Abstract
Acetylation of nucleosomal histones is a major regulatory step during activation of eukaryotic gene expression. Among the known acetyltransferase (AT) families, the structure–function relationship of the GNAT superfamily is the most well understood. In contrast, less information is available regarding mechanistic and regulatory aspects of p300/CBP AT function. In this paper, we investigate in closer detail the structure and sequence requirements for p300/CBP enzymatic activity. Unexpectedly, we find that the PHD finger of p300, but not of CBP, is dispensable for AT activity. In order to identify residues involved in substrate or acetyl-coenzyme A (acetyl-CoA) recognition, we have introduced 19 different amino acid substitutions in segments that are highly conserved between animal and plant p300/CBP proteins. By performing acetylation reactions with histones, a p53 peptide or the AT domain itself, we define several residues required for histone and p53 substrate recruitment but not for acetyl-CoA binding. Finally, we show that identical mutations in the p300 and CBP AT domain impair AT activity differently. This latter result combined with the finding of a differential requirement for the PHD finger provides evidence for structural differences between p300 and CBP that may in part underlie a previously reported functional specialization of the two proteins.
INTRODUCTION
Gene activation in eukaryotes is regulated at multiple levels. One regulatory level is acetylation of core histones, which frequently accompanies transcriptionally active chromosomal loci (1). Histone acetylation alone, however, is often not sufficient for gene activation (2,3). The cloning of nuclear histone acetyltransferases (HATs) allowed investigators to directly assess and dissect the contribution of each enzyme to transcriptional activation (4). Nuclear and cytoplasmic HATs (5) catalyze the transfer of an acetyl group from acetyl-coenzyme A (acetyl-CoA) to the ɛ-amino group of specific lysine residues in the N-terminal tails of core histones. This modification leads to the partial neutralization of the positively charged histone tail and is believed to destabilize the nucleosomal packaging of the DNA, thereby facilitating the access of other components required for transcription. In general, active chromatin (euchromatin) is associated with hyperacetylated histones, whereas heterochromatin is associated with hypoacetylated histones (reviewed in 6,7). However, exceptions to this rule have been reported in yeast, where specific acetylation of histone H4 has been observed in transcriptionally silenced chromatin (8). Based on sequence similarities and on the presence of common motifs, nuclear acetyltransferases (ATs) are grouped into distinct families. These include the GNAT (Gcn5-related N-acetyltransferase) superfamily (9), the MYST family (10), CBP/p300 (11,12), the TBP-associated factor TAFII 250 (13), members of the steroid receptor co-activators such as SRC-1 (14), and ACTR (15) and the gene-specific transcription factors ATF-2 (16) and CIITA (17). The best-characterized group of ATs is the GNAT superfamily whose members share sequence similarity in four conserved motifs—C, D, A (the most highly conserved region also present in the MYST family) and B—which represents the catalytic core of this large enzyme family. Together with mutational studies (18,19), the crystal structure of several members of the GNAT superfamily bound to acetyl-CoA alone (20–25) or to a substrate (26) has been determined. Based on the recently solved crystal structure of the first MYST family member, Esa1 (27), it was proposed that a structurally conserved central core domain mediates acetyl-CoA binding and catalysis in all ATs, whereas the sequence variability within a structurally related framework of the N- and C-terminal domains may determine substrate binding specificity.
p300 and CBP are transcriptional co-regulators that harbor multiple functional domains and regulate gene expression at several levels (reviewed in 28). In addition, they are essential for embryonic development (29–31). The AT activity of p300/CBP is able to modify not only histones but also other chromosomal proteins and sequence-specific transcription factors. A close correlation between AT activity and activation of transcription has been observed in the analysis of the wild-type or mutated CBP AT domain fused to the Gal4 DNA binding domain (32). The AT domain of p300/CBP has been mapped to a region that extends from the beginning of the cysteine/histidine-rich domain 2 (C/H2) to the N-terminal part of the cysteine/histidine-rich domain 3 (C/H3) (11,12). This region is evolutionarily conserved in p300/CBP orthologs ranging from animals to plants (33). To date, no crystal structure determination of the AT domain of p300/CBP has been reported and thus the mechanism of catalysis remains unknown.
The C/H2 region harbors in its N-terminal part a Cys4–His–Cys3 zinc binding module originally identified in the maize homeodomain protein ZMHOX1 (34) and its Arabidopsis relative HAT3.1 (35) and hence has been named plant homeodomain (PHD) finger. PHD fingers are primarly found in nuclear proteins presumed to regulate transcription in a chromatin environment.
In this paper, we have examined the AT domain of p300 by introducing progressive truncations or amino acid substitutions and measuring the resulting enzymatic activity. Our data show that both the PHD finger and residues belonging to the C/H3 region are not required for AT activity, confining the catalytic core domain to a segment of 385 amino acids in length. Mutations in this domain affect acetylation of histones and of p53 to a comparable degree, suggesting that both substrates are recognized in a similar way. Most mutations that strongly reduced AT activity are localized to a highly conserved fragment of 70 amino acid residues in length. Autoacetylation of the AT domain was the least affected activity as only two mutations abolished it. Surprisingly, in contrast to p300, the enzymatic activity of CBP is severely reduced by removal of the PHD finger or by some of the mutations that only marginally affected p300 activity. We therefore suggest that the AT domains of p300 and CBP are not equivalent and likely encompass elements of common as well as distinct structure.
MATERIALS AND METHODS
Plasmids
For the construction of plasmids allowing expression of glutathione S-transferase (GST)–AT fusion proteins in bacteria, cDNA fragments encoding the p300 and CBP protein segments shown in Figures 2A and 3A, respectively, or the AT domain of the short version (36) of human GCN5 (amino acids 8–477) were amplified by PCR and cloned into the pGEX-KG plasmid (37). The 19 mutations of the p300 AT domain, the eight mutations of the CBP AT domain and the four mutations in the PHD finger of p300 and CBP, respectively, were introduced using the QuickChange site-directed mutagenesis kit (Stratagene). The sequence of the mutagenesis primers is available upon request. The mutated plasmids were sequenced on both strands to verify that they contain the correct sequence. The p300 AT domain mutations of Figure 6 were introduced into the p300 full-length context by transferring an XbaI–XmaI fragment from the respective pGEX-p300-AT plasmid into the CMVβ-p300-CHA plasmid, which expresses p300 carrying a C-terminal HA-tag (38).
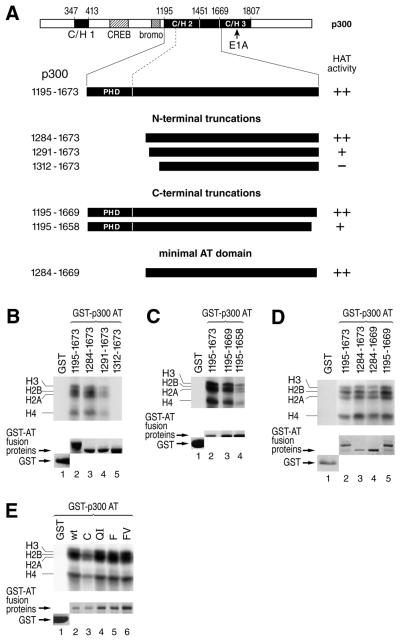
Figure 2.
The p300 AT core domain encompasses a segment of 385 residues (amino acids 1284–1669) and does not include the PHD finger and the C/H3 region. (A) Schematic representation of human p300 and N- and C-terminal truncations of the originally identified p300 AT domain (amino acids 1195–1673). The indicated fragments were purified as GST fusion proteins, incubated with core histones and [14C]acetyl-CoA and analyzed by SDS–PAGE. The HAT activity exhibited by each fragment is summarized on the right: ++, wild-type activity; +, reduced activity compared with wild-type; –, inactive. (B–D) HAT activity of the indicated p300 AT fragments. (E) The four mutations C(1201)A, FV(1244–1245)AA, QI(1256–1257)AA and F(1270)A were each introduced into the PHD finger of the p300 AT domain protein (amino acids 1195–1673) (see Fig. 4 for location of mutations) and HAT assays were performed. In all cases, acetylated core histones were detected following autoradiography of the SDS–PAGE gel (top panels). (B–E) The bottom panels show a Coomassie Blue staining of the gel displayed in the respective upper panel, revealing expression levels of the different GST–p300 AT fusion proteins.
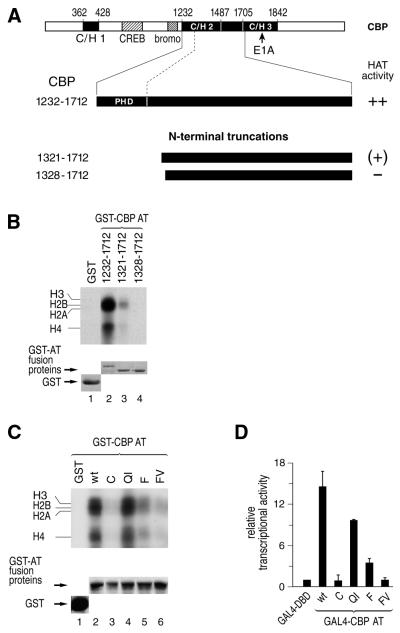
Figure 3.
The PHD finger of CBP is essential for enzymatic activity. (A) Schematic representation of mouse CBP and N-terminal truncations of the originally identified CBP AT domain (amino acids 1232–1712) are shown. The indicated fragment were purified as GST fusion proteins, incubated with core histones and [14C]acetyl-CoA and analyzed by SDS–PAGE. The HAT activity exhibited by each fragment is summarized to the right: ++, wild-type activity; (+), strongly reduced activity compared with wild-type; –, inactive. (B) HAT activity of the indicated progressively truncated CBP AT fragments. (C) The four mutations C(1237)A, FV(1280–1281)AA, QI(1292–1293)AA and F(1306)A were introduced into the PHD finger of mouse GST-CBP AT expression plasmids (see Fig. 4 for location of mutations) and HAT activity was determined with purified GST–CBP AT proteins. (B and C) Wild-type and mutant proteins were expressed as GST–AT fusion proteins and standard HAT assays were performed. Acetylated core histones were viewed following autoradiography of the SDS–PAGE gel (top panel). The bottom panels show Coomassie Blue staining of the gels displayed in the upper panels, revealing expression levels of the different GST–CBP AT fusion proteins. (D) U2OS cells were transiently transfected with a GAL4-dependent luciferase reporter gene along with the indicated wild-type and mutant GAL4-CBP AT expression plasmids. Luciferase activity was determined and normalized to a co-transfected internal control. The activity of the GAL4-DBD alone was set to 1.0 and the activity of the other effectors is expressed relative to it.
Figure 6.
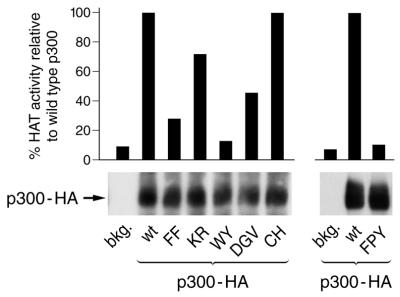
Effect of selected AT mutations in the full-length p300 context. Human U2OS cells were transiently transfected with CMV-p300 plasmids expressing wild-type or mutant, HA-tagged full-length p300 and expression levels of proteins were monitored by western blot (bottom panel). In parallel, HA-tagged p300 proteins were immunoprecipitated and assayed for HAT activity in the presence of H4 peptide and [14C]acetyl-CoA (top panel). The activity of the mutants is indicated as percentage of activity relative to that of wild-type p300, which was set to 100%.
Purification of recombinant proteins
Recombinant GST fusion proteins were expressed in Escherichia coli and purified using glutathione–Sepharose (Pharmacia) as described previously (33). In order to assay for the sensitivity of the enzymatic reaction catalyzed by the p300 AT domain to the zinc chelating agent 1,10-phenanthroline (Fig. 7), bacteria were lyzed in NETN buffer (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5% NP-40 supplemented with the indicated amount of 1,10-phenanthroline) by sonication and GST–AT domain fusion proteins were maintained throughout purification in NETN buffer supplemented with the indicated amount of phenanthroline. HAT assays were performed in the presence of the indicated concentration of 1,10-phenanthroline.
Figure 7.
Sensitivity of AT reaction catalyzed by p300 AT domain to 1,10-phenanthroline. The indicated AT domain fragments derived from human p300 or human GCN5 proteins were purified from E.coli as GST fusion proteins in the presence of the indicated 1,10-phenanthroline concentration. Standard HAT assays with core histones were performed in the presence of the same 1,10-phenanthroline concentration used during the purification. The fragment of human GCN5 (residues 8–477) used in this experiment exhibits a relaxed substrate specificity for histone acetylation and, therefore, all core histone are acetylated to a comparable degree. The expression level of the various GST fusion proteins was analyzed by staining the gel with Coomassie Blue (bottom panel) and acetylated histones were detected by autoradiography (top panel).
HAT assays
Standard HAT assays were performed with purified GST fusion proteins using a modification of the assay described previously (39). Enzyme samples were incubated at room temperature for 7.5 min in 30 µl of HAT assay buffer [50 mM Tris–HCl pH 8.0, 10% (v/v) glycerol, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM sodium butyrate] containing, unless otherwise specified, 25 nCi [14C]acetyl-CoA (55 mCi/mmol; Hartmann) and 5 µg of calf thymus histones (Sigma, type IIA). The reaction products were separated on SDS–polyacrylamide gels, which were treated with Na-salycilate for fluorography. Signals corresponding to acetylated histones and autoacetylation activity were quantified by scanning the gel with a densitometer (Molecular Dynamics).
HAT assays with H4 or p53 peptides as substrate were performed as reported (40). Purified GST fusion proteins or immunoprecipitated HA-tagged p300 proteins were incubated in 30 µl of HAT assay buffer containing 30 µM of either a biotinylated p53 (41) or H4 peptide (Chiron) and 0.25 nCi of [14C]acetyl-CoA (55 mCi/mmol; Hartmann) at room temperature for 7.5 min. After centrifugation, the supernatants were incubated in 500 µl HAT buffer with 20 µl of pre-washed streptavidine beads (Sigma) for 20 min at 4°C on a rotating wheel. The beads were then washed twice with lysis buffer, mixed with 3 ml of scintillation liquid (Hionic, Packard) and counted using a β-counter (BETAmatic).
Cell culture and transient transfections
U2OS cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (ICN). Transient transfection assays were carried out using the calcium–phosphate precipitation method (42). For the immunoprecipitation experiments, U2OS cells were transfected with 10 µg of CMVβ-p300-CHA plasmid encoding either wild-type HA-tagged p300 or one of the mutated variants (WY, FF, FPY, DGV, CH or KR), washed the following day and harvested 36 h after transfection. Cells were lyzed in IPH buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.5% desoxycholate, 0.1 mM PMSF) for 30 min at 4°C on a rotating wheel. Half of the cleared lysate was first incubated with HA.11 monoclonal antibody (BAbCO) for 1 h at 4°C followed by another 30 min with the secondary rabbit anti-mouse IgG antibody (Sigma). Immunocomplexes were captured on 30 µl of protein A–Sepharose beads (Pharmacia) for 40 min at 4°C. The beads were washed three times with IPH buffer and H4 peptide HAT assays were performed as described above. Western blots with a PCAF antibody showed that under the conditions employed, PCAF does not co-precipitate with p300 (data not shown). In parallel, the other half of the cell extract was separated on a 5% SDS–PAGE gel and HA-tagged p300 proteins were detected in western blots with the HA.11 monoclonal antibody.
In order to measure the transactivation potential linked to the CBP AT activity, 3 µg of CMV-promoter driven GAL4-CBP AT plasmid (32), expressing either wild-type CBP AT domain or one harboring one of the mutations C, FV, F or QI in the PHD finger of CBP, were transfected together with 7 µg of 5× GAL4-AdML-(firefly)luciferase reporter gene and 50 ng of pRL-CMV Renilla luciferase control reporter vector as internal standard. Cells were washed the following day and harvested 36 h after transfection. Lysis of cells and reporter gene assays were performed using the Dual-Luciferase Reporter Assay System (Promega) followed by measurement of luciferase activity in a luminometer (EG&G Berthold Lumat LB 9507).
RESULTS
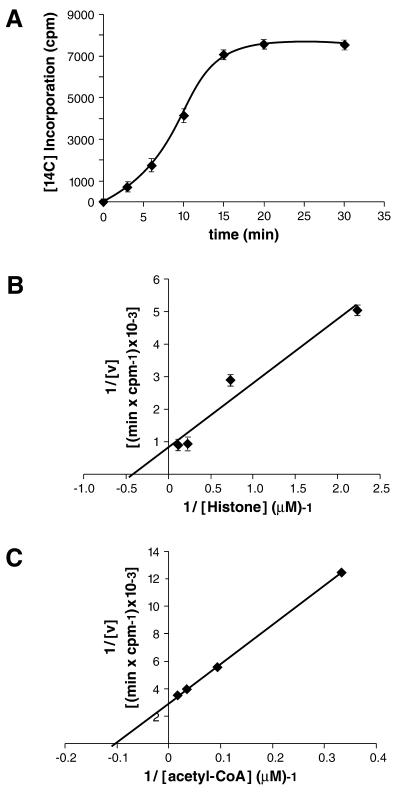
The AT domain of human p300 has been mapped previously to residues 1195–1673 (12). We have expressed this segment of p300 as a GST fusion protein in E.coli and first performed a kinetic analysis of the acetylation reaction using saturating amounts of core histones and [14C]acetyl-CoA. This analysis was carried out in order to determine suitable parameters for subsequent enzymatic reactions. The acetylation reaction was in the linear range between 3 and 15 min of incubation time (Fig. 1A). As a result, an incubation time of 7.5 min was used in all subsequent experiments. In order to determine the apparent Km for histone H4 acetylation, acetylation reactions with varying concentrations of histone H4 (0.45–9 µM) were carried out. Incorporated radioactivity (H4 acetylation) was plotted against H4 concentration in a double reciprocal graph (Fig. 1B). Taking data from three independent experiments, the apparent Km of the p300 AT domain for histone H4 was determined to be 2.7 µM. In similar experiments using radioactive acetyl-CoA and a full-length histone, the apparent Km of yeast GCN5 for histone H3 has been determined to be 28 µM (43) or 10 µM (44). Measurements of p300 HAT enzyme reactions with varying acetyl-CoA and constant histone concentrations suggested an apparent Km for acetyl-CoA of 9.8 µM (Fig. 1C), compared with the apparent Km (acetyl-CoA) of 20 µM for yGCN5. We conclude that the bacterially expressed human p300 AT domain binds histones and acetyl-CoA at least as efficiently as yGCN5.
Figure 1.
Kinetic analysis of the AT reaction catalyzed by the p300 AT domain. (A) Determination of the linear range of HAT enzymatic reaction. Purified GST–p300 AT fusion protein containing the AT domain (amino acids 1195–1673) of hp300 was assayed in 30 µl assay reactions for the indicated time points. The rate of incorporation of [14C]acetyl groups was measured using saturating amounts of core histones (20 µg) and acetyl-CoA [0.5 µCi (55 mCi/mmol)]. Points on the graph represent an average of three independent experiments. (B) Determination of the apparent Km for histone H4 acetylation. Standard HAT reactions were carried out with varying concentrations (0.45–9 µM) of histone H4. Assays were performed in triplicate and the apparent Km for p300 AT domain was calculated to be 2.7 µM. (C) Determination of the apparent Km for acetyl-CoA. Standard HAT assay was performed with varying acetyl-CoA concentrations (3.6–12.5 µM). Data obtained were represented in a double reciprocal plot yielding an apparent Km for acetyl-CoA of 9.8 µM.
The previously mapped AT domain of human p300 includes C/H2 in its entirety, as well as an intervening sequence and the N-terminal part of C/H3 (12). To refine the borders of the enzymatic domain, we have prepared a series of N- and C-terminal truncations of the p300 AT domain which are schematically depicted in Figure 2A. The truncated versions of the AT domain were again expressed in bacteria as GST fusion proteins and HAT assays using core histones as substrate were performed according to the conditions determined in Figure 1. The HAT activity of the N-terminally truncated GST–p300 AT fusion proteins is shown in Figure 2B. Deletion of the first 89 residues containing the PHD finger did not compromise AT activity (Fig. 2B, compare lanes 2 and 3). However, further truncation of an additional seven residues resulted in a protein (amino acids 1291–1673) with diminished AT activity (lane 4). The deletion of a further 21 amino acids resulted in a complete loss of AT activity (Fig. 2B, lane 5). In order to assess the requirement of the C/H3 domain for AT activity, we analyzed C-terminal deletions. A p300 fragment (amino acids 1195–1669) lacking the C/H3 region retained full enzymatic activity (Fig. 2C, compare lanes 3 and 2). Removal of an additional 11 residues resulted in a protein (amino acids 1195–1658) displaying a 4-fold reduced AT activity (Fig. 2C, lane 4). In order to identify the minimal p300 AT domain showing full activity, we expressed a GST–AT fusion protein combining the maximal N- and C-terminal truncations, which still exhibited wild-type activity in the above assays. This protein (residues 1284–1669) was as active as the originally defined AT domain (Fig. 2D, compare lanes 4 and 2). Finally, to confirm the independence of p300 AT activity from PHD finger sequences, we have introduced four different mutations in conserved residues of the PHD finger of p300 (residues C, QI, F and FV were individually replaced by alanine; see Fig. 4 for location of mutations). Residue C, a cysteine predicted to ligate zinc, and residues F are likely to be essential for the structural integrity of the PHD finger (45). In HAT assays, mutant C showed a 2–3-fold reduced activity, probably due to a collapse of the PHD zinc finger, whereas the other three mutants exhibited wild-type activity (Fig. 2E). This result supports the notion that the PHD finger of p300 is not essential for p300 AT activity. From this analysis, we conclude that the minimal region of p300 conferring wild-type enzymatic activity encompasses 385 residues (1284–1669), which is at the N-terminus shorter than previously defined. In particular, the PHD finger is dispensable for the in vitro AT activity of p300. In accordance with previous studies (12,46), we also show that the AT core domain does not extend into the C/H3 region.
Figure 4.
Sequence conservation of the C/H2 domain of animal and plant p300/CBP proteins and mutations introduced in this study. A sequence alignment of the segment encompassing the C/H2 region of human p300 and CBP, Drosophila and Caenorhabditis elegans CBP and the two Arabidopsis thaliana orthologs of p300/CBP PCAT1 and PCAT2 (33) is shown. Multiple sequence analysis was performed with the clustalx software (50) and shading was done using the standard settings of Boxshade 3.21 program (www.ch.embnet.org/software/BOX_doc.html). Asterisks above the aligned sequences indicate AT domain or PHD finger residues that were mutated in p300 and CBP, respectively. The PHD finger is located between residues 1201 and 1272 of human p300 and residues 1237–1281 of human CBP, respectively. The effect of the AT mutations on the activity of p300/CBP are summarized in Table 1 and illustrated in Figures 5 and 6. GST fusion proteins carrying mutations in the PHD finger were assayed in Figures 2 and 3.
We next examined whether the PHD finger of CBP is similarly dispensable for AT activity. Unexpectedly, deletion of the PHD finger resulted in a strong reduction of CBP enzymatic activity (Fig. 3B, compare lanes 2 and 3). Removal of an additional seven N-terminal residues completely abolished CBP AT activity (lane 4). The introduction of four mutations identical to those analyzed for p300 in the CBP PHD finger led to a strong reduction in CBP AT activity for mutations C, F and FV whereas mutation of residues QI did not show any effect (Fig. 3C). The data were confirmed by transactivation assays. Indeed, transactivation by a Gal4–CBP AT fusion protein whose activation potential is strictly dependent on its intrinsic AT activity (32) mirrored the results obtained in the in vitro acetylation assays (Fig. 3D). We therefore conclude that the CBP AT domain, unlike that of p300, requires the presence of intact PHD finger sequences for enzymatic activity.
To identify residues within the p300 AT domain that are essential for AT activity, double and triple amino acid substitution mutations were introduced into the evolutionarily most conserved segments of this domain. Similar scanning strategies have been applied previously to yGCN5 (18,19) and CBP (32). The mutated p300 AT residues are highlighted in Figure 4 by asterisks above the sequence alignment of p300/CBP proteins from different species. Wild-type and mutant AT domains were expressed in bacteria as GST fusion proteins and assayed for their ability to acetylate either core histones, a p53 peptide or the AT domain itself (autoacetylation). Assaying proteins expressed in bacteria rules out that the enzymatic activity of other mammalian HATs binding to p300 contributes to the measured activity.
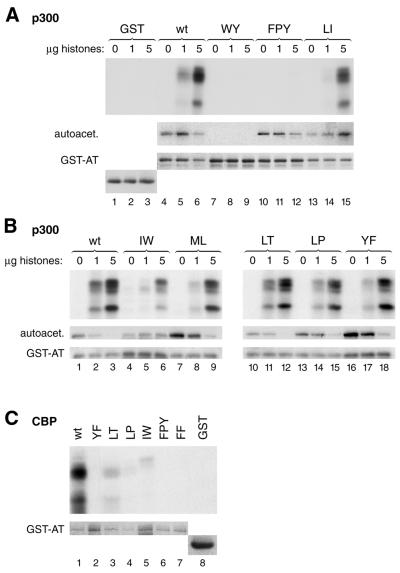
The p300 AT mutants were named according to the wild-type residues that were altered. A summary of the activity exhibited by the mutants is given in Table 1. For all three substrates tested, the activity of the wild-type enzyme was set to 100% and the activity of the mutated proteins was expressed relative to wild-type levels. In addition, Figure 5 shows histone and autoacetylation of the key mutants. Overall, there is a good correlation between the histone and p53 acetylation activity of most mutants. Based on the enzymatic activity, two major groups of mutants can be defined. The first one is represented by mutations that have either no or only a minor effect (<2-fold) on the AT activity of p300. Mutations CH, YF, ML, LT, KR, LI, EY, LP and DGV belong to this group (see top part of Table 1). In contrast, a second group of mutations including VY, DDY, YL and FF showed considerably reduced autoacetylation as well as a reduced ability to acetylate histones and non-histone substrates such as p53. In particular, one mutation, WY, was completely defective in acetylation of all three substrates tested (Fig. 5A, lanes 7–9). Mutations FPY and IW showed strongly reduced AT activity but no (FPY), or only a mild (IW), reduction in autoacetylation (Fig. 5A and B, lanes 10–12 and 4–6, respectively). These results suggest that mutation of p300 residues IW and FPY might primarily impair the ability of the AT domain to bind substrates such as histones and p53, while leaving the catalytic core function of the enzyme largely intact. In contrast, mutations that strongly impair the acetylation of substrates as well as autoacetylation (particularly YL, FF and WY) are likely to interfere with acetyl-CoA and substrate binding (see Discussion and Table 1). Mutations ML and YF stimulated autoacetylation activity 3–4-fold (Fig. 5B, lanes 7–9 and 16–18), confirming that the catalytic function is not reduced by these mutants. One possible explanation for such elevated autoacetylation levels might be that these two mutations increase the accessibility of lysine residues, which are otherwise buried and, therefore, normally not modified by the autoacetylation activity.
Table 1. Summary of mutations introduced into the AT domain of human p300 and their relative AT activity.
| |
Histone acetylation (% wild-type) |
p53 acetylation (% wild-type) |
Autoacetylation |
Suggested defect |
| Wild-type | 100 | 100 | 100 | |
| First group of mutations (mild ones) | ||||
| CH(1450–1451)AA | 99 ± 11 | 98 ± 15 | 133 ± 12 | |
| YF(1503–1504)AA | 96 ± 13 | nd | 380 ± 21 | |
| ML(1470–1471)AA | 94 ± 16 | nd | 377 ± 42 | |
| LT(1495–1496)AA | 91 ± 15 | nd | 92 ± 9 | |
| KR(1461–1462)AA | 91 ± 9 | 91 ± 4 | 66 ± 8 | |
| EY(1423–1424)AA | 80 ± 7 | nd | 82 ± 5 | |
| LP(1501–1503)AA | 84 ± 9 | nd | 110 ± 3 | |
| DGV(1367–1369)AAA | 70 ± 5 | 85 ± 14 | 74 ± 4 | |
| LI(1418–1419)AA | 65 ± 18 | 86 ± 11 | 80 ± 10 | |
| Second group of mutations (severe ones) | ||||
| IW(1435–1436)AA | 18 ± 5 | nd | 50 ± 4 | S |
| VY(1413–1414)AA | 15 ± 4 | 6 ± 3 | 12 ± 3 | A/S |
| FPY(1353–1355)AAA | 5 ± 2 | 2 ± 2 | 121 ± 11 | S |
| DDY(1444–1446)AAQ | 5 ± 1 | nd | 11 ± 1 | A/S |
| YL(1397–1398)AA | 5 ± 1 | 3 ± 0 | 6 ± 2 | A/S |
| FF(1403–1404)AA | 2.1 ± 0.3 | 4 ± 1 | 1.8 ± 0.3 | A/S |
| WY(1466–1467)AS | 1.8 ± 0.3 | 1.0 ± 0.5 | 1.9 ± 0.3 | A/S |
| LT/YF | 15 ± 3 | nd | 115 | S |
| LP/YF | 19 ± 1 | nd | nd | |
| LT/LP/YF | 4 ± 1 | nd | 124 | S |
| Background | 1.7 ± 0.2 | 1.3 ± 0.2 | 2.0 ± 0.2 | |
The top part of the table [‘first group of mutations (mild ones)’] summarizes the activity exhibited by mutants, which showed <2-fold reduced AT activity. The middle part [‘second group of mutations (severe ones)’] shows mutants with >2-fold diminished AT activity and the third section (LT/YF, LP/YF, LT/LP/YF) shows the results obtained by combining two or three mutations. The last column labeled ‘Suggested defect’ indicates whether a given mutation is impaired in substrate (S) or acetyl-CoA and/or substrate (A/S) binding. Comparable amounts of GST–p300 AT proteins (∼100 ng) were used to carry out HAT assays in the presence of 5 µg of core histones. Autoacetylation reactions were performed with the GST–p300 AT fusion protein alone. Proteins were analyzed by SDS–PAGE followed by Coomassie blue staining and autoradiography of the gel. Signals corresponding to acetylated histones or autoacetylation were quantified by scanning the gel with a densitometer. p53 acetylation was assayed using a p53 peptide as substrate. Incorporated radioactivity was measured in a scintillation counter. Background activity associated with core histones or p53 was determined by performing the assays in the absence of the enzyme. Background activity of autoacetylation was determined by measuring the signal associated with GST alone. The activity is expressed in percentage relative to wild-type. Most of the assays were performed in triplicate and in these cases the standard deviations are indicated. nd, not determined
Figure 5.
Substrate titration experiments of key p300 and CBP AT mutant proteins. Wild-type human p300 AT domain (amino acids 1195–1673) and the indicated mutant proteins were expressed as GST fusion proteins. The recombinant proteins were incubated with 0, 1 or 5 µg of core histones, in the presence of [14C]acetyl-CoA and analyzed by SDS–PAGE. Coomassie Blue staining of the gel [lower panels in (A) and (B)] show expression levels of the wild-type and mutant recombinant proteins. Acetylated histones and autoacetylation of GST–p300 AT fusion proteins are shown in the middle and bottom panels, respectively (A and B). (C) Wild-type and mutant mouse CBP AT domain (amino acids 1232–1712) fragments were expressed as GST–AT fusion proteins and standard HAT assays were performed. The top panel shows acetylation levels of core histones incubated with the indicated CBP mutant protein. The bottom panel shows a Coomassie Blue stained section of the SDS–PAGE revealing expression levels of the GST–CBP AT fusion proteins.
Although amino acid residues altered in the first group are highly conserved throughout evolution, they are not essential for the AT activity of p300. Of particular interest are the results of mutations LT, LP and YF as comparable mutations have previously been introduced into the CBP AT domain. While our results indicate little interference with p300 catalytic function (Fig. 5B, lanes 10–18), the corresponding point mutations of mouse CBP residues L(1532), L(1538), Y(1540) and F(1541) were reported to strongly reduce CBP AT activity (32). Based on sequence similarity between PCAF and CBP, Martinez-Balbas et al. (32) have suggested that the above CBP residues, 1532–1541, harbor a catalytically important motif, which is similar to motif B of GNAT enzymes (9). To examine more thoroughly the potential role of residues LT, LP and YF in catalytic activity, we have constructed GST–p300 AT proteins bearing combinations of these mutations. Combination of LT/YF and LT/LP/YF mutations resulted in a p300 AT domain exhibiting strongly reduced HAT activity while still retaining autoacetylation activity (Table 1, third section). Taken together, these results suggest that residues LT, LP, YF of p300 may be involved in substrate recognition rather than catalysis or acetyl-CoA binding (see Discussion).
In order to clarify the role of residues LT, LP and YF for AT activity of p300 and CBP, these residues were also individually mutated in a GST–CBP AT protein. In contrast to p300 and in agreement with previous results of a single amino acid substitution analysis (32), mutation of CBP residues LT, LP and YF led to a severe reduction of CBP AT activity (Fig. 5C, lanes 2–4). Therefore, an identical mutation can lead to a differential impairment of p300 and CBP AT activity, indicating that the structure of the two enzyme domains is not entirely identical. This is not true for all mutations because mutation of CBP residues IW, FPY and FF led to a result comparable with that obtained with the corresponding p300 mutations. CBP mutants FPY and FF showed no enzymatic activity (Fig. 5C, lanes 6 and 7) and the CBP IW mutant protein showed a 10-fold reduced AT activity. Interestingly, mutation of residues IW in p300 and CBP selectively affected acetylation of histone H3 the least if 5 µg of histones were used (Fig. 5B and C, lanes 6 and 5, respectively). This result suggests that p300 and CBP may be folded similarly in the regions containing residues FF, FPY and IW.
We introduced the six most interesting p300 AT mutations into the context of the full-length protein to examine whether there is a difference in enzymatic activity between full-length p300 and the AT domain alone fused to GST. Human U2OS osteosarcoma cells were transiently transfected with p300-HA expression plasmids encoding one of the six AT mutations or wild-type p300. A western blot showed that the seven p300 proteins were expressed at comparable levels (Fig. 6, lower panel), allowing us to compare their relative HAT activities. The HA-tagged p300 was immunoprecipitated and subjected to a HAT assay using a histone H4 peptide as substrate. Stringent immunoprecipitation conditions were used to ensure that no other HAT proteins binding to p300, such as PCAF (36) or SRC-1/ACTR (15), were co-immunoprecipitated. In keeping with the results from the GST–AT protein assays, mutant WY did not exhibit a significant AT activity and that of mutants FF and FPY was severely reduced (Fig. 6, bar diagram). Mutatation of residues CH and KR failed to affect AT activity (CH) or only modestly diminished it (KR). We conclude that the AT activity of the full-length p300 proteins expressed in mammalian cells is affected by the mutations in a manner comparable with that of the AT domain alone.
The p300/CBP AT domain contains several cysteine and histidine residues, which may coordinate zinc or form disulfide bridges. Some of these residues are located within the PHD finger, which was shown above to be dispensable for AT activity. Three cysteine and two histidine residues, however, are located C-terminal of the PHD finger and are conserved in all known p300/CBP orthologs. It is of interest to note that MYST AT family members also contain a zinc finger motif in the midst of their AT domain. This motif was shown to be essential for enzymatic activity (47,48). To examine the role of the cysteine and histidine residues in enzyme structure or activity we exposed GST–p300 AT proteins to a reducing agent (DTT) or to a zinc-chelating agent (1,10-phenanthroline). HAT reactions were not inhibited by DTT concentrations ranging from 1 to 125 mM (data not shown). The insensitivity of the enzyme towards high DTT concentrations speaks against the existence of cysteine disulfide bridges essential for p300 catalytic activity. In order to probe for the presence of zinc ions, GST–AT fusion proteins were isolated from E.coli in the presence of 0, 5, 10 or 15 mM 1,10-phenanthroline and and the subsequent HAT assays were performed in the presence of the same phenanthroline concentrations. As it is known that the PHD finger coordinates zinc, its chelation may lead to a collapse of the finger thereby impairing the activity of the neighboring AT domain. Therefore, we carried out this experiment with GST fusion proteins including and excluding the PHD finger. Figure 7 shows that the enzymatic activity of the GST–p300 fusion protein both in the presence (Fig. 7, lanes 1–4) and absence (Fig. 7, lanes 5–8) of the PHD finger was diminished by the presence of 5 mM phenanthroline and abrogated at 10 mM phenanthroline. As expected, at both phenanthroline concentrations, the HAT activity of the catalytic domain of human GCN5 protein, which is known not to chelate zinc (24,48), was not impaired (Fig. 7, lanes 10 and 11). This result suggests that the activity of the p300 AT domain depends on zinc chelation by the AT core domain itself rather than that of the PHD finger.
DISCUSSION
In this study, we have carried out the first detailed mutational analysis of the AT domain of p300 and have compared the AT domain of p300 and CBP. Four major conclusions have been reached. First, the PHD finger of p300 is dispensable for AT activity, whereas that of CBP is essential. Indeed, our mapping data indicate that the minimal AT domain spans residues 1284–1669 of human p300. Secondly, histone and p53 acetylation are affected by the AT mutations to a comparable degree, suggesting that these two substrates are most likely to be recognized in a similar way. Thirdly, our substitution mutations define residues that are involved in substrate recognition and separate them in part from those probably involved in acetyl-CoA binding (see Table 1 for summary). Since the latter residues do not show any similarity to motifs known in GNAT family members to participate in acetyl-CoA binding, we suggest that p300 and CBP belong to a separate group of ATs. Fourthly, several, but not all, identical mutations introduced in p300 and CBP differentially impair their respective AT activity, indicating that the two enzymes have in part distinct structures.
In order to examine the relationship between evolutionarily conserved sequences and p300 AT activity, we have performed scanning mutagenesis, substituting in total 16 conserved amino acid pairs or triplets. Comparison of the ability of the 16 simple and the three combined AT mutants to acetylate core histones leads to two major conclusions. First, more than half of the introduced simple mutations (nine in total) only reduced AT activity modestly, suggesting that these residues are not essential for p300 AT function. Some of these mutations, however, such as ML, LP or YF, concern residues strictly conserved in all known p300/CBP species. It is possible that these residues have a conserved role in functions other than AT activity that remain to be determined. Secondly, seven mutations completely abolished or severely compromised AT function. With the exception of the FPY mutation, these mutations are all located within a 70 amino acid segment, encompassing residues 1397–1467 of human p300. This segment, which is located in the middle of the p300/CBP AT domain, is part of the evolutionarily best-conserved subregion. Based on the drastic phenotype of these mutations for HAT activity, this 70 amino acid segment is likely to be intimately involved in both acetyl-CoA and substrate binding. However, not all residues within this segment are absolutely required for AT activity as mutations LI, EY, CH and KR alter residues within this region without severely decreasing enzymatic activity. It is likely that these residues are not directly exposed to the enzyme surface, thus not interacting directly with substrate and/or acetyl-CoA.
The above conclusions can be extended by taking into account the autoacetylation activity of each mutant protein. Autoacetylation is mainly dependent on the ability of the catalytic domain to bind acetyl-CoA and does not require substrate recruitment as the acetyl-group is transferred to one or several lysine residues within the AT domain. Therefore, if a given mutant is active in autoacetylation, it is likely to be able to recruit acetyl-CoA. Accordingly, the pronounced defect of mutation FPY in histone acetylation (5% of wild-type activity), while autoacetylation is intact, is probably not due to a defect in acetyl-CoA binding because autoacetylation is intact, but rather due to a specific defect in substrate recruitment. In contrast, mutations VY, DDY, YL, FF and WY, which are not only severely impaired in histone acetylation but also in autoacetylation, are likely to exhibit a defect in acetyl-CoA binding. This conclusion, however, does not rule out that the above five mutations are also impaired in substrate recruitment. In fact, this is likely to be the case as acetyl-CoA and substrate have to be brought in close and precise juxtaposition in order to achieve specificity in lysine acetylation of histones (22).
In a previous study, a series of single amino acid substitutions was introduced into the CBP AT domain and three regions sharing limited homology with GNAT motifs A, B and D in PCAF were described within this domain (32). Thus, a comparison of the results of the two studies is of interest. Four mutations introduced into a CBP segment proposed to correspond to motif B of GNATs can directly be compared with three p300 mutations because they affect residues identical in the two proteins. CBP mutations L(1532), L(1538), Y(1540) and F(1541) are matched by p300 mutations LT(1495–1496), LP(1501–1503) and YF(1503–1504) (the matching residues are highlighted in bold). All four single residue CBP mutations were reported to almost abolish AT activity. In keeping with this result, mutation of CBP residues LT, LP and YF in this study indeed essentially eliminates CBP AT activity. Interestingly, as the identical mutations in the p300 AT domain only marginally compromise enzyme activity, p300 and CBP AT domains clearly exhibit a differential sensitivity towards identical mutations. This observation indicates that, despite a high degree of similarity in their primary protein sequence, subtle structural differences are likely to exist between p300 and CBP AT domains.
Based on our results with p300, we believe that residues LT, LP and YF may be part of enzyme structures involved in substrate interactions. Since in p300 none of the three double amino acid substitution mutants LT, LP or YF showed strongly reduced p300 AT activity (see Table 1 and Fig. 5B), it is unlikely that these residues participate in acetyl-CoA recognition, a function that is probably highly conserved among p300/CBP family members. Moreover, the triple mutation LT/LP/YF displayed almost no HAT activity but wild-type autoacetylation activity, implying that acetyl-CoA binding was indeed intact. This finding strongly suggests that the integrity of p300 residues LT, LP and YF is not required for acetyl-CoA binding, a function linked to GNAT motifs A and B, but rather contributes to substrate binding. We also note that several of our p300 mutations showing strongly reduced AT activity, such as YL, FF, VY and WY (this study and 33), are actually located outside of one of the GNAT motifs proposed by Martinez-Balbas et al. (32). In addition, if one compares p300/CBP proteins from different species, the assignment of GNAT motif A is located in a region of CBP that is not very well conserved within p300/CBP family members (see for example Fig. 4, residues 1281–1293 of PCAT2). In summary, these observations lead to the suggestion that the AT domain of p300/CBP does not belong to the GNAT superfamily, but rather constitutes a family on its own.
Our deletion analysis of the AT domain of p300 and CBP revealed that the PHD finger, located in the N-terminal half of the C/H2 region, is required for CBP but not for p300 enzymatic activity. In accordance with this finding is the observation that three of the four amino acid substititions in the CBP PHD finger greatly reduced CBP AT activity while three of the four identical substitution mutations in the p300 PHD finger had no effect on p300 AT function. Therefore, the relative arrangement of PHD finger and core AT domain is different in the two proteins. This view is further supported by the observation that Gal4–CBP AT fusion proteins (including the PHD finger) efficiently activate transcription of reporter plasmids in vivo (32), whereas an identical Gal4–p300 AT fusion protein is incapable of triggering any transcriptional activation (data not shown). This observation also points to significant structural differences between p300 and CBP AT domains, which in this particular case may prevent acetylation by p300, but not by CBP, of a key protein whose modification is essential for transcriptional activity of the Gal4 binding site-dependent promoter. Moreover, a report investigating p300 and CBP function in F9 cells has suggested that in vivo the two proteins fulfill partially distinct functions (49). The identification in the present report of a differential sensitivity of p300 and CBP AT activity towards identical mutations in the PHD finger or AT domain provides the first experimental evidence for subtle structural differences between the two proteins, which may translate into significant functional differences in vivo.
ACKNOWLEGEMENTS
We are most grateful to Drs Yoshihiro Nakatani and Xiang-Jiao Yang for the gift of the short version of the human GCN5 cDNA. We thank Drs Jeanne-Françoise Roth and Noriko Shikama for critical reading of the manuscript. This work was supported by a START fellowship and a grant from the Swiss National Science Foundation to R.E.
References
- 1.Allfrey V.G., Faulkner,R. and Mirsky,A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebbes T.R., Thorne,A.W. and Crane-Robinson,C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebbes T.R., Thorne,A.W., Clayton,A.L. and Crane-Robinson,C. (1992) Histone acetylation and globin gene switching. Nucleic Acids Res., 20, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- 5.Kleff S., Andrulis,E.D., Anderson,C.W. and Sternglanz,R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem., 270, 24674–24677. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98, 1–4. [DOI] [PubMed] [Google Scholar]
- 7.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuwald A.F. and Landsman,D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci., 22, 154–155. [DOI] [PubMed] [Google Scholar]
- 10.Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- 12.Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 13.Mizzen C.A., Yang,X.J., Kokubo,T., Brownell,J.E., Bannister,A.J., Owen-Hughes,T., Workman,J., Wang,L., Berger,S.L., Kouzarides,T., Nakatani,Y. and Allis,C.D. (1996) The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- 14.Spencer T.E., Jenster,G., Burcin,M.M., Allis,C.D., Zhou,J., Mizzen,C.A., McKenna,N.J., Onate,S.A., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature, 389, 194–198. [DOI] [PubMed] [Google Scholar]
- 15.Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H., Schiltz,L., Chiu,R., Itakura,K., Taira,K., Nakatani,Y. and Yokoyama,K.K. (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature, 405, 195–200. [DOI] [PubMed] [Google Scholar]
- 17.Raval A., Howcroft,T.K., Weissman,J.D., Kirshner,S., Zhu,X.S., Yokoyama,K., Ting,J. and Singer,D.S. (2001) Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell, 7, 105–115. [DOI] [PubMed] [Google Scholar]
- 18.Kuo M.-H., Zhou,J., Jambeck,P., Churchill,M.E.A. and Allis,D.C. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Liu,L. and Berger,S.L. (1998) Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev., 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutnall R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1998) Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell, 94, 427–438. [DOI] [PubMed] [Google Scholar]
- 21.Wolf E., Vassilev,A., Makino,Y., Sali,A., Nakatani,Y. and Burley,S.K. (1998) Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell, 94, 439–449. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y., Fletcher,C.M., Zhou,J., Allis,C.D. and Wagner,G. (1999) Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature, 400, 86–89. [DOI] [PubMed] [Google Scholar]
- 23.Trievel R.C., Rojas,J.R., Sterner,D.E., Venkataramani,R.N., Wang,L., Zhou,J., Allis,C.D., Berger,S.L. and Marmorstein,R. (1999) Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl Acad. Sci. USA, 96, 8931–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clements A., Rojas,J.R., Trievel,R.C., Wang,L., Berger,S.L. and Marmorstein,R. (1999) Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J., 18, 3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angus-Hill M.L., Dutnall,R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1999) Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol., 294, 1311–1325. [DOI] [PubMed] [Google Scholar]
- 26.Rojas J.R., Trievel,R.C., Zhou,J., Mo,Y., Li,X., Berger,S.L., Allis,C.D. and Marmorstein,R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature, 401, 93–98. [DOI] [PubMed] [Google Scholar]
- 27.Yan Y., Barlev,N.A., Haley,R.H., Berger,S.L. and Marmorstein,R. (2000) Crystal structure of yeast esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell, 6, 1195–1205. [DOI] [PubMed] [Google Scholar]
- 28.Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- 29.Yao T.-P., Oh,S.P., Fuchs,M., Zhou,N.-D., Ch′ng,L.-E., Newsome,D., Bronson,R.T., Livingston,D.M. and Eckner,R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- 30.Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.E., Sieff,C.A., Livingston,D.M. and Yao,T.P. (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- 31.Akimaru H., Chen,Y., Dai,P., Hou,D.X., Nonaka,M., Smolik,S.M., Armstrong,S., Goodman,R.H. and Ishii,S. (1997) Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature, 386, 735–738. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Balbas M.A., Bannister,A.J., Martin,K., Haus-Seuffert,P., Meisterernst,M. and Kouzarides,T. (1998) The acetyltransferase activity of CBP stimulates transcription. EMBO J., 17, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordoli L., Netsch,M., Luthi,U., Lutz,W. and Eckner,R. (2001) Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res., 29, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellmann R. and Werr,W. (1992) Zmhox1a, the product of a novel maize homeobox gene, interacts with the Shrunken 26 bp feedback control element. EMBO J., 11, 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler U., Beckmann,H. and Cashmore,A.R. (1993) HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J., 4, 137–150. [DOI] [PubMed] [Google Scholar]
- 36.Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- 37.Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- 38.Eckner R., Ewen,M.E., Newsome,D., Gerdes,M., DeCaprio,J.A., Lawrence,J.B. and Livingston,D.M. (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- 39.Brownell J.E. and Allis,C.D. (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl Acad. Sci. USA, 92, 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ait-Si-Ali S., Ramirez,S., Robin,P., Trouche,D. and Harel-Bellan,A. (1998) A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res., 26, 3869–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 42.Chen C. and Okayama,H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol., 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner K.G., Trievel,R.C., Kuo,M.-H., Howard,R.M., Berger,S.L., Allis,C.D., Marmorstein,R. and Denu,J.M. (1999) Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem., 274, 18157–18160. [DOI] [PubMed] [Google Scholar]
- 44.Syntichaki P. and Thireos,G. (1998) The Gcn5-Ada complex potentiates the histone acetyltransferase activity of Gcn5. J. Biol. Chem., 273, 24414–24419. [DOI] [PubMed] [Google Scholar]
- 45.Capili A.D., Schultz,D.C., Rauscher,I.F. and Borden,K.L. (2001) Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J., 20, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraus W.L., Manning,E.T. and Kadonaga,J.T. (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol., 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takechi S. and Nakayama,T. (1999) Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem. Biophys. Res. Commun., 266, 405–410. [DOI] [PubMed] [Google Scholar]
- 48.Akhtar A. and Becker,P.B. (2001) The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep., 2, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawasaki H., Eckner,R., Yao,T.-P., Taira,K., Chiu,R., Livingston,D.M. and Yokoyama,K.K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- 50.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL:X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]