Abstract
The biotype El Tor of serogroup O1 and most of the non-O1/non-O139 strains of Vibrio cholerae can produce an extracellular pore-forming toxin known as cholera hemolysin (HlyA). Expression of HlyA has been previously reported to be regulated by the quorum sensing (QS) and the regulatory proteins HlyU and Fur, but lacks the direct evidence for their binding to the promoter of hlyA. In the present work, we showed that the QS regulator HapR, along with Fur and HlyU, regulates the transcription of hlyA in V. cholerae El Tor biotype. At the late mid-logarithmic growth phase, HapR binds to the three promoters of fur, hlyU, and hlyA to repress their transcription. At the early mid-logarithmic growth phase, Fur binds to the promoters of hlyU and hlyA to repress their transcription; meanwhile, HlyU binds to the promoter of hlyA to activate its transcription, but it manifests direct inhibition of its own gene. The highest transcriptional level of hlyA occurs at an OD600 value of around 0.6–0.7, which may be due to the subtle regulation of HapR, Fur, and HlyU. The complex regulation of HapR, Fur, and HlyU on hlyA would be beneficial to the invasion and pathogenesis of V. cholerae during the different infection stages.
Keywords: Vibrio cholerae, regulation, HlyA, quorum sensing, HapR, Fur, HlyU
Introduction
Vibrio cholerae, a Gram-negative and curved bacterium, is the causative agent of the diarrheal disease cholera (Clemens et al., 2017). This pathogen expresses various key virulence factors, including major ones, such as cholera toxin (CT), toxin co-regulated pilus (TCP), flagellum, and cholera hemolysin (HlyA; Almagro-Moreno et al., 2015; Benitez and Silva, 2016; Clemens et al., 2017). HlyA, an extracellular pore-forming toxin, is expressed in the El Tor biotype and most of the non-O1/non-O139 isolates (Yamamoto et al., 1990a; Singh et al., 2001; Diep et al., 2015). It possesses various biological activities including hemolytic activity, lethality, cardiotoxicity, cytotoxicity, and enterotoxicity (Ichinose et al., 1987; Benitez and Silva, 2016). HlyA has been recognized as a virulence determinant in the infant mouse cholera model (Fallarino et al., 2002). Purified HlyA can induce fluid accumulation and a histological change in the mucosa when injected into rabbit ileal loops (Ichinose et al., 1987; Debellis et al., 2009). In vitro studies showed that HlyA induces cell vacuolation and apoptosis in cultured mammalian cells (Coelho et al., 2000; Mitra et al., 2000; Figueroa-Arredondo et al., 2001; Chakraborty et al., 2011). HlyA was also strongly suggested to be responsible for lethality, developmental delay, and intestinal vacuoles formation in Caenorhabditis elegans during V. cholerae infection (Cinar et al., 2010; Sahu et al., 2012).
Expression of HlyA was highly induced when V. cholerae was cultured in rabbit ileal loops (Xu et al., 2003). It has also been found to be positively regulated by HlyU, a member of SmtB/ArsR family of transcriptional repressors (Saha and Chakrabarti, 2006). Deletion of hlyU decreased HlyA production but increased LD50 in the infant mouse cholera model (Williams et al., 1993). HlyU acts as a dimer that binds to the promoter of hlyA to activate its transcription (Mukherjee et al., 2015). In addition, the quorum sensing (QS) master regulator HapR was shown to be involved in repressing HlyA expression at both the transcriptional and posttranscriptional levels (Tsou and Zhu, 2010). Repression of hlyA by HapR at the transcriptional level was achieved through direct binding of HapR to the hlyA promoter, while that at posttranscriptional level was mediated via the metalloprotease HapA (Tsou and Zhu, 2010). However, as a virulence factor, expression of hlyA should be under the tight control of multiple regulators.
The ferric uptake regulator Fur is a metal-dependent DNA-binding protein that regulates multiple genes related to metabolism and virulence in V. cholerae (Occhino et al., 1998; Davis et al., 2005; Mey et al., 2005; Wyckoff et al., 2007; Davies et al., 2011). A 19 bp palindromic sequence was previously described as the DNA binding box of Fur in V. cholerae (Goldberg et al., 1990). However, the palindromic sequence cannot explain all of the DNA-binding characteristics of Fur. Thus, an enhanced V. cholerae Fur box with a 21 bp palindromic sequence was constructed based on the ChIP-seq-identified binding sites, but it shares an identical span of bases with the previously predicted (Davies et al., 2011). One Fur box-like sequence, TGAATATCAGTAATTGTTATT, was found within the upstream DNA region of hlyA, suggesting that its transcription might be under the direct control of Fur. In the present study, we showed that the highest transcription of hlyA occurs at early mid-logarithmic growth phase due to the collective and elaborate regulation of HapR, Fur, and HlyU, suggesting that HlyA would only function during the early mid-logarithmic growth phase in V. cholerae. The complex regulatory actions of HapR, Fur, and HlyU on hlyA transcription would be beneficial to the invasion and pathogenesis of V. cholerae.
Materials and Methods
Construction of the Mutants and Complementary Mutants
Vibrio cholerae O1 El Tor strain C7258 (Peru, 1991) was used as the wild type (WT) in this study. The deletion mutants of hapR, fur, and hlyU (designated as ΔhapR, Δfur, and ΔhlyU, respectively) were constructed from WT using the suicide plasmid pWM91 by allelic exchange, which was similarly performed as previously described (Wu et al., 2015). To construct the complementary mutants, the entire coding region of each deleted gene was cloned into the pBAD24 vector harboring an arabinose PBAD promoter and an ampicillin resistance gene (Guzman et al., 1995; Sun et al., 2014). After being verified by DNA sequencing, the complementary plasmid for each deleted gene was transferred into the corresponding mutant, yielding the complementary mutant strain ΔhapR/pBAD24-hapR, Δfur/pBAD24-fur, or ΔhlyU/pBAD24-hlyU. In order to counteract the effects of arabinose and ampicillin on bacterial growth, the empty vector pBAD24 was introduced into WT or each mutant to generate WT/pBAD24, ΔhapR/pBAD24, Δfur/pBAD24, or ΔhlyU/pBAD24, respectively (Sun et al., 2014). All the primers used are listed in Table 1.
Table 1.
Oligonucleotide primers used in this study.
| Target | Primers (forward/reverse, 5’-3’) |
|---|---|
| Construction of mutants | |
| hapR | GCGGGATCCCCAGCAATACATCTTTACC/GTGCTGCCCAAGAAAAGGGGTATATCCTTGCC |
| GGCAAGGATATACCCCTTTTCTTGGGCAGCAC/GCGACTAGTAACTCACCAAAACCTTC | |
| GCGGGATCCCCAGCAATACATCTTTACC/GCGACTAGTAACTCACCAAAACCTTC | |
| fur | CGGGATCCTTCGTGTAAGGCAGCAGTAATC/CAGAGCGTAAAGCCTATGGATACTTTCCTGTTGATGTTC |
| GAACATCAACAGGAAAGTATCCATAGGCTTTACGCTCTG/GGACTAGTAGATGAAGATGGTGTGGGAAAC | |
| CGGGATCCTTCGTGTAAGGCAGCAGTAATC/GGACTAGTAGATGAAGATGGTGTGGGAAAC | |
| hlyU | GCGGGATCCCCAGGCAGTCGAACCGCA/TACCTTTTTTTCGACCACCTTTAATTCCAACCCATTCATTC |
| GAATGAATGGGTTGGAATTAAAGGTGGTCGAAAAAAAGGTA/GGACTAGTGAAAGGATAAGAATGTCATAG | |
| GCGGGATCCCCAGGCAGTCGAACCGCA/GGACTAGTGAAAGGATAAGAATGTCATAG | |
| Construction of complemented mutants | |
| hapR | GATTCTAGAAGGAGGAATTCACCATGGACGCATCAATCGAAAAAC/GCGAAGCTTCTAGTTCTTATAGATACACAG |
| fur | GATTCTAGAAGGAGGAATTCACCATGTCAGACAATAACCAAG/GCGAAGCTTTTATTTCTTCGGCTTGTGAG |
| hlyU | GATTCTAGAAGGAGGAATTCACCATGCCGTATTTAAAGG/GCGAAGCTTCTACTGATTCGCCTGAC |
| Protein expression | |
| hapR | GCGGGATCCATGGACGCATCAATCGAAAAAC/GCGAAGCTTCTAGTTCTTATAGATACACAG |
| fur | GCGGGATCCATGTCAGACAATAACCAAG/GCGAAGCTTTTATTTCTTCGGCTTGTGAG |
| hlyU | GCGGGATCCATGCCGTATTTAAAGG/GCGAAGCTTCTACTGATTCGCCTGAC |
| qRT-PCR | |
| hapR | AAACGCAAACTACAACTGATGG/AGCACATCGTCAACCAAGTC |
| fur | AGCCAGAGTGCCAACATATTAG/AATACTGACTTGCCGCCTTC |
| hlyU | CTCAGCCAATCTGCTCTT/AGTTCAATCATCGCCTTC |
| hlyA | CGTTAGATGCCTATTTCCG/CTCCACTGACTTCCACCC |
| Luminescence assay | |
| hapR | GCGGAGCTCCCAGCAATACATCTTTACC/GCGACTAGTTGAGGCGATAGCCGAGTT |
| fur | GCGGAGCTCGCATCAAGGCATAAACGG/GCGACTAGTATACTTTCCTGTTGATGTTC |
| hlyU | GCGGAGCTCTGTTAGTTCCAGGCAGTC/GCGACTAGTTTTAATTCCAACCCATTC |
| hlyA | GCGGAGCTCCAATCTATGCTTATACGG/GCGACTAGTGCAACGATTGAGTTTTGG |
| Primer extension | |
| hlyU | /TTTGCAGTCGCCGCTCATTG |
| hlyA | /TCATGGGTTACCCTCGTC |
| DNase I footprinting | |
| fur | GTAAAACGACGGCCAGTGCATCAAGGCATAAACGG/CAGGAAACAGCTATGACGAGGCAAATCACTGAACAAA |
| hlyU | GTAAAACGACGGCCAGTCGTGTTTATGGCTCCCTC/CAGGAAACAGCTATGACGATGTCGTAATTCGGTTG |
| hlyA | GTAAAACGACGGCCAGTCTTATGTGTAAGCGTATTG/CAGGAAACAGCTATGACCGGATCACAGATTTTAGC |
| M13 | (FAM)GTAAAACGACGGCCAGT/(HEX)CAGGAAACAGCTATGAC |
Growth Conditions
The LB broth (1% tryptone, 0.5% yeast extract, and 1% NaCl) was used for V. cholerae cultivation. Overnight bacterial cultures were diluted 1:50 into 15 ml of fresh LB broth, and grown under 37°C with shaking at 200 rpm to reach an OD600 value of 1.0, and then diluted 1:100 into 15 ml of fresh LB broth for the third-round growth, and were harvested at required cell densities. When necessary, the LB broth was supplemented with 100 μg/ml ampicillin, 5 μg/ml chloramphenicol, 100 μg/ml kanamycin, or 0.1% arabinose.
Hemolytic Activity Assay
The hemolytic activity of V. cholerae strains were tested using the method previously described with slight modifications (Zhang et al., 2017b); 5 μl of the third-round bacterial cultures were transferred onto LB agar containing 5% sheep blood erythrocytes, 100 μg/ml ampicillin, and 0.1% arabinose. The LB blood plates were incubated at 37°C for 20 h.
Luminescence Assay
For the lux activity assay (Xu et al., 2010), the promoter DNA region of each target gene was PCR amplified and cloned into the corresponding restriction endonuclease sites of pBBRlux vector harboring a promoterless luxCDABE reporter gene and a chloramphenicol resistance gene. The resulting plasmid was then transferred into WT and mutant strains, respectively. The V. cholerae strains transformed with recombinant plasmids were cultivated completely in LB broth at 37°C, and harvested at the required cell densities. The luminescence was measured using an Infinite® 200 Pro NanoQuant (Tecan, Switzerland). The lux activity was calculated as light units/OD600.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted using the TRIzol Reagent (Invitrogen, United States). The cDNAs were generated by using 12 μg of total RNAs and 3 μg of random hexamer primers. The quantitative real-time PCR (qRT-PCR) assay was performed and analyzed as previously described (Gao et al., 2011). The relative mRNA levels were determined based on the standard curve of recA (reference gene) expression for each RNA preparation.
Preparation of 6× His-Tagged Proteins
The entire coding region of hapR, fur, and hlyU was amplified and cloned into the plasmid pET28a (Novagen, United States), respectively. The resulting plasmids encoding His-tagged proteins were transferred into Escherichia coli BL21λDE3 cells for protein expression (Kleber-Janke and Becker, 2000). The methods for purification of His-tagged proteins were done as previously described (Gao et al., 2008; Zhang et al., 2012). The eluted His-tagged proteins were dialyzed and then concentrated to a final concentration of 0.3–0.6 mg/ml. The purity of purified proteins was analyzed by SDS-PAGE.
DNase I Footprinting
The procedures for DNase I footprinting assay and DNA sequencing were carried out as previously described (Gao et al., 2017; Osei-Adjei et al., 2017; Zhang et al., 2017b). Briefly, upon being incubated with the increasing amounts of His-tagged protein, the FAM (or HEX)-labeled DNA probes were digested by the optimized RQ1 RNase-Free DNase I (Promega). The digested DNA fragments were then analyzed using an ABI 3500XL DNA Genetic analyzer with GeneMarker software 2.2, while the DNA sequencing products were surveyed with Sequence Scanner software v1.0.
Primer Extension Assay
The primer extension assay was performed as previously described with slight modifications (Gao et al., 2011). Briefly, 12 μg of total RNA was annealed with 1 pmol of 5′-HEX-labeled reverse oligonucleotide primer to generate cDNAs using the Primer Extension System (Promega) according to the manufacturer’s instructions. The primer extension products and sequencing materials were analyzed using the same methods as that of the DNase I footprinting assay.
Experimental Replicates and Statistical Methods
The presented data of hemolytic activity assay, DNase I footprinting, and primer extension were done at least two independent times. The luminescence assay and qRT-PCR were performed with at least three independent bacterial cultures, and the values were expressed as the mean ± SD. Paired Student’s t-test was used to calculate statistically significant differences, and p < 0.01 was considered to indicate statistical significance.
Results
HapR and Fur Represses the Hemolytic Activity of Vibrio cholerae
The hemolytic activities against sheep blood erythrocytes were compared between WT/pBAD24, mutant (ΔhapR/pBAD24, Δfur/pBAD24, or ΔhlyU/pBAD24), and the complementary mutant (ΔhapR/pBAD24-hapR, Δfur/pBAD24-fur, or ΔhlyU/ pBAD24-hlyU) strains (Figure 1). The results showed that the WT/pBAD24 and all of the complementary mutants exhibit α-type hemolysis, but ΔhapR/pBAD24 and Δfur/pBAD24 manifest β-type hemolysis. Thus, both HapR and Fur strongly inhibited the hemolytic activity of V. cholerae. Although the ΔhlyU/pBAD24 strain also exhibited α-type hemolysis, its colony was much dull than that of WT/pBAD24 or ΔhlyU/pBAD24-hlyU, which is consistent with HlyU activation of hlyA transcription. Taken together, these results indicated that the transcription of hlyA would be under the negative control of HapR and Fur, but under the positive regulation of HlyU.
FIGURE 1.
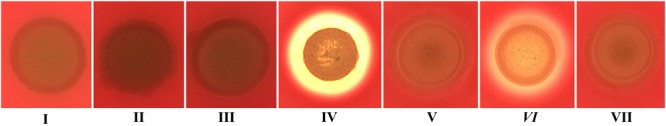
Hemolytic activity of different Vibrio cholerae strains on LB-5% sheep blood agar. After being incubated at 37°C for 48 h, the LB blood plates were photographed. I, II, III, IV, V, VI, and VII represent WT/pBAD24, ΔhlyU/pBAD24, ΔhlyU/pBAD24-hlyU, ΔhapR/pBAD24, ΔhapR/pBAD24-hapR, Δfur/pBAD24, and Δfur/pBAD24-fur, respectively.
Transcription of hapR, fur, hlyU, and hlyA Were All Cell Density-Dependent
The luminescence reporter assay was employed to measure the transcription changes of hapR, fur, hlyU, and hlyA during the growth periods of the strains (Figure 2). The transcription levels of all of the four genes were increased but then reduced with the increase of cell density. In addition, the transcriptional activity of each of the four genes could be detected at all cell densities. However, the highest transcription of hapR occurred at an OD600 value of around 1.0, while that of fur, hlyU, and hlyA appeared at an OD600 value of around 0.6–0.7. Thus, the V. cholerae cells were harvested at the OD600 value of about 1.0 and 0.6 for characterizing HapR- and Fur/HlyU-mediated gene regulation, respectively. Moreover, the cell density-dependent transcription of fur and hlyU suggests that their expression would be under the control of QS.
FIGURE 2.
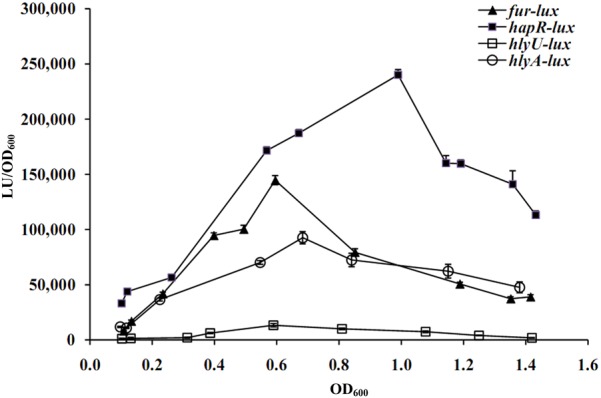
Cell density-dependent expression of target genes. The promoter DNA region of each target gene was cloned into the pBBRlux vector and then transferred into WT to determine the luminescence activity under various OD600 values. The bacteria were cultivated completely in LB broth containing the appropriate antibiotics and grown with shaking at 37°C.
HlyU Activates hlyA Transcription but Represses Its Own Gene
As determined by the luminescence assay (Figure 3A), the promoter activity of hlyA in ΔhlyU was much lower relative to that in WT, whereas that of luminescence under the control of hlyU promoter in ΔhlyU was much higher than that in WT, suggesting the positive and negative regulation of hlyA and hlyU by HlyU, respectively. The qRT-PCR assay further confirmed the positive correlation between HlyU and hlyA transcription in V. cholerae (Figure 3B). The DNase I footprinting assay showed that His-HlyU protected a single region, i.e., –563...–627 or –99...–155, for hlyA or hlyU promoter, respectively, against DNase I digestion in a dose-dependent manner (Figure 3C). Taken together, these results suggested that HlyU acts as a transcriptional activator of hlyA but serves as a repressor of its own gene in V. cholerae.
FIGURE 3.
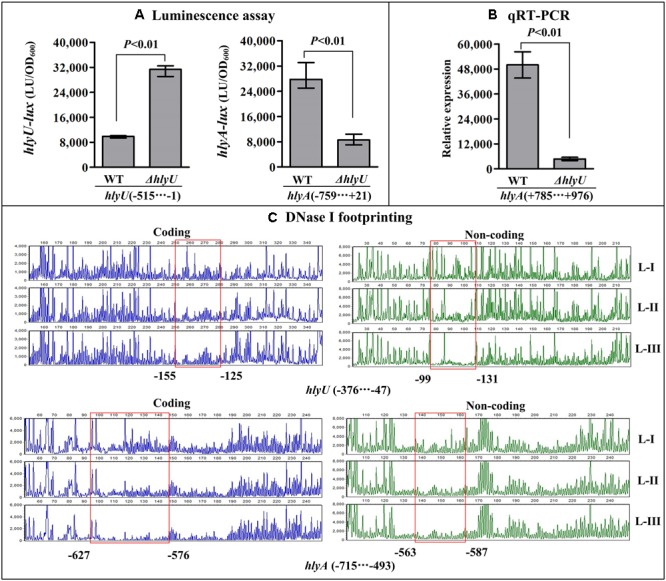
Regulation of hlyU and hlyA by HlyU. The luminescence assay (A) was done as Figure 2. (B) qRT-PCR. The relative mRNA level of fur was compared between ΔhlyU and WT. (C) DNase I footprinting. The promoter fragment of each target gene was labeled with FAM or HEX, incubated with increasing amounts of purified His-HlyU (Lanes-I, II, and III contain 0, 4.06, and 12.12 pmol, respectively), and then subjected to DNase I footprinting assay. The results were analyzed using an ABI 3500XL DNA analyzer. The protected regions are boxed and marked with positions. The negative and positive numbers indicate the nucleotide positions relative to the translation start site (+1) of each target genes, respectively.
Negative Regulatory Actions of HapR and Fur on hlyU and hlyA
The results of luminescence assay showed that the promoter activities of hlyU and hlyA in both ΔhapR and Δfur were much higher relative to that in WT (Figures 4A, 5A). The qRT-PCR assay indicated that the transcriptional levels of hlyU and hlyA significantly increased in ΔhapR and Δfur relative to WT (Figures 4B, 5B). The DNase I footprinting assay disclosed that His-HapR protected two different DNA regions for each promoter against DNase I digestion (Figure 4C), while His-Fur only protected a single region for each promoter (Figure 5C). Taken together, both HapR and Fur repressed the transcription of hlyU and hlyA in a direct manner.
FIGURE 4.
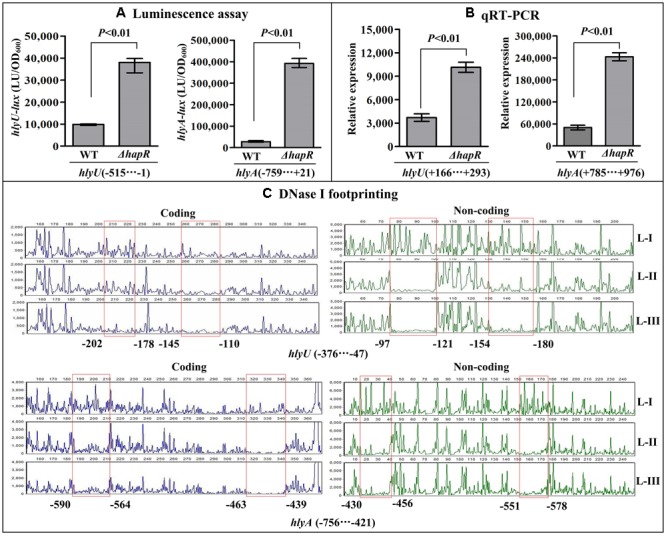
Negatively regulation of hlyU and hlyA by HapR. The luminescence assay (A) was done as Figure 2. The qRT-PCR (B) and DNase I footprinting assay (C) were done as Figure 3. Lanes-I, II, and III contain 0, 2.31, and 6.92 pmol His-HapR, respectively.
FIGURE 5.
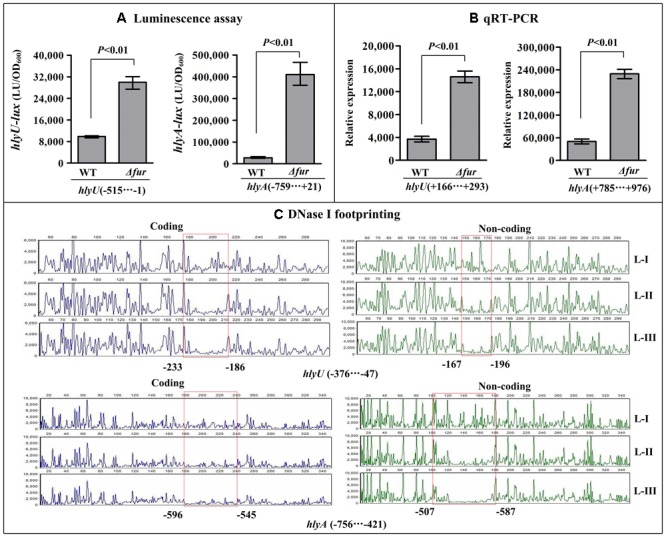
Negatively regulation of hlyU and hlyA by Fur. The luminescence assay (A) was done as Figure 2. The qRT-PCR (B) and DNase I footprinting assay (C) were done as Figure 3. Lanes-I, II, and III contain 0, 2.95, and 8.85 pmol His-Fur, respectively.
Identification of the Transcription Start Sites for hlyA and hlyU
Two transcriptions start sites of fur have been previously reported in V. cholerae (also seen in Figure 8; Litwin et al., 1992; Lam et al., 1994). In the present work, the primer extension assay was employed to map the transcription start sites of hlyA and hlyU. The assay detected only one transcription start site for each gene located at 287 bp upstream of hlyA and 432 bp upstream of hlyU, respectively (Figure 6).
FIGURE 8.
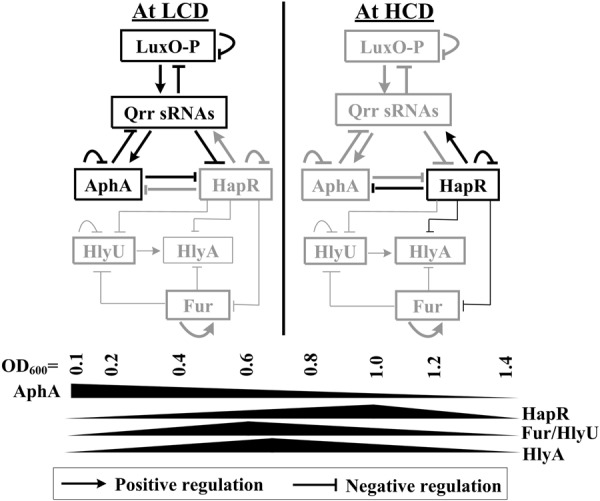
Regulatory circuit. The regulatory actions between LuxO, Qrr sRNAs, AphA, and HapR were previously described in Vibrio cholerae and closely related vibrios (Henke and Bassler, 2004; Tu et al., 2010; Rutherford et al., 2011; Sun et al., 2012; Zhang et al., 2012). AphA and HapR are the two master QS regulators operating at LCD and HCD, respectively. Fur and HlyU, which are transcribed highly at an OD600 value of around 0.6, coordinate with HapR to tightly regulate hlyA transcription, leading to the high expression of HlyA at the early mid-logarithmic growth phase. Positive autoregulation of Fur has been established in V. vulnificus (Lee et al., 2007); this mechanism would be conserved between V. vulnificus and Vibrio cholerae.
FIGURE 6.
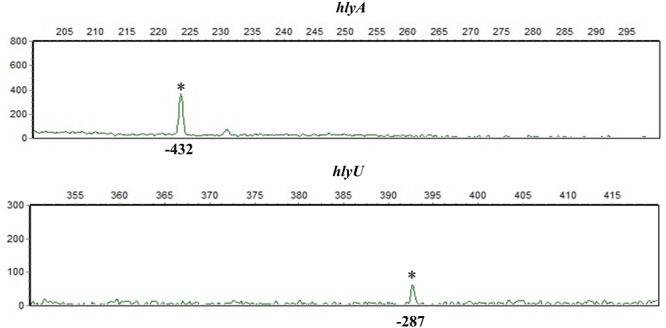
Transcription start sites of hlyA and hlyU. A 5’-HEX-labeled oligonucleotide primer was designed to be complementary to the RNA transcript of each target gene. The primer extension products were analyzed with an ABI 3500XL DNA Genetic analyzer. The transcription start sites are marked with asterisks and positions.
HapR Represses fur Transcription
The recombinant pBBRlux plasmid that contains the promoter-proximal region of fur and a promoterless luxCDABE reporter gene was transferred into ΔhapR and WT, respectively, to test the action of HapR on the promoter activity of fur. As shown in Figure 7A, the promoter activity of fur in ΔhapR was much higher relative to that in WT, indicating the negative correlation of HapR and fur transcription in V. cholerae. As further determined by the qRT-PCR assay (Figure 7B), the mRNA level of fur was significantly enhanced in ΔhapR relative to WT. The results of in vitro DNase I footprinting (Figure 7C) demonstrated that His-HapR protected a single region from 238 to 558 bp upstream of fur against DNase I digestion in a dose-dependent manner. Taken together, HapR directly and negatively regulates the transcription of fur in V. cholerae.
FIGURE 7.
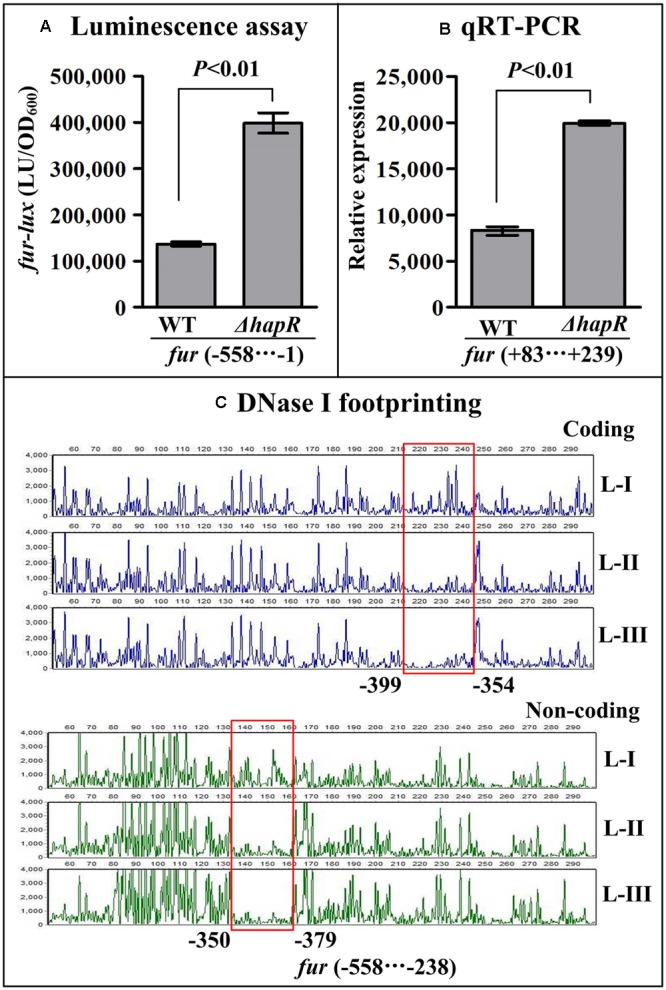
Transcription of fur was negatively regulated by HapR. The luminescence assay (A) was done as Figure 2. The qRT-PCR (B) and DNase I footprinting assay (C) were done as Figure 3.
Discussion
Although iron-, Fur-, HapR-, and HlyU-dependent expression of hlyA has been previously reported in V. cholerae (Stoebner and Payne, 1988; Williams et al., 1993; Tsou and Zhu, 2010; Mukherjee et al., 2015), the detailed regulatory mechanisms need to be further illustrated. In this study, we showed that the transcription of hlyA was regulated coordinately by HlyU, HapR, and Fur in V. cholerae El Tor biotype (Figure 8). At the late mid-logarithmic growth phase (OD600 ≈ 1.0), the highly expressed HapR bound to the promoters of fur, hlyU, and hlyA to repress their transcription. At the early mid-logarithmic growth phase (OD600 ≈ 0.6), the highly expressed Fur bounds to the promoters of hlyU and hlyA to repress their transcription; meanwhile, HlyU bounds to the promoters of hlyA and hlyU to activate and inhibit their transcription, respectively. The highest transcriptional level of hlyA occurred at an OD600 value of about 0.7 due to the tight regulation of HapR, Fur, and HlyU, suggesting that HlyA would function at the early mid-logarithmic growth phase in V. cholerae.
Vibrio vulnificus secretes a potent hemolysin (VvhA) sharing homologous regions with HlyA (Yamamoto et al., 1990b). VvhA exhibits strongly cytolytic and hemolytic activities and may contribute to the bacterial invasion and causes vasodilatation (Kim et al., 1993; Kook et al., 1996; Elgaml and Miyoshi, 2017). Vibrio vulnificus SmcR, a HapR homolog, directly represses the expression of hlyU and vvhA, while HlyU directly activates vvhA transcription (Shao et al., 2011; Wen et al., 2012). Deletion of hlyU resulted in the loss of cytotoxicity and reduced VvhA production in the smcR mutant (Shao et al., 2011). The double mutant of smcR and hlyU regained cytotoxicity and hemolytic activity when hns was further deleted (Shao et al., 2011). HlyU seems act as an anti-repressor of H-NS in the regulation of the virulence genes in V. vulnificus (Liu et al., 2009). In addition, it has been shown that iron represses vvhA transcription via Fur, which represses vvsA transcription in the presence of iron through the protein–promoter DNA association (Kim et al., 2009). The binding site of Fur overlaps with that of SmcR but with a higher affinity than SmcR (Wen et al., 2012). Moreover, Fur has been shown to be involved in the regulation of smcR transcription in V. vulnificus (Kim et al., 2013; Wen et al., 2016). However, V. cholerae Fur seems to have no regulatory activity on hapR transcription (data not shown), suggesting that the regulation of the QS regulator gene by Fur may depend on the bacterial growth conditions. Nevertheless, the conservative regulatory mechanisms might be employed to tightly control of the HlyA production in V. vulnificus and V. cholerae.
The transcription of hlyA, fur, and hlyU was stimulated at the early mid-logarithmic growth phase but repressed at both low cell density (LCD) and high cell density (HCD; Figure 2), suggesting that some unknown regulators can repress their transcription at LCD. AphA has been considered as the bottom master regulator of QS operating at LCD (Ng and Bassler, 2009; Ball et al., 2017). The DNA binding box of AphA has been identified as an inverted repeat of ATATGC with a 6-nt centered spacer, i.e., ATATGCA-N6-TGCATAT (Sun et al., 2012). An AphA box-like sequence (ATACTCCTCTTTAATCTCAT) was detected within the promoter of hlyU but was not found in the other two promoters (Figure 9). Thus, the transcription of hlyU would be under the direct control of AphA. The asymmetric production of AphA and HapR orthologs coupled with their combined inhibition of downstream targets has been observed in other vibrios (Van Kessel et al., 2013; Zhang et al., 2017a). For example, in Vibrio harveyi, both AphA and LuxR bound to the promoters of the type III secretion system (T3SS) genes to repress their transcription, resulting in the highest expression levels of T3SS occurring at LCD-to-HCD transition (Van Kessel et al., 2013); in Vibrio parahaemolyticus, ToxR coordinates with AphA and OpaR to repress T6SS1, leading to the highest expression of T6SS1 occurring at the mid-logarithmic growth phase (Zhang et al., 2017a). However, the detailed regulatory mechanisms of AphA or other additional factors on hlyA transcription in V. cholerae need to be further investigated.
FIGURE 9.
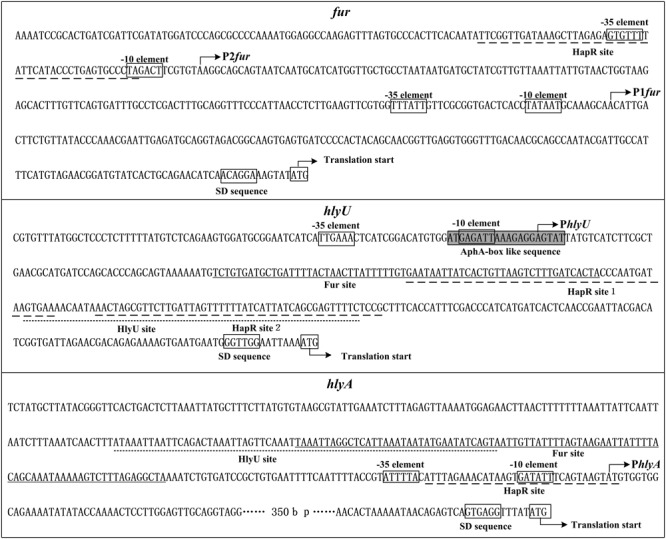
Structural organization of target promoters. The DNA sequence was derived from Vibrio cholerae El Tor C7258. The transcription/translation start sites are indicated by bent arrows. Shine-Dalgarno (SD) box and –10/–35 elements are enclosed in boxes. The HapR sites are underlined with broken lines, the Fur sites are underlined with solid lines, while the HlyU sites are underlined with dotted lines.
The organization of fur, hlyU, and hlyA promoters was reconstructed herein, by collecting the data of translation/transcription start sites, promoter -10 and -35 elements, HapR/Fur/HlyU binding sites, AphA box-like sequence, and Shine-Dalgarno (SD) sequences (ribosomal binding sites; Figure 9). One HapR binding site was detected in the upstream of fur and hlyA, respectively, and each overlaps the core -10 element; two HapR binding sites for hlyU were detected and both of them were located downstream of the transcription start site. Thus, the binding of HapR would block the entry or elongation of the RNA polymerase to repress the transcription of the target genes. Both the Fur and HlyU sites for hlyU are located downstream of the transcription start site, indicating that the repression mechanisms of hlyU by Fur and HlyU would be similar to that by HapR. Notably, the Fur site for hlyU overlaps the HapR site 1, while HlyU site overlaps the HapR site 2. Thus, there may be a competitive binding activity between HapR and Fur or HlyA in the binding of the hlyU promoter. Although the binding site of Fur to hlyA is located upstream of the transcription start site, it overlaps with that of HlyU. Thus, the binding of HapR would block the binding of HlyU, which acts as a transcriptional activator of hlyA in V. cholerae.
Conclusion
This work reports that QS coordinates with HlyU and Fur to regulate hlyA transcription in V. cholerae, leading to the highest transcription of hlyA occurring at the early mid-logarithmic growth phase when bacteria cells are grown in LB broth. Therefore, we propose that at the early or middle stage of infection, V. cholerae produces high amount of HlyA in the small intestine, which promotes the bacterial invasion and pathogenesis, and contributes to the watery diarrhea; at the end of the infectious cycle, since HapR is highly expressed, it activates the protease production and inhibits the biofilm formation, to detach the mutual aggregation of V. cholerae cells in the initial infection sites in the intestine. Meanwhile, the expression of CT decreases, and therefore the shift to lower expression of HlyA may have a similar response of CT.
Author Contributions
HG, JX, YZ, and BK conceived the study and designed experimental procedures. XL, JLi, JLo, HZ, BD, and QS performed the experiments and carried out data analysis. HG, YZ, and BK wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer RW and handling Editor declared their shared affiliation.
Acknowledgments
We thank Dr. George Osei-Adjei from the Republic of Ghana for language editing the manuscript.
Footnotes
Funding. This study was supported by grants from the Science Foundation for the State Key Laboratory for Infectious Disease Prevention and Control from China (Grant No. 2015SKLID509) and the National Natural Science Foundation of China (Grant Nos. 81471917 and 81401715).
References
- Almagro-Moreno S., Pruss K., Taylor R. K. (2015). Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog. 11:e1004787. 10.1371/journal.ppat.1004787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball A. S., Chaparian R. R., Van Kessel J. C. (2017). Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J. Bacteriol. 199:e00105-17. 10.1128/JB.00105-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez J. A., Silva A. J. (2016). Vibrio cholerae hemagglutinin(HA)/protease: an extracellular metalloprotease with multiple pathogenic activities. Toxicon 115 55–62. 10.1016/j.toxicon.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D. C., Mukherjee G., Banerjee P., Banerjee K. K., Biswas T. (2011). Hemolysin induces Toll-like receptor (TLR)-independent apoptosis and multiple TLR-associated parallel activation of macrophages. J. Biol. Chem. 286 34542–34551. 10.1074/jbc.M111.241851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar H. N., Kothary M., Datta A. R., Tall B. D., Sprando R., Bilecen K., et al. (2010). Vibrio cholerae hemolysin is required for lethality, developmental delay, and intestinal vacuolation in Caenorhabditis elegans. PLoS One 5:e11558. 10.1371/journal.pone.0011558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J. D., Nair G. B., Ahmed T., Qadri F., Holmgren J. (2017). Cholera. Lancet 390 1539–1549. 10.1016/S0140-6736(17)30559-7 [DOI] [PubMed] [Google Scholar]
- Coelho A., Andrade J. R., Vicente A. C., Dirita V. J. (2000). Cytotoxic cell vacuolating activity from Vibrio cholerae hemolysin. Infect. Immun. 68 1700–1705. 10.1128/IAI.68.3.1700-1705.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. W., Bogard R. W., Mekalanos J. J. (2011). Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl. Acad. Sci. U.S.A. 108 12467–12472. 10.1073/pnas.1107894108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. M., Quinones M., Pratt J., Ding Y., Waldor M. K. (2005). Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187 4005–4014. 10.1128/JB.187.12.4005-4014.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellis L., Diana A., Arcidiacono D., Fiorotto R., Portincasa P., Altomare D. F., et al. (2009). The Vibrio cholerae cytolysin promotes chloride secretion from intact human intestinal mucosa. PLoS One 4:e5074. 10.1371/journal.pone.0005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep T. T., Nguyen N. T., Nguyen T. N., An H. K., Nguyen T. Q., Nguyen V. H., et al. (2015). Isolation of New Delhi metallo-beta-lactamase 1-producing Vibrio cholerae non-O1, non-O139 strain carrying ctxA, st and hly genes in southern Vietnam. Microbiol. Immunol. 59 262–267. 10.1111/1348-0421.12248 [DOI] [PubMed] [Google Scholar]
- Elgaml A., Miyoshi S. I. (2017). Regulation systems of protease and hemolysin production in Vibrio vulnificus. Microbiol. Immunol. 61 1–11. 10.1111/1348-0421.12465 [DOI] [PubMed] [Google Scholar]
- Fallarino A., Attridge S. R., Manning P. A., Focareta T. (2002). Cloning and characterization of a novel haemolysin in Vibrio cholerae O1 that does not directly contribute to the virulence of the organism. Microbiology 148(Pt 7) 2181–2189. 10.1099/00221287-148-7-2181 [DOI] [PubMed] [Google Scholar]
- Figueroa-Arredondo P., Heuser J. E., Akopyants N. S., Morisaki J. H., Giono-Cerezo S., Enríquez-Rincón F., et al. (2001). Cell vacuolation caused by Vibrio cholerae hemolysin. Infect. Immun. 69 1613–1624. 10.1128/IAI.69.3.1613-1624.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhang L., Osei-Adjei G., Yang W., Zhou D., Huang X., et al. (2017). Transcriptional regulation of cpsQ-mfpABC and mfpABC by CalR in Vibrio parahaemolyticus. Microbiologyopen 6:e00470. 10.1002/mbo3.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhang Y., Yang L., Liu X., Guo Z., Tan Y., et al. (2011). Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol. 11:40. 10.1186/1471-2180-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhou D., Li Y., Guo Z., Han Y., Song Y., et al. (2008). The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190 3063–3075. 10.1128/JB.01910-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. B., Boyko S. A., Calderwood S. B. (1990). Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J. Bacteriol. 172 6863–6870. 10.1128/jb.172.12.6863-6870.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke J. M., Bassler B. L. (2004). Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186 6902–6914. 10.1128/JB.186.20.6902-6914.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose Y., Yamamoto K., Nakasone N., Tanabe M. J., Takeda T., Miwatani T., et al. (1987). Enterotoxicity of El Tor-like hemolysin of non-O1 Vibrio cholerae. Infect. Immun. 55 1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. M., Chung Y. Y., Shin S. H. (2009). Iron differentially regulates gene expression and extracellular secretion of Vibrio vulnificus cytolysin-hemolysin. J. Infect. Dis. 200 582–589. [DOI] [PubMed] [Google Scholar]
- Kim H. R., Rho H. W., Jeong M. H., Park J. W., Kim J. S., Park B. H., et al. (1993). Hemolytic mechanism of cytolysin produced from V. vulnificus. Life Sci. 53 571–577. 10.1016/0024-3205(93)90714-E [DOI] [PubMed] [Google Scholar]
- Kim I. H., Wen Y., Son J. S., Lee K. H., Kim K. S. (2013). The fur-iron complex modulates expression of the quorum-sensing master regulator, SmcR, to control expression of virulence factors in Vibrio vulnificus. Infect. Immun. 81 2888–2898. 10.1128/IAI.00375-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber-Janke T., Becker W. M. (2000). Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Expr. Purif. 19 419–424. 10.1006/prep.2000.1265 [DOI] [PubMed] [Google Scholar]
- Kook H., Lee S. E., Baik Y. H., Chung S. S., Rhee J. H. (1996). Vibrio vulnificus hemolysin dilates rat thoracic aorta by activating guanylate cyclase. Life Sci. 59 L41–L47. 10.1016/0024-3205(96)00292-5 [DOI] [PubMed] [Google Scholar]
- Lam M. S., Litwin C. M., Carroll P. A., Calderwood S. B. (1994). Vibrio cholerae fur mutations associated with loss of repressor activity: implications for the structural-functional relationships of fur. J. Bacteriol. 176 5108–5115. 10.1128/jb.176.16.5108-5115.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Bang S. H., Lee K. H., Park S. J. (2007). Positive regulation of fur gene expression via direct interaction of fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 189 2629–2636. 10.1128/JB.01791-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin C. M., Boyko S. A., Calderwood S. B. (1992). Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J. Bacteriol. 174 1897–1903. 10.1128/jb.174.6.1897-1903.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Naka H., Crosa J. H. (2009). HlyU acts as an H-NS antirepressor in the regulation of the RTX toxin gene essential for the virulence of the human pathogen Vibrio vulnificus CMCP6. Mol. Microbiol. 72 491–505. 10.1111/j.1365-2958.2009.06664.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A. R., Wyckoff E. E., Kanukurthy V., Fisher C. R., Payne S. M. (2005). Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73 8167–8178. 10.1128/IAI.73.12.8167-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R., Figueroa P., Mukhopadhyay A. K., Shimada T., Takeda Y., Berg D. E., et al. (2000). Cell vacuolation, a manifestation of the El tor hemolysin of Vibrio cholerae. Infect. Immun. 68 1928–1933. 10.1128/IAI.68.4.1928-1933.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Pal A., Chakravarty D., Chakrabarti P. (2015). Identification of the target DNA sequence and characterization of DNA binding features of HlyU, and suggestion of a redox switch for hlyA expression in the human pathogen Vibrio cholerae from in silico studies. Nucleic Acids Res. 43 1407–1417. 10.1093/nar/gku1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43 197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhino D. A., Wyckoff E. E., Henderson D. P., Wrona T. J., Payne S. M. (1998). Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29 1493–1507. 10.1046/j.1365-2958.1998.01034.x [DOI] [PubMed] [Google Scholar]
- Osei-Adjei G., Gao H., Zhang Y., Zhang L., Yang W., Yang H., et al. (2017). Regulatory actions of ToxR and CalR on their own genes and type III secretion system 1 in Vibrio parahaemolyticus. Oncotarget 8 65809–65822. 10.18632/oncotarget.19498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T., Van Kessel J. C., Shao Y., Bassler B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 25 397–408. 10.1101/gad.2015011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R. P., Chakrabarti P. (2006). Molecular modeling and characterization of Vibrio cholerae transcription regulator HlyU. BMC Struct. Biol. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu S. N., Lewis J., Patel I., Bozdag S., Lee J. H., LeClerc J. E., et al. (2012). Genomic analysis of immune response against Vibrio cholerae hemolysin in Caenorhabditis elegans. PLoS One 7:e38200. 10.1371/journal.pone.0038200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C. P., Lo H. R., Lin J. H., Hor L. I. (2011). Regulation of cytotoxicity by quorum-sensing signaling in Vibrio vulnificus is mediated by SmcR, a repressor of hlyU. J. Bacteriol. 193 2557–2565. 10.1128/JB.01259-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. V., Matte M. H., Matte G. R., Jiang S., Sabeena F., Shukla B. N., et al. (2001). Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67 910–921. 10.1128/AEM.67.2.910-921.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebner J. A., Payne S. M. (1988). Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56 2891–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhang Y., Qiu Y., Yang H., Yang W., Yin Z., et al. (2014). H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front. Microbiol. 5:675. 10.3389/fmicb.2014.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Zhang Y., Wang L., Yan X., Tan Y., Guo Z., et al. (2012). Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One 7:e44210. 10.1371/journal.pone.0044210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou A. M., Zhu J. (2010). Quorum sensing negatively regulates hemolysin transcriptionally and posttranslationally in Vibrio cholerae. Infect. Immun. 78 461–467. 10.1128/IAI.00590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu K. C., Long T., Svenningsen S. L., Wingreen N. S., Bassler B. L. (2010). Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell. 37 567–579. 10.1016/j.molcel.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel J. C., Rutherford S. T., Shao Y., Utria A. F., Bassler B. L. (2013). Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J. Bacteriol. 195 436–443. 10.1128/JB.01998-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Kim I. H., Kim K. S. (2016). Iron- and quorum-sensing signals converge on small quorum-regulatory RNAs for coordinated regulation of virulence factors in Vibrio vulnificus. J. Biol. Chem. 291 14213–14230. 10.1074/jbc.M116.714063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Kim I. H., Son J. S., Lee B. H., Kim K. S. (2012). Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J. Biol. Chem. 287 26727–26739. 10.1074/jbc.M112.374165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. G., Attridge S. R., Manning P. A. (1993). The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol. Microbiol. 9 751–760. 10.1111/j.1365-2958.1993.tb01735.x [DOI] [PubMed] [Google Scholar]
- Wu R., Zhao M., Li J., Gao H., Kan B., Liang W. (2015). Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in Vibrios. Sci. Rep. 5:14921. 10.1038/srep14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E. E., Mey A. R., Payne S. M. (2007). Iron acquisition in Vibrio cholerae. Biometals 20 405–416. 10.1007/s10534-006-9073-4 [DOI] [PubMed] [Google Scholar]
- Xu Q., Dziejman M., Mekalanos J. J. (2003). Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U.S.A. 100 1286–1291. 10.1073/pnas.0337479100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Stern A. M., Liu Z., Kan B., Zhu J. (2010). Virulence regulator AphB enhances toxR transcription in Vibrio cholerae. BMC Microbiol. 10:3. 10.1186/1471-2180-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Ichinose Y., Shinagawa H., Makino K., Nakata A., Iwanaga M., et al. (1990a). Two-step processing for activation of the cytolysin/hemolysin of Vibrio cholerae O1 biotype El Tor: nucleotide sequence of the structural gene (hlyA) and characterization of the processed products. Infect. Immun. 58 4106–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Wright A. C., Kaper J. B., Morris J. G., Jr. (1990b). The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae E1 Tor hemolysin gene. Infect. Immun. 58 2706–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao H., Osei-Adjei G., Zhang Y., Yang W., Yang H., et al. (2017a). Transcriptional regulation of the Type VI Secretion System 1 Genes by Quorum Sensing and ToxR in Vibrio parahaemolyticus. Front. Microbiol. 8:2005. 10.3389/fmicb.2017.02005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qiu Y., Tan Y., Guo Z., Yang R., Zhou D. (2012). Transcriptional regulation of opaR, qrr2-4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One 7:e34622. 10.1371/journal.pone.0034622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Y., Gao H., Zhang L., Yin Z., Huang X., et al. (2017b). Vibrio parahaemolyticus CalR down regulates the thermostable direct hemolysin (TDH) gene transcription and thereby inhibits hemolytic activity. Gene 613 39–44. 10.1016/j.gene.2017.03.001 [DOI] [PubMed] [Google Scholar]


