Rice exocyst subunit OsSEC3A interacts with OsSNAP32, a SNAP25-type SNARE protein, and disruption of OsSEC3A causes a lesion-mimic phenotype and enhanced defense response
Keywords: Defense responses, exocyst complex, lesion-mimic phenotype, Oryza sativa, pathogenesis, salicylic acid, SEC3A
Abstract
The exocyst, an evolutionarily conserved octameric protein complex involved in exocytosis, has been reported to be involved in diverse aspects of morphogenesis in Arabidopsis. However, the molecular functions of such exocytotic molecules in rice are poorly understood. Here, we examined the molecular function of OsSEC3A, an important subunit of the exocyst complex in rice. The OsSEC3A gene is expressed in various organs, and OsSEC3A has the potential ability to participate in the exocyst complex by interacting with several other exocyst subunits. Disruption of OsSEC3A by CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) caused dwarf stature and a lesion-mimic phenotype. The Ossec3a mutant exhibited enhanced defense responses, as shown by up-regulated transcript levels of pathogenesis- and salicylic acid synthesis-related genes, increased levels of salicylic acid, and enhanced resistance to the fungal pathogen Magnaporthe oryzae. Subcellular localization analysis demonstrated that OsSEC3A has a punctate distribution with the plasma membrane. In addition, OsSEC3A interacted with rice SNAP25-type t-SNARE protein OsSNAP32, which is involved in rice blast resistance, via the C-terminus and bound to phosphatidylinositol lipids, particularly phosphatidylinositol-3-phosphate, through its N-terminus. These findings uncover the novel function of rice exocyst subunit SEC3 in defense responses.
Introduction
Upon attacks by pathogens, host plant cells activate a wide range of defense responses. Endocytic and exocytic vesicle trafficking is required for diverse aspects of defense responses ranging from regulation of early signaling to deployment of immune responses at later stages (Robatzek, 2007; Žárský et al., 2013; Inada and Ueda, 2014). Recently, plant soluble SNARE complexes, which are involved in vesicle trafficking during exocytosis, were shown to be involved in immune signaling. HvSNAP34, a SNAP25 (soluble N-ethylmaleimide-sensitive factor attachment protein 25)-type SNARE protein in barley, inhibits pathogen penetration into host cells and is required for resistance to powdery mildew (Collins et al., 2003). In rice, the SNAP25-type SNARE protein OsSNAP32 is involved in rice blast resistance (Luo et al., 2016). PEN1/SYP121, a Qa-SNARE protein in Arabidopsis, is believed to function by forming a ternary SNARE complex with SNAP33, vesicle-associated membrane protein (VAMP) 721, and VAMP722 (Kwon et al., 2008).
Before SNARE-mediated membrane fusion, the exocyst mediates tethering of vesicles to the plasma membrane in the early stage of exocytosis (Guo et al., 2000). The exocyst, an evolutionarily conserved protein complex composed of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84, was initially discovered in yeast (TerBush et al., 1996; Liu and Guo, 2012). Yeast Sec3 localized to the bud tip independent of the actin cytoskeleton is a landmark protein of polarized exocytosis that is involved in recruitment of the other exocyst subunits residing in the cytoplasm to the plasma membrane (Finger et al., 1998; Wiederkehr et al., 2003). The polarized localization of Sec3 to the plasma membrane is mediated by its interaction with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] in the inner leaflet of the plasma membrane in yeast and mammalian cells (He et al., 2007; Liu et al., 2007; Zhang et al., 2008). In yeast, Sec3 is a downstream effector of Cdc42 and Rho; interaction of Sec3 with Cdc42 and Rho is important for its function in exocytosis and cell polarity (Guo et al., 2001; Zhang et al., 2008).
In terrestrial plants, the exocyst exists as a subcomplex that primarily functions in regulating polarized exocytosis during cell expansion (Žárský et al., 2009). While budding yeast, fungi, and most animals have only a single copy of each exocyst subunit, expansion of exocyst subunit genes, especially Exo70 genes, in plants has resulted in the presence of several copies of some exocyst subunits. For example, Arabidopsis has 23 Exo70 genes, whereas rice (Oryza sativa) has 47 Exo70 genes; the functions of these copies are largely unknown (Chong et al., 2010). Corresponding to the complexity of plant exocyst subunits, orthologs of exocyst subunits display varying specialized developmental functions. Sec5, Sec6, Sec8, and Sec15 subunits encoded by one or two copies of each gene function in pollen germination, pollen tube growth (Hála et al., 2008), and secretory processes during cytokinesis (Fendrych et al., 2010). Exo70A1 in Arabidopsis has been well characterized and functions in specialized developmental processes (Synek et al., 2006; Samuel et al., 2009; Fendrych et al., 2010; Kulich et al., 2010; Drdova et al., 2013). Few rice exocyst subunits have been studied in detail, although rice exocyst subunit OsEXO70A1 is known to be required for normal vascular bundle differentiation and primary mineral nutrient assimilation (Tu et al., 2015).
Recent reports demonstrated that some plant EXO70 proteins are involved in plant immunity. For example, Arabidopsis Exo70B2 and Exo70H1 are up-regulated in response to pathogens and necessary for immune responses (Pečenková et al., 2011). EXO70B2 is a target of plant U-box-type ubiquitin ligase 22 and negatively regulates PAMP (pathogen-associated molecular pattern)-triggered responses (Stegmann et al., 2012). Mutation of EXO70B1 leads to activation of a defense pathway with an intracellular immune system receptor TIR-NBS2 (Zhao et al., 2015). In addition, EXO70B1 is involved in autophagy-related membrane trafficking to vacuoles (Kulich et al., 2013). In Nicotiana benthamiana, exocyst subunit SEC5 is required for secretion of pathogenesis-related protein PR-1 and plays a role in CRN2-induced cell death (Du et al., 2015). Although little is known about the function of the exocyst complex in rice, OsExo70F3 was reported to be involved in AVR-Pii-triggered immunity to Magnaporthe oryzae by forming an AVR-Pii–OsExo70F2/3 complex (Fujisaki et al., 2015).
The role of exocyst subunit SEC3 in plants has been widely investigated. This subunit is required for root hair elongation, embryogenesis, and pollen germination (Elias et al., 2003; Wen et al., 2005; Zhang et al., 2013; Bloch et al., 2016). However, the function of SEC3 in rice has not been elucidated. Although SEC3 has been reported to have molecular characteristics similar to those of EXO70, namely anchoring the exocyst to the plasma membrane (Wiederkehr et al., 2003), there is no evidence linking SEC3 with plant immunity. In this study, we confirmed the ability of OsSEC3A to participate in the exocyst complex and obtained OsSEC3A mutants (Ossec3a) using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) technology. Our results showed that lesion spots that resembled the phenotype of lesion-mimic mutants (LMMs) appeared in Ossec3a, along with induced cell death and defense responses. As clues to the mechanism underlying the observed defense responses, we show that OsSEC3A interacted with rice SNAP25-type SNARE protein OsSNAP32 and phosphatidylinositol-3-phosphate [PI(3)P], which are involved in plant disease resistance, through the C-terminus and N-terminus, respectively. These results provide new insight into our understanding of SEC3 in the context of its participation in plant defense, which is a role that has not been identified in previous studies.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa L. japonica var. Kitaake) and N. benthamiana plants were used in this study. All rice plants used in this study were grown in paddy fields of the Institute of Crop Science in Beijing, paddy fields of the Institute of Crop Science in Sanya (China), or a greenhouse at 25–28 °C with 16 h of light and 8 h of darkness every day.
Sequence analysis and prediction of protein structure
Sequence analysis was performed using SMART (simple modular architecture research tool) (http://smart.embl-heidelberg.de/). The protein sequences used for the phylogenetic analysis and sequence alignment were obtained from the NCBI (http://www.ncbi.nlm.nih.gov/pmc/) and assessed using BLASTp software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was conducted with the DNAMAN software package (Lynnon BioSoft, Canada).
Gene expression analysis
RNA extraction and quantitative reverse transcription–PCRs (qRT–PCRs) were performed as previously described (Ma et al., 2016). mRNA was prepared using RNeasy plant mini kits (Qiagen, http://www.qiagen.com/). A 1 µg aliquot of mRNA was reverse-transcribed to cDNA using a QuantiTect reverse transcription kit (Qiagen, German). All qRT–PCR assays were carried out using the SYBR Premix Ex TaqKit (TaKaRa, Japan) with a 7900HT Real-time PCR System (Applied Biosystems, USA). Expression levels were calculated in three technical replicates consisting of two biological replicates each, with the rice ubiquitin gene serving as an internal control. The primers used in this study are listed in Supplementary Table S1 at JXB online.
For the β-glucuronidase (GUS) assay, 2.5 kb of the OsSEC3A promoter sequence was amplified using OsSEC3A-Pro-F/R, and the resulting construct was transformed into cv. Kitaake by Agrobacterium tumefaciens. Histochemical GUS staining was performed as previously described (Jefferson, 1987).
Subcellular localization of OsSEC3A in rice roots and N. benthamiana
To obtain green fluorescent protein (GFP) transgenic plants, the OsSEC3A cDNA fragment was amplified using 1305OsSEC3A-GFP-F/R and cloned into the pCAMBIA1305-GFP vector (produced by insertion of the GFP backbone of pAN580 into the pCAMBIA1305 vector at the SacI–SalI sites) to generate the pCAMBIA1305-D35S-OsSEC3A-GFP-NOS vector. This construct was introduced into wild-type and Ossec3a mutant plants to generate transgenic plants. For brefeldin A (BFA) treatment, leaf epidermal cells of N. benthamiana were observed following incubation in 100 µg ml–1 BFA for periods of 20 min and 40 min. For transient expression analysis in N. benthamiana, the binary vector pCAMBIA1305-OsSEC3A-GFP (described above) was introduced into Agrobacterium strain EHA105, which was used to infiltrate N. benthamiana leaves as described previously; N. benthamiana protoplasts were isolated using the same method used for Arabidopsis (Ren et al., 2014). Confocal images were acquired using a Leica TCS SP5 laser scanning confocal microscope.
Transient expression analysis of rice protoplasts
Wild-type OsSEC3A cDNA fragments were amplified with primer pair OsSEC3A-GFP-F/R (Supplementary Table S1) and inserted into pAN transient expression vectors (http://www.bio.utk.edu/cellbiol/markers/vectors.htm) to produce OsSEC3A–GFP fusion proteins. The recombinant plasmid was co-transformed into rice protoplasts with plasma membrane markers. Transient expression using rice protoplasts from rice leaf sheaths was achieved as described previously (Shen et al., 2014). Confocal images were acquired at 12 h after transformation using a Leica TCS SP5 system. For each experiment, >20 individual cells were imaged. For GFP, the excitation maximum was 488 nm, while the emission maximum was 530 nm. For monomeric red fluorescent protein (mRFP), the excitation maximum was 587 nm, while the emission maximum was 610 nm. Sequential imaging of both channels was employed. A single confocal slice was presented.
Vector construction for the CRISPR/Cas9 system
The single guide RNA (sgRNA) design method for the CRISPR/Cas9 system in plants was described previously (Lei et al., 2014). Here, a web application tool, CRISPR-P (http://cbi.hzau.edu.cn/crispr), was used to select sgRNAs targeting the third and 10th exon of the OsSEC3A gene. Plasmid construction was conducted as previously described. The Cas9 destination vector was driven by the maize ubiquitin promoter for expression in rice, and sgRNA expression was driven by the pol III type promoter of U3 sgRNA. Gateway recombination was used to mobilize sgRNA, after which the binary T-DNA vectors for co-expression of Cas9 and sgRNA were transformed into rice calli.
Measurement of salicylic acid
Samples of seedling and leaf sheath tissue weighing ~0.3 g (FW) were prepared. Extraction and quantification of salicylic acid (SA) was performed as described previously, with minor modifications (Chen et al., 1995).
Pathogen infection assay
Magnaportje oryzae isolate HLJ07-45-1-2 (collected from Heilongjiang Province, China) was evaluated for virulence in Kitaake and the Ossec3a mutants. Four-week-old rice seedlings were spray-inoculated following a previously described procedure (Li et al., 1998). The number and size of lesions in ~35 inoculated seedlings from each line were evaluated 7 days post-inoculation (dpi). Some of the seedlings were sampled at 0, 1, 2, 3, 4, and 5 dpi for qPCR. To quantify the disease symptoms, the disease severity, which reflects the percentage of the leaf area exhibiting necrosis/spots, was analyzed. Using ImageJ software, we quantified the disease severity (1–100%) of the first, second, third, and fourth fully expanded leaves for lines #5, #7, and #11. For the visualization of dead cells, the leaves of plants at the tillering stage were observed by trypan blue staining as previously described (Tang et al., 2011). Hydrogen peroxide (H2O2) accumulation was detected by 3,3-diaminobenzidine (DAB) staining. H2O2 content was measured with a Hydrogen Peroxide Assay Kit (Beyotime, S0038) according to the manufacturer’s protocol.
Assays for protein–protein interactions
For the yeast two-hybrid (Y2H) assay, the coding regions of OsSEC3A and various domain deletion variants of OsSEC3A were cloned into Y2H bait vector pGBKT7 (Clontech). The coding regions of other exocyst subunit genes were cloned into prey construct pGADT7 (Clontech) (see Supplementary Table S1 for primer sequences). Bait and prey constructs were co-transformed into Saccharomyces cerevisiae strain AH109. Positive clones grown on SD/-Leu/-Trp plates at 28 °C for 3 d were incubated in liquid media, after which a 10 μl drop of the culture dilution (OD600=0.5 with sterile water) was plated on SD/-Ade/-His/-Leu/-Trp plates to test the interaction between bait and prey.
For the in vitro pull-down assays, the coding regions of OsSEC3A, OsSEC3AC (amino acids 310–888), OsSNAP32, OsExo70A1, and OsExo70B1 were inserted into pMAL-c2X and pGEX4T-1 vectors to generate maltose-binding protein (MBP)–OsSEC3A, MBP–OsSEC3C (amino acids 310–888), glutathione S-trasferase (GST)–OsSNAP32, GST–OsExo70A1, and GST–OsExo70B1 plasmids, respectively (see Supplementary Table S1 for primer sets). Amylose resin (for MBP purification; New England Biolabs) and GST-binding resin (for GST purification; Merck) were used to purify proteins, including fusions and empty tags. The pull-down analyses were performed as previously described (Zhou et al., 2013). Products were detected with an enhanced chemiluminescence reagent (GE Healthcare).
For the luciferase (LUC) reporter assay, the OsSEC3A_C, full-length OsSNAP32, OsSEC5, OsSEC6, and OsSEC15bN sequences were PCR amplified and inserted into 35S:NLuc or 35S:CLuc plasmids to construct LUC vectors as described previously (Chen et al., 2008). These constructs were transiently introduced into N. benthamiana by infiltration with A. tumefaciens strain GV3101. After 3 d, Agrobacterium-infiltrated leaves were placed into a 96-well microtiter plate with a hole punch and kept in the dark for 5 min to allow detection of luminescence. Relative LUC activity was measured with the Dual-Luciferase Reporter Assay System (Promega).
Lipid binding assay
The coding sequence (CDS) of OsSEC3A and OsSEC3AN (amino acids 1–450) were amplified using primers (see Supplementary Table S1) and inserted into vector pMAL-c2X tagged with MBP using an infusion cloning kit (Clontech). Protein expression was induced with 200 µM isopropyl-β-d-thiogalactopyranoside at 16 °C overnight. Amylose affinity chromatography (New England Biolabs) was used to purify MBP-tagged proteins. The PIP strips/arrays (P-6001/ 6100; Echelon Biosciences) were blocked with 1% non-fat dry milk in phosphate-buffered saline (PBS) for 1 h and incubated in 10 μg ml–1 MBP-tagged recombinant protein in blocking buffer for 1 h at room temperature with gentle agitation. The blots were washed three times with PBS plus 0.1% Tween-20 (pH 7.4) and soaked in this buffer supplemented with 1:1000 murine horseradish peroxidase-conjugated anti-MBP antibodies (New England Biolabs) at room temperature for 1 h. After thorough washing, the signals were detected following the ECL Plus immunoblot method. As a negative control, we also incubated the membrane with the MBP tag only, and proceeded with the same experimental protocol.
Results
SEC3A is an active ortholog in rice
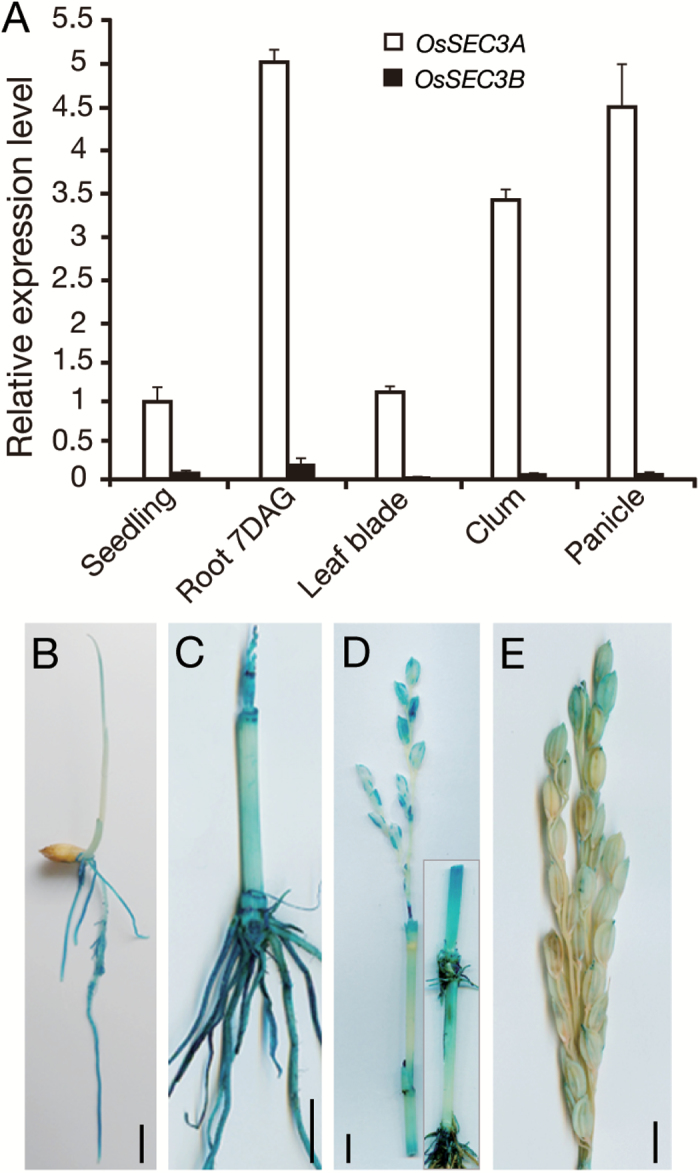
Rice exocyst subunit SEC3 has two orthologous genes, OsSEC3A (LOC_Os03g42750) and OsSEC3B (LOC_Os11g17600), which share 64% sequence identity at the protein level (Cvrčková et al., 2012). The expression patterns of OsSEC3A and OsSEC3B were determined by qRT–PCR. OsSEC3A was expressed in various organs, including the roots, culms, panicles, and leaves, with the highest expression in the seedling roots, whereas almost no OsSEC3B transcript was detected (Fig. 1A). GUS expression driven by the OsSEC3A promoter confirmed the constitutive expression pattern detected by qRT–PCR and further indicated that GUS staining was higher in young roots (Fig. 1B, C), shoot apices (Fig. 1C), the basal parts of internodes (divisional and elongating zones) originating from the intercalary meristem (Fig. 1D), and young glumes (Fig. 1D, E). The broadly high expression of OsSEC3A in organs examined in our analysis indicated that OsSEC3A is the major SEC3 paralog in rice. Next, we focused our work mainly on rice exocyst subunit OsSEC3A.
Fig. 1.
Expression analysis of OsSEC3A. (A) qRT–PCR expression patterns of OsSEC3A and OsSEC3B in various organs. Values are the means ±SE of three independent experiments. (B–E) Histochemical staining of young seedlings 4 d after germination (B), at 4 weeks (C), 1 week before heading (D), and at the heading stage (E). The image inserted in (D) is another part of the same transgenic plant. Scale bars=1 cm in (B)–(E).
OsSEC3A is a member of the exocyst complex
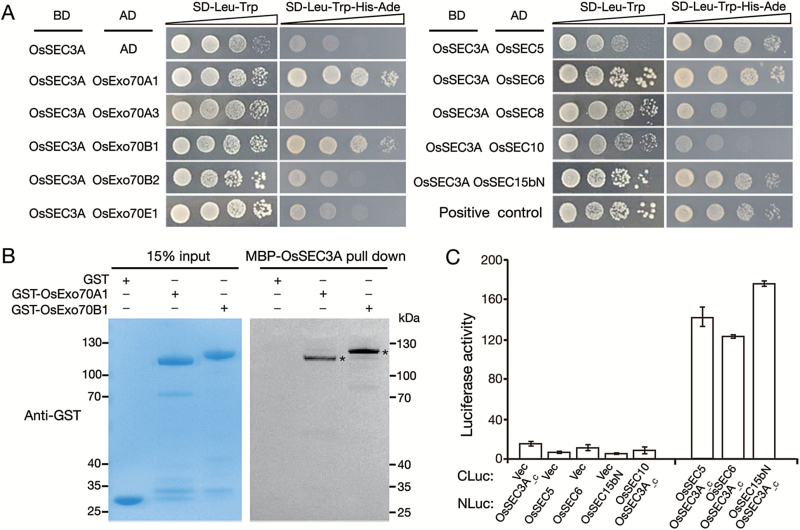
To gain further insight into whether exocyst subunit OsSEC3A functions as a component of the exocyst complex, we performed Y2H assays with a group of subunits that do not display auto-activation (Supplementary Fig. S1). OsSEC3A interacted with OsExo70A1 and OsExo70B1. There was no interaction between OsSEC3A and OsExo70A3, OsExo70B2, or OsExo70E1 (Fig. 2A). In addition, our in vitro pull-down assay verified that MBP–OsSEC3A pulled down GST–OsExo70A1 and GST–OsExo70B1 fusion proteins, but not GST itself (Fig. 2B). For other exocyst subunits, OsSEC3A interacted with OsSEC5, OsSEC6, and OsSEC15bN in Y2H assays (Fig. 2A). We further used a split LUC complementation assay based on firefly LUC (Chen et al., 2008) to confirm this interaction. Consistently, enhanced LUC activity was detected in N. benthamiana leaves (Fig. 2C). Taken together, these findings show that OsSEC3A might have the ability to participate in exocyst complexes.
Fig. 2.
Interaction of OsSEC3A with other exocyst subunits. (A) Y2H assay in S. cerevisiae. Full-length OsSEC3A was ligated into pGBKT7 to generate the OsSEC3A-BD plasmid. Several exocyst constructs were fused with the activation doamin (AD), displaying no auto-activation. (B) In vitro pull-down assay of recombinant GST–OsExo70A1, GST–OsExo70B1, and GST using resins containing MBP–OsSEC3A. Asterisks indicate GST–OsExo70A1 and GST–OsExo70B1. (C) Quantification of luciferase activity in N. benthamiana leaves expressing: OsSEC3A_C (amino acids 521–888)-NLuc and CLuc-OsSEC5; OsSEC3A_C-NLuc and CLuc-OsSEC10; OsSEC3A_C-NLuc and CLuc-OsSEC6; or OsSEC3A_C-NLuc and CLuc-OsSEC15BN. The data shown are representative of three independent experiments.
Mutation of OsSEC3A causes dwarfism and lesion spots
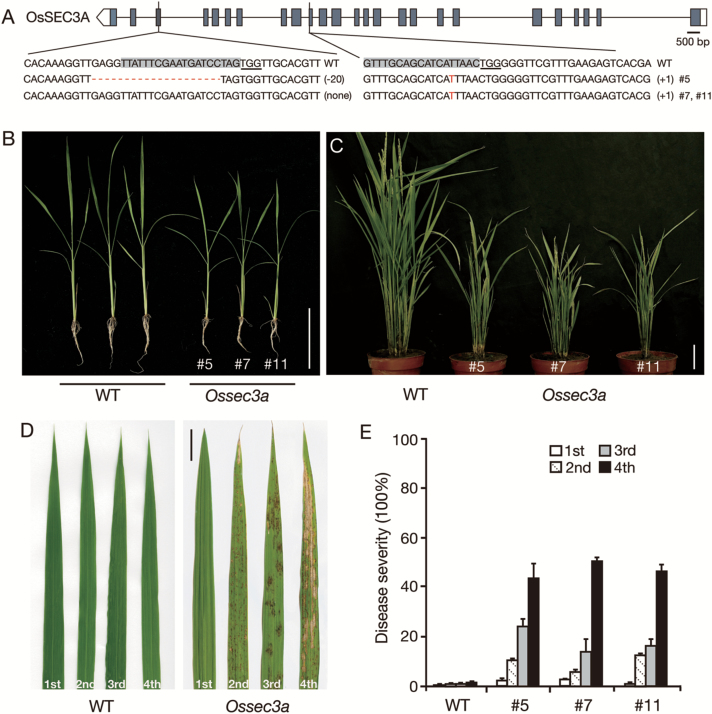
To decipher the functional role of OsSEC3A in plant development, CRISPR/Cas9 technology, the application of which has been demonstrated to achieve efficient targeted mutagenesis in transgenic rice (Miao et al., 2013), was used to disrupt specifically the OsSEC3A gene. To enhance mutation efficiency, we designed sgRNAs targeting the third and 10th exon of the OsSEC3A gene and transformed them into O. japonica var. Kitaake simultaneously (see the Materials and methods). Twenty independent transgenic lines for the sgRNA construct were obtained, after which DNA sequencing analysis of these lines was conducted using gene-specific primers. The sequencing analysis showed either loss of peak/gain of peak or overlapping peaks around the target site in the sequencing chromatograms of five lines: #2, #5, #7, #11, and #18 (Fig. 3A; Supplementary Fig. S2), which confirmed mutations in the OsSEC3A gene. In addition, qRT–PCR analyses showed severely reduced abundance of OsSEC3A as expected (Supplementary Fig. S3). The occurrence of the detected mutation upstream of the protospacer adjacent motif (PAM) indicated that the CRISPR/Cas9 system cleaved the target sequence.
Fig. 3.
Phenotypes of field-grown wild-type (WT) and Ossec3a mutant plants. (A) Targeted mutagenesis of the OsSEC3A gene. The positions of target sites are shown on the gene structure. The protospacer adjacent motif (PAM) sequence is underlined, while the target sequence is shaded in gray. The sequencing results of mutant alleles are aligned to the reference genome sequence. The indels are shown in red letters or dashes. The net change in length is noted to the right of each sequence (+, insertion; –, deletion). (B and C) Phenotype of WT and Ossec3a mutant plants at the seedling (B) and heading stage (C). (D) Phenotype of the first, second, third, and fourth fully expanded leaves from the tip to the base of the main tiller of plants at the heading stage. (E) Quantification of disease symptoms for leaves of different ages. Disease severity reflects the percentage of the leaf area exhibiting necrosis/spots. Scale bars=10 cm in (B, C) and 5 cm in (D).
Detailed phenotype analysis showed that each independent transgenic line (lines #5, #7, and #11) exhibited similarly altered growth in comparison with that of wild-type plants (Fig. 3). The progeny of transgenic line #5 were selected for the following studies and designated Ossec3a. In the seedling stage, Ossec3a plants were smaller than wild-type seedlings (Fig. 3B). The length of the main roots of Ossec3a plants was reduced in comparison with those of wild-type plants (Supplementary Fig. S4A). Lesions appeared on Ossec3a leaf blades and leaf sheaths from the tillering stage to the ripening stage (Fig 3C; Supplementary Fig. S4B). The disease severity of the lesions formed on Ossec3a mutants appeared to follow a developmental pattern and was correlated with the age of the affected leaf blades. No spots were present on newly emerging flag leaves, whereas the second leaf blades from the top of each main tiller had larger and more numerous spots, while the third and fourth leaves showed some withering (Fig 3C–E). However, the corresponding leaf blades of wild-type plants showed no spots in all developmental stages (Fig 3C–E). When the last emerging flag leaf was filled with spots, the Ossec3a spots began to senesce far earlier than those of the wild-type plants (Fig 3C, D). In addition to an obvious leaf blade phenotype, several agricultural traits, including plant height, panicle length, tiller number, thousand-grain weight, and spikelet fertility, were decreased/impaired in the Ossec3a mutants in comparison with the wild-type plants (Supplementary Fig. S5).
The lesion phenotype of Ossec3a mutant plants was fully recovered when the full-length OsSEC3A cDNA was introduced into Ossec3a mutant line #5 under the control of the 35S promoter (Supplementary Fig. S4C). This result indicates that the lesion phenotype was due to a loss of function of OsSEC3A. The results of the experiments with OsSEC3A RNAi (T3 homozygous) showed a lesion phenotype similar to that of the Ossec3a mutant plants (Supplementary Fig. S4D). qRT–PCR analyses showed severely reduced abundance of OsSEC3A (~5% of the normal transcript level), whereas OsSEC3B and OsSEC5 transcript levels were not affected (Supplementary Fig. S4E), demonstrating that the RNAi vector was specific for OsSEC3A.
Ossec3a displays induced cell death
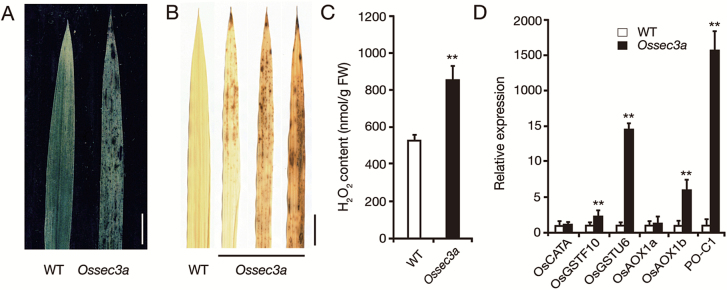
The appearance of lesion spots in the Ossec3a mutant resembled that of LMMs, which usually exhibit immunity-mediated cell death. Trypan blue staining revealed that the lesions of Ossec3a mutants contained large areas of dead cells, whereas no blue staining was observed in wild-type leaves (Fig. 4A). To confirm this result, we analyzed production of H2O2, another marker of cell death. We observed intense staining in Ossec3a leaves following treatment with DAB, whereas no DAB signal was found in wild-type leaves (Fig. 4B). Consistently, the H2O2 content in Ossec3a leaves was 1.6-fold (852.3 ± 83.5 for the wild type; 529.4 ± 28.7 for Ossec3) higher than that of wild-type leaves (Fig. 4C). In addition, the expression level of hydrogen peroxidase PO-C1 (Wally et al., 2009) was significantly increased in Ossec3a mutants. Transcript levels of OsAOX1b (LI et al., 2013), OsGSTU6, and OsGSTF10 (Jardim-Messeder et al., 2015), which have been shown to be induced by H2O2, were also up-regulated in Ossec3a mutants, consistent with H2O2 accumulation (Fig. 4D). These results demonstrate that cell death is induced in Ossec3a mutants in a manner similar to that observed in LMMs.
Fig. 4.
Ossec3a displays induced cell death. (A) Staining of the leaf blades of wild-type (WT) and Ossec3a mutant plants with trypan blue. (B) DAB staining in WT and Ossec3a leaf blades. Scale bars=5 cm in (A, B). (C) Quantitative measurements of H2O2 in the leaves of WT and Ossec3a mutant plants. (D) qRT–PCR analysis of antioxidant enzyme genes in WT and Ossec3a plants. Significance was assessed using the Student’s t-test (**P<0.01). Error bars indicate the SD.
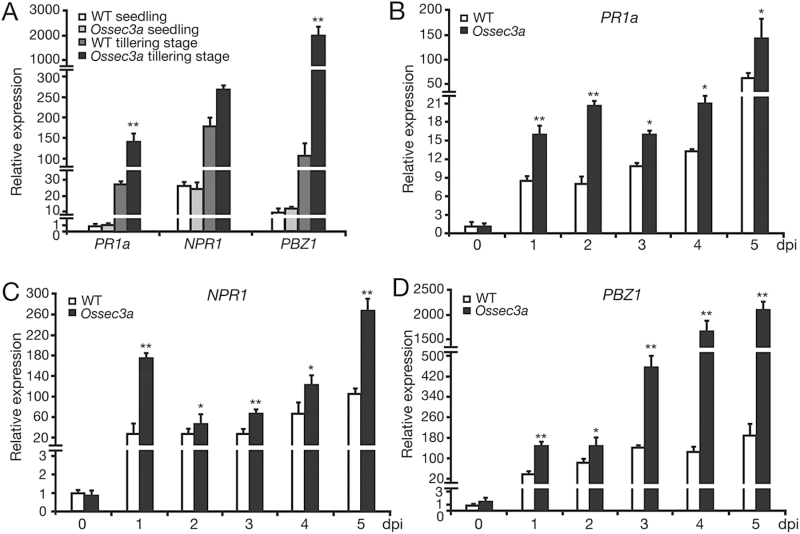
Ossec3a mutants display an enhanced defense response
It has been reported that defense responses are activated during lesion formation in rice LMMs (Manosalva et al., 2011). To address this possibility in Ossec3a mutant plants, we measured the transcript levels of pathogenesis-related (PR) genes, including PR1a, NPR1, and PBZ1. At the seedling stage, when lesions had not developed, transcript levels of PR genes were very low and no difference was detected between Ossec3a and wild-type leaf blades. However, 3 d after the appearance of lesions of the tillering stage, PR1a and PBZ1 were up-regulated significantly in the leaf blades of Ossec3a mutants in comparison with corresponding wild-type leaf blades (Fig. 5A). We also examined PR gene expression at various time points from 0 to 5 dpi with M. oryzae in Ossec3a mutants and wild-type plant seedlings. After spray-inoculation, the relative transcript levels of PR genes were increased and found to be much higher in Ossec3a mutants in comparison with those of wild-type plants; in particular, transcript abundance of PBZ1 was increased markedly in Ossec3a mutants at 4 and 5 dpi in comparison with that of wild-type plants (Fig. 5B–D).
Fig. 5.
Expression of pathogenesis-related genes in wild-type (WT) and Ossec3a mutant plants. (A) qRT–PCR of pathogenesis-related genes in WT and Ossec3a plants. Samples were detached from leaf blades at the seedling stage and tillering stage when lesions appeared. (B–D) qRT–PCR of pathogenesis-related genes PR1a (B), NPR1 (C), and PBZ1 (D) using 4-week-old WT and Ossec3a plants at various time points from 0 to 5 dpi (days post-inoculation) with Magnaporthe oryzae. Values are presented as the mean ±SE of three independent experiments. Significant differences were determined with the Student’s t-test (*0.01<P<0.05; ** P<0.01).
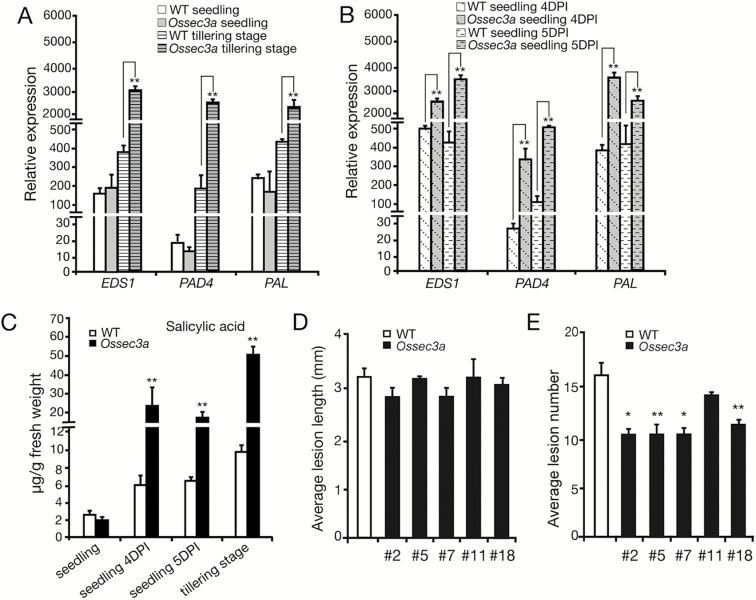
As a molecular marker for defense responses, SA accumulates in leaves and activates PR genes (Durrant and Dong, 2004). To investigate this role of SA, we examined transcript levels of SA synthesis-related genes EDS1 (enhanced disease susceptibility 1), PAD4 (phytoalexin deficient 4), and PAL (phenylalanine ammonia-lyase) (Du et al., 2009). At the seedling stage, no differences between Ossec3a and wild-type leaves were detected. However, after inoculation with M. oryzae, SA synthesis-related genes were up-regulated significantly in Ossec3a seedlings in comparison with those of wild-type seedlings (Fig. 6A, B). The same up-regulation of these genes was also observed in the Ossec3a mutants at the tillering stage (Fig. 6A). We also measured the abundance of free SA using HPLC. As expected, no difference was observed in the abundance of SA in Ossec3a and wild-type seedlings. However, at the tillering stage and after inoculation with M. oryzae, Ossec3a seedlings had ~3.0- to 5.5-fold higher free SA in comparison with wild-type seedlings (Fig. 6C). The observed up-regulation of defense response-related genes and accumulation of SA in response to pathogen inoculation indicated that plant defense responses were enhanced in Ossec3a mutant plants.
Fig. 6.
Accumulation of salicylic acid (SA) and enhanced disease resistance to Magnaporthe oryzae in the Ossec3a mutant plants. (A and B) qRT–PCR of SA synthesis-related genes EDS1, PAD4, and PAL in Ossec3a seedlings or tillering plants without (A) and with (B) M. oryzae inoculation. (C) Levels of free SA in Ossec3a and wild-type (WT) plants with and without M. oryzae inoculation. (D and E) Evaluation of resistance to M. oryzae in WT and Ossec3a mutant plants. The average lesion length (D) and number (E) in WT and Ossec3a mutant (line #2, #5, #7, #11, and #18) leaves at 7 dpi are shown. The bars represent the SE of three replicates. Significance was determined with the Student’s t-test (*0.01<P<0.05; **P<0.01).
Another molecular marker commonly known to participate in plant defense responses is jasmonic acid (JA) (Glazebrook, 2001). Therefore, we explored the expression patterns of genes related to JA biosynthesis and signaling. The results were consistent with those of our experiments assessing expression of SA-related genes. Significant up-regulation of JA biosynthesis gene OsLOX2.2 and JA signaling genes OsJAZ1 and OsJAZ2 was observed in Ossec3a leaves at the tillering stage, as well as in Ossec3a seedlings with inoculation of M. oryzae (Supplementary Fig. S6). No changes were detected between wild-type and Ossec3a seedlings without inoculation (Supplementary Fig. S6). These results indicated activation of JA synthesis and the JA signaling pathway, which is consistent with the idea that Ossec3a mutants have an enhanced defense response.
To test whether Ossec3a plants show actual resistance to pathogens, we spray-inoculated Ossec3a and wild-type plants with an M. oryzae isolate at the seedling stage. One week after inoculation, Ossec3a mutants and wild-type plants showed typical lesions of similar length (Fig. 6D). However, the leaves of Ossec3a mutants had fewer lesions than did the wild-type seedlings (Fig. 6E), suggesting that Ossec3a mutant plants exhibited enhanced disease resistance against M. oryzae.
OsSEC3A localizes with the plasma membrane
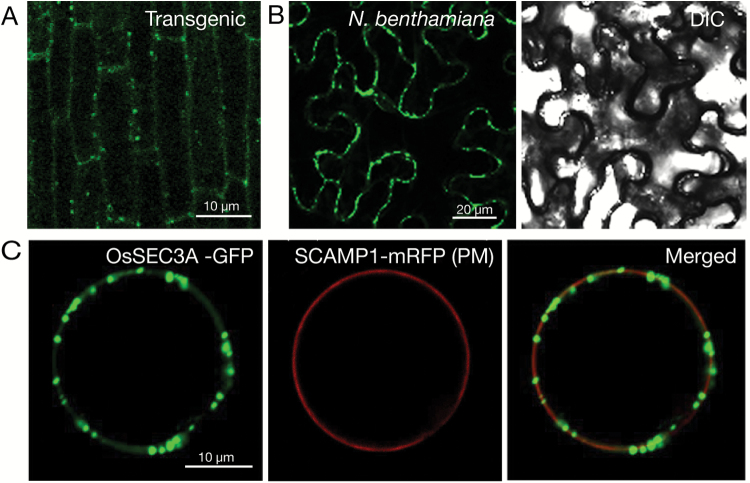
To elucidate the subcellular localization of OsSEC3A, a 35S:OsSEC3A:GFP construct, with the C-terminus of OsSEC3A connected with the N-terminus of GFP, was expressed under the control of the 35S promoter and transformed into rice calluses. Under a fluorescence microscope, we observed strong GFP signals in punctate structures in the periphery of root epidermal cells in OsSEC3A transgenic seedlings (Fig. 7A). A similar subcellular localization pattern was observed in leaf epidermal cells of N. benthamiana (Fig. 7B). To confirm further that these punctate structures accumulated in the plasma membrane, we transiently transformed OsSEC3A–GFP into rice protoplasts with a plasma membrane marker (SCAMP–mRFP; Lam et al., 2007). The punctate OsSEC3A–GFP signals were localized within the plasma membrane signal (Fig. 7C). These results indicated that OsSEC3A exhibited a punctate distribution and was most probably localized within, or linked to, the plasma membrane.
Fig. 7.
Subcellular localization of OsSEC3A–GFP. (A) Confocal microscopy of OsSEC3A–GFP in the root epidermal cells of OsSEC3A transgenic seedlings. (B) Confocal microscopy of OsSEC3A–GFP in the leaf epidermal cells of N. benthamiana. (C) Confocal micrograph of the distributions of OsSEC3A–GFP and the indicated markers (SCAMP1–mRFP; plasma membrane) 12 h after transformation.
A previous study reported plasma membrane localization of OsSNAP32, a SNARE protein that mediates membrane fusion, in rice (Bao et al., 2008). In agreement with these findings, our results provide solid evidence for interaction between OsSEC3A and OsSNAP32 (see below), in accordance with the idea that OsSEC3A localizes with the plasma membrane.
OsSEC3A binds to phosphatidylinositols through its N-terminus
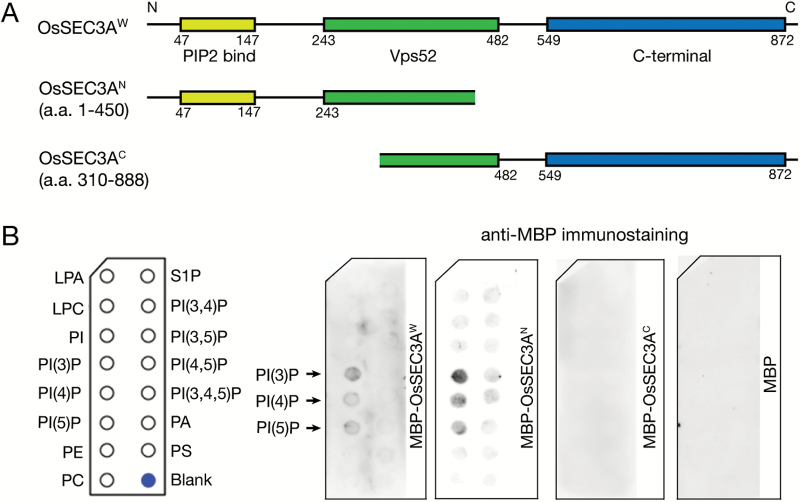
In yeast and humans, the Sec3 subunit anchors the exocyst complex to the plasma membrane via interaction with phosphatidylinositol (He et al., 2007; Liu et al., 2007; Zhang et al., 2008). The pleckstrin homology (PH) domain in the N-terminus of the Sec3 subunit plays a critical role in binding to the plasma membrane (He et al., 2007; Liu et al., 2007). To test whether OsSEC3A works in a similar manner, we performed a bioinformatics analysis on the encoding gene. The results of this analysis showed that OsSEC3A encodes a protein composed of 888 amino acid residues, containing a PIP2-binding domain (amino acids 47–147, corresponding to the PH domain), a Vps52 domain (amino acids 243–482), and the C-terminal region (amino acids 549–872) (Fig. 8A). Amino acid sequence alignment analysis showed that the key amino acids responsible for membrane binding are highly conserved (Supplementary Fig. S7). This implies that the N-terminus of OsSEC3A should bind phospholipids as demonstrated in previous studies (Bloch et al., 2016). Therefore, we implemented a lipid binding assay using purified recombinant MBP–OsSEC3AW protein (whole protein), MBP–OsSEC3AN (amino acids. 1–450, N-terminal half), and MBP–OsSEC3AC (amino acids 310–888, C-terminal half). The results of this assay showed obvious binding of MBP–OsSEC3AW and MBP–OsSEC3AN to phosphatidylinositol, whereas MBP–OsSEC3AC showed no binding signal (Fig. 8B). These results suggest that OsSEC3A, similarly to the SEC3 subunit in humans and yeast, has the ability to bind phospholipids through its N-terminus, which supports our observation that OsSEC3A localizes in or at the plasma membrane.
Fig. 8.
Lipid binding assays using the full-length OsSEC3A protein and different domains. (A) Schematic structure of the full-length OsSEC3A protein, including three predicted domains and two segmentations, used for purifying recombinant proteins. (B) Purified recombinant proteins with the MBP tag were used to probe PIP strips. Interactions are indicated by arrows. Lipid binding was assayed with three biological replicates. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine.
OsSEC3A interacts with OsSNAP32 through its C-terminus
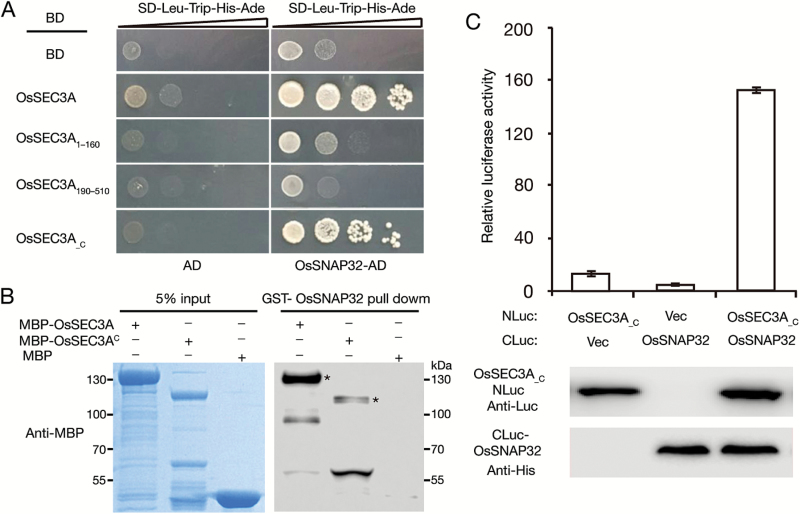
To explore how and why OsSEC3A participates in plant immunity and defense responses, we assessed whether OsSEC3A interacts with a group of known immunity- and defense-related protein factors, including SPL11, SPL28, OsVAMP721, OsSNAP32, OsPUB51, and ORPM1 (Zeng et al., 2004, 2008; Qiao et al., 2010). Our Y2H assays revealed interaction between OsSNAP32 and OsSEC3A (Fig. 9A; Supplementary Fig. S8), whereas no interaction was detected between OsSEC3A and any of the other tested proteins (Supplementary Fig. S8). OsSNAP32 is a SNAP25-type SNARE protein involved in resistance to rice blast (Luo et al., 2016). Considering that SNAREs play important roles in plant responses to various biotic and abiotic stresses (Collins et al., 2003; Kwon et al., 2008), and Ossec3a plants exhibited enhanced defense response, including resistance to rice blast (Fig. 5; Supplementary Fig. S6), we boldly speculated that OsSEC3A might function in a SNARE complex-mediated pathway. Furthermore, our deletion analysis revealed that the C-terminus, OsSEC3A_C (amino acids 521–888), displayed a specific interaction with OsSNAP32 (Fig. 9A), whereas the other domains, OsSEC3A1–160 (including the PH domain) and OsSEC3A190–510 including Vps52), failed to interact with OsSNAP32 (Fig. 9A). These results indicate that the C-terminal domain of OsSEC3A was essential for mediating the interaction between OsSEC3A and OsSNAP32. We further performed in vitro pull-down assays and split LUC complementation assays to verify this conclusion. The results of the in vitro pull-down assays confirmed that OsSNAP32–GST pulled down MBP–OsSEC3A and MBP–OsSEC3AC (amino acids 310–888) (Fig. 9B). In the split LUC complementation assay, we showed that LUC activity in N. benthamiana leaves that co-expressed Pro35S:OsSEC3A_C-NLuc and Pro35S:CLuc-OsSNAP32 was ~15-fold greater than that of leaves expressing two negative vector controls (Pro35S:OsSEC3A_C-NLuc and Pro35S:CLuc or Pro35S:NLuc and Pro35S:CLuc-OsSNAP32) (Fig. 9C). These results confirmed the interaction between OsSNAP32 and OsSEC3A through the C-terminal region (OsSEC3A_C; amino acids 521–888).
Fig. 9.
OsSEC3A physically interacts with OsSNAP32. (A) Y2H assay showing that full-length OsSEC3A and OsSEC3A_C (amino acids 521–888) interact with OsSNAP32, while two other predicted domains, OsSEC3A1–160 (including the PH domain) and OsSEC3A190–510 (including Vps52) failed to interact with OsSNAP32. (B) In vitro pull-down assay of recombinant MBP–OsSEC3A, MBP–OsSEC3AC (amino acids 310–888, C-terminal half), and MBP using resins containing GST–OsSNAP32. Asterisks indicate MBP–OsSEC3A and MBP–OsSEC3AC proteins. (C) Quantification of luciferase (LUC) activity in N. benthamiana leaves expressing OsSEC3A_C-NLuc and CLuc-OsSNAP32. The western blot shows the expression levels of CLuc and NLuc fusion proteins. OsSEC3A_C derivatives were detected by anti-firefly LUC antibodies, whereas CLuc-OsSNAP32 was detected by anti-His antibodies. The staining shows equal loading of protein in each lane. Data were collected 3 d after infiltration. The data shown are representative of three independent experiments.
Discussion
OsSEC3A participates in plant defense responses
SEC3 is a subunit of the exocyst complex that recruits other subunits residing in the cytoplasm to the plasma membrane (Finger et al., 1998; Wiederkehr et al., 2003). Previous studies in maize and Arabidopsis reveal that mutations in SEC3 result in failure to achieve correct root hair elongation (Wen et al., 2005) and defective pollen tube growth (Bloch et al., 2016). These observations of compromised polarized growth in plants with mutated SEC3 indicate that SEC3 plays a role in polar exocytosis. The novelty of this study is that we show for the first time that SEC3 has a role in the plant defense response in rice. Although the Ossec3a mutant rice plants showed slower growth of the main roots in comparison with that of wild-type plants, no specific inhibition of root hair growth was observed (Fig. 3B). Moreover, defects in pollen tube growth were not observed because the homozygous Ossec3a mutant plants produced normal seeds. Taking these results into account, it is likely that OsSEC3A may not be required for polarized growth. Instead, the Ossec3a mutants in our study presented a phenotype resembling that of LMMs. Lesions appeared on Ossec3a leaf blades and leaf sheaths from the tillering stage to the ripening stage (Fig. 3; Supplementary Fig. S4), and cell death was induced in Ossec3a mutants in a manner similar to that observed in LMMs (Fig. 4). In addition, Ossec3a mutants also showed signs of enhanced immunity, including enhanced SA content, up-regulation of defense response-related genes, and enhanced disease resistance against M. oryzae (Figs 5, 6).
Recently, ~15 LMM mutants showing enhanced immunity and spontaneous cell death were identified in rice (Zhu et al., 2016). The well-studied proteins responsible for the LMM phenotype are E3 ubiquitin ligase SPL11 and AAA ATPase LRD-6. The former protein negatively regulates plant defense by associating with SPIN6 and OsRac1, and the latter probably inhibits immunity by regulating multivesicular body-mediated vesicular trafficking (Zeng et al., 2004; Zhu et al., 2016). In this study, Ossec3a mutants showed spontaneous lesions and enhanced resistance to disease, which are the most typical phenotypes of LMMs. This finding implies a possible interaction between OsSEC3A and the molecular pathway of LMM immunity.
Localization of OsSEC3A and interaction with other exocyst subunits
In Arabidopsis, SEC3A, as well as other exocyst subunits such as SEC6, SEC8, and EXO70A1, have been shown to localize at the plasma membrane with a punctate distribution (Žárský et al., 2013; Zhang et al., 2013). Our results showed almost the same localization pattern for OsSEC3A (Fig. 7). The distribution of SEC3A and other exocyst subunits in Arabidopsis can occur in a manner dependent on (Zhang et al., 2013) or independent of (Fendrych et al., 2013) secretory vesicles. In this study, using the inhibitor BFA, which causes aggregation of the trans-Golgi network and inhibits exocytosis (Wang et al., 2016), we showed that, upon BFA treatment, the punctate-localized OsSEC3A–GFP signal maintained the same pattern as that observed in untreated plants, which differed from that of the positive control Man1–mRFP (cis-Golgi marker), which was completely redirected to the endoplasmic reticulum (Supplementary Fig. S9). This finding suggests that localization of OsSEC3A is independent of the presence of secretory vesicles, in agreement with a previous study of Arabidopsis (Fendrych et al., 2013).
Previous studies demonstrate that SEC3A functions as a subunit of the exocyst complex (Zhang et al., 2013). Membership of a protein complex such as the exocyst is usually determined by assessing protein–protein interactions with the other subunits in the complex. In Arabidopsis, SEC3A interacts with EXO70A1 and SEC5A in the exocyst complex (Zhang et al., 2013). In rice, however, we showed that OsSEC3A interacts with EXO70B1, SEC5A, SEC6, and SEC15B (Fig. 2). Notably, Arabidopsis SEC3A does not interact with SEC6 or SEC15B (Zhang et al., 2013). Our results showed that OsSEC3A also failed to interact with EXO70A1 (Fig. 2). We were unable to locate any further information about the subunits that interact with SEC3A in other plant species. Thus, our results provide some initial insight into exocyst assembly in plants. The additional subunits that were observed to interact with OsSEC3A, SEC6, and SEC15 (besides EXO70 and SEC5 in Arabidopsis) may reflect the existence of exocyst subcomplexes in rice.
Possible links between OsSEC3A and plant defense
In plants, the function of the exocyst is linked to normal development and defense response (Žárský et al., 2013; Fujisaki et al., 2015; Zhao et al., 2015). Among the subunits that comprise the exocyst complex, EXO70B1 is a unique subunit of the exocyst complex, whose mutant shows the lesion-mimic phenotype and enhanced disease resistance (Stegmann et al., 2013; Zhao et al., 2015). In our study, we found that mutation of OsSEC3A caused spontaneous leaf lesions resembling those caused by hypersensitive response (HR) in the absence of pathogen infection. This phenotype of Ossec3a mutants is very similar to that of exo70B1 mutants. It is interesting that mutations of SEC3A in other plants species, such as maize and Arabidopsis, do not produce a similar phenotype (Wen et al., 2005; Zhang et al., 2013). To explore how and why OsSEC3A is involved in plant immunity and defense responses, we examined the possibility that OsSEC3A interacts with a group of known immunity- and defense-related protein factors. We discovered that OsSEC3A interacts directly with OsSNAP32 (Fig. 9), a SNAP25-type SNARE protein that influences the resistance of rice to disease (Luo et al., 2016). This finding provides a molecular link between OsSEC3A and plant immunity and defense responses. In Arabidopsis, AtSNAP33, an OsSNAP32 homolog, is involved in immune responses by playing a role in the formation of papilla below infection sites (Kwon et al., 2008; Yun et al., 2013). Moreover, AtSNAP33 mutants show a lesion-mimic phenotype. According to these findings, beyond our initial report of the involvement of SEC3A in plant immunity, we hypothesized that SNARE complex-mediated signaling might be critical for this function.
Using lipid binding assays, we showed that OsSEC3A binds to PI(3)P; the N-terminus of OsSEC3A was essential for this binding (Fig. 8). Strikingly, this type of binding is different from that through which PI(4,5)P2 interacts with Sec3 in yeast (Zhang et al., 2008). The latter type of lipid binding to PI(4,5)P2 has also been observed in Arabidopsis, although the sequence similarity of SEC3A in Arabidopsis and rice is high (Bloch et al., 2016). It is notable that external PI(3)P is reported to mediate the entry of pathogen effectors into plant host cells (Kale et al., 2010; Helliwell et al., 2016). This may suggest another link between OsSEC3A and the defense response. In addition, the difference in phosphatidylinositol binding may reflect differences in the manner in which SEC3As participate in developmental processes; Arabidopsis SEC3A probably participates in pollen tube growth and embryo development, whereas OsSEC3A probably participates in plant defense.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Several exocyst subunits fused with the activation domain displayed no auto-activation in our experiments.
Fig. S2. Alignment of mutations in OsSEC3A from chromatograms to the reference genome sequence.
Fig. S3. qRT–PCR analysis of OsSEC3A in wild-type and Ossec3a plants (#5, #7, #11).
Fig. S4. Performance of whole plants (wild-type and transgenic lines) at different developmental stages.
Fig. S5. Agronomic traits (plant height, panicle length, tiller number, thousand-grain weight, and spikelet fertility) of wild-type and Ossec3a mutant plants.
Fig. S6. qRT–PCR of jasmonic acid synthesis-related genes (OsLOX2.2 and OsJAR1;2) and signaling pathway genes (OsJAZ1 and OsJAZ2) in wild-type and Ossec3a plants.
Fig. S7. Evolutionary analysis of OsSEC3A.
Fig. S8. Interaction of OsSEC3A with a group of known immunity- and defense-related protein factors.
Fig. S9. BFA treatment of leaf epidermal cells co-expressing OsSEC3A–GFP and Man1–mRFP.
Table S1. Sequences of DNA oligonucleotides used in this study
Acknowledgements
We thank Professor Liumin Fan (Peking University) for providing the 35S:NLuc and 35S:CLuc plasmids. We thank Xin Zhang and Xiuping Guo (Chinese Academy of Agricultural Sciences) for technical assistance with the transformation of rice calluses. This work was supported by the Ministry of Agriculture of China for Transgenic Research (2014ZX0800938B) and the National Program on Key Basic Research Project (973 Program) of China (2013CB126905).
Glossary
Abbreviations
- BFA
brefeldin A
- DAB
3,3′-diaminobenzidine
- dpi
days post-inoculation
- H2O2
hydrogen peroxide
- LUC
luciferase
- PH
pleckstrin homology
- PI(3)P
phosphatidylinositol-3-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PR
pathogenesis-related
- SA
salicylic acid
- SNAP
soluble N-ethylmaleimide-sensitive factor attachment protein
- VAMP
vesicle-associated membrane protein
- Y2H
yeast two-hybrid.
References
- Bao YM, Wang JF, Huang J, Zhang HS. 2008. Molecular cloning and characterization of a novel SNAP25-type protein gene OsSNAP32 in rice (Oryza sativa L.). Molecular Biology Reports 35, 145–152. [DOI] [PubMed] [Google Scholar]
- Bloch D, Pleskot R, Pejchar P, Potocký M, Trpkošová P, Cwiklik L, Vukašinović N, Sternberg H, Yalovsky S, Žárský V. 2016. Exocyst SEC3 and phosphoinositides define sites of exocytosis in pollen tube initiation and growth. Plant Physiology 172, 980–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. 2008. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Malamy J, Henning J, Conrath U, Sánchez-Casas P, Silva H, Ricigliano J, Klessig DK. 1995. Induction, modification, and transduction of the salicylic acid signal in plant defense responses. Proceedings of the National Academy of Sciences, USA 92, 4134–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, Goring DR. 2010. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytologist 185, 401–419. [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC. 2003. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Cvrčková F, Grunt M, Bezvoda R, Hála M, Kulich I, Rawat A, Zárský V. 2012. Evolution of the land plant exocyst complexes. Frontiers in Plant Science 3, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drdova EJ, Synek L, Pečenková T, Hala M, Kulich I, Fowler JE, Murphy AS, Žárský V. 2013. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. The Plant Journal 73, 709–719. [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B. 2009. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proceedings of the National Academy of Sciences, USA 106, 22163–22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Mpina MH, Birch PR, Bouwmeester K, Govers F. 2015. Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiology 169, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. 2004. Systemic acquired resistance. Annual Review of Phytopathology 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Elias M, Drdova E, Ziak D, Bavlnka B, Hala M, Cvrckova F, Soukupova H, Zarsky V. 2003. The exocyst complex in plants. Cell Biology International 27, 199–201. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pecenková T, Drdová EJ, Sekeres J, de Rycke R, Nowack MK, Zársky V. 2013. Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Molecular Biology of the Cell 24, 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pečenková T, Toupalová H, Cole R, Drdová E, Nebesářová J, Šedinová M, Hála M, Fowler JE. 2010. The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. The Plant Cell 22, 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Hughes TE, Novick P. 1998. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell 92, 559–571. [DOI] [PubMed] [Google Scholar]
- Fujisaki K, Abe Y, Ito A et al. 2015. Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. The Plant Journal 83, 875–887. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. 2001. Genes controlling expression of defense responses in Arabidopsis—2001 status. Current Opinion in Plant Biology 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. 2000. Protein complexes in transport vesicle targeting. Trends in Cell Biology 10, 251–255. [DOI] [PubMed] [Google Scholar]
- Guo W, Tamanoi F, Novick P. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nature Cell Biology 3, 353–360. [DOI] [PubMed] [Google Scholar]
- Hála M, Cole R, Synek L et al. 2008. An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. The Plant Cell 20, 1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xi F, Zhang X, Zhang J, Guo W. 2007. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO Journal 26, 4053–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell EE, Vega-Arreguín J, Shi Z, Bailey B, Xiao S, Maximova SN, Tyler BM, Guiltinan MJ. 2016. Enhanced resistance in Theobroma cacao against oomycete and fungal pathogens by secretion of phosphatidylinositol-3-phosphate-binding proteins. Plant Biotechnology Journal 14, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N, Ueda T. 2014. Membrane trafficking pathways and their roles in plant–microbe interactions. Plant and Cell Physiology 55, 672–686. [DOI] [PubMed] [Google Scholar]
- Jardim-Messeder D, Caverzan A, Rauber R, Souza Ferreira E, Margis-Pinheiro M, Galina A. 2015. Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytologist 208, 776–789. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405. [Google Scholar]
- Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, Rumore A, Arredondo FD, Hanlon R, Fudal I, Rouxel T. 2010. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295. [DOI] [PubMed] [Google Scholar]
- Kulich I, Cole R, Drdová E, Cvrcková F, Soukup A, Fowler J, Zárský V. 2010. Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytologist 188, 615–625. [DOI] [PubMed] [Google Scholar]
- Kulich I, Pečenková T, Sekereš J, Smetana O, Fendrych M, Foissner I, Höftberger M, Žárský V. 2013. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14, 1155–1165. [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H. 2008. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L. 2007. Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular–vesicular structures as an early endosome in tobacco BY-2 cells. The Plant Cell 19, 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. 2014. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Molecular Plant 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li P, Zhai H, Zhang H, Lu Z, Chen Z, Wang F. 1998. Inheritance of blast resistance in two japonica rice landraces from Taihu Lake area. ZhongGuo ShuiDao KeXue 13, 11–14. [Google Scholar]
- Liu J, Guo W. 2012. The exocyst complex in exocytosis and cell migration. Protoplasma 249, 587–597. [DOI] [PubMed] [Google Scholar]
- Liu J, Zuo X, Yue P, Guo W. 2007. Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Molecular Biology of the Cell 18, 4483–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Zhang H, He W, Zhang Y, Cao W, Zhang H, Bao Y. 2016. OsSNAP32, a SNAP25-type SNARE protein-encoding gene from rice, enhanced resistance to blast fungus. Plant Growth Regulation 80, 37–45. [Google Scholar]
- Ma J, Cheng Z, Chen J, Shen J, Zhang B, Ren Y, Ding Y, Zhou Y, Zhang H, Zhou K. 2016. Phosphatidylserine synthase controls cell elongation especially in the uppermost internode in rice by regulation of exocytosis. PLoS One 11, e0153119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva PM, Bruce M, Leach JE. 2011. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. The Plant Journal 68, 777–787. [DOI] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. 2013. Targeted mutagenesis in rice using CRISPR–Cas system. Cell Research 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pečenková T, Hála M, Kulich I, Kocourková D, Drdová E, Fendrych M, Hana T, Žárský V. 2011. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant–pathogen interaction. Journal of Experimental Botany 62, 2017–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Jiang W, Lee J et al. 2010. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytologist 185, 258–274. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Liu F, Zhou K, Ding Y, Zhou F, Wang Y, Liu K, Gan L, Ma W. 2014. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. The Plant Cell 26, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S. 2007. Vesicle trafficking in plant immune responses. Cellular Microbiology 9, 1–8. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. 2009. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. The Plant Cell 21, 2655–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Fu J, Ma J, Wang X, Gao C, Zhuang C, Wan J, Jiang L. 2014. Isolation, culture, and transient transformation of plant protoplasts. Current Protocols in Cell Biology 17, 2.8.1–2.8.8. [DOI] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M. 2012. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. The Plant Cell 24, 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Westphal L, Rosahl S, McDowell JM, Trujillo M. 2013. The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signaling and Behavior 8, e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L, Schlager N, Eliáš M, Quentin M, Hauser MT, Žárský V. 2006. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. The Plant Journal 48, 54–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Zhu X, Wang Y, Liu L, Xu B, Li F, Fang J, Chu C. 2011. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. The Plant Journal 66, 996–1007. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. 1996. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO Journal 15, 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tu B, Hu L, Chen W et al. 2015. Disruption of OsEXO70A1 causes irregular vascular bundles and perturbs mineral nutrient assimilation in rice. Scientific Reports 5, 18609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wally O, Jayaraj J, Punja Z. 2009. Comparative resistance to foliar fungal pathogens in transgenic carrot plants expressing genes encoding for chitinase, β-1, 3-glucanase and peroxidise. European Journal of Plant Pathology 123, 331–342. [Google Scholar]
- Wang Y, Liu F, Ren Y et al. 2016. GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. The Plant Cell 28, 2850–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS. 2005. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiology 138, 1637–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Du Y, Pypaert M, Ferro-Novick S, Novick P. 2003. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Molecular Biology of the Cell 14, 4770–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun HS, Kwaaitaal M, Kato N, Yi C, Park S, Sato MH, Schulze-Lefert P, Kwon C. 2013. Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Molecules and Cells 35, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zárský V, Cvrcková F, Potocký M, Hála M. 2009. Exocytosis and cell polarity in plants—exocyst and recycling domains. New Phytologist 183, 255–272. [DOI] [PubMed] [Google Scholar]
- Zárský V, Kulich I, Fendrych M, Pečenková T. 2013. Exocyst complexes multiple functions in plant cells secretory pathways. Current Opinion in Plant Biology 16, 726–733. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. 2004. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL. 2008. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Molecular Plant 1, 800–815. [DOI] [PubMed] [Google Scholar]
- Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. 2008. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. Journal of Cell Biology 180, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Immink R, Liu CM, Emons AM, Ketelaar T. 2013. The Arabidopsis exocyst subunit SEC3A is essential for embryo development and accumulates in transient puncta at the plasma membrane. New Phytologist 199, 74–88. [DOI] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, Tang D. 2015. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genetics 11, e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L et al. 2013. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yin J, Liang S, Liang R, Zhou X, Chen Z, Zhao W, Wang J, Li W, He M. 2016. The multivesicular bodies (MVBs)-localized AAA ATPase LRD6-6 inhibits immunity and cell death likely through regulating MVBs-mediated vesicular trafficking in rice. PLoS Genetics 12, e1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.