Abstract
The application of viral and cellular metagenomics to natural environments has expanded our understanding of the structure, functioning, and diversity of microbial and viral communities. The high diversity of many communities, e.g., soils, surface ocean waters, and animal-associated microbiomes, make it difficult to establish virus-host associations at the single cell (rather than population) level, assign cellular hosts, or determine the extent of viral host range from metagenomics studies alone. Here, we combine single-cell sequencing with environmental metagenomics to characterize the structure of virus–host associations in a Yellowstone National Park (YNP) hot spring microbial community. Leveraging the relatively low diversity of the YNP environment, we are able to overlay evidence at the single-cell level with contextualized viral and cellular community structure. Combining evidence from hexanucelotide analysis, single cell read mapping, network-based analytics, and CRISPR-based inference, we conservatively estimate that >60% of cells contain at least one virus type and a majority of these cells contain two or more virus types. Of the detected virus types, nearly 50% were found in more than 2 cellular clades, indicative of a broad host range. The new lens provided by the combination of metaviromics and single-cell genomics reveals a network of virus–host interactions in extreme environments, provides evidence that extensive virus–host associations are common, and further expands the unseen impact of viruses on cellular life.
Introduction
For most natural environments, we lack a comprehensive inventory of both viruses, their microbial hosts and the virus–host networks they form [1, 2]. A comprehensive understanding is necessary because viruses likely play a central role in controlling microbial community structure and function [3–6]. Culture-based assays have revealed complex networks of infection between bacteriophage and bacterial hosts where a single bacteriophage may be able to infect multiple bacterial species, and each bacterial species may be a host for multiple different phage types [7–10]. Comparative genomics of bacterial and archaeal strains also identified the presence of many different proviral elements [11–13]. However, culture-based infection assays and host range determination are limited in scope by the small number of microbial species and their viruses that can presently be cultured.
In recent years, several culture-independent methods have been developed to investigate host–virus associations (reviewed by [14]). These include analysis by metaviromics [15, 16], CRISPR spacer sequences [17–19], phageFISH [20], viral tagging [21, 22], microfluidic digital PCR [23], and single-cell genomics (SCG) [24–27]. Of these methods, SCG has provided some of the most detailed in situ insights into virus–host associations. For example, analysis of 58 single-cell amplified genomes (SAGs) from marine surface bacterioplankton showed that 20 of the SAGs contained viral sequences, some of which were shown to be actively replicating [28]. As a second example, analysis of 127 uncultivated SUP05 bacterial SAGs from an oxygen minimum zone revealed that ~1/3 were infected and that viruses reshaped core cellular metabolism [25]. Yet, few studies combine methods to provide a comprehensive inventory of virus–host associations for the entire microbial community.
Microbial communities in high temperature (>80 °C) acidic hot springs (pH < 4) are typically composed of only a limited number of bacterial and archaeal cell types and their viruses, in contrast to natural environments with higher resident diversity, like the surface oceans or the human gut. We have previously used community metagenomics to establish that the Yellowstone Nymph Lake 01 (NL01) hot spring microbial community is comprised of only 110 dominant viral types and 8 archaeal cellular species [16], and that this microbial community is relatively stable over a period of years [29]. The majority of virus and cellular types identified in NL01 remain uncultured nor has it been possible to characterize virus–host interactions from the metagenomic sequence data alone. Here, we combine SCG, CRISPR, and hexanucelotide analysis with community and viral metagenomics analysis to detect rampant and broad virus-host associations within the NL01 microbial community.
Materials and methods
Sample site
Water samples (1 mL) were collected from the Nymph Lake 01 (NL01) hot spring in Yellowstone National Park (YNP, Supplemental Fig. 1). At the time of sampling, the hot spring conditions were 83.3 °C, pH 2.45, and 1.085 mS conductivity. Samples were preserved on site with 5% glycerol and immediately flash frozen in a dry ice–ethanol bath. Samples were provided to the Bigelow Single Cell Genomics Center (East Boothbay, ME).
Single cell genome sequencing
Flow cytometric separation of individual cells and whole genome amplification were performed at the Bigelow Single Cell Genomics Center using previously described methods [30, 31]. Based on effective MDA amplification of genetic material, a 384-well plate was selected for low coverage shotgun sequencing with an Ilumina end-paired HiSeq. The obtained reads were trimmed with trimmomatic v0.32 [32], normalized with kmernorm 1.05 (https://sourceforge.net/projects/kmernorm/), and assembled with SPAdes version 3.0.0 [33]. All contigs over 2.2 kb were used to estimate genome size and completeness using CheckM [34].
Cellular classification
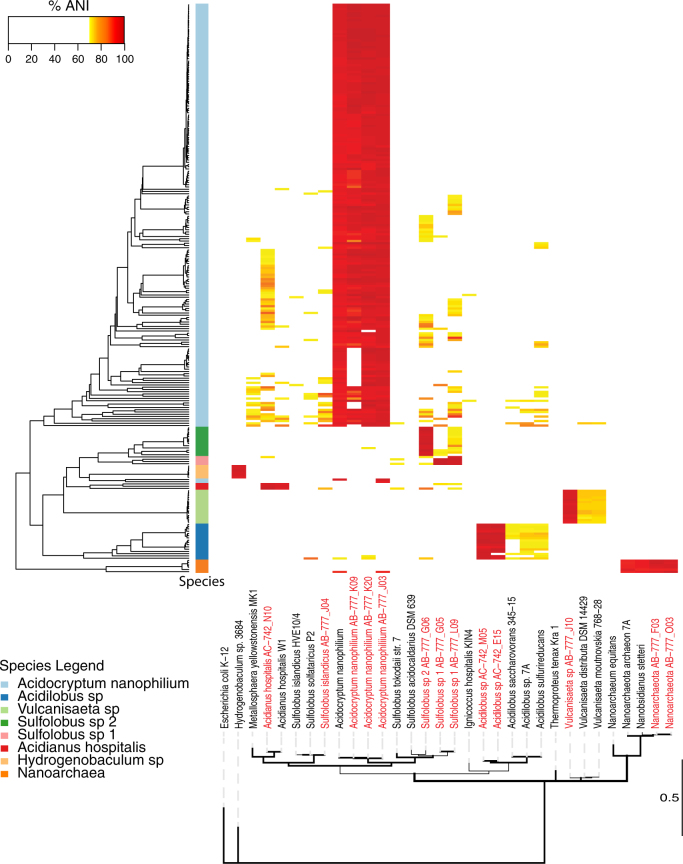
Cells were classified using a script (https://github.com/chjp/ANI) that measures sequence similarities between the contigs and reference genomes in terms of the average nucleotide identity (ANI). All cells were compared against the reference panel consisting of previously sequenced single-cell genomes from the same hot spring [29] as well as 18 thermophile reference genomes (Supplemental Table 1). ANI scores were combined with the percent of SAG base pairs to generate an ANI bar code for every SAG against the 32 reference genomes (https://github.com/speng32/SAG_hot_spring_YNP). All ANI matches covering <5% of the SAG genome were discarded. SAGs with two or more species present at ≥91% ANI were examined for the presence of double cells. SAGs with less than 1/3 reference genomes shared were considered as double cells. Twelve SAGs showed evidence of having two cells present. Eight of these SAGs were classified as double cells and the remaining 4 were unclassified and removed from further analysis. SAGs with only a single species present at ≥95% ANI using at least 30% of the SAG genome were classified as belonging to the same species as the reference genome(s). SAGs that failed to meet the above categories (≥95% ANI and ≥30% coverage) were classified as likely single cells (≥95% ANI and <30% coverage) (14 SAGs) or unclassifiable (28 SAGs) and removed from further analysis. ANI results were clustered hierarchically and a heatmap of ANI (Fig. 1) and bp coverage (Supplemental Fig. 2) was generated for every classified SAG against every reference genome. 16S rRNA sequences were identified in 8 SAGs and compared to the reference genomes as a means to evaluate the accuracy of ANI-based taxonomic identification.
Fig. 1.
Cellular classification of SAGs. Heatmap of the average nucleotide identity (ANI) of 253 classified single cell SAGs sequenced in this study compared against 32 reference genomes including 13 SAGs previously sequenced at high coverage from the same hot spring [29] (red text). SAGs were hierarchically clustered using complete linkage (left hierarchical dendogram). The column directly to the right of the hierarchical dendogram indicates classified cell species (color key provided) for all SAGs classified as single cells. Partial length 16S rRNA sequences from the 32 reference genomes were used to construct a maximum likelihood phylogenetic tree and nodes with greater than 95% posterior probability are bolded. The E. coli strain served as the outgroup. The scale bar is in substitutions per site
Hexamer frequency analysis
The contigs from SAGs classified as the same species were grouped together for hexamer frequency analysis. The hexamer frequency distribution of the grouped SAGs as well as a dataset of the viral types present in the NL01 hot spring [16] were generated using VirusHostMatcher [35]. The virus–host pair with the lowest hexamer distance was calculated by d2* [35] and pairs with a distance value <0.3 were used as an indication of a potential virus–host pair.
Viral sequence identification
All sequence reads obtained from SAG sequencing were used as the query of a BLASTn search against the viral database previously described [16]. Reads with a significant match (e-value <1.0−10) to the viral database were filtered and classified as having a viral origin if they matched at >95% nucleotide identity over 100 bp. Identified viral reads were subsequently mapped back to their viral group previously established using network analytics [16] using a custom script. Reads that mapped to multiple viral groups were assigned to the viral group with the most reads from that individual SAG to reduce false positives. To test if this mapping protocol resulted in false identification of viruses, controls were performed where the same SAG reads were mapped to the contigs from the Tara Oceans Virome (TOV) datasets (18SUR 66 Mbp and 18DCM 99 Mbp) [15] and a virome from the human gut (6 Mbp) [36] all of which were not expected to contain viruses found in hot spring environments. Additionally, sequence reads from 25 publically available SAGs generated from non-hot spring environments from the JGI IMG (http://jgi.doe.gov/) representing ten bacterial and two archaeal phyla (703.7 million total reads) were compared against the viral database at the same stringency described above.
We used the following rationale to establish a threshold criteria for identifying virus–host associations within an individual SAG dataset. Since the estimated genome completeness for each SAG varied, we first determined the ratio of identified unique viral sequence reads (average of 150 bp in length) to the total unique host base pairs for each SAG. The number of unique viral base pairs was determined by mapping SAG reads to the NL01 viral dataset using BLASTn and removing any overlap to the reference viral genomes. The unique number of host base pairs was calculated using the ANI-based composition statistic [37, 38] for each SAG with respect to the 32 reference genomes, minus the unique viral base pairs. These ratios were compared to expected ratios using an average viral genome size of 30 kb, a host genome size from 1.5–3.0 MB, and assuming no sequencing bias towards either virus or host or a 2× bias towards virus or host (arbitrarily chosen to account for variation in amplification). Using this rationale, we determined that a minimum of 2–5 unique 150 bp viral sequence reads should be present in an individual SAG dataset if that SAG were in fact associated with a virus whose genome was present at the same copy number as the cellular genome.
After determining the profile of viral content in each individual SAG, the dataset was treated as a bipartite network. The BiMat algorithm [39] was applied to the bipartite viral–host network for modularity analysis. The binary network was generated using a minimum cutoff of 2 or 5 unique viral sequence reads from a SAG to the 110 viral groups previously identified in the NL01 hot spring [16].
CRISPR spacer sequence identification
CRISPR spacer sequences were identified in SAG contigs using Piler-CR [40]. Identified CRISPR spacer sequences were extracted and compared against the viral database with virus–host associations assigned to CRISPR spacer sequences that match ≥90% identity over the entire spacer length. Contigs with CRISPR matches were selected and the viral group they belonged to was identified using a custom python script. As controls for the false identification of CRISPR spacer–virus associations, a CRISPR spacer dataset of 966 unique spacers from a human gut microbial community was analyzed against the NL01 viral database. In addition, the SAG CRISPR spacer sequences were compared to the viral dataset of the human gut bacterial community [36] under the same conditions described above.
Statistical test for contamination
To identify the possibility of sample contamination within adjacent wells on the 384-well plates during sample preparation, a statistical approach was used to evaluate the correlation between the physical distance and the sequence similarity between adjacent wells. First, the physical distance between two neighboring wells from the same row or the same column as a unit was defined. A distance matrix with all pairwise distances was computed based on the Euclidean distance between any two wells. Second, the sequence similarity between two wells was calculated based on the number of unique and shared viral groups of the two wells. The Jaccard index of a given pair of wells A and B was calculated as , where SA denotes the set of viral groups in SAG A and SB denotes the set of detected viral groups in SAG B. Third, the Spearman’s rank correlation was calculated to evaluate the relationship between physical distances of the wells and the Jaccard index. A series of distance cutoffs between 1.5 and 3 were used to calculate the Spearman’s correlation of two wells to focus on the cross contamination in nearby wells. Finally, to evaluate the statistical significance of the observed Spearman’s correlation coefficients at different distance cutoffs, a permutation test was performed to obtain the null distribution of the Spearman’s coefficients. For the permutation test, the plate layout was randomly shuffled 100 times and the Spearman’s correlation coefficients were re-calculated at corresponding distance cutoffs. The observed Spearman’s correlation coefficients were then compared with the null distributions.
Results and discussion
In this study, we combined single-cell genomics and community metagenomics to characterize virus–host interactions. Single cells were randomly isolated directly from hot spring samples, their genomes amplified and sequenced. 109,930,697 total paired end reads were produced from 307 single amplified genomes (SAGs, average ~358,000 reads per cell) with a maximum of 2,015,593 and a minimum of 3823 reads per SAG (Supplemental Table 2). A total of 34.1 Mbp was assembled ranging from a minimum total bp of 7806 to a maximum of 380,184 with an average total assembled length of 110,997 bp per cell. This correlates to an average genome completeness of approximately 9% but ranges from <1 to 44% complete based on CheckM analysis.
In order to determine the cellular identity of each SAG a multistep process was developed (Supplemental Fig. 3). First, the average nucleotide identity (ANI) [37, 38] for all contigs greater than 2 kb for each SAG was calculated with respect to 32 reference genomes. The reference genomes consisted of a combination of SAGs previously sequenced at high depth (17–90% genome completeness) from the same hot spring and other complete or near complete thermophilic archaeal and bacterial reference genomes from the NCBI database (WGS release 212, February, 2016). Second, the percentage of sequence homology between a SAG and the reference genomes were determined. SAGs were hierarchically clustered and assigned to their closest cellular species based on ANI score in combination with the percentage of sequence homology between the SAG and its closest reference genome (Fig. 1, Supplemental Table 3). We utilized an ANI score of 95% in combination with 30% sequence coverage to classify the majority of SAGs (253/307 SAGs). The 54 SAGs that were not classified were either double cells of the symbiont Nanoarchaea with its Acidocryptum host (8 examples), or 46 SAG cells that failed to meet our classification criteria. These 54 SAGs were removed from further analysis. To further support cellular identification, all SAGs were examined for 16S rRNA gene sequences. 16S rRNA sequences were present in only 8 SAGs and cellular classification based on their 16S rRNA was determined by alignment to reference genomes. In all 8 cases, the 16S rRNA gene and ANI classifications produced the same result.
The classification of SAGs revealed a low-diversity microbial community consisting of 8 cellular clades, dominated by Archaea (Fig. 1), consistent with our previous studies [29]. The 253 SAGs classified to one of 8 cellular clades. Of these, 247 were classified as one of 7 clades of Archaea (97.6%), 6 were classified as members of a single clade of Bacteria (2.4%), and none were classified as Eukaryotic. The vast majority (98%) of the Archaeal cells are members of the Crenarchaeota (241/247 SAGs) while Nanoarchaeota (6) make up the remaining 2.0%. The only bacterial species detected belonged to the Aquificales. The NL01 microbial community structure was nearly identical to the community structure determined by 16S rRNA amplicon sequencing from a sample taken 12 months previously. Overall, 6 of the 8 clades identified in this study have not been cultured to date, and these 6 uncultured clades comprise 96% of the SAGs in this study (244/253 SAGs).
As a first step in characterizing virus-host associations, we generated a distance matrix based on hexamer nucleotide analysis using the d2* metric [35] of the 8 cellular clades against the 110 viral types previously determined to be present in the hot spring [16] (Supplemental Table 4). If the smallest measured d2* between a cell type and a virus type was <0.3 it was used as indication of a possible virus–host association. Previous studies have indicated that hexamer nucleotide analysis can be a useful predictor of virus–host associations, given a cutoff of <0.3 as a conservative identification of possible virus-host pairs [35]. Hexamer nucleotide analysis indicated that 61 virus types were potentially associated with the 7 archaeal cell types. The number of virus types associated with a particular archaeal cell type ranged from 28 virus types for the Acidilobus clade to 1 for the Sulfolobus sp 1, clade. Controls consisting of 75 bacterial genomes unlikely to serve as hosts for the hot spring viruses along with the grouped sequences from the 8 SAG cellular clades of this study, found no false virus–host associations to the bacterial genomes (Supplemental Table 4). A limitation of hexanucleotide analysis is that it only suggests a possible virus–host association and does not indicate viral host range [35]. Moreover, hexanucelotide analysis lacks resolution when closely related cellular species/strains are compared [35]. Therefore, this analysis provides an indication of possible virus–host associations and not definitive proof of the association.
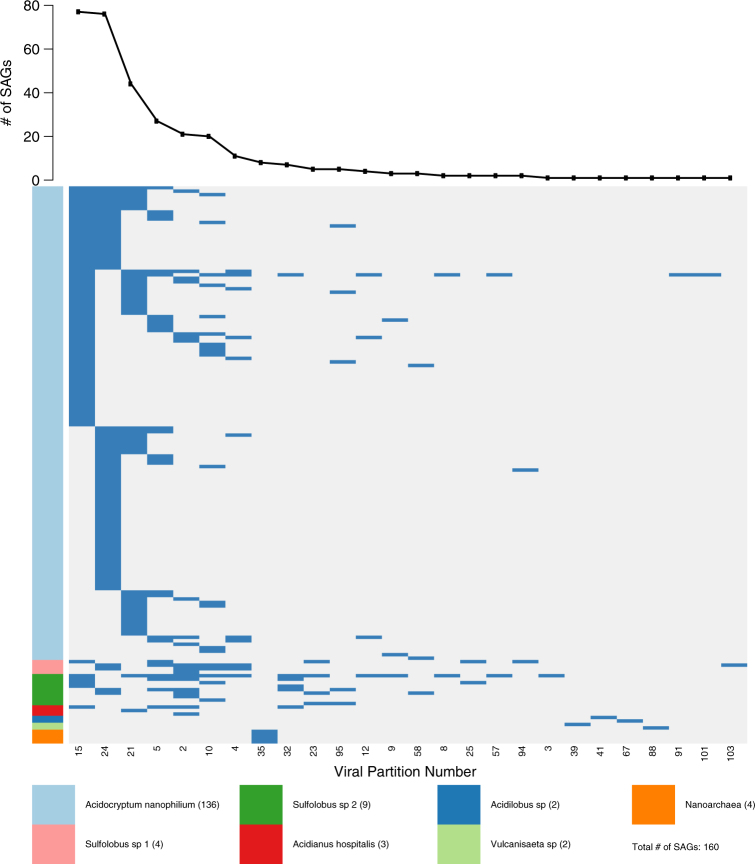
Further identification of individual virus types within each SAG was accomplished by mapping sequencing reads from individual SAGs to the 110 viral types present in NL01 previously established by network-based analytics using time-series community viromics data [16]. We first established a rationale for how many viral base pairs would be expected to be detected in given SAGs given the low level of genome completeness obtained (average host genome completion was 9%). This was accomplished by determining the ratio of viral sequence to host base pairs for each SAG (Supplemental Figure 4) and comparing observed ratios to expectations (see Methods). We estimate that finding two or more unique SAG viral sequences (at least 300 bp) represents a reasonable minimum for detecting virus–host associations. A conservative threshold for virus–host association assumes a two-fold bias in sequence amplification, suggesting a threshold of five or more unique sequence reads (at least 750 bp) to a given viral group in a SAG. Using the more conservative requirement of ≥5 SAG viral reads (750 bp) matching a virus type, viral sequences were detected in 160 of the 253 classified single cell SAGs (63% of SAGs) (Fig. 2, Supplemental Table 5), virus–host associations identified using the lower value of ≥2 viral reads (300 bp) matching a virus type are provided in Supplemental Table 5). Viral sequences were detected in all cellular groups except for Hydrogenobaculum. Of the 110 viral types, 26, were detected (24% of total vial types) in the 253 SAGs. For example, over 49,851 reads mapped to 34.5 kb of continuous sequence represented on the entirety of 3 contigs assembled from a single Acidocryptum nanophilum SAG (AD-903-K19). This 34.5 kb segment likely represents the near-full length genome of a new archaeal virus.
Fig. 2.
Detection of viral types in 160 SAGs. 26 of 110 virus types were detected by BLASTn identification of SAG sequencing reads to NL01 viral community [16]. Viral group numbers are taken from [16]. Blue indicates the detection of a viral group in a SAG and white indicates that a viral group was not detected in a SAG. SAGs are grouped by cell type (vertical axis, a color key for cell the type is provided) and viral groups (horizontal axis) are ordered by detection frequency (top graph)
Next, we examined the number of virus types found in each infected SAG. Surprisingly, more than one viral type was detected in a majority of the cells. Of the 160 SAGs where viral reads were detected, 95 (59%) had ≥750 bp sequence reads from 2 or more viral types, with an average of 2 viral types detected per cell (Fig. 2). This data suggests that co-infection may be common in the hot spring environment. Indeed, 63% of cells randomly sampled by SAG analysis had evidence of virus association. Given the low depth of average SAG genome coverage (approx. 9%), we anticipate that actual association levels are much higher, suggesting that (nearly) all cells in the hot spring interact with viruses. This work extends the scope of virus associations measured in previous reports in marine environments where viral sequences were found in 30–50% of cells [25, 28].
Several lines of evidence indicate that the detected virus-host associations are biologically relevant and not a consequence of random associations. First, no sequencing reads from any of the 307 SAGs were recruited onto two much larger marine viral metagenomic or a human gut viral metagenomic datasets using the identical mapping stringency conditions (Supplemental Table 6). Additionally, sequencing reads from 25 publically available non-hot spring associated SAGs from the JGI IMG (https://img.jgi.doe.gov/) representing 10 bacterial and two archaeal phyla were compared against the viral database used in this study. These SAG’s isolated from other environments, totaling 703.7 million reads, did not match any of the 110 viral groups used in this study at the same stringency settings (Supplemental Table 7). These controls support the conclusion that the conditions used in this study strike a balance between viral detection sensitivity and stringency sufficient to detect biologically relevant virus–host associations in individual SAGs. Future targeted virus RTqPCR analysis on single cells should clarify if the detected viruses are actively replicating.
Analysis of CRISPR spacer sequences were used to detect additional virus–host associations. CRISPR spacer sequences were extracted from SAGs and mapped to the 110 viral types (Supplemental Table 8). A total of 2321 unique CRISPR spacer sequences were detected in 135 SAGs. Spacer sequences were found in all cell types except for the Nanobsidianus. Previous studies had also failed to identify CRISPR sequences in Nanobsidianus sp from YNP hot springs [29, 41]. CRISPR spacer-virus matches were found for 695 (30%) spacer sequences to 38 of the 110 viral types from 121 SAGs (90% of spacer-containing SAGs). The majority of spacers with matches were found in Acidocryptum cells (541/695). Twenty-two viral types were identified by both read mapping and by CRISPR spacer matching to the same cellular species. As expected, controls of comparing 966 non-relevant CRISPR spacer sequences derived from the human gut microbial community to the 110 hot springs viral types failed to detect any virus–host associations under the same conditions. Overall, 47 of the 110 viral types (42%) were detected by either mapping of SAG reads or by SAG CRISPR spacer matching. Furthermore, 18 of these 47 virus types were predicted by hexamer distance analysis to the same host. Taken together, these three independent measures support the conclusion that virus–host associations are a common feature in this hot spring environment.
It is worthwhile to retrospectively consider how useful it is to rely on hexanucleotide analysis to accurately connect viruses to potential hosts. In this work, we have the advantage of having internal standards of viral sequences present within individual SAGs to compare against hexanucleotide analysis at different threshold cut offs. We observe that the hexanucleotide cut off values of <0.3 balance the need to reduce false positives while maintaining the detection of meaningful host-virus pairs (Supplemental Figure 5).
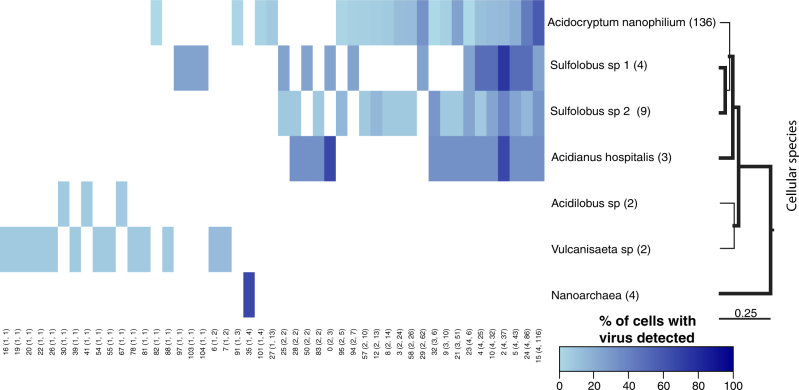
The contextualized virus–host associations (Fig. 3) and CRISPR spacer analysis (Fig. 3, Supplemental Tables 8) provide complementary information on the realized and potential host range of viruses, respectively. By combining these two lines of evidence we asked: what is the host range of individual virus types? Twenty-four viruses infected only a single cellular clade. In contrast, 23 virus types were detected in >2 host genera within the Sulfolobaceae family. Every previously characterized virus detected was found in at least one new host species. For example, STIV previously shown to infect S. solfataricus [42], was also detected in Acidocryptum cells. These results demonstrate that culture-independent approaches can be used to investigate the host range of uncultured viruses across the entire microbial community. Despite finding multiple new associations, it is important to recognize that reported host ranges remain lower bounds, i.e., increased depth of sampling could reveal even more virus types within classified SAGs.
Fig. 3.
Ubiquitous interaction of multiple viruses with cells. The heatmap indicates the detection frequency of 47 viral groups detected by BLASTn analysis or the matching of CRISPR spacer sequences. Viral groups are arranged from least frequently detected to the most frequently detected. Numbers below the heatmap are viral group numbers taken from [16] and numbers in parenthesis indicate the number of species and cells that a group was detected in. The number after the species name on the right hand side is the number of cells classified as members of that species. Partial length 16S sequences from representative genomes were used to make a ML tree and nodes with greater than 0.95 posterior probability are bolded. The scale bar is in substitutions per base. Detected viral groups with described members are: group 0 = SIRV1,2, group 23 = ASV1, SSV1,2, 4–9, group 26 = ATV, group 28 = AFV1, group 29 = STIV1,2 and group 32 = STST1,2 and ARSV1
The inference methods in the present analysis are made possible by network-based analytics that determine viral groups but also limited by relatively low SAG coverage (∼9%). As a consequence, we cannot easily distinguish actively replicating viruses within individual SAGs, define their viral lifestyles (lytic, lysogenic, or chronic) or define individual viruses at the species level. Despite these limitations, it is remarkable that we detect in situ the majority of host and viral types—currently identifiable from whole community sequencing projects—and their associations within a relatively low number of SAGs.
This work shows the benefits of combining single-cell genomics with metagenomics to establish a comprehensive understanding of virus-host associations in a focal environment. Unlike previous studies of virus–microbe interactions, we are able to contextualize virus–host infection networks and link the identity of viruses found in different cells. Guided by the knowledge of the overall virus community, the incorporation of SAG analysis—including contextualized community network mapping and CRISPR detection—allows for the identification of individual hosts and the host range of an individual virus type in a culture-independent fashion. This study shows that (nearly) all cells in the NL01 hot spring interact with viruses, that multiple, concurrent interactions are common, and that a broad spectrum of virus types from specialists to generalists coexist in a relatively low-diversity community. These results should encourage the development of more robust empirical methods and theoretical models to assess the relevance of superinfection and a diversity of viral lifestyles in shaping natural communities.
Electronic supplementary material
Supplemental Table 2. SAG sequencing and assembly information
Supplemental Table 3. Average nucelotide identity
Supplemental Table 4. Virus host hexamer distance matrix
Supplemental Table 5. SAG classification & Identification of viral reads
Supplemental Table 6. Recruitment hot spring SAGs to viromes from other environments
Supplemental Table 7. SAGs from other envrionemnt recruited to NL virome
Acknowledgements
We thank Jennifer Wirth and Ross Hartman for critical review of this work, and three reviewers for their feedback on the manuscript. This study was supported by National Science Foundation grant DEB-4W4596 to RJW, JSW, and MJY. This research was conducted in Yellowstone National Park under the conditions of permit YELL-2013-SCI-5090.
Author contributions
RJW, JSW, and MJY conceived the study. JHMM, RS, SP, and SD performed the experiments and analysis. JHMM, SP, RJW, JSW, and MY prepared the manuscript with input from all authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0071-7) contains supplementary material, which is available to authorized users.
References
- 1.Fuhrman JA, Schwalbach M. Viral influence on aquatic bacterial communities. Biol Bull. 2003;204:192–5. doi: 10.2307/1543557. [DOI] [PubMed] [Google Scholar]
- 2.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 3.Needham DM, Chow CET, Cram JA, Sachdeva R, Parada A, Fuhrman JA. Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J. 2013;7:1274–85. doi: 10.1038/ismej.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–12. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan MB, Weitz JS, Wilhelm S. Viral ecology comes of age. Environ Microbiol Rep. 2017;9:33–5. doi: 10.1111/1758-2229.12504. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–48. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 7.Comeau M, Buenaventura E, Suttle CA. A persistent, productive, and seasonally dynamic vibriophage population within Pacific Oysters (Crassostrea gigas) Appl Environ Microbiol. 2005;71:5324–31. doi: 10.1128/AEM.71.9.5324-5331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Statistical structure of host–phage interactions. Proc Natl Acad Sci. 2011;108:288–97. doi: 10.1073/pnas.1101595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malki K, Kula A, Bruder K, Sible E, Hatzopoulos T, Steidel S, et al. Bacteriophages isolated from Lake Michigan demonstrate broad host-range across several bacterial phyla. Virol J. 2015;12:164–8. doi: 10.1186/s12985-015-0395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21:82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40:258–72. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci. 2009;106:8605–10. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winstanley C, Langille MGI, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of pseudomonas aeruginosa. Genome Res. 2009;19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat Rev Microbiol. 2015;13:147–59. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 15.Brum JR, Ignacio-espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:121498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 16.Bolduc B, Wirth JF, Mazurie A, Young MJ. Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis. ISME J. 2015;9:2162–77. doi: 10.1038/ismej.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson RE, Brazelton WJ, Baross JA. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol. 2011;77:120–33. doi: 10.1111/j.1574-6941.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 18.Snyder JC, Bateson MM, Lavin M, Young MJ. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol. 2010;76:7251–8. doi: 10.1128/AEM.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg Miller ME, Yeoman CJ, Chia N, Tringe SG, Angly FE, Edwards Ra, et al. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14:207–27. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 20.Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Barrero-Canosa J, et al. Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol. 2013;15:2306–18. doi: 10.1111/1462-2920.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. Contrasting life strategies of viruses that infect photo-and heterotrophic bacteria, as revealed by viral tagging. MBio. 2012;3:e00373–12. doi: 10.1128/mBio.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513:242–5. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 23.Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria or viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labonté JM, Field EK, Lau M, Chivian D, Van Heerden E, Wommack KE et al. Single cell genomics indicates horizontal gene transfer and viral infections in a deep subsurface Firmicutes population. Front Microbiol. 2015;6:349. [DOI] [PMC free article] [PubMed]
- 25.Roux S, Hawley AK, Torres Beltran M, Scofield M, Schwientek P, Stepanauskas R et al. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. Elife. 2014;3:e03125. [DOI] [PMC free article] [PubMed]
- 26.Stepanauskas R. Single cell genomics: an individual look at microbes. Curr Opin Microbiol. 2012;15:613–20. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Wilson WH, Gilg IC, Moniruzzaman M, Field EK, Koren S, Lecleir GR, et al. Genomic exploration of individual giant ocean viruses. Int Soc Microb Ecol J. 2017;11:1736–45. doi: 10.1038/ismej.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ, et al. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015;9:2386–99. doi: 10.1038/ismej.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munson-McGee JH, Field EK, Bateson M, Rooney C, Stepanauskas R, Young MJ. Nanoarchaeota, their sulfolobales host, and nanoarchaeota virus distribution across Yellowstone National Park hot springs. Appl Environ Microbiol. 2015;81:7860–8. doi: 10.1128/AEM.01539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci. 2007;104:9052–7. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–9. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 32.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bankevich A, Nurk S, Antipov D, Gurevich Aa, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. Alignment-free d2∗ oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017;45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci. 2016;113:10400–5. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neubeck MVon, Huptas C, Glu C, Krewinkel M, Stoeckel M, Stressler T, et al. Pseudomonas helleri sp. nov. and Pseudomonas weihenstephanensis sp. nov., isolated from raw cow’s milk. Int J Syst Evol Microbiol. 2016;66:1163–73. doi: 10.1099/ijsem.0.000852. [DOI] [PubMed] [Google Scholar]
- 38.Richter M, Rossello R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106:19126–31. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flores CO, Poisot T, Valverde S, Weitz JS. BiMat: a MATLAB package to facilitate the analysis of bipartite networks. Methods Ecol Evol. 2015;7:127–32. doi: 10.1111/2041-210X.12458. [DOI] [Google Scholar]
- 40.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. [DOI] [PMC free article] [PubMed]
- 41.Podar M, Makarova KS, Graham DE, Wolf YI, Koonin E V, Reysenbach A-L. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct. 2013;8:9. [DOI] [PMC free article] [PubMed]
- 42.Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci. 2004;101:7716–20. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 2. SAG sequencing and assembly information
Supplemental Table 3. Average nucelotide identity
Supplemental Table 4. Virus host hexamer distance matrix
Supplemental Table 5. SAG classification & Identification of viral reads
Supplemental Table 6. Recruitment hot spring SAGs to viromes from other environments
Supplemental Table 7. SAGs from other envrionemnt recruited to NL virome