DWARF1 functions as a brassinosteroid C-24 reductase that converts 24-methylene brassinosteroids to 24-methyl brassinosteroids to regulate the endogenous level of an active brassinosteroid, castasterone, to control growth and development in Arabidopsis.
Keywords: Arabidopsis thaliana, biosynthetic connections, brassinosteroids, BR C-24 reductase, DWARF1
Abstract
DWARF1 (DWF1) is a sterol C-24 reductase that catalyses the conversion of 24-methylenecholesterol (24-MCHR) to campesterol (CR) in Arabidopsis. A loss-of-function mutant, dwf1, showed similar phenotypic abnormalities to brassinosteroid (BR)-deficient mutants. These abnormalities were reversed in the wild-type phenotype by exogenous application of castasterone (CS) and brassinolide (BL), but not dolichosterone (DS). Accumulation of DS and decreased CS were found in quantitative analysis of endogenous BRs in dwf1. The enzyme solution prepared from dwf1 was unable to convert 6-deoxoDS to 6-deoxoCS and DS to CS, as seen in either wild-type or 35S:DWF1 transgenic plants. This suggests that DWF1 has enzyme activity not only for a sterol C-24 reductase, but also for a BR C-24 reductase that catalyses C-24 reduction of 6-deoxoDS to 6-deoxoCS and of DS to CS in Arabidopsis. Overexpression of DWF1 in a BR-deficient mutant (det2 35S:DWF1) clearly rescued abnormalities found in det2, indicating that DWF1 functions in biosynthesis of active BRs in Arabidopsis. Expression of DWF1 is down-regulated by application of CS and BL and in a BR-dominant mutant, bes1-D. E-boxes in the putative promoter region of DWF1 directly bind to a BR transcription factor, BES1, implying that DWF1 expression is feedback-regulated by BR signaling via BES1. Overall, biosynthesis of 24-methylene BR is an alternative route for generating CS, which is mediated and regulated by DWF1 in Arabidopsis.
Introduction
Plant growth and development are controlled by chemical signals synthesized in plants (Santner et al., 2009; Wolters and Jürgens, 2009). Brassinosteroids (BRs) are endogenous steroid signals that regulate diverse phenomena at very low concentrations (>10−8 M) in processes related to the growth and development of plants, such as root development, stem elongation, leaf development, photomorphogenesis, vascular and floral development, and senescence (Clouse and Sasse, 1998; Fujioka and Yokota, 2003; Choe, 2004; Wu et al., 2008; Kim and Wang, 2010). In addition, BRs are important factors in stress modulation and defense in plants (Krishna, 2003; Bari and Jones, 2009). Since brassinolide (BL) (Fig. 1) and castasterone (CS) were identified from pollen of Brassica napus and insect galls of Castanea sativa, over 50 plant steroids structurally related to them have been characterized from many plant tissues, ranging from lower to higher plants (Bishop and Yokota, 2001; Bajguz and Tretyn, 2003; Fujioka and Yokota, 2003). Therefore, BRs are regarded as essential plant hormones for normal growth and development.
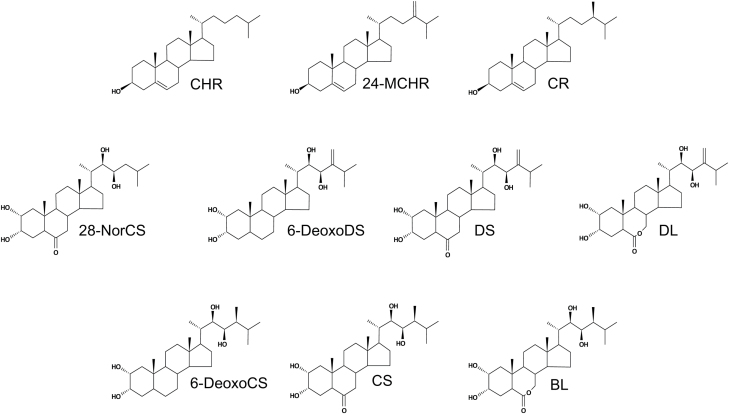
Fig. 1.
Structure of frequently mentioned phytosterols and BRs in the study. BL, brassinolide; CHR, cholesterol; CS, castasterone; CR, campesterol; 6-DeoxoCS, 6-deoxocastasterone; 6-DeoxoDS, 6-deoxodolichosterone; DL, dolicholide; DS, dolichosterone; 24-MCHR, 24-methylenecholesterol; 28-NorCS, 28-norcastasterone.
Naturally occurring BRs can be classified into C27-, C28-, and C29-BRs based on the presence of alkyl groups occupying the C-24 position in 5α-cholestane carbon skeleton side chain (Yokota, 1997; Bajguz and Tretyn, 2003). C27-BRs carry no alkyl group at C-24 (28-norBRs). C28-BRs contain 24-methylene, 24S- and 24R-methyl BRs. C29-BRs include 24-ethylidene, 24-ethyl and 24-methylene-25-methyl BRs. The carbon skeletons of C27-, C28-, and C29-BRs are similar to those of 4-demethylsterols, which are abundant in plants. This implies that BRs are biosynthesized from 4-demethylsterols in plants (Fujioka and Yokota, 2003). Based on this speculation, multiple biosynthetic pathways that generate diverse kinds of BRs are operant in plants (Fujioka et al., 1997; Yokota, 1997; Joo et al., 2009).
To date, CS and BL have been the BRs most frequently identified from plants (Fujioka, 1999; Bajguz and Tretyn, 2003). Given their stronger biological activity and wider distribution in the plant kingdom than any other types of BRs, CS and BL are also considered the most important bioactive BRs in plants (Bajguz and Tretyn, 2003). CS and BL possess 24S-oriented methyl groups at C-24 positions, suggesting that they can be biosynthesized from campesterol (CR). Feeding experiments using CR and downstream intermediates in Catharanthus roseus indicated the presence of two parallel biosynthetic pathways for CS and BL generation, the early and late C-6 oxidation pathways (Suzuki et al., 1993). Early and/or late C-6 oxidation have also been demonstrated in other higher plants, such as Arabidopsis, tomato, rice, maize, and Phaseolus vulgaris, as well as in lower plants such as Marchantia polymorpha (Kim et al., 2001; Fujioka and Yokota, 2003; Kim et al., 2004a, b, c; Kim et al., 2005c). This suggests that early and late C-6 oxidation pathways are common biosynthetic routes for BRs in the plant kingdom. However, some evidence has accumulated that is contrary to the notion that early C-6 oxidation plays a role in CS and BL biosynthesis (Bishop et al., 1999; Kim et al., 2000; Noguchi et al., 2000). The first reaction in the pathway is C-6 oxidation of campestanol (CN) to 6-oxoCN, which was established in Catharanthus crown gall cells (Fujioka et al., 2000). Since that study, C-6 oxidation of CN to 6-oxoCN has not been confirmed in other plants. C-22 hydroxylation of 6-oxoCN to cathasterone (CT) has not been confirmed in Catharanthus crown gall cells, even though CT is endogenous to such cells (Fujioka et al., 1995). Neither the conversion of 6-oxoCN to CT nor the presence of CT has been demonstrated in other plants (Joo et al., 2002). Additionally, CYP90B1 (DWF4) hydroxylates CN to 6-deoxoCT, but does not convert 6-oxoCN to CT (Fujita et al., 2006). These results indicate that the early C-6 oxidation pathway is commonly interrupted in plant tissues.
BRs are highly oxidized steroids. It is thus thought that the incorporation of oxygen atoms into phytosterols is catalysed by cytochrome P450 (CYP) monooxygenase in BR biosynthesis. Functional studies using heterogeneously expressed Arabidopsis CYPs in Escherichia coli, yeast, and insect cells have revealed that CYP90B1 (DWF4) catalyses C-22 hydroxylation of CR to 22-hydroxyCR (Supplementary Fig. S1A at JXB online), rather than catalysing C-22 hydroxylation of 6-oxoCN to CT or CN to 6-deoxoCT in the early and late C-6 oxidation pathways (Fujita et al., 2006). CYP90A1 (CPD) mediates C-3 oxidation of 3β-hydroxylated intermediates to their corresponding 3-dehydro derivatives such as 22-hydroxyCR to 22-hydroxy-campesta-4-en-3-one, 6-deoxoCT to 22-hydroxy-campesta-3-one and 6-deoxo-teasterone (6-deoxoTE) to 6-deoxo-3-dehydroteasterone (6-deoxo3DT) (Ohnishi et al., 2012). CYP90C1 and CYP90D1 have similar enzyme activities for C-23 hydroxylation in 6-deoxoCT to 6-deoxoTE, 22-hydroxy-campesta-3-one to 3-dehydro-6-deoxoTE, and 3-epi-6-deoxoCT to 6-deoxo-typhasterol (6-deoxoTY) (Kim et al., 2005a, Ohnishi et al., 2006). Although CYP85A2 has stronger enzyme activity than CYP85A1, both molecules catalyse C-6 oxidation of 6-deoxoTE, 3-dehydro-6-deoxoTE, 6-deoxoTY, and 6-deoxoCS to teasterone (TE), 3-dehydroTE, typhasterol (TY), and CS, respectively (Kim et al., 2005b). Finally, CYP85A2 has been shown to convert CS to BL, which suggests that CYP85A2 is a bifunctional enzyme, not only a BR C-6 oxidase but also a BL synthase, in Arabidopsis plants (Kim et al., 2005b).
Together with the presence of a biosynthetic sequence, CS and BL are biosynthesized by a new pathway called the CN-independent or early CR C-22 hydroxylation pathway (Supplementary Fig. S1A) in Arabidopsis (Fujita et al., 2006). The pathway conversion diagram is as follows: CR→22-hydroxy-CR→22-hydroxy-campesta-4-en-3-one→22-hydroxy-campesta-3-one→6-deoxoCT and 3-epi-dexoxoCT. In the CN-independent pathway, conversion of 22-hydroxy-campesta-4-en-3-one to 22-hydroxy-campesta-3-one is catalysed by DET2, originally identified as a 5α-reductase for CR to CN conversion in plants (Fujita et al., 2006).
DS is a C28-BR that carries an exo-methylene group at a C-24 side chain (Baba et al., 1983). To date, DS and its biosynthetically related BRs, such as 6-deoxoDS and dolicholide (DL), have been identified from four dicots (Arabidopsis, Phaseolus vulgaris, Dolichos purpreus and Vicia faba), two monocots (rice and maize) and a pteridophyte (Equisetum arrense), suggesting that these 24-methylene BR molecules are common in the plant kingdom (Bajguz and Tretyn, 2003). In all plants from which DS has been identified, CS has also been identified. This suggests that the biosynthetic pathways that synthesize the two compounds might operate simultaneously in plants (Bajguz and Tretyn, 2003; Hwang et al., 2009; Lee et al., 2010). Recently, a crude enzyme solution prepared from Arabidopsis successfully mediated conversion of 6-deoxoDS to CS, intermediated by DS (Hwang et al., 2009; Lee et al., 2010). This suggests successful biosynthesis of 24-methylene BRs to 24-methyl BRs in plants. However, the enzyme responsible for C-24 reduction of 24-methylene BRs to 24-methyl BRs, a BR C-24 reductase, has not yet been identified in plants. This prompted us to identify a BR C-24 reductase in Arabidopsis. Here, biological and biochemical evidence for DWF1 as a BR C-24 reductase in Arabidopsis is presented. In addition, molecular regulation of DWF1 expression in the plant is described.
Materials and methods
Plant materials and growth conditions
Seeds of Col-0, En-2, dwf1, det2, 35S:DWF1, det2 35S:DWF1, bes1-D, and bzr1-1D were each surface-sterilized in ethanol–water (70:30, v/v), rinsed in distilled water (DW), cold-treated at 4 °C for 2 d and plated on 0.5× Murashige and Skoog (Duchefa, Haarlem, the Netherlands) medium containing 1% sucrose and 0.7% agar. Plates were kept in the light (120 μmol m−2 s−1) at 22 °C for 16 h and in the dark for 8 h in a growth chamber (Sanyo, Osaka, Japan).
Analysis of BRs and sterols in Arabidopsis
Arabidopsis plants (30 kg) grown for 6 weeks on soil were harvested and extracted three times with 3 liters of 90% methanol. Evaporated extracts were partitioned three times between water (3 liters) and chloroform (3 liters). The chloroform-soluble fractions were concentrated and partitioned three times between 80% methanol (3 liters) and n-hexane (3 liters). The concentrated 80% methanol extracts were repartitioned three times between ethyl acetate (3 liters) and phosphate buffer (pH 7.8, 3 liters). Ethyl acetate-soluble residues were subjected to silica gel chromatography. Columns were eluted with 150 ml of chloroform containing 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, 50, or 100% (v/v) methanol. The 3–7% (v/v) methanol fractions were combined, concentrated and subsequently purified using a Sep-Pak C18 cartridge column (Waters, Milford, MA, USA) eluted with 0, 50, and 100% methanol (20 ml each). The 100% methanol fraction obtained from the Sep-Pak column was dried, dissolved in a small amount of methanol and then subjected to reversed-phase HPLC (Senshu-Pak C18, 10 × 150 mm). The column was eluted at a flow rate of 2.5 ml min−1 using different percentages of acetonitrile (MeCN) in water for different lengths of time: 0–20 min, 45% MeCN; 20–40 min, 45–100% MeCN; or 40–70 min, 100% MeCN. Fractions were collected every minute. Under the same HPLC condition, synthetic 6-deoxoDS, DS, and DL was detected in fractions 34–37, 14–15 and 9–10, respectively. These fractions were analysed by capillary GC-MS after bismethanboronation. Endogenous amounts of 6-deoxoDS and DS were calculated by a calibration curve established with a molecular ion (m/z 496 and m/z 510 for 6-deoxoDS and DS, respectively) in GC–selective ion monitoring (SIM) analysis. The n-hexane-soluble fraction was extracted with methanol:chloroform (4:1, v/v). Extracts were then concentrated and solvent-partitioned between chloroform and water. D7-Cholesterol (D7-CHR), 0.5 μg, was added to the chloroform-soluble fraction as an internal standard. The fraction extracted with n-hexane after alkaline hydrolysis was purified on a Sep-Pak C18 cartridge column and subjected to GC-MS analysis after trimethylsilylation. Endogenous amounts of 24-MCHR and CR were measured based on relative ratios of 24-MCHR and CR against endogenous amounts of cholesterol (CHR) with D7-CHR as an internal standard.
Enzyme assay
Three-week-old light-grown plants (20 g) were homogenized with 0.1 M sodium phosphate buffer (pH 7.4) containing 1 mM EDTA, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 15 mM 2-mercaptoethanol, 15% glycerol, 250 mM sucrose, 40 mM ascorbate, and 1% insoluble polyvinyl-polypyrrolidone, and centrifuged for 10 min at 8000 g to remove cell debris. Resulting supernatants were re-centrifuged for 30 min at 20 000 g. Following the addition of cold acetone (final volume 40%), precipitates were suspended in 0.1 M sodium phosphate buffer (pH 7.4) containing 1.5 mM 2-mercaptoethanol and 30% glycerol in order to create a crude enzyme solution. The protein concentration of the enzyme solution was estimated with a Bradford assay (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as a standard.
Enzyme assays for metabolism of 6-deoxoDS and DS were initiated by the addition of substrates (5 μg each) to the enzyme solution (4–5 mg protein ml−1) in the presence of NADPH. Following incubation at 37 °C for 30 min, metabolites of enzyme reactions were extracted with ethyl acetate (1.2 ml) and concentrated. Ethyl acetate-soluble fractions were loaded onto Sep-Pak C18 cartridge columns and washed with aqueous methanol (in 5 ml 50% methanol, 5 ml of 60% and 5 ml of 80% methanol). The 80% methanol-eluted fractions were concentrated, dissolved in 50 μl of methanol and then subjected to reversed-phase HPLC (Senshu-Pak C18, 10 × 150 mm). Fractions were then eluted at a flow rate of 2.5 ml min−1 with the following MeCN-water gradient: 0–20 min, 45% MeCN; 20–40 min, 45–100% MeCN; and 40–70 min, 100% MeCN. Fractions were collected every minute. The HPLC fractions corresponding to authentic 6-deoxoCS, DS and CS in the same HLPC conditions were collected and analysed via GC-MS or GC-SIM.
GC-MS and GC-SIM analysis
GC-MS and GC-SIM analyses were carried out on a Hewlett-Packard 5973 mass spectrometer (Electron impact ionization, 70 eV) coupled to an Agilent 6890 gas chromatograph (Palo Alto, CA, USA) fitted with a fused silica capillary column (HP-5, 0.25 mm×30 m, 0.25 μm film, Agilent). The oven temperature was maintained at 175 °C for 2 min, raised to 280 °C at a rate of 40 °C min−1 and then maintained at 280 °C. Helium was used as the carrier gas at a flow rate of 1 ml min−1 and samples were introduced using the on-column injection mode. Methaneboronation was carried out by heating samples dissolved in pyridine-containing methaneboronic acid (2 mg ml−1) at 70 °C for 30 min.
RNA isolation of qRT-PCR
Total RNAs were extracted using TRI reagent (Sigma-Aldrich, St Louis, MO, USA) in accordance with the manufacturer’s instructions. For RT-PCR, 2 μg of total RNA was reverse-transcribed by M-MLV RT (Promega, Madison, WI, USA); 2 μl of the RT product was employed as a PCR template. Quantitative RT-PCR (qRT-PCR) was performed using iQ SYBR Green SuperMix and the iCycler iQTM RT-PCR Detection System (Bio-Rad). PCR conditions consisted of denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 15 s and extension at 72 °C for 30 s. A dissociation curve was generated at the end of each cycle to verify amplification of a single product. mRNA levels were quantified using the 2−ΔΔCTmethod (Livak and Schmittgen 2001). Expression level of the target gene was normalized relative to the expression of the housekeeping gene UBIQUITIN 5 (UBQ5). Gene-specific primers are described in Supplementary Table S1.
Selection of dwf1, 35S:DWF1, and det2 35S:DWF1
An Arabidopsis dwf1 knockout mutant (Salk006932) with a transfer (T)-DNA insertion in the DWF1 gene’s first exon was obtained. T-DNA insertion was confirmed using T-DNA LB primer (5′-CTTTGACG TTGGAGTCCACGTTCTTTAATA-3′) and DWF1 reverse primer (Supplementary Table S1). For overexpression, DWF1 full-length cDNA was amplified by PCR and cloned into pGEM-T Easy vectors (Promega), followed by sub-cloning into a binary vector (pBI121) driven by a constitutive 35S promoter (35S:DWF1). Transgene constructs were confirmed by sequencing and subsequently transformed into wild-type (T0) and BR-deficient mutant, det2, by the floral-dip method (Clough and Bent, 1998). T1 seeds with cold pretreatment were screened on 0.5× MS medium containing 50 μg l−1 kanamycin, 1% sucrose, and 0.7% agar. Homozygous lines resistant to kanamycin were obtained at the T3 generation.
Expression level of DWF1 in wild-type, dwf1, 35S:DWF1, and det2 35S:DWF1 was measured by semi-quantitative RT-PCR (semi-qRT-PCR) using DWF1 forward and reverse primers (Supplementary Table S1). Cycling conditions were as follows: 5 min at 95 °C, 35 cycles of 15 s at 95 °C, 15 s at 59 °C, and 15 s at 72 °C.
Rescue experiments
For rescue experiments for dwf1 and det2, DS, CS and BL were sprayed onto 21-day-old dwf1 and det2 plants 3–5 times at 5-day intervals. In hormone stocks, 5 μM of BRs were dissolved in ethanol containing 1% DMSO and 0.5% Tween-20 in ethanol. For seedlings, dwf1 and det2 were planted and grown in the dark on 0.5× MS medium containing 10–9 M CS, DS and BL, 1% sucrose, and 0.7% agar. Hypocotyl length was measured after 7 d.
Electrophoretic mobility shift assay
Maltose binding protein (MBP) and MBP–BES1 proteins were expressed and affinity-purified from E. coli (BL21-DE3) using amylose resins (New England Biolabs, Ipswitch, MA, USA). The DWF1 promoter fragments (WT probes) and mutated probes (MT probes, CANNTG to AAAAAA) used for electrophoretic mobility shift assay (EMSA) were prepared using primers described in Supplementary Table S1. Double-stranded DNA probes, 100 ng, were labeled with [32P]dATP using T4 polynucleotide kinase using a 5′ end labeling system (‘hot probes’). Unlabeled double-stranded DNA probes were used as ‘cold probes’. Binding reactions were carried out in 2 μl of 5× binding buffer (50 mM Tris–HCl pH 7.5, 250 mM NaCl, 2.5 mM EDTA, 2.5 mM DTT, 5 mM MgCl2, and 20% glycerol), 4 μl of 3 μg MBP-BES1 proteins and 3 μl of DW with a 1 μl probe. After 30 min of incubation at room temperature, reactions were resolved using 4% native polyacrylamide gels with 0.5× TBE buffer (containing 5.39 g l−1 Tris, 2.75 g l−1 boric acid and 0.37 g l−1 EDTA, pH 8.0) and exposed to a phosphor imaging screen.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on 10-day-old 35S:YFP and BES1-YFP overexpression transgenic lines. Seedlings (1 g) were cross-linked with 50 ml of 1% formaldehyde in a vacuum for 5 min. A total of 2.5 ml of 2 M glycine was added to quench cross-linking. After rinsing seedlings with DW, tissues were ground with liquid nitrogen and resuspended in cold extraction buffer I (0.4 M sucrose, 10 mM Tris–HCl, pH 8, 10 mM MgCl2, 5 mM 2-mercaptoethanol, 0.1 mM PMSF, and 1× protease inhibitor (Roche Diagnostics, Indianapolis, IN, USA)). Filtrates filtered through Miracloth (Calbiochem, Darmstadt, Germany) were centrifuged at 1800 g for 10 min at 4 °C. The pellet was resuspended in 1ml of extraction buffer II (0.25 M sucrose, 10 mM Tris–HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM 2-mercaptoethanol, 1 mM PMSF, and 1× protease inhibitor) and centrifuged at 14 000 g for 10 min at 4 °C. The pellet was resuspended in 600 μl of extraction buffer III (1.7 M sucrose, 10 mM Tris–HCl, pH 8, 0.15% Triton X-100, 2 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM PMSF, and 1× protease inhibitor) and centrifuged at 14 000 g for 1 h at 4 °C. The crude chromatin pellet was resuspended in 250 μl of nuclear lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 200 mM NaCl, 0.5% Triton X-100, 1 mM PMSF and 1× protease inhibitor) and sonicated with a Branson sonifier (VWR) to achieve an average fragment size of ∼0.5–1.0 kb. The solution obtained was centrifuged at 14 000 g for 10 min at 4 °C and supernatant was transferred to a new tube. Chromatin solution was diluted with an equal volume of DW and 50 μl was taken out to use as input. After pre-clearing with protein A-sepharose beads (Sigma-Aldrich), 5 μl of with yellow fluorescent protein (YFP)-specific monoclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA) was added to chromatin solution and incubated overnight with rotating at 4 °C. The immunocomplexes were extracted by incubating with 50 μl of protein A beads for 1 h at 4 °C. The extract was washed with 250 μl of wash buffer (1% SDS and 0.1 M NaHCO3) and then reverse cross-linked with a final concentration of 200 mM NaCl at 65 °C for overnight. Extracted DNA using a gel elution kit (MP) was eluted by 50 μl of TE and used for PCR analysis. ChIP-PCR was performed using primers specific for DWF1 and SAUR-AC1 promoter regions (Supplementary Table S1). Cycling conditions were as follows: 3 min at 95 °C, 35 cycles of 10 s at 95 °C, 10 s at 52 °C, and 10 s at 72 °C. The ChIP experiments were performed three independent times.
Results
Identification and metabolism of 6-deoxoDS in Arabidopsis
The presence of DS was recently demonstrated with a large quantity (30 kg) of Arabidopsis (Lee et al., 2010). The presence of DS analogs, especially 6-deoxoDS and DL, in Arabidopsis extract was investigated in order to understand the biosynthesis of DS. Following reversed-phase HPLC, the fractions corresponding to synthetic 6-deoxoDS and DL were collected and analysed by capillary GC-MS/SIM after derivatization to bismethaneboronates (BMBs). BMBs, thought to be a form of 6-deoxoDS, exhibited characteristic molecular ions for 6-deoxoDS-BMB at m/z 496 (M+), 356, 342, 327, 273, 153, and 124 in HPLC fractions at GC retention times similar to authentic 6-deoxoDS-BMB, verifying that the compound is 6-deoxoDS (Table 1). However, BMBs in compounds of HPLC fractions equivalent to synthetic DL failed to show any characteristic molecular ions for DL-BMB, suggesting that DL was not present in Arabidopsis or that endogenous DL levels in Arabidopsis were too low to be detected by GC-MS/SIM. Endogenous levels of 6-deoxoDS and DS in Arabidopsis were measured by GC-SIM-based calibration curves using molecular ions. Arabidopsis contains approximately 0.13 and 0.01 ng g−1 fresh weight of 6-deoxoDS and DS, respectively (Table 1).
Table 1.
HPLC and GC data for 6-DeoxoDS and DS in Arabidopsis
| Compound | Rtb on HPLC | RRtc on GC | Prominent ions | Endogenous amountd |
|---|---|---|---|---|
| Arabidopsis | ||||
| 6-DeoxoDSa | 34–37 | 0.721 | 496 (M+, 26), 356 (1), 342 (1), 327 (1), 273 (44), 153 (67), 124 (100) | 0.13 |
| DSa | 14–15 | 1.002 | 510 (M+, 22), 495 (16), 411 (16), 387 (15), 355/356 (23), 327 (96), 287 (9), 153 (69), 124 (100) | 0.01 |
| Authentic | ||||
| 6-DeoxoDSa | 34–37 | 0.721 | 496 (M+, 24), 356 (1), 342 (1), 327 (1), 273 (46), 153 (87), 124 (100) | — |
| DSa | 14–15 | 1.002 | 510 (M+,25), 495 (13), 411 (12), 387 (18), 355/356 (20), 327 (95), 287 (10), 153 (72), 124 (100) | — |
aThe sample was analysed as bismethanboronate.
bRetention time.
cRelative retention time to CS (30.10 min).
dAmount is denoted as ng g−1 fresh weight.
6-DeoxoDS is thought to be a direct biosynthetic precursor of DS in Arabidopsis (Lee et al., 2010). To confirm this, metabolism of 6-deoxoDS was examined using a crude enzyme solution prepared from Arabidopsis. Following enzyme assay, products were purified by reversed-phase HPLC, and the obtained HPLC fractions were tested in rice lamina inclination assays. In addition to HPLC fraction 34–37 for 6-deoxoDS, HPLC fractions 14–15, 19–21, and 40–42 showed biological activities. The HPLC fractions were derivatized to BMBs and analysed by capillary GC-MS. As summarized in Table 2, DS, CS, and 6-deoxoCS were present in HPLC fractions 14–15, 19–21, and 40–42, respectively. Coupled with identification of CS as an enzyme product of DS, this strongly suggests that partial biosynthetic sequences, 6-deoxoDS→DS→CS and 6-deoxoDS→6-deoxoCS→CS, are operating in Arabidopsis plants.
Table 2.
In vitro conversion of 6-deoxoDS and DS in enzyme solution obtained from Arabidopsis
| Substrate | Product | Rtb on HPLC | RRtc on GC | Prominent ions |
|---|---|---|---|---|
| 6-DeoxoDS | 6-DeoxoCSa | 40–42 | 0.705 | 498 (M+, 35), 483 (15), 332 (4), 273 (100), 213 (4), 155 (37) |
| DSa | 14–15 | 1.002 | 510 (M+, 26), 495 (15), 411 (15), 387 (17), 355/356 (20), 327 (97), 287 (10), 153 (72), 124 (100) | |
| CSa | 19–21 | 1 | 512 (M+, 81), 358 (35), 327 (10), 287 (29), 155 (100) | |
| DS | CSa | 19–21 | 1 | 512 (M+, 80), 358 (33), 327 (12), 287 (32), 155 (100) |
aThe sample was analysed as bismethanboronate.
bRetention time.
cRelative retention time to CS (30.10 min).
DWF1 exhibits BR C-24 reductase activity in Arabidopsis
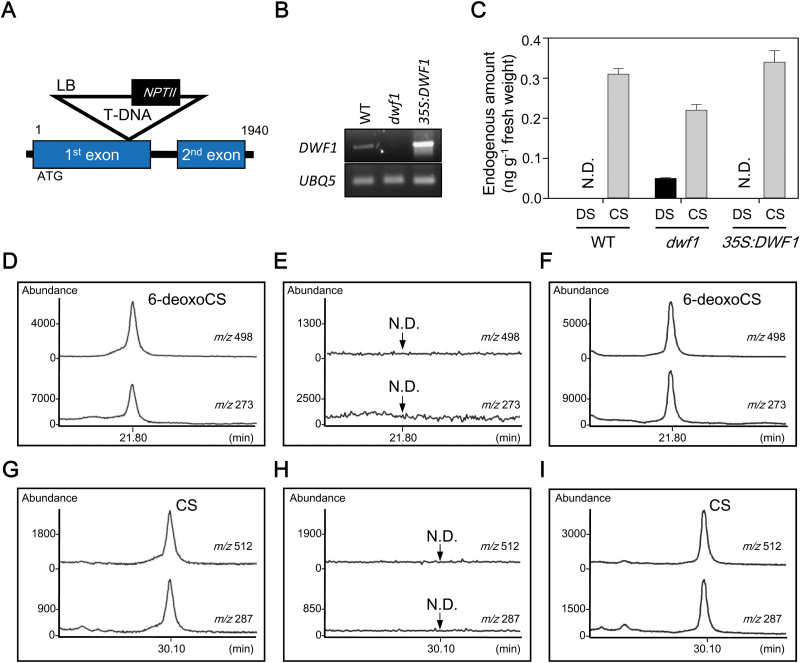
Arabidopsis DWF1 loss-of-function mutant dwf1 was selected from SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress) mutant pools. Sequence analysis of genomic DNA flanking the T-DNA insertion sites revealed that dwf1 contained a T-DNA insertion at the first exon in chromosome 3 (Fig. 2A). Semi-qRT-PCR analysis using RNA isolated from the homozygous mutant and from the wild-type (Col-0) showed that dwf1 was a null allele (Fig. 2B; Supplementary Fig. S2A).
Fig. 2.
Biochemical analysis of DWF1 enzyme activity as a BR C-24 reductase in Arabidopsis. (A) Schematic diagram of T-DNA insertion in dwf1 mutant. Exons and intron are indicated by boxes and a line, respectively. (B) Semi-qRT-PCR analysis of DWF1 expression in the 40-day-old dwf1 and 35S:DWF1 plants. (C) Endogenous amounts of DS and CS in dwf1, 35S:DWF1, and wild-type (50 g each), which represent the average value obtained from two independent quantitative analyses (+SE). DS was quantified by a GC-SIM-based calibration curve using molecular ion at m/z 510. CS was quantified via GC-SIM using an internal standard (D6-labeled CS). (D–I) In vitro enzymatic conversion of 6-deoxoDS to 6-deoxoCS and DS to CS in wild-type (D, G), dwf1 (E, H), and 35S:DWF1 (F, I). In GC-SIM analysis, ions at m/z 498 (M+) and m/z 273 were monitored for 6-deoxoCS-BMB. Ions at m/z 512 (M+) and m/z 287 were monitored for CS-BMB. N.D., not detected.
DWF1 is a sterol C-24 reductase that catalyses C-24 reduction of 24-MCHR to CR in Arabidopsis plants (Choe et al., 1999). In dwf1 mutants, endogenous levels of 24-MCHR are greatly increased compared with wild-type (Table 3). On the other hand, endogenous levels of CR are significantly reduced compared with those in wild-type. Therefore, loss-of-function of DWF1 as a sterol C-24 reductase was biochemically verified in the dwf1 mutant.
Table 3.
Endogenous amount of 24-MCHR and CR in dwf1 and wild-type (Col-0) Arabidopsis
| Compound | Wild-type (Col-0) | dwf1 | ||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |
| 24-MCHR (µg g−1 fresh weight) | 5260 | 5560 | 15 591 | 13 940 |
| CR (µg g−1 fresh weight) | 44 369 | 43 990 | 668 | 673 |
Conversion of 6-deoxoDS to 6-deoxoCS and of DS to CS occurs via the same C-24 reduction mechanism as conversion of 24-MCHR to CR, implying that DWF1 can also mediate C-24 reduction of 6-deoxoDS to 6-deoxoCS and DS to CS in Arabidopsis. Endogenous levels of DS and CS were compared among wild-type (Col-0), dwf1, and 35S:DWF1 (DWF1 overexpression line; Fig. 2B; Supplementary Fig. S2A), each with 50 g of Arabidopsis plants. As shown in Fig. 2C, Col-0 contained 0.31 ng g−1 fresh weight CS, but no detectable amount of DS. In 35S:DWF1, CS was slightly increased (0.34 ng g−1 fresh weight) compared with Col-0, but no DS was detected, indicating that overexpression of DWF1 can generate increased CS in 35S:DWF1. In dwf1, CS was reduced (0.22 ng g−1 fresh weight) relative to Col-0, and 0.05 ng g−1 fresh weight of DS was detected. To demonstrate BR C-24 reductase activity of DWF1, conversion of 6-deoxoDS to 6-deoxoCS and DS to CS was examined in enzyme solutions prepared from dwf1 and 35S:DWF1 plants. In GC-SIM analysis, C-24 reduction of 6-deoxoDS to 6-deoxoCS and DS to CS was shown in the enzyme solution prepared from 35S:DWF1, but not in the enzyme solution prepared from dwf1, compared with those in wild-type (Fig. 2D–I). This suggests that DS accumulates in dwf1 due to loss-of-function in the enzymatic activity for conversion of 6-deoxoDS to 6-deoxoCS and DS to CS in the mutant. In other words, DWF1 exhibits a BR C-24 reductase activity to convert 6-deoxoDS to 6-deoxoCS and DS to CS as well as a sterol C-24 reductase in Arabidopsis.
Overexpression of DWF1 enhances growth and development in Arabidopsis
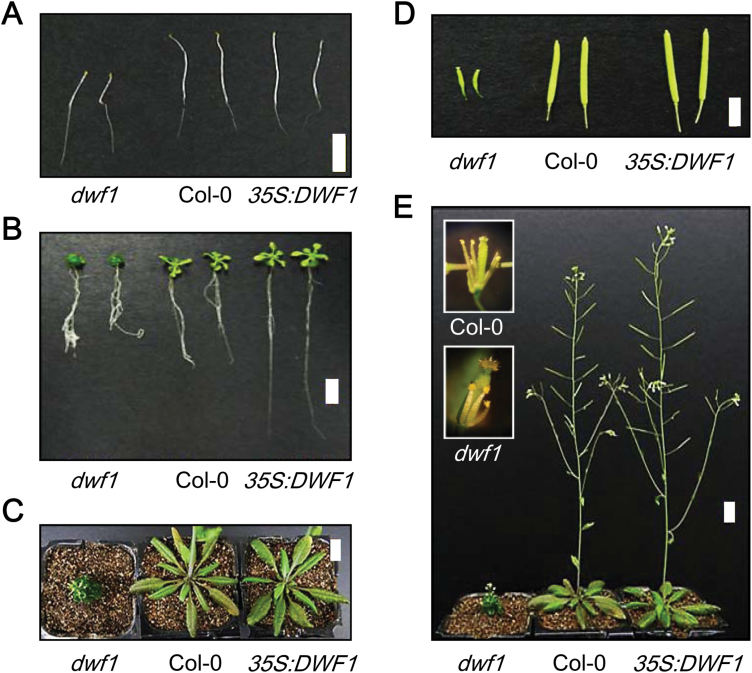
Abnormal growth and development in dwf1 have been reported by several authors (Takahashi et al., 1995, Klahre et al., 1998, Choe et al., 1999). As shown in Fig. 3, similar abnormalities were found in dwf1 in this study. Phenotypic alterations in 35S:DWF1 were investigated over the entire developmental growth stage of transgenic plants. In the dark, 35S:DWF1 exhibited almost no differences in seedling growth compared with wild-type seedlings (Fig. 3A). Under long-day conditions (16 h light and 8 h dark), however, 12-day-old 35S:DWF1 seedlings showed more developed leaves and longer roots compared with wild-type plants (Fig. 3B). In the rosette plant stage, 35S:DWF1 showed similar sized leaves, but their shapes were more expanded than those in wild-type plants (Fig. 3C). In intact plants, 35S:DWF1 showed stems with slightly increased fluorescence as well as an increased number of siliques (Fig. 3D, E). These phenotypic alterations in 35S:DWF1 imply that overexpression of DWF1 yields positive effects on growth and development of Arabidopsis under light conditions.
Fig. 3.
Phenotype of Arabidopsis wild-type (Col-0), dwf1, and 35S:DWF1 plants. (A) Comparison of 5-day-old dark-grown seedlings and (B) 7-day-old light-grown seedlings on MS media for dwf1, 35S:DWF1, and wild-type. (C) Rosette leaves of 27-day-old dwf1, 35S:DWF1, and wild-type plants. (D) Siliques and (E) adult phenotype of the dwf1, 35S:DWF1, and wild-type. Scale bar=1 cm.
DWF1 is important for biosynthesis of active BRs in Arabidopsis
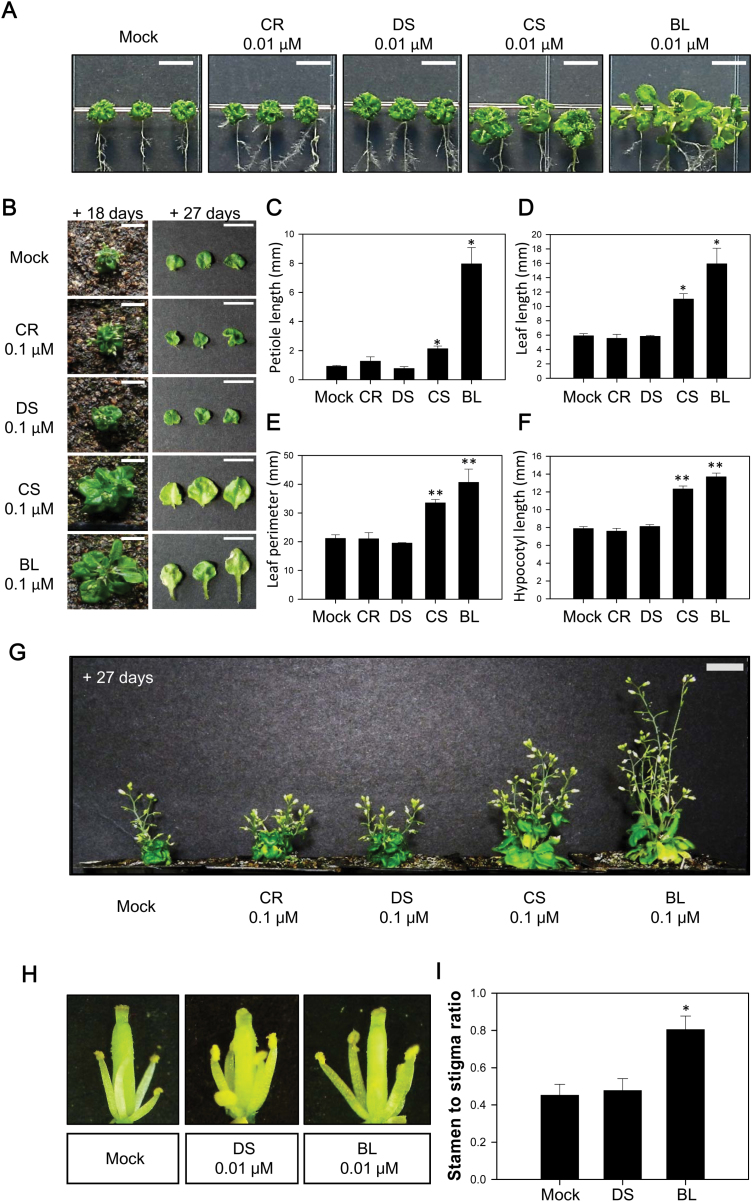
Restoration of phenotypic abnormalities in dwf1 was examined by application of CR, DS, CS, and BL. In light-grown seedlings and rosette plants, application of CR and DS did not improve abnormal leaf size, leaf shape or petiole growth, while application of CS and BL almost completely restored leaf size, leaf shape and petiole length to that observed in wild-type plants (Fig. 4A–E). In dark-grown seedlings, application of CS and BL greatly increased hypocotyl length compared with CR and DS application (Fig. 4F; Supplementary Fig. S3A). In intact plants, abnormalities found in dwf1 such as unexpanded and less-developed dark green rosette leaves, reduced fluorescence in stems, and less-developed stamen were not restored by application of CR and DS. Abnormal stem growth and development of rosette leaves and reproductive organs in dwf1 were clearly reversed by application of CS and BL (Fig. 4G–I). These findings indicate that abnormalities shown in dwf1 are likely caused by insufficient endogenous levels of CS and BL.
Fig. 4.
Reversal of abnormalities in dwf1 through application of BRs. (A) Restoration of shoot growth in 7-day-old dwf1 seedlings after application of 0.01 µM CR, DS, CS, and BL. Scale bar=1 cm. (B–E) Restoration of shoot growth in adult dwf1 plants after application of 0.1 µM CR, DS, CS, and BL. BRs were sprayed 3–5 times a day onto dwf1 for 27 d at an interval of 3 d. A representative result from six individual dwf1 adult plants is shown in (B). Comparison of petiole length (C), leaf length (D), and leaf perimeter (E) after application of 0.01 µM CR, DS, CS, and BL. Each column represents the mean (+SE) of the individual measurements (n>20). (F) Restoration of hypocotyl growth in dwf1 mutant seedlings after application of 0.1 µM CR, DS, CS and BL. Hypocotyl length was measured after 7 d. Each column represents the mean (+SE) of the individual measurements (n>20). Asterisks indicate the statistical significance by Student’s t-test. *P<0.05, **P<0.01 compared with Mock control. (G) Restoration of abnormal stem growth in dwf1 adult plants after application of 0.1µM CR, DS, CS, and BL. BRs were sprayed 3–5 times a day onto dwf1 for 27 d at an interval of 3 d. A representative result from six individual dwf1 intact plants is shown. (H, I) Restoration of stamen development of dwf1 after 0.01 µM DS and BL treatment. After artificial fertilization, BRs were sprayed 3–5 times onto dwf1 flower every day. A representative result from five individual dwf1 flowers is shown in (H). Hormonal effects on phenotypic rescue of less-developed stamen in dwf1 are presented as stigma to stamen length ratio (I). Each column represents the mean (+SE) of the individual measurements (n=5).
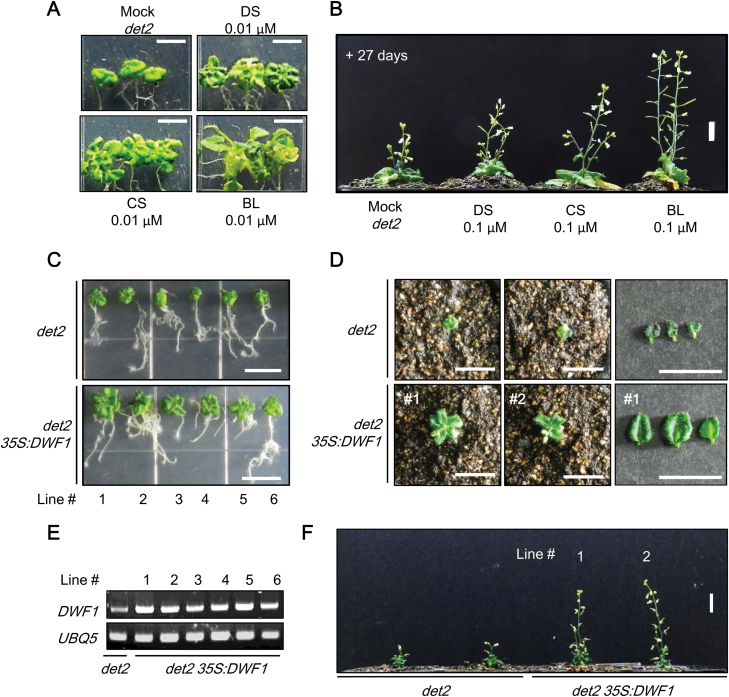
Restoration of abnormalities in a BR-deficient mutant, det2, was tested by application of BRs. Exogenously applied DS as well as CS and BL successfully rescued abnormalities in det2 (Fig. 5A, B; Supplementary Fig. S3B). Next, DWF1 was overexpressed in det2, and the recovery of the phenotype in det2 35S:DWF1 was examined. As shown in Fig. 5C–F, abnormalities in det2 were clearly reversed in det2 35S:DWF1 at the seedling, rosette, and intact plant stages, which demonstrates a significant role of DWF1 in BR biosynthesis in Arabidopsis plants.
Fig. 5.
Reversal of abnormalities in det2 via application of BRs and overexpression of DWF1. (A) Restoration of shoot growth in det2 mutant seedlings after application of 0.01 µM DS, CS, and BL. Seedlings were grown on vertically oriented MS plates for 7 d in the presence or absence of BRs. Scale bar=1 cm. (B) Restoration of shoot growth in det2 adult plants after application of 0.1 µM DS, CS, and BL. BRs were sprayed 3–5 times a day onto det2 for 27 d at an interval of 3 d. A representative result from 10 individual det2 adult plants is shown in (B). (C–F) Reversal of the det2 abnormal phenotype in det2 35S:DWF1 at the seedling (C), rosette (D), and intact plant (F) stages. Expression levels of DWF1 in each det2 35S:DWF1 transgenic line were confirmed by semi-qRT-PCR (E).
DWF1 expression is down-regulated by BR signaling in Arabidopsis
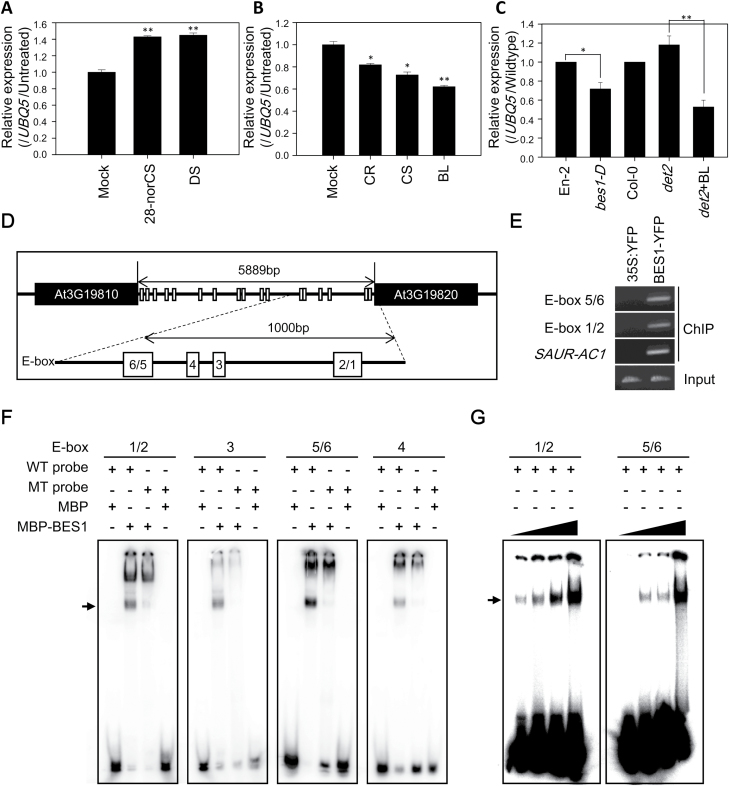
Molecular regulation of DWF1 expression in BR biosynthesis was examined in Arabidopsis. Exogenously applied DS and 28-norCS as a potent biosynthetic precursor of DS increased DWF1 expression (Fig. 6A), implying that DWF1 expression is up-regulated by the substrate and its biosynthetic precursor. In contrast, exogenously applied CR, CS, and BL inhibited expression of DWF1 (Fig. 6B), which suggests that DWF1 expression is down-regulated by the products. In a BR-deficient mutant (det2), expression of DWF1 was increased compared with that in wild-type (Col-0), but this increased DWF1 expression was greatly reduced when BL was applied to det2 (Fig. 6C). Compared with wild-type (En-2), expression of DWF1 was inhibited in the BR-dominant mutant, bes1-D (Fig. 6C), while expression of DWF1 was not changed in the gain-of function mutant of the BZR1, bzr1-1D (Supplementary Fig. S4). These results suggest that feedback regulation of DWF1 by the products mainly occurs through BR signaling via a transcription factor, BES1 in Arabidopsis.
Fig. 6.
Feedback regulation of DWF1 by BR signaling via BES1 in Arabidopsis. (A) Comparison of DWF1 expression by application of 28-norCS and DS. (B) Comparison of DWF1 expression by application of CR, CS, and BL. Seedlings grown in 0.5× MS medium for 7 d were transferred to DW containing 0.1 µM BRs and incubated for 2 h. (C) Expression of DWF1 in BR-deficient (det2) and dominant (bes1-D) mutants. Data are presented as a relative value (mean ±SE) relative to Mock control (A, B) and wild-type (C). Asterisks indicate the statistical significance by Student’s t-test. *P<0.05, **P<0.01 compared with mock or wild-type. (D) Schematic diagram of DWF1 promoter region. White rectangles indicate E-boxes (CANNTG) on the intergenic region. The six E-boxes were denoted as E-box 1 to E-box 6. E-box 1/2 and 5/6 contain two overlapped E-boxes. (E) ChIP assay results for binding of BES1 to DWF1 promoter in vivo (lanes 1 and 2; Input, lanes 3–8; ChIP with anti-YFP antibody prepared from 10-day-old light-grown 35S:YFP (negative control) and BES1-YFP transgenic seedlings). ChIP-PCR was performed with primers from indicated E-box positions at the DWF1 promoter and SAUR-AC1 (positive control). Each assay was repeated three times. (F, G) EMSA results for binding of BES1 to DWF1 promoter in vitro (F). 32P-labeled DWF1 probes were incubated with or without MBP-tagged BES1 protein. The absence or presence of MBP-tagged BES1 protein is indicated by – or +. MBP proteins and mutated probes (MT probes) were used as a negative control (G). 32P-labeled DWF1 probes were incubated at an increasing concentration (0.5, 1, 1.5, and 2 µg) of MBP-tagged BES1 protein.
Seventeen cis-regulatory elements (E-boxes, CANNTG) for BES1 binding sites were located on the intergenic region of DWF1 (at 5889 bp) in the Arabidopsis genome (Fig. 6D). In particular, six E-boxes were present in the region 1000-bp upstream from the start codon of DWF1, a putative promoter region for DWF1. The six E-boxes were denoted E-box 1 to E-box 6, which were respectively nearest to and farthest from the start codon of DWF1. Using BES1–YFP transgenic plants, a ChIP assay was carried out. As shown in Fig. 6E, BES1–YFP proteins successfully bind to both E-box 1/2 and E-box 5/6 in Arabidopsis plants (Supplementary Fig. S2B). Maltose binding protein (MBP)–BES1 was obtained from E. coli using a construct in which MBP was fused to the N-terminus of full-length BES1, and an EMSA was also conducted. As shown in Fig. 6F, MBP, used as a negative control, was not bound to six E-boxes, but MBP–BES1 was directly bound to the E-boxes (Fig. 6F; Supplementary Fig. S2C). Two overlapped probes in particular containing E-box 1/2 and E-box 5/6 were strongly bound to MBP–BES1 in a concentration-dependent manner (Fig. 6G; Supplementary Fig. S2D). The binding of MBP–BES1 was diminished by mutation of E-boxes (CANNTG to AAAAAA) and excess unlabeled probes (Supplementary Figs S2E and S5). These results demonstrate that BES1 binds to the E-boxes on the promoter region of DWF1 both in vitro and in vivo.
Discussion
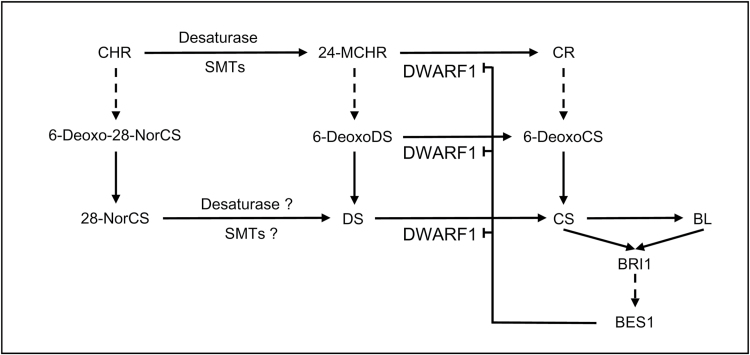
In many plants, the biosynthetic end products of C27-, C28-, and C29-BRs co-exist, suggesting that multiple biosynthetic pathways function within the plants (Fujioka et al., 1997; Yokota, 1997; Sakurai, 1999; Bajguz and Tretyn, 2003; Joo et al., 2009). In tomato plants, endogenous levels of a C27-BR, 28-norCS, are comparable to that of CS (Yokota et al., 1997). In vitro conversion experiments using enzyme solutions prepared from tomato plants revealed that 28-norCS is biosynthesized from cholesterol (CHR) via the same late C-6 oxidation pathway as has been established in CS and BL biosynthesis (Kim et al., 2004b). In Arabidopsis, endogenous 28-norCS can also be biosynthesized from CHR via the late C-6 oxidation pathway. However, the early C-6 oxidation pathway for 28-norCS is interrupted between 6-oxocholestanol (6-oxoCHN) and 28-norTE, as it is intermediated by 28-norCT (Joo et al., 2012). In the biosynthetic pathway of 28-norCS, involvement of DET2, CYP90B1, CYP85A1, and CYP85A2 has been demonstrated. Although the presence of biosynthetic sequences, such as 22-hydroxy-CHR→22-hydroxy-cholesta-4-en-3-one→22-hydroxy-cholesta-3-one→6-deoxo-28-norCT and 3-epi-6-deoxo-28-norCT, and the involvement of CYP90C1 and CYP90D1 for C-23 hydroxylation of 22-hydroxy-cholesta-3-one, 6-deoxo-28-norCT, and 3-epi-6-deoxo-28-norCT to 6-deoxo-28-norTE, 3-dehydro-6-deoxo-28-norTE, and 6-deoxo-28-norTY, respectively, have not yet been demonstrated, these suggest that 28-norCS is biosynthesized through a cholestanol (CHN)-independent pathway via the same biosynthetic reactions that have been established in CN-independent pathways for the biosynthesis of CS and BL in Arabidopsis (Supplementary Fig. S1). Enzyme solutions prepared from both tomato and Arabidopsis successfully mediate conversion of 28-norCS to CS, providing that biosynthesis of 28-norCS, a C27-BR, biosynthesis is an alternative route to produce a biologically active BR, CS, in both plants (Fig. 7, Kim et al., 2004b; Joo et al., 2012).
Fig. 7.
Connections of three BR biosynthetic pathways that function in Arabidopsis. Three biosynthetic pathways are funneled into CS. DWF1 is feedback-regulated through BR signaling via a transcription factor, BES1, in the plant. Solid arrows indicate a single biosynthetic step. Dotted arrows indicate multiple biosynthetic steps. SMT, sterol methyltransferase.
In the current study, we identified 6-deoxoDS in Arabidopsis. Coupled with the presence of DS and 24-MCHR (Fujita et al., 2006), identification of 6-deoxoDS suggests that 24-methylene BR biosynthesis took place in Arabidopsis plants. CYP90B1 can catalyse 22-hydroxylation of 24-MCHR to 22-hydroxy-24-MCHR (Fujita et al., 2006). Previous results from our group demonstrated that C-6 oxidation of 6-deoxoDS to DS is primarily mediated by CYP85A2 (Lee et al., 2010). Although the presence of biosynthetic sequences from 22-hydroxy-24-MCHR to 6-deoxoDS and the involvement of CYP90B1, CYP90C1/D1, and DET2 in 24-methylene BR biosynthesis have not yet been established, the results of this study imply that 24-methylene BRs are biosynthesized via a similar biosynthetic route to that in the biosynthesis of 28-norCS and CS. DL was not found even in large quantities of Arabidopsis. In addition, enzyme prepared from Arabidopsis was unable to catalyse the conversion of DS to DL. Therefore, the end product of 24-methylene BR biosynthesis in Arabidopsis plants seems to be DS rather than DL.
The crude enzyme solution prepared from P. vulgaris and Arabidopsis successfully mediated conversion of 6-deoxoDS to CS via DS. In the study where we demonstrated this conversion, we also demonstrated conversion of 6-deoxoDS to CS via 6-deoxoCS in Arabidopsis plants. This evidence suggests that biosynthesis of 24-methylene BRs and 24-methyl BRs involves a connection between 6-deoxoDS and 6-deoxoCS and between DS and CS in Arabidopsis. We recently demonstrated in Arabidopsis that biosynthetic precursors for 28-norCS such as 28-norTE, 28-nor-3-dehydroTE, and 28-norTY can be converted into biosynthetic precursors for CS such as TE, 3-dehydroTE and TY, respectively (Joo et al., 2012). Therefore, the biosynthetic pathways for 28-norCS, DS and CS are likely to be connected in Arabidopsis not only between end products, but also between their biosynthetic precursors. With complicated connections, three biosynthetic pathways for BRs are funneled into CS to maintain biologically active BRs, CS and BL in plants (Fig. 7).
In Arabidopsis, dwf1, diminuto (dim), and cabbage1 (cbb1) alleles are associated with typical BR-deficient phenotypes such as abnormal root and leaf development, reduced stem elongation (dwarfism), failed stamen formation, reduced silique and seed formation, and delayed senescence. All mutants exhibit accumulation of side chain unsaturated phytosterols at C-24, such as 24-MCHR and isofucosterol, whereas they show reductions in C-24 saturated phytosterols, such as CR and sitosterol (Takahashi et al., 1995, Klahre et al., 1998, Choe et al., 1999). Similarly, a pea lkb mutant shows the same phenotypes as the BR-deficient mutant, while 24-MCHR and isofucosterol accumulate to levels comparable to those in dim, suggesting that the LKB gene is an ortholog of DWF1 (Nomura et al., 1997; Nomura et al., 1999). Accompanied by feeding experiments using isotope-labeled substrates, these results suggest that in plants, DWF1 catalyses conversion of 24-MCHR and isofucosterol to CR and sitosterol, respectively, as a sterol C-24 reductase (Klahre et al., 1998; Choe et al., 1999). Nevertheless, exogenous application of CR cannot reverse abnormalities in dwf1 to their original states in wild-type plants, while the application of CS, BL, and their biosynthetic precursors can reverse these abnormalities (Klahre et al., 1998). Although the reason why exogenously applied CR could not restore dwf1 phenotype is still unknown, the conversion of campesterol to CS and BL seems to be inefficient or ineffective for rescuing the dwf1 phenotype. In this study, we also found that abnormalities in dwf1 cannot be rescued by CR, but this can be achieved by CS and BL (Fig. 4). Therefore, the phenotype found in dwf1 is likely to originate from deficiency of active BRs and not from phytosterol deficiency in Arabidopsis.
Based on GC-SIM quantification with 30 kg of Arabidopsis, endogenous amounts of DS were calculated to be approximately 0.01 ng g−1 fresh weight in Arabidopsis (Table 1). When 50 g of Arabidopsis was used for quantitative analysis, endogenous levels of DS were not detectable on GC-SIM analysis, which resulted in a lack of DS in wild-type plants and in 35S:DWF1 mutants (Fig. 2C). However, approximately 0.05 ng g−1 fresh weight of DS was detected in the dwf1 mutant. In contrast, an endogenous amount of CS at 0.31 ng g−1 fresh weight in wild-type was reduced to 0.22 ng g−1 fresh weight in the dwf1 mutant. Therefore, defects in DWF1 caused accumulation of DS and reduction of CS via blockage of conversion of DS to CS in the dwf1 mutant. We previously reported that an approximately 20% reduction in CS can cause severe abnormalities in cyp85a2 (Kim et al., 2005b), suggesting that the abnormal phenotype in dwf1 is caused by a 29% reduction in CS in the mutant. In 35S:DWF1, DS was not detected by GC-SIM analysis, but endogenous levels of CS in 35S:DWF1 were slightly increased at 0.34 ng g−1 fresh weight. This may have enhanced the formation of BL, the most biologically active BR in Arabidopsis, resulting in slightly increased stem elongation, silique formation, and altered root development in the 35S:DWF1 mutant. Collectively, changes in endogenous CS level seem to induce phenotypic alternations in dwf1 and 35S:DWF1, which suggests that 24-methylene BR biosynthesis to generate CS catalysed by DWF1 is required for homeostasis of active BRs to control the growth and development of Arabidopsis plants.
In Arabidopsis, endogenous CS is extremely reduced (to <10%) in BR-deficient dwarf mutants such as dwf4, det2, and cyp85a1/cyp85a2 (Fujioka et al., 1997; Choe et al., 2001; Kim et al., 2005b). In contrast, sterol-deficient dwarf mutants such as smt1/2, dwf7, and dwf5 contain a fair amount (>50%) of CS, suggesting that the biosynthetic pathway to produce CS may be fairly operant with small amounts of sterols in sterol-deficient mutants (Choe et al., 1999, 2000, 2001; Diener et al., 2000; Carland et al., 2010). In this study, we determined that a small amount of CR can be generated in dwf1 in spite of DWF1 deficiency (Table 3). We also found that expression of biosynthetic genes for BRs such as DWF4, CPD, and CYP85A2 is clearly up-regulated in dwf1 (Supplementary Fig. S6). Therefore, endogenous CS (79% of CS in wild-type) can be effectively synthesized from a low level of CR by the activated biosynthesis of BRs in dwf1.
BR-deficient dwarf2 (brd2), found in rice, is a homolog of Arabidopsis dwf1 (Hong et al., 2005). Like dwf1, brd2 leads to accumulated 24-MCHR and DS, but reduced CR and CS. Exogenous application of DS to brd2 exhibits activity similar to CS: sheath elongation, inhibited root growth, and inclined lamina, suggesting that DS is an alternative bioactive BR in rice. In this study, we also examined the extent to which application of DS, CS, and BL led to restoration in dwf1 mutants. As shown in Fig. 4, application of DS led to little reversal of dwf1 abnormalities. On the other hand, applied CS and BL led to complete reversal of abnormalities in Arabidopsis. This indicates that DS is not a biologically active BR, but rather a biosynthetic precursor to the generation of bioactive BRs, CS and BL in Arabidopsis.
DWF1 is a flavin adenine dinucleotide (FAD)-dependent oxidoreductase (Choe et al., 1999). In CR biosynthesis, DWF1 exhibits enzyme activity for C-24 isomerization and for reduction of 24-MCHR (Klahre et al., 1998). We previously proposed that C-24 methylation of 28-norCS to CS occurs via three steps: (i) desaturation of 28-norCS to form Δ24-28-norCS by a desaturase, (ii) S-adenosylmethionine-dependent methylation of Δ24-28-norCS to form DS by sterol methyltransferase1 (SMT1), and (iii) reduction of DS by NADPH to form CS by DWF1 (Joo et al., 2012). In this study, we concretely demonstrate that the reduction of DS to CS is catalysed by DWF1. This reduction consists of isomerization and reduction, implying that DWF1 has both isomerase and reductase activities in the reduction in Arabidopsis. Successful conversion of 28-norCS to CS via DS indicates that DS can be biosynthesized from not only 24-MCHR, but also CHR via 28-norCS in Arabidopsis. Nevertheless, endogenous levels of DS are quite low in Arabidopsis, suggesting that DWF1 enzyme activity in the reduction of DS to CS can be high in plants. Expression of DWF1 was enhanced by 28-norCS and DS (Fig. 6A). This feed-forward activation of DWF1 by each 28-norCS and DS seems to be another reason that endogenous levels of DS are low in Arabidopsis.
Exogenously applied, biologically active BRs, primarily CS and BL, down-regulate expression of BR biosynthetic genes, but up-regulate BR signaling genes in Arabidopsis (Goda et al., 2002; Tanaka et al., 2005; Gruszka, 2013). This suggests that increased levels of active BRs trigger enhanced BR signaling towards down-regulation of biosynthetic genes in plants (Tanaka et al., 2005). In the current study, we found DWF1 expression to be down-regulated by active BRs (CS and BL) in Arabidopsis (Fig. 6B). In addition, application of BL led to decreased DWF1 expression in det2, such that expression became much lower than that of the wild-type state (Fig. 6C). These findings indicate that DWF1 expression was negatively regulated by endogenous levels of active BRs. DWF1 expression was also down-regulated by CR. However, the down-regulation of DWF1 by CR was weaker than that by CS and BL, suggesting that feedback regulation of DWF1 is more effective in downstream BR biosynthesis rather than in upstream phytosterol biosynthesis with regard to maintaining physiological levels of active BRs in Arabidopsis.
In bes1-D, DWF1 was down-regulated (Fig. 6C), implying that regulation of DWF1 may have occurred through feedback in BR-induced signal transduction pathways in Arabidopsis (Fig. 7). In the intergenic region of DWF1, 17 E-boxes for BES1 binding sites were present, but no BR response element (BRRE) sequence for the BZR1 binding site were found. Furthermore, DWF1 was not down-regulated in bzr1-1D (Supplementary Fig. S4). Although BZR1 can bind to E-boxes, this suggests that BES1 is a BR transcription factor that regulates DWF1 transcription in plants. In fact, EMSA and ChIP showed direct binding of BES1 to E-boxes (Fig. 6E, F), more strongly to two overlapped E-boxes, 1/2 and 5/6, both located on the potent DWF1 promoter region (Fig. 6F, G). Therefore, it is likely that down-regulation of DWF1 occurs via direct binding of BES1 to the DWF1 promoter in Arabidopsis. Feedback regulation of BR biosynthetic genes occurs mainly by BZR1 (Wang et al., 2002, He et al., 2005). From this viewpoint, this study provides a good example of BR biosynthetic genes also acting in feedback regulated by BES1 to maintain appropriate concentrations of endogenous BRs in Arabidopsis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. CN-independent (A) and CHN-independent (B) pathways for biosynthesis of BRs in Arabidopsis.
Fig. S2. Original images for all of the gels and blots presented in this report. (A) Fig. 2B, (B) Fig. 6E, (C) Fig. 6F, (D) Fig. 6G, (E) Supplementary Fig. S5.
Fig. S3. Hypocotyl length assay in 5-day-old seedlings of dwf1 (A) and det2 (B) mutant after application of 0.1 µM CR, DS, CS, and BL.
Fig. S4. Expression level of DWF1 and DWF4 in BR-dominant mutants bes1-D and bzr1-1D.
Fig. S5. Competitive EMSA results for binding of MBP-BES1 to DWF1 promoter in vitro.
Fig. S6. Alternation of expression of BR biosynthetic genes in dwf1 and wild-type (Col-0).
Table S1. Sequences of DNA probes and primers used in this study.
Acknowledgements
This work was supported by grants from Next-Generation BioGreen 21 Program (PJ01178601 to JHY, PJ01114901 to TWK and PJ0132082018 to SKK), in Korea.
Glossary
Abbreviations:
- BL
brassinolide
- BMB
bismethaneboronate
- BR
brassinosteroid
- CHN
cholestanol
- CHR
cholesterol
- CN
campestanol
- CR
campesterol
- CS
castasterone
- CT
cathasterone
- CYP
cytochrome P450
- DL
dolicholide
- DS
dolichosterone
- DT
dehydroteasterone
- 24-MCHR
24-methylenecholesterol
- TE
teasterone
- TY
typhasterol.
References
- Baba J, Yokota T, Takahashi N. 1983. Brassinolide-related new bioactive steroids from Dolichos lablab seed. Agricultural and Biological Chemistry 47, 659–661. [Google Scholar]
- Bajguz A, Tretyn A. 2003. The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62, 1027–1046. [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y. 1999. The tomato dwarf enzyme castalyses C-6 oxidation in brassinosteroid biosynthesis. Proceedings of the National Academy of Sciences, USA 96, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T. 2001. Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant & Cell Physiology 42, 114–120. [DOI] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Nelson T. 2010. The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiology 153, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S. 2004. Brassinosteroid biosynthesis and metabolism. In: Davies PJ, ed. Plant hormones: biosynthesis, signal transduction, action! Dordrecht: Kluwer Academic Publishers, 156–178. [Google Scholar]
- Choe S, Dilkes BP, Gregory BD et al. . 1999. The Arabidopsis dwf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiology 119, 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. 2001. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. The Plant Journal 26, 573–582. [DOI] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. 2000. Lesions in the sterol delta reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. The Plant Journal 21, 431–443. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. 1998. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annual Review of Plant Physiology and Plant Molecular Biology 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. 2000. Sterol methyltransferase 1 controls the level of cholesterol in plants. The Plant Cell 12, 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S. 1999. Natural occurrence of brassinosteroids in the plant kingdom. In: Sakurai A, Yokota T, Clouse SD, eds. Brassinosteroids: Steroidal plant hormones. Tokyo: Springer-Verlag, 21–45. [Google Scholar]
- Fujioka S, Inoue T, Takatsuto S, Yanagisawa T, Yokota T, Sakurai A. 1995. Identification of a new brassinosteroid, cathasterone, in cultured cells of Catharanthus roseus as a biosynthetic precursor of teasterone. Bioscience, Biotechnology, and Biochemistry 59, 1543–1547. [Google Scholar]
- Fujioka S, Li J, Choi YH et al. . 1997. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. The Plant Cell 9, 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Watanabe T, Takatsuto S, Yoshida S. 2000. Biosynthesis of brassinosteroids in cultured cells of Catharanthus roseus. Phytochemistry 53, 549–553. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yokota T. 2003. Biosynthesis and metabolism of brassinosteroids. Annual Review of Plant Biology 54, 137–164. [DOI] [PubMed] [Google Scholar]
- Fujita S, Ohnishi T, Watanabe B, Yokota T, Takatsuto S, Fujioka S, Yoshida S, Sakata K, Mizutani M. 2006. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. The Plant Journal 45, 765–774. [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. 2002. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiology 130, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka D. 2013. The brassinosteroid signaling pathway—new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. International Journal of Molecular Sciences 14, 8740–8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. 2005. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. 2005. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. The Plant Cell 17, 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Joo SH, Park CH, Lee SC, Kim SK. 2009. Substrate specificity for cytochrome P450 85A1 and 85A2 in brassinosteroids biosynthesis. Bulletin of the Korean Chemical Society 30, 293–294. [Google Scholar]
- Joo SH, Hwang JY, Park CH, Lee SC, Kim SK. 2009. Biosynthetic connection of 24-methylene- and 24-methyl-brassinosteroids in Phaseolus vulgaris. Bulletin of the Korean Chemical Society 30, 502–504. [Google Scholar]
- Joo SH, Kim TW, Son SH, Lee WS, Yokota T, Kim SK. 2012. Biosynthesis of a cholesterol-derived brassinosteroid, 28-norcastasterone, in Arabidopsis thaliana. Journal of Experimental Botany 63, 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo SH, Yun HS, Kim TW, Kim YS, Kim SK. 2002. Identification and transformation of campestanol in cultured cells of Phaseolus vulgaris.Bulletin of the Korean Chemical Society 23, 1035–1038. [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H. 2005a CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. The Plant Journal 41, 710–721. [DOI] [PubMed] [Google Scholar]
- Kim TW, Chang SC, Lee JS, Hwang B, Takatsuto S, Yokota T, Kim SK. 2004a Cytochrome P450-catalyzed brassinosteroid pathway activation through synthesis of castasterone and brassinolide in Phaseolus vulgaris. Phytochemistry 65, 679–689. [DOI] [PubMed] [Google Scholar]
- Kim TW, Chang SC, Lee JS, Takatsuto S, Yokota T, Kim SK. 2004b Novel biosynthetic pathway of castasterone from cholesterol in tomato. Plant Physiology 135, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK. 2005b Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. The Plant Cell 17, 2397–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Park HH, Kim SK. 2004c Cell-free conversion of castasterone in cultured cells of Phaseolus vulgaris and Marchantia polymorpha. Bulletin of the Korean Chemical Society 25, 955–956. [Google Scholar]
- Kim TW, Park SH, Han KS, Choo J, Lee JS, Hwang S, Kim SK. 2000. Occurrence of teasterone and typhasterol, and their enzymatic conversion in Phaseolus vulgaris. Bulletin of the Korean Chemical Society 21, 373–374. [Google Scholar]
- Kim TW, Park SH, Joo SH, Kim YS, Choo J, Kim SK. 2001. Metabolism of typhasterol, a brassinosteroid, in suspension cultured cells of Marchantia polymorpha. Bulletin of the Korean Chemical Society 22, 651–654. [Google Scholar]
- Kim TW, Wang ZY. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annual Review of Plant Biology 61, 681–704. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim TW, Kim SK. 2005c Brassinosteroids are inherently biosynthesized in the primary roots of maize, Zea mays L. Phytochemistry 66, 1000–1006. [DOI] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. 1998. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. The Plant Cell 10, 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. 2003. Brassinosteroid-mediated stress responses. Journal of Plant Growth Regulation 22, 289–297. [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang JY, Joo SH, Son SH, Youn JH, Kim SK. 2010. Biosynthesis and metabolism of dolichosterone in Arabidopsis thaliana. Bulletin of the Korean Chemical Society 31, 3475–3478. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔC(T) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Yoshida S, Feldmann KA. 2000. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiology 124, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T. 1999. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of Pea. Plant Physiology 119, 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. 1997. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in Pisum sativum.Plant Physiology 113, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Godza B, Watanabe B, Fujioka S, Hategan L, Ide K, Shibata K, Yokota T, Szekeres M, Mizutani M. 2012. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. The Journal of Biological Chemistry 287, 31551–31560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari AM, Watanabe B et al. . 2006. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. The Plant Cell 18, 3275–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A. 1999. Brassinosteroid biosynthesis. Plant Physiology and Biochemistry 37, 351–361. [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M. 2009. Plant hormones are versatile chemical regulators of plant growth. Nature Chemical Biology 5, 301–307. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A. 1993. Biosynthesis of brassinolide from castasterone in cultured cells of Catharanthus roseus. Journal of Plant Growth Regulation 12, 101–106. [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH. 1995. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes & Development 9, 97–107. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. 2005. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiology 138, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J et al. . 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Wolters H, Jürgens G. 2009. Survival of the flexible: hormonal growth control and adaptation in plant development. Nature Reviews. Genetics 10, 305–317. [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P et al. . 2008. Brassinosteroids regulate grain filling in rice. The Plant Cell 20, 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T. 1997. The structure, biosynthesis and function of brassinosteroids. Trends in Plant Science 2, 137–143. [Google Scholar]
- Yokota T, Nomura T, Nakayama M. 1997. Identification of brassinosteroids that appear to be derived from campesterol and cholesterol in tomato shoots. Plant Cell Physiology 38, 1291–1294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.