Abstract
Changes in developmental gene regulatory networks enable evolved changes in morphology. These changes can be in cis regulatory elements that act in an allele-specific manner, or changes to the overall trans regulatory environment that interacts with cis regulatory sequences. Here we address several questions about the evolution of gene expression accompanying a convergently evolved constructive morphological trait, increases in tooth number in two independently derived freshwater populations of threespine stickleback fish (Gasterosteus aculeatus). Are convergently evolved cis and/or trans changes in gene expression associated with convergently evolved morphological evolution? Do cis or trans regulatory changes contribute more to gene expression changes accompanying an evolved morphological gain trait? Transcriptome data from dental tissue of ancestral low-toothed and two independently derived high-toothed stickleback populations revealed significantly shared gene expression changes that have convergently evolved in the two high-toothed populations. Comparing cis and trans regulatory changes using phased gene expression data from F1 hybrids, we found that trans regulatory changes were predominant and more likely to be shared among both high-toothed populations. In contrast, while cis regulatory changes have evolved in both high-toothed populations, overall these changes were distinct and not shared among high-toothed populations. Together these data suggest that a convergently evolved trait can occur through genetically distinct regulatory changes that converge on similar trans regulatory environments.
Author summary
Convergent evolution, where a similar trait evolves in different lineages, provides an opportunity to study the repeatability of evolution. Convergent morphological evolution has been well studied at multiple evolutionary time scales ranging from ancient, to recent, such as the gain in tooth number in freshwater stickleback fish. However, much less is known about the accompanying evolved changes in gene regulation during convergent evolution. Here we compared evolved changes in gene expression in dental tissue of ancestral low-toothed marine fish to fish from two independently derived high-toothed freshwater populations. We also partitioned gene expression changes into those affecting a gene’s regulatory elements (cis), and those affecting the overall regulatory environment (trans). Both freshwater populations have evolved similar gene expression changes, including a gain of expression of putative dental genes. These similar gene expression changes are due mainly to shared changes to the trans regulatory environment, while the cis changes are largely population specific. Thus, during convergent evolution, overall similar and perhaps predictable transcriptome changes can evolve despite largely different underlying genetic bases.
Introduction
Development is controlled by a complex series of interlocking gene regulatory networks. Much of this regulation occurs at the level of transcription initiation, where trans acting factors bind to cis regulatory elements to control their target gene’s expression [1,2]. Evolved changes in an organism's morphology are the result of changes in this developmental regulatory landscape. It has been proposed that the genetic bases of many of these evolved changes are mutations within the cis-regulatory elements of genes [3–5]. Indeed, recent work in evolutionary genetics suggests the molecular bases of a diverse array of traits from Drosophila wing spots [6] to mouse pigmentation [7] to stickleback armored plate number [8,9] and size [10] are changes in the activity of cis-regulatory elements.
Evolved changes in gene expression can be divided into two broad regulatory classes. Cis regulatory changes can occur within the proximal promoter [11], distal enhancer [12], or the gene body itself [13], and result in allele-specific gene expression differences in hybrid diploids [14]. Trans regulatory changes modify the overall regulatory environment [15,16], but are usually genetically unlinked to the expression change, and do not result in allele-specific expression in hybrid diploids. For any gene with an evolved expression difference, the total evolved gene expression difference can be partitioned into changes in cis and trans by quantifying expression differences between two populations and also testing for expression differences between alleles in F1 hybrids between the two populations [14]. As both alleles in F1 hybrids animals are exposed to the same regulatory environment, any difference in their expression must be due to a cis-regulatory change. Several studies have attempted to characterize evolved cis and trans-regulatory changes at a transcriptome-wide level [17–21]. Though the relative contribution of cis and trans regulatory changes varies extensively among studies, cis changes have been found to dominate [17,18,21] or at least be approximately equivalent to trans changes [19,20,22]. Additionally, compensatory changes (cis and trans changes in opposing directions) have been found to be enriched over neutral models [17,18], showing evidence for selection for stable gene expression levels. However, none of these studies examined contribution of cis and trans gene expression changes during convergent morphological evolution.
Populations evolve new traits following a shift to a novel environment, due to a mixture of drift and selection. Truly adaptive traits can often be repeatedly observed in multiple populations following a similar ecological shift. Threespine sticklebacks are an excellent system for the study of evolved changes in phenotypes, including gene expression [23–27]. Marine sticklebacks have repeatedly colonized freshwater lakes and streams along the coasts of the Northern hemisphere [28]. Each of these freshwater populations has independently adapted to its new environment; however, several morphological changes, including a loss in armored plates and a gain in tooth number, are shared among multiple newly derived populations [29,30]. The repeated evolution of lateral plate loss is due to repeated selection of a standing variant regulatory allele of the Eda gene within marine populations [8,9] and genome sequencing studies found over a hundred other shared standing variant alleles present in geographically diverse freshwater populations [31]. These studies suggest the genetic basis of freshwater adaptation might typically involve repeated reuse of the same standing variants to evolve the same adaptive freshwater phenotype.
However, more recent evidence has shown that similar traits have also evolved through different genetic means in freshwater stickleback populations. A recent study which mapped the genetic basis of a gain in pharyngeal tooth number in two independently derived freshwater populations showed a largely non-overlapping genetic architecture [30]. Another study using three different independently derived benthic (adapted to the bottom of a lake) populations showed that, even when adapting to geographically and ecologically similar environments, the genetic architecture of evolved traits is a mix of shared and unique changes [32]. Even in cases where the same gene is targeted by evolution in multiple populations (the loss of Pitx1 expression resulting in a reduction in pelvic spines), the individual mutations are often independently derived [33,34]. All of these genomic scale studies have looked at the genetic control of morphological changes, while the extent and nature of genome-wide gene expression changes has been less studied. It remains an open question as to whether similar gene expression patterns evolve during the convergent evolution of morphology, and if so, to what extent those potential shared gene expression changes are due to shared cis or trans changes.
Teeth belong to a class of vertebrate epithelial appendages (including mammalian hair) that develop from placodes, and have long served as a model system for studying organogenesis and epithelial-mesenchymal interactions in vertebrates [35]. Odontogenesis is initiated and controlled by complex interactions between epithelial and mesenchymal cell layers, and involves several deeply conserved signaling pathways [36–38]. Sticklebacks retain the ancestral jawed vertebrate condition of polyphyodonty, or continuous tooth replacement, and offer an emergent model system for studying tooth replacement. Previous work has supported the hypothesis that two independently derived freshwater stickleback populations have evolved an increase in tooth replacement rate, potentially mediated through differential odontogenic stem cell dynamics [30]. Recent studies have found teeth and taste bud development to be linked, with one study supporting a model where teeth and taste buds are copatterned from a shared oral epithelial source [39], and another study supporting a model where teeth and taste buds share a common progenitor stem cell pool [40].
We sought to examine the evolution of the regulatory landscape controlling stickleback tooth development and replacement. Using high-throughput RNA sequencing (RNA-seq) in parental non-hybrid fish, we found that two independently derived high-toothed freshwater populations display highly convergent gene expression changes, especially in orthologs of known tooth-expressed genes in other vertebrates, likely reflecting the convergently evolved tooth gain phenotype and the deep homology of teeth across all jawed vertebrates. We also quantitatively partitioned these evolved gene expression changes into cis and trans regulatory changes [14,19] in both populations at a transcriptome-wide level using RNA-seq on F1 marine-freshwater hybrids. We found that trans regulatory changes predominate evolved changes in gene expression in dental tissue. Additionally, we found that the trans regulatory changes are more likely to be shared between the freshwater populations than the cis regulatory changes. Thus, similar downstream transcription networks controlling tooth development and replacement have convergently evolved largely through different upstream genetic regulatory changes.
Results
Convergent evolution of tooth gain in two freshwater populations
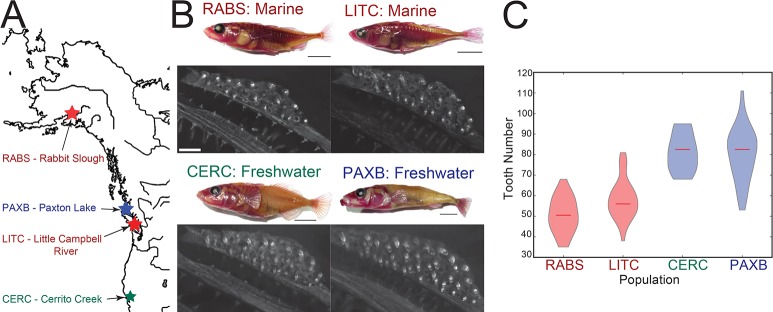
To test whether multiple freshwater populations have evolved increases in tooth number compared to multiple ancestral marine populations [30,41], we quantified total ventral pharyngeal tooth number of lab reared sticklebacks from four distinct populations: (1) a marine population from the Little Campbell river (LITCM) in British Columbia, Canada, (2) a second marine population from Rabbit Slough (RABSM) in Alaska, USA, (3) a benthic freshwater population from Paxton Lake (PAXBFW) in British Columbia, Canada, and (4) a second freshwater population from Cerrito Creek (CERCFW) in California, USA (Fig 1A and 1B). Freshwater fish from both populations had more pharyngeal teeth than marine fish at this 35-50mm standard length (SL) stage, consistent with previous findings [30,41] of increases in tooth number in freshwater sticklebacks (Fig 1B and 1C, S1 Table).
Fig 1. Evolved tooth gain in two freshwater populations.
(A) Stickleback population locations. (B) Representative Alizarin red stained adult lab-reared sticklebacks (top, scale bars = 1 cm) and dissected ventral pharyngeal tooth plates (scale bars = 100μm). (C) Total ventral pharyngeal tooth number of 35–50 millimeter standard length lab-reared adult fish from each population. N = 44,52,12,32 for RABSM, LITCM, CERCFW, and PAXBFW, respectively.
To estimate the genomic relatedness of these populations, we resequenced the genomes of three marine and six freshwater sticklebacks from the four different populations (S2 Table). We aligned the resulting reads (mean of ~53 million reads per sample, see Methods and S2 Table) to the stickleback reference genome [31] using Bowtie2 [42], and called 8.3 million (see Methods) variants using the Genome Analysis Toolkit (GATK) [43–45]. As it has been previously shown that Pacific marine stickleback populations are an outgroup to freshwater populations from Canada (PAXBFW) and California (CERCFW) [31], we hypothesized the two high-toothed populations would be more related to each other genomically than either marine population. A phylogeny constructed using a down-sampled set of 67.5 thousand genome-wide variants (see Methods) cleanly separated freshwater populations from each other and from marine fish (S1A Fig). Principal component analysis using 1.7 million filtered genome-wide variants (see Methods) revealed that the first principle component explains nearly half (41.4%) of the overall variance and separates PAXBFW sticklebacks from both CERCFW and marine fish (S1B Fig), representing the independent evolution of PAXBFW genomes. The second principal component separated both freshwater populations from marine populations, showing partially shared freshwater genome evolution. These results further support the model that populations of freshwater sticklebacks used a combination of shared and independent genetic changes [31,32] when evolving a set of similar morphological changes in response to a new environment.
Convergent evolution of gene expression
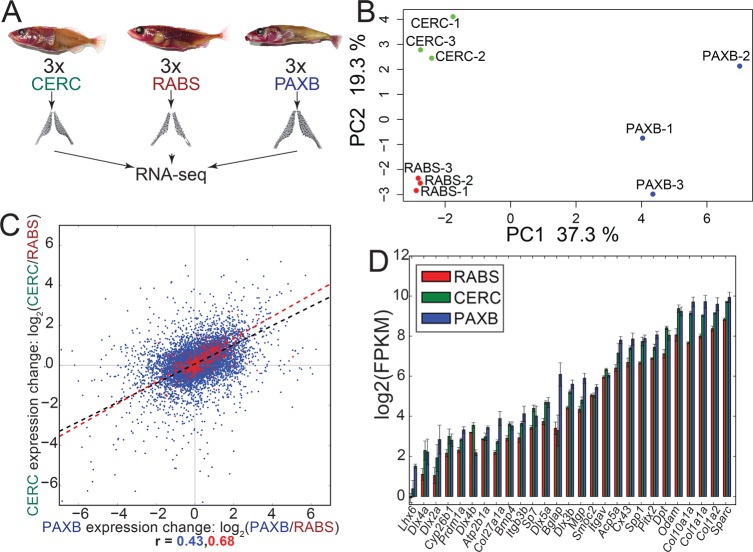
As morphological changes are often the result of changes in gene expression patterns and levels, we sought to identify evolved changes in gene expression during tooth development at stages soon after the evolved differences emerge [41]. We quantified gene expression in ventral pharyngeal dental tissue for three females each from the two high-toothed freshwater (PAXBFW and CERCFW) and Alaskan (RABSM) low-toothed marine populations using RNA-seq (Fig 2A, S3 and S4 Tables). Principal component (PC) analysis of the resulting gene expression matrix showed a clustering of gene expression by population, with the first PC separating PAXBFW samples, and the second PC separating both PAXBFW and CERCFW samples from marine, similar to the PC analysis of the genome-wide variants (Fig 2B) [46].
Fig 2. Convergent evolution of gene expression in dental tissue.
(A) Ventral pharyngeal tooth plates from three different populations were dissected and gene expression quantified by RNA-seq. (B) Principal component analysis of dental tissue gene expression shows population specific expression profiles. (C) Freshwater dental tissue exhibited correlated gene expression changes for all genes (blue), with increased correlation observed for orthologs of genes known to be expressed during mammalian tooth development (red). (D) Expression of genes annotated as expressed in zebrafish teeth (zfin.org) which were significantly upregulated in one or both freshwater populations.
Given the convergently evolved morphological change of increases in tooth number, we hypothesized that convergent evolution has occurred at the gene expression level in freshwater dental tissue. To test this hypothesis, we performed a differential expression analysis, defining evolved changes in gene expression as changes found to be significant in a differential expression analysis using cuffdiff2 [47]. We compared evolved change in gene expression in PAXBFW dental tissue (PAXBFW expression vs marine) to the evolved change in CERCFW dental tissue (CERCFW expression vs marine). We found 6,693 and 3,501 genes (out of a total of 22,442) with significant (as determined by cuffdiff2 [47], see Methods) evolved expression changes in PAXBFW and CERCFW respectively. Of these genes with evolved expression changes, 2,223 were called differentially expressed in both populations, with 1,898 (85%) showing expression changes in the same direction relative to marine.
At a genome-wide level, correlated changes in gene expression levels have evolved in the two high-toothed freshwater populations (Fig 2C, Spearman's r = 0.43). We next asked if orthologs of genes implicated in tooth development in other vertebrates showed an increase in correlated evolved expression changes. We compared the gene expression changes of stickleback orthologs of genes in the BiteIt (http://bite-it.helsinki.fi/) [48] or ToothCODE (http://compbio.med.harvard.edu/ToothCODE/) [36] databases (hereafter referred to as the “BiteCode” gene set, S5 Table), two databases of genes implicated in mammalian tooth development. Consistent with the conserved roles of gene regulatory networks regulating mammalian and fish teeth [49–52] and the major evolved increases in tooth number in both freshwater populations (Fig 1C), these predicted dental genes showed an increase in their correlated evolved gene expression change (Fig 2C red points, Spearman's r = 0.68), and tended to have an overall increase in gene expression (S2 Fig, P = 7.36e-6, GSEA, see methods). This correlation coefficient was higher than any observed in over 100,000 bootstrapped (sampled with replacement) gene sets of the same size from the same gene expression matrix. We also examined the expression levels of genes whose orthologs are annotated as being expressed in zebrafish pharyngeal teeth (www.zfin.org). Within this gene set, 27 of 40 genes were significantly more highly expressed in at least one freshwater population, with no genes expressed significantly higher (as determined by cuffdiff2 [47,53–55], see Materials and Methods) in marine samples than either freshwater population (Fig 2D).
Increased freshwater expression of stem cell maintenance genes
Tooth development is controlled by several deeply conserved developmental signaling pathways [50,52]. To test whether expression changes in the components of specific developmental signaling pathways have evolved in the two high-toothed freshwater populations, we next analyzed the expression levels of stickleback orthologs of genes implicated in mammalian tooth development and annotated as components of different signaling pathways [36]. When comparing gene expression levels in freshwater dental tissue to marine dental tissue, genes annotated as part of the TGF-ß signaling pathway displayed significantly increased expression in freshwater dental tissue (S3A–S3F Fig).
Since these two freshwater populations have a largely different developmental genetic basis for their evolved tooth gain [30], we next asked whether any pathways were upregulated or downregulated specifically in one freshwater population. When comparing the expression of genes in PAXBFW dental tissue to expression in CERCFW or marine dental tissue, genes not only in the TGF-ß pathway, but also in the WNT signaling pathway, displayed significantly increased expression, consistent with the differing genetic basis of tooth gain in these populations (S3B Fig). In contrast, no significant pathway differences were found comparing CERCFW to PAXBFW or marine (S3C Fig).
We next asked whether any pathways, regardless of previous implication in tooth development, were significantly upregulated in either or both freshwater transcriptomes. Genes upregulated in freshwater dental tissue were enriched for Gene Ontology (GO) terms involved in anatomical structure development, signaling, and regulation of cell proliferation (S4A Fig, S6 Table). Genes upregulated in PAXBFW dental tissue over marine were enriched for GO terms involved in cell proliferation, division and cell cycle regulation, as well as DNA replication (S4B Fig, S7 Table), while genes upregulated in CERCFW over marine were enriched for GO terms involved in cell locomotion, movement, and response to lipids (S4C Fig, S8 Table). 204 of the 454 and 432 GO terms that were enriched in genes upregulated in PAXBFW and CERCFW relative to marine, respectively, were shared, further supporting the convergent gain of freshwater gene expression.
As teeth are constantly being replaced in polyphyodont adult fish, potentially due to the action of dental stem cells [40], we hypothesized that genes involved in stem cell maintenance have evolved increased expression in freshwater tooth plates, given the higher rate of newly forming teeth previously found in adults [30], and the possibly greater number of stem cell niches in high-toothed fish. We further hypothesized that since teeth are developmentally homologous to hair, perhaps an ancient genetic circuit regulating vertebrate placode replacement controls both fish tooth and mammalian hair replacement. For example, the Bmp6 gene, previously described as expressed in all stickleback teeth [41] was significantly upregulated in CERCFW fish, consistent with the evolved major increases in tooth number in this population (S4 Table). In contrast, no such significant upregulation was observed in the expression of PAXBFW Bmp6 (S4 Table), consistent with the observed evolved cis-regulatory decrease in PAXBFW Bmp6 expression [41]. Further supporting this hypothesis, the expression of the stickleback orthologs of a previously published set of mouse hair follicle stem cell (HFSC) signature genes [56] were significantly upregulated in freshwater dental tissue (S3A Fig), with 84 and 75 out of 254 genes displaying significant increases in expression in PAXBFW and CERCFW, respectively. CERCFW dental tissue displayed a small but significant increase in expression of this set of HFSC orthologs relative to both PAXBFW and marine samples (S3C Fig).
In cichlid fish, pharmacology experiments revealed that reductions in tooth density can be accompanied by concomitant increases or decreases in taste bud density [39]. To begin to test whether derived high-toothed stickleback populations have also evolved significantly altered levels of known taste bud marker gene expression, we examined the expression levels of known taste bud markers Calbindin2 and Phospholipase Beta 2 [57], as well as taste receptors such as Taste 1 Receptor Member 1, Taste 1 Receptor Member 3, and Polycystin 2 Like 1 [58]. Although four of these five genes had detectable significant expression changes between different populations, no consistent freshwater upregulation or downregulation of taste bud marker genes was seen (S5 Fig).
Cis and trans regulatory changes in gene expression
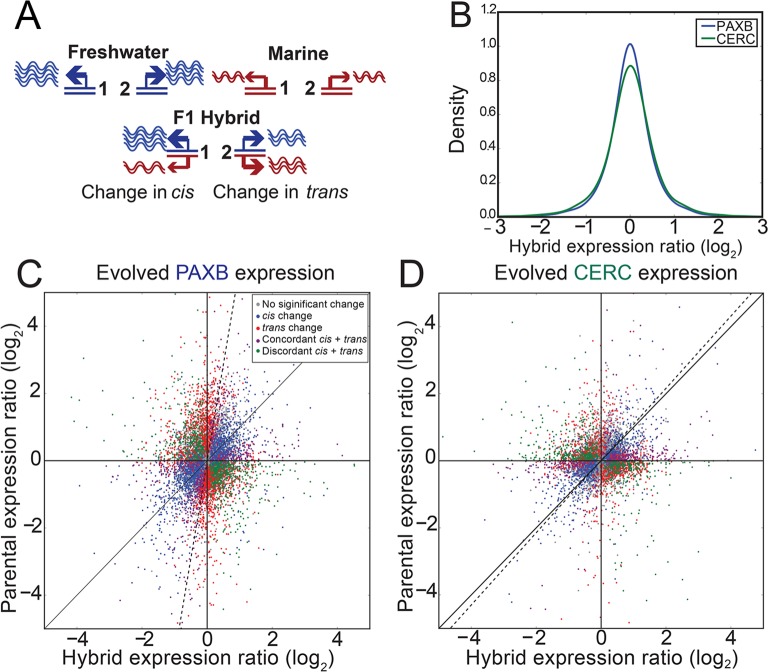
Evolved changes in gene expression are due to a combination of cis acting changes that are linked to the genes they act on, and trans acting changes which usually are genetically unlinked to the gene or genes they regulate. Since the genetic basis of freshwater tooth gain mapped to largely non-overlapping intervals in these two populations [30], we hypothesized that the observed shared freshwater gene expression changes were the result of a similar trans environment, but a largely different set of cis changes. To test this hypothesis, we measured evolved cis expression changes in marine-freshwater F1 hybrids, which have marine and freshwater alleles present in the same trans environment. We raised both CERCFW-marine and PAXBFW-marine F1 hybrids to the late juvenile stage, dissected their ventral pharyngeal tooth plates, then generated and sequenced five barcoded RNA-seq libraries per population (10 total). We then quantified the cis expression change as the ratio of the number of reads mapping uniquely to the freshwater allele of a gene to the number of uniquely mapping marine reads (Fig 3A, S9–S11 Tables). Trans expression changes were calculated by factoring the cis change out from the overall parental expression change [19].
Fig 3. Evolved changes in cis-regulation.
(A) Cartoon showing the two different regulatory changes detectable by our F1 hybrid system. Both genes 1 and 2 show an evolved increase of expression in freshwater fish, but the freshwater allele of gene 1 but not gene 2 is expressed more highly in F1 hybrids. Therefore, gene 1 has evolved its increased gene expression through cis-regulatory changes, while gene 2 was modulated by trans regulatory changes. (B) Density plot showing the measured cis-regulatory changes. Neither population displayed a significant allelic bias, as measured by a Wilcoxon signed-rank test. (C-D) Gene expression changes in both parental and hybrid dental tissue–genes are color-coded based on the role of cis and/or trans change in PAXBFW (C) or CERCFW (D) dental tissue. Dashed line indicates the first principal component axis.
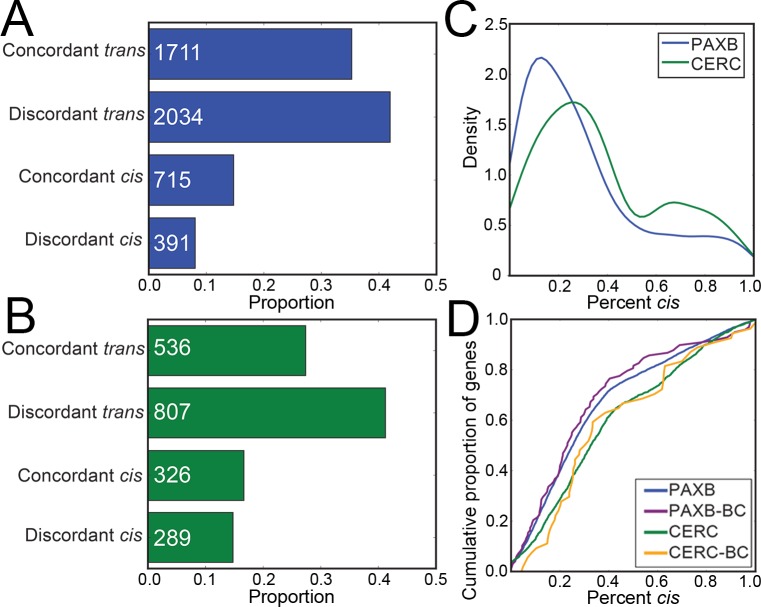
We found 11,832 and 8,990 genes in PAXBFW and CERCFW F1 hybrids, respectively, that had a fixed marine-freshwater sequence difference which had more than 20 total reads mapping to it. We observed no significant bias towards either the marine or freshwater allele in either set of F1 hybrids (Fig 3B). We next classified genes into one of four categories (cis change only, trans change only, concordant cis and trans changes, discordant cis and trans changes). We found 1640 and 1116 PAXBFW (Fig 3C) and CERCFW (Fig 3D) genes, respectively, with only significant cis changes, and 1873 and 1048 genes, respectively, with only significant trans changes. We also found 478 and 359 genes with significant cis and trans changes in the same direction, which we term concordant changes in gene expression. Conversely, we found 772 and 607 genes with significant cis and trans changes in opposing directions, which we termed discordant changes. Discordant cis and trans changes were more common in both populations, suggesting selection for stable levels of gene expression.
Trans regulatory changes dominate
We next wanted to determine the relative contribution of cis and trans gene expression changes to evolved changes in gene expression. We restricted our analysis to differentially expressed genes (as determined by cuffdiff2 [47]) to examine only genes with a significant evolved difference in gene expression and quantifiable (i.e. genes with transcripts containing a polymorphic variant covered by at least 20 reads) cis and trans expression changes. When evolving a change in gene expression, the cis and trans regulatory basis for this change can be concordant (cis and trans effects both increase or decrease expression) or discordant (cis effects increase and trans decrease or vice versa). We hypothesized that genes would tend to display more discordant expression changes, as stabilizing selection has been found to buffer gene expression levels [17,22,59]. To test this hypothesis, we binned differentially expressed genes into a 2x2 contingency table, with genes classified as cis or trans based on which effect controlled the majority of the evolved expression change, and discordant or concordant based on the direction of the cis and trans changes (Fig 4A and 4B). In the CERCFW population, significantly more discordant changes than expected by a neutral model (P = 1.35e-7, binomial test) have evolved. In both populations, we found increased discordant changes when the trans effect is larger than the cis effect (P = 1.29e-7, 1.44e-13, PAXBFW and CERCFW respectively, binomial test). In both populations, we observe the opposite (an enrichment of concordant changes) when the cis effect is stronger, relative to the ratio when the trans effect is dominant (P = 1.34e-36, 8.2e-11 PAXBFW and CERCFW respectively, binomial test). When considering all (not just differentially expressed) genes with quantifiable cis and trans expression changes, discordant changes dominated regardless of the relative strength of the cis effect (S6 Fig).
Fig 4. Trans changes predominate evolved dental gene expression changes.
(A-B) Proportion of differentially expressed genes displaying opposing and concordant cis and trans changes in PAXBFW (A) or CERCFW (B) dental tissue. Genes whose expression differences were mostly explained by cis changes tended to be more concordant (P = 5.0e-17, 0.002 for PAXBFW and CERCFW, respectively) than those mostly explained by trans changes. (C) Density of the relative percentage of gene expression differences which are explained by cis changes in PAXBFW and CERCFW dental tissue. (D) Cumulative percentage of percentage of gene expression due to cis changes. Genes in CERCFW samples display a higher percentage cis change than genes in PAXBFW samples (P = 1.25e-22, Mann-Whitney U test).
If all gene expression changes were due to changes only in cis, we would expect to see the measured cis ratios in the hybrids match the parental expression ratios. Instead, in both cases of evolved change, we saw parental expression ratios of a greater magnitude than F1 hybrid ratios, indicating a stronger contribution of trans changes to overall gene expression changes (Fig 3C and 3D). Indeed, when we examined the overall percentage of expression changes of differentially expressed genes that were due to changes in cis, we observed median per gene values of only 25.2% and 32.5% of PAXBFW and CERCFW gene expression changes, respectively (Fig 4C). Comparing the expression levels of orthologs of known dentally expressed genes from the BiteIt [48] and ToothCODE [36] databases revealed a similarly small number of gene expression changes explained by changes in cis, relative to the genome-wide average (Fig 4D). Evolved changes in CERCFW gene expression were more due to changes in cis than PAXBFW genes (Fig 4D, P = 1.25e-22, Mann-Whitney U test). Thus, trans effects on gene expression dominate the evolved freshwater gene expression changes.
Trans regulatory changes are more likely to be shared between freshwater populations
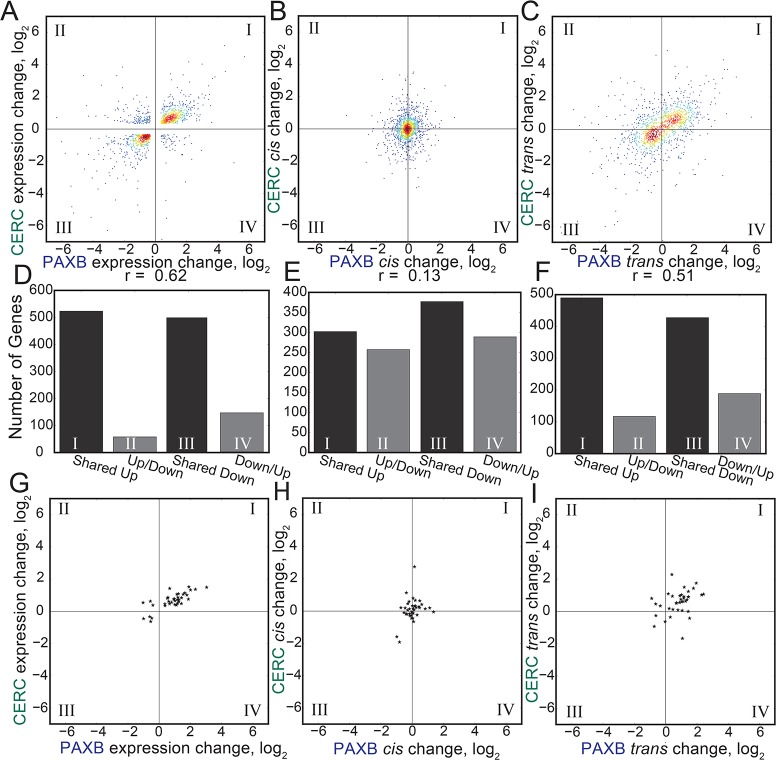
We next wanted to test the hypothesis that the shared freshwater gene expression changes were primarily due to shared trans changes, rather than shared cis changes. We first compared the overall expression levels of genes called differentially expressed between PAXBFW and marine as well as CERCFW and marine. We restricted our analysis to differentially expressed genes whose cis-regulatory change we were able to measure in our F1 hybrids, including genes without a significant cis change. Similar to the genome-wide comparison, we found a highly significant non-parametric correlation coefficient (Spearman's r = 0.62, P = 1.2e-132) for the expression change of these shared differentially expressed genes (Fig 5A). When comparing the PAXBFW cis changes of these genes to the CERCFW cis changes, however, we found a much lower (though still significant) correlation coefficient (Spearman's r = 0.13, P = 5.1e-6) (Fig 5B). We calculated trans changes for each of these differentially expressed genes, defined as the difference between the expression change in the freshwater parent relative to marine and the freshwater allele relative to the marine in the F1 hybrid [18,19,60]. When comparing the calculated trans changes for these shared differentially expressed genes, we observed much higher correlation coefficient (Spearman's r = 0.51, P = 1.2e-80) (Fig 5C). When comparing all, not just differentially expressed, genes, trans changes are still likely to be more shared than cis (S7 Fig). Additionally, 35/38 of the shared differentially expressed putative dental genes have shared regulatory increases or decreases in both freshwater populations relative to marine in overall expression difference. 32/38 of these gene show regulatory changes in the same direction in trans, but only 25/38 in cis (Fig 5G–5I). Thus, the trans effects on evolved gene expression are more likely to be shared by both freshwater populations than the cis changes.
Fig 5. Trans changes are more likely to be shared across populations.
(A) Genes with significantly different evolved expression in both freshwater populations relative to marine fish, showing significantly correlated changes in gene expression in PAXBFW and CERCFW dental tissue. (B) Freshwater dental tissue had a significant but small number of shared cis-regulatory changes. (C) Freshwater dental tissue showed significantly correlated changes in trans expression changes. A-C show genes with significant expression changes between populations and quantifiable (i.e. genes with transcripts containing a polymorphic SNP covered by at least 20 reads) cis-regulatory changes in both populations. Density (color) was estimated with a Gaussian kernal density estimator. (D-F) Bar graphs show the number of genes with shared or divergent expression patterns from the above panels. (G-I) Similar to (A-C), but showing only genes in the BiteCode gene set.
Discussion
We sought to test the relative contribution of cis and trans gene regulatory changes during convergent evolution of tooth gain, as well as to ask whether the same or different regulatory changes underlie evolved changes in gene expression during this case of convergent evolution. We quantified the overall regulatory divergence, as well as the specific contribution of cis and trans changes, between ancestral low-toothed marine and two different independently derived populations of high-toothed freshwater sticklebacks. Similar overall changes in gene expression have evolved in both freshwater populations, especially in orthologs of known dental regulators in mammals. In this system, trans-regulatory changes play a larger role than cis changes in both populations. Furthermore, trans acting changes were much more likely to be shared between freshwater populations than cis changes, suggesting the two high-toothed populations evolved their similar gene expression patterns through independent genetic changes.
Convergent evolution of dental gene expression
Convergent evolution at the gene expression level occurs when similar gene expression levels evolve in different populations. Both the PAXBFW and CERCFW stickleback populations have adapted from an ancestral marine form to their current freshwater environments. The genomic nature of their derived changes appears largely divergent, with major axis of variation separating PAXBFW genomes from the geographically proximal marine populations (LITCM), as well as the more distant marine (RABSM) and CERCFW populations. However, when looking at the gene expression basis of their convergently evolved gain in tooth number, orthologs of genes implicated in mammalian dental development showed strong correlated freshwater gains in expression. This correlation suggests both that sticklebacks deploy conserved genetic circuits regulating tooth formation during tooth replacement, but also that both populations have convergently evolved changes to similar downstream transcriptional circuits resulting in a gain of tooth number.
Though both freshwater populations showed strongly correlated changes in evolved gene expression at the trans regulatory level, the cis changes were largely not shared across populations. This was especially true for putative dentally expressed genes with evolved expression changes–the vast majority of the trans but not cis expression changes were shared between both freshwater populations. This suggests that the similar freshwater gene expression patterns evolved through independent genetic changes. It is possible that the small number of shared cis changes are sufficient to drive the observed changes to the overall trans regulatory environments. However previous work has shown that the genetic basis of tooth gain in these two populations is largely distinct [30], and it seems parsimonious that the genetic basis of a gain in dental gene expression is also mostly independent. Thus, convergent freshwater gene expression changes appear to be largely due to distinct, independent population-specific regulatory changes. This finding suggests that there are many regulatory alleles that are accessible during the evolution of an adaptive trait.
Trans effects dominate
Other studies have used RNA-seq to compare the relative contribution of cis and trans-regulatory changes in the evolution of gene expression in a multitude of species and tissues. In mice, evolved gene expression changes in the liver [18] and the retina [61] were driven primarily by cis-regulatory changes. In Drosophila, work on organismal-wide evolved gene expression changes on the genome-wide level has shown the opposite, with trans-regulatory effects playing a larger role in the evolution of gene expression [19,22]. Other studies have found trans effects contribute more to intraspecific comparisons, while cis effects contribute more to interspecific comparisons [17,20,60]. Consistent with this, we observe trans effects dominating in both of our intraspecific comparisons.
Another key distinction could be that cis-regulatory effects dominate when looking at more cellularly homogenous tissues, while trans-regulatory effects dominate when looking at more heterogeneous tissues. Stickleback tooth plates likely fall into an intermediate category, less heterogenous in cell type composition than a full adult fly or fly head, but more heterogeneous than a specialized tissue such as the mouse retina. Overall, freshwater tooth plates are more morphologically similar to each other than marine, with freshwater tooth plates possessing a larger area, increased tooth number, and decreased intertooth spacing [30,41]. Freshwater tooth plates likely have more similar cell type abundances and compositions (e.g. more developing tooth germs with inner and outer dental epithelia, and odontogenic mesenchyme) compared to each other than to marine tooth plates. Similar cell types tend to have similar gene expression patterns, even when compared across different species [62]. Much of the shared freshwater increase in dental gene expression could be due to an increase in dental cell types in both freshwater populations. As other evolved changes to stickleback morphology have been shown to be due to cis regulatory changes to key developmental regulatory genes [8,33,41,63], this trans regulatory increase in cell type abundance could be due to a small number of cis regulatory changes. These initially evolved developmental regulatory changes could result in similar downstream changes in the developmental landscape, resulting in the shared increase in dental cell types. Consistent with this interpretation, stickleback orthologs of genes known to be expressed during mammalian tooth development were found here to have a much greater incidence of convergently evolved increase in trans regulatory gene expression.
Compensatory cis and trans
Previous studies [17,18] have shown compensatory cis and trans changes are essential for the evolution of gene expression. These findings are consistent with the idea that the main driving force in the evolution of gene expression is stabilizing selection [59] where compensatory changes to regulatory elements are selected for to maintain optimal gene expression levels. In both PAXBFW and CERCFW dental tissue, when considering all genes with a quantifiable (i.e. polymorphic and covered by ~20 reads, see Methods) cis effects, discordant compensatory cis and trans changes were far more common than concordant ones. This trend could be driven by some initial selection on pleiotropic trans changes, followed by selection for compensatory cis changes to restore optimal gene expression levels [17,18,22]. However, the trans, but not the cis, evolved changes in gene expression were highly shared among the two freshwater populations. Thus, collectively our data support a model where two independently derived populations have convergently evolved both similar genome-wide expression levels as well as ecologically relevant morphological changes through different genetic means.
Potential parallels between teeth and hair regeneration
PAXBFW and CERCFW sticklebacks have an increased rate of new tooth formation in adults relative to their marine ancestors [30]. In constantly replacing polyphyodonts, it has been proposed that teeth are replaced through a dental stem cell intermediate [37,38]. A strong candidate gene underlying a large effect PAXBFW tooth quantitative trait locus (QTL) is the secreted ligand Bone Morphogenetic Protein 6 (Bmp6) [41], which is also a key regulator of stem cells in the mouse hair follicle [56]. Freshwater dental tissue displayed significantly increased expression of known signature genes of mouse hair follicle stem cells, perhaps reflecting more stem cell niches supporting the higher tooth numbers in freshwater fish. Genes upregulated in freshwater dental tissue also were significantly enriched for GO terms involved in the cell cycle and cell proliferation. Together these findings suggest that both freshwater populations have evolved an increased tooth replacement rate through an increased activity or abundance of their dental stem cells, and also suggest the genetic circuitry regulating mammalian hair and fish tooth replacement might share an ancient, underlying core gene regulatory network.
Materials and methods
Ethics statement
Experiments were approved by the Institutional Animal Care and Use Committee of the University of California-Berkeley (protocol # R330).
Stickleback husbandry
Fish from all populations were raised in 110L aquaria in brackish water (3.5g/L Instant Ocean salt, 0.217mL/L 10% sodium bicarbonate) at 18°C in 8 hours of light per day. Young fry [standard length (SL) < 10 millimeters (mm)] were fed a diet of live Artemia, early juveniles (SL ~10–20 mm) a combination of live Artemia and frozen Daphnia, and older juveniles (SL > ~20 mm) and adults a combination of frozen bloodworms and Mysis shrimp.
Skeletal staining and imaging
Sticklebacks were fixed in 10% neutral buffered formalin overnight at 4°C. Fish were washed once with water and then stained in 1% KOH, 0.008% Alizarin Red for 24 hours. Following a water rinse, fish were cleared in 0.25% KOH, 50% glycerol for 2–3 weeks. Branchial skeletons were dissected as previously described [64]. Pharyngeal teeth were quantified with fluorescent illumination using a TX2 filter on a Leica DM2500 microscope. Representative tooth plates were created using montage z-stacks on a Leica M165 FC using the RhodB filter. Adult fish were imaged using a Canon Powershot S95. Some tooth count data from the CERCFW, RABSM, and PAXBFW populations; n = 11, 13, 29, respectively, (see S1 Table) have been previously published [30].
DNA preparation and genome resequencing
Caudal fin tissue was placed into 600μl tail digestion buffer [10mM Tris pH 8.0, 100mM NaCl, 10mM EDTA, 0.05% SDS, 2.5μl ProK (Ambion AM2546)] for 12 hours at 55°C. Following addition of 600 μl of 1:1 phenol:chloroform solution and an aqueous extraction, DNA was precipitated with the addition of 1ml 100% ethanol, centrifuged, washed with 75% ethanol, and resuspended in water. 50ng of purified genomic DNA was used as input for the Nextera Library prep kit (Illumina FC-121-1031), and barcoded libraries were constructed following the manufacturer’s instructions. Library quality was verified using an Agilent Bioanalyzer. Libraries were pooled and sequenced on an Illumina HiSeq 2000 (see S2 Table for details), resulting in a mean of 52.8 million reads per sample, with a max of 70.3 million reads and a minimum of 39 million reads (S2 Table).
RNA purification and creation of RNA-seq libraries
Late juvenile stage female sticklebacks (SL ~40mm) were euthanized in 0.04% Tricaine. Dissected [64] bilateral ventral pharyngeal tooth plates were placed into 500μl TRI reagent, then incubated at room temperature for 5 minutes. Following addition of 100μl of chloroform, a further 10 minute incubation and centrifugation, the aqueous layer was extracted. Following addition of 250μl isopropyl alcohol and 10 minute incubation, RNA was precipitated by centrifugation, washed with 75% EtOH, and dissolved in 30ul of DEPC-treated water. RNA integrity was assayed by an Agilent Bioanalyzer. 500ng of RNA from each fish was used as input to the Illumina stranded TruSeq polyA RNA kit (Illumina RS-122-2001), and libraries were constructed following the manufacturer’s instructions. Library quality was analyzed on an Agilent Bioanalyzer, and libraries were pooled and sequenced on an Illumina HiSeq2000 (see S3 Table). We obtained a mean of 84.1 million reads among the parental samples, with a max of 91.0 million and a minimum of 78.6 million (S3 Table).
Gene expression quantification and analysis
RNA-seq reads were mapped to the stickleback reference genome [31] using the STAR aligner [65] (version 2.3, parameters = —alignIntronMax 100000—alignMatesGapMax 200000—outFilterMultimapNmax 20—outFilterMismatchNmax 999—outFilterMismatchNoverLmax 0.04—outFilterType BySJout), using ENSEMBL genes release 85 as a reference transcriptome. The resulting SAM files were sorted and indexed using Samtools version 0.1.18 [66], PCR duplicates were removed, read groups added and mate pair information fixed using Picard tools (version 1.51) (http://broadinstitute.github.io/picard/) with default settings. Gene expression was quantified with the Cufflinks suite (v 2.2.1) [47,53–55] using ENSEMBL genes as a reference transcriptome, with gene expression quantified with cuffquant (-u—library-type fr-firststrand) and normalized with cuffnorm. Differentially expressed genes were found using cuffdiff2, with parameters (-u—FDR .1—library-type fr-firststrand, using the reference genome for bias correction). Genes with a mean expression less than 0.1 FPKM were filtered from further analysis.
Gene set and gene ontology enrichment
The BiteCode gene set was generated by combining all genes in the BiteIt (http://bite-it.helsinki.fi/) or ToothCODE (http://compbio.med.harvard.edu/ToothCODE/) [36] databases. Stickleback orthologs or co-orthologs were found using the annotated names of ENSEMBL stickleback genes. Gene set expression change statistical enrichment was done as previously described [67]. Briefly, a t-test was performed for each gene to test for a difference in mean expression between the two treatments. The resulting t-values were subject to a 1-sample t-test, with the null model that the mean of the t-values was 0. Cutoffs were validated using 10,000 bootstrapped replicate gene sets drawn from the same gene expression matrix. Stickleback orthologs of mouse or human genes were determined using annotated ENSEMBL orthologs. Sorted lists of genes, ranked by log2 expression change in PAXBFW dental tissue relative to marine, CERCFW relative to marine, or the mean of CERCFW and PAXBFW relative to marine, were generated using the measured gene expression data. Gene Ontology enrichment was done using Gorilla [68,69], and results were visualized using REVIGO [70].
Detection of genomic and transcriptomic variants
Genomic resequencing reads were aligned to the stickleback reference genome [31] using the bwa aln and bwa sampe modules of the Burrows-Wheeler Alignment tool (v 0.6.0-r85) [71]. Resulting SAM files were converted to BAM files, sorted and indexed by Samtools version 0.1.18 [66], with PCR duplicates removed by Picard tools. GATK's (v3.2–2) IndelRealigner (parameter: '-LOD 0.4'), BaseRecalibrator, and PrintReads were used on the resulting BAM files. BAM files from the above RNA-seq alignment were readied for genotype calling using GATK's SplitNCigarReads, BaseRecalibrator, and PrintReads. Finally, the UnifiedGenotyper was used to call variants from the RNA-seq and DNA-seq BAM files, with parameters (-stand_call_conf 30 -stand_emit_conf 30 -U ALLOW_N_CIGAR_READS—genotype_likelihoods_model BOTH) [43,45]. This analysis identified a set of 8,341,326 variants.
Principal components analysis of the genome-wide set of variants was performed by first filtering all multiallelic variants or variants with a missing genotype, resulting in a set of 1,690,729 variants. PCA was performed using FactoMiner [46] and a set of custom R scripts. Phylogenetic trees were constructed using the set of variants, downsampled to 67,507 SNPs (no indels) for use with BEAST and SNAPP [72,73]. We constructed phylogenies using SNAPP, estimating substitution rate and proportion invariant from the data, and ran 1 million generations of MCMC simulations. The best tree was picked with TreeAnnotator and visualized with FigTree.
To accurately phase RNA-seq data from F1 hybrids, pseudo-transcriptomes were created for each hybrid. The pseudo-transcriptomes consist of the predicted sequence for each allele within an F1 hybrid, with all predicted splicing variants of a gene collapsed to a single transcript. A variant was added to the pseudo-transcriptome if and only if it was homozygous in the sequenced parents (or parent’s sibling in the case of the RABSM parent of the CERCFW x RABSM F1 hybrids) and called heterozygous in the F1 hybrid.
Cis and trans regulatory divergence quantification
RNA-seq reads from F1 hybrid sticklebacks were aligned to the individual’s pseudo-transcriptome using STAR (v 2.3) with the parameters:—outFilterMultimapNmax 1 and—outFilterMultimapScoreRange 1. By only looking at uniquely aligning reads, we ensured we only considered reads which overlapped a heterozygous variant site. Counting these unique reads minimizes double counting a single read that supports two different variant positions. Total cis divergence in each F1 hybrid was quantified by comparing the number of reads mapping uniquely to each allele in the pseudo-transcriptome.
Following cis divergence quantification in all F1 hybrids, we considered the overall cis change in the different freshwater populations. Genes which only had 20 or fewer uniquely mapping reads across all replicates were filtered from further analysis. We filtered 28 genes that had >32 fold expression changes that included genes that either had zero reads from one allele and thus infinite expression differences (20 genes), were highly repetitive (2 genes), or mitochondrial (2 genes). Reported cis ratios were calculated by comparing the ratio of uniquely mapped freshwater reads to uniquely mapped marine reads. Evolved trans changes were quantified as the difference between the log of the overall gene expression change between the freshwater and marine parents and the log of measured cis freshwater expression change. Percent cis change was calculated as the absolute value of the log of the cis change divided by the sum of the absolute value of the log of the cis change and the absolute value of the log of the trans change. Statistical significance of cis changes was determined by a binomial test comparing overall reads mapping to the freshwater allele to a null model of no cis divergence, with a false discovery rate of 1% applied using the Benjamini-Hochberg method. Statistical significance of trans changes was determined by a G-test, comparing the expected (based on the measured cis change) and observed ratios of marine and freshwater, with a 1% false discovery rate.
Supporting information
(A) Genome-wide phylogeny created from genomic resequencing data. Wild-caught fish are non-italicized. All nodes have 100% posterior probability. Scale bar shows 3% sequence divergence at variant positions. (B) Principal component analysis of genome-wide genotypes separates marine and CERCFW populations from the PAXBFW lake population, with the 2nd PC separating marine and freshwater populations.
(TIF)
(A) PAXBFW upregulation of BiteCode genes (282 expressed orthologs, P = 9.8e-3, GSEA). (B) CERCFW upregulation of BiteCode genes (P = 2.1e-5, GSEA). (C) PAXBFW and CERCFW upregulation of BiteCode genes (P = 5.1e-6, GSEA).
(TIF)
(A-F) Changes in gene expression changes of genes annotated as components of the indicated signaling pathways (BMP, FGF, SHH, WNT, ACT, TGFB, NOTCH, or EDA, containing 59, 60, 28, 75, 19, 11, 12, and 6 expressed orthologs, respectively) [36] or orthologs of a described set of mouse hair follicle stem cell signature genes (HFSC, containing 254 expressed orthologs) [56]. Violin plots show the mean expression change of genes in the pathway. (A) Change in freshwater (PAXBFW + CERCFW) relative to marine. (B) PAXBFW specific changes (PAXBFW relative to CERCFW + marine). In the WNT and TGFB pathway, 22/75 and 6/11 genes had significantly increased expression respectively (C) CERCFW specific changes (CERCFW relative to PAXBFW + marine). (D) PAXBFW evolved changes (PAXBFW relative to marine) (E) CERCFW evolved changes (CERCFW relative to marine) (F) PAXBFW vs CERCFW changes (PAXBFW relative to CERCFW).
(TIF)
(A-C) GO enrichment of genes upregulated in freshwater (A), PAXBFW (B), or CERCFW (C). GO analysis was performed using Gorilla [68], with the results visualized with Revigo [70].
(TIF)
Expression levels of known taste bud marker genes in marine, PAXBFW and CERCFW tooth plates as assayed by RNA-seq. * indicates differentially expressed genes. Error bars are standard error of the mean.
(TIF)
(A-B) Proportion of genes with quantifiable (i.e. genes with transcripts containing a polymorphic SNP covered by at least 20 reads) hybrid expression displaying opposing and concordant cis and trans changes in PAXBFW (A) or CERCFW (B) dental tissue. Similar to Fig 5, but here showing all genes, not just genes with significantly different expression levels compared to marine. Trans regulatory changes predominate, as do opposing over concordant changes. (C) Density plot of the percentage of gene expression changes explained by cis-regulatory changes.
(TIF)
(A) Expression changes of genes with quantifiable (i.e. genes with transcripts containing a polymorphic SNP covered by at least 20 reads) hybrid expression in both freshwater populations relative to marine fish, showing significantly correlated changes in gene expression in PAXBFW and CERCFW tooth plates. (B) cis regulatory changes of genes with quantifiable hybrid expression in freshwater dental tissue overall do not display correlated evolved changes. (C) trans regulatory changes of genes with quantifiable hybrid expression in freshwater dental tissue. Density (color) was estimated with a Gaussian kernel density estimator. (D-F) Similar to A-C, but showing only genes in the BiteCode gene set, revealing that these orthologs have evolved highly convergent changes in the two freshwater populations (D), despite non-convergent cis regulatory changes (E).
(TIF)
For each fish, the population, ecotype (freshwater or marine), total ventral pharyngeal tooth number (TVTP), total length (TL), standard length (SL), and whether data has been published [30] is shown.
(XLSX)
For each fish, population and biological replicate number (Fish), the total number of barcoded reads from each fish (reads), and number of reads that mapped and passed all filters (final mapped) is listed.
(XLSX)
For each fish, population of parents and biological replicate number (sample), standard length (SL), total reads (generated by HiSeq2000 over two different runs (run1 and run2)), mapped reads (reads that mapped to the genome), and final reads (excludes reads filtered due to low quality or PCR duplication) is listed.
(XLSX)
Estimated abundance in in fragments per kilobases per million reads (FPKM) of ENSEMBL genes (rows) in ventral pharyngeal dental tissue from three individual fish from three populations (in columns). Mean expression (in FPKM) is shown after the 3 replicates. Log2(Pop1/Pop2) shows the fold-change in log2 of the estimated mean expression between the two populations. IsSig(Pop1/Pop2) indicates whether the difference was significant as reported by cuffdiff2.
(XLSX)
A list of stickleback orthologs in the BiteIt [48] (http://bite-it.helsinki.fi/) or ToothCODE (http://compbio.med.harvard.edu/ToothCODE/) [36] databases.
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in freshwater dental tissue reported by GOrilla [68].
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in PAXBFW dental tissue reported by GOrilla [68].
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in CERCFW dental tissue reported by GOrilla [68].
(XLSX)
For each ventral pharyngeal tooth plate (VTP), population of parents and biological replicate number (sample), standard length (SL), total reads (generated by HiSeq2000), mapped reads (reads that mapped to the genome), final reads (excludes reads filtered due to low quality or PCR duplication), and unique reads (reads that mapped uniquely to one haplotype) is listed.
(XLSX)
Estimated gene expression change in cis in log2, PAXBFW vs marine. Name is the reported ENSEMBL gene name. Log2(F/M) is the log2 of the ratio of freshwater vs marine reads mapping uniquely to the gene.
(XLSX)
Estimated gene expression change in cis in log2, CERCFW vs marine. Name is the reported ENSEMBL gene name. Log2(F/M) is the log2 of the ratio of freshwater vs marine reads mapping uniquely to the gene.
(XLSX)
Acknowledgments
We thank Nikunj Donde for collecting a subset of tooth count data, and Michael Nachman and Dan Rokhsar for helpful discussions.
Data Availability
All sequencing reads are available on the Sequence Read Archive (SRP142616). All scripts used for analysis are available on GitHub (https://github.com/trahsemaj/Conv_Evo_Gene_Exp). A subset of the tooth count data (see S1 Table) is available from Ellis et al., 2015, Development, PMID 26062935
Funding Statement
This work was supported by National Institutes of Health R01 DE021475 (to CTM), National Institutes of Health genomics training grant 5T32HG000047-15 (to JCH), National Science Foundation Graduate Research Fellowship (to NAE), and an Howard Hughes Medical Institute investigator award (to MBE). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by National Institutes of Health S10 Instrumentation Grants S10RR029668 and S10RR027303. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27: 299–308. [DOI] [PubMed] [Google Scholar]
- 2.Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12: 283–293. doi: 10.1038/nrg2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- 4.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. The Quarterly Review of Biology. 1971;46: 111–138. [DOI] [PubMed] [Google Scholar]
- 5.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188: 107–116. [DOI] [PubMed] [Google Scholar]
- 6.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433: 481–487. doi: 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- 7.Manceau M, Domingues VS, Mallarino R, Hoekstra HE. The developmental role of agouti in color pattern evolution. Science. 2011;331: 1062–1065. doi: 10.1126/science.1200684 [DOI] [PubMed] [Google Scholar]
- 8.O’Brown NM, Summers BR, Jones FC, Brady SD, Kingsley DM. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. eLife. 2015;4: e05290 doi: 10.7554/eLife.05290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307: 1928–1933. doi: 10.1126/science.1107239 [DOI] [PubMed] [Google Scholar]
- 10.Indjeian VB, Kingman GA, Jones FC, Guenther CA, Grimwood J, Schmutz J, et al. Evolving new skeletal traits by cis-regulatory changes in bone morphogenetic proteins. Cell. 2016;164: 45–56. doi: 10.1016/j.cell.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schor IE, Degner JF, Harnett D, Cannavò E, Casale FP, Shim H, et al. Promoter shape varies across populations and affects promoter evolution and expression noise. Nature Genetics. 2017;49: 550 doi: 10.1038/ng.3791 [DOI] [PubMed] [Google Scholar]
- 12.McCoy RC, Wakefield J, Akey JM. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168: 916–927.e12. doi: 10.1016/j.cell.2017.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritter DI, Dong Z, Guo S, Chuang JH. Transcriptional enhancers in protein-coding rxons of vertebrate developmental genes. PLOS ONE. 2012;7: e35202 doi: 10.1371/journal.pone.0035202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430: 85–88. doi: 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- 15.Gibbons TC, Metzger DCH, Healy TM, Schulte PM. Gene expression plasticity in response to salinity acclimation in threespine stickleback ecotypes from different salinity habitats. Mol Ecol. 2017;26: 2711–2725. doi: 10.1111/mec.14065 [DOI] [PubMed] [Google Scholar]
- 16.Yao C, Joehanes R, Johnson AD, Huan T, Liu C, Freedman JE, et al. Dynamic role of trans regulation of gene expression in relation to complex traits. The American Journal of Human Genetics. 2017;100: 571–580. doi: 10.1016/j.ajhg.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack KL, Campbell P, Nachman MW. Gene regulation and speciation in house mice. Genome Res. 2016; gr.195743.115. doi: 10.1101/gr.195743.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goncalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, et al. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012;22: 2376–2384. doi: 10.1101/gr.142281.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20: 816–825. doi: 10.1101/gr.102491.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmon ZH, Bukowski R, Sun Q, Doebley JF. The role of cis regulatory evolution in maize domestication. PLOS Genet. 2014;10: e1004745 doi: 10.1371/journal.pgen.1004745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osada N, Miyagi R, Takahashi A. cis- and trans-regulatory effects on gene expression in a natural population of Drosophila melanogaster. Genetics. 2017; 117.201459. doi: 10.1534/genetics.117.201459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. Tempo and mode of regulatory evolution in Drosophila. Genome Res. 2014;24: 797–808. doi: 10.1101/gr.163014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peichel CL, Marques DA. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Phil Trans R Soc B. 2017;372: 20150486 doi: 10.1098/rstb.2015.0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard VL, Viitaniemi HM, McCairns RJS, Merilä J, Nikinmaa M, Primmer CR, et al. Regulatory architecture of gene expression variation in the threespine stickleback Gasterosteus aculeatus. G3. 2017;7: 165–178. doi: 10.1534/g3.116.033241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa A, Kusakabe M, Yoshida K, Ravinet M, Makino T, Toyoda A, et al. Different contributions of local- and distant-regulatory changes to transcriptome divergence between stickleback ecotypes. Evolution. 2017;71: 565–581. doi: 10.1111/evo.13175 [DOI] [PubMed] [Google Scholar]
- 26.Di Poi C, Bélanger D, Amyot M, Rogers S, Aubin-Horth N. Receptors rather than signals change in expression in four physiological regulatory networks during evolutionary divergence in threespine stickleback. Mol Ecol. 2016;25: 3416–3427. doi: 10.1111/mec.13690 [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Chain FJJ, Panchal M, Eizaguirre C, Kalbe M, Lenz TL, et al. Transcriptome profiling of immune tissues reveals habitat-specific gene expression between lake and river sticklebacks. Mol Ecol. 2016;25: 943–958. doi: 10.1111/mec.13520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell MA, Foster SA. The evolutionary biology of the threespine stickleback Oxford University Press; 1994. [Google Scholar]
- 29.Hagen DW, Gilbertson LG. Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America. Evolution. 1972;26: 32–51. doi: 10.1111/j.1558-5646.1972.tb00172.x [DOI] [PubMed] [Google Scholar]
- 30.Ellis NA, Glazer AM, Donde NN, Cleves PA, Agoglia RM, Miller CT. Distinct developmental genetic mechanisms underlie convergently evolved tooth gain in sticklebacks. Development. 2015;142: 2442–2451. doi: 10.1242/dev.124248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484: 55–61. doi: 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson PA, Glazer AM, Killingbeck EE, Agoglia RM, Baek J, Carsanaro SM, et al. Partially repeatable genetic basis of benthic adaptation in threespine sticklebacks: repeatable evolution in benthic sticklebacks. Evolution. 2016;70: 887–902. doi: 10.1111/evo.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327: 302–305. doi: 10.1126/science.1182213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428: 717 doi: 10.1038/nature02415 [DOI] [PubMed] [Google Scholar]
- 35.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262: 195–205. [DOI] [PubMed] [Google Scholar]
- 36.O’Connell DJ, Ho JWK, Mammoto T, Turbe-Doan A, O’Connell JT, Haseley PS, et al. A WNT-BMP feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci Signal. 2012;5: ra4 doi: 10.1126/scisignal.2002414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jernvall J, Thesleff I. Tooth shape formation and tooth renewal: evolving with the same signals. Development. 2012;139: 3487–3497. doi: 10.1242/dev.085084 [DOI] [PubMed] [Google Scholar]
- 38.Tucker AS, Fraser GJ. Evolution and developmental diversity of tooth regeneration. Seminars in Cell & Developmental Biology. doi: 10.1016/j.semcdb.2013.12.013 [DOI] [PubMed] [Google Scholar]
- 39.Bloomquist RF, Parnell NF, Phillips KA, Fowler TE, Yu TY, Sharpe PT, et al. Coevolutionary patterning of teeth and taste buds. PNAS. 2015;112: E5954–E5962. doi: 10.1073/pnas.1514298112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin KJ, Rasch LJ, Cooper RL, Metscher BD, Johanson Z, Fraser GJ. Sox2+ progenitors in sharks link taste development with the evolution of regenerative teeth from denticles. Proc Natl Acad Sci. 2016;113: 14769–14774. doi: 10.1073/pnas.1612354113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, et al. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. PNAS. 2014;111: 13912–13917. doi: 10.1073/pnas.1407567111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9: 357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20: 1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43: 491–498. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43: 11.10.1–33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sébastien Lê Julie Josse, Husson François. FactoMineR: an R package for multivariate analysis | Lê | Journal of Statistical Software. Journal of Statistical Software. 2008;25 Available: https://www.jstatsoft.org/article/view/v025i01 [Google Scholar]
- 47.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotech. 2013;31: 46–53. doi: 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gene expression in tooth (WWW database), http://bite-it.helsinki.fi. Developmental Biology Programme of the University of Helsink; 1996.
- 49.Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7: e1000031 doi: 10.1371/journal.pbio.1000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fraser GJ, Bloomquist RF, Streelman JT. Common developmental pathways link tooth shape to regeneration. Dev Biol. 2013;377: 399–414. doi: 10.1016/j.ydbio.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock DW. Zebrafish dentition in comparative context. J Exp Zool B Mol Dev Evol. 2007;308: 523–549. doi: 10.1002/jez.b.21187 [DOI] [PubMed] [Google Scholar]
- 52.Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274: 139–157. doi: 10.1016/j.ydbio.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 53.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28: 511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011; btr355. doi: 10.1093/bioinformatics/btr355 [DOI] [PubMed] [Google Scholar]
- 55.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biology. 2011;12: R22 doi: 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kandyba E, Leung Y, Chen Y-B, Widelitz R, Chuong C-M, Kobielak K. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci USA. 2013;110: 1351–1356. doi: 10.1073/pnas.1121312110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuoka A, Aihara Y, Matsumoto I, Abe K. Phospholipase C-beta 2 as a mammalian taste signaling marker is expressed in the multiple gustatory tissues of medaka fish, Oryzias latipes. Mechanisms of Development. 2004;121: 985–989. doi: 10.1016/j.mod.2004.03.009 [DOI] [PubMed] [Google Scholar]
- 58.Bachmanov AA, Beauchamp GK. Taste Receptor Genes. Annu Rev Nutr. 2007;27: 389–414. doi: 10.1146/annurev.nutr.26.061505.111329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends in Genetics. 2006;22: 456–461. doi: 10.1016/j.tig.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 60.Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40: 346–350. doi: 10.1038/ng.77 [DOI] [PubMed] [Google Scholar]
- 61.Shen SQ, Turro E, Corbo JC. Hybrid mice reveal parent-of-origin and cis- and trans-regulatory effects in the retina. PLOS ONE. 2014;9: e109382 doi: 10.1371/journal.pone.0109382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478: 343–348. doi: 10.1038/nature10532 [DOI] [PubMed] [Google Scholar]
- 63.Chen H, Capellini TD, Schoor M, Mortlock DP, Reddi AH, Kingsley DM. Heads, shoulders, elbows, knees, and toes: modular Gdf5 enhancers control different joints in the vertebrate skeleton. PLOS Genetics. 2016;12: e1006454 doi: 10.1371/journal.pgen.1006454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellis NA, Miller CT. Dissection and flat-mounting of the threespine stickleback branchial skeleton. Journal of Visualized Experiments. 2016; doi: 10.3791/54056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012; bts635. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18: 565–575. doi: 10.1177/0962280209351908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10: 48 doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLOS Comput Biol. 2007;3: e39 doi: 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE. 2011;6: e21800 doi: 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLOS Computational Biology. 2014;10: e1003537 doi: 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring Species Trees Directly from Biallelic Genetic Markers: Bypassing Gene Trees in a Full Coalescent Analysis. Mol Biol Evol. 2012;29: 1917–1932. doi: 10.1093/molbev/mss086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Genome-wide phylogeny created from genomic resequencing data. Wild-caught fish are non-italicized. All nodes have 100% posterior probability. Scale bar shows 3% sequence divergence at variant positions. (B) Principal component analysis of genome-wide genotypes separates marine and CERCFW populations from the PAXBFW lake population, with the 2nd PC separating marine and freshwater populations.
(TIF)
(A) PAXBFW upregulation of BiteCode genes (282 expressed orthologs, P = 9.8e-3, GSEA). (B) CERCFW upregulation of BiteCode genes (P = 2.1e-5, GSEA). (C) PAXBFW and CERCFW upregulation of BiteCode genes (P = 5.1e-6, GSEA).
(TIF)
(A-F) Changes in gene expression changes of genes annotated as components of the indicated signaling pathways (BMP, FGF, SHH, WNT, ACT, TGFB, NOTCH, or EDA, containing 59, 60, 28, 75, 19, 11, 12, and 6 expressed orthologs, respectively) [36] or orthologs of a described set of mouse hair follicle stem cell signature genes (HFSC, containing 254 expressed orthologs) [56]. Violin plots show the mean expression change of genes in the pathway. (A) Change in freshwater (PAXBFW + CERCFW) relative to marine. (B) PAXBFW specific changes (PAXBFW relative to CERCFW + marine). In the WNT and TGFB pathway, 22/75 and 6/11 genes had significantly increased expression respectively (C) CERCFW specific changes (CERCFW relative to PAXBFW + marine). (D) PAXBFW evolved changes (PAXBFW relative to marine) (E) CERCFW evolved changes (CERCFW relative to marine) (F) PAXBFW vs CERCFW changes (PAXBFW relative to CERCFW).
(TIF)
(A-C) GO enrichment of genes upregulated in freshwater (A), PAXBFW (B), or CERCFW (C). GO analysis was performed using Gorilla [68], with the results visualized with Revigo [70].
(TIF)
Expression levels of known taste bud marker genes in marine, PAXBFW and CERCFW tooth plates as assayed by RNA-seq. * indicates differentially expressed genes. Error bars are standard error of the mean.
(TIF)
(A-B) Proportion of genes with quantifiable (i.e. genes with transcripts containing a polymorphic SNP covered by at least 20 reads) hybrid expression displaying opposing and concordant cis and trans changes in PAXBFW (A) or CERCFW (B) dental tissue. Similar to Fig 5, but here showing all genes, not just genes with significantly different expression levels compared to marine. Trans regulatory changes predominate, as do opposing over concordant changes. (C) Density plot of the percentage of gene expression changes explained by cis-regulatory changes.
(TIF)
(A) Expression changes of genes with quantifiable (i.e. genes with transcripts containing a polymorphic SNP covered by at least 20 reads) hybrid expression in both freshwater populations relative to marine fish, showing significantly correlated changes in gene expression in PAXBFW and CERCFW tooth plates. (B) cis regulatory changes of genes with quantifiable hybrid expression in freshwater dental tissue overall do not display correlated evolved changes. (C) trans regulatory changes of genes with quantifiable hybrid expression in freshwater dental tissue. Density (color) was estimated with a Gaussian kernel density estimator. (D-F) Similar to A-C, but showing only genes in the BiteCode gene set, revealing that these orthologs have evolved highly convergent changes in the two freshwater populations (D), despite non-convergent cis regulatory changes (E).
(TIF)
For each fish, the population, ecotype (freshwater or marine), total ventral pharyngeal tooth number (TVTP), total length (TL), standard length (SL), and whether data has been published [30] is shown.
(XLSX)
For each fish, population and biological replicate number (Fish), the total number of barcoded reads from each fish (reads), and number of reads that mapped and passed all filters (final mapped) is listed.
(XLSX)
For each fish, population of parents and biological replicate number (sample), standard length (SL), total reads (generated by HiSeq2000 over two different runs (run1 and run2)), mapped reads (reads that mapped to the genome), and final reads (excludes reads filtered due to low quality or PCR duplication) is listed.
(XLSX)
Estimated abundance in in fragments per kilobases per million reads (FPKM) of ENSEMBL genes (rows) in ventral pharyngeal dental tissue from three individual fish from three populations (in columns). Mean expression (in FPKM) is shown after the 3 replicates. Log2(Pop1/Pop2) shows the fold-change in log2 of the estimated mean expression between the two populations. IsSig(Pop1/Pop2) indicates whether the difference was significant as reported by cuffdiff2.
(XLSX)
A list of stickleback orthologs in the BiteIt [48] (http://bite-it.helsinki.fi/) or ToothCODE (http://compbio.med.harvard.edu/ToothCODE/) [36] databases.
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in freshwater dental tissue reported by GOrilla [68].
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in PAXBFW dental tissue reported by GOrilla [68].
(XLSX)
Gene Ontology (GO) term category and name are given in GO term and description, with the p-value, q-value, and relative enrichment within genes upregulated in CERCFW dental tissue reported by GOrilla [68].
(XLSX)
For each ventral pharyngeal tooth plate (VTP), population of parents and biological replicate number (sample), standard length (SL), total reads (generated by HiSeq2000), mapped reads (reads that mapped to the genome), final reads (excludes reads filtered due to low quality or PCR duplication), and unique reads (reads that mapped uniquely to one haplotype) is listed.
(XLSX)
Estimated gene expression change in cis in log2, PAXBFW vs marine. Name is the reported ENSEMBL gene name. Log2(F/M) is the log2 of the ratio of freshwater vs marine reads mapping uniquely to the gene.
(XLSX)
Estimated gene expression change in cis in log2, CERCFW vs marine. Name is the reported ENSEMBL gene name. Log2(F/M) is the log2 of the ratio of freshwater vs marine reads mapping uniquely to the gene.
(XLSX)
Data Availability Statement
All sequencing reads are available on the Sequence Read Archive (SRP142616). All scripts used for analysis are available on GitHub (https://github.com/trahsemaj/Conv_Evo_Gene_Exp). A subset of the tooth count data (see S1 Table) is available from Ellis et al., 2015, Development, PMID 26062935