Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is associated with a wide range of clinical presentations, from asymptomatic or mildly ill to severe respiratory illness including death. We describe isolation of infectious MERS-CoV from the upper respiratory tract of a mildly ill 27-year-old female in Saudi Arabia 15 days after illness onset.
Keywords: asymptomatic, MERS, mild, prolonged detection, virus isolation
Since its emergence in 2012, Middle East respiratory syndrome coronavirus (MERS-CoV) transmission has been associated with both direct exposure to dromedary camels [1] and with exposure to markedly symptomatic MERS patients, usually in households or health care settings [2–4]. Other potential sources of infection are less clear, but unrecognized transmission associated with infected individuals who are mildly ill or asymptomatic has been suspected [5, 6]. Although MERS-CoV RNA has been detected for prolonged periods from respiratory specimens of confirmed patients who were mildly ill or asymptomatic [7–9], isolation of live MERS-CoV has not been previously documented among this group.
During October 2015, the Saudi Arabia Ministry of Health (MoH) reported a cluster of MERS-CoV infections identified in female janitors working at a university in Riyadh [10, 11]. We summarize below the clinical course and concomitant laboratory investigation of a case identified early in this cluster.
INITIAL MERS DIAGNOSIS
A 27-year-old expatriate female was tested for MERS-CoV on October 10, 2015, following diagnosis of her roommate with MERS the previous day. She was found to be positive by MERS-CoV-specific real-time reverse transcriptase polymerase chain reaction (rRT-PCR) of a nasopharyngeal (NP) swab and was subsequently admitted to a MERS referral hospital in Riyadh on October 10 for isolation and monitoring. Based on public health investigation at that time, she reported the onset of mild upper respiratory symptoms on September 30, 2015, 10 days before detection and hospitalization (Figure 1); the following findings are presented according to this date, as days post–illness onset.
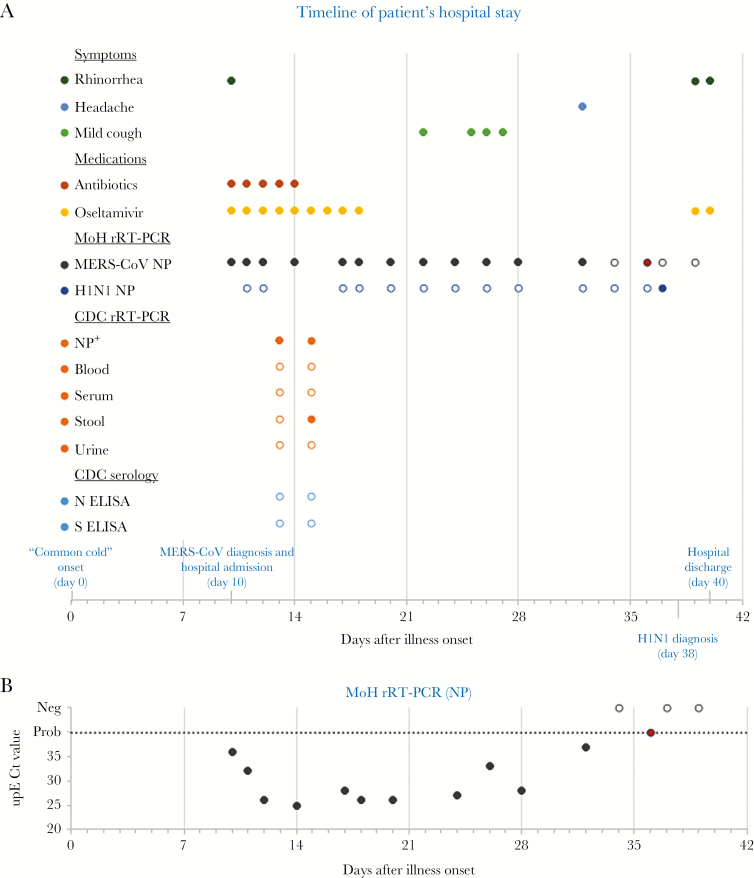
Figure 1.
Clinical, laboratory, and hospital findings. A, Timeline of events during the patient’s hospital stay, by days after illness onset. Patient-reported illness onset (represented as day 0) was on September 30, 2015. Hospital admission and discharge were on days 10 and 40 after illness onset, respectively. Reported symptoms, treatments, and laboratory tests are depicted. The patient’s medical chart was systematically reviewed for administration of medications and for mention of the following symptoms on each day of hospitalization: fever, cough (with description if available), headache, sore throat, dyspnea, chest pain, muscle ache, chills, abdominal pain, vomiting, diarrhea, and other; rhinorrhea was reported under other symptoms. A solid circle means that the medication was administered or the symptom was reported in the chart. Symptoms not depicted were not reported throughout hospitalization. Temperature was also recorded from regular vital sign measurements. We defined fever as a measured oral temperature of >38.0°C or a measured axillary temperature of >37.5°C; she remained afebrile throughout. For test results, a solid circle represents a positive result and an open circle represents a negative result; the red circle at day 36 of the Saudi Arabia Ministry of Health (MoH) real-time reverse transcriptase polymerase chain reaction (rRT-PCR) results represents a probable positive result (only 1 of 2 rRT-PCR assays was positive; cycle threshold (Ct) values not available). Nasopharyngeal (NP+): live virus was also isolated from the 2 NP specimens submitted to the Centers for Disease Control and Prevention (CDC). B, Middle East respiratory syndrome coronavirus (MERS-CoV) upE rRT-PCR Ct values obtained from MoH diagnostic testing by days after illness onset. The dashed line represents the limit of detection, above which specimens were considered MERS-CoV-negative (open circle). Probable positive (red circle) results were assigned a value of 40 for graphing purposes. A Ct value was not available for the specimen collected 22 days after illness. Abbreviations: ELISA, enzyme-linked immunosorbent assay; N, nucleocapisd; S, spike.
CLINICAL PRESENTATION AND HOSPITAL COURSE
According to a retrospective review of her medical chart, the patient had rhinorrhea at the time of admission and denied cough, shortness of breath, chest pain, gastrointestinal symptoms, or fever (Figure 1A). She reported no underlying medical conditions. White blood cell counts with differential and blood chemistry analyses were within normal limits throughout hospitalization (Supplementary Table 1). A chest x-ray was obtained 13 days after illness onset (on October 13) and was interpreted as unremarkable. She received oseltamivir, ceftriaxone, and azithromycin empirically beginning at admission. Following her hospital admission, no respiratory symptoms were reported until 22 days after illness onset (on October 22), when the patient developed a mild cough, which was noted for 6 days (Figure 1A); she remained on room air throughout. No hypoxia was identified during hospitalization, and her oral temperature peaked at 37.6°C. Just before discharge, she reported rhinorrhea and was started on oseltamivir following a new diagnosis of influenza. She was discharged 40 days after illness onset (on November 9).
LABORATORY INVESTIGATION
Clinical Testing in Respiratory Specimens
To assess MERS-CoV status, we reviewed hospital records for serial diagnostic testing of respiratory specimens, performed by MoH using rRT-PCR assays targeting MERS-CoV upE and ORF1a genes [12]. The same diagnostic specimens were also routinely tested for influenza A virus (H1N1), and these results were reviewed if available.
Based on hospital records, 16 NP swabs were collected for MERS-CoV diagnostic testing (Figure 1). Twelve NP swabs collected during 10–32 days after illness onset (between October 10 and November 1) were confirmed positive (Figure 1A). Cycle threshold (Ct) values of MERS-CoV upE rRT-PCR were available for 11 of the 12 positive specimens and ranged from 25 to 37 (Figure 1B). On day 10 after onset, the upE Ct value was 37, indicating a low viral load; by day 14, the Ct value was much lower, at 25, indicating a notable rise in viral load. An NP swab collected 36 days after illness onset was probable for MERS-CoV, meaning that 1 target was detected but the other was not; target-specific results were not available. The final 2 NP swabs collected 37 and 39 days after illness onset tested negative. The swab collected on day 37 tested positive for influenza A virus (H1N1) by rRT-PCR. Among the 14 NP swabs collected during days 10–37, 12 tested negative for H1N1, and results for the remaining 2 were not available (Figure 1A).
MERS-CoV Detection and Sequencing
For additional molecular and serologic analysis at the US Centers for Disease Control and Prevention (CDC), 2 (of each) NP, whole blood, sera, stool, and urine specimens were collected on days 13 and 15 after illness onset (on October 13 and 15); NP swabs were collected in 2 mL of viral transport medium. All specimens were frozen and shipped on dry ice. At the CDC, stool suspensions were prepared in phosphate-buffered saline (10% weight/volume). RNA was extracted from 200-µL sample aliquots on a NucliSens EasyMAG (BioMerieux), recovering 100 µL of total nucleic acid. Testing was performed by MERS-CoV upE and N2 and/or N3 rRT-PCR assays [13]. All specimen extracts were tested neat; stool extracts were also diluted 1:5 in nuclease-free water to remove potential stool inhibitors. Genome sequencing was attempted on positive specimens with Ct values ≤36, as described previously [14]. Nucleotide sequences were aligned using Clustal X, version 1.83, implemented in BioEdit, version 7.2.5. Phylogenetic analyses were performed using MrBayes 3.2.6 under a GTR model of nucleotide substitution with 4 categories of γ-distributed rate heterogeneity and a proportion of invariant sites (GTR + 4 + I).
MERS-CoV RNA was detected in the 2 NP swabs, and 1 stool specimen (at 1:5 dilution) was collected 15 days after illness onset. Estimated viral loads, based on the upE assay, were 4.8 × 106 and 9.6 × 105 genome copies/mL in the NP specimens collected on days 13 and 15, respectively, and 1.0 × 104 genome copies/gm of stool from day 15. Whole-genome sequences were obtained from the 2 sequential NP swabs and were 100% identical (accession numbers: MG520075-MG520076). They showed 99.9% similarity and closest phylogenetic clustering with sequences from 4 severely ill MERS patients who were linked to a hospital outbreak in Riyadh in August 2015 (accession numbers: KU851860-851862 and KU851864) [15].
MERS-CoV Isolation
The 2 MERS-CoV RNA-positive NP specimens submitted to the CDC (collected on days 13 and 15) were serially diluted 10-fold in DMEM in a 96-well plate, and subsequently used to inoculate Vero cell suspensions. The cells were observed daily between days 3 and 7 postinoculation; cytopathic effect (CPE) was observed under inverted scope 3 days postinoculation. Any wells that exhibited CPE were harvested and passaged in a 24-well plate. RNA was extracted from 50 μL of the potential virus lysate. Both lysates were confirmed positive for MERS-CoV by N2 rRT-PCR assay; Ct values were 12 (day 13) and 15 (day 15). The isolated viruses (accession numbers: MG546330-MG546331) were subsequently sequenced using Fluidigm Access Array PCR and MiSeq amplicon sequencing [16]. Whole-genome sequences of the 2 isolated viruses were identical to their clinical specimens, except the day 13 isolated virus, which had mixed bases at positions 23 364 and 25 861 (nucleotide location based on accession number JX869059.2). The variant alleles of the isolate would predict amino acid changes (N>D in spike [S] protein and V>L in ORF4a) that may reflect adaptation in culture. Virus isolation was not attempted for the MERS-CoV RNA-positive stool specimen because of low virus load.
Antibody Responses
Sera collected at days 13 and 15 after onset were examined for the presence of MERS-CoV-specific antibodies against the nucleocapisd (N) and S proteins, as previously described [17]. Both specimens were below the limit of detection for both N and S enzyme-linked immunosorbent assays (ELISAs). No sera were available after day 15.
DISCUSSION
We describe isolation of live MERS-CoV from the upper respiratory tract of a mildly ill patient in Saudi Arabia. The ability of a mildly ill MERS patient to shed live virus has not previously been documented and fills an important gap in our understanding of MERS-CoV natural history.
The patient’s illness began with reports of upper respiratory tract symptoms, or “a common cold.” Rhinorrhea was noted 10 days later, when she was admitted to the hospital for isolation, after which no symptoms were reported for a further 12 days. A mild cough then began 22 days after illness onset and lasted for 6 days. Despite this mild cough, she was noted to be without hypoxia, dyspnea, chest pain, or other signs/symptoms of respiratory illness throughout hospitalization. MERS-CoV RNA was detected in her stool specimen (at day 15) and up to 32 days after illness onset in upper respiratory specimens; low virus load in the stool was coincident with the highest virus loads measured in the upper respiratory tract and may reflect shedding from that compartment. Most notably, live virus was isolated from the NP swabs collected 13 and 15 days after illness onset, when she was reported to be asymptomatic. N and S ELISA antibody responses were not detected by day 15 of illness. We were unable to test subsequent serum specimens.
The patient was admitted to the hospital for isolation following MERS-CoV detection. Home isolation may be considered for asymptomatic or mildly ill patients who test positive for MERS-CoV [18], but this case patient resided in shared accommodation, and home isolation was not practical. Although the role of mildly ill patients in transmission is not fully understood [19], the ability of these patients to shed infectious MERS-CoV, as demonstrated here, should inform home isolation considerations.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Jennifer Harcourt for gamma irradiation of serum and Shifaq Kamili for specimen coordination.
Financial support. This work was supported by the Ministry of Health in Saudi Arabia and the US Centers for Disease Control and Prevention.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Disclosure. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Ethical approval. The patient provided informed written consent. The study was reviewed and approved by the Saudi Arabia Ministry of Health Institutional Review Board before initiation.
References
- 1. Alraddadi BM, Watson JT, Almarashi A, et al. . Risk factors for primary Middle East respiratory syndrome coronavirus illness in humans, Saudi Arabia, 2014. Emerg Infect Dis 2016; 22:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Assiri A, McGeer A, Perl TM, et al. ; KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med 2013; 369:407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drosten C, Meyer B, Müller MA, et al. . Transmission of MERS-coronavirus in household contacts. N Engl J Med 2014; 371:828–35. [DOI] [PubMed] [Google Scholar]
- 4. Oboho IK, Tomczyk SM, Al-Asmari AM, et al. . 2014 MERS-CoV outbreak in Jeddah—a link to health care facilities. N Engl J Med 2015; 372:846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Omrani AS, Matin MA, Haddad Q, et al. . A family cluster of Middle East respiratory syndrome coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int J Infect Dis 2013; 17:e668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alfaraj SH, Al-Tawfiq JA, Altuwaijri TA, et al. . Middle East respiratory syndrome coronavirus transmission among health care workers: implication for infection control. Am J Infect Control 2018; 46:165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al Hosani FI, Pringle K, Al Mulla M, et al. . Response to emergence of Middle East respiratory syndrome coronavirus, Abu Dhabi, United Arab Emirates, 2013–2014. Emerg Infect Dis 2016; 22:1162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Gethamy M, Corman VM, Hussain R, et al. . A case of long-term excretion and subclinical infection with Middle East respiratory syndrome coronavirus in a healthcare worker. Clin Infect Dis 2015; 60:973–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J 2015; 12:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saudi Arabia Ministry of Health. MOH: 4 workers at PNU quarantined, no MERS-CoV cases reported amongst students. 2015. Available at: https://www.moh.gov.sa/en/Ministry/MediaCenter/News/Pages/news-2015-10-15-002.aspx. Accessed 19 October 2017. [Google Scholar]
- 11. World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia. 2015. Available at: http://www.who.int/csr/don/22-october-2015-mers-saudi-arabia/en/. Accessed 19 October 2017. [Google Scholar]
- 12. Corman VM, Ölschläger S, Wendtner CM, et al. . Performance and clinical validation of the RealStar MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J Clin Virol 2014; 60:168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu X, Whitaker B, Sakthivel SK, et al. . Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol 2014; 52:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Assiri AM, Midgley CM, Abedi GR, et al. . Epidemiology of a novel recombinant Middle East respiratory syndrome coronavirus in humans in Saudi Arabia. J Infect Dis 2016; 214:712–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Assiri AM, Biggs HM, Abedi GR, et al. . Increase in Middle East respiratory syndrome-coronavirus cases in Saudi Arabia linked to hospital outbreak with continued circulation of recombinant virus, July 1–August 31, 2015. Open Forum Infect Dis 2016; 3:ofw165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yusof MF, Queen K, Eltahir YM, et al. . Diversity of Middle East respiratory syndrome coronaviruses in 109 dromedary camels based on full-genome sequencing, Abu Dhabi, United Arab Emirates. Emerg Microbes Infect 2017; 6:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trivedi S, Miao C, Al-Abdallat MM, et al. . Inclusion of MERS-spike protein ELISA in algorithm to determine serologic evidence of MERS-CoV infection. J Med Virol 2018; 90:367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. World Health Organization. Management of asymptomatic persons who are RT-PCR positive for middle east respiratory syndrome coronavirus (MERSCoV). World Health Organization; 2018. Available at: http://www.who.int/csr/disease/coronavirus_infections/management_of_asymptomatic_patients/en/. Accessed 6 January 2018. [Google Scholar]
- 19. Moon SY, Son JS. Infectivity of an asymptomatic patient with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis 2017; 64:1457–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.