Abstract
Extensive utilization of polybrominated diphenyl ethers (PBDEs) as flame retardants since the 1960s in a variety of commercial products has resulted in ubiquitous environmental distribution of commercial PBDE mixtures. Dangers posed to biological populations became apparent after the discovery of elevated levels of PBDEs in biota, most notably in human breast milk and tissues. Environmental persistence of PBDEs results in significant transboundary displacement, threatening fragile ecosystems globally. Despite efforts to curtail usage of PBDEs, public concern remains about the effects of legacy PBDEs contamination and continued discharge of PBDEs in regions lacking restrictions on usage and manufacture. Among available technologies for remediation of PBDEs such as ex-situ soil washing, electrokinetic degradation, and biodegradation, this review focuses on bioremediation by microbes under anaerobic conditions. Bioremediation is generally preferred as it is less disruptive to contaminated ecosystems, is cost-effective, and can be implemented at sites that may be inaccessible to more traditional ex-situ methods. The aims of this review are to (1) summarize current knowledge of anaerobic microbes that debrominate PBDEs and their associated synergistic partnerships with non-dehalogenating microbes; (2) explore current understandings of the metabolic reductive debromination of PBDE congeners; (3) discuss recent discoveries on dehalogenase genes involved in debromination of PBDEs.
Keywords: flame retardants, organohalides, polybrominated diphenyl ethers (PBDEs), reductive debromination, debromination pathways, reductive dehalogenase genes
Introduction
Polybrominated diphenyl ethers (PBDEs) have been widely used as flame retardant additives in a variety of manufactured products, from paints and plastics to textiles and televisions since 1960s. Deposition of anthropogenic PBDEs has subsequently been identified in air, soils, and water across the world (McGrath et al., 2017), even in remote areas including isolated mountaintop sediments, the Faroe Islands, and the Antarctic (Lindström, 1999; Gallego et al., 2007; Wild et al., 2015). In recent years, PBDEs concentrations as high as 10,000 ng/g soil have been detected at manufacturing and e-recycling sites (Alabi et al., 2012; Labunska et al., 2013; Li et al., 2015; Deng et al., 2016). Notably, PBDEs have the tendency to accumulate in biota (de Boer et al., 1998; Boon et al., 2002; Norstrom et al., 2002; Zhu and Hites, 2004) and have diverse toxicological effects including endocrine disruption, metabolic disorders in biological populations, as well as neurological and developmental disorders in human (Siddiqi et al., 2003; Windham et al., 2015; ATSDR, 2017). Growing concerns over risks to human and environmental health have resulted in hepta- through tetra-BDEs being listed as persistent organic pollutants (POPs) on the United Nations Stockholm Convention in 2009 (United Nations Environment Programme, 2009).
Though restrictions and bans on manufacture and usage of PBDEs have been in place for several years, these legislations have no effect on the release of PBDEs from existing products or from recycled materials containing PBDEs. Environmental deposition of PBDEs can occur via release during manufacture and use of consumer products, improper disposal, and recycling of PBDEs containing products, volatilization during incineration, and discharge from wastewater treatment facilities. PBDEs are highly lipophilic, have low water solubility and low vapor pressures. These physical characteristics are largely dictated by the number and arrangement of bromine atoms on the molecule, with highly brominated PBDEs being more lipophilic, less soluble and less volatile (Table 1). Hence, highly brominated PBDEs adsorb readily to soils and sediments, where they persist and act as reservoirs leaching into other compartments over time (O'Driscoll et al., 2016). PBDEs also tend to bind to the organic fraction of particulate matter, soils, vegetation, and sediments following environmental deposition. Thus, PBDEs have a tendency to be exchanged between the atmosphere and surface, allowing passive transport over long distances.
Table 1.
Physical properties of PBDE mixtures.
| Property | Penta-BDE | Octa-BDE | Deca-BDE |
|---|---|---|---|
| Molecular weight | Mixture | Mixture | 959.22 |
| Color | Clear, amber to pale yellow | Off-white | Off-white |
| Physical state | Highly viscous liquid | Powder | Powder |
| Melting point | −7 to −3°C (commercial) | 85–89°C (commercial); 200°C (range, 167–257); 79–87°C; 170–220°C | 290–306°C |
| Boiling point | >300°C (decomposition starts above 200°C) | Decomposes at >330°C (commercial) | Decomposes at >320, >400, and 425°C |
| Density (g/mL) | 2.28 at 25°C; 2.25–2.28 | 2.76; 2.8 (commercial) | 3.0; 3.25 |
| Solubility: | |||
| – Water | 13.3 μg/L (commercial); | < 1 ppb at 25°C (commercial); | < 0.1 μg/L |
| 2.4 μg/L (pentabromodiphenyl ether component); | 1.98 μg/L (heptabromodiphenyl ether component) | ||
| 10.9 μg/L (tetrabromodiphenyl ether component) | |||
| – Organic solvent(s) | 10 g/kg methanol; miscible in toluene | Acetone (20 g/L); benzene (200 g/L); methanol (2 g/L)—all at 25°C | d-limonene (0.1823 g/100 g solvent); n-propanol (0.1823 g/100 g solvent)—all at 20°C * |
| Partition coefficients: | |||
| – Log Kow | 6.64–6.97; 6.57 (commercial) | 6.29 (commercial) | 6.265 |
| – Log Koc | 4.89–5.10 | 5.92–6.22 | 6.8 |
| Vapor pressure | 2.2 × 10−7−5.5 × 10−7 mm Hg at 25°C; 3.5 × 10−7 mm Hg (commercial) | 9.0 × 10−10–1.7 × 10−9 mm Hg at 25°C; 4.9 × 10−8 mm Hg at 21°C (commercial) | 3.2 × 10−8 mm Hg |
| Henry's Law constant (atm-m3/mole) | 1.2 × 10−5; 1.2 × 10−6; 3.5 × 10−6 | 7.5 × 10−8; 2.6 × 10−7 | 1.62 × 10−6; 1.93 × 10−8; 1.2 × 10−8; 4.4 × 10−8 |
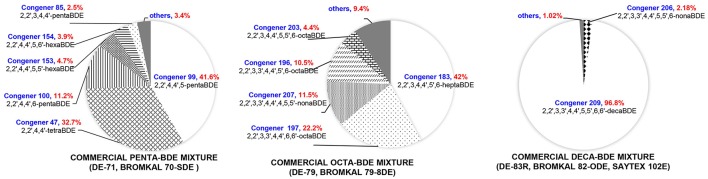
PBDE formulations have been primarily manufactured as three different technical mixtures (Figure 1): penta-BDEs, octa-BDEs, and deca-BDEs. Although BDE-209 has historically been the most widely used PBDE congener, BDE −47, −99, −100, and −153 are the most commonly observed PBDEs in the environment (Darnerud et al., 2001), indicating that environmental transformation of highly brominated BDEs is an important pathway contributing to the dispersal and environmental impact of PBDEs. The partial debromination of deca- and octa-BDEs is particularly worrisome because the less brominated metabolites like tetra- and penta-BDEs are of higher toxicity (Palm et al., 2002). Both abiotic and biotic processes are responsible for the breakdown of PBDEs in the environment. Photolysis, which can occur in the atmosphere and at soil surfaces, of lesser brominated BDEs (BDE-28, BDE-47, BDE-99, BDE-100, BDE-153, and BDE-183) (Rayne et al., 2006; Fang et al., 2008) and of BDE-209 (Stapleton and Dodder, 2008) has been demonstrated under laboratory conditions, although there is only weak evidence that atmospheric photodegradation is a major contributor to environmental attenuation of PBDEs (Schenker et al., 2008). Anaerobic and anoxic sediments and soils are major sinks and environmental reservoirs for PBDEs, making anoxic debromination by microorganisms an important route for eliminating PBDEs in the environment.
Figure 1.
Compositions of representative penta-, octa-, and deca-BDE mixtures. Congeners below 2% (w/w) are considered as others. As a result of the chemical process used to generate PBDE mixtures, the exact congener composition remains undefined and may vary significantly between different manufacturers and production batches (La Guardia et al., 2006).
Debromination of a variety of PBDEs, including the three primary technical mixtures, has been observed in soil, sewage sludge, and estuarine and marine sediments under different environmental conditions (redox, pH, available electron donors, etc.), although the process is typically slow and often results in incomplete debromination of PBDEs. Despite these apparent limitations of anaerobic microbial degradation of PBDEs, bioremediation is considered the most environmentally friendly technology to remediate PBDEs. Although bioremediation requires extensive site characterization to be successful and is often much slower than traditional ex-situ treatment strategies, it is less expensive, less disruptive to sites, and can be more complete in degrading hazardous compounds. Considering the fragility and inaccessibility of many of the environments and ecosystems plagued by PBDEs contamination, bioremediation via anaerobic microbes is the best available option for eliminating PBDEs from contaminated areas.
This review aims to summarize current knowledge of microbial reductive debromination of PBDEs under anaerobic conditions, the debromination pathways involved, and dehalogenase genes identified so far. Investigation of PBDEs debrominating microbes and exploration of the underlying mechanisms of debromination will enable more effective tracking the fate of PBDEs in the environment.
Microorganisms involved in reductive debromination of PBDEs
The hydrophobicity of PBDEs impedes bioavailability, which results in low biomass of debrominating microorganisms. This low abundance is a critical challenge to enrichment, isolation, and characterization of PBDE debrominating bacteria. After pioneering efforts identified debromination of PBDEs in previously isolated organochlorine dehalogenating members of the genera Sulfurospirillum and Dehalococcoides (He et al., 2006) as well as Desulfitobacterium and Dehalobacter (Robrock et al., 2008), later studies successfully enriched and isolated PBDEs debrominating bacteria from multiple environmental sources (Lee et al., 2011; Ding et al., 2013, 2017) and confirmed the role of members of the genera Dehalococcoides and Acetobacterium in environmental debromination of PBDEs (Figure 2, Table 2).
Figure 2.
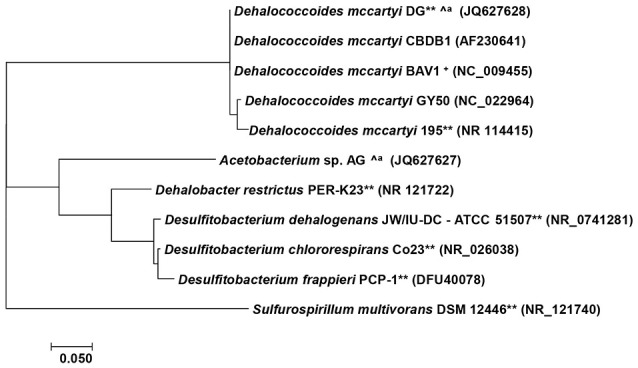
Phylogenetic analysis of PBDE debrominating bacteria. The tree was constructed with MEGA 7 (Kumar et al., 2016) using Neighbor-joining method in Kimura 2-parameter mode (Kimura, 1980). ∧a, derived from same culture, **require auxiliary substrate, +only in mixed culture.
Table 2.
PBDE debrominating cultures.
| Sample ID | Source | Auxiliary substrate | deca-BDEs | octa-BDEs | penta-BDEs | Remarks | Duration | Citation | |
|---|---|---|---|---|---|---|---|---|---|
| Pure cultures | Dehalococcoides mccartyi 195 | Anaerobic sewage digestor sludge | TCE | No | hepta- through tetra- | N.A. | 6 months | He et al., 2006 | |
| Dehalococcoides mccartyi DG | Isolate from G | TCE | N.A. | penta- tetra- | tetra- | Minimal conversion; Lost octa-BDE debromination activity of parent culture, G, which can only be resumed by co-cultivating AG and DG; | Octa-BDEs 5 months; penta-BDEs 1 month | Ding et al., 2013 | |
| Acetobacterium sp. AG | Isolate from G | Lactate | N.A. | penta- tetra- | penta- through di- | ||||
| Dehalococcoides mccartyi CBDB1/n-ZVI | Saale river sediment | n-ZVI | 20% conversion to diphenyl ether | N.A. | N.A. | No debromination of deca-BDE by CBDB1 without addition of n-ZVI; the role of CBDB1 in this study is uncertain due to pH and unbalanced hydrogen consumption. | 1 month | Xu et al., 2014 Yang, 2017 | |
| Dehalococcoides mccartyi GY50 | Isolate from co-culture, GY2, sand, and silt near Lianjiang River | No | N.A. | N.A. | Diphenyl ether | GY2 share the same debromination activity with its isolate GY50; Complete debromination; Ortho-removal preferred; identified PBDE RDases, PbrA1, PbrA2, PbrA3 | 2 weeks | Lee et al., 2011; Ding et al., 2017 | |
| Dehalococcoides mccartyi GY52 | Variant of GY50 after consecutive transfer in TCE | No | N.A. | N.A. | Di- | A genome island where pbrA1 and pbrA2 locate was deleted from GY50 | 2 weeks | ||
| Desulfitobacterium chlororespirans Co23 | 2,3-CP dehalogenating compost soil | 3-chloro-4-hyxdroxybenzoate | Similar debromination profile as ANAS195 (data not shown) | N.A. | Robrock et al., 2008 | ||||
| Desulfitobacterium halogenans JW/IU-DC | Freshwater sediments-pond | 3-chloro-4-hydroxphenylacetate | Similar debromination profile as ANAS195 (data not shown) | N.A. | Robrock et al., 2008 | ||||
| Desulfitobacteriun frappieri PCP-1 | A mixture of pentachlorophenol contaminated soil and anaerobic sewage sludge | pentachlorophenol | N.A. | hepta- hexa- penta- | penta 99 to tetra, tri, di; tetra 47 to tri and di | Debromination pathways were identified by spiking individual congeners; para- and meta-removal is preferred; debromination for higher brominated BDEs are slower; | 3 months | Robrock et al., 2008 | |
| Dehalobacter restrictus PER-K23 | Anaerobic Rhine river sediment and ground anaerobic granular sludge | PCE | N.A. | hepta- hexa- penta- | penta 99 to tetra-; tetra47 to tri and di | 3 months | Robrock et al., 2008 | ||
| Sulfurospirillum multivorans DSM12446 | Activated sludge | TCE | Octa- Hepta- | No | N.A. | 2 months | He et al., 2006 | ||
| Mixed Cultures | Culture G | Soil samples from a river bank in Wisconsin | TCE | N.A. | hexa-, penta-, tetra-(dominant), tri- | penta- through di- | Para- and meta-removal preferred in octa-BDEs debromination; Penta-BDEs debromination similar as AG, strictly para-removal | octa-BDEs 5 months; penta-BDEs 1 month | Ding et al., 2013 |
| EC195 | Highly enriched 195 containing autotrophic culture | TCE | No | hepta- hexa- penta- tetra- di- | N.A. | Additional one hexa and two penta, as well as tetra and di congeners produced compared with strain 195 | 3 months | He et al., 2006 | |
| ANAS195 | An enrichment culture with strain 195 | TCE | No | hepta- through di- | N.A. | Faster and more extensive debromination compared with strain195 | 3 months | He et al., 2006 | |
| EC195+BAV1 | EC195 with strain BAV1 | TCE | No | tetra- tri- di- | N.A. | BAV1 shows no debromination on deca- or octa-BDEs. More extensive debromination compared with EC195 suggests BAV1 can debrominate lesser brominated PBDEs | 3 months | He et al., 2006 | |
| Microcosms | Plug-flow bioreactor with colonization water from wetland near munition dump | No | N.A. | N.A. | N.A. | Di-BDE 15 to mono-BDE 3 and diphenyl ether | 12% and 61% conversion with HRT 3.4 h and 6.8 h, respectively | Rayne et al., 2003 | |
| Sewage sludge from mesophilic digester in Dubendorf, Switzerland | 4-bromobenzoic acid, 2,6-dibromobiphenyl, tetrabromobisphenyl A, hexabromocyclododecan, and decabromobiphenyl | Nona- Octa- | N.A. | N.A. | Para- and meta-removal preferred | 238 days | Gerecke et al., 2005 | ||
| Loam sediment from Celey Bog Park, West Lafayette, IN, USA | Methanol and dextrose | Nona- through Hexa- | N.A. | Tetra- Tri- | Para-removal preferred | 8 months for penta- and tetra-BDEs, 3.5 years for deca-BDE | Tokarz et al., 2008 | ||
| A few soils and sediments from multiple locations in China, Singapore, and US | No | N.A. | Hexa- Penta- Tetra- | N.A. | Dehalococcoides exist in 11 out of 14 active microcosms; more extensive debromination was observed with TCE as auxiliary substrate | 2 months | Lee and He, 2010 | ||
| TCE | N.A. | Hexa- through di- | N.A. | ||||||
| Sediments at the riverside of Lianjiang Rriver, Guiyu, E-recycling town in China | No | Nona- through tri-BDE | N.A. | N.A. | Too ambitious to correlate Pseudomonas with PBDE debromination simply by DGGE study in original microcosm | 3 months | Qiu et al., 2012 | ||
| A few sediment slurries from mangrove, fresh water ponds, and marine subsurface sediments, Hongkong SAR | No | No | N.A. | Hexa 153 to hexa-, penta-, tetra-, tri-, and di-; tetra 47 to tri- | Para- removal preferred, followed by meta- and ortho-removal | 90 days for tetra 47; 7.6 to 165 days for hexa 153 | Zhu et al., 2014 | ||
| River sediment from Erren River, Taiwan (heavily contaminated rivers) | No | Nono-through mono-BDE | N.A. | N.A. | 6 months | Huang et al., 2014 | |||
| Wastewater sludge samples, Hrade Kralove and Brno | No | A relative distribution of individual congeners changed with a significance increase of tetra 49 | A mixture of mono-through hepta BDEs was spiked. | 15 months | Stiborova et al., 2015 | ||||
| E-waste contaminated soils | Lactate | Deca-209 and tetra 47 decrease while penta-99, hexa-154, 153, and hepta 183 increase at significant levels | PBDEs contamination already exist in soils samples; iron-reducing conditions | ||||||
| Mangrove sediment from Guandu and Bali, Taiwan | No | Nona-through di- | N.A. | N.A. | Para- removal preferred, followed by meta- and ortho-removal; Debrominating populations were tried to be identified through analysis of microbial communities; ZVI can enhance debromination | 75 days | Yang et al., 2017 | ||
| Subsurface sediment from mature mangrove forest in Maipo, Hongkong SAR | No | N.A. | N.A. | Tetra- 47 to tri, di- | Biochar accelerates PBDEs' reductive debromination in electron transfer among microbial populations. Abundance of dehalogenating populations were enriched | 20 weeks | Chen et al., 2018 | ||
Studies evaluating debromination potential with either technical PBDE mixtures or environmentally relevant BDE congeners in microcosms established from soils and sediments collected from various locations and environments have reported differences in both the rate and extent of PBDE debromination after long-term incubation (Table 2). When the primary goal is identification of functional bacteria, rather than developing enrichment cultures for fundamental studies, high-throughput sequencing and quantitative real-time PCR have been used to detect changes in microbial composition and identify specific populations whose increase correlates with debromination. Chen et al used this strategy to identify organohalide respiring Dehalobacter, Dehalococcoides, Dehalogenimonas, and Desulfitobacterium populations in a microcosm during reductive debromination of tetra-BDE 47 (Chen et al., 2018). Though some studies have purported to identify debrominating populations in microbial communities using less-sensitive molecular techniques [e.g., denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP), Qiu et al., 2012; Huang et al., 2014], these results must be viewed skeptically. Organohalide respiring bacteria are typically minor populations, even within enrichment cultures, whose presence can be masked by more dominant non-debrominating populations, thus large changes in detected abundance that appear to be related to debromination often only represent small relative changes in abundance of the population—which is difficult to discern by DGGE or T-RFLP. In such cases, researchers may make erroneous conclusions about the identity of debrominating populations in the microbial community.
PBDEs debromination has been reported by bacteria of at least six different genera, and can be broadly divided as either metabolic process—energy from debromination to support cell growth, or co-metabolic process—not supporting cell growth (Tiehm and Schmidt, 2011). Co-metabolic dehalogenation typically requires supplementation with auxiliary substrates, e.g., other types of organohalides. Several studies of PBDEs debrominating microbial consortia and isolates have reported debromination only in the presence of additional halogenated electron acceptors (He et al., 2006; Robrock et al., 2008; Lee and He, 2010). Carbon sources may also act as auxiliary substrates, as in the case of the lactate, pyruvate or H2-CO2 dependent co-metabolic degradation of PBDEs identified in Acetobacterium sp. strain AG (Ding et al., 2013). The use of benign, non-halogenated auxiliary substrates in co-metabolic debromination makes strain AG a particularly interesting candidate for in-situ bioremediation. Co-metabolic reductive debromination of PBDEs is currently more frequently reported than metabolic debromination. However, metabolic reductive debromination is generally favored for site remediation because it does not require auxiliary substrates and has higher energy-utilization efficiency. Thus far, only Dehalococcoides mccartyi strains GY50 and GY52, both isolated from co-culture GY2 (Lee et al., 2011), have been shown to metabolically debrominate PBDEs (Ding et al., 2017). Strain GY50 is of particular interest as it completely debrominates penta-BDE mixtures to diphenyl ether, rather than producing partially debrominated end-products.
Synergistic interactions in microbial reductive debromination of PBDEs
Synergistic metabolic interactions between organohalide respiring bacteria and other bacterial populations can increase the robustness of dehalogenation in microbial consortia by providing certain growth factors (He et al., 2007; Men et al., 2011). Although the exact nature of most of these synergisms is unknown, faster and extensive debromination in mixed microbial communities has been observed. Co-metabolic debromination in co-culture G consisting of D. mccartyi strain DG and Acetobacterium sp. strain AG, with presence of auxiliary substrate, TCE, is an example of this synergistic metabolic interactions (Ding et al., 2013). Culture G, a co-culture originating from a river bank, was found to reductively debrominate octa- and penta-BDE mixtures to less brominated congeners ranging from penta- to di-BDEs. Debromination of the BDE mixtures by strain DG was slower and also less extensive than that in the parent culture, by producing only trace amounts of penta- and tetra-BDEs after 6 weeks' incubation. Strain AG had the same penta-BDEs debromination capacity as culture G, albeit more slowly and less extensively than its parent culture, and had an octa-BDE debrominating profile similar to that of strain DG. Debromination of octa-BDE could be rescued in co-cultures of strain AG and strain DG, indicating a synergistic relationship between these two populations. The authors speculated that the Acetobacterium provides certain growth factors, such as vitamin B12 in the form of cobalamin, which are essential for PBDEs debromination by strain DG.
Besides supplying essential nutrients during synergistic interactions, another benefit of synergetic microbial interactions can be seen in the more extensive debromination of PBDEs by culture EC195 plus strain BAV1 compared to culture EC195 alone, as direct participants in debromination processes (He et al., 2006). The highly enriched autotrophic culture EC195 produced hepta- through di-BDEs from an octa-BDE mixture and D. mccartyi strain BAV1 did not debrominate octa-BDEs in pure culture. However, addition of strain BAV1 to EC195 resulted in further debromination, with generation of only tetra- through di-BDEs from the octa-BDE mixture. The hexa- and penta-BDE debromination by EC195 plus strain BAV1 suggests a role for BAV1 in synergistic debromination of lesser brominated PBDEs.
However, interactions among populations in mixed microbial communities do not necessarily improve debromination activity. For example, inhibition of methanogenic bacteria in culture GY-T-2 by the addition of BES increased the rate and extent of the debromination of PBDEs (Lee and He, 2010). It is possible that methanogens in GY-T-2 compete with the PBDE debrominating populations in the community for limiting factors, such as hydrogen, thereby inhibiting debromination.
Reductive debromination of PBDEs and functional genes
Anaerobic reductive debromination of PBDEs was first observed in a bioreactor where di-BDE 15 was converted to mono-BDE 3 and diphenyl ether (Rayne et al., 2003). Later investigations of biological transformation of PBDE mixtures and some individual congeners (e.g., tetra-BDE 47, penta-BDE 99) identified and characterized anaerobic microbial reductive debromination of PBDEs in sewage sludge treatments as well as in terrestrial, marine, and estuarial soils and sediments, and in microbial consortia and isolates derived from these sources (Gerecke et al., 2005; He et al., 2006; Robrock et al., 2008; Tokarz et al., 2008; Lee and He, 2010; Lee et al., 2011; Xu et al., 2014; Zhu et al., 2014; Stiborova et al., 2015).
Debromination of PBDEs is partial in almost all observed systems, yielding lesser brominated metabolites, and typically occurs slowly and at nanomolar concentrations. Gerecke et al. reported microbial debromination of 5% of 11.2 nM deca-BDE 209 to nona- and octa-BDEs after 238 days in a microcosm established from sewage sludge (Gerecke et al., 2005). A separate study utilizing a D. mccartyi-containing microbial consortia, ANAS195, observed production of 500 nM hepta- to di-BDEs from 1.3 μM of an octa-BDE mixture with the presence of TCE after 6 months' incubation (He et al., 2006). Similarly, partial debromination of deca-BDE 209 and tetra-BDE 47 was reported after 90 days incubation by the autochthonous microbial community in e-waste contaminated soil containing a range of deca- to tri-BDEs (Song et al., 2015). Though uncommon, complete debromination of tetra- and penta-BDEs to diphenyl ether was demonstrated by Lee et al in a co-culture, GY2 (Lee et al., 2011), from which a novel D. mccartyi strain, GY50, that debrominated ~1,180 nM tetra-BDE 47, penta-BDEs 99 and 100 to diphenyl ether in 2 weeks was isolated (Ding et al., 2017).
There is a marked variation in observed debromination rates, pathways, and relative abundance of daughter products among different studies of anaerobic microbial debromination. This is most likely due to regional differences in nutrient availability and bioavailability of PBDEs as well as site-specific variations in microbial community composition. However, it is generally true that more highly substituted PBDEs (deca- and octa-BDEs) are debrominated by fewer organisms and more slowly than lesser substituted congeners (penta- and tetra-BDEs), which is a common trend in microbial dehalogenation of aromatic organohalides [i.e., PBDEs, polychlorinated biphenyl ethers (PCBs), polychlorinated dibenzo-p-dioxins and polychlorinated dibenzofurans (PCDD/Fs), etc.]. This is thought to be a result of the increased hydrophobicity of more highly halogenated aromatics and of the reduced reactivity of more highly substituted aromatic rings resulting from changes in electron density (Fagervold et al., 2007; Cooper et al., 2015; Zhang et al., 2017, 2018). Instances of this phenomenon can be seen in several studies in which prevalence of octa-BDEs debromination was observed in pure cultures and defined mixed cultures than debromination of deca-BDEs (He et al., 2006; Robrock et al., 2008; Xu et al., 2014). Similarly, studies of the debromination potential of municipal sewage sludge (Shin et al., 2010) and various sediment slurries (Zhu et al., 2014) have noted debromination of hexa- to tetra-BDEs but no debromination of deca- or hepta-BDEs.
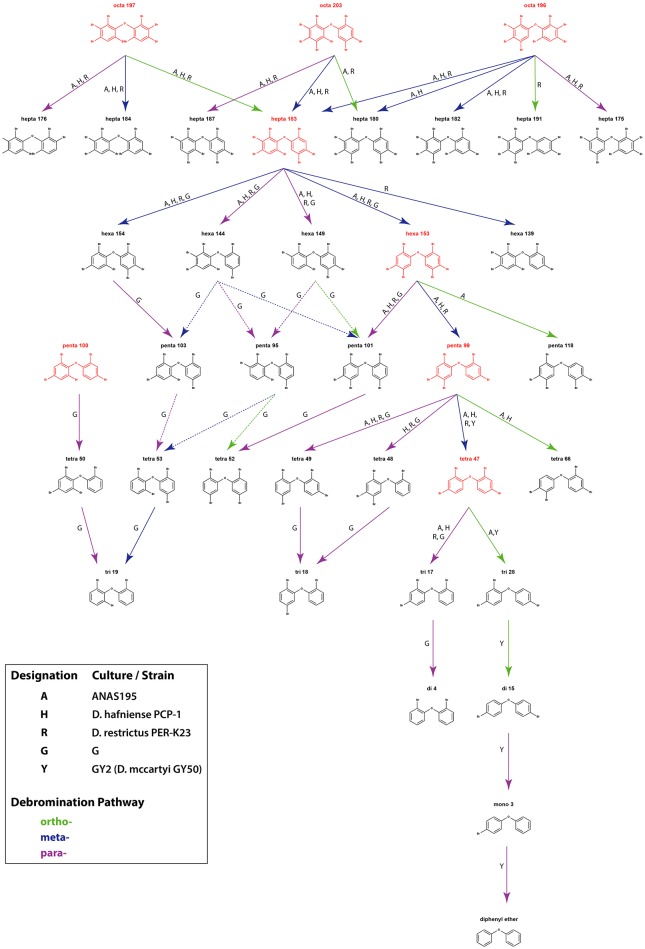
Attempts to identify the products of anaerobic microbial debromination have also revealed preferential removal of para and meta bromine substituents (Figure 3), a trend which is also present in microbial dehalogenation of aromatic organohalides, such as PCBs (Wang and He, 2013; Wang et al., 2014). This preference appears to exist regardless of the degree of bromine saturation, and has been observed in debromination of deca-BDE 209 (Gerecke et al., 2005) as well as in debromination of octa-BDEs (BDE 196, 203, and 197), hepta-BDE 183, hexa-BDE 153, penta-BDE 99, and tetra-BDE 47 (Robrock et al., 2008). The 2008 study by Robrock et al. also suggests preferential removal of double-flanked bromine moieties. Strict para and para-dominant debromination patterns in penta- and octa-BDE mixtures are also found in culture G with the presence of TCE, although meta- and ortho-bromine substitution were suggested as minor pathways in octa-BDE debromination (Ding et al., 2013). The only exception thus far identified not following preferential para and meta substitution is the predominance of ortho-bromine removal in co-culture GY2, as well as in D. mccartyi strain GY50 (derived from culture GY2), in stepwise conversion of penta-BDE 100 to di-BDE 15 via tetra-BDE 47 and tri-BDE 28 (Lee et al., 2011).
Figure 3.
Debromination pathways of PBDEs in mixed and pure microbial cultures.
The preference on para-, meta-, or ortho-bromine substitution could be determined by the reductive dehalogenases present in the debrominating bacteria. Identification and characterization of reductive dehalogenases responsible for dehalogenation of specific compounds allow researchers to investigate the mechanisms of organohalide respiration and provide targets that can be used to monitor populations of organohalide respiring bacteria in laboratory and field-scale bioremediation studies. Reductive dehalogenases responsible for dehalogenation of a wide variety of halogenated compounds have been reviewed in Hug et al. (2013). Identification of PBDEs reductive dehalogenases is impeded by the co-metabolic nature of PBDEs debromination in most mixed cultures and isolates, as the presence of auxiliary substrates makes it more challenging to determine single gene products responsible for observed activities, and by marginal cell yield of PBDEs debrominating populations in mixed cultures.
The only PBDE reductive dehalogenases characterized to date, PbrA1, PbrA2, and PbrA3, were identified in D. mccartyi strain GY50 using a combination of transcriptomics and proteomics (Lee et al., 2011; Ding et al., 2017). The fortuitous emergence of two variant strains that exhibited distinct dehalogenation profiles to strain GY50 allowed for functional characterization of the three PBDE dehalogenases. The deletion of a genomic island containing both PbrA1 and PbrA2 in the genome of strain GY52 and the lack of di-BDE debromination in this strain provided strong evidence for the role of PbrA3 in debromination of penta- and tetra-BDEs to di-BDE (BDE 15), while simultaneously implicating PbrA1 and PbrA2 in removal of unflanked para-bromines from BDE 15 to mono-BDE (BDE 4) and diphenyl ether. Several hundred putative reductive dehalogenase homologous genes have been identified in the genomes of different Dehalococcoides strains and have been categorized into more than 50 orthologous groups based on amino acid similarity (Hug et al., 2013). The three PBDE reductive dehalogenases, PbrA1, PbrA2, and PbrA3, which catalyze different debromination pathways and attack bromine moieties at different positions, are phylogenetically distinct—sharing < 40% amino acid sequence similarity with each other (Figure 4). It is not uncommon for reductive dehalogenase genes with similar functionality to exhibit significant disparities in nucleotide sequence, but the phylogenetic similarity of the PBDE reductive dehalogenases to enzymes which catalyze dehalogenation of other poly-halogenated aromatic compounds may reveal some structural aspect that is common among enzymes mediating catalysis of these types of organohalides. While neither nucleotide nor amino acid sequence similarity among reductive dehalogenases is predictive of substrate range (Hug, 2016), the clustering of known reductive dehalogenase into orthologous groups at least provides a starting point for putatively identifying other PBDE reductive dehalogenases. For example, debromination of PBDEs and other brominated benzenes (Wagner et al., 2012) and phenols (Yang et al., 2015) by strain CBDB1 contains one reductive dehalogenase gene that clusters within the same ortholog group as PbrA3, though no functional genes were implicated in the original study. More information about the functions of uncharacterized dehalogenases is necessary before meaningful comparative analyses of these orthologs can be performed. The emergence and characterization of the GY52 variant facilitated description of the regiospecificity of these three dehalogenases and demonstrated a strategy by which other PBDE dehalogenases can be investigated.
Figure 4.
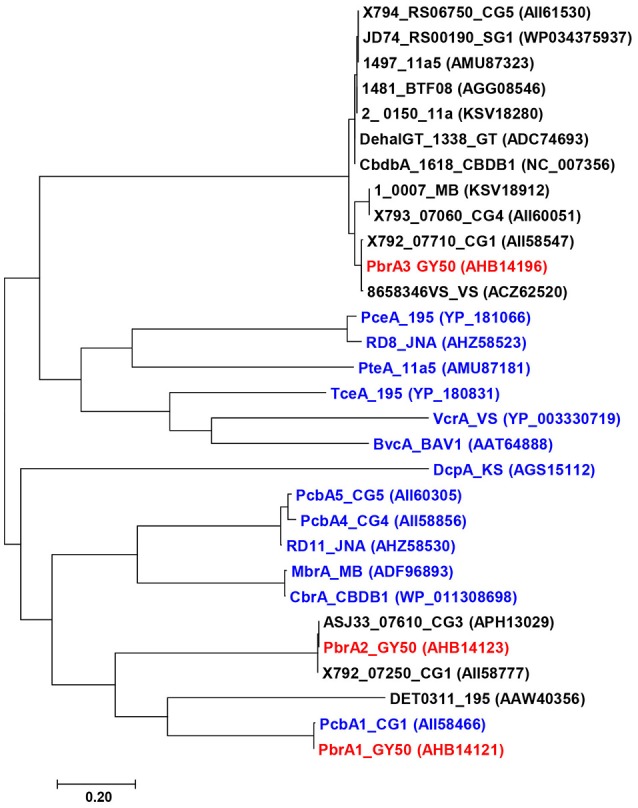
Phylogenetic analysis of functionally characterized RDases (blue color) in Dehalococcoides mccartyi including orthologs (black color) of PBDE reductive dehalogenases (red color). The tree was constructed with MEGA 7 (Kumar et al., 2016) using maximum likelihood method in JTT matrix-based model (Jones et al., 1992).
Outlook
Though production and usage of PBDE mixtures have declined after implementation of bans and voluntarily cessation of manufacture, the environmental persistence and potential for transboundary disbursement make PBDEs a continuing threat to biological populations around the globe. The degree of the debromination of PBDEs not only affects physical and chemical properties, but also toxicity and potential for bioaccumulation. Highly substituted deca-, nona-, and octa-BDEs are thermally labile and are partially degraded to lesser brominated congeners in the environment resulting in an increase in the risk presented by the original contamination. Because PBDEs preferentially sorb to organic matter, they tend to accumulate in anaerobic and anoxic soils and sediments. Harnessing the metabolic potential of anaerobic microbes that can detoxify PBDEs by removing bromine substituents has the potential to be a cost effective and efficient approach to remediate PBDEs in the environment.
Microbial debromination of PBDEs must overcome several obstacles before it can be considered a viable technology for bioremediation. Most of the bacteria that are currently known to debrominate PBDEs do so co-metabolically and partially. This incomplete debromination may often cause additional problems in-situ and the requirement for auxiliary substrates can severely limit the rate and extent of debromination. The only anaerobic microbe that can completely detoxify PBDEs and couple cell growth is D. mccartyi GY50 which can metabolize penta- and tetra-BDEs to produce diphenyl ether as an end-product. However, since deca- and octa-BDE mixtures also represent the majority of PBDEs production and pollution globally, further study is necessary to find other bacterial isolates and mixed cultures that can metabolize these highly brominated congeners. In general, co-cultures could likely be a promising solution to completely debrominate higher brominated BDEs to diphenyl ether via intermediates such as penta- and tetra-BDEs.
The slow growth rate and low cell yields associated with debrominating bacteria have impeded efforts to elucidate the mechanisms of microbial PBDE degradation. The isolation of D. mccartyi GY50 has revealed functional PBDE reductive dehalogenases for the first time, which may facilitate identification of additional PBDE dehalogenases in other Dehalococcoides. Discovery of enzymes responsible for the observed debromination of higher brominated deca- and octa-BDEs would be particularly valuable to the advancement of PBDEs bioremediation efforts. Recent advances in heterologous expression and purification of functional reductive dehalogenases will facilitate investigations into the dehalogenation potential of uncharacterized dehalogenases and may make it possible to establish a platform for in-vitro production of specific dehalogenases for bioremediation.
In summary, the innate capacity of some anaerobic microbes to detoxify different PBDEs can potentially be exploited as a tool to remediate contaminated soils and sediments. Application of these microbes in-situ has been hindered by the slow rate of cell growth and associated debromination of target compounds. Combining organohalide respiring bacteria with other physical and chemical processes to increase the rate and extent of anaerobic debromination of PBDEs has been investigated with varying degrees of success. Recent descriptions of PBDE debrominating isolates and defined microbial consortia have shed light on genes responsible for some, but not all, of the natural attenuation of PBDEs that has been observed. Future investigations to elucidate and characterize additional PBDEs dehalogenases in anaerobic systems may provide a clearer picture of the mechanisms responsible for the partial degradation of highly substituted PBDEs in the environment and pave the way for development of new strategies to address the persistent threat that PBDEs pose to biological populations.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author contributions
SZ, MR, and JH developed the structure of the article. SZ and MR drafted the article. SZ, MR, CD, and JH revised the article. JH did the final approval of the version to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for financial support by the Ng Teng Fong Charitable Foundation fund under project No.: R-302-000-198-720.
References
- Alabi O. A., Bakare A. A., Xu X., Li B., Zhang Y., Huo X. (2012). Comparative evaluation of environmental contamination and DNA damage induced by electronic-waste in Nigeria and China. Sci. Total Environ. 423, 62–72. 10.1016/j.scitotenv.2012.01.056 [DOI] [PubMed] [Google Scholar]
- ATSDR (2017). Toxicological Profile for Polybrominated Diphenyl Ethers (PBDEs), Public Health Service, Department of Health and Human Services. Atlanta, GA: Agency for Toxic Substances and Disease Registry. [PubMed] [Google Scholar]
- Boon J. P., Lewis W. E., Tjoen-A-Choy M. R., Allchin C. R., Law R. J., De Boer J., et al. (2002). Levels of polybrominated diphenyl ether (PBDE) flame retardants in animals representing different trophic levels of the North Sea food Web. Environ. Sci. Technol. 36, 4025–4032. 10.1021/es0158298 [DOI] [PubMed] [Google Scholar]
- Chen J., Wang C., Pan Y., Farzana S. S., Tam N. F. (2018). Biochar accelerates microbial reductive debromination of 2,2',4,4'-tetrabromodiphenyl ether (BDE-47) in anaerobic mangrove sediments. J. Hazard. Mater. 341, 177–186. 10.1016/j.jhazmat.2017.07.063 [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao X., Linag S., Yu M., Liu W., et al. (2016). The research on the solubility of decabromodiphenyl ether based on recycling of waste electronic plastic by solvent process. Proc. Environ. Sci. 31, 827–831. 10.1016/j.proenv.2016.02.083 [DOI] [Google Scholar]
- Cooper M., Wagner A., Wondrousch D., Sonntag F., Sonnabend A., Brehm M., et al. (2015). Anaerobic microbial transformation of halogenated aromatics and fate prediction using electron density modeling. Environ. Sci. Technol. 49, 6018–6028. 10.1021/acs.est.5b00303 [DOI] [PubMed] [Google Scholar]
- Darnerud P. O., Eriksen G. S., Jóhannesson T., Larsen P. B., Viluksela M. (2001). Polybrominated diphenyl ethers: occurrence, dietary exposure, and toxicology. Environ. Health Perspect. 109, 49–68. 10.1289/ehp.01109s149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J., Wester P. G., Klamer H. J., Lewis W. E., Boon J. P. (1998). Do flame retardants threaten ocean life? Nature 394, 28–29. [DOI] [PubMed] [Google Scholar]
- Deng C., Chen Y., Li J., Li Y., Li H. (2016). Environmental pollution of polybrominated diphenyl ethers from industrial plants in China: a preliminary investigation. Environ. Sci. Pollut. Res. 23, 7012–7021. 10.1007/s11356-015-5902-8 [DOI] [PubMed] [Google Scholar]
- Ding C., Chow W. L., He J. (2013). Isolation of Acetobacterium sp. strain AG, which reductively debrominates octa- and pentabrominated diphenyl ether technical mixtures. Appl. Environ. Microbiol. 79, 1110–1117. 10.1128/AEM.02919-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Rogers M. J., Yang K. L., He J. (2017). Loss of the ssrA genome island led to partial debromination in the PBDE respiring Dehalococcoides mccartyi strain GY50. Environ. Microbiol. 19, 2906–2915. 10.1111/1462-2920.13817 [DOI] [PubMed] [Google Scholar]
- Fagervold S. K., May H. D., Sowers K. R. (2007). Microbial reductive dechlorination of aroclor 1260 in baltimore harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl. Environ. Microbiol. 73, 3009–3018. 10.1128/AEM.02958-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Huang J., Yu G., Wang L. (2008). Photochemical degradation of six polybrominated diphenyl ether congeners under ultraviolet irradiation in hexane. Chemosphere 71, 258–267. 10.1016/j.chemosphere.2007.09.041 [DOI] [PubMed] [Google Scholar]
- Gallego E., Grimalt J. O., Bartrons M., Lopez J. F., Camarero L., Catalan J., et al. (2007). Altitudinal gradients of PBDEs and PCBs in fish from European high mountain lakes. Environ. Sci. Technol. 41, 2196–2202. 10.1021/es062197m [DOI] [PubMed] [Google Scholar]
- Gerecke A. C., Hartmann P. C., Heeb N. V., Kohler H. P., Giger W., Schmid P., et al. (2005). Anaerobic degradation of decabromodiphenyl ether. Environ. Sci. Technol. 39, 1078–1083. 10.1021/es048634j [DOI] [PubMed] [Google Scholar]
- He J., Holmes V. F., Lee P. K., Alvarez-Cohen L. (2007). Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73, 2847–2853. 10.1128/AEM.02574-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Robrock K. R., Alvarez-Cohen L. (2006). Microbial reductive debromination of polybrominated diphenyl ethers (PBDEs). Environ. Sci. Technol. 40, 4429–4434. 10.1021/es052508d [DOI] [PubMed] [Google Scholar]
- Huang H.-W., Chang B.-V., Lee C.-C. (2014). Reductive debromination of decabromodiphenyl ether by anaerobic microbes from river sediment. Int. Biodeterior. Biodegrad. 87, 60–65. 10.1016/j.ibiod.2013.10.011 [DOI] [Google Scholar]
- Hug L. A. (2016). Diversity, evolution, and environmental distribution of reductive dehalogenase genes, in Organohalide-Respiring Bacteria, eds Adrian L., Löffler F. E. (Berlin; Heidelberg: Springer Berlin Heidelberg; ), 377–393. [Google Scholar]
- Hug L. A., Maphosa F., Leys D., Loffler F. E., Smidt H., Edwards E. A., et al. (2013). Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. B Biol. Sci. 368:20120322 10.1098/rstb.2012.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunska I., Harrad S., Santillo D., Johnston P., Brigden K. (2013). Levels and distribution of polybrominated diphenyl ethers in soil, sediment and dust samples collected from various electronic waste recycling sites within Guiyu town, southern China. Environ. Sci. Process Impacts 15, 503–511. 10.1039/c2em30785e [DOI] [PubMed] [Google Scholar]
- La Guardia M. J., Hale R. C., Harvey E. (2006). Detailed polybrominated diphenyl ether (PBDE) congener composition of the widely used penta-, octa-, and deca-PBDE technical flame-retardant mixtures. Environ. Sci. Technol. 40, 6247–6254. 10.1021/es060630m [DOI] [PubMed] [Google Scholar]
- Lee L. K., Ding C., Yang K.-L., He J. (2011). Complete debromination of tetra- and penta-brominated diphenyl ethers by a coculture consisting of Dehalococcoides and Desulfovibrio species. Environ. Sci. Technol. 45, 8475–8482. 10.1021/es201559g [DOI] [PubMed] [Google Scholar]
- Lee L. K., He J. (2010). Reductive debromination of polybrominated diphenyl ethers by anaerobic bacteria from soils and sediments. Appl. Environ. Microbiol. 76, 794–802. 10.1128/AEM.01872-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Niu S., Hai R., Li M. (2015). Concentrations and distribution of polybrominated diphenyl ethers (PBDEs) in soils and plants from a deca-BDE manufacturing factory in China. Environ. Sci. Pollut. Res. Int. 22, 1133–1143. 10.1007/s11356-014-3214-z [DOI] [PubMed] [Google Scholar]
- Lindström G., Wingfors H., Dam M., Bavel B. V. (1999). Identification of 19 polybrominated diphenyl ethers (PBDEs) in long-finned pilot whale (Globicephala melas) from the Atlantic. Arch. Environ. Contam. Toxicol. 36, 355–363. 10.1007/s002449900482 [DOI] [PubMed] [Google Scholar]
- McGrath T. J., Ball A. S., Clarke B. O. (2017). Critical review of soil contamination by polybrominated diphenyl ethers (PBDEs) and novel brominated flame retardants (NBFRs); concentrations, sources and congener profiles. Environ. Pollut. 230, 741–757. 10.1016/j.envpol.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Men Y., Feil H., VerBerkmoes N. C., Shah M. B., Johnson D. R., Lee P. K. H., et al. (2011). Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6, 410–421. 10.1038/ismej.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norstrom R. J., Simon M., Moisey J., Wakeford B., Weseloh D. V. (2002). Geographical distribution (2000) and temporal trends (1981-2000) of brominated diphenyl ethers in Great Lakes hewing gull eggs. Environ. Sci. Technol. 36, 4783–4789. 10.1021/es025831e [DOI] [PubMed] [Google Scholar]
- O'Driscoll K., Robinson J., Chiang W.-S., Chen Y.-Y., Kao R.-C., Doherty R. (2016). The environmental fate of polybrominated diphenyl ethers (PBDEs) in western Taiwan and coastal waters: evaluation with a fugacity-based model. Environ. Sci. Pollut. Res. 23, 13222–13234. 10.1007/s11356-016-6428-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm A., Cousins I. T., Mackay D., Tysklind M., Metcalfe C., Alaee M. (2002). Assessing the environmental fate of chemicals of emerging concern: a case study of the polybrominated diphenyl ethers. Environ. Pollut. 117, 195–213. 10.1016/S0269-7491(01)00276-7 [DOI] [PubMed] [Google Scholar]
- Qiu M., Chen X., Deng D., Guo J., Sun G., Mai B., et al. (2012). Effects of electron donors on anaerobic microbial debromination of polybrominated diphenyl ethers (PBDEs). Biodegradation 23, 351–361. 10.1007/s10532-011-9514-9 [DOI] [PubMed] [Google Scholar]
- Rayne S., Ikonomou M. G., Whale M. D. (2003). Anaerobic microbial and photochemical degradation of 4,4'-dibromodiphenyl ether. Water Res. 37, 551–560. 10.1016/S0043-1354(02)00311-1 [DOI] [PubMed] [Google Scholar]
- Rayne S., Wan P., Ikonomou M. (2006). Photochemistry of a major commercial polybrominated diphenyl ether flame retardant congener: 2,2′, 4,4′, 5,5′-hexabromodiphenyl ether (BDE153). Environ. Int. 32, 575–585. 10.1016/j.envint.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Robrock K. R., Korytar P., Alvarez-Cohen L. (2008). Pathways for the anaerobic microbial debromination of polybrominated diphenyl ethers. Environ. Sci. Technol. 42, 2845–2852. 10.1021/es0720917 [DOI] [PubMed] [Google Scholar]
- Schenker U., Soltermann F., Scheringer M., Hungerbühler K. (2008). Modeling the environmental fate of polybrominated diphenyl ethers (PBDEs): the importance of photolysis for the formation of lighter PBDEs. Environ. Sci. Technol. 42, 9244–9249. 10.1021/es801042n [DOI] [PubMed] [Google Scholar]
- Shin M., Duncan B., Seto P., Falletta P., Lee D.-Y. (2010). Dynamics of selected pre-existing polybrominated diphenylethers (PBDEs) in municipal wastewater sludge under anaerobic conditions. Chemosphere 78, 1220–1224. 10.1016/j.chemosphere.2009.12.057 [DOI] [PubMed] [Google Scholar]
- Siddiqi M. A., Laessig R. H., Reed K. D. (2003). Polybrominated diphenyl ethers (PBDEs): new pollutants–old diseases. Clin. Med. Res. 1, 281–290. 10.3121/cmr.1.4.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. K., Luo C. L., Li F. B., Jiang L. F., Wang Y., Zhang D. Y., et al. (2015). Anaerobic degradation of polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs), and microbial community dynamics of electronic waste-contaminated soil. Sci. Total Environ. 502, 426–433. 10.1016/j.scitotenv.2014.09.045 [DOI] [PubMed] [Google Scholar]
- Stapleton H. M., Dodder N. G. (2008). Photodegradation of decabromodiphenyl ether in house dust by natural sunlight. Environ. Toxicol. Chem. 27, 306–312. 10.1897/07-301R.1 [DOI] [PubMed] [Google Scholar]
- Stiborova H., Vrkoslavova J., Pulkrabova J., Poustka J., Hajslova J., Demnerova K. (2015). Dynamics of brominated flame retardants removal in contaminated wastewater sewage sludge under anaerobic conditions. Sci. Total Environ. 533, 439–445. 10.1016/j.scitotenv.2015.06.131 [DOI] [PubMed] [Google Scholar]
- Tiehm A., Schmidt K. R. (2011). Sequential anaerobic/aerobic biodegradation of chloroethenes—aspects of field application. Curr. Opin. Biotechnol. 22, 415–421. 10.1016/j.copbio.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Tokarz J. A., Ahn M. Y., Leng J., Filley T. R., Nies L. (2008). Reductive debromination of polybrominated diphenyl ethers in anaerobic sediment and a biomimetic system. Environ. Sci. Technol. 42, 1157–1164. 10.1021/es071989t [DOI] [PubMed] [Google Scholar]
- United Nations Environment Programme (2009). Stockholm Convention on Persistent Organic Pollutans (POPs). Geneva, Switzerland. [Google Scholar]
- Wagner A., Cooper M., Ferdi S., Seifert J., Adrian L. (2012). Growth of Dehalococcoides mccartyi strain CBDB1 by reductive dehalogenation of brominated benzenes to benzene. Environ. Sci. Technol. 46, 8960–8968. 10.1021/es3003519 [DOI] [PubMed] [Google Scholar]
- Wang S., Chng K. R., Wilm A., Zhao S., Yang K.-L., Nagarajan N., et al. (2014). Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc. Natl. Acad. Sci. U.S.A. 111, 12103–12108. 10.1073/pnas.1404845111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., He J. (2013). Phylogenetically distinct bacteria involve extensive dechlorination of Aroclor 1260 in sediment-free cultures. PLoS ONE 8:e59178. 10.1371/journal.pone.0059178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild S., McLagan D., Schlabach M., Bossi R., Hawker D., Cropp R., et al. (2015). An Antarctic research station as a source of brominated and perfluorinated persistent organic pollutants to the local environment. Environ. Sci. Technol. 49, 103–112. 10.1021/es5048232 [DOI] [PubMed] [Google Scholar]
- Windham G. C., Pinney S. M., Voss R. W., Sjödin A., Biro F. M., Greenspan L. C., et al. (2015). Brominated flame retardants and other persistent organohalogenated compounds in relation to timing of puberty in a longitudinal study of girls. Environ. Health Perspect. 123, 1046–1052. 10.1289/ehp.1408778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wang J., Lu M. (2014). Complete debromination of decabromodiphenyl ether using the integration of Dehalococcoides sp. strain CBDB1 and zero-valent iron. Chemosphere 117, 455–461. 10.1016/j.chemosphere.2014.07.077 [DOI] [PubMed] [Google Scholar]
- Yang C. (2017). Anaerobic Transformation of Brominated Aromatic Compounds by Dehalococcoides mccartyi Strain CBDB1. Doctoral Thesis.
- Yang C., Kublik A., Weidauer C., Seiwert B., Adrian L. (2015). Reductive dehalogenation of oligocyclic phenolic bromoaromatics by Dehalococcoides mccartyi strain CBDB1. Environ. Sci. Technol. 49, 8497–8505. 10.1021/acs.est.5b01401 [DOI] [PubMed] [Google Scholar]
- Yang C. W., Huang H. W., Chang B. V. (2017). Microbial communities associated with anaerobic degradation of polybrominated diphenyl ethers in river sediment. J. Microbiol. Immunol. Infect. 50, 32–39. 10.1016/j.jmii.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Zhang S., Adrian L., Schuurmann G. (2018). Interaction mode and regioselectivity in vitamin B12-dependent dehalogenation of aryl halides by Dehalococcoides mccartyi strain CBDB1. Environ. Sci. Technol. 52, 1834–1843. 10.1021/acs.est.7b04278 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wondrousch D., Cooper M., Zinder S. H., Schuurmann G., Adrian L. (2017). Anaerobic dehalogenation of chloroanilines by Dehalococcoides mccartyi strain CBDB1 and Dehalobacter Strain 14DCB1 via different pathways as related to molecular electronic structure. Environ. Sci. Technol. 51, 3714–3724. 10.1021/acs.est.6b05730 [DOI] [PubMed] [Google Scholar]
- Zhu H. W., Wang Y., Wang X. W., Luan T. G., Tam N. F. Y. (2014). Intrinsic debromination potential of polybrominated diphenyl ethers in different sediment slurries. Environ. Sci. Technol. 48, 4724–4731. 10.1021/es4053818 [DOI] [PubMed] [Google Scholar]
- Zhu L. Y., Hites R. A. (2004). Temporal trends and spatial distributions of brominated flame retardants in archived fishes from the Great Lakes. Environ. Sci. Technol. 38, 2779–2784. 10.1021/es035288h [DOI] [PubMed] [Google Scholar]