology. 2011;20:595–606. doi: 10.1007/s10646-011-0615-3. [DOI] [PubMed] [Google Scholar]
- Chiari Y, Glaberman S, Serén N, Carretero MA, Capellini I. Phylogenetic signal in amphibian sensitivity to copper sulfate relative to experimental temperature. Ecol Appl. 2015;25:596–602. doi: 10.1890/14-0439.1. [DOI] [PubMed] [Google Scholar]
- Coats JR. Mechanisms of toxic action and structure-activity relationships for organochlorine and synthetic pyrethroid insecticides. Environ Health Perspect. 1990;87:255–262. doi: 10.1289/ehp.9087255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission. Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation. Authorisation and Restriction of Chemicals (REACH); 2006. [Google Scholar]
- Felsenstein J. Phylogenies and the Comparative Method. Am Nat. 1985;125:1–15. [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic Analysis and Comparative Data: A Test and Review of Evidence. Am Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Fukuto TR. Mechanism of Action of Organophosphorus and Carbamate Insecticides. Environ Health Perspect. 1990;87:245–254. doi: 10.1289/ehp.9087245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Bennett AF, Rezende EL. Phylogenetic approaches in comparative physiology. J Exp Biol. 2005;208:3015–3035. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- Groh KJ, Carvalho RN, Chipman JK, Denslow ND, Halder M, Murphy CA, Roelofs D, Rolaki A, Schirmer K, Watanabe KH. Development and application of the adverse outcome pathway framework for understanding and predicting chronic toxicity: I. Challenges and research needs in ecotoxicology. Chemosphere. 2015;120:764–777. doi: 10.1016/j.chemosphere.2014.09.068. [DOI] [PubMed] [Google Scholar]
- Guénard G, von der Ohe PC, de Zwart D, Legendre P, Lek S. Using phylogenetic information to predict species tolerances to toxic chemicals. Ecol Appl. 2011;21:3178–3190. [Google Scholar]
- Guénard G, von der Ohe PC, Walker SC, Lek S, Legendre P. Using phylogenetic information and chemical properties to predict species tolerances to pesticides. Proc R Soc Lond B Biol Sci. 2014;281:20133239. doi: 10.1098/rspb.2013.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JI, Jones DK, Stephens PR, Relyea RA. Phylogeny meets ecotoxicology: evolutionary patterns of sensitivity to a common insecticide. Evol Appl. 2012;5:593–606. doi: 10.1111/j.1752-4571.2011.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford; New York: Oxford University Press; 1991. [Google Scholar]
- Hooper DU, Adair EC, Cardinale BJ, Byrnes JEK, Hungate BA, Matulich KL, Gonzalez A, Duffy JE, Gamfeldt L, O’Connor MI. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature. 2012;486:105–108. doi: 10.1038/nature11118. [DOI] [PubMed] [Google Scholar]
- van den Hurk P, Gerzel LE, Calomiris P, Haney DC. Phylogenetic signals in detoxification pathways in Cyprinid and Centrarchid species in relation to sensitivity to environmental pollutants. Aquat Toxicol. 2017;188:20–25. doi: 10.1016/j.aquatox.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Knöbel M, Busser FJM, Rico-Rico Á, Kramer NI, Hermens JLM, Hafner C, Tanneberger K, Schirmer K, Scholz S. Predicting Adult Fish Acute Lethality with the Zebrafish Embryo: Relevance of Test Duration, Endpoints, Compound Properties, and Exposure Concentration Analysis. Environ Sci Technol. 2012;46:9690–9700. doi: 10.1021/es301729q. [DOI] [PubMed] [Google Scholar]
- Köhler HR, Triebskorn R. Wildlife Ecotoxicology of Pesticides: Can We Track Effects to the Population Level and Beyond? Science. 2013;341:759–765. doi: 10.1126/science.1237591. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Lyons D, Helgen HW, Robinson SL, Swintek JA, Saari TW, Ankley GT. Editor’s Highlight: Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A Web-Based Tool for Addressing the Challenges of Cross-Species Extrapolation of Chemical Toxicity. Toxicol Sci. 2016;153:228–245. doi: 10.1093/toxsci/kfw119. [DOI] [PubMed] [Google Scholar]
- Malaj E, von der Ohe PC, Grote M, Kühne R, Mondy CP, Usseglio-Polatera P, Brack W, Schäfer RB. Organic chemicals jeopardize the health of freshwater ecosystems on the continental scale. Proc Natl Acad Sci. 2014;111:9549–9554. doi: 10.1073/pnas.1321082111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millennium Ecosystem Assessment. Ecosystems and human well-being: biodiversity synthesis. Washington, DC: World Resources Institute; 2005. [Google Scholar]
- Muller EB, Nisbet RM, Berkley HA. Sublethal toxicant effects with dynamic energy budget theory: model formulation. Ecotoxicology. 2010;19:48. doi: 10.1007/s10646-009-0385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD. OECD Guidelines for the Testing of Chemicals, Section 2: Effects on Biotic Systems 2017 [Google Scholar]
- Orme D. The caper package: comparative analysis of phylogenetics and evolution in R 2013 [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zool Scr. 1997;26:331–348. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pool R, Rusch E. Identifying and reducing environmental health risks of chemicals in our society: workshop summary. Washington, DC: The National Academies Press; 2014. [PubMed] [Google Scholar]
- Poteat MD, Buchwalter DB. Calcium uptake in aquatic insects: influences of phylogeny and metals (Cd and Zn) J Exp Biol. 2014;217:1180–1186. doi: 10.1242/jeb.097261. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, Chang J, Alfaro ME. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat Commun. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- Raimondo S, Lilavois CR, Barron MG. Web-based Interspecies Correlation Estimation (Web-ICE) for Acute Toxicity: User Manual. 2015 Version 3.3, EPA/600/R-15/192. [Google Scholar]
- Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. Comparative thresholds for acetylcholinesterase inhibition and behavioral impairment in coho salmon exposed to chlorpyrifos. Environ Toxicol Chem. 2005;24:136–145. doi: 10.1897/04-195r.1. [DOI] [PubMed] [Google Scholar]
- Scholz NL, Truelove NK, French BL, Berejikian BA, Quinn TP, Casillas E, Collier TK. Diazinon disrupts antipredator and homing behaviors in chinook salmon (Oncorhynchus tshawytscha) Can J Fish Aquat Sci. 2000;57:1911–1918. [Google Scholar]
- Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, von Gunten U, Wehrli B. The Challenge of Micropollutants in Aquatic Systems. Science. 2006;313:1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Secretariat of the Convention on Biological Diversity. Global Biodiversity Outlook 3 (Montréal) 2010 [Google Scholar]
- Stark JD, Banks JE, Vargas R. How risky is risk assessment: The role that life history strategies play in susceptibility of species to stress. Proc Natl Acad Sci U S A. 2004;101:732–736. doi: 10.1073/pnas.0304903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strempel S, Scheringer M, Ng CA, Hungerbühler K. Screening for PBT Chemicals among the “Existing” and “New” Chemicals of the EU. Environ Sci Technol. 2012;46:5680–5687. doi: 10.1021/es3002713. [DOI] [PubMed] [Google Scholar]
- U.S. EPA. TSCA Chemical Substance Inventory 2017a [Google Scholar]
- U.S. EPA. Series 850 - Ecological Effects Test Guidelines 2017b [Google Scholar]
- Vörösmarty CJ, McIntyre PB, Gessner MO, Dudgeon D, Prusevich A, Green P, Glidden S, Bunn SE, Sullivan CA, Liermann CR, et al. Global threats to human water security and river biodiversity. Nature. 2010;467:555–561. doi: 10.1038/nature09440. [DOI] [PubMed] [Google Scholar]
- Wheeler JR, Leung KMY, Morritt D, Sorokin N, Rogers H, Toy R, Holt M, Whitehouse P, Crane M. Freshwater to saltwater toxicity extrapolation using species sensitivity distributions. Environ Toxicol Chem. 2002;21:2459–2467. [PubMed] [Google Scholar]
- Wilson MP, Schwarzman MR. Toward a New U.S. Chemicals Policy: Rebuilding the Foundation to Advance New Science, Green Chemistry, and Environmental Health. Environ Health Perspect. 2009;117:1202–1209. doi: 10.1289/ehp.0800404. [DOI] [PMC free article] [PubMed] [Google Scholar]


