Abstract
Background
Virus is detected in about 80% of upper respiratory tract infections (URTIs) in children and is also detectable in the nasopharynx of 30% of asymptomatic children. The effect of asymptomatic viral infection on the dynamics of bacterial density and colonization of the nasopharynx has not been reported. The current study was performed
to assess the presence and density of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the nasopharynx of 4–7-year-old children during URTI and when well.
Methods
Nasal samples were obtained during 4 surveillance periods when children were asymptomatic and whenever they had symptoms of URTI. Respiratory viruses and bacterial pathogens were identified and quantified using polymerase chain reaction.
Results
The proportion of children colonized with all 3 bacteria was higher during visits for acute URTI than during asymptomatic surveillance visits. Mean bacterial densities were significantly higher at all visits for all 3 pathogens when a virus was detected. The differences between the means were 1.0, 0.4, and 0.7 log10 colony-forming unit equivalents per milliliter for S. pneumoniae, H. influenzae, and M. catarrhalis, respectively, compared with visits in which virus was not detected. The percentage of children colonized and density were also higher at asymptomatic visits in which virus was detected than at visits in which virus was not detected.
Conclusion
The density and frequency of colonization with S. pneumoniae, H. influenzae, and M. catarrhalis in nasal wash samples increase during periods of both symptomatic and asymptomatic viral infection. Increases in bacterial colonization observed during asymptomatic viral infection were nearly the same magnitude as when children were symptomatic.
Keywords: Upper respiratory infections, Streptococcus pneumoniae, Haemophilus influenza, Moraxella, colonization
The densities of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in nasal wash samples increase during both symptomatic and asymptomatic viral infection. Increases in bacterial colonization observed during asymptomatic viral infection were nearly the same magnitude as during symptomatic periods.
Upper respiratory tract infection (URTI) is the most common infectious disease of childhood and one of the most frequent indications for medical care [1]. Although URTIs are caused by respiratory viruses, the common local bacterial complications of URTI, acute otitis media, acute bacterial sinusitis and pneumonia, result from pathogenic bacteria found in the nose and nasopharynx. The current understanding of these complications is that viral URTI precedes bacterial infection and that alterations in mucosal conditions facilitate proliferation of bacteria in the respiratory tract and the development of secondary bacterial infections. Previous studies, performed using semiquantitative culture techniques, have supported this theory [2–4]. A virus is detected in about 80% of URTIs in children, but notably ≥30% of asymptomatic children will also have a detectable virus in the nasopharynx, depending on the population studied [5–9]. The effect of asymptomatic viral infection on the dynamics of bacterial colonization of the nasopharynx has not been reported. The objective of the current study was to prospectively investigate the frequency and density of bacterial colonization during periods of symptomatic and asymptomatic respiratory viral infection in an observational cohort of 4–7-year-old children.
PATIENTS AND METHODS
Enrollment and Inclusion Criteria
Healthy children aged 48–96 months were recruited throughout the study period from 2 pediatric practices in Madison, Wisconsin, and followed up for 1 year. Children were excluded if they had an underlying condition, reported by the parent or noted in the medical record, that was likely to alter the natural history of URTI, including asthma, congenital or acquired immunodeficiency, craniofacial abnormalities, cystic fibrosis, allergic rhinitis, or a history of chronic sinusitis. Written, informed consent was secured, and assent was obtained from children ≥7 years of age. Subjects received a small stipend for participation. The study was approved by the University of Wisconsin Institutional Review Board.
Procedures
Nasal samples were obtained at entry and during 4 surveillance visits (February, April, September, and December) when children were asymptomatic as verified by study nurses. If respiratory symptoms were present, the surveillance visit was postponed for 2 weeks. Parents were instructed to call the study nurse at the first sign of a URTI, which was defined as ≥48 hours of respiratory symptoms, including nasal congestion, nasal discharge, or cough. Nasal samples were obtained on day 2–3 (range, day 1–9) of illness by the study nurse. and a recovery sample was obtained at about day 15. However, because of variability in scheduling, the recovery swab sample was obtained 10–42 days after onset of URTI. A clinical assessment at the time of the visit assured that symptoms reflected illness confined to the upper respiratory tract.
Collection of Samples
Samples of nasal mucus were obtained using an established nose-blowing technique [10–12]. Saline nasal spray was sprayed into each of the child’s nostrils. The study nurse then held a transparent plastic bag to the nose and occluded one of the nostrils, taking care to not touch the inside of the bag. The child was then instructed to blow their nose forcefully into the plastic bag. This procedure was then repeated with the other nostril. If children were unable to blow their nose, a sufficient sample was obtained by allowing the sprayed saline to drip into the plastic bag. The sample was placed in M4RT transport medium (Remel) and transported on ice to the processing laboratory.
Virus Identification
Diagnostic virology was performed on nasal samples by multiplex polymerase chain reaction (PCR; Respiratory Multicode Assay [EraGen Biosciences] or Respiratory Viral Panel [Luminex]) to test for the following viruses: respiratory syncytial virus (RSV; groups A and B), rhinovirus (RV; approximately 160 known types), parainfluenza (1, 2, 3, 4a and 4b), influenza (A, B and C), adenovirus (B, C and E), coronavirus (229E, NL63, OC43, HK, and severe acute respiratory syndrome), enterovirus, human bocavirus, and human metapneumovirus (A and B). Nasal specimens were also analyzed by means of partial sequencing to determine which RV types were present and differentiate closely related enterovirus from RV [13].
Bacterial Polymerase Chain Reaction
Nasal samples were also analyzed for Streptococcus pneumoniae, H. influenzae, and Moraxella catarrhalis by using quantitative real-time PCR. DNA was extracted with the BiOstic Bacteremia DNA Isolation Kit (Mo Bio Laboratories). Quantitative LytA PCR for the detection of S. pneumoniae [14] was combined with hpd PCR for the detection of H. influenzae [15] and copB PCR for the detection of M. catarrhalis [16], as described elsewhere. All primers and probes were obtained from Applied Biosystems, and the real-time PCR assay was performed with a 7300 Applied Biosystems instrument. Standard curves consisted of bacterial DNA extracted from known quantities of clinical isolates of each bacterium obtained from the University of Wisconsin Hospital Clinical Microbiology Laboratory, and PCR results were expressed as colony-forming unit equivalents (CFUe) per milliliter. The same standards were used throughout the study and were internally consistent. Colonization was defined as any detectable bacteria by PCR (≥1 CFUe/mL) in nasal samples.
Statistical Methods
Demographic characteristics were summarized in terms of means and standard deviation or percentages. The proportion of subjects colonized was analyzed using the generalized estimating equation approach, with a logit link function to account for multiple visits. Proportions were summarized along with the corresponding 95% confidence intervals, stratified by visit type and virus detection status. Tukey’s honestly significant difference method was used to control the type I error when conducting multiple comparisons between visit types. Bacterial density values (in CFUe per milliliter) were log (base 10) transformed and analyzed using a linear mixed-effects model with subject-specific random effects. A compound symmetry correlation structure was used to account for correlations between repeated visits within the same subject. Residual plots were examined to verify model assumptions. All reported P values are 2 sided, and P < .05 was used to define statistical significance. Statistical analyses were conducted using SAS software (version 9.4; SAS Institute).
RESULTS
Invitations to participate in this study were sent to families of 4516 patients from 2 pediatric clinics after screening 6394 possible participants; 1878 subjects were not eligible owing to an underlying medical condition (Figure 1). From February 2012 to September 2015, a total of 279 subjects were enrolled. One subject was excluded after enrollment because nasal samples could not be obtained, 205 were followed up for an entire year, and the remaining 73 subjects had ≥1 surveillance sample taken. The demographic characteristics of enrolled subjects are shown in Table 1.
Figure 1.
Enrollment of subjects. Abbreviation: URTI, upper respiratory tract infection.
Table 1.
Subject Demographics (n = 278)
| Demographic Variable | Subjects, %a |
|---|---|
| Age, mean (range), y | 5.2 (4.0–7.7) |
| Female sex | 46 |
| Race | |
| American Indian or Alaska Native | 1 |
| Asian | 5 |
| African American | 7 |
| Other | 6 |
| Unknown or not reported | 1 |
| White | 80 |
| Hispanic ethnicity | 7 |
| Attends day care | 83 |
| Tobacco exposure | 3 |
| Maternal education level | |
| Graduate/professional | 36 |
| College degree | 37 |
| Some college | 14 |
| Vocational/technical training | 5 |
| High school or less | 6 |
| Not reported | 1 |
| Public insurance (Medicaid) | 18 |
aData represent No. (%) of subjects unless otherwise specified.
There were 847 surveillance, 363 acute URTI, and 298 recovery visits (Figure 1) A virus was detected in 34% of nasal samples obtained during surveillance visits, 81% of those obtained during acute URTI visits, and 32% of those obtained during recovery visits. The rates of detection of individual viruses at each visit type are detailed in Table 2. As expected, RV was the most common virus detected for all 3 visit types.
Table 2.
Virus Identification at Study Visits
| Virus | Samples with Virus Detected, (%) | ||
|---|---|---|---|
| Surveillance Visits (n = 847) | Acute URTI Visits (n = 363) | Recovery Visits (n = 298) | |
| AdV | 2 (<1) | 0 (0) | 2 (1) |
| hBoV | 12 (1) | 2 (1) | 3 (1) |
| CoV | 24 (3) | 24 (7) | 11 (4) |
| EV | 4 (<1) | 7 (2) | 1 (<1) |
| FluA | 2 (<1) | 5 (1) | 0 (0) |
| FluB | 2 (<1) | 7 (2) | 0 (0) |
| MPV | 6 (2) | 11 (3) | 3 (1) |
| PIV | 8 (1) | 21 (6) | 3 (1) |
| RSV | 3 (<1) | 15 (4) | 2 (1) |
| RV | 205 (25) | 160 (44) | 59 (20) |
| Mixed | 20 (1) | 43 (12) | 10 (3) |
| Negative | 559 (66) | 68 (19) | 204 (68) |
Abbreviations: AdV, adenovirus; CoV, coronavirus; EV, enterovirus; FluA, influenza A virus; FluB, influenza B virus; hBoV, human bocavirus; MPV, metapneumovirus, PIV, parainfluenza virus; RSV, respiratory syncytial virus; RV, rhinovirus; URTI, upper respiratory tract infection.
The proportion of children colonized was determined at the initial surveillance visit for each bacterial pathogen, and these proportions are shown in Table 3. The proportion of children colonized was significantly higher for S. pneumoniae (54%) than for Haemophilus influenzae (26%) or M. catarrhalis (33%) (P < .001) and higher for M. catarrhalis than for H. influenzae (P = .04). Next we compared the proportion of children colonized at surveillance visits where virus was detected compared with visits where a virus was not detected. The proportion colonized was significantly higher when virus was detected than when not detected for all 3 respiratory pathogens (62% vs 47% for S. pneumoniae [P < .001], 28% vs 20% for H. influenzae [P = .007], and 43% vs 36% for M. catarrhalis [P = .03]; Table 4). Patients were often colonized with >1 bacterial pathogen. The proportions of children colonized with ≥1 pathogen at surveillance and acute visits are shown in Table 5. Colonization with any 1 of the 3 bacteria was more common during acute visits than during surveillance visits (P < .001), as was the detection of any 2 bacteria (P < .001) or all 3 bacteria in the same sample (P = .05).
Table 3.
Colonization Frequency at Initial Surveillance Visits
| Bacterial Pathogen | Children Colonized, No. (%; 95% CI) (n = 278) | Comparison | P Valuea |
|---|---|---|---|
| Streptococcus pneumoniae | 150 (54; 48–60) | S. pneumoniae vs Haemophilus influenzae | <.001 |
| H. influenzae | 72 (26; 21–31) | S. pneumoniae vs Moraxella catarrhalis | <.001 |
| M. catarrhalis | 93 (33; 28–39) | H. influenzae vs M. catarrhalis | .04 |
Abbreviation: CI, confidence interval.
aBased on generalized estimating equation analysis.
Table 4.
Colonization Frequency at Surveillance Visits: Virus Versus No Virus
| Bacterial Pathogen | Detectable Colonization, No. (%; 95% CI) | P Valuea | |
|---|---|---|---|
| Virus Detected (n = 288) | No Virus Detected (n = 559) | ||
| Streptococcus pneumoniae | 178 (62; 55–68) | 254 (47; 43–52) | <.001 |
| Haemophilus influenzae | 82 (28; 23–35) | 111 (20; 16–24) | .007 |
| Moraxella catarrhalis | 125 (43; 38–49) | 200 (36; 31–41) | .03 |
Abbreviation: CI, confidence interval.
a P values based on generalized estimating equation analysis
Table 5.
Bacteria Detected
| Bacterial Pathogens Detected | Proportion Colonized, % (95% CI) | P Valuea | |
|---|---|---|---|
| Surveillance Visits (n = 847) | Acute Visits (n = 363) | ||
| None | 29 (25–33) | 16 (12–20) | <.001 |
| Single pathogen | |||
| Total | 37 (33–41) | 35 (29–41) | .49 |
| S. pneumoniae | 21 (18–24) | 17 (12–21) | .13 |
| Haemophilus influenzae | 4 (2–5) | 3 (1–5) | .73 |
| Moraxella catarrhalis | 13 (10–15) | 15 (11–19) | .26 |
| 2 Pathogens | |||
| Total | 25 (22–29) | 36 (31–41) | <.001 |
| S. pneumoniae and H. influenzae | 8 (6–10) | 12 (8–15) | .04 |
| S. pneumoniae and Moraxella catarrhalis | 15 (12–17) | 19 (14–24) | .1 |
| H. influenzae and M. catarrhalis | 2 (1–4) | 5 (3–8) | .04 |
| 3 Pathogens: S. pneumoniae, H influenzae, and M. catarrhalis | 9 (7–11) | 13 (9–17) | .05 |
Abbreviation: CI, confidence interval.
a P values based on generalized estimating equation analysis.
Bacterial densities in nasal samples obtained at study visits are shown in Figure 2. Densities were significantly higher at acute URTI visits for S. pneumoniae, H. influenzae and M. catarrhalis, demonstrating nearly a log increase in density compared with surveillance visits. In addition, bacterial densities for all 3 bacteria were significantly decreased at recovery visits compared with acute URTI visits. Bacterial densities at recovery visits dropped to near baseline surveillance levels for S. pneumoniae and H. influenzae but remained significantly above those for surveillance visits for M. catarrhalis (P < .05).
Figure 2.
Bacterial density for Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis at surveillance visits (n = 847), acute upper respiratory tract infection (URTI) visits (n = 363), and recovery visits (n = 298). *P < .001 for surveillance versus acute URTI visits; †P < .01 for acute URTI versus recovery visits; ‡P = .03 for recovery versus surveillance visits. Error bars show 95% confidence intervals. Abbreviation: CFUe, colony-forming unit equivalents.
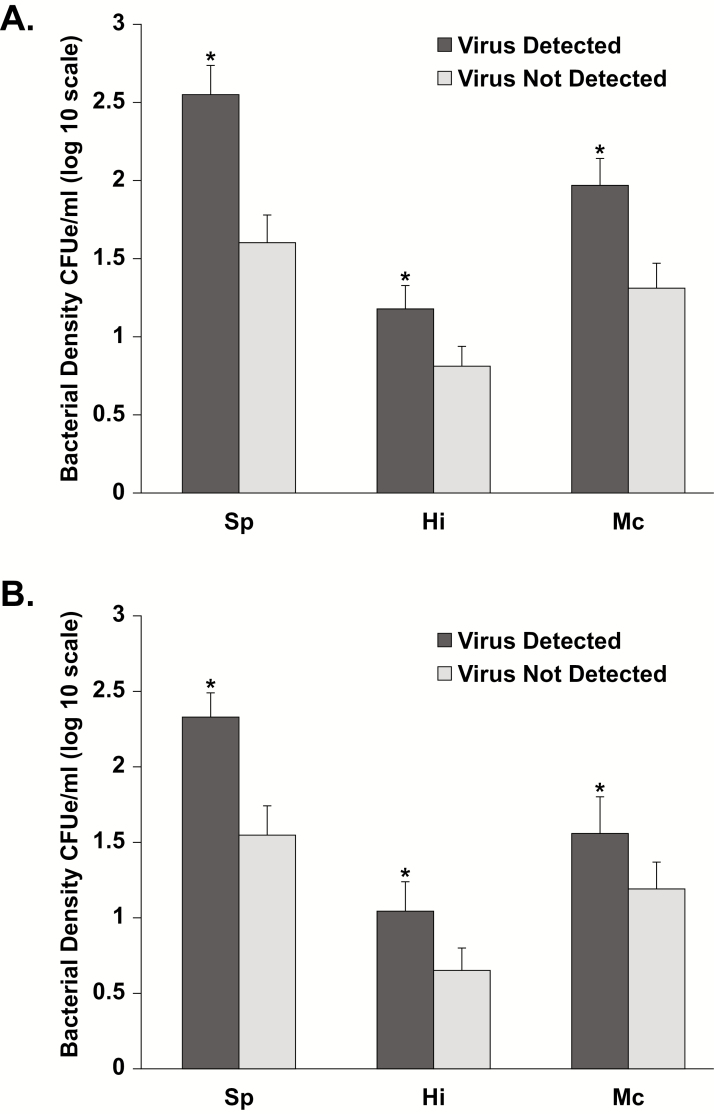
To isolate the effect of viral URTIs on bacterial density, the mean densities of bacteria were compared between visits at which a virus was detected and those at which a virus was not detected. Mean densities were significantly higher at all visits for all 3 bacterial pathogens (Figure 3A), when a virus was detected, with mean differences in density of 1.0, 0.4, and 0.7 log10 CFUe/mL for S. pneumoniae, H. influenzae, and M. catarrhalis, respectively, compared with visits at which a virus was not detected (P < .001). The effect of viral infection on increasing bacterial density occurred regardless of whether symptoms were present. Figure 3B shows bacterial densities for asymptomatic visits at which a virus was detected compared with those at which a virus was not detected. At surveillance visits, the mean differences in density between these visits were 0.8, 0.4, and 0.4 log10 CFUe/mL for S. pneumoniae, H. influenzae, and M. catarrhalis, respectively (P < .01).
Figure 3.
Bacterial density for Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis for all visits (n = 1508; A) and surveillance visits (n = 847; B) at which a virus was detected versus not detected. *P < .01 for virus detected versus not detected. Abbreviation: CFUe, colony-forming unit equivalents.
Bacterial densities were analyzed for individual respiratory viruses (Table 6). Samples that had >1 virus detected were labeled as mixed. Samples in which RV was detected had significantly higher densities for all 3 bacterial pathogens than samples without detection of a virus. Samples with mixed virus detection showed significantly higher densities of S. pneumoniae and M. catarrhalis but not H. influenzae. Comparison between other individual respiratory viruses was not possible because of low numbers.
Table 6.
Adjusted Mean Bacterial Density By Virus for All Visitsa
| Virus | Streptococcus pneumoniae | Haemophilus influenzae | Moraxella catarrhalis | |||
|---|---|---|---|---|---|---|
| No. | Bacterial Density, Mean (95% CI), Log10 CFUe/mL | No. | Bacterial Density, Mean (95% CI), Log10 CFUe/mL | No. | Bacterial Density, Mean (95% CI), Log10 CFUe/mL | |
| AdV | 4 | 0.72 (−1.4 to 2.85) | 3 | 2.12 (.36–3.89) | 3 | 2.54 (.49–4.58) |
| hBoV | 17 | 1.46 (.4–2.51) | 7 | 1.32 (.45–2.2) | 8 | 1.22 (.21–2.23) |
| CoV | 59 | 1.79b (1.22–2.36) | 8 | 0.67 (.19–1.14) | 37 | 2.44c (1.9–2.98) |
| FluA | 7 | 2.17 (.57–3.77) | 5 | 2.6 (1.32–3.98) | 4 | 3.1 (1.55–4.64) |
| FluB | 9 | 2.18 (.80–3.57) | 2 | 0.34 (−.82 to 1.5) | 7 | 3.41 (2.06–4.76) |
| RV | 424 | 2.57c (2.33–2.81) | 125 | 1.24c (1.04–1.44) | 189 | 1.82c (1.60–2.04) |
| MPV | 20 | 2.58 (1.64–3.51) | 6 | 1.14 (.36–1.92) | 8 | 1.36 (.45–2.27) |
| RSV | 20 | 2.84 (1.91–3.78) | 7 | 1.3 (.52–2.08) | 12 | 2.64 (1.73–3.55) |
| PIV | 32 | 2.87 (2.13–3.62) | 4 | 0.63 (0–1.25) | 12 | 1.41 (.69–2.13) |
| Mixed | 73 | 3.05c (2.54–3.56) | 25 | 1.17 (.74–1.6) | 46 | 2.68c (2.19–3.17) |
| EV | 12 | 3.17 (1.96–4.38) | 5 | 1.65 (.65–2.66) | 3 | 1.38 (.21–2.56) |
| Negative | 73 | 1.61 (1.43–1.79) | 198 | 0.81 (.67–.96) | 318 | 1.31 (1.15–1.47) |
Abbreviations: AdV, adenovirus; CI, confidence interval; CoV, coronavirus; EV, enterovirus; FluA, influenza A virus; FluB, influenza B virus; hBoV, human bocavirus; MPV, metapneumovirus,; PIV, parainfluenza virus; RSV, respiratory syncytial virus; RV, rhinovirus.
aBased on a linear mixed-effects model.
b P = .04 for comparison with mixed results.
c P < .02 for comparison with negative results..
To further determine the role of viral infection in affecting bacterial colonization and density, we compared 4 groups: surveillance visits with no virus detected (SN), surveillance visits with virus detected (SV), acute URTI visits with no virus detected (AN), and acute URTI visits with virus detected (AV). Differences were adjusted for multiple comparisons. For S. pneumoniae (Figure 4A), the proportions of children colonized were similar for SV and AV visits, and both were significantly higher than for SN visits. The density of each bacterial species was grouped into 4 levels (1–100, 100–1000, 1000-10000, or >10000 CFUe/mL) for each of the visit types (SN, SV, AN, and AV). For S. pneumoniae, when a virus was detected, the greatest increase in the proportion of samples occurred in the group with >10000 CFUe/mL. For H. influenzae (Figure 4B) colonization frequency was significantly higher for SV, AN, and AV visits than for SN visits. Again the greatest change in bacterial density occurred in the proportion of samples with >10000 CFUe/mL. For M. catarrhalis (Figure 4C), the density of colonization was higher for both AN and AV visits than for SN visits, also with the greatest change in the >10000 CFUe/mL group.
Figure 4.
Bacterial colonization rates for Streptococcus pneumoniae (A), Haemophilus influenzae (B), and Moraxella catarrhalis (C) at surveillance visits with no virus detected (SN), surveillance visits with virus detected (SV), acute upper respiratory tract infection (URTI) visits with no virus detected (AN), and acute URTI visits with virus detected (AV). *P < .05 adjusted for multiple comparisons by Tukey’s honestly significant difference method. Error bars represent 95% confidence intervals. CFUe, colony-forming unit equivalents. Abbreviations: Hi, haemophilus influenzae; Mc, moraxella catarrhalis; Sp, streptococcus pneumoniae.
Bacterial density for all 3 pathogens was tracked over the time course of a URTI. The mean observed duration for bacterial density to return to baseline levels was 13.6 days (range, 5–40 days). Overall 223 of 295 or 75% of URTIs (95% confidence interval, 70%–80%) showed a return to baseline of bacterial density by day 14 of illness for all 3 bacterial pathogens. The rate of return to baseline density did not differ between URTIs by virus type detected (data not shown).
DISCUSSION
In this study, dynamic changes in upper respiratory bacterial flora associated with viral infection were demonstrated. The density of all 3 of the common respiratory bacterial pathogens in nasal samples, S. pneumoniae, H. influenzae, and M. catarrhalis, increased by nearly a factor of 10 between surveillance periods and episodes of symptomatic URTI. In addition, it has been shown for the first time that bacterial detection and density of all 3 sinus pathogens increase during asymptomatic viral infection. In general, bacterial density was lowest for asymptomatic visits at which virus was not detected and highest for acute URTI visits at which virus was detected. Bacterial densities decreased to near-baseline levels during recovery. The impact of virus detection on bacterial detection and density lasted about 14 days from the onset of symptoms.
The effect of virus on increasing bacterial density and colonization was greatest for S. pneumoniae. This large effect of virus detection on S. pneumoniae was also observed during asymptomatic viral infections. Interestingly, the proportion colonized for all 3 pathogens was similar between surveillance visits with virus detected and acute URTI visits with no virus detected. Furthermore, there were no differences in the frequency of bacterial colonization between episodes of acute URTI with virus and those without virus. Accordingly, it is possible that the upper respiratory tract symptoms in acute episodes of URTI without virus are in fact caused by a virus not in our PCR panel or present in undetectable amounts.
For all 3 pathogens, bacterial density (but not colonization) was higher for symptomatic URTI visits at which a virus was detected than for surveillance visits at which a virus was detected. This suggests that increased density of pathogenic bacteria may itself be a cause of respiratory symptoms without viral coinfection or contribute to symptoms (not only complications) when virus is present. A similar observation was made by Kloepfer et al [17] for S. pneumoniae and M. catarrhalis but not H. influenzae in a group of children with or without asthma who were followed up for 5 weeks in the autumn season. These differences (in bacterial species) may be explained by season or age, but the suggestion that bacterial species may cause respiratory symptoms is the same and may have therapeutic implications.
The proportion of children colonized in our study was higher than expected for S. pneumoniae, whereas H. influenzae and M. catarrhalis were detected at frequencies similar to those previously reported [4, 18–20]. A study done in infants demonstrated a higher risk of URTI when subjects were colonized with M. catarrhalis and S. pneumoniae but not H. influenzae [20]. In contrast, our study did not demonstrate an increased risk of URTI with colonization with any of the bacteria tested. This difference may be due to the ages of populations studied and the methods used to detect colonization (culture vs PCR).
The results of our study affirm previous important insights into the pathogenesis of the bacterial complications of viral URTIs. Proliferation of S. pneumoniae, H. influenzae, and M. catarrhalis is thought to be a necessary step in the development of these conditions. Faden et al demonstrated an increase in the carriage rate of otopathogens in the nasopharynx of children with acute otitis media during URTI, compared with well children [21, 22]. Other more recent studies, using culture and quantitative PCR, have shown an increase in pneumococcal carriage [23] and density during acute URTI [24–26]. Xu et al [27] also recently reported that nasopharyngeal colonization rates of otopathogens during health were significantly lower than during URTI. Studies of otitis media and sinusitis have demonstrated that the presence of pathogenic bacteria in the nasopharynx markedly increases the risk of these complications [2–4, 28–31]. During URTI, there are complex interactions between bacteria and viruses [32, 33]. These observations suggest that the combination of virus and bacteria is necessary but not sufficient for the development of bacterial complications of URTI.
Detection of viruses during surveillance, when patients are asymptomatic, is not a new finding [6, 7, 34]. However, the implications of the finding that bacterial densities increase substantially during periods of asymptomatic viral infection are important and may have clinical relevance. These data allow us to understand the development of cases of acute otitis media, acute bacterial sinusitis, and their complications (mastoiditis and subperiosteal orbital cellulitis) in children without respiratory symptoms or whose respiratory symptoms are extremely modest [35]. In nearly 50% of children presenting with complications of sinusitis, the complication is the first signal of the underlying sinus infection. The current findings demonstrate that, even in the absence of symptoms, viral infection enhances bacterial colonization and density. We can speculate that this enhancement of colonization, especially with S. pneumoniae, predisposes the patient to the complications of URTI.
The mechanism by which viral URTI increases bacterial colonization and density is not well understood. A cotton rat model of RSV infection demonstrated a higher frequency of colonization of nontypable H. influenzae than non-RSV infected rats during acute RSV infection. This was not associated with changes in bacterial adherence to epithelial cells or to changes in production of specific antibacterial antibody [36]. In vitro studies have shown that RV facilitates the binding, translocation, and persistence of bacteria by disrupting epithelial barrier function [37]. In addition, increased mucus production and host cell debris may enhance the nutritional milieu for bacterial growth during infection with influenza virus [38].
This study has several unique characteristics and possible limitations that should be considered. The longitudinal follow-up of subjects for 1 year allowed the avoidance of seasonal variation in viral infections. Sensitive PCR techniques permitted identification of viruses and quantitation of bacterial colonization. Obtaining nasal samples during surveillance and illness permitted new observations of increased bacterial density even when subjects had asymptomatic viral infection. Samples were obtained with a nasal blow technique rather than by nasopharyngeal swab sample for patient comfort and to improve adherence. This method may seem to be prone to inconsistency, given the potential for variations in dilution. However, this technique is well established and has been used in many other studies of viral URTI [11, 12, 17, 39–43].
Although specimens obtained during URTI could theoretically have higher concentrations of mucus, and thus higher concentrations of bacteria, similar densities of bacteria were detected during asymptomatic viral infections. This latter observation suggests that an increased yield of mucous during URTI did not affect these results. In addition, our overall colonization proportions were higher than those previously reported, supporting the contention that the use of nasal wash samples did not underestimate the detection of bacterial pathogens. It is also notable that, despite the potential for dilution, the increases in bacterial density and colonization were very consistently found during viral URTI in this study and in others [17].
In conclusion, we have shown that the density and frequency of colonization with the bacterial pathogens S. pneumoniae, H. influenzae, and M. catarrhalis in nasal wash samples increase during periods of symptomatic and asymptomatic viral infection. In the current study, episodes of symptomatic respiratory infection with or without virus identification were similar with regard to bacterial colonization, suggesting that virus may be present even when undetected. Furthermore, the frequency of bacterial colonization observed during asymptomatic viral infection, especially for S. pneumoniae, was nearly of the same magnitude as that observed when children were symptomatic. These observations provide important insights into the pathogenesis of the bacterial complications of URTI, particularly in patients without respiratory symptoms. Further investigations that determine the precise mechanism of virus-bacterial interaction are needed.
Notes
Financial support. This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NAIAD), National Institutes of Health (NIH; grant R01 AI097172).
Potential conflicts of interest. S. V. L. reports grants from Broad Foundation, Janssen, Sloan Foundation, Pfizer, Gilead Sciences, and the NIH/NAIAD during the conduct of the study. She received personal fees from Janssen, Boston Consulting Group, and Regeneron, outside the submitted work. S. V. L. also has the following patents issued: Stan449-PRV Reductive Prodrug Cancer Chemothera, Combination Antibiotic and Antibody Therapy for the Treatment of Pseudomonas aeruginosa Infection (licensed and royalties paid by KaloBios), Therapeutic Microbial Consortium for Induction of Immune Tolerance (licensed by Siolta Therapeutics), Systems and Methods for Detecting Antibiotic Resistance (WO 2012027302 A3), Nitroreductase Enzymes (US 7687474 B2), Sinusitis Diagnostics and Treatments (WO 2013155370 A1), and Methods and Systems for Phylogenetic Analysis (US 20120264637 A1). S. V. L. cofounded and is currently a board member and paid consultant for Siolta Therapeutics, and owns 25% of its stock. J. C. E. reports a consultancy relationship with American Congress of Rehabilitative Medicine and grants received and pending from Sanofi. G. P. D. reports that his institution has received grant money from the NIH. He has also received money from the NIH, which was used for travel to present the results of the study at a national meeting. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med 2003; 163:487–94. [DOI] [PubMed] [Google Scholar]
- 2. Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y;. Tonawanda/Williamsville Pediatrics. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis 1997; 175:1440–5. [DOI] [PubMed] [Google Scholar]
- 3. Faden H, Stanievich J, Brodsky L, Bernstein J, Ogra PL. Changes in nasopharyngeal flora during otitis media of childhood. Pediatr Infect Dis J 1990; 9:623–6. [PubMed] [Google Scholar]
- 4. Harabuchi Y, Kodama H, Faden H. Outcome of acute otitis media and its relation to clinical features and nasopharyngeal colonization at the time of diagnosis. Acta Otolaryngol 2001; 121:908–14. [PubMed] [Google Scholar]
- 5. Advani S, Sengupta A, Forman M, Valsamakis A, Milstone AM. Detecting respiratory viruses in asymptomatic children. Pediatr Infect Dis J 2012; 31:1221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byington CL, Ampofo K, Stockmann C et al. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin Infect Dis 2015; 61:1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chonmaitree T, Alvarez-Fernandez P, Jennings K et al. Symptomatic and asymptomatic respiratory viral infections in the first year of life: association with acute otitis media development. Clin Infect Dis 2015; 60:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jansen RR, Wieringa J, Koekkoek SM et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol 2011; 49:2631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeMuri GP, Gern JE, Moyer SC, Lindstrom MJ, Lynch SV, Wald ER. Clinical features, virus identification, and sinusitis as a complication of upper respiratory tract illness in children ages 4-7 years. J Pediatr 2016; 171:133–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olenec JP, Kim WK, Lee WM et al. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J Allergy Clin Immunol 2010; 125:1001–6.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Powell KR, Shorr R, Cherry JD, Hendley JO. Improved method for collection of nasal mucus. J Infect Dis 1977; 136:109–11. [DOI] [PubMed] [Google Scholar]
- 12. Hayden FG, Herrington DT, Coats TL et al. ; Pleconaril Respiratory Infection Study Group Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin Infect Dis 2003; 36:1523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bochkov YA, Grindle K, Vang F, Evans MD, Gern JE. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol 2014; 52:2461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wessels E, Schelfaut JJ, Bernards AT, Claas EC. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J Clin Microbiol 2012; 50:1171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X, Mair R, Hatcher C et al. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int J Med Microbiol 2011; 301:303–9. [DOI] [PubMed] [Google Scholar]
- 16. Greiner O, Day PJ, Altwegg M, Nadal D. Quantitative detection of Moraxella catarrhalis in nasopharyngeal secretions by real-time PCR. J Clin Microbiol 2003; 41:1386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kloepfer KM, Lee WM, Pappas TE et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol 2014; 133:1301–7, 7.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brook I, Gober AE. Frequency of recovery of pathogens from the nasopharynx of children with acute maxillary sinusitis before and after the introduction of vaccination with the 7-valent pneumococcal vaccine. Int J Pediatr Otorhinolaryngol 2007; 71:575–9. [DOI] [PubMed] [Google Scholar]
- 19. Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J 2010; 29:304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chonmaitree T, Trujillo R, Jennings K et al. Acute otitis media and other complications of viral respiratory infection. Pediatrics 2016; 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernstein JM, Faden HF, Dryja DM, Wactawski-Wende J. Micro-ecology of the nasopharyngeal bacterial flora in otitis-prone and non-otitis-prone children. Acta Otolaryngol 1993; 113:88–92. [DOI] [PubMed] [Google Scholar]
- 22. Faden H, Waz MJ, Bernstein JM, Brodsky L, Stanievich J, Ogra PL. Nasopharyngeal flora in the first three years of life in normal and otitis-prone children. Ann Otol Rhinol Laryngol 1991; 100:612–5. [DOI] [PubMed] [Google Scholar]
- 23. Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis 2001; 184:451–9. [DOI] [PubMed] [Google Scholar]
- 24. Hanke CR, Grijalva CG, Chochua S et al. Bacterial density, serotype distribution and antibiotic resistance of pneumococcal strains from the nasopharynx of Peruvian children before and after pneumococcal conjugate vaccine 7. Pediatr Infect Dis J 2016; 35:432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Syrjänen RK, Herva EE, Mäkelä PH et al. The value of nasopharyngeal culture in predicting the etiology of acute otitis media in children less than two years of age. Pediatr Infect Dis J 2006; 25:1032–6. [DOI] [PubMed] [Google Scholar]
- 26. Fan RR, Howard LM, Griffin MR et al. Nasopharyngeal pneumococcal density and evolution of acute respiratory illnesses in young children, Peru, 2009–2011. Emerg Infect Dis 2016; 22:1996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Q, Wischmeyer J, Gonzalez E, Pichichero ME. Nasopharyngeal polymicrobial colonization during health, viral upper respiratory infection and upper respiratory bacterial infection. J Infect 2017; 75:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marom T, Alvarez-Fernandez PE, Jennings K, Patel JA, McCormick DP, Chonmaitree T. Acute bacterial sinusitis complicating viral upper respiratory tract infection in young children. Pediatr Infect Dis J 2014; 33:803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Revai K, Mamidi D, Chonmaitree T. Association of nasopharyngeal bacterial colonization during upper respiratory tract infection and the development of acute otitis media. Clin Infect Dis 2008; 46:e34–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaikh N, Hoberman A, Colborn DK et al. Are nasopharyngeal cultures useful in diagnosis of acute bacterial sinusitis in children?Clin Pediatr (Phila) 2013; 52:1118–21. [DOI] [PubMed] [Google Scholar]
- 31. Syrjänen RK, Herva EE, Mäkelä PH et al. The value of nasopharyngeal culture in predicting the etiology of acute otitis media in children less than two years of age. Pediatr Infect Dis J 2006; 25:1032–6. [DOI] [PubMed] [Google Scholar]
- 32. Pettigrew MM, Gent JF, Pyles RB, Miller AL, Nokso-Koivisto J, Chonmaitree T. Viral-bacterial interactions and risk of acute otitis media complicating upper respiratory tract infection. J Clin Microbiol 2011; 49:3750–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis 2008; 14:1584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Zalm MM, Wilbrink B, van Ewijk BE, Overduin P, Wolfs TF, van der Ent CK. Highly frequent infections with human rhinovirus in healthy young children: a longitudinal cohort study. J Clin Virol 2011; 52:317–20. [DOI] [PubMed] [Google Scholar]
- 35. Kristo A, Uhari M. Timing of rhinosinusitis complications in children. Pediatr Infect Dis J 2009; 28:769–71. [DOI] [PubMed] [Google Scholar]
- 36. Patel J, Faden H, Sharma S, Ogra PL. Effect of respiratory syncytial virus on adherence, colonization and immunity of non-typable Haemophilus influenzae: implications for otitis media. Int J Pediatr Otorhinolaryngol 1992; 23:15–23. [DOI] [PubMed] [Google Scholar]
- 37. Sajjan U, Wang Q, Zhao Y, Gruenert DC, Hershenson MB. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am J Respir Crit Care Med 2008; 178:1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siegel SJ, Roche AM, Weiser JN. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 2014; 16:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lemanske RF Jr, Jackson DJ, Gangnon RE et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol 2005; 116:571–7. [DOI] [PubMed] [Google Scholar]
- 40. Lee WM, Grindle K, Pappas T et al. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol 2007; 45:2626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jackson DJ, Gangnon RE, Evans MD et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med 2008; 178:667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gern JE, Pappas T, Visness CM et al. Comparison of the etiology of viral respiratory illnesses in inner-city and suburban infants. J Infect Dis 2012; 206:1342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee WM, Lemanske RF Jr, Evans MD et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 2012; 186:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]