Abstract
The expression levels of estrogen receptor 1 (ESR1), arylamine N-acetyltransferase 1 (NAT1), and arylamine N-acetyltransferase 2 (NAT2) are implicated in breast cancer; however, their co-expression profiles in normal breast tissue, primary breast tumors and established breast cancer cell lines are undefined. NAT1 expression is widely reported to be associated with ESR1 expression and is frequently investigated in breast cancer etiology. Furthermore, the NAT2 phenotype has been reported to modify breast cancer risk in molecular epidemiological association studies. Understanding the relationships between the expression levels of these genes is essential to understand their role in breast cancer etiology and treatment. In the present study, NAT1, NAT2 and ESR1 expression data were accessed from repositories of RNA-Seq data covering 57 breast cancer cell lines, 1,043 primary breast tumors and 99 normal breast tissues. The relationships between gene expression, and between NAT1 activity and RNA expression in breast cancer cell lines were evaluated using non-parametric statistical analyses. Differences in gene expression in each dataset, as well as gene expression differences in normal breast tissue compared to primary breast tumors, and stratification by estrogen receptor status were determined. NAT1 and NAT2 mRNA expression were detected in normal and primary breast tumor tissues; NAT1 expression was much higher than NAT2. NAT1 and ESR1 expression were strongly associated, whereas NAT2 and ESR1 expression were not. Although NAT1 and NAT2 expression were associated, the magnitude was moderate. NAT1, NAT2, and ESR1 expression were increased in primary breast tumor tissue compared with normal breast tissue; however, the magnitude and significance of the differences were lower for NAT2. Analysis of NAT1, NAT2, and ESR1 expression in normal and primary breast tissues and breast cancer cell lines suggested that NAT1 and NAT2 expression are regulated by distinctive mechanisms, whereas NAT1 and ESR1 expression may have overlapping regulation. Defining these relationships is important for future investigations into breast cancer prevention.
Keywords: NAT1, NAT2, ESR1, breast cancer, TCGA, CCLE
Introduction
Human arylamine N-acetyltransferase 1 (NAT1) and arylamine N-acetyltransferase 2 (NAT2) are cytosolic phase II xenobiotic metabolizing isozymes, which catalyze the acetylation of a wide range of aromatic and heterocyclic amines via a ping-pong bi-bi reaction mechanism (1,2). This acetylation can ultimately lead to bioactivation and/or deactivation of various substrates, including breast cancer carcinogens (3–5). In addition to metabolizing xenobiotics, NAT1, but not NAT2, can catalyze the hydrolysis of acetyl-CoA using folate as a cofactor (6,7). NAT1 and NAT2 are encoded by two separate loci in close proximity on chromosome 8p22 (8,9) and each consist of an intronless open reading frame of 870 base pairs (10). Although NAT1 and NAT2 share ~87% nucleotide sequence identity and 81% deduced amino acid homology, they exhibit differing tissue localizations, and distinct but overlapping substrate specificities (11). In addition, NAT1 and NAT2 expression vary inter-individually from single nucleotide polymorphisms (SNPs) (2,12–17).
Although NAT1 and NAT2 catalyze N-acetylation, their roles in breast cancer etiology may differ. Numerous studies have investigated possible roles for NAT1 in breast cancer etiology and progression (18–24), given the association between increased expression of NAT1 and estrogen receptor (ER)-positive breast cancers (25–31). Notably, NAT1 expression is not directly regulated by estrogens or dihydrotestosterone (32), thus suggesting that there may be a common regulatory element between NAT1 and ESR1. Furthermore, congenic rats expressing higher NAT2 activity (orthologous to human NAT1) have been reported to exhibit greater carcinogen-induced mammary tumor susceptibility independent of carcinogen metabolism (22). In addition, SNPs in NAT2 have been well described and have been revealed to influence acetylation rates of many known carcinogens; an association between NAT2 genotype with breast cancer risk among smokers has been reported (33). Since NAT1 and NAT2 may have different roles in breast cancer, it is important to analyze relationships between the expression levels of these isozymes.
The mRNA expression levels of NAT1 and NAT2 have been detected by reverse transcription-polymerase chain reaction (RT-PCR) in human mammary tissue (34,35). NAT1 N-acetylation activity has been widely reported in normal breast tissue and breast tumor tissue (34,36–40), whereas NAT2 N-acetylation activity has not been observed as consistently; when NAT2 activity is observed the activity is much lower than NAT1 activity (34,38,39). In addition, since NAT1 and NAT2 have overlapping substrate specificities, activity studies of the two isozymes can be complex. For example, Deitz probed human mammary tissue samples for NAT1 and NAT2 activities with p-aminobenzoic acid (PABA; selective for NAT1) and sulfamethazine (SMZ; selective for NAT2), and reported that SMZ was acetylated by NAT1 at very low levels (40). By normalizing the SMZ N-acetylation activity to NAT1 activity, Deitz demonstrated that the SMZ N-acetylation activity was most likely catalyzed by NAT1 rather than NAT2. NAT1 and NAT2 activities have also been reported in rat mammary tissues (41).
Wakefield et al profiled NAT1 expression and activity in seven breast cancer cell lines (MCF-7, T47D, ZR-75-1, Cal51, MDA-MB-231, MDA-MB-437 and MDA-MB-453) and detected NAT1 mRNA expression and activity in all seven cell lines (28); however, NAT2 expression and activity were not investigated. In addition, NAT2 mRNA has been detected in MCF-7 breast cancer cells at very low levels (35); however, NAT1 was not measured at the same time preventing a direct comparison of expression between the two isozymes. Bradshaw et al detected NAT1 and NAT2 by western blotting in the ER-positive breast cancer cell line MCF-7; however, the expression levels were not compared between the two proteins (42).
Limited studies have investigated the expression profiles of these isozymes together in breast tissues. Based on limited data, it has been hypothesized that NAT2 expression is very low in breast tissue and negligible in comparison to NAT1 expression; however, previous investigations have not addressed this hypothesis rigorously or comprehensively. To gain a better understanding of the relationship between NAT1 and NAT2 in breast tissues the present study evaluated the RNA expression levels of each in breast cancer cell lines, breast tumor tissue, and normal breast tissue. In addition, this study evaluated the extent to which established breast cancer cell lines reflect the NAT expression profile observed in primary breast tumors and normal breast tissue. Since NAT1 and NAT2 are so similar in terms of sequence, structure and substrates, and the association between NAT1 and ESR1 has been well established, the present study also evaluated the relationship between NAT2 and ESR1 expression in breast tissues. Since inhibition of NAT1 activity is under investigation for breast cancer prevention and treatment, understanding the relationships between NAT1, NAT2 and ESR1 is of great importance.
Materials and methods
Acquisition of publicly available data from the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) data repositories
RNA expression (RNA-Seq) data for ESR1, NAT1 and NAT2 in established breast cancer cell lines were accessed on 8/11/17 (n=57) from the CCLE (43); RNA expression values were reported in reads per kilobase of transcript per million mapped reads (RPKM). A total of 15 breast cancer cell lines had no detectable NAT2 gene expression. Data from TCGA (44) for the breast invasive carcinoma (BRCA) cohort were accessed on 2/4/18 (primary breast tumor tissue, n=1,043; normal breast tissue, n=99) via FirebrowseR (45), an R client to the Broad Institute’s RESTful Firehose Pipeline; RNA expression values were reported in RNA-Seq by Expectation-Maximization (RSEM). A total of 59 of the breast tumor samples and seven of the normal tissue samples did not have gene expression data for NAT2.
Established breast cancer cell lines analyzed
The following breast cancer cell lines were analyzed in this study: AU565, BT-20, BT-474, BT-483, BT-549, CAL-120, CAL-148, CAL-51, CAL-85-1, CAMA-1, DU4475, EFM-19, EFM-192A, HCC1143, HCC1187, HCC1395, HCC1419, HCC1428, HCC1500, HCC1569, HCC1599, HCC1806, HCC1937, HCC1954, HCC202, HCC2157, HCC2218, HCC38, HCC70, HDQ-P1, HMC-1-8, HMEL, Hs 274.T, Hs 281.T, Hs 343.T, Hs 578.T, Hs 606.T, Hs 739.T, Hs 742.T, JIMT-1, KPL-1, MCF-7, MDA-MB-134-VI, MDA-MB-157, MDA-MB-175-VII, MDA-MB-231, MDA-MB-361, MDA-MB-415, MDA-MB-436, MDA-MB-453, MDA-MB-468, SK-BR-3, T-47D, UACC-812, UACC-893, ZR-75-1 and ZR-75-30.
Statistical analyses
Shapiro-Wilk tests were conducted to determine if the expression of the genes under study were approximately normally distributed. Significant evidence of departures from approximate normality was observed; therefore, non-parametric statistical techniques were employed for subsequent analyses. Spearman’s correlation was used to evaluate the RNA expression levels between gene pairs (i.e. ESR1 and NAT1, ESR1 and NAT2, NAT1 and NAT2). Differences in the mRNA expression levels of NAT1 and NAT2 in each dataset, and differences in RNA expression between primary breast tumor samples and normal breast tissue samples, were evaluated using the Wilcoxon rank-sum test; median values were compared to determine fold-differences.
RNA expression data for each gene were stratified by ER status (+ or −) as defined in the literature (46–48) for the CCLE data, or as determined by immunohistochemistry during sample collection and cataloging for TCGA data. Differences in gene expression following stratification were evaluated using Wilcoxon rank-sum tests for each gene; median values were compared to determine fold-differences. A total of 10 of the breast cancer cell lines had either conflicting or unknown ER status in the literature.
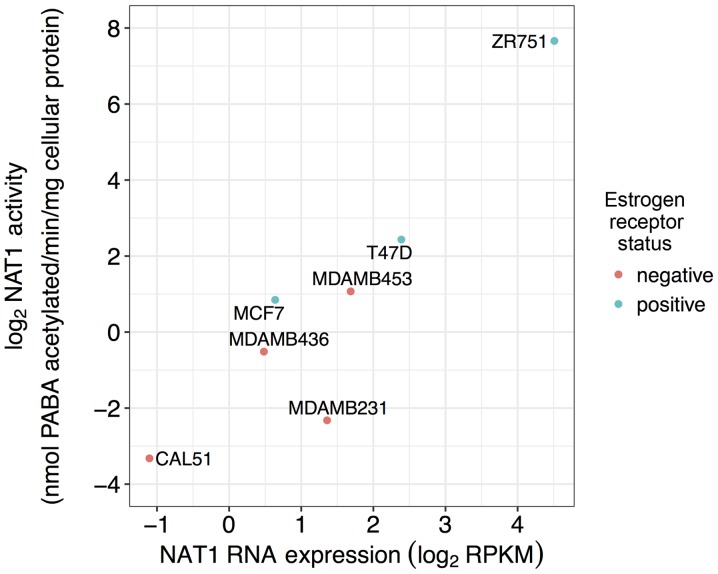
Wakefield et al published the NAT1 PABA N-acetylation activities of seven (ZR-75-1, T47D, MCF-7, MDA-MB-453, MDA-MB-436, MDA-MB-231 and CAL-51) of the 57 breast cancer cell lines included in the present study (28). The association between previously reported NAT1 activity and NAT1 RNA expression data for the same cell lines in the CCLE repository was evaluated. All statistical analyses were performed in R: A Language and Environment for Statistical Computing, version 3.4.2 (49).
Results
Association between NAT1 and ESR1, NAT2 and ESR1, and NAT1 and NAT2
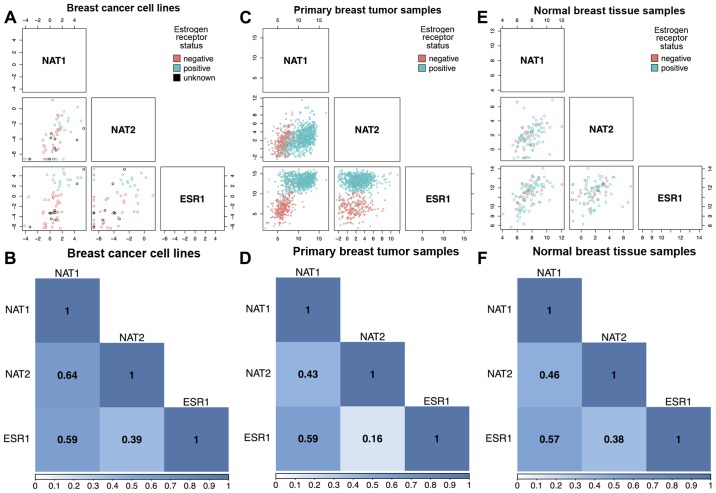
NAT1 RNA and ESR1 RNA were significantly correlated (P<0.0001 for all) at moderately high magnitudes in breast cancer cell lines (Spearman rho ρ=0.59; Fig. 1A and B), human primary breast tumors (ρ=0.59; Fig. 1C and D) and normal breast tissue (ρ=0.57; Fig. 1E and F). A significant (P<0.005 for all) association between ESR1 and NAT2 expression was observed, although the magnitude of the association was low and varied across datasets. The primary breast tumor dataset exhibited the weakest association (ρ=0.16; Fig. 1C and D), whereas the normal breast tissue (ρ=0.38; Fig. 1E and F) and breast cancer cell line (ρ=0.39; Fig. 1A and B) datasets exhibited similar, albeit low, association. Strong evidence of an association (P<0.0001 for all) between NAT1 RNA and NAT2 RNA levels was observed in all three datasets, with moderately high magnitude in the breast cancer cell lines (ρ=0.64; Fig. 1A and B). The primary breast tumor and normal breast tissue datasets exhibited interdependence similar to each other (ρ= 0.43 and 0.46, respectively; Figs. 1C–F); however, the association was lower than that observed in the breast cancer cell lines.
Figure 1.
Scatterplot and correlation matrices for NAT1, NAT2, and ESR1. Associations between NAT1, NAT2, and ESR1 RNA expression were analyzed in breast cancer cell lines, primary breast tumor tissue, and normal breast tissue using the Spearman method. In the scatterplot matrices, each open circle represents a single sample and is color-coded according to ER status; pink circles, ER− samples; blue circles, ER+ samples; black circles, samples with unknown ER status. In the association matrices, boxes are labeled with the Spearman correlation coefficient (ρ) for each comparison and color reflects strength of association; dark blue represents high association, light blue represents low association, and white represents no association. (A) Scatterplot matrix of the association between NAT1, NAT2 and ESR1 RNA expression in breast cancer cell lines (n=57). (B) Correlation matrix between NAT1, NAT2 and ESR1 RNA expression in breast cancer cell lines (n=57). (C) Scatterplot matrix of the association between NAT1, NAT2 and ESR1 RNA expression in primary breast tumor samples (n=1,043 for NAT1 vs. ESR1, n=984 for NAT1 vs. NAT2 and NAT2 vs. ESR1). (D) Correlation matrix between NAT1, NAT2, and ESR1 RNA expression in primary breast tumor samples (n=1,043 for NAT1 vs. ESR1, n=984 for NAT1 vs. NAT2 and NAT2 vs. ESR1). (E) Scatterplot matrix of the association between NAT1, NAT2 and ESR1 RNA expression in normal breast tissue samples (n=99 for NAT1 vs. ESR1, n=92 for NAT1 vs. NAT2 and NAT2 vs. ESR1). (F) Correlation matrix between NAT1, NAT2 and ESR1 RNA expression in normal breast tissue samples (n=99 for NAT1 vs. ESR1, n=92 for NAT1 vs. NAT2 and NAT2 vs. ESR1). ER, estrogen receptor; ESR1, estrogen receptor 1; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2.
Comparison of NAT1 and NAT2 expression
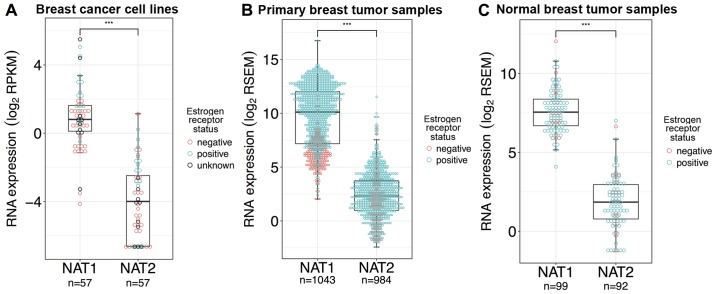
NAT1 RNA expression in breast cancer cell lines, primary breast tumors, and normal breast tissue was significantly higher compared with NAT2 expression by 33-, 222- and 52-fold, respectively (P<0.0001 for all; Fig. 2A–C). NAT1 expression was higher than NAT2 expression in all 57 breast cancer cell lines tested, with the exception of the UACC-893 cell line, which expressed the highest NAT2 RNA of any of the breast cancer cell lines analyzed. A total of 15 of the 57 breast cancer cell lines (MDA-MB-134-IV, CAL-120, DU4475, MCF-7, JIMT-1, Hs 281.T, KPL-1, Hs 606.T, HCC70, EFM-19, CAL-148, HCC1569, HMC-1-8, HCC1599 and HCC1395) had no reported NAT2 RNA expression, whereas all 57 reported NAT1 RNA expression. The KPL-1 breast cancer cell line has been reported to be contaminated/misidentified and to be an MCF-7 derivative (50).
Figure 2.
NAT1 and NAT2 RNA expression in breast cancer cell lines, primary breast tumor samples, and normal breast tissue samples. Differences in gene expression between NAT1 and NAT2 in breast cancer cell lines, primary breast tumor tissue and normal breast tissue were statistically evaluated by Wilcoxon rank-sum test; ***P<0.001. Each dot represents a single sample and is color-coded according to ER status; pink dots, ER− samples; blue dots, ER+ samples; black dots, samples with unknown ER status. In the boxplots, the solid black line represents the median, the upper hinge represents the 75th quartile and the lower hinge represents the 25th quartile. The upper whisker represents the largest observation less than or equal to the upper hinge + 1.5 x IQR, the lower whisker represents the smallest observation greater than or equal to the lower hinge - 1.5 x IQR. (A) NAT1 RNA expression was significantly higher than NAT2 RNA expression in the breast cancer cell lines. (B) NAT1 RNA expression was significantly higher than NAT2 RNA expression in the primary breast tumor samples. (C) NAT1 RNA expression was significantly higher than NAT2 RNA expression in the normal breast tissue samples. ER, estrogen receptor; IQR, interquartile range; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; RPKM, reads per kilobase of transcript per million mapped reads; RSEM, RNA-Seq by Expectation-Maximization.
In TCGA dataset, normal breast tissue samples were collected from patients in which primary breast tumor samples were also collected (but only for 99 individuals), allowing comparison of gene expression between normal breast tissue and primary tumor breast tissue within single individuals. In the primary breast tumor samples, only nine of the 984 samples had higher NAT2 RNA expression than NAT1; of those nine samples, two were ER+ and seven were ER−, and only one sample had a corresponding normal breast tissue sample. Notably, in that individual’s normal breast tissue sample, NAT2 RNA expression was not higher than NAT1 RNA expression. In the normal breast tissue samples, only one of the 92 samples had higher NAT2 RNA expression than NAT1; the corresponding primary breast tumor sample from the same patient had lower NAT2 than NAT1.
Comparison of gene expression between ER+ and ER− samples
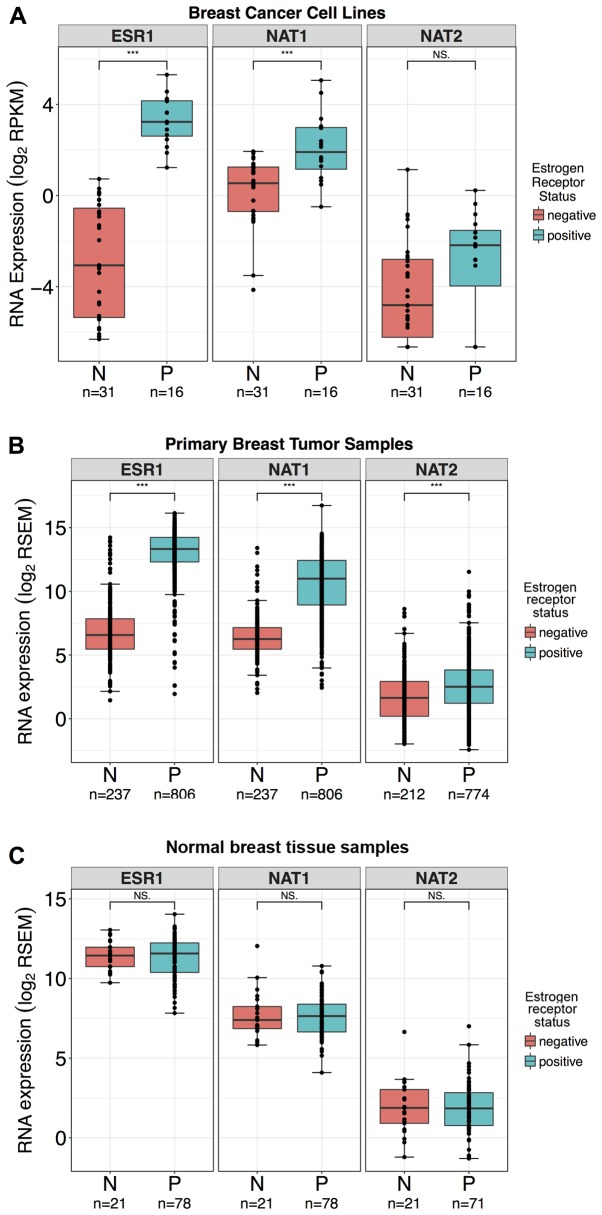
ESR1 and NAT1 gene expression were significantly increased, 86- and 2.6-fold, respectively, in ER+ breast cancer cell lines (P<0.0001 for both; Fig. 3A), whereas NAT2 gene expression did not significantly vary between ER+ and ER− breast cancer cell lines (P>0.05; Fig. 3A). Of the breast cancer cell lines with ER status defined in the literature (46–48), a connection between ESR1 RNA expression and the reported ER status has been observed. According to the literature, samples in the dataset with ESR1 RNA expression <1.7 RPKM were defined as ER−, whereas samples with ESR1 expression >2.3 RPKM were defined as ER+. The expression levels of all three genes were significantly higher in ER+ primary breast tumor samples (P<0.0001 for all; Fig. 3B); however. the fold-change between NAT2 expression in ER+ and ER− samples was smaller (1.8-fold difference) than for NAT1 and ESR1. In comparison, ESR1 and NAT1 were ~108- and 27-fold higher, respectively. The expression levels of genes were not significantly different between ER+ and ER− normal breast tissue samples (P>0.05 for all; Fig. 3C). Most of the breast cancer cell lines were ER−, whereas most of the primary breast tumor and normal breast tissue samples were ER+.
Figure 3.
ESR1, NAT1 and NAT2 RNA expression in breast cancer cell lines, primary breast tumor samples, and normal breast tissue stratified by ER status. Differences in the expression levels of ESR1, NAT1 and NAT2 genes in breast cancer cell lines, primary breast tumor tissue, and normal breast tissue stratified by ER status were evaluated by Wilcoxon rank-sum test; ***P<0.001; NS, not significant. Boxplots are color-coded according to ER status; pink boxplots, ER− samples; blue boxplots, ER+ samples. In the boxplots, the solid black line represents the median, the upper hinge represents the 75th quartile and the lower hinge represents the 25th quartile. The upper whisker represents the largest observation less than or equal to the upper hinge + 1.5 x IQR, the lower whisker represents the smallest observation greater than or equal to the lower hinge - 1.5 x IQR. (A) ESR1 and NAT1 RNA expression were significantly higher in ER+ breast cancer cell lines compared with ER− breast cancer cell lines. NAT2 RNA expression was not significantly different in ER+ breast cancer cell lines compared with ER− breast cancer cell lines. A total of 10 cell lines had either conflicting reports or no available data for ER status in the literature and were excluded from the analysis. (B) ESR1, NAT1 and NAT2 RNA expression were significantly higher in ER+ samples compared with ER− samples in the primary breast tumor dataset. (C) ESR1, NAT1 and NAT2 RNA expression levels were not significantly different in ER+ samples compared with ER− samples in the normal breast tissue dataset. ER, estrogen receptor; IQR, interquartile range; ESR1, estrogen receptor 1; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; RPKM, reads per kilobase of transcript per million mapped reads; RSEM, RNA-Seq by Expectation-Maximization.
Comparison of NAT1, NAT2, and ESR1 gene expression between normal breast tissue and primary breast tumors
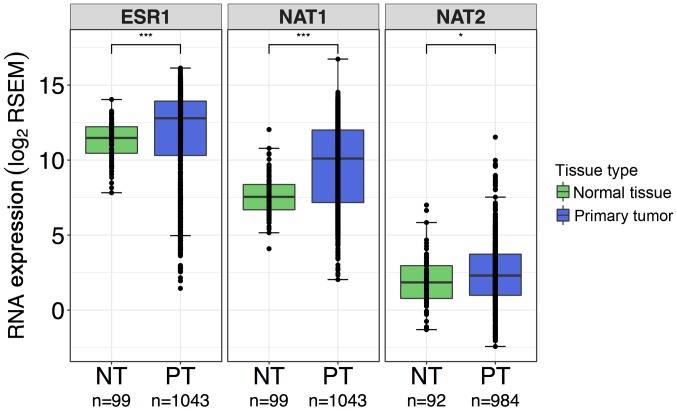
Differences in gene expression between normal breast tissue and primary breast tumor tissue were evaluated for each gene, ESR1, NAT1, and NAT2. More spread was observed in the primary breast tumor samples compared with the normal breast tissue samples for each gene. ESR1 and NAT1 gene expression were significantly elevated 2.5- and 5.9-fold, respectively, in primary breast tumor samples compared with normal breast tissue samples (P<0.0001 for both; Fig. 4). NAT2 expression was also significantly higher in primary breast tumor samples compared with normal breast tissue samples, but at a lower significance and fold-change (1.4-fold) than ESR1 and NAT1 (P<0.05; Fig. 4).
Figure 4.
Comparison of ESR1, NAT1 and NAT2 RNA expression in normal breast tissue and primary breast tumor samples. Differences in gene expression of ESR1, NAT1 and NAT2 in normal breast tissue and primary breast tumor tissue were evaluated by Wilcoxon rank-sum test; ***P<0.001; *P<0.05. Boxplots are color-coded according to tissue type; green boxplots, normal breast tissue samples; blue boxplots, primary breast tumor samples. In the boxplots, the solid black line represents the median, the upper hinge represents the 75th quartile and the lower hinge represents the 25th quartile. The upper whisker represents the largest observation less than or equal to the upper hinge + 1.5 x IQR, the lower whisker represents the smallest observation greater than or equal to the lower hinge - 1.5 * IQR. For all genes, more spread was observed in data from the primary breast tumor samples compared with the normal breast tissue samples. ESR1 and NAT1 gene expression were significantly elevated in primary tumor tissue compared with normal breast tissue. NAT2 expression was also significantly higher in primary tumor tissue compared with normal breast tissue, but at a lower significance than ESR1 and NAT1. IQR, interquartile range; ESR1, estrogen receptor 1; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; RSEM, RNA-Seq by Expectation-Maximization.
Relationship between previously reported NAT1 N-acetylation activity and NAT1 RNA expression
NAT1 N-acetylation activity previously reported in the literature and NAT1 RNA expression in seven of the 57 breast cancer cell lines were significantly associated (P<0.05) with a high magnitude (ρ=0.89; Fig. 5).
Figure 5.
Association between NAT1 RNA expression and previously reported NAT1 N-acetylation activity in seven established breast cancer cell lines. NAT1 RNA expression from Cancer Cell Line Encyclopedia and previously reported NAT1 N-acetylation activity (28) in seven breast cancer cell lines were significantly associated (P<0.05; ρ=0.89). Dots represent a single cell line and are color-coded according to ER status: Pink dots, ER− samples; blue dots, ER+ samples. ER, estrogen receptor; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; PABA, p-aminobenzoic acid; RPKM, reads per kilobase of transcript per million mapped reads.
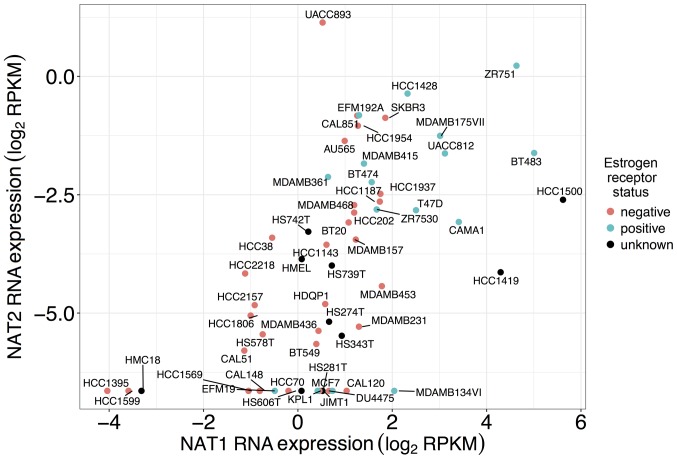
Co-expression of NAT1 and NAT2 RNA expression in established breast cancer cell lines
Co-expression profiles of NAT1 and NAT2 RNA for each established breast cancer cell line included in this study are presented in Fig. 6. Of all the cell lines included in the present study, the UACC-893 cell line expressed the highest level of NAT2 RNA, whereas the HCC1500 cell line expressed the highest level of NAT1 RNA. The ZR-75-1 cell line expressed high levels of both NAT1 and NAT2 RNA, whereas the HCC1395 cell line expressed low levels of both.
Figure 6.
NAT1 and NAT2 association in breast cancer cell lines. Association between NAT1 and NAT2 RNA expression was analyzed in breast cancer cell lines (each labeled in this figure). Each dot represents a single breast cancer cell line and is color-coded according to ER status; pink dots, ER− samples; blue dots, ER+ samples; black dots, samples with unknown or conflicting ER status in the literature (46–48). NAT1 and NAT2 RNA expression was significantly associated in breast cancer cell lines (P<0.0001, ρ= 0.64).
Discussion
The present study analyzed established breast cancer cell lines and samples from patients with breast cancer to evaluate the extent to which breast cancer cell lines serve as appropriate models for NAT1, NAT2 and ESR1 expression in breast tumors. Overall, the present findings demonstrated a strong association between NAT1 and ESR1 expression, which is in agreement with previous reports that NAT1 and ESR1 are positively associated (25–31), and this association was observed in all three sample types at approximately the same magnitude. These findings suggested that breast cancer cell lines may accurately reflect this relationship and provide a useful model for further research into the relationship. It is well known that ESR1 expression is frequently altered in breast cancer; therefore, the decrease in association between NAT2 and ESR1 in primary breast tumors compared with normal breast tissue samples and established breast cancer cell lines may be due to more dysregulation of ESR1 than NAT2 in primary breast tumors. The results of an analysis between NAT2 and ESR1 expression suggested that, while NAT2 and ESR1 are associated, the magnitude is low.
Interdependence between NAT1 and NAT2 expression was moderately high in the breast cancer cell line dataset, but substantially lower in the primary breast tumor and normal breast tissue datasets. Additionally, the strength of the association between NAT1 and NAT2 in the breast cancer cell line dataset was similar to the strength of the association observed between NAT1 and ESR1 in that dataset; however, in the primary breast tumor and normal breast tissue datasets, the association between NAT1 and NAT2 was lower. These findings suggested that breast cancer cell lines may over-represent the interdependence between NAT1 and NAT2, and not fully replicate the relationship observed in primary breast tumors or normal breast tissue.
In the breast cancer cell line data there appears to be a cut-off (between 1.7 and 2.3 RPKM) linking ESR1 RNA expression and the reported ER status of the breast cancer cell lines. This may provide a method to predict the ER status of breast cancer cell lines that currently have conflicting or unknown ER status in the literature. Using that method, it may be predicted that the HCC1500 and HCC1419 cell lines are ER+, whereas the HMC-1-8, Hs 742.T, Hs 343.T, Hs 739.T, HMEL, Hs 274.T, Hs 281.T and Hs 606.T cell lines are ER−. Notably, although 67–82% of breast cancers are ER+ (51) and most of the primary breast tumor samples were ER+, the majority of established breast cancer cell lines are ER−.
NAT1 and NAT2 RNA expression were reported in almost all samples included in the present study, which concurs with previously published results that have detected NAT1 and NAT2 mRNA by RT-PCR in human mammary tissue in smaller cohorts (34–36). NAT1 RNA expression was significantly higher than NAT2 RNA expression in the breast cancer cell lines, primary breast tumor samples and normal breast tissue. In addition, with only a few exceptions, NAT1 RNA expression was always higher than NAT2 RNA expression in matched samples from the breast cancer cell line, primary breast tumor sample and normal breast tissue sample datasets, thus supporting previous findings that indicated NAT1 transcripts were 2- to 3-fold higher than NAT2 transcripts in human mammary tissues (52). The UACC-893 cell line, the only breast cancer cell line observed in this study to express higher NAT2 RNA than NAT1 RNA, is an ER− and progesterone receptor-negative cell line that has a ~20-fold amplification of the human epidermal growth factor receptor 2/neu oncogene sequence. Further study of this cell line may aid in the identification of additional regulatory mechanisms of NAT1 and/or NAT2, since it expresses a unique profile of NAT1 and NAT2 compared with the other breast cancer cell lines.
While NAT1 expression was reported in all 57 breast cancer cell lines in the present study, 15 of those breast cancer cell lines had no reported NAT2 RNA expression (Fig. 6). The cell lines with no detected NAT2 RNA are plotted at ~−6.6 log2 RPKM NAT2. One of those 15 cell lines, MCF-7, has previously been reported to express NAT2 RNA expression (35,53) albeit at very low levels. One reason for the difference in observation between this study and the previous studies may be that the detection threshold for NAT2 was higher when measured by RNA-Seq for the CCLE dataset than in the previous studies. Additionally, in the previous studies that detected NAT2 RNA in the MCF-7 breast cancer cell line, NAT1 RNA was not measured at the same time; therefore, direct comparisons of the isozymes was not possible. To the best of our knowledge, NAT2 RNA expression has not been investigated in any of the other 56 breast cancer cell lines until this study. The results of this study indicated that NAT2 may be expressed in breast tissues and expression should be considered when studying NAT1, due to their overlapping substrate specificities and the high degree of structural similarity.
In normal breast tissue samples no significant difference in gene expression for ESR1, NAT1 and NAT2 was observed when data was stratified by ER status. However, in the primary breast tumor samples and in the breast cancer cell lines, ESR1 and NAT1 exhibited increased expression in the ER+ samples compared with in the ER− samples. NAT2 RNA expression did not significantly vary in breast cancer cell lines when comparing ER+ and ER− samples, but was significantly increased in ER+ primary breast tumor samples compared with in ER− primary breast tumor samples, although the difference was small. This finding suggested that the dysregulation of NAT1 and ESR1 during tumorigenesis may share similar mechanisms; however, NAT2 does not.
ESR1, NAT1 and NAT2 RNA expression were each increased in primary breast tumor samples compared with normal breast tissue samples although the significance and fold-change of NAT2 were smaller than that of ESR1 and NAT1. Additionally, for all genes, more widely spread expression was observed in the primary breast tumor samples compared with normal breast tissue. These data suggested that expression of all three genes may become modified during breast cancer tumorigenesis; however, the expression of NAT1 and ESR1 appear to be dysregulated to a greater extent. As recently reviewed (54), the role of NAT2 in breast cancer etiology is considered to be due to its effects on carcinogen metabolism. The present study suggested that the role of NAT2 in breast cancer is less likely a product of cell transformation, as the expression levels of NAT2 between normal and tumor tissues exhibited smaller variance than the expression levels of NAT1 and ESR1.
NAT1 N-acetylation activity has been reported in normal breast tissue and breast tumor tissue (34,36–40), whereas NAT2 N-acetylation activity has not been observed as consistently; when NAT2 activity is observed the activity is much lower than that of NAT1 activity (34,38,39). Wakefield et al profiled NAT1 expression and activity in seven breast cancer cell lines (MCF-7, T47D, ZR-75-1, Cal51, MDA-MB-231, MDA-MB-437 and MDA-MB-453); NAT1 mRNA and activity was observed in all seven cell lines (28); however, NAT2 expression and activity were not co-investigated. The high degree of association between the previously reported NAT1 N-acetylation activity and the NAT1 RNA expression of the same seven breast cancer cell lines suggested that NAT1 RNA expression is highly reflective of NAT1 N-acetylation activity. Gene expression is not always predictive of enzyme activity, due to the numerous regulatory mechanisms that can occur between RNA expression and protein function; however, these results suggested that RNA expression of NAT1 may serve as an appropriate predictor of NAT1 N-acetylation activity. Further studies with an increased number of breast cancer cell lines in which NAT1 N-acetylation activity has been measured are required to confirm this hypothesis. Additionally, further studies are required to determine association between NAT2 RNA expression and NAT2 N-acetylation activity.
The CCLE and TCGA repositories offer a wealth of publicly available data. The present study utilized this data to analyze and annotate the previously undefined relationships between NAT1, NAT2 and ESR1 in breast cancer cell lines, primary breast tumors and normal breast tissue. The results demonstrated that NAT1 and NAT2 RNA were expressed in normal breast tissue and primary breast tumor tissue; however, NAT1 RNA expression was much higher than NAT2. The expression of NAT1 and NAT2 were found to be associated; however, the magnitude was lower than that observed between NAT1 and ESR1 in the primary breast tumors and normal breast tissue. Additionally, although the association between NAT1 and NAT2 was slightly exaggerated in the breast cancer cell lines dataset, the cell lines generally reflected the NAT1 and NAT2 expression profiles of the primary breast tumors investigated. The present study demonstrated that while NAT1 and ESR1 expression were moderately associated in all datasets included in this study, NAT2 and ESR1 expression were associated at a lower magnitude, particularly in the primary breast tumor samples.
NAT1 and ESR1 expression were increased in primary breast tumor samples compared with normal breast tissue samples, and were increased in ER+ primary breast tumors compared with ER− primary breast tumors. NAT2 expression was slightly increased in primary breast tumor samples compared with normal breast tissue samples and in ER+ primary breast tumors compared with ER− primary breast tumors. Although NAT1 and NAT2 are both implicated in breast cancer, the majority of previous breast cancer studies have investigated each isozyme individually. The present study suggested that both isozymes should be considered in each study, since both are expressed in breast tissues. Defining the association between NAT1, NAT2 and ESR1 is of great importance, as modification of NAT1 is currently being studied for breast cancer prevention (20,21,55,56).
Acknowledgments
The results published here are in whole or part based upon data generated by TCGA, managed by the National Cancer Institute and the National Human Genome Research Institute, and the CCLE, a collaboration between the Broad Institute and the Novartis Institutes for Biomedical Research and its Genomics Institute of the Novartis Research Foundation. Information about TCGA and CCLE can be found at http://cancergenome.nih.gov and https://portals.broadinstitute.org/ccle, respectively. Those who carried out the original analysis and collection of the TCGA and CCLE data bear no responsibility for the further analysis or interpretation of it presented in this manuscript. Results of this study represent partial fulfillment of Samantha M. Carlisle’s PhD in Pharmacology and Toxicology from the University of Louisville. The authors thank Mr. Patrick J. Trainor, MS, MA (Department of Medicine, University of Louisville, Louisville, KY, USA) for his careful review of the manuscript and insights.
Abbreviations
- NAT1
arylamine N-acetyltransferase 1
- NAT2
arylamine N-acetyltransferase 2
- SNPs
single nucleotide polymorphisms
- ESR1
estrogen receptor 1
- CCLE
Cancer Cell Line Encyclopedia
- TCGA
The Cancer Genome Atlas
- ER
estrogen receptor
- SMZ
sulfamethazine
- PABA
p-aminobenzoic acid
- RPKM
reads per kilobase of transcript per million mapped reads
- RSEM
RNA-Seq by Expectation-Maximization
- ρ
Spearman correlation coefficient
Funding
The present study was partially supported by US Public Health Service grants (grant nos. T32-ES011564 and R25-CA134283).
Availability of data and materials
The CCLE data have been deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) using accession number GSE36139 and are also available at http://www.broadinstitute.org/ccle. TCGA data portal can be accessed at https://portal.gdc.cancer.gov/ or via FirebrowseR, an R client to the Broad Institute’s RESTful Firehose Pipeline.
Authors’ contributions
SMC designed the study, retrieved and analyzed all data, and prepared all figures in partial fulfilment of her PhD dissertation carried out under the direction of DWH. Both authors drafted the manuscript. DWH reviewed, modified and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hein DW. Molecular genetics and function of NAT1 and NAT2: Role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002:506–507. 65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 2.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 3.Bendaly J, Zhao S, Neale JR, Metry KJ, Doll MA, States JC, Pierce WM, Jr, Hein DW. 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline-induced DNA adduct formation and mutagenesis in DNA repair-deficient Chinese hamster ovary cells expressing human cytochrome 4501A1 and rapid or slow acetylator N-acetyltransferase 2. Cancer Epidemiol Biomarkers Prev. 2007;16:1503–1509. doi: 10.1158/1055-9965.EPI-07-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millner LM, Doll MA, Cai J, States JC, Hein DW. NATb/NAT1*4 promotes greater arylamine N-acetyltransferase 1 mediated DNA adducts and mutations than NATa/NAT1*4 following exposure to 4-aminobiphenyl. Mol Carcinog. 2012;51:636–646. doi: 10.1002/mc.20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hein DW. Acetylator genotype and arylamine-induced carcinogenesis. Biochim Biophys Acta. 1988;948:37–66. doi: 10.1016/0304-419x(88)90004-2. [DOI] [PubMed] [Google Scholar]
- 6.Stepp MW, Mamaliga G, Doll MA, States JC, Hein DW. Folate-dependent hydrolysis of acetyl-Coenzyme A by recombinant human and rodent arylamine N-acetyltransferases. Biochem Biophys Rep. 2015;3:45–50. doi: 10.1016/j.bbrep.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurieri N, Dairou J, Egleton JE, Stanley LA, Russell AJ, Dupret JM, Sim E, Rodrigues-Lima F. From arylamine N-acetyltransferase to folate-dependent acetyl CoA hydrolase: Impact of folic acid on the activity of (HUMAN)NAT1 and its homologue (MOUSE)NAT2. PLoS One. 2014;9:e96370. doi: 10.1371/journal.pone.0096370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matas N, Thygesen P, Stacey M, Risch A, Sim E. Mapping AAC1, AAC2 and AACP, the genes for arylamine N-acetyltransferases, carcinogen metabolising enzymes on human chromosome 8p22, a region frequently deleted in tumours. Cytogenet Cell Genet. 1997;77:290–295. doi: 10.1159/000134601. [DOI] [PubMed] [Google Scholar]
- 9.Hickman D, Risch A, Buckle V, Spurr NK, Jeremiah SJ, McCarthy A, Sim E. Chromosomal localization of human genes for arylamine N-acetyltransferase. Biochem J. 1994;297:441–445. doi: 10.1042/bj2970441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant DM, Blum M, Demierre A, Meyer UA. Nucleotide sequence of an intronless gene for a human arylamine N-acetyltransferase related to polymorphic drug acetylation. Nucleic Acids Res. 1989;17:3978. doi: 10.1093/nar/17.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamura A, Graham J, Mushtaq A, Tsiftsoglou SA, Vath GM, Hanna PE, Wagner CR, Sim E. Eukaryotic arylamine N-acetyltransferase. Investigation of substrate specificity by high-throughput screening. Biochem Pharmacol. 2005;69:347–359. doi: 10.1016/j.bcp.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Vatsis KP, Weber WW. Structural heterogeneity of Caucasian N-acetyltransferase at the NAT1 gene locus. Arch Biochem Biophys. 1993;301:71–76. doi: 10.1006/abbi.1993.1116. [DOI] [PubMed] [Google Scholar]
- 13.Weber WW, Vatsis KP. Individual variability in p-aminobenzoic acid N-acetylation by human N-acetyltransferase (NAT1) of peripheral blood. Pharmacogenetics. 1993;3:209–212. doi: 10.1097/00008571-199308000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Grant DM, Hughes NC, Janezic SA, Goodfellow GH, Chen HJ, Gaedigk A, Yu VL, Grewal R. Human acetyltransferase polymorphisms. Mutat Res. 1997;376:61–70. doi: 10.1016/S0027-5107(97)00026-2. [DOI] [PubMed] [Google Scholar]
- 15.Cloete R, Akurugu WA, Werely CJ, van Helden PD, Christoffels A. Structural and functional effects of nucleotide variation on the human TB drug metabolizing enzyme arylamine N-acetyltransferase 1. J Mol Graph Model. 2017;75:330–339. doi: 10.1016/j.jmgm.2017.04.026. [DOI] [PubMed] [Google Scholar]
- 16.McDonagh EM, Boukouvala S, Aklillu E, Hein DW, Altman RB, Klein TE. PharmGKB summary: Very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics. 2014;24:409–425. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butcher NJ, Boukouvala S, Sim E, Minchin RF. Pharmacogenetics of the arylamine N-acetyltransferases. Pharmacogenomics J. 2002;2:30–42. doi: 10.1038/sj.tpj.6500053. [DOI] [PubMed] [Google Scholar]
- 18.Tiang JM, Butcher NJ, Minchin RF. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015;4:565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiang JM, Butcher NJ, Cullinane C, Humbert PO, Minchin RF. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One. 2011;6:e17031. doi: 10.1371/journal.pone.0017031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiang JM, Butcher NJ, Minchin RF. Small molecule inhibition of arylamine N-acetyltransferase Type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun. 2010;393:95–100. doi: 10.1016/j.bbrc.2010.01.087. [DOI] [PubMed] [Google Scholar]
- 21.Stepp MW, Doll MA, Carlisle SM, States JC, Hein DW. Genetic and small molecule inhibition of arylamine N-acetyltransferase 1 reduces anchorage-independent growth in human breast cancer cell line MDA-MB-231. Mol Carcinog. 2018;57:549–558. doi: 10.1002/mc.22779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stepp MW, Doll MA, Samuelson DJ, Sanders MA, States JC, Hein DW. Congenic rats with higher arylamine N-acetyltransferase 2 activity exhibit greater carcinogen-induced mammary tumor susceptibility independent of carcinogen metabolism. BMC Cancer. 2017;17:233. doi: 10.1186/s12885-017-3221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlisle SM, Trainor PJ, Yin X, Doll MA, Stepp MW, States JC, Zhang X, Hein DW. Untargeted polar metabolomics of transformed MDA-MB-231 breast cancer cells expressing varying levels of human arylamine N-acetyltransferase 1. Metabolomics. 2016;12:111. doi: 10.1007/s11306-016-1056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witham KL, Minchin RF, Butcher NJ. Role for human arylamine N-acetyltransferase 1 in the methionine salvage pathway. Biochem Pharmacol. 2017;125:93–100. doi: 10.1016/j.bcp.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, Langerød A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D, Børresen-Dale AL. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)70933-8. [DOI] [PubMed] [Google Scholar]
- 28.Wakefield L, Robinson J, Long H, Ibbitt JC, Cooke S, Hurst HC, Sim E. Arylamine N-acetyltransferase 1 expression in breast cancer cell lines: A potential marker in estrogen receptor-positive tumors. Genes Chromosomes Cancer. 2008;47:118–126. doi: 10.1002/gcc.20512. [DOI] [PubMed] [Google Scholar]
- 29.Tozlu S, Girault I, Vacher S, Vendrell J, Andrieu C, Spyratos F, Cohen P, Lidereau R, Bieche I. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 30.Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bièche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:R252–R263. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Carlisle SM, Doll MA, Martin RCG, States JC, Klinge CM, Hein DW. High N-acetyltransferase 1 expression is associated with estrogen receptor expression in breast tumors, but is not under direct regulation by estradiol, 5α-androstane-3β,17β-diol, or dihydrotestosterone in breast cancer cells. J Pharmacol Exp Ther. 2018;365:84–93. doi: 10.1124/jpet.117.247031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrosone CB, Kropp S, Yang J, Yao S, Shields PG, Chang-Claude J. Cigarette smoking, N-acetyltransferase 2 genotypes, and breast cancer risk: Pooled analysis and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:15–26. doi: 10.1158/1055-9965.EPI-07-0598. [DOI] [PubMed] [Google Scholar]
- 34.Sadrieh N, Davis CD, Snyderwine EG. N-acetyltransferase expression and metabolic activation of the food-derived heterocyclic amines in the human mammary gland. Cancer Res. 1996;56:2683–2687. [PubMed] [Google Scholar]
- 35.Husain A, Zhang X, Doll MA, States JC, Barker DF, Hein DW. Identification of N-acetyltransferase 2 (NAT2) transcription start sites and quantitation of NAT2-specific mRNA in human tissues. Drug Metab Dispos. 2007;35:721–727. doi: 10.1124/dmd.106.014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Chung JG, Lai JM, Levy GN, Weber WW. Kinetics of arylamine N-acetyltransferase in tissues from human breast cancer. Cancer Lett. 1997;111:39–50. doi: 10.1016/S0304-3835(96)04491-6. [DOI] [PubMed] [Google Scholar]
- 37.Husain A, Barker DF, States JC, Doll MA, Hein DW. Identification of the major promoter and non-coding exons of the human arylamine N-acetyltransferase 1 gene (NAT1) Pharmacogenetics. 2004;14:397–406. doi: 10.1097/01.fpc.0000114755.08559.6e. [DOI] [PubMed] [Google Scholar]
- 38.Geylan YS, Dizbay S, Güray T. Arylamine N-acetyltransferase activities in human breast cancer tissues. Neoplasma. 2001;48:108–111. [PubMed] [Google Scholar]
- 39.Geylan-Su YS, Isgör B, Coban T, Kapucuoglu N, Aydintug S, Iscan M, Iscan M, Güray T. Comparison of NAT1, NAT2 and GSTT2-2 activities in normal and neoplastic human breast tissues. Neoplasma. 2006;53:73–78. [PubMed] [Google Scholar]
- 40.Deitz AC. PhD dissertation. University of North Dakota; 1999. N-acetyltransferase genetic polymorphisms and breast cancer risk. [Google Scholar]
- 41.Jefferson FA, Xiao GH, Hein DW. 4-Aminobiphenyl downregulation of NAT2 acetylator genotype-dependent N- and O-acetylation of aromatic and heterocyclic amine carcinogens in primary mammary epithelial cell cultures from rapid and slow acetylator rats. Toxicol Sci. 2009;107:293–297. doi: 10.1093/toxsci/kfn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradshaw TD, Chua MS, Orr S, Matthews CS, Stevens MF. Mechanisms of acquired resistance to 2-(4-aminophenyl)benzothiazole (CJM 126, NSC 34445) Br J Cancer. 2000;83:270–277. doi: 10.1054/bjoc.2000.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng M, Bragelmann J, Kryukov I, Saraiva-Agostinho N, Perner S. FirebrowseR: an R client to the Broad Institute’s Firehose Pipeline. Database (Oxford) 2017. 2017:baw160. doi: 10.1093/database/baw160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.American Type Culture Collection (ATCC©) Breast cancer and normal cell lines. 2013 https://www.atcc.org/~/media/PDFs/Cancer%20and%20Normal%20cell%20lines%20tables/Breast%20cancer%20and%20normal%20cell%20lines.ashx. Accessed Feb 2, 2018.
- 48.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. http://www.R-project.org/. Accessed Dec 15, 2017. [Google Scholar]
- 50.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 51.Thorpe SM. Estrogen and progesterone receptor determinations in breast cancer. Technology, biology and clinical significance Acta Oncol. 1988;27:1–19. doi: 10.3109/02841868809090312. [DOI] [PubMed] [Google Scholar]
- 52.Williams JA, Stone EM, Fakis G, Johnson N, Cordell JA, Meinl W, Glatt H, Sim E, Phillips DH. N-Acetyltransferases, sulfotransferases and heterocyclic amine activation in the breast. Pharmacogenetics. 2001;11:373–388. doi: 10.1097/00008571-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 53.AbuHammad S, Zihlif M. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics. 2013;101:213–220. doi: 10.1016/j.ygeno.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Hein DW. N-acetyltransferase 2 polymorphism and human urinary bladder and breast cancer risks. In: Laurieri N, Sim E, editors. Arylamine N-Acetyltransferases in Health and Disease: From Pharmacogenetics to Drug Discovery and Diagnostics. World Scientific Publishing; Singapore: 2018. pp. 327–349. [DOI] [Google Scholar]
- 55.Butcher NJ, Minchin RF. Arylamine N-acetyltransferase 1: A novel drug target in cancer development. Pharmacol Rev. 2012;64:147–165. doi: 10.1124/pr.110.004275. [DOI] [PubMed] [Google Scholar]
- 56.Laurieri N, Egleton JE, Russell AJ. Human N-acetyltransferase type 1 and breast cancer. In: Laurieri N, Sim E, editors. Arylamine N-Acetyltransferases in Health and Disease: From Pharmacogenetics to Drug Discovery and Diagnostics. World Scientific Publishing; Singapore: 2018. pp. 351–384. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CCLE data have been deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) using accession number GSE36139 and are also available at http://www.broadinstitute.org/ccle. TCGA data portal can be accessed at https://portal.gdc.cancer.gov/ or via FirebrowseR, an R client to the Broad Institute’s RESTful Firehose Pipeline.