Abstract
Iron is an essential nutrient for many bacteria. Since the metal is highly sequestered in host tissues, bound predominantly to heme, pathogenic bacteria often take advantage of heme uptake and degradation mechanisms to acquire iron during infection. The most common mechanism of releasing iron from heme is through oxidative degradation by heme oxygenases (HOs). In addition, an increasing number of proteins that belong to two distinct structural families have been implicated in aerobic heme catabolism. Finally, an enzyme that degrades heme anaerobically was recently uncovered, further expanding the mechanisms for bacterial heme degradation. In this analysis, we cover the spectrum and recent advances in heme degradation by infectious bacteria. We briefly explain heme oxidation by the two groups of recognized HOs to ground readers before focusing on two new types of proteins that are reported to be involved in utilization of heme iron. We discuss the structure and enzymatic function of proteins representing these groups, their biological context, and how they are regulated to provide a more complete look at their cellular role.
Keywords: pathogenic bacteria, heme oxygenase, heme degradation, heme binding, iron regulation
Introduction
In the late 1960s, Tenhunen and colleagues isolated the first heme oxygenase, HO-1 (Tenhunen et al., 1968, 1969). While the enzyme was discovered in rats, HO-1 is highly conserved in mammals. The discovery of HO-1 provided valuable insights into the pathway that reduces hemoglobin to bile (Ostrow et al., 1962). Previous to this discovery, research showed that heme is cleaved to generate the linear tetrapyrrole biliverdin, which is subsequently reduced to bilirubin by biliverdin reductase. Yet, the mechanism by which heme was converted to biliverdin was not understood. Almost 30 years after discovering HO-1, the first prokaryotic HO, HmuO, was described in the Gram-positive Corynebacterium (Schmitt, 1997b; Wilks and Schmitt, 1998). Schmitt and colleagues screened a genomic library of Corynebacterium diphtheriae in Corynebacterium ulcerans mutants that were unable to grow in medium containing heme or hemoglobin as iron sources. The clone selected based on its ability to restore growth carried a gene (hmuO) with high homology to HO-1. Biochemical characterization revealed that, in the presence of a redox partner, HmuO catalyzes in vitro the oxidative cleavage of heme to produce biliverdin IXα (Wilks and Schmitt, 1998; Kunkle and Schmitt, 2007). Two more bacterial HOs, HO-1 and HemO, were then cloned from Synechocystis spp. PCC 6803 of the Cyanobacteria phylum, and the Gram-negative Proteobacterium, Neisseria meningitides respectively (Cornejo et al., 1998; Zhu et al., 2000). Both HOs also share overall structural and mechanical similarities with the mammalian HO-1. The discovery of bacterial enzymes that are comparable to the mammalian counterparts promoted the initial notion that heme degradation represents an evolutionarily conserved mechanism. The mechanism these bacterial and mammalian HOs use to cleave the heme ring is referred to as “canonical heme oxidation” (Wilks and Heinzl, 2014). The discovery of IsdG and IsdI, two paralogous HOs from Staphylococcus aureus, and MhuD, a similar enzyme from Mycobacterium tuberculosis, gave rise to the second family of HOs due to their unique structure and novel mechanism for cleaving the protoporphyrin ring (Skaar et al., 2004a; Chim et al., 2010; Nambu et al., 2013). These enzymes demonstrated that oxidative heme catabolism could be achieved by more than one class of bacterial enzymes.
Since the discovery of HmuO, heme degradation in bacteria has become an active research area. Currently HOs are divided into the canonical HOs, which share structure and mechanistic similarities to HO-1, and the IsdG-like group of HOs (Wilks and Heinzl, 2014). Many potential HOs have also been identified based on their ability to carry out aerobic heme degradation in vitro. However, the acceptance of these proteins as HOs has met with some skepticism over the reaction mechanism and whether these proteins catalyze ring opening in vivo (Wilks and Ikeda-Saito, 2014). Non-enzymatic degradation of heme to verdoheme can be achieved by the addition of hydrogen peroxide (H2O2) to the solution (known as coupled oxidation). Coupled oxidation can take place in the presence of molecular oxygen (O2) and a reducing agent that converts the heme iron to ferrous iron (Fe+2) and H2O2. Various hemoproteins (e.g., myoglobin) and cytochrome mutants, in which the axial ligand was removed or replaced with a weaker ligand, readily exhibit this reaction (Avila et al., 2003). While oxidative cleavage by a HO or breakdown by coupled oxidation is achieved by two separate mechanisms, they cannot be discriminated based on their products alone as they both yield CO and verdoheme (which is hydrolyzed to biliverdin under acidic or alkali conditions). Because of this complexity and since proteins that exhibit heme degradation in vitro (in the presence of a reductant) may have another function under physiological conditions, it was suggested that a comprehensive approach that combines genetics, mechanistic enzymology, and metabolite profiling is necessary for the characterization these proteins before terming them HOs (Wilks and Ikeda-Saito, 2014). Such extensive studies are not yet available for all cases but in this review, we attempt to compel what information exists. We briefly explain the HO-1 and the IsdG-like HOs before focusing on two new types of bacterial proteins that are reported to be involved in iron gain from heme. We discuss the structure and enzymatic function of these proteins, the biological context, and regulation mode. For this paper, we reviewed the published literature as well as structural information. Protein structures were generated by downloading the Protein Data Bank file into PyMol (Table 1) and sequence comparisons were generated through NIH Protein BLAST.
Table 1.
Proteins and their corresponding Protein Data Bank Identity (PDB) used for bioinformatical analysis.
Lastly, many bacteria degrade heme to obtain the iron. Here we focus on infectious bacteria, where heme catabolism is particularly important since it allows pathogens to leverage on the host heme pools. Nevertheless, it is important to note that non-pathogenic bacteria also use HOs and that these enzymes can be involved in additional functions. For example, in the photosynthetic Cyanobacteria, HO-1 catalyzes the first step in the production of phycocyanobilin, the tetrapyrrole chromophore the bacteria use to harvest solar energy (Cornejo et al., 1998).
Heme is a key iron source for pathogenic bacteria within the host environment
Although iron is a common element on Earth, very little of it is free in host tissues, partially because iron is insoluble under physiological conditions (Sánchez et al., 2017). Additionally, free ferric ions can interact with H2O2 through the Haber-Weiss reaction to generate free radicals (Valko et al., 2005). This can lead to cell death due to oxidative stress, which injures tissues through lipid peroxidation and can damage genetic material. Sequestration of iron is vital for the host because iron is an essential nutrient for most invading microorganisms. Iron overload is linked to increase in both the quantity of incidents and the severity of bacterial infections in patients as can be seen in hemodialysis patients (Ishida and Johansen, 2014). Evidences linking iron availability to increased susceptibility were also obtained in murine infection models with multiple bacterial pathogens including Vibrio vulnificus, Listeria monocytogenes, Yersinia pestis, and Yersinia pseudotuberculosis (Sword, 1966; Quenee et al., 2012; Arezes et al., 2015; Miller et al., 2016). For these reasons, the host iron is tightly sequestered in proteins.
Most mammalian iron is bound to heme which is predominately in complex with hemoglobin (67% of the total iron) but also found in myoglobin and cytochromes (3.5 and 3% of the total iron, respectively). A significant but smaller fraction of the body iron (25%) is stored in the cellular compartment in ferritin or hemosiderin. The plasma contains only 0.1% of the iron in the body, where it is mobilized as ferric ion bound to transferrin, a glycoprotein that is maintained under physiological conditions only at 30–40% saturation level. Hemoglobin and heme that are liberated by lysis are captured by the plasma proteins haptoglobin and hemopexin, respectively. The resulting complexes are then cleared by macrophages. Ferric iron is cleared from secretions (e.g., tears, saliva, etc.) by lactoferrin. Therefore, under physiological conditions the majority of a host's iron is bound to proteins and held in the intracellular compartment. In response to infection, the innate immune system orchestrates a systemic reduction in the iron levels and bioavailability by inhibiting the dietary uptake of iron and its export into circulation, increasing the level of heme- and ferric- binding proteins in the serum, and releasing apo-lactoferrin into the infection site (Johnson and Wessling-Resnick, 2012; Ganz and Nemeth, 2015; Knutson, 2017).
As a result, pathogenic bacteria have developed mechanisms that compete for the host's iron during infection (Carver, 2018). A significant fraction of the strategies used by bacteria to obtain iron from the host is dedicated to heme acquisition (Sheldon et al., 2016; Huang and Wilks, 2017). Many bacteria secrete hemolysins that can rupture erythrocytes and other blood cell types. Some intracellular pathogens, such as Salmonella, activate hemophagocytic macrophages to take up erythrocytes (Silva-Herzog and Detweiler, 2008). Heme is not only readily taken up and used by numerous bacterial pathogens, it serves as the preferred iron source for important pathogens such as S. aureus and Streptococcus pyogenes (Eichenbaum et al., 1996; Skaar et al., 2004b). Bacteria employ soluble and/or surface proteins that can seize heme from hemoproteins (e.g., hemoglobin, hemopexin etc.). Heme is then shuttled across the outer-membrane and/or the cell wall onto dedicated transporters for import across the cytoplasmic membrane. Inside the bacterial cell, heme is incorporated directly into proteins or it is degraded to release the iron (Sheldon and Heinrichs, 2015). Oxidative degradation by HOs is the most common mechanism used to liberate iron from heme, although other mechanisms have been discovered.
The structure and function of heme degrading enzymes
Canonical heme oxygenases
The first discovered group of HOs are usually referred to as canonical HOs, and include mammalian-HO-1, HmuO, and PigA/HemO. They are α-only proteins and typically contain nine to ten helixes. Their catalytic sites are highly conserved, especially the proximal α-helix (Figure 2A) (Hirotsu et al., 2004). Comparative studies between apo- and holo-HO-1 and HmuO show that heme binding induces a conformational change where the proximal α-helix twists in on itself, tightening the helix and shortening it. This results in the proximal α-helix moving toward the heme molecule. Concurrently, the distal α-helix swings toward the heme, causing the proximal and distal α-helix to open and close around the heme-binding pocket.
This group of HOs degrade heme to α-biliverdin (a linear tetrapyrrole), CO and free ferrous iron (Fe+2) by three successive oxygenation steps and consumes three O2 and seven electrons (Figure 1A). This mechanism has been extensively reviewed in several articles (Wilks and Heinzl, 2014; Wilks and Ikeda-Saito, 2014). Briefly, the reaction is initiated by the reduction of the heme ferric iron (Fe+3) and O2 binding. The coordinated oxygen (Fe+2-O2) is then reduced to peroxide (Fe+3-OOH), a reactive species that hydroxylates the heme on the α-meso carbon. In the second step, the α-meso-hydroxyheme is oxidized to α-verdoheme while releasing CO. In the last step, the verdoheme ring is cleaved, generating Fe+3-biliverdin complex that is reduced to release the iron (Fe+2). This differs from coupled oxidation in the first step, were peroxide from the solvent (H2O2) instead of coordinated peroxide (Fe+3-OOH) reacts with the heme iron to form the meso-hydroxyheme (which in turn progress to yield verdoheme). Hence, unlike HO catalysis, coupled oxidation is inhibited by the H2O2 scavenging enzyme, catalase.
Figure 1.
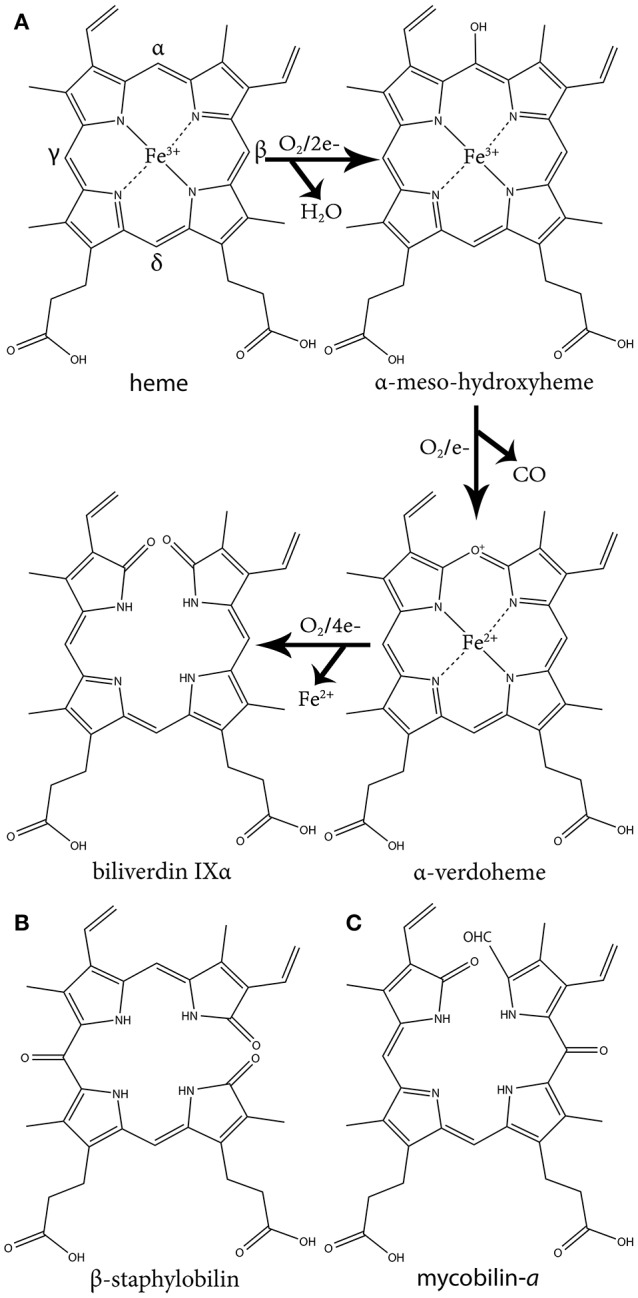
The three oxidative steps of canonical heme oxidation and the chemical structure of staphylobilin and mycobilin. (A) The three oxidative steps of canonical heme oxidation. In the first step heme is oxidized to ferric-hydroperoxide, then it is self-hydroxylated to α-meso-hydroxyheme. Next it is oxidized to α-verdoheme, which is ultimately oxidized to α-biliverdin. (B) One of the two isomers of staphylobilin produced by IsdG/I. (C) One of the two isomers of mycobilin produced by MhuD.
Regioselectivity is the preference of a specific chemical bond over other possible bonds during a reaction. For this group of HOs the regioselectivity is for the α-meso carbon over the β-, γ-, or δ-meso carbons. Regioselectivity is controlled during the hydroxylation step due to steric hindrance of the distal helix interacting with the protoporphyrin ring and allowing ring opening only at the α-meso carbon (Figure 2A) (Unno et al., 2013). The distal helix also bends, bringing the reactive iron dioxygen species closer to the α-meso carbon. The interactions between the distal helix and the protoporphyrin ring are stabilized through an extensive hydrogen bond network that is supplied by water molecules. This network also serves as the proton relay network required for oxygen activation (Unno et al., 2007, 2013).
Figure 2.
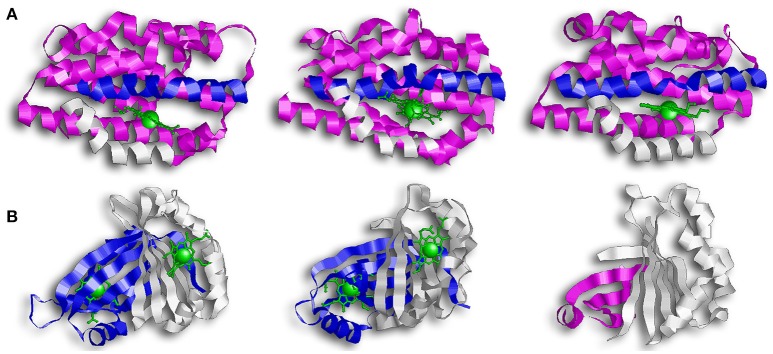
Overall structure of canonical and IsdG-like heme oxygenases. (A) From left to right: HO-1, HmuO, and PigA. This family of enzymes are commonly referred to as the canonical HOs. They are α-only proteins and have been colored so that the proximal helix is in white and the distal helix is in blue. These helixes take an open conformation when the binding pocket is empty but tighten and close around the heme molecule. Note how the propionate groups in PigA are rotated compared to HO-1 and HmuO. (B) From left to right: IsdG, MhuD, Isd-LmHde. These enzymes represent the second group of HOs, the IsdG-like HOs. This group consists of α/β proteins that dimerize across their β-sheets. Both IsdG and MhuD have been colored so that one monomer is in white and the second monomer is in blue. The Isd-LmHde structure has been colored so that the N-terminal is in magenta and the C-terminal is in white.
While the regioselectivity of the α-meso carbon is a defining characteristic of this group of bacterial HOs, a notable exception exists in PigA/HemO. Interestingly, PigA/HemO produces a mixed product of β- and δ-biliverdin in a 3:7 ratio (Friedman et al., 2004). A structure study of heme bound PigA/HemO shows that the heme is rotated about 100° in the binding pocket as compared to HO-1 and HmuO (Figure 2A). This heme rotation is believed to result in the favoring of the δ-meso carbon over the α-meso carbon in PigA/HemO during the hydroxylation step (Friedman et al., 2004; Fujii et al., 2004). Mutagenic analysis of HO-1 and HmuO show that mutations to the amino acids that interact directly with the propionates or through solvent interactions are important for proper placement of the heme in the binding site (Unno et al., 2007). In PigA/HemO, substituting Lys34 and/or Lys132 with alanine residues allows the production of α-biliverdin by the mutant enzymes in addition to β- and δ-biliverdin (Fujii et al., 2004). It was suggested that these residues interact with the propionate of the rotated heme molecule and that when the interactions are lost, it releases the ring rotation allowing the positioning the of α-meso carbon at the oxidation site. In addition, the ratio of β- and δ-biliverdin in PigA/HemO is greatly influenced by the amino acid identity at position 189, which is occupied by phenylalanine. A F189W mutant was found to produce mostly β-biliverdin (~90%) during heme catalysis. These observations suggest that steric interactions bewteen the amino acid residue at position 189 with the heme impact the enzyme's topology and hence the reaction regiospecifificy.
The IsdG family of heme oxygenases
The second group of bacterial HOs are commonly referred to as both non-canonical and IsdG-like HOs (Wilks and Heinzl, 2014). The founder of this group, IsdG, and its paralog, IsdI (64% sequence identity and 79% sequence similarity), were discovered in S. aureus, and degrade heme to staphylobilin and formaldehyde instead of biliverdin and CO (Figure 1B) (Skaar et al., 2004a; Reniere et al., 2010; Matsui et al., 2013). Following the discovery of IsdG/I, MhuD was discovered in M. tuberculosis. During heme degradation, MhuD retains its meso-carbon as an aldehyde and generates mycobilin as its final product (Figure 1C) (Nambu et al., 2013). MhuD shares 24% sequence identity and 46% sequence similarity with IsdG. Both of these enzymes are α+β class of protein and are part of the dimeric α- and β-barrel superfamily (Figure 2B). MhuD and IsdG/I contain a ferredoxin-like fold, consisting of three α-helices and four β-strands arranged in a βαββααβ pattern, in their ABM domain (PF03992) (Figure 2B). These enzymes also have similar active site structures. In IsdG, enzymatic activity is supplied by the catalytic triad Asn7, Trp67, and His77 (Wu et al., 2005). His77 serves as the axial ligand, Trp67 induces heme ruffling, and Asn7, which is critical for catalytic activity, acts to stabilize the reaction intermediates and contributes to heme distortion. Mutagenesis of key residues in MhuD, showed that its heme ligand is a conserved histidine residue (His75) and that a mutation of Asn7 to alanine resulted in decreased enzymatic activity (Nambu et al., 2013).
Advancements in time-resolved in proteo mass spectrometry have allowed for the characterization of the heme-derived species formed during IsdG mediated heme degradation (Streit et al., 2016). Using this process, Streit and colleagues observed the production of an intermediate species with the same molecular weight as mycobilin (a formyloxobilin with α regiospecificity) (Figure 1C). Further analysis revealed this intermediate consists of mixed β/δ-formyloxobilin isomers which is consistent with the reported production of both β- and δ-staphylobilin by IsdG (Reniere et al., 2010). Overall, the data suggests that in the IsdG reaction, an initial rapid conversion of heme to meso-hydroxyheme is followed by the formation of formyloxobilin, which resolves into staphylobilin by releasing CH2O. More research is needed to understand why MhuD does not release formaldehyde from its formyloxobilin product. Interestingly, while a non-catalytic conversion of formyloxobilin to staphylobilin occurs when the product is extracted from the enzyme under aerobic conditions, in proteo MS analysis revealed that IsdG converts only a fraction of the produced formyloxobilins to staphylobilins (Streit et al., 2016). Hence, it seems possible that the remained formyloxobilin population might have a biological function.
The difference in regioselectivity of staphylobilin and mycobilin may be explained by a 90° in-plane rotation of the heme in the catalytic site which results in alternative forms of heme ruffling (Figure 3). Heme ruffling occurs when steric forces from the protein cause the usually planar heme molecule to bend. In IsdG/I, this results in the β- and δ-meso carbons being pushed toward the distal ligand (Lee et al., 2008; Takayama et al., 2015). This is distinct from the MhuD-induced heme ruffling, which leads to the α- and γ-meso carbons being exposed to reactive iron-dioxygen species (Graves et al., 2014). However, since the final product for MhuD are all α-formyloxobilins, other interactions must also be important for its regiospecificity. This differs from HmuO catalyzed heme oxidation, where water molecules associated with the HO push the α-meso carbon toward the reactive iron dioxygen species.
Figure 3.
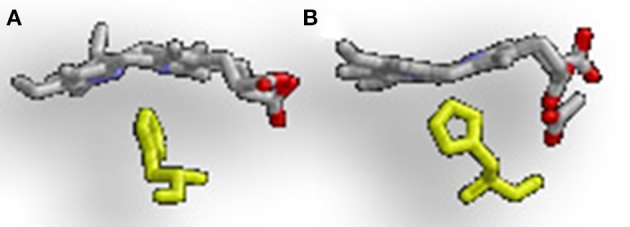
Alternative heme ruffling in IsdG and MhuD. (A) Represents the heme ruffling in IsdG and (B) Represents the heme ruffling in MhuD. Notice in (A), how the IsdG bends the heme so that the β- and δ-meso carbons are pushed away from the histidine, causing the α-meso carbon to move toward the ligand. In (B), MhuD bends the heme so that the β- and δ-meso carbons are closer to the histidine ligand.
In addition to a putative isdG ortholog, the food-born pathogen L. monocytogenes codes for a second protein, Isd-LmHde, which shares structurally similarity with IsdG (Figure 2B) (Duong et al., 2014). Isd-LmHde is a monomer, and the full-length protein forms a β-barrel that is highly similar to the dimmers formed by IsdG/I and MhuD. The C-terminal region of Isd-LmHde shares 58% sequence similarity and 26% sequence identity with the IsdG monomer, but Isd-LmHde has an additional domain at the protein amino terminus. The N-terminal domain of Isd-LmHde consists of a pseudo-ferredoxin-like fold, arranged βαβββ, while the C-terminal domain of the enzyme has of the same ferredoxin-like fold found in the IsdG-like HOs (βαββααβ). Interestingly, an isolated C-terminal fragment of Isd-LmHde forms a homodimer in solution similar to the IsdG protein (Duong et al., 2014). Both the full-length protein and isolated C-terminal domain of Isd-LmHde degrade heme in vitro (in the presence of catalase, favoring enzymatic degradation). However, activity by the truncated enzyme is significantly reduced compared to the full-length protein, demonstrating that both protein regions in Isd-LmHde are necessary for full function.
Enzymes with HemS motifs
This group consists of heme-binding and degrading proteins, intracellular heme shuttling proteins, and possibly have other functions. We discuss these proteins together due to overall structural similarity. The group's founder member, HemS, is associated with the heme-specific transport system in Yersinia enterocolitica (Stojiljkovic and Hantke, 1994). HemS alleviates heme toxicity in Escherichia coli and is required for Y. enterocolitica to utilize heme as an iron source (Stojiljkovic and Hantke, 1992; Schneider et al., 2006). It also contains tandem repeats of a domain termed the HemS motif (PF05171). This motif is also found in the heme shuttling protein PhuS of Pseudomonas aeruginosa, and in HmuS from the related Y. pseudotuberculosis and ChuS from E. coli O157:H7, both of which degrade heme in vitro.
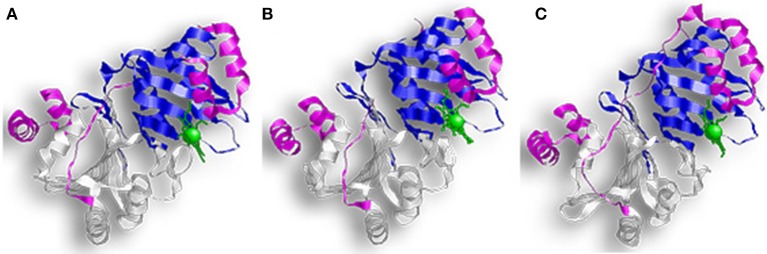
HemS and ChuS function as monomers; while PhuS crystalized as a dimer, the monomeric form is more stable and the dominant form in solution (Figure 4) (Schneider and Paoli, 2005; Suits et al., 2006; Block et al., 2007; Tripathi et al., 2013). HemS shares 35% sequence identity and 51% similarity to PhuS, and 34% sequence identity and 49% similarity to ChuS. Looking at the structure, ChuS consists of 10 α-helices and 18 β-strands arranged so that two β-sheets of nine antiparallel β-strands form the core of the enzyme. The core is flanked asymmetrically by three α-helices arranged in an α-loop-α-loop-α motif on the C-terminal side and a pair of parallel α-helices on the N-terminal. This configuration is conserved in the structure of HemS, although HemS is symmetrical, with three α-helices arranged in an α-loop-α-loop-α on each side. For ChuS the carboxy and amino termini share limited primary sequence homology, but they represent structural duplication in its tertiary structure (Suits et al., 2005). The two halves appear to be associated across a central β-sheet, with key residues being conserved between both halves (i.e., Arg29/209, Phe127/304, and Tyr138/315). In the crystalized form, the PhuS dimers align across the β-sheets, forming a structure highly similar to ChuS. The C-terminal side of PhuS consists of three α-helices arranged in an α-loop-α-loop-α motif and a pair of parallel α-helices on the N-terminal side (Figure 4C).
Figure 4.
Overall structure of proteins with HemS motifs. (A) Represents ChuS, (B) Represents HemS, and (C) Represents PhuS. In all representations the N-terminal HemS domain is in white and the C-terminal HemS domain is in blue. Looking at the structure, ChuS consists of a mix of α-helices and β-strands arranged so that the β-sheets form the core of the enzyme and are flanked by three α-helices on one side and two on the other side. This configuration is conserved in the structure of HemS, although HemS is symmetrical, with three α-helices on each side.
The binding pocket in PhuS and HemS is more open than in ChuS, where the C-terminal α-helices are closer to the binding site (Schneider and Paoli, 2005; Lee et al., 2014). This supports studies showing that PhuS acts in vivo as an intracellular heme transport protein (Bhakta and Wilks, 2006). PhuS shuttles heme to PigA/HemO, a HO in P. aeruginosa (O'neill and Wilks, 2013). Upon binding heme, the C-terminal domain of PhuS rearranges to facilitate PhuS and PigA/HemO interactions, and heme transfer (Deredge et al., 2017). Heme-induced conformational changes were also reported for HemS, which alters between an apo-form that is open to a closed, heme-bound state (Schneider et al., 2006).
The heme binding residues for these proteins are located on their α7 helix with a conserved histidine acting as the iron axial ligand (His196 for HemS, His193 for ChuS, and His209 for PhuS) (Schneider and Paoli, 2005; Suits et al., 2006; Tripathi et al., 2013). Studies of a PhuS H209A mutant revealed that it can alternatively bind heme using His212 and that His210 helped stabilize these interactions (O'neill et al., 2012). Further analysis showed that the H209A mutant could still form a complex with the PigA/HemO, although to a less extend than the wild type protein. In comparison, while H212A and H210A mutants bound heme, they lost the ability to interact with PigA/HemO. Therefore, the current model suggests His209 and His212 of holo-PhuS act as a ligand switch for PigA/HemO (O'neill et al., 2012; Tripathi et al., 2013).
ChuS completes several cycles of aerobic heme degradation in vitro when either ascorbic acid or cytochrome P450 reductase (CPR)/NADPH are used as electron sources (Suits et al., 2005, 2006). Interestingly, both the N-terminal and the C-terminal halves of ChuS are tandem repeats that retained enzymatic activity, but the activity of the isolated domains varies depending on which reductant is used in the reaction (Suits et al., 2005). Catalase was found to inhibit this reaction in a concentration dependent manner, indicating that H2O2 is required for activity. While the exact mechanism is not fully understood, UV/Visual stop flow spectroscopy, NMR spectroscopy, and EPR analysis have provided insights into the reaction intermediates and products (Ouellet et al., 2016). Ouellet and colleges suggest that the first intermediate, α-meso hydroxyheme generated by H2O2 attack of ChuS-bound heme, is a short-lived intermediate that progresses to ferric verdoheme. A subsequent ring cleavage releases Fe+3, hematic acid (a monopyrrole moiety) and a tripyrrole product (containing both vinyl groups and one propionate). The tetrapyrrole ring cleavage in this reaction is different from simple hydrolysis of verdoheme, which produces a mixture of the four biliverdin isomers. This reaction also takes place in the presence of ascorbate or CPR/NADPH without externally added H2O2. The enzyme accelerates the production of H2O2 by ascorbate, suggesting that ChuS promotes H2O2-coupled heme degradation. The chuS gene is associated with heme-uptake machinery and is regulated due to iron availability, implying a function in supplying the bacteria with heme iron. The ability to release iron from heme under a relatively mild peroxide concentration, and the absence of known HOs in E. coli that degrade heme under aerobic conditions, are consistent with this proposal. Nevertheless, genetic analysis and addition biochemical characterization are required to test this hypothesis, and the possibility that ChuS serves alternative roles (e.g., protection from oxidative stress).
An ortholog of HemS from Bartonella henselae's was tested for heme degradation in vivo using the E. coli strain FB8.27 (Liu M. et al., 2012). This strain is engineered to take up heme, but it cannot release the iron from the porphyrin ring due to inactivation of the enzymes efeB and yfeX. Therefore, FB8.27 cannot grow using externally added heme as the sole source of iron. A recombinant B. henselae HemS protein restored growth of FB8.27 using heme iron, suggesting that HemS promotes the release of iron from heme in vivo. Currently the degradation products have not been characterized, and while it degrades heme in the presence of ascorbate or CPR/NADPH, HemS's sensitivity to catalase has not been investigated (Liu M. et al., 2012). Interestingly, a reduction of hemS gene expression does not impact growth, although it did increase the bacterial sensitivity to H2O2, implicating this enzyme in oxidative stress protection.
HmuS, a close HemS ortholog from Y. pseudotuberculosis, degrades heme to mixed biliverdin β- and δ-isomers in vitro (Onzuka et al., 2017). Heme degradation using either ascorbate or ferredoxin/NADPH both with and without catalase produced biliverdin while consuming molecular oxygen and releasing CO. The spectral readings of reactions performed with and without catalase remained the same. Although the reaction was slower with catalase, the heme was eventually converted to biliverdin. When HmuS was incubated with only H2O2, verdoheme accumulated as the final product. Heme degradation (but not binding) was abolished when the proposed axial ligand (His196) or a distal arginine (Arg102) was mutated to alanine. Together the data is consistent with enzymatic degradation of heme, but direct evidence that supports its function in heme catabolism in vivo is still missing.
In summary, evolution of the HemS family may reflect individual adaptation of different bacteria to their growth environment. While some proteins from this family (e.g., PhuS) serve in intracellular heme shuttling, others may contribute to protection from oxidative stress and/or iron supply (e.g., B. henselae HemS and ChuS). In vitro, many of the family members are reported to exhibit heme degradation activity, yet biliverdin was confirmed only with HmuS from Y. pseudotuberculosis. Non-enzymatic cleavage was not ruled out in all cases and at least with ChuS, heme cleavage by H2O2-driven reaction is suggested, albeit by a unique enzymatic mechanism that seems possible under physiological conditions. Hence more investigations, specifically with whole bacteria, are required to gain a better understanding of the function of proteins from this family.
Enzymes with FMN-binding domains
The FMN binding-like superfamily is divided into the NADH FMN oxidoreductase-like subfamily and the pyridoxamine 5'-phosphate oxidase (PNPOx)-like subfamily (PF01243). Several proteins from the PNPOx-like subfamily bind and/or degrade heme in vitro. Based on this proliferation, Hu et al. (2011) recommend adding a third subfamily to the FMN superfamily that includes these heme-binding proteins (Hu et al., 2011). HutZ (Vibrio cholerae), ChuZ (Campylobacter jejuni), HugZ (Helicobacter pylori), and HupZ (S. pyogenes) are examples of heme-binding proteins with PNPOx-like domains that could belong to this third subfamily. While they all contain FMN-like binding domain, neither HupZ nor HutZ bound FMN in vitro (HupZ data not published) (Liu X. et al., 2012).
All four of these enzymes crystallized as dimers, and size-exclusion assays for HugZ, ChuZ, and HutZ confirmed that they purify as dimers (Hu et al., 2011; Zhang et al., 2011; Uchida et al., 2012). Looking at their genetic sequence, the C-terminal regions are more conserved than the N-terminal region. HugZ and ChuZ have the highest level of similarity (54% sequence identity and 68% sequence similarity). Their N-terminal regions contain a simple α/β-type domain arranged in a three-stranded anti-parallel β-sheet, stacked against two α-helices in HugZ and three α-helices in ChuZ (Figure 5A). Their C-terminal regions contain the PNPOx-like domain and consist of eight anti-parallel β-stands and four α-helices. In ChuZ, the β-barrel takes on a distorted form with β10 and β11 extending out on one end of the barrel. HutZ and HupZ are smaller than HugZ and ChuZ, and share more similarity to the C-terminal region of HugZ and ChuZ than the N-terminal region (Figure 5B). In HutZ and HupZ, the antiparallel split-barrels are formed by six β-strands and helices α1 and α2 block the open ends of the β-barrel. HutZ has two more α-helices that are packed against the side of the β-barrel and HupZ, the smallest of all, has only one additional α-helix.
Figure 5.
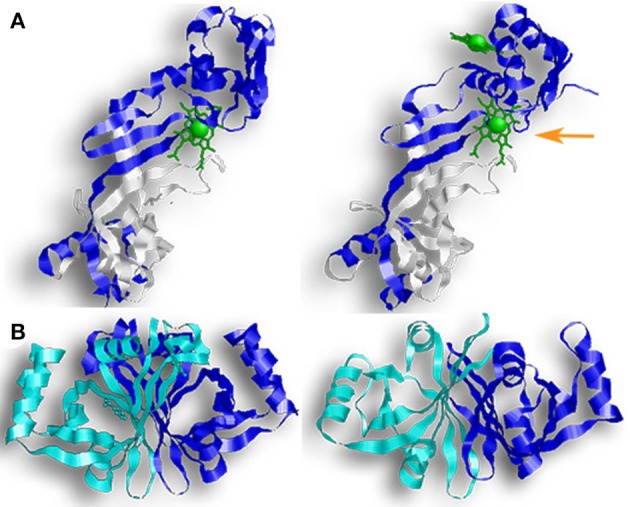
Enzymes with FMN-binding domains. Proteins from the PNPOx-like subfamily contain a β-barrel with a Greek key topology (a protein structure consisting of four antiparallel strands connected by three hairpin loops that is named for a common pattern in Grecian decoration). The pictured enzymes also contain FMN-binding domains, although neither HutZ or HupZ can bind FMN. (A) (from left to right) a monomer of HugZ and a monomer of ChuZ. In both representations, the PNPOx domain is in white. In ChuZ, the canonical heme binding pocket is indicated by the orange arrow. (B) (from left to right) HutZ and HupZ. In both representations one monomer is colored blue and the other monomer is colored teal.
Dimerization in these proteins occurs across their β-sheets with one monomer packed at about 90° from the second monomer. Crystal structures of holo-HugZ and ChuZ show that the heme-binding pocket lies near the dimerized β-sheets and that heme binding is stabilized through interactions with residues from both monomers (Figure 5A). In ChuZ, the heme is bound between neighboring dimers using hydrophobic interactions between the protoporphyrin ring and the side-chains of His9 and His14 on different dimers (Zhang et al., 2011). The heme molecule binds with the propionic groups pointed toward the center of the dimer (Figure 5A). This is distinctive from canonical HOs where the propionic group is parallel to the enzyme (Figure 2A). Site directed mutagenesis of HugZ confirmed that His245 is involved in heme iron coordination and Arg166 interacts with the carboxylate group of the propionates (Hu et al., 2011). In ChuZ, His245 is also involved in heme iron coordination but Arg166 is not conserved and instead Lys166 interacts with the carboxylate group (Zhang et al., 2011). Currently only apo-crystal structures exist for HutZ and HupZ, but analyses using single site substitution mutagenesis of HutZ have shown that His63 and Arg92 (corresponding to His245 and Arg166 in HugZ) interact with the propionate due to shifts in heme-bound spectra (Liu X. et al., 2012). HupZ, however, lacks His245 and contains a glycine instead of Arg166, making it difficult to determine the heme coordinating residues based on sequence analysis alone (Sachla et al., 2016). Interestingly, ChuZ crystallized not only with the expected heme binding site, but with an additional surface bound heme molecule (Zhang et al., 2011). If ChuZ can also bind heme via the surface pocket in vivo, it may serve as a mechanism for preventing heme toxicity.
While all four proteins bind heme and mediate heme degradation in vitro, their physiological role is still under debate. A recent biophysical analysis of HutZ demonstrated that in the presence of exogenically added H2O2, heme is degraded to verdoheme (possibly β- or δ-isomer) and CO (Uchida et al., 2012). In the presence of ascorbate, the HutZ reaction is suggested to produce ferric biliverdin based on the appearance of broad featureless absorption in the visible region but this reaction is inhibited by catalase, which suggests coupled oxidation over enzymatic activity (Wyckoff et al., 2004). The hutZ gene is encoded by the iron-regulated hutWXZ operon that is divergently expressed from a heme import system in V. cholera. This operon compliments the growth of hmuO mutants in Cyanobacteria on heme iron, although complementation requires an intact hutZ gene (but not hutW or X). In addition, a V. cholera mutant that lost hutZ could not efficiently use heme as a source of iron. Since the hutZ mutant is not more sensitive to heme toxicity, it's phenotype is likely related to a defect in acquisition of heme iron (Wyckoff et al., 2004). In vivo studies implicate HutZ in heme catabolism, but more research is required.
In vitro degradation of heme by HugZ, ChuZ, and HupZ was observed in the presence of catalase (Ridley et al., 2006; Guo et al., 2008; Sachla et al., 2016). All three proteins degrade heme to produce CO and a chromophore that absorbs at 660 nm (in the presence of ascorbate or CPR/NADH). HPLC chromatography has confirmed that HugZ produce δ-biliverdin (Guo et al., 2008). While the chromophore produced by the ChuZ and HupZ reactions were not identified, the stability of HupZ's chromophore under acidic conditions suggests it is not verdoheme (Sachla et al., 2016). The iron-regulated expression of all three enzymes is consistent with a role in iron acquisition. Genetic studies are available only for chuZ where inactivation of this gene impaired the ability of C. jejuni to use heme or hemoglobin as a source of iron. Notably, the loss of chuZ had no impact on the bacterial sensitivity to oxidative stress (Ridley et al., 2006). In summary, similarly to the proteins with HemS domains, heme-binding proteins from the PNPOx-like subfamily might have evolved to play separate roles in different bacterial species, and more comprehensive investigations are required to describe the reaction catalyzed by these proteins and to determine their role in bacterial physiology.
The genetic link to iron metabolism in pathogenic bacteria
Bacteria tightly regulate iron levels to prevent overload and the resulting oxidative stress. Accordingly, expression of genes involved in heme catabolism are often repressed by iron and activated by heme, sometimes in competing manners. Here we discuss several examples of metal-dependent transcriptional regulation, as well as heme regulation by a two-component system and a post-translational modification.
Many enzymes, including isdG and isdI, are regulated by the Ferric Uptake Regulator, or Fur (Carpenter et al., 2009; Troxell and Hassan, 2013). The isdG gene is part of a chromosomal locus that encodes a key heme uptake pathway and it is the terminal gene of the isdCDEF/srtB/isdG operon. This operon also encodes a membrane heme transporter (isdCDEF) and a dedicated heme import sortase (srtB) (Reniere and Skaar, 2008). Transcription of this operon is derived from the isdC promoter, divergently from the heme-receptor genes isdA and isdB. The isdI gene is the second gene in a bicistronic operon located elsewhere in the genome (Reniere and Skaar, 2008). The promoters controlling the expression of isdG and isdI each contain a Fur box. In the wild type strain, IsdG and IsdI levels increase when medium iron levels are decreased, however the enzymes levels remain stationary in a null fur mutant (Reniere and Skaar, 2008). This shows that expression is repressed in the presence of iron, although IsdG is also stabilized in the presence of heme (discussed below). Of note, isdG and isdI double knock-out mutants still exhibit growth with heme as the only iron source, suggesting S. aureus contains a third heme degrading mechanism (Reniere and Skaar, 2008).
Similarly to S. aureus, P. aeruginosa also encodes two HOs, PigA/HemO and BphO; however, only PigA/HemO is involved in iron metabolism. The pigA/hemO gene, which is part of a Fur regulated gene cluster, is required for efficient use of heme iron by P. aeruginosa and can complement the phenotype of hemO mutants in N. meningitidis (Ratliff et al., 2001). The second HO, BphO, catalyzes the production of biliverdin used for the assembling the photoreceptor phytochrome, BphP (Wegele et al., 2004). BphO is transcribed upstream from BphP in a bicistronic operon that is regulated by cell density and expressed independently of the iron status. PhuS, however, only transfers heme to PigA/HemO. In addition to studies demonstrating heme transfer from holo-PhuS to PigA/HemO, 13C-heme isotopic labeling studies showed that a ΔphuS mutant degrades externally provided heme using both of its HOs, PigA/HemO and BphO (Barker et al., 2012). Since the wild type strain uses only PigA/HemO to degrade externally provided heme, these experiments support the role of PhuS as intracellular heme chaperon that shuttles the heme specifically to PigA/HemO. Interestingly, these studies also implicated PhuS as a modulator of heme uptake since the mutant strain demonstrated altered expression of the heme uptake machinery.
The expression of the chuZ gene, an enzyme with a PNPOx-like domain found in C. jejuni, is also regulated by Fur. The chuZ gene, is located directly upstream of the chuABCD genes, which together code for a heme ABC transporter (Ridley et al., 2006). Mutants that lack either chuZ or chuA are unable to utilize heme as an iron source, but mutants that lack chuB, chuC, or chuD are only partly affected, suggesting an additional heme uptake system. Two fur boxes lie in between chuZ and chuA. Using transcriptional fusions and competitive EMSA assays, Ridley and colleagues demonstrated that both promoters are expressed in an iron-dependent manner and bind to Fur protein in vitro (Ridley et al., 2006).
The Diphtheria Toxin Repressor (DtxR) is mechanistically similar to Fur and controls the expression of genes involved in metal homeostasis and pathogenesis of many Gram-positive bacteria (Nobles and Maresso, 2011; Merchant and Spatafora, 2014). The transcription of hmuO from C. diphtheriae is repressed in the presence of iron by DtxR (and activated by heme, as will be described below) (Schmitt, 1997a). EMSA and DNase I footprinting experiments showed that DtxR binds in a metal-dependent manner in the promoter region of hmuO in C. diphtheriae (Schmitt, 1997a). The expression of hmuO in C. ulcerans appears to be controlled differently (Kunkle and Schmitt, 2007). Although the hmuO promoter in C. ulcerans contains a putative DtxR like box, promoter fusion studies revealed that it is only weakly repressed by iron. The basal expression of hmuO grown in the absence of heme is significantly higher in C. ulcerans than in C. diphtheriae, although the mechanism is unclear. A null hmuO C. ulcerans mutant cannot survive with heme as the only iron source while the phenotype of C. diphtheriae mutants vary, exhibiting moderate to no growth attenuation depending on the strain (Kunkle and Schmitt, 2007). Together these observations suggest that hmuO is important for the extraction of iron from heme in C. ulcerans and in some strains of C. diphtheriae, and that C. diphtheriae likely encodes additional factor(s) that allow the use of heme iron.
A DtxR-like protein named MtsR regulates the transcription of hupZ, a recently described enzyme in S. pyogenes (Bates et al., 2005; Sachla et al., 2016). MtsR binds to DNA in the presence of either iron or manganese. It was first recognized as the repressor of the sia operon (shr/shp/siaABCDEFG) which encodes a central heme import pathway and of the mtsABC metal transporter (Hanks et al., 2006; Sachla et al., 2016). Global transcription analysis of null mtsR mutants established MtsR as a global regulator that controls the expression of many S. pyogenes genes, including iron and heme uptake, metal homeostasis and important virulence factors (Toukoki et al., 2010). A two-gene cluster encoding a putative surface receptor and a cytoplasmic enzyme, later named HupZ, was identified in the MtsR regulon (Sachla et al., 2016). The in vitro activity of HupZ and its repression by MtsR implicate this enzyme in the utilization of heme as an iron source, but this role has yet to be established in vivo.
The expression of some heme associated genes is dependent on heme availability. The heme dependent-regulation of hmuO from C. diphtheriae is the first case where bacteria were shown to use two-component systems to modulate the expression of HOs. Out of the eleven two-component systems encoded by C. diphtheriae, two (ChrAS and HrrAS) modulate heme homeostasis. Both regulatory systems activate the hmuO promoter in the presence of heme, but the ChrAS system is the dominant one (Bibb et al., 2007; Bibb and Schmitt, 2010). ChrAS also activates the transcription of hrtAB, an ABC transporter that functions in heme detoxification. While ChrAS and HrtAB both repress hemA, a heme biosynthesis gene. Fluorescence staining experiments demonstrated that the sensor kinase, ChrS, autophosphorylates then phosphorylates ChrA, the response regulator (Burgos and Schmitt, 2012). A conserved aspartate residue in ChrA (Asp50) was implicated as the phosphorylation site and shown to be important for ChrA binding to hmuO promoter. DNase I footprint experiments identified two 10-bp segments (designated as S2ho and S3ho) that protected by ChrA and are conserved in other ChrA regulated promoters. S3ho partially overlaps the DtxR binding box, suggesting that ChrA and DtxR compete for binding during growth in the presence of heme and iron. Mutagenesis and promoter fusion experiments confirm the importance of the both S2ho and S3ho in the regulated expression of hmuO (Burgos and Schmitt, 2012).
Heme availability also regulates IsdG, but via a post-translational modification (Reniere and Skaar, 2008; Reniere et al., 2011). As was previously mentioned, IsdG is the terminal gene of the isd operon. While the entire operon is repressed by Fur in the presence of iron, only the concentration of IsdG is enhanced in the presence of heme. A pulse-chase and immunoprecipitation study of IsdG (expressed from a constitutive promoter) found that the half-life of IsdG increased 2.5 fold in medium containing heme, suggesting that heme stabilizes the protein (Reniere and Skaar, 2008). The stability of a catalytically inactive IsdG was similarly increased by heme, indicating that it is the heme and not the reaction product (staphylobilin) that facilitates stability. Analysis of a collection of chimeric proteins consisting of different IsdI and IsdG segments implicated an internal, β-strands containing, fragment in IsdG degradation. Further investigations identified a flexible loop near the heme-binding site that is required (but not sufficient) for proteolysis in the absence of heme (Reniere et al., 2011). It was hypothesized that the differential regulation of IsdG/I allows S. aureus to specifically adjust the expression of its HOs to the different host environment. Interestingly, while both enzymes are required for full pathogenesis, isdG mutants are more impaired in heart and kidney colonization models in comparison to isdI mutants (Reniere and Skaar, 2008).
Future direction
Advances in the fields of biochemistry and bioinformatics are improving the understanding of bacterial heme degradation. There is, however, still more to learn about these enzymes, such as why MhuD retains an aldehyde while IsdG does not and whether proteins with HemS or PNPOx domains degrade heme or protect from oxidative stress in vivo. Additionally, several species of bacteria lack proteins with homology to reported heme degrading enzymes and several enzymes were discovered that do not use oxidation to open the heme ring. One such protein is the recently discovered ChuW, a radical S-adenosylmethionine methyltransferase from E. coli O157-H7, which appears to degrade heme use a primary carbon radical to initiate a methyl transfer instead of binding O2 (Lamattina et al., 2016). In E. coli K12 the enzymes YfeX and EfeB also liberate iron from heme without breaking the protoporphyrin ring (Létoffé et al., 2009).
More research is needed to understand the mechanism for many of these proteins and would allow better understanding of their cellular role. Oxidative heme degradation requires an electron supply in addition to molecular oxygen and the electron donor for most bacterial HOs are unknown. Mammalian HOs use CPR and in the absence of a known physiological electron donor, in vitro heme degradation is achieved using CPR or an alternative reducing agent (e.g., trolox or ascorbate). Often, different reducing agents produce different degradation kinetics and can halt the reaction at different stages, therefore they give a limited view of the in vivo reaction. In P. aeruginosa, the NADPH-dependent ferredoxin reductase, Fpr, was implicated as the physiological electron donor for PigA/HemO due to its ability to sustain catalytic activity during in vitro studies (Wang et al., 2007). The two likely reductase partners for the Staphylococcal HOs are IruO (a pyridine nucleotide-disulfide oxidoreductase) and NtrA (a nitroreductase) (Hannauer et al., 2015). Severe defects in heme usage are exhibited by an iruO ntrA double mutant (but not with either of the single gene mutants) implicated these enzymes in the process of heme catabolism. The search for native electron donors for bacterial HOs is important to develop the understanding of heme catalysis at the molecular level. New advances in our ability to monitor heme degradations as they are carried out in the cell are likely to enhance this important area of bacterial physiology.
Understanding the downstream effects of heme degradation products, such as biliverdin, staphylobilin, and CO is another area of research that could be expanded. In mammals, biliverdin is released from HO-1 and reduced to bilirubin by the action of biliverdin reductase. In bacteria, the destiny of the bilin produced during heme catalysis and its role in the cell physiology remains largely unknown. A recent mouse study showed that over expression of the E. coli ChuS leads to attenuation of the innate immune system (Maharshak et al., 2015). This immunomodulation was achieved by decreasing the expression of the pro-inflammatory cytokine, IL-12 p40, and increasing expression of the anti-inflammatory cytokine, IL-10. Researchers speculated that cross-talk between mammalian HO-1 and enteric bacteria HOs might be a key component of maintaining healthy intestinal microbiota, making it an important area for farther study.
Since heme iron is used by many pathogenic bacteria, targeting heme uptake and degradation provides attractive targets for therapeutic inhibition. A direct role between HOs and virulence was established in only a few cases. Mice challenged with mutant S. aureus that lacked either isdG, isdI, or both genes, showed decreased bacterial loads in heart and kidney tissues compared to WT indicating that they are necessary for full pathogeneses (Reniere and Skaar, 2008). Deleting the hemO gene in Leptosria interorgans, which is required for growth on hemoglobin iron, resulted in a 50% increase of survival rate of infected hamsters (Murray et al., 2009). A better understanding of HOs function and the mechanisms that facilitate heme degradation during infection may lay the ground for new therapeutic designs.
Conclusion
In many pathogenic bacteria, heme catabolism is carried out by HOs similar to either HO-1 or IsdG. Other bacteria encode enzymes that bind and degrade heme in vitro. These proteins fall into two families based on structure: enzymes that contain HemS motif and those with PNPOx domains. Proteins from these families are regulated by iron, support bacterial growth on heme iron, and/or aid in tolerance of heme and oxidative stress. It seems possible that proteins from these families might have evolved to serve separate functions in different bacteria. Already, PhuS was shown in vivo to shuttle heme to the HO, PigA/HemO. While in vitro degradation and additional investigations link proteins from these families to iron/heme metabolism, more work is required to determine their physiological role. Genetic studies suggest that even in bacteria with recognized HOs there are additional (likely non-homologous) enzymes that aid in heme degradation. This redundancy emphasizes the importance of this mechanism for bacterial physiology. For 50 years, this area has challenged both researchers and technology. While much has been learned about how pathogenic bacteria obtain iron, the complexity and diverse strategies employed by the bacteria could easily fascinate researchers for many more years.
Author contribution
KL generated the figures and the sequence comparisons, and wrote the manuscript; ZE critically revised the manuscript. Both authors have made substantial, direct and intellectual contribution to the work, and have approved it for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MV and handling Editor declared their shared affiliation.
Footnotes
Funding. This work was supported by American Heart Association Greater Southeast Affiliate Grant-in-Aid 15GRNT25600006 (ZE).
References
- Arezes J., Jung G., Gabayan V., Valore E., Ruchala P., Gulig P. A., et al. (2015). Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 17, 47–57. 10.1016/j.chom.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila L., Huang H.-W., Damaso C. O., Lu S., Moënne-Loccoz P., Rivera M. (2003). Coupled oxidation vs heme oxygenation: insights from axial ligand mutants of mitochondrial cytochrome b5. J. Am. Chem. Soc. 125, 4103–4110. 10.1021/ja029311v [DOI] [PubMed] [Google Scholar]
- Barker K. D., Barkovits K., Wilks A. (2012). Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J. Biol. Chem. 287, 18342–18350. 10.1074/jbc.M112.359265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates C. S., Toukoki C., Neely M. N., Eichenbaum Z. (2005). Characterization of MtsR, a new metal regulator in Group A Streptococcus, involved in iron acquisition and virulence. Infect. Immun. 73, 5743–5753. 10.1128/IAI.73.9.5743-5753.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta M. N., Wilks A. (2006). The mechanism of heme transfer from the cytoplasmic heme binding protein phus to the δ-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry 45, 11642–11649. 10.1021/bi060980l [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb L. A., Kunkle C. A., Schmitt M. P. (2007). The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 75, 2421–2431. 10.1128/IAI.01821-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb L. A., Schmitt M. P. (2010). The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 192, 4606–4617. 10.1128/JB.00525-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D. R., Lukat-Rodgers G. S., Rodgers K. R., Wilks A., Bhakta M. N., Lansky I. B. (2007). Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein phus from Pseudomonas aeruginosa and their relevance to function. Biochemistry 46, 14391–14402. 10.1021/bi701509n [DOI] [PubMed] [Google Scholar]
- Burgos J. M., Schmitt M. P. (2012). The ChrA response regulator in Corynebacterium diphtheriae controls hemin-regulated gene expression through binding to the hmuO and hrtAB promoter regions. J. Bacteriol. 194, 1717–1729. 10.1128/JB.06801-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B. M., Whitmire J. M., Merrell D. S. (2009). This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601. 10.1128/IAI.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver P. (2018). The battle for iron between humans and microbes. Curr. Med. Chem. 25, 85–86. 10.2174/0929867324666170720110049 [DOI] [PubMed] [Google Scholar]
- Chim N., Iniguez A., Nguyen T. Q., Goulding C. W. (2010). Unusual diheme conformation of the heme degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395, 595–608. 10.1016/j.jmb.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo J., Willows R. D., Beale S. I. (1998). Phytobilin biosynthesis: cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 15, 99–107. [DOI] [PubMed] [Google Scholar]
- Deredge D. J., Huang W., Hui C., Matsumura H., Yue Z., Moenne-Loccoz P., et al. (2017). Ligand-induced allostery in the interaction of the Pseudomonas aeruginosa heme binding protein with heme oxygenase. Proc. Natl. Acad. Sci. U.S.A. 114, 3421–3426. 10.1073/pnas.1606931114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., Park K., Kim T., Kang S. W., Hahn M. J., Hwang H. Y., et al. (2014). Structural and functional characterization of an Isd-type haem-degradation enzyme from Listeria monocytogenes. Acta Crystallogr. D Biol. Crystallogr. 70, 615–626. 10.1107/S1399004713030794 [DOI] [PubMed] [Google Scholar]
- Eichenbaum Z., Muller E., Morse S. A., Scott J. R. (1996). Acquisition of iron from host proteins by the Group A Streptococcus. Infect. Immun. 64, 5428–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Lad L., Li H., Wilks A., Poulos T. L. (2004). Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry 43, 5239–5245. 10.1021/bi049687g [DOI] [PubMed] [Google Scholar]
- Fujii H., Zhang X., Yoshida T. (2004). Essential amino acid residues controlling the unique regioselectivity of heme oxygenase in Pseudomonas aeruginosa. J. Am. Chem. Soc. 126, 4466–4467. 10.1021/ja031791i [DOI] [PubMed] [Google Scholar]
- Ganz T., Nemeth E. (2015). Iron homeostasis in host defence and inflammation. Nat. rev. Immunol. 15, 500–510. 10.1038/nri3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves A. B., Morse R. P., Chao A., Iniguez A., Goulding C. W., Liptak M. D. (2014). Crystallographic and spectroscopic insights into heme degradation by Mycobacterium tuberculosis MhuD. Inorg. Chem. 53, 5931–5940. 10.1021/ic500033b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Guo G., Mao X., Zhang W., Xiao J., Tong W., et al. (2008). Functional identification of HugZ, a heme oxygenase from Helicobacter pylori. BMC Microbiol. 8:226. 10.1186/1471-2180-8-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks T. S., Liu M., Mcclure M. J., Fukumura M., Duffy A., Lei B. (2006). Differential regulation of iron- and manganese-specific MtsABC and heme-specific HtsABC transporters by the metalloregulator MtsR of Group A Streptococcus. Infect. Immun. 74, 5132–5139. 10.1128/IAI.00176-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannauer M., Arifin A. J., Heinrichs D. E. (2015). Involvement of reductases IruO and NtrA in iron acquisition by Staphylococcus aureus. Mol. Microbiol. 96, 1192–1210. 10.1111/mmi.13000 [DOI] [PubMed] [Google Scholar]
- Hirotsu S., Chu G. C., Unno M., Lee D. S., Yoshida T., Park S. Y., et al. (2004). The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae. J. Biol. Chem. 279, 11937–11947. 10.1074/jbc.M311631200 [DOI] [PubMed] [Google Scholar]
- Hu Y., Jiang F., Guo Y., Shen X., Zhang Y., Zhang R., et al. (2011). Crystal structure of HugZ, a novel heme oxygenase from Helicobacter pylori. J. Biol. Chem. 286, 1537–1544. 10.1074/jbc.M110.172007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Wilks A. (2017). Extracellular heme uptake and the challenge of bacterial cell membranes. Annu. Rev. Biochem. 86, 799–823. 10.1146/annurev-biochem-060815-014214 [DOI] [PubMed] [Google Scholar]
- Ishida J. H., Johansen K. L. (2014). Iron and infection in hemodialysis patients. Semin. Dial. 27, 26–36. 10.1111/sdi.12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. E., Wessling-Resnick M. (2012). Iron metabolism and the innate immune response to infection. Microbes Infect. 14, 207–216. 10.1016/j.micinf.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson M. D. (2017). Iron Transport proteins: gateways of cellular and systemic iron homeostasis. J. Biol. Chem. 292, 12735–12743. 10.1074/jbc.R117.786632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle C. A., Schmitt M. P. (2007). Comparative analysis of hmuO function and expression in Corynebacterium species. J. Bacteriol. 189, 3650–3654. 10.1128/JB.00056-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina J. W., Nix D. B., Lanzilotta W. N. (2016). Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U.S.A. 113, 12138–12143. 10.1073/pnas.1603209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. J., Schep D., Mclaughlin B., Kaufmann M., Jia Z. (2014). Structural analysis and identification of PhuS as a heme-degrading enzyme from Pseudomonas aeruginosa. J. Mol. Biol. 426, 1936–1946. 10.1016/j.jmb.2014.02.013 [DOI] [PubMed] [Google Scholar]
- Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. (2008). Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283, 30957–30963. 10.1074/jbc.M709486200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009). Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. U.S.A. 106, 11719–11724. 10.1073/pnas.0903842106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Boulouis H. J., Biville F. (2012). Heme degrading protein HemS is involved in oxidative stress response of Bartonella henselae. PLoS ONE 7:e37630. 10.1371/journal.pone.0037630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gong J., Wei T., Wang Z., Du Q., Zhu D., et al. (2012). Crystal structure of HutZ, a heme storage protein from Vibrio cholerae: a structural mismatch observed in the region of high sequence conservation. BMC Struct. Biol. 12:23. 10.1186/1472-6807-12-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharshak N., Ryu H. S., Fan T.-J., Onyiah J. C., Schulz S., Otterbein S. L., et al. (2015). Escherichia coli heme oxygenase modulates host innate immune responses. Microbiol. Immunol. 59, 452–465. 10.1111/1348-0421.12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Nambu S., Ono Y., Goulding C. W., Tsumoto K., Ikeda-Saito M. (2013). Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry 52, 3025–3027. 10.1021/bi400382p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A. T., Spatafora G. A. (2014). A role for the DtxR family of metalloregulators in gram-positive pathogenesis. Mol. Oral Microbiol. 29, 1–10. 10.1111/omi.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. K., Schwiesow L., Au-Yeung W., Auerbuch V. (2016). Hereditary hemochromatosis predisposes mice to Yersinia pseudotuberculosis infection even in the absence of the type III secretion system. Front. Cell. Infect. Microbiol. 6:69. 10.3389/fcimb.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray G. L., Srikram A., Henry R., Puapairoj A., Sermswan R. W., Adler B. (2009). Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 11, 311–314. 10.1016/j.micinf.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Nambu S., Matsui T., Goulding C. W., Takahashi S., Ikeda-Saito M. (2013). A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem. 288, 10101–10109. 10.1074/jbc.M112.448399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles C. L., Maresso A. W. (2011). The theft of host heme by Gram-positive pathogenic bacteria. Metallomics 3, 788–796. 10.1039/c1mt00047k [DOI] [PubMed] [Google Scholar]
- O'neill M. J., Bhakta M. N., Fleming K. G., Wilks A. (2012). Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc. Natl Acad. Sci. U.S.A. 109, 5639–5644. 10.1073/pnas.1121549109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'neill M. J., Wilks A. (2013). The, P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem. Biol. 8, 1794–1802. 10.1021/cb400165b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onzuka M., Sekine Y., Uchida T., Ishimori K., Ozaki S.-I. (2017). HmuS from Yersinia pseudotuberculosis is a non-canonical heme-degrading enzyme to acquire iron from heme. Biochim. Biophys. Acta 1861, 1870–1878. 10.1016/j.bbagen.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Ostrow J. D., Jandl J. H., Schmid R. (1962). The formation of bilirubin from hemoglobin in vivo. J. Clin. Invest. 41, 1628–1637. 10.1172/JCI104620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet Y. H., Ndiaye C. T., Gagne S. M., Sebilo A., Suits M. D., Jubinville E., et al. (2016). An alternative reaction for heme degradation catalyzed by the Escherichia coli O157:H7 ChuS protein: release of hematinic acid, tripyrrole and Fe(III). J. Inorg. Biochem. 154, 103–113. 10.1016/j.jinorgbio.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Quenee L. E., Hermanas T. M., Ciletti N., Louvel H., Miller N. C., Elli D., et al. (2012). Hereditary hemochromatosis restores the virulence of plague vaccine strains. J. Infect. Dis. 206, 1050–1058. 10.1093/infdis/jis433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff M., Zhu W., Deshmukh R., Wilks A., Stojiljkovic I. (2001). Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403. 10.1128/JB.183.21.6394-6403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Haley K. P., Skaar E. P. (2011). The flexible loop of Staphylococcus aureus IsdG is required for its degradation in the absence of heme. Biochemistry 50, 6730–6737. 10.1021/bi200999q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Skaar E. P. (2008). Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 69, 1304–1315. 10.1111/j.1365-2958.2008.06363.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Ukpabi G. N., Harry S. R., Stec D. F., Krull R., Wright D. W., et al. (2010). The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75, 1529–1538. 10.1111/j.1365-2958.2010.07076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley K. A., Rock J. D., Li Y., Ketley J. M. (2006). Heme utilization in Campylobacter jejuni. J. Bacteriol. 188, 7862–7875. 10.1128/JB.00994-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachla A. J., Ouattara M., Romero E., Agniswamy J., Weber I. T., Gadda G., et al. (2016). in vitro heme biotransformation by the HupZ enzyme from Group A Streptococcus. Biometals 29, 593–609. 10.1007/s10534-016-9937-1 [DOI] [PubMed] [Google Scholar]
- Sánchez M., Sabio L., Gálvez N., Capdevila M., Dominguez-Vera J. M. (2017). Iron chemistry at the service of life. IUBMB Life 69, 382–388. 10.1002/iub.1602 [DOI] [PubMed] [Google Scholar]
- Schmitt M. (1997a). Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65, 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. (1997b). Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S., Paoli M. (2005). Crystallization and preliminary X-ray diffraction analysis of the haem-binding protein HemS from Yersinia enterocolitica. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 802–805. 10.1107/S1744309105023523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S., Sharp K. H., Barker P. D., Paoli M. (2006). An induced fit conformational change underlies the binding mechanism of the heme transport proteobacteria-protein HemS. J. Biol. Chem. 281, 32606–32610. 10.1074/jbc.M607516200 [DOI] [PubMed] [Google Scholar]
- Sheldon J. R., Heinrichs D. E. (2015). Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39, 592–630. 10.1093/femsre/fuv009 [DOI] [PubMed] [Google Scholar]
- Sheldon J. R., Laakso H. A., Heinrichs D. E. (2016). Iron acquisition strategies of bacterial pathogens. Microbiol. Spectr. 4, 1–32. 10.1128/microbiolspec.VMBF-0010-2015 [DOI] [PubMed] [Google Scholar]
- Silva-Herzog E., Detweiler C. S. (2008). Intracellular microbes and haemophagocytosis. Cell. Microbiol. 10, 2151–2158. 10.1111/j.1462-5822.2008.01192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P., Gaspar A. H., Schneewind O. (2004a). IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443. 10.1074/jbc.M307952200 [DOI] [PubMed] [Google Scholar]
- Skaar E. P., Humayun M., Bae T., Debord K. L., Schneewind O. (2004b). Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628. 10.1126/science.1099930 [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I., Hantke K. (1992). Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11, 4359–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I., Hantke K. (1994). Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13, 719–732. 10.1111/j.1365-2958.1994.tb00465.x [DOI] [PubMed] [Google Scholar]
- Streit B. R., Kant R., Tokmina-Lukaszewska M., Celis A. I., Machovina M. M., Skaar E. P., et al. (2016). Time-resolved studies of IsdG protein identify molecular signposts along the non-canonical heme oxygenase pathway. J. Biol. Chem. 291, 862–871. 10.1074/jbc.M115.666560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suits M. D., Jaffer N., Jia Z. (2006). Structure of the Escherichia coli O157:H7 heme oxygenase ChuS in complex with heme and enzymatic inactivation by mutation of the heme coordinating residue His-193. J. Biol. Chem. 281, 36776–36782. 10.1074/jbc.M607684200 [DOI] [PubMed] [Google Scholar]
- Suits M. D., Pal G. P., Nakatsu K., Matte A., Cygler M., Jia Z. (2005). Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. U.S.A. 102, 16955–16960. 10.1073/pnas.0504289102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sword C. (1966). Mechanisms of pathogenesis in Listeria monocytogenes infection. J. Bacteriol. 92, 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S. J., Loutet S. A., Mauk A. G., Murphy M. E. (2015). A ferric-peroxo intermediate in the oxidation of heme by IsdI. Biochemistry 54, 2613–2621. 10.1021/acs.biochem.5b00239 [DOI] [PubMed] [Google Scholar]
- Tenhunen R., Marver H. S., Schmid R. (1968). The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc. Natl. Acad Sci. U.S.A. 61, 748–755. 10.1073/pnas.61.2.748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhunen R., Marver H. S., Schmid R. (1969). Microsomal heme oxygenase: characterization of the enzyme. J. Biol. Chem. 244, 6388–6394. [PubMed] [Google Scholar]
- Toukoki C., Gold K. M., Mciver K. S., Eichenbaum Z. (2010). MtsR is a dual regulator that controls virulence genes and metabolic functions in addition to metal homeostasis in GAS. Mol. Microbiol. 76, 971–989. 10.1111/j.1365-2958.2010.07157.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S., O'neill M. J., Wilks A., Poulos T. L. (2013). Crystal structure of the Pseudomonas aeruginosa cytoplasmic heme binding protein, Apo-PhuS. J. Inorg. Biochem. 128, 131–136. 10.1016/j.jinorgbio.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Hassan H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T., Sekine Y., Matsui T., Ikeda-Saito M., Ishimori K. (2012). A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem. Commun. 48, 6741–6743. 10.1039/c2cc31147j [DOI] [PubMed] [Google Scholar]
- Unno M., Ardevol A., Rovira C., Ikeda-Saito M. (2013). Structures of the substrate-free and product-bound forms of HmuO, a heme oxygenase from corynebacterium diphtheriae: x-ray crystallography and molecular dynamics investigation. J. Biol. Chem. 288, 34443–34458. 10.1074/jbc.M113.486936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M., Matsui T., Ikeda-Saito M. (2007). Structure and catalytic mechanism of heme oxygenase. Nat. Prod. Rep. 24, 553–570. 10.1039/b604180a [DOI] [PubMed] [Google Scholar]
- Valko M., Morris H., Cronin M. T. D. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. 10.2174/0929867053764635 [DOI] [PubMed] [Google Scholar]
- Wang A., Zeng Y., Han H., Weeratunga S., Morgan B. N., Moënne-Loccoz P., et al. (2007). Biochemical and structural characterization of Pseudomonas aeruginosa Bfd and FPR: ferredoxin NADP+ Reductase and not ferredoxin is the redox partner of heme oxygenase under iron-starvation conditions. Biochemistry 46, 12198–12211. 10.1021/bi7013135 [DOI] [PubMed] [Google Scholar]
- Wegele R., Tasler R., Zeng Y., Rivera M., Frankenberg-Dinkel N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. 10.1074/jbc.M408303200 [DOI] [PubMed] [Google Scholar]
- Wilks A., Heinzl G. (2014). Heme oxygenation and the widening paradigm of heme degradation. Arch. Biochem. Biophys. 544, 87–95. 10.1016/j.abb.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A., Ikeda-Saito M. (2014). Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res. 47, 2291–2298. 10.1021/ar500028n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A., Schmitt M. P. (1998). Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273, 837–841. 10.1074/jbc.273.2.837 [DOI] [PubMed] [Google Scholar]
- Wu R., Skaar E. P., Zhang R., Joachimiak G., Gornicki P., Schneewind O., et al. (2005). Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280, 2840–2846. 10.1074/jbc.M409526200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E. E., Schmitt M., Wilks A., Payne S. M. (2004). HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186, 4142–4151. 10.1128/JB.186.13.4142-4151.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Zhang J., Guo G., Mao X., Tong W., Zhang Y., et al. (2011). Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode. Biochem. Biophys. Res. Commun. 415, 82–87. 10.1016/j.bbrc.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Zhu W., Wilks A., Stojiljkovic I. (2000). Degradation of heme in gram-negative bacteria: the product of the hemo gene of neisseriae is a heme oxygenase. J. Bacteriol. 182, 6783–6790. 10.1128/JB.182.23.6783-6790.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]