ABSTRACT
During the peptidoglycan (PG) maturation of mycobacteria, the glycan strands are interlinked by both 3-3 (between two meso-diaminopimelic acids [meso-DAPs]) and 4-3 cross-links (between d-Ala and meso-DAP), though there is a predominance (60 to 80%) of 3-3 cross-links. The dd-carboxypeptidases (dd-CPases) act on pentapeptides to generate tetrapeptides that are used by ld-transpeptidases as substrates to form 3-3 cross-links. Therefore, dd-CPases play a crucial role in mycobacterial PG cross-link formation. However, the physiology of dd-CPases in mycobacteria is relatively unexplored. In this study, we deleted two dd-CPase genes, msmeg_2433 and msmeg_2432, both individually and in combination, from Mycobacterium smegmatis mc2155. Though the single dd-CPase gene deletions had no significant impact on the mycobacterial physiology, many interesting functional alterations were observed in the double-deletion mutant, viz., a predominance in PG cross-link formation was shifted from 3-3 cross-links to 4-3, cell surface glycopeptidolipid (GPL) expression was reduced, and susceptibility to β-lactams and antitubercular agents was enhanced. Moreover, the survival rate of the double mutant within murine macrophages was higher than that of the parent. Interestingly, the complementation with any one of the dd-CPase genes could restore the wild-type phenotype. In a nutshell, we infer that the altered ratio of 4-3 to 3-3 PG cross-links might have influenced the expression of surface GPLs, colony morphology, biofilm formation, drug susceptibility, and subsistence of the cells within macrophages.
IMPORTANCE The glycan strands in mycobacterial peptidoglycan (PG) are interlinked by both 3-3 and 4-3 cross-links. The dd-CPases generate tetrapeptides by acting on the pentapeptides, and ld-transpeptidases use tetrapeptides as substrates to form 3-3 cross-links. In this study, we showed that simultaneous deletions of two dd-CPases alter the nature of PG cross-linking from 3-3 cross-links to 4-3 cross-links. The deletions subsequently decrease the expression of glycopeptidolipids (significant surface lipid present in many nontuberculous mycobacteria, including Mycobacterium smegmatis) and affect other physiological parameters, like cell morphology, growth rate, biofilm formation, antibiotic susceptibility, and survival within murine macrophages. Thus, unraveling the physiology of dd-CPases might help us design antimycobacterial therapeutics in the future.
KEYWORDS: peptidoglycan, dd-carboxypeptidase, glycopeptidolipids, biofilm, macrophage, DacB2, MSMEG_2433, MSMEG_2432
INTRODUCTION
The cell wall biosynthetic components of mycobacteria are among the prime targets of many antibiotics. The mycobacterial cell wall is covalently linked with three polymers, namely, mycolic acid, arabinogalactan, and peptidoglycan (PG), collectively known as the MAP complex (1, 2). PG is the innermost component of the MAP complex, and its synthesis requires cross-link formation between the stem peptides (l-Ala1–d-iso-Glu2–meso-diaminopimelic acid3 [meso-DAP3]–d-Ala4–d-Ala5) of the glycan chains. In mycobacteria, the glycan chains are interlinked by 3-3 cross-links between the two meso-DAP3s and 4-3 cross-links between d-Ala4 and meso-DAP3 of the stem peptides (3–6). It is important to note that the prevalence of 3-3 cross-links in mycobacteria is higher than that of 4-3 cross-links. The abundance of 3-3 cross-links in various growth phases of Mycobacterium tuberculosis cells is 60 to 80% (4, 6), whereas it is 64 to 73% in Mycobacterium abscessus throughout its growth phases (5). In contrast, only 3 to 10% of 3-3 cross-links are present in Escherichia coli (7, 8). The classical dd-transpeptidases (penicillin-binding proteins [PBPs]) catalyze the 4-3 cross-link formation (9), whereas ld-transpeptidases catalyze 3-3 cross-link formation (10). ld-Transpeptidase was originally detected in Enterococcus faecium and acts on tetrapeptides generated from pentapeptides by the action of dd-carboxypeptidases (dd-CPases) (11). The dd-CPases are low-molecular-mass (LMM) PBPs that remove the terminal d-Ala5 from the pentapeptides linked to the glycan chains in the PG layer and play an essential role in the formation of PG cross-links in mycobacteria. There are multiple established and/or putative dd-CPases; for example, E. coli has 4 (PBP4, PBP5, PBP6, and DacD) (12), M. tuberculosis has 3 (DacB2 [Rv2911], Rv3330, and Rv3627c), and Mycobacterium smegmatis contains 4 (MSMEG_1661, MSMEG_2432, MSMEG_2433, and MSMEG_6113) (13). However, the physiology of dd-CPases in mycobacteria has been relatively unexplored. To date, only MSMEG_2433 of M. smegmatis has been characterized as a dual-activity enzyme (dd-CPase and β-lactamase) (14) and DacB2 of M. tuberculosis as a dd-CPase (6). These two proteins are ectopically expressed in heterologous systems and have been shown to have certain impacts on physiology and cellular morphology (14, 15). The dacB2 mutant is reported as a better intracellular survivor in THP-1 cells during infection than the parent and complemented strains (15).
Glycopeptidolipids (GPLs), comprising nearly 85% of the total surface lipids, are found in many nontuberculous mycobacteria, including Mycobacterium smegmatis (16). GPLs play a significant role in maintaining cell wall permeability, sliding motility, and biofilm formation in M. smegmatis (16, 17). The structure of GPLs is well elaborated (18–20). Among the known GPL-linked proteins, tmtpC (MmpL family protein) and gap (GPL addressing protein) are the only described GPL transporters (21–23) (see Fig. S1 in the supplemental material). Recently, it has been shown that a GPL-deficient M. smegmatis strain has a stronger ability to survive in murine macrophages than the GPL-restored phenotype, though the underlying mechanism is unknown (24). Despite several advances in knowledge, the biosynthetic and export pathways of GPLs in mycobacteria are poorly understood.
In this study, we aimed to investigate the physiological roles of two dd-CPase homologs of M. smegmatis. Accordingly, we deleted two dd-CPase genes, msmeg_2433 and msmeg_2432, individually and in combination from M. smegmatis mc2155. We studied the impact of both single (resulting in strains Δms32 and Δms33) and double (resulting in strain Δms33/32) deletions on the bacterial physiology, such as the nature of PG cross-link formation, surface GPL expression, antibiotic susceptibility, and survival within murine macrophages. Based on the results obtained, we hypothesize that alteration in PG cross-linking in the double dd-CPase mutant affects the expression of surface GPLs, directly or indirectly, and simultaneously influences other physiological parameters, such as growth rate, colony morphology, biofilm formation, antibiotic susceptibility, and survival within murine macrophages. These additional insights on the varied roles of dd-CPases in mycobacterial physiology might be helpful in devising better therapeutic strategies to combat mycobacterial infections.
RESULTS
Construction of single and double knockouts of msmeg_2433 and msmeg_2432.
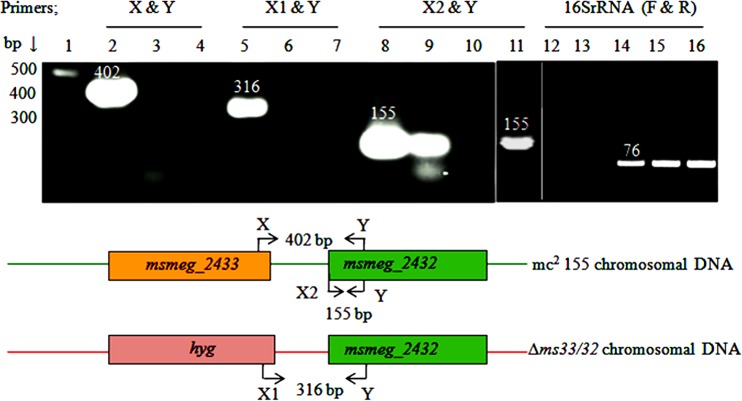
To study the physiology of dd-CPases in mycobacteria, we deleted msmeg_2433 from the M. smegmatis mc2155 chromosome (yielding Δms33/32 [Fig. S2]). The genes msmeg_2433 and msmeg_2432 are placed in tandem in the mc2155 genome, though separated by 69 nucleotides. As the genes are situated in close proximity, it was essential to check whether they are expressed from a single promoter and whether the deletion of msmeg_2433 had any polar effect on msmeg_2432 (another dd-CPase homolog), which is downstream. Using semiquantitative reverse transcription-PCR (sqRT-PCR), we confirmed that these two genes do not share an mRNA transcript, as the primer pairs X and Y and X1 and Y (Fig. 1; see also Table S1) could not amplify the products from the cDNA templates of both mc2155 and the Δms33/32 mutant (Fig. 1, lanes 3 and 7). The primer pair X2 and Y, designed to amplify a 155-bp product from the msmeg_2432 open reading frame, could amplify the product from the cDNA template of mc2155 but not from Δms33/32 (Fig. 1, lanes 9 and 10). The results revealed that both the genes are nonoperonic, which is also supported by the computational operon prediction database ProOpDB (25), and showed that the deletion of msmeg_2433 had a polar effect on msmeg_2432. Integration of the hyg cassette inactivated the promoter machinery of msmeg_2432, thus making the gene functionally inactive. Next, to understand the individual roles of both genes, we constructed two separate single deletions of msmeg_2433 (Δms33) and msmeg_2432 (Δms32) in the mc2155 chromosome. The polar effect of msmeg_2433 on msmeg_2432 was tested again. This time the deletion had no polar effect, as an amplicon of the expected size (155 bp) was generated using primers X2 and Y (Fig. 1, lane 11).
FIG 1.
Analysis of the polar effect of msmeg_2433 deletion on msmeg_2432, and transcriptional uncoupling between the two, by semiquantitative RT-PCR (sqRT-PCR). Lane 1, 100-bp DNA ladder; lanes 2, 8, and 14, PCR amplicons amplified from the mc2155 genomic DNA (gDNA) template and used as positive controls; lane 5, amplicon from the mutant (Δms33/32) gDNA template; lanes 3, 6, and 9 and lanes 4, 7, and 10, amplicons from the cDNA templates of mc2155 and Δms33/32, respectively. The PCR product in lane 11 was amplified from Δms33 cDNA. The coupled primers X and Y represent the forward and reverse primers designed from the msmeg_2433 and msmeg_2432 regions that can amplify 402-bp products, including the intermediate region, from the parent gDNA (shown in the schematic representation at the bottom). Similarly, in the second primer set X1 and Y, the forward primer, X1, was designed from the hygromycin cassette and can amplify a 316-bp product, including the intermediate region, from the Δms33/32 gDNA. The primer pairs X2 and Y were designed from the msmeg_2432 region. The primers F and R, for the internal positive control 16S rRNA, could not amplify the 76-bp product from the RNA templates of mc2155 and Δms33/32 (lanes 12 and 13), whereas they amplified it from their respective cDNA templates (lanes 15 and 16), overcoming the possibility of genomic DNA contamination in the sample.
Complementation studies and confirmation of β-lactamase activity.
Our study was focused on three dd-CPase homologs, namely, those encoded by msmeg_2433, msmeg_2432, and dacB2. All three proteins are LMM PBPs and possess the conserved signature motifs of PBPs, i.e., STIK, SGN, and KTG (Fig. S3). To study the effect of the genes, the single- and double-deletion mutants were complemented with msmeg_2433, msmeg_2432, and dacB2 or the Rv2911 gene (corresponding homolog in M. tuberculosis). The genes were cloned in the tetracycline-inducible expression vector pMIND. The transformants obtained were named Δms33/32::pM33, Δms33/32::pM32, and Δms33/32::pM11, respectively.
All the aforementioned LMM PBPs were ectopically expressed and the presence of the expressed proteins in the membrane fraction was confirmed by binding the cell extracts with Bocillin FL (a commercially available fluorescent tagged penicillin V that binds to PBPs) (Fig. S4A). The presence of DacB2 in the culture filtrates has been reported earlier (26); however, in this study, we could simultaneously locate it in the membrane fractions as well. Interestingly, the band intensities of the expressed proteins observed in the Coomassie brilliant blue (CBB)-stained gels did not correlate with Bocillin FL binding (Fig. S4A). There are two possible explanations for the reduced band intensities upon Bocillin FL binding: either the proteins had different binding affinities for CBB and Bocillin FL or they had β-lactamase activity (that cleaves the bound Bocillin FL). As the first possibility is less likely because the binding intensities of both the proteins were reduced, we tested the latter, i.e., the β-lactamase activity of the proteins. The β-lactamase activity of MSMEG_2433 is already established, as Glu (E) in the Ω-like loop has been shown to play a crucial role (Fig. S3) (14). As MSMEG_2432 and DacB2 share distinct amino acid identities with MSMEG_2433, 58% and 70%, respectively, there was a possibility that both MSMEG_2432 and DacB2 also possess β-lactamase activity. This hypothesis was further supported by their ability to hydrolyze nitrocefin (Fig. S4B and C).
Deletion of both msmeg_2433 and msmeg_2432 hampers the growth of the organism.
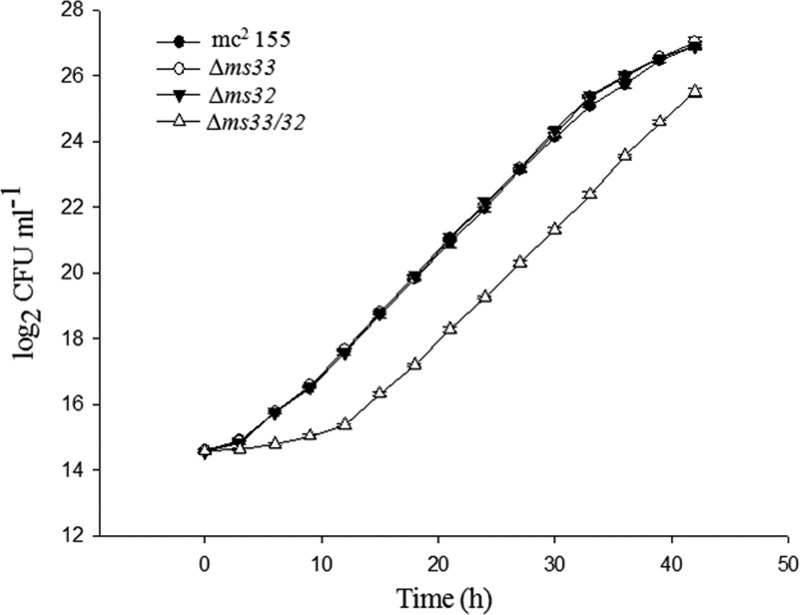
The effects of the deletions on the growth rate of the cells were investigated for 42 h by calculating the CFU per milliliter at 3-h intervals. We observed a 16% lower growth rate (0.21 ± 0.003 h−1) and a 16% longer doubling time (3.26 ± 0.05 h) in the double mutant than the growth rate (0.25 ± 0.001 h−1) and doubling time (2.80 ± 0.19 h) of the parent strain. However, the single mutants Δms33 and Δms32 had approximately the same growth rate and doubling time as the parent (Fig. 2). The results indicate that simultaneous deletion of two dd-CPases affects the growth rate of the bacteria, whereas the single dd-CPase deletions do not.
FIG 2.
Growth analysis of M. smegmatis mc2155 and knockout mutants (two single mutants [Δms33 and Δms32] and one double mutant [Δms33/32]). The strains were inoculated (McFarland number 0.5) into Middlebrook 7H9 medium (supplemented with OADC enrichment and appropriate antibiotics), and the CFU per milliliter were counted every 3 h for 42 h. The graph was plotted on semilogarithmic base 2 scales (y axis). The values are means ± SDs from three independent experiments. The growth rate and doubling time were calculated for each replicate using the linear part of the log phase.
Peptidoglycan cross-linking is altered in the double mutant, with a predominance of 4-3 cross-links.
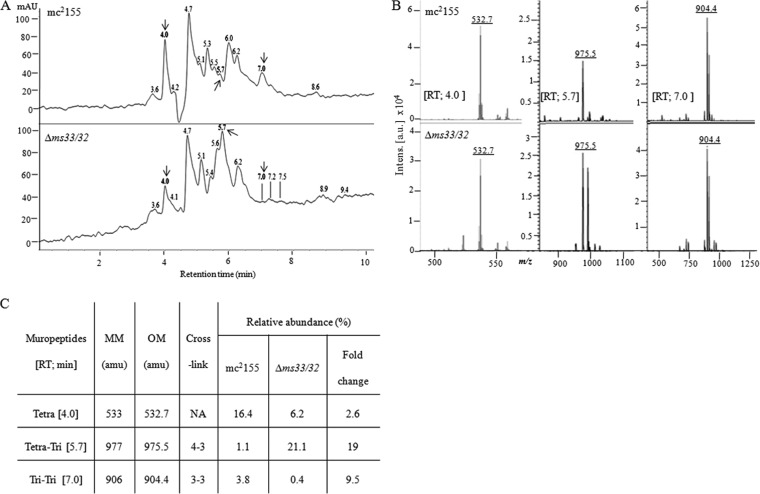
The dd-CPases play a vital role in PG remodeling. They facilitate the process of 3-3 cross-link formation via ld-transpeptidases by providing the tetrapeptide substrates (10). As the double deletion had a significant impact on the bacterial growth rate, we decided to further analyze the PG components in the double-knockout strain, Δms33/32. We isolated PG from mc2155 and Δms33/32 and analyzed it by reverse-phase high-pressure liquid chromatography (RP-HPLC). We collected the individual muropeptides and noted their relative abundances (peak area) (Table S2). The muropeptides of mycobacterial PG have been studied previously. Therefore, we chose not to report all the peaks but selected only those showing the same retention time but a ≥2.5-fold difference in their relative abundances. Thereafter, three important peaks with retention time of 4.0, 5.7, and 7.0 min were analyzed through matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Upon MS analysis, the masses obtained for the above-mentioned peaks were 532.7, 975.6, and 904.4 atomic mass units (amu), respectively (Fig. 3), which were close to the monoisotopic masses of tetrapeptide (533 amu), tetra-tri dimer (4-3; 977 amu), and tri-tri dimer (3-3; 906 amu), respectively (Fig. S5). The masses obtained differed by ∼2 amu from the actual monoisotopic masses of mycobacterial PG-muropeptides, which were possibly due to the amidation of the free carboxyl groups of the glutamate and DAP residues, regardless of the cross-link status (6, 27). We observed a 2.6-fold reduction in the production of tetrapeptides in Δms33/32, which indicated that the mutant strain did not possess enough dd-CPases to produce tetrapeptides. Such a decrease was further reflected in their 3-3 cross-link formation, where a drastic reduction (9.5-fold) was observed in Δms33/32 compared to the parent strain. Surprisingly, we observed a noticeably large (19-fold) amount of 4-3 cross-link formation in Δms33/32 (Fig. 3). Accordingly, we hypothesize that the absence of dd-CPases reduces the production of tetrapeptides, consequently affecting the ld-transpeptidation that prevents 3-3 cross-link formation and ultimately forces the bacteria to form more 4-3 cross-links.
FIG 3.
Comparative analysis of the muropeptides from the parent (mc2155) and the double-knockout (Δms33/32) strain of M. smegmatis by RP-HPLC and MALDI-TOF MS. (A) RP-HPLC profile of the PG muropeptides extracted from the mid-log phase (optical density at 600 nm [OD600] ∼ 0.6) culture. Absorbance (milli-absorbance units [mAU] × 103) was recorded at 210 nm. Numbers above the peaks represent the retention times. Arrows indicate the peaks with similar retention times and the >2.5-fold difference in the relative abundance (peak area) (see Table S2) that were chosen for further analysis. (B) MALDI-TOF MS analysis of the selected peaks. (C) Summary of the muropeptides analyzed through RP-HPLC and MALDI-TOF MS. MM and OM, monoisotopic and observed masses, respectively; amu, atomic mass units; RT, retention time.
Expression of surface GPL is reduced in the double-deletion mutant.
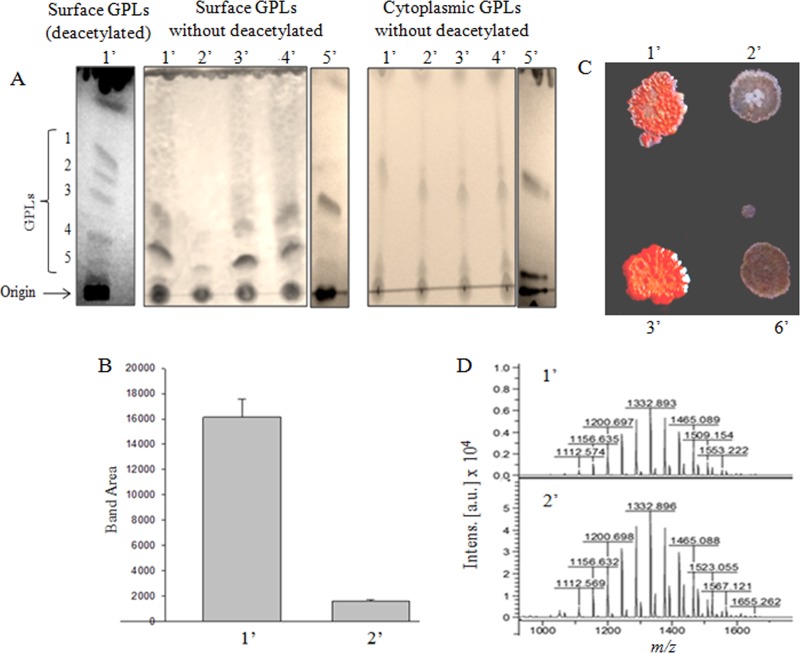
Mycobacteria have a thick cell wall made up of the MAP complex. It is possible that any change in the PG cross-link formation is likely to affect the other cell surface components. Therefore, we analyzed the effect of dd-CPase deletions on expression of glycopeptidolipid (GPL), which is a major type of cell surface lipid. We isolated the GPLs from the parent and the mutants, and half of GPLs were subjected to mild deacetylation. Deacetylation removes the ester-linked fatty acids and the entire O-acetyl group, which improves the resolution of GPLs on thin-layer chromatography (TLC) (17, 28). We observed five GPL bands (1 to 5; shown only for mc2155) on the TLC plate (Fig. 4A). As the deacetylation process could mask some of the natural characteristics of the isolated GPLs, we chose to work with the unmodified (nondeacetylated) GPLs. We observed that the surface GPL content of the double-knockout mutant, Δms33/32, was drastically reduced (∼10-fold) compared to that of the parent strain (Fig. 4B). Interestingly, the effect was restored upon complementation with either of the dd-CPases (MSMEG_2433, MSMEG_2432, or DacB2), which shows that multiple copies of any one of the dd-CPases could rectify the loss of surface GPLs.
FIG 4.
Analysis of surface and cytoplasmic GPLs of the M. smegmatis strains by TLC, MALDI-TOF MS, and Congo red assay. (A) (From left to right) TLC of deacetylated surface GPLs (1′ to 5′), without deacetylated surface and cytoplasmic GPLs, resolved on a silica gel plate using the GPL-specific solvent system of chloroform-methanol-water (90:10:1) (refer to Materials and Methods for details). Numbers 1′ to 6′ (see panel C) represent strains mc2155, Δms33/32, Δms33/32::pM33, Δms33/32::pM11, Δms33/32::pM32, and Δms33/32::pMIND (Δms33/32 transformed with empty pMIND vector), respectively. (B) Quantitative analysis (by ImageJ) of surface GPL (total bands/lane) extracted from 1′ and 2′. Error bars show SDs calculated from three replicates. (C) Colony appearance of the indicated strains in the Congo red assay. (D) MALDI-TOF MS analysis of the scraped cytoplasmic GPLs from mc2155 and Δms33/32, showing similar peak patterns for both the strains. Here m/z is the mass/charge ratio of the fragments, and the intensities of the peaks are shown in arbitrary units (a.u.).
Further, we wanted to validate the lack of surface GPLs in Δms33/32 by the Congo red (CR) assay. CR is a hydrophobic, planar diazo dye that binds to glycolipids and lipoproteins, which are abundant on the mycobacterial cell surface (29). Interestingly, we observed the appearance of dry white colonies for Δms33/32 and Δms33/32::pMIND strains but a typical smooth reddish colony of the parent strain, mc2155, on the CR-containing plates, indicating the absence of surface GPLs in the mutants. However, the complemented strain, Δms33/32::pM33, was able to restore the parental phenotype (Fig. 4C).
The reduced expression of GPLs on the cell surface could be due to the suppression of GPL biosynthesis. If the biosynthesis is affected, the cytoplasmic GPL content would also be reduced. To address this issue, we assessed the cytoplasmic GPL levels in the respective strains. Surprisingly, there was no difference in the cytoplasmic GPL levels between the parent and the mutant strains (Fig. 4A), indicating that the biosynthesis of GPLs was not affected. Next, we tried to analyze the nature of GPLs in these strains. We scraped the cytoplasmic GPLs of Δms33/32 and mc2155 from the TLC plate and examined their masses using MALDI-TOF MS. We observed a similar pattern of the peaks in both the parent and the double mutant. The results indicated that though the surface expression of GPL was affected, the compositions of GPL were similar in the two strains (Fig. 4D). Therefore, the results point out that the absence of dd-CPases changes the expression of surface GPLs without affecting GPL biosynthesis, though the mechanism behind the process is as yet unknown.
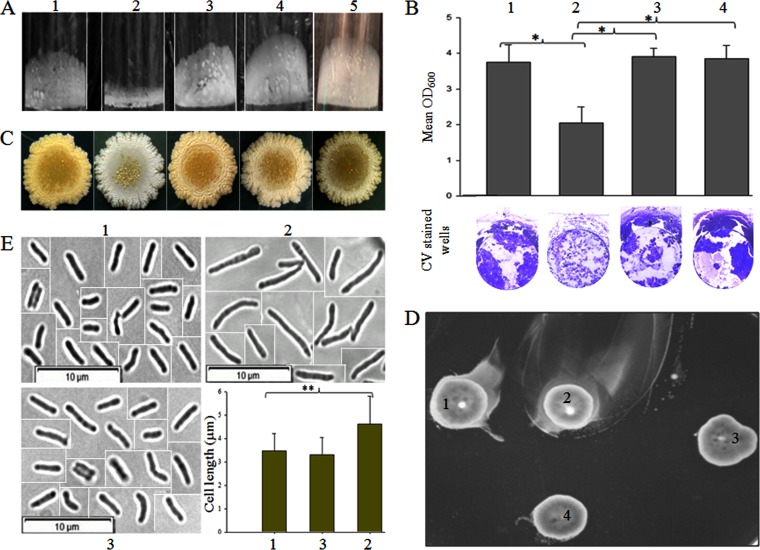
Double dd-CPase deletion results in reduced biofilm/pellicle formation, altered colony morphology, hypermotility, and cell elongation.
We observed that dd-CPases play an essential role in PG cross-link formation and surface GPL expression. The latter is also reported to play a significant role in maintaining bacterial colony morphology, biofilm formation, and sliding motility (21). Therefore, we tried to assess the impact of the double dd-CPase deletion on the phenotypic properties of M. smegmatis. We observed that the double-knockout strain was unable to adhere properly to the wells, which resulted in reduced pellicle formation as well as reduced biofilm at the air-liquid interface (Fig. 5A and B). A distinct change in the colony morphology was also observed. The parent strain showed a pale yellow, smooth appearance, whereas the mutant had an unusual dry white colony appearance on LB agar plates (Fig. 5C). Based on the difference in the sliding motility (as observed by measuring the zone diameter), where the double mutant (4.3 ± 0.61 cm) had a value nearly 2-fold higher than that of the parent (2.2 ± 0.17 cm), it can be hypothesized that the motility of the double-deletion mutant was enhanced significantly, which we refer to as hypermotility (Fig. 5D). We also examined the cell shape of Δms33/32 using phase-contrast microscopy. Although we could not see any difference in the cell shape, we observed that the average cell length (n = 100) of Δms33/32 (4.6 ± 1.2 μm) was significantly greater (P < 0.001) than that of the parent (3.4 ± 0.7 μm) and complemented (3.3 ± 0.7 μm) strains (Fig. 5E). It is important to mention that we also tested the impact of the single deletions on surface GPL expression, pellicle formation, colony morphology, and motility, using single mutants Δms33 and Δms32, but no significant difference from mc2155 was observed (Fig. S6). All these oddities of Δms33/32 were restored in the complemented strains. Overall, the results confirm that the absence of these two dd-CPases has a significant impact on the physiology and cellular morphology of mycobacteria.
FIG 5.
Effect of double deletion on the phenotype of M. smegmatis mc2155. Numbers 1 to 5 represent strains mc2155, Δms33/32, Δms33/32::pM33, Δms33/32::pM11, and Δms33/32::pM32, respectively. (A) Pellicle formation of the different strains. (B) Quantitative analysis of the biofilm formation from six replicates (means ± SDs). The lower panel shows the representative images of the crystal violet (CV)-stained biofilms in the well. (C) Colony morphology of the strains on an LB agar plate. (D) Motility of the strains on an M63 plate containing 0.3% agarose. We refer the motility of the double-knockout mutant, Δms33/32, as hypermotility. (E) Cell shape and cell length analysis by phase-contrast microscopy (picked from the three different focuses) at a magnification of ×100. As all the cells were not present in the same field during microscopy, the cropped and separate images of the individual cells are presented here together with demarcated boundaries. The bar graph shows the cell length measurement (mean ± SD; 100 cells/strain). The asterisks in both the bar graphs (B and E) indicate the statistical significance (P < 0.001), calculated using the Bonferroni t test of one-way ANOVA.
The sensitivity of the double mutant to antitubercular and β-lactam drugs is enhanced.
The proteins MSMEG_2433, MSMEG_2432, and DacB2 had β-lactamase activities, as revealed by their ability to hydrolyze nitrocefin (Fig. S4B and C). To further validate this result, we assessed the antibiotic susceptibilities of the double-knockout mutant and the complemented strains to various front-line antitubercular and β-lactam drugs by determining their MIC values using the broth microdilution assay. The sensitivity of the double mutant to antitubercular drugs was increased 2- to 4-fold, whereas a 2- to 8-fold susceptibility enhancement was observed with β-lactam antibiotics. Among the β-lactams tested, Δms33/32 showed a notable (8-fold) lower resistance against imipenem. A similar result was obtained in a previous study, in which M. tuberculosis lacking a functional copy of LdtMt5 (an ld-transpeptidase) was found to be more susceptible to some of the carbapenem antibiotics (30). Complementation with any of the three genes msmeg_2433, msmeg_2432, and dacB2 was able to overcome the loss of resistance in Δms33/32 (Table 1). For ampicillin, the parent strain showed resistance, with an MIC of 512 μg ml−1, whereas Δms33/32 showed an MIC of 256 μg ml−1. However, in combination with clavulanic acid (CA), a known β-lactamase inhibitor, ampicillin showed an MIC of 4 μg ml−1 for both of the strains. The sensitization of the double mutant to β-lactams, which was the result of the absence of both MSMEG-2433 and MSMEG-2432, is likely due to the loss of their β-lactamase-like property or certain changes in cell permeability.
TABLE 1.
Susceptibilities of M. smegmatis strains to β-lactam and antitubercular drugsa
| Antibiotic | MIC (μg ml−1) for indicated strain |
||||
|---|---|---|---|---|---|
| mc2155 | Δms33/32 | Δms33/32::pM33 | Δms33/32::pM32 | Δms33/32::pM11 | |
| Ethambutol | 312 | 156 | 312 | 312 | 312 |
| Rifampin | 10 | 2.5 | 10 | 10–20 | 5–10 |
| Isoniazid | 500 | 125 | 250–500 | 500 | 250–500 |
| Imipenem | 12 | 1.56 | ND | 6–12 | ND |
| Oxacillin | 500 | 125 | 250–500 | 500 | 250 |
| Penicillin G | 500 | 125 | 500 | 250–500 | 500 |
| Ampicillin | 512 | 256 | 512 | 512 | 512 |
| Ampicillin-CA | 4 | 4 | 4–8 | 4–8 | 4 |
Ampicillin was used in combination with clavulanic acid (CA) in a 2:1 ratio. ND, test not done.
The double mutant survives longer in the murine macrophage line RAW 264.7.
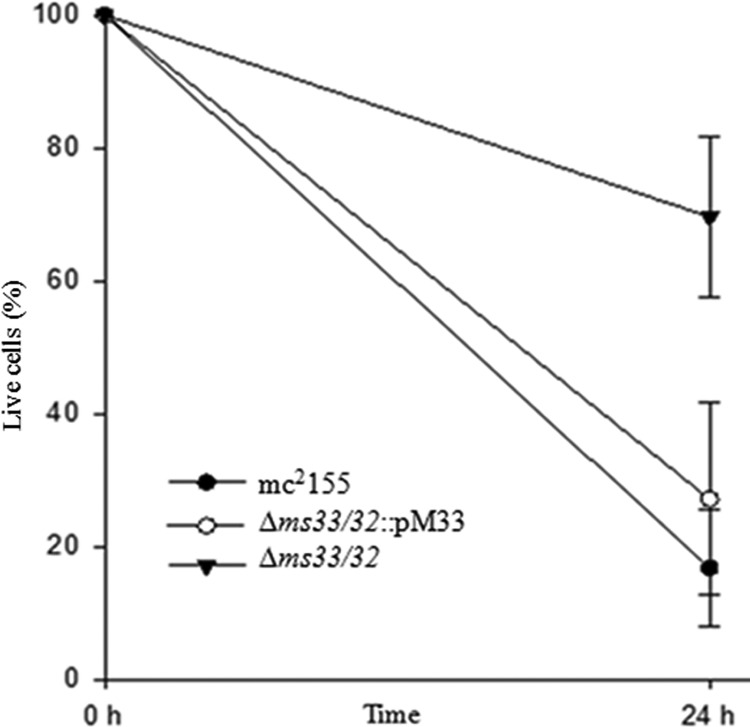
Bacterial cell surface components influence the survival of the organism in human and murine macrophages (24, 31, 32). In this study, an enormous loss of surface GPLs was observed in the double-knockout mutant Δms33/32, so we wanted to study its effect on the survival of the mycobacterium in the murine macrophage line RAW 264.7. We monitored the organism within macrophages for 24 h and observed that the number of viable cells was significantly higher (∼70%) in the double dd-CPase GPL-deficient strain than in the parent strain (∼20%). However, the level of the MSMEG_2433-complemented mutants was reduced to ∼25% (Fig. 6). From this result, we infer that surface GPLs may sensitize a nonpathogenic bacterium to killing by RAW 264.7 cells, though the decreased sensitivity of the double mutant could also be due to some other effects that arose because of the dd-CPase deletions.
FIG 6.
Existence of mycobacterial strains in murine macrophage line RAW 264.7. The macrophages were infected with an approximate MOI of 10:1 (refer to Materials and Methods for details). The net CFU per milliliter of the mycobacterial strains before lyses (considered 100%) as well as postlyses (after 24 h of infection) are plotted. The data represent the results from six replicates; error bars show SDs. Statistical significance was calculated by the Bonferroni t test using one-way ANOVA. The comparison showed a significant difference between Δms33/32 and mc2155 or Δms33/32::pM33 (P < 0.001) but an insignificant difference between mc2155 and Δms33/32::pM33 (P = 0.156).
DISCUSSION
To understand the roles of msmeg_2433 and msmeg_2432 in mycobacterial physiology, in this study we deleted these two dd-CPase genes both singly and in combination. The double deletion had a drastic impact on the physiology of the organism, whereas no significant effect was visible upon single deletions, indicating that MSMEG_2433 and MSMEG_2432 might play compensatory roles in the bacterium. Though there is a possibility of the existence of other analogous proteins, their compensatory roles are probably not enough to support the loss of two dd-CPases. This is further confirmed by ectopic-complementation studies, where we observed that the expression of any one of the orthologous genes, namely, msmeg_2433, msmeg_2432, and dacB2, is capable of overcoming all the oddities observed in the double-knockout mutant Δms33/32.
The most significant phenotypic changes that resulted from the double deletion included reduced surface GPLs, reduced biofilm/pellicle formation, and hypermotility. Previously, it has been reported that dacB2-deficient M. tuberculosis and DacB2-overexpressing M. smegmatis have altered physiological properties compared to those of the parent strain; the authors speculated that the change in the cell surface components and PG structure were the possible reasons for such alterations (15). Interestingly, here we report that PG cross-link formation was drastically affected by the absence of these two dd-CPases, resulting in a predominance of 4-3 cross-links over the regular 3-3 cross-links that are present in the parent. In M. tuberculosis, the 3-3 cross-links are essential for normal colony morphology (30, 33). We observed that the colony appearance of M. smegmatis was changed to dry whitish in the double mutant, in contrast to the parent, which originally had a regular smooth shape and pale yellow color. The exact reason for these changes is not understood. However, we speculate that as PG is an essential component of the MAP complex, the modified PG cross-link formation might have an impact on the MAP complex organization or plays a role in the destabilization of the MAP complex (28), which possibly influences the colony appearance, either directly or indirectly. We have noticed that the cell surface GPL expression is reduced in the double mutant without affecting its level in the cytoplasm, suggesting that either the cytoplasmic GPLs in Δms33/32 have undergone repression, possibly by a negative-feedback mechanism, or the GPLs are degraded. It is well known that a lack of surface GPLs reduces the formation of pellicle and biofilm at the air-liquid interface (17). The role of GPLs in sliding motility of M. smegmatis has also been reported (34). However, the surface GPLs might not be the sole reason for such changes in the phenotypes, as seen in the case of an lsr2 mutant of M. smegmatis which has normal surface GPLs but lacks a mycolate-containing apolar lipid (mycolyl-diacylglycerols), resulting in reduced surface hydrophobicity and pellicular-biofilm formation. This lsr2 mutant also showed hypermotility (21, 28, 35). Interestingly, in our study, hypermotility was observed in the GPL-deficient strain. These variations indicate that these phenotypic characteristics are not solely dependent on surface GPLs or cell surface hydrophobicity, as reported earlier, but that other cellular mechanisms might also influence it. Therefore, we hypothesize that the oddities in the double dd-CPase mutant might be due to the synergistic effect of surface GPL deficit and destabilization of the MAP complex.
Furthermore, MSMEG_2433, MSMEG_2432, and DacB2 have shown the ability to hydrolyze nitrocefin, suggesting that they have β-lactamase activity (see Fig. S4B and C in the supplemental material). The reduction in the antibiotic resistance further supports the same property observed in the mutants. In other words, the deletion of both the dd-CPases lowered the intrinsic β-lactam resistance of mycobacteria, whereas the lost resistance was regained upon complementing the double mutant by any of the cloned genes for dd-CPase homologs, namely, msmeg_2433, msmeg_2432, and dacB2 of M. tuberculosis. However, a lower resistance to the ampicillin-clavulanic acid combination (4 μg ml−1 [Table 1]) was observed in all the strains. This shows an apparent β-lactam sensitization in Δms33/32 that might be due to the loss of β-lactamase activity, because msmeg_2433 and msmeg_2432 appear to be inhibited by clavulanic acid. Loss of the surface GPLs could also impact the permeability of the cell wall. It can eventually sensitize the cell by increasing the intake of antibiotics in the double mutant, and a similar possibility has also previously been shown, where an M. smegmatis mutant lacking surface GPLs substantially accumulated chenodeoxycholate more effectively than did its parent strain (16).
The mycobacterial cell surface components play a significant role in immunogenicity (36). In the M. tuberculosis complex, a large number of virulence factors have evolved as a response to the host immune reaction that mostly includes cell envelope proteins, lipids, mycolic acid, etc. (36). The role of GPLs is hypothesized to mask the interaction of phosphatidyl-myo-inositol mannosides with Toll-like receptor 2 (37) and elicit the antibody response in M. avium complex-related infections (38). Previously, it has been shown that a dacB2 deletion strain of M. tuberculosis and a GPL-deficient M. smegmatis J15cs strain can survive longer in human and murine macrophages, respectively (15, 24). Our results correlate well with these findings, as we have also shown better survival of Δms33/32 cells in the RAW 264.7 cell line than that of the parent strain, though the mechanism of involvement of surface GPLs and/or dd-CPases in immune modulation remains unclear.
Hence, the results suggest that the altered ratio of 4-3 and 3-3 cross-links in the PG layer as a consequence of double dd-CPase deletion hampers surface GPL expression (directly or indirectly), affects its phenotype, and enhances the survival of the bacteria within macrophages. Nevertheless, the precise mechanisms by which the altered PG-cross-links influence the phenotype and the expression of surface GPLs and modulate the immune response remain unclear.
MATERIALS AND METHODS
Bacterial strains and growth media.
Mycobacterial strains (Table 2) were grown in Middlebrook (MB) 7H9 medium supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; Difco, MD), 0.05% (vol/vol) Tween 80, and glycerol. For solid media, 7H11 and Luria-Bertani (LB) agar were used with appropriate antibiotics. E. coli cells were grown in LB broth and agar. M63 minimal medium (39) along with MgSO4·7H2O (1 mM), glycerol (0.2%), and vitamin B1 (1 μg ml−1) was used for biofilm and pellicle formation assays. M63 with 0.8% and 0.3% agarose was used for colony morphology and motility experiments, respectively. Antibiotics were used at the following concentrations: hygromycin at 150 μg ml−1 (Invitrogen, CA), kanamycin at 30 μg ml−1, chloramphenicol at 20 μg ml−1, and tetracycline at 0.02 μg ml−1 (HiMedia, Mumbai, India). Unless otherwise indicated, all the enzymes for genetic modifications were purchased from New England BioLabs (MA) and all the reagents and chemicals were from Sigma-Aldrich (St. Louis, MO).
TABLE 2.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype or relevant features | Source or reference |
|---|---|---|
| Strains | ||
| mc2155 | Wild-type strain of Mycobacterium smegmatis | ATCC |
| Δms33 | msmeg_2433 deletion strain of M. smegmatis; Hygr | This study |
| Δms32 | msmeg_2432 deletion strain of M. smegmatis; Hygr | This study |
| Δms33/32 | msmeg_2433/32 deletion strain of M. smegmatis; Hygr | This study |
| Δms33/32::pM33 | Δms33/32 complemented with msmeg_2433 (pM33); Hygr Kanr | This study |
| Δms33/32::pM32 | Δms33/32 complemented with msmeg_2432 (pM32); Hygr Kanr | This study |
| Δms33/32::pM11 | Δms33/32 complemented with dacB2/rv2911 (pM11); Hygr Kanr | This study |
| Δms33/32::pMIND | Δms33/32 transformed with empty vector pMIND; Hygr Kanr | This study |
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pSMT100 | Mycobacterial suicide vector; sacB Hygr | 40 |
| pSD33-2 | pSMT100 with right and left flanking regions of msmeg_2433; Hygr | This study |
| pMIND | Tetracycline-inducible vector; Kanr | Bose Institute, Kolkata, India |
| pM33 | pMIND harboring msmeg_2433; Kanr | This study |
| pM32 | pMIND harboring msmeg_2432; Kanr | This study |
| pM11 | pMIND harboring dacB2/rv2911 of M. tuberculosis; Kanr | This study |
Genetic manipulations.
Inactivation of the msmeg_2433 gene was done by following the homologous-recombination technique as described previously (40). Briefly, the left and right flanking regions of msmeg_2433 were amplified with primers (see Table S1 in the supplemental material) from the mc2155 chromosomal DNA template using a thermocycler (Mastercycler personal; Eppendorf, Hamburg, Germany). The amplicon was sequentially cloned into the mycobacterial suicide vector pSMT100 to generate pSD33-2 (Fig. S2) and electroporated (Eporator; Eppendorf) into electrocompetent mc2155. The transformants were selected on 7H11 plates containing hygromycin and 10% (wt/vol) sucrose. As pSMT100 harbors the sacB gene cassette, only double-crossover mutants can grow on a sucrose-hygromycin plate (41). The deletion was further confirmed by amplifying the DNA product using various combinations of primers (Table S1). We tested the polar effect of msmeg_2433 deletion on the downstream gene msmeg_2432 using semiquantitative RT-PCR. The total RNA was isolated (from ab A600 of ∼0.6) using an RNASure minikit from Nucleo-pore (New Delhi, India). The purity of the RNA was tested by PCR, and DNA contamination, if any, was further removed by DNase I treatment. The integrity of the RNA was ensured by 2% agarose gel electrophoresis by visualizing 23S and 16S RNA bands. The cDNA was synthesized using equal amounts of RNA and the respective 3′ primers for each gene, using a reverse transcriptase (murine leukemia virus [MuLV]) kit. The gene-specific primers (Table S1) were used for PCR amplification of the products from the appropriate templates.
Single-deletion mutants (without any polar effect) for msmeg_2433 (Δms33) and msmeg_2432 (Δms32) were also created. The deletion of msmeg_2432 was carried out using the process mentioned above (unpublished data), but for msmeg_2433, we chose to follow the one-step deletion protocol (42), with some modifications. Briefly, the primers for the hygromycin cassette were designed in such a way that the forward and reverse primers would contain 50-bp extensions, matching the promoter region and the open reading frame of msmeg_2433, respectively, with a gap of 200 bp, including the start codon. The amplicons were ligated to pSMT100 at the BamHI and PstI sites before transformation. Double-crossover mutants were selected on sucrose-hygromycin plates. The mutants were further confirmed by amplifying and sequencing the entire fragments.
Complementation.
The double mutant Δms33/32 was complemented with msmeg_2433, msmeg_2432, and its ortholog from M. tuberculosis, dacB2 (Fig. S3), cloned in a tetracycline-inducible vector, pMIND. The nucleotide sequences of msmeg_2433 and msmeg_2432 were extracted from MycoBrowser smegmaList, and that of dacB2 was extracted from tubercuList. The genes were amplified using the respective gene-specific primers (Table S1), cloned into the pMIND vector, and electroporated into Δms33/32. The transformants were screened on 7H11 plates containing hygromycin and kanamycin and named Δms33/32::pM33, Δms33/32::pM32 and Δms33/32::pM11, respectively. The empty pMIND vector was also transformed into Δms33/32 to create Δms33/32::pMIND, which was used as a control.
Ectopic expression and membrane localization studies using Bocillin FL.
The cultures were grown in 7H9 medium containing appropriate antibiotics. The strains harboring pMIND plasmids were grown up to an A600 of ∼0.2 and induced with 20 ng ml−1 of tetracycline. The membrane proteins were isolated from the cells grown to an A600 of ∼1.0. The cells were harvested and ruptured by sonication. The cell debris was removed by spinning at 18,000 × g for 10 min at 4°C, followed by spinning the supernatant at 50,000 × g for 3 h at 4°C. Membrane proteins (pellet) were dissolved and allowed to bind with Bocillin FL (Invitrogen, Carlsbad, CA) at 30°C for 1 h. The proteins were resolved by 15% sodium dodecyl sulfate (SDS)-PAGE, and the Bocillin FL-labeled proteins were detected in Typhoon FLA 7000 (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) at an excitation wavelength of 488 nm and an emission wavelength of 526 nm.
Growth curve analysis.
Primary inocula were diluted to a density equivalent to McFarland standard 0.5 and inoculated in 100 ml of MB 7H9 medium with appropriate antibiotics. The culture was grown at 37°C, and the CFU per milliliter were calculated from 0 to 42 h at a growth interval of 3 h. The experiment was done in triplicate and the graph was plotted on semilog base 2 scale (y axis; CFU per milliliter). The growth rate and generation time of each replicate were calculated from the linear part of the log phase in the growth curve.
Preparation of peptidoglycan.
To prepare the PG, previously published protocols (3, 43) were followed, with slight modifications. The mycobacterial strains were grown in 500 ml of nutrient broth (HiMedia) to mid-log phase and harvested, and the pellet was washed twice with phosphate-buffered saline (PBS). The pellet was resuspended in 10 mM NH4HCO3 containing 1 mM protease inhibitor and phenylmethylsulfonyl fluoride (PMSF) and sonicated intermittently for 30 cycles (with a 60-s burst followed by 60 s of cooling). The sonicated cell suspension was treated with DNase and RNase (10 μg ml−1 each) for 1 h at 4°C before centrifugation at 27,000 × g for 30 min. The pellet was resuspended in PBS with 4% SDS and incubated at 100°C for 30 min. The pellet was further treated with pronase (200 μg ml−1 in 1 ml of 10 mM Tris-HCl [pH 7.4]) and trypsin (200 μg ml−1 in 1 ml of 20 mM phosphate buffer [pH 7.8]) for 18 h at 37°C. The pellet was washed twice with water (20 ml) and digested overnight at 37°C with lysozyme and mutanolysin (200 μg ml−1 each) prepared in 1 ml of 25 mM phosphate buffer, pH 6.0, containing 0.1 mM MgCl2. Both of the enzymes were inactivated (at 100°C for 3 min), and the soluble disaccharide peptides were recovered by ultracentrifugation at 100,000 × g for 30 min at 20°C. The lactoyl peptide PG fragments were prepared from the disaccharide peptides under alkaline conditions. To the solution of 200 μl of disaccharide peptides was added 30% ammonium hydroxide (68 μl), and the reaction mixture was incubated at 37°C for 5 h. The reaction was neutralized with 61 μl of acetic acid, lyophilized, and dissolved in water containing 0.05% trifluoroacetic acid (TFA).
Purification of PG fragments by reverse-phase high-pressure liquid chromatography (RP-HPLC).
The lactoyl peptides were separated by an Agilent 1260 Infinity HPLC system (Agilent Technologies, CA) using a C18 column (ZORBAX SB-C18, 5 μm, 4.6 by 250 mm) and a diode array detector (DAD) as described previously (3), with little modification. Lactoyl peptides (50 μl) were applied on the C18 column and the muropeptides were eluted in 30% acetonitrile (containing 0.05% TFA) at a flow rate of 0.5 ml min−1. The elution was monitored at 210 nm. The relative abundances of the muropeptides were estimated as a percentage of the total integrated peak area. The materials eluted in the individual peaks were collected separately, lyophilized, and dissolved in water for MALDI-TOF analysis.
Extraction of GPLs and TLC analysis.
The surface and cytoplasmic glycopeptidolipids (GPLs) were extracted by following previously published protocols (23, 44, 45), with few modifications. Briefly, the cells were grown and harvested and the pellet was washed twice with cold PBS (1 ml) and mixed with chloroform-methanol (C-M; ratio of chloroform to methanol, 2:1) by shaking vigorously. The insoluble materials were removed by centrifugation. The supernatant was mixed with 0.9% NaCl and centrifuged (2,000 × g for 5 min), and the GPL-containing organic phase was evaporated to dryness. The dried extracts were dissolved in C-M (2:1) before being subjected to thin-layer chromatography (TLC). For deacetylation (46), one half of the extracted GPLs was mixed with an equal amount of 0.2 N NaOH (in methanol) and incubated at 37°C for 1 h. The solution was neutralized (with 2.5 μl of glacial acetic acid), evaporated to dryness, and dissolved in a mixture of 500 μl of C-M (2:1) and 100 μl of water. The mixture was spun down and the lower organic phase was restored, dried, and dissolved in C-M (2:1). TLC was performed on silica gel 60 F254 (Merck, Germany) using a previously established combination of solvent mix and C-M-water (90:10:1) (17). The GPL bands were developed by spraying 0.1% orcinol (in 40% sulfuric acid) and charring them at 120°C. As and when required, the GPL bands were also marked using an iodine chamber and UV light.
MALDI-TOF MS analysis.
The HPLC-purified muropeptides and GPLs were analyzed by MALDI-TOF MS using an UltrafleXtreme ToF/ToF (Bruker Daltonics, MA). The sample preparation was done as described previously (47). Briefly, the acetylated GPLs were run on the TLC plate and the GPL bands were scraped and dissolved in C-M (2:1). After incubation for 4 h at room temperature, the samples were mixed and the clear upper phase was collected for MALDI-TOF MS analysis. The matrix, 2,5-dihydrobenzoic acid (DHB; 10 mg ml−1 in methanol), was mixed with equal amounts of GPLs and muropeptides (1 μl each). The samples were spotted and dried before the analysis. For the analysis, regular standard operating protocol, i.e., Reflectron mode with an accelerating voltage in positive mode at 20 kV, and the manufacturer's instructions were followed.
CR binding assay.
Actively growing mycobacterial cultures were washed twice with sterile water, and 3 μl was spotted onto LB agar plates (without NaCl) supplemented with Congo red (CR) and Coomassie brilliant blue (CBB) in 40- and 20-μg ml−1 final concentrations, respectively. Plates were incubated at 37°C for 5 days, after which the effect of CR binding on the colony morphology was analyzed (48).
Determination of pellicle and biofilm formation, motility, and colony morphology.
To determine the pellicle and biofilm formation ability of the strains, previously published protocols (17, 28, 49, 50) were followed. For pellicle formation, cultures containing ∼1.5 × 108 cells were grown without shaking in 10 ml of M63 medium (devoid of Tween 80) in sterile glass tubes at 25°C, and the results were analyzed 1 week postinoculation. The biofilm formation assay was performed with 24-well cell culture plates with 1 ml of M63 medium containing an inoculum of ∼1.5 × 108 cells/well. Strains were incubated at 37°C for 48 h under static conditions. Wells were carefully washed with sterile water to remove the planktonic cells/nonadherent bacteria, and the biofilms were stained with 0.1% crystal violet (CV). The bound CV was extracted from the biofilms in 1 ml of 33% acetic acid and measured at A600 to quantify the biofilm formation. Six replicates of the experiment were performed. For colony morphology, 3 μl of actively growing culture at an A600 of ∼1.0 was spotted onto LB agar plates and M63 plates with 0.8% agarose (containing appropriate antibiotics and inducers) and incubated at 37°C. Results were analyzed 5 days postinoculation. For the motility assay the procedure was the same as described above except that 0.3% agarose was used in the M63 plates.
Phase-contrast microscopy.
Cells were grown to mid-log phase, harvested by spinning at 4,000 × g for 5 min at 4°C, washed twice with cold PBS (pH 7.0), and spotted onto poly-lysine-coated glass slides. Live cells were subsequently visualized using a 1X71 phase-contrast microscope (Olympus Corporation, Tokyo, Japan) with a 100× oil immersion objective (51).
Determination of drug susceptibilities via MIC.
The MIC values for the β-lactam antibiotics and front-line antitubercular drugs were determined by using the broth microdilution method as described previously (52, 53). Briefly, the assay was performed in 96-well microtiter plates with 7H9 broth containing primary inocula of 1.5 × 108 cells/well. The plates were incubated at 37°C for 3 days, and growth was assessed at A600 in a Multiskan Spectrum-1500 spectrophotometer (Thermo Scientific, Nyon, Switzerland). Six replicates of the experiments were performed.
Studies of survival in RAW 264.7 cells.
The RAW 264.7 murine macrophage line, procured from the National Centre for Cell Sciences (NCCS; Pune, India), was maintained in 5% CO2 at 37°C in Dulbecco's modified Eagle medium (DMEM; HiMedia) supplemented with 10% fetal bovine serum (FBS; Gibco, Invitrogen Life Technologies) and 1% penicillin-streptomycin solution (Sigma-Aldrich). The infection studies were performed as detailed earlier (24), with some modifications. Briefly, the RAW 264.7 cells were scraped from the culture plate, suspended in fresh DMEM, and allowed to adhere to the surfaces of 6-well flat-bottom polystyrene plates at a count of 2.5 × 105. Actively growing mycobacterial cells were washed, and a single-cell suspension was prepared and inoculated into the RAW 264.7 cells at an approximate multiplicity of infection (MOI) of 10:1. The cocultivation was allowed to proceed for 4 h at 37°C and 5% CO2 and washed twice with sterile PBS to remove noninfecting bacteria. Thereafter, fresh DMEM was added and the coculturing was allowed for a period of 24 h. Next, the macrophages were lysed with 0.1% Triton X-100 by resuspension for 15 min with intermittent shaking. The lysates of the respective samples were serially diluted, plated on LB agar, and incubated at 37°C for 5 days, and the viable bacteria were enumerated in the form of CFU per milliliter. Six replicates of the experiments were performed, and statistical significance was calculated by the Bonferroni t test of one-way analysis of variance (ANOVA).
Supplementary Material
ACKNOWLEDGMENTS
S.D.P. and A.S.G. thank Science and Engineering Research Board (SERB), Government of India, for funding this work under the NPDF vide grant no. PDF/2015/000011.
S.D.P. and A.S.G. designed the experiments and analyzed the results. S.D.P. performed most of the experiments, S.P. did the biofilm assay, A.B. did the MIC experiments, G.K.N. performed the cell shape analysis, and S.M. did the macrophage studies.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00760-17.
REFERENCES
- 1.McNeil M, Daffe M, Brennan PJ. 1990. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J Biol Chem 265:18200–18206. [PubMed] [Google Scholar]
- 2.McNeil M, Daffe M, Brennan PJ. 1991. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J Biol Chem 266:13217–13223. [PubMed] [Google Scholar]
- 3.Mahapatra S, Crick DC, McNeil MR, Brennan PJ. 2008. Unique structural features of the peptidoglycan of Mycobacterium leprae. J Bacteriol 190:655–661. doi: 10.1128/JB.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol 193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HI, Barry CE III. 2012. Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol 86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glauner B, Holtje JV, Schwarz U. 1988. The composition of the murein of Escherichia coli. J Biol Chem 263:10088–10095. [PubMed] [Google Scholar]
- 8.Pisabarro AG, de Pedro MA, Vazquez D. 1985. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol 161:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem 280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 10.Sanders AN, Wright LF, Pavelka MS Jr. 2014. Genetic characterization of mycobacterial L,D-transpeptidases. Microbiology 160:1795–1806. doi: 10.1099/mic.0.078980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. 2000. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem 275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh AS, Chowdhury C, Nelson DE. 2008. Physiological functions of D-alanine carboxypeptidases in Escherichia coli. Trends Microbiol 16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Machowski EE, Senzani S, Ealand C, Kana BD. 2014. Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol 14:75. doi: 10.1186/1471-2180-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bansal A, Kar D, Murugan RA, Mallick S, Dutta M, Pandey SD, Chowdhury C, Ghosh AS. 2015. A putative low-molecular-mass penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characteristics of DD-carboxypeptidase and beta-lactamase. Microbiology 161:1081–1091. doi: 10.1099/mic.0.000074. [DOI] [PubMed] [Google Scholar]
- 15.Bourai N, Jacobs WR Jr, Narayanan S. 2012. Deletion and overexpression studies on DacB2, a putative low molecular mass penicillin binding protein from Mycobacterium tuberculosis H(37)Rv. Microb Pathog 52:109–116. doi: 10.1016/j.micpath.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont MA, Daffe M. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089–3100. doi: 10.1099/00221287-148-10-3089. [DOI] [PubMed] [Google Scholar]
- 17.Recht J, Kolter R. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J Bacteriol 183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schorey JS, Sweet L. 2008. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18:832–841. doi: 10.1093/glycob/cwn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan PJ, Goren MB. 1979. Structural studies on the type-specific antigens and lipids of the Mycobacterium avium·Mycobacterium intracellulare·Mycobacterium scrofulaceum serocomplex. J Biol Chem 254:4205–4211. [PubMed] [Google Scholar]
- 20.Chatterjee D, Khoo KH. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell Mol Life Sci 58:2018–2042. doi: 10.1007/PL00000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Recht J, Martinez A, Torello S, Kolter R. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol 182:4348–4351. doi: 10.1128/JB.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshayes C, Laval F, Montrozier H, Daffe M, Etienne G, Reyrat JM. 2005. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J Bacteriol 187:7283–7291. doi: 10.1128/JB.187.21.7283-7291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sondén B, Kocincova D, Deshayes C, Euphrasie D, Rhayat L, Laval F, Frehel C, Daffe M, Etienne G, Reyrat JM. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol Microbiol 58:426–440. doi: 10.1111/j.1365-2958.2005.04847.x. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara N, Ohara N, Ogawa M, Maeda S, Naka T, Taniguchi H, Yamamoto S, Ayata M. 2015. Glycopeptidolipid of Mycobacterium smegmatis J15cs affects morphology and survival in host cells. PLoS One 10:e0126813. doi: 10.1371/journal.pone.0126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. 2012. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res 40:D627–D631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Målen H, Softeland T, Wiker HG. 2008. Antigen analysis of Mycobacterium tuberculosis H37Rv culture filtrate proteins. Scand J Immunol 67:245–252. doi: 10.1111/j.1365-3083.2007.02064.x. [DOI] [PubMed] [Google Scholar]
- 27.Mahapatra S, Piechota C, Gil F, Ma Y, Huang H, Scherman MS, Jones V, Pavelka MS Jr, Moniz-Pereira J, Pimentel M, McNeil MR, Crick DC. 2013. Mycobacteriophage Ms6 LysA: a peptidoglycan amidase and a useful analytical tool. Appl Environ Microbiol 79:768–773. doi: 10.1128/AEM.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JM, German GJ, Alexander DC, Ren H, Tan T, Liu J. 2006. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J Bacteriol 188:633–641. doi: 10.1128/JB.188.2.633-641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortalo-Magné A, Lemassu A, Laneelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffe M. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol 178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brammer Basta LA, Ghosh A, Pan Y, Jakoncic J, Lloyd EP, Townsend CA, Lamichhane G, Bianchet MA. 2015. Loss of a functionally and structurally distinct ld-transpeptidase, LdtMt5, compromises cell wall integrity in Mycobacterium tuberculosis. J Biol Chem 290:25670–25685. doi: 10.1074/jbc.M115.660753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sut A, Sirugue S, Sixou S, Lakhdar-Ghazal F, Tocanne JF, Laneelle G. 1990. Mycobacteria glycolipids as potential pathogenicity effectors: alteration of model and natural membranes. Biochemistry 29:8498–8502. doi: 10.1021/bi00488a042. [DOI] [PubMed] [Google Scholar]
- 32.Villeneuve C, Etienne G, Abadie V, Montrozier H, Bordier C, Laval F, Daffe M, Maridonneau-Parini I, Astarie-Dequeker C. 2003. Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. J Biol Chem 278:51291–51300. [DOI] [PubMed] [Google Scholar]
- 33.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. 2010. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez A, Torello S, Kolter R. 1999. Sliding motility in mycobacteria. J Bacteriol 181:7331–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arora K, Whiteford DC, Lau-Bonilla D, Davitt CM, Dahl JL. 2008. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J Bacteriol 190:4291–4300. doi: 10.1128/JB.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, Cataldi AA, Bigi F. 2013. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhoades ER, Archambault AS, Greendyke R, Hsu FF, Streeter C, Byrd TF. 2009. Mycobacterium abscessus glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-alpha by preventing interaction with TLR2. J Immunol 183:1997–2007. doi: 10.4049/jimmunol.0802181. [DOI] [PubMed] [Google Scholar]
- 38.Matsunaga I, Komori T, Ochi A, Mori N, Sugita M. 2008. Identification of antibody responses to the serotype-nonspecific molecular species of glycopeptidolipids in Mycobacterium avium infection. Biochem Biophys Res Commun 377:165–169. doi: 10.1016/j.bbrc.2008.09.091. [DOI] [PubMed] [Google Scholar]
- 39.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey SD, Choudhury M, Yousuf S, Wheeler PR, Gordon SV, Ranjan A, Sritharan M. 2014. Iron-regulated protein HupB of Mycobacterium tuberculosis positively regulates siderophore biosynthesis and is essential for growth in macrophages. J Bacteriol 196:1853–1865. doi: 10.1128/JB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelicic V, Reyrat JM, Gicquel B. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol 178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, Blanot D, Brouard JP, Arthur M. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in Gram-positive bacteria. J Biol Chem 279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- 44.Billman-Jacobe H, McConville MJ, Haites RE, Kovacevic S, Coppel RL. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol Microbiol 33:1244–1253. doi: 10.1046/j.1365-2958.1999.01572.x. [DOI] [PubMed] [Google Scholar]
- 45.Tatham E, Sundaram Chavadi S, Mohandas P, Edupuganti UR, Angala SK, Chatterjee D, Quadri LE. 2012. Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family. BMC Microbiol 12:118. doi: 10.1186/1471-2180-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennan PJ, Souhrada M, Ullom B, McClatchy JK, Goren MB. 1978. Identification of atypical mycobacteria by thin-layer chromatography of their surface antigens. J Clin Microbiol 8:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naka T, Nakata N, Maeda S, Yamamoto R, Doe M, Mizuno S, Niki M, Kobayashi K, Ogura H, Makino M, Fujiwara N. 2011. Structure and host recognition of serotype 13 glycopeptidolipid from Mycobacterium intracellulare. J Bacteriol 193:5766–5774. doi: 10.1128/JB.05412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jonas K, Tomenius H, Kader A, Normark S, Romling U, Belova LM, Melefors O. 2007. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol 7:70. doi: 10.1186/1471-2180-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallant CV, Daniels C, Leung JM, Ghosh AS, Young KD, Kotra LP, Burrows LL. 2005. Common beta-lactamases inhibit bacterial biofilm formation. Mol Microbiol 58:1012–1024. doi: 10.1111/j.1365-2958.2005.04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar A, Mallik D, Pal S, Mallick S, Sarkar S, Chanda A, Ghosh AS. 2015. Escherichia coli O8-antigen enhances biofilm formation under agitated conditions. FEMS Microbiol Lett 362:fnv112. doi: 10.1093/femsle/fnv112. [DOI] [PubMed] [Google Scholar]
- 51.Nelson DE, Young KD. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182:1714–1721. doi: 10.1128/JB.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown BA, Wallace RJ Jr, Onyi GO. 1992. Activities of clarithromycin against eight slowly growing species of nontuberculous mycobacteria, determined by using a broth microdilution MIC system. Antimicrob Agents Chemother 36:1987–1990. doi: 10.1128/AAC.36.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarkar SK, Dutta M, Chowdhury C, Kumar A, Ghosh AS. 2011. PBP5, PBP6 and DacD play different roles in intrinsic beta-lactam resistance of Escherichia coli. Microbiology 157:2702–2707. doi: 10.1099/mic.0.046227-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.