Abstract
Objective
Osteoarthritis (OA) chondrocytes have impaired autophagy, one arm of the proteostasis network that coordinates proteome and organelle quality control and degradation. Deficient proteostasis impacts differentiation and viability, and inflammatory processes in aging and disease. Studying OA chondrocytes, we assessed ubiquitin proteasome system proteasomal function.
Methods
We evaluated human knee cartilages by immunohistochemistry, and assessed proteasomal function, levels of proteasomal core subunits and chaperones, and autophagy in cultured chondrocytes. Assays included Western blotting, quantitative RT-PCR, proteasomal protease activity and cell immunofluorescence.
Results
Human knee OA cartilages demonstrated polyubiquitin accumulation, with increased ubiquitin K48-linked polyubiquitinated proteins in situ, suggesting proteasomal impairment. Cultured OA chondrocytes demonstrated accumulation of K48 polyubiquitinated proteins, significantly decreased 20S proteasome core protease activity, and decreased levels of phosphorylated FOXO4 and PSMD11, a FOXO4-inducible promoter of proteasomal activation. Levels of proteasomal core subunits PSMB3, 5, 6, and assembly chaperone PSMG1 were not decreased in OA chondrocytes. In normal chondrocytes, PSMD11 siRNA knockdown stimulated certain autophagy machinery elements, elevated nitric oxide (NO) release, and decreased levels of chondrocytic master transcription factor SOX9 protein (and mRNA) and aggrecan (AGC1) mRNA. PSMD11 gain of function by transfection increased proteasomal function, raised levels of SOX9-induced AGC1 mRNA, stimulated elements of the autophagic machinery, and inhibited IL-1-induced NO and MMP13 release in OA chondrocytes.
Conclusion
Deficient PSMD11, associated with less phosphorylated FOXO4, promotes impaired proteasomal function in OA chondrocytes, dysregulated chondrocytic homeostasis, and decreased levels of SOX9 mRNA, SOX9 protein, and AGC1 mRNA. Chondrocyte proteasomal impairment may be a therapy target for OA.
Keywords: Osteoarthritis, Proteostasis, Autophagy, Cartilage, Sox9, Aggrecan, MMP13
INTRODUCTION
OA is a highly prevalent disease of the whole joint, in which aging, biomechanical injury, inflammation, metabolism, and other processes, converge to promote altered articular chondrocyte differentiation and function, and ultimate cartilage failure (1). Multiple hallmarks of tissue aging (2) are detected in OA articular chondrocytes (1). These include deficient activities of cell homeostasis signaling by Forkhead Box O (FOXO) transcription factor and AMP-activated protein kinase (AMPK))(3,4), mitochondrial dysfunction (5,6), and dysregulated proteostasis (5,7–11).
The proteostasis network, which includes the unfolded protein response (UPR), autophagy, and the ubiquitin-proteasome system (UPS), coordinates proteome synthesis and folding, and employs chaperones to direct proteins to loci for degradation (12–14). UPR activation alleviates stress from accumulation of damaged proteins in the ER lumen, but, including in experimental OA, dysregulated UPR activity can contribute to inflammation, tissue damage and degeneration (14–16). Conversely, distinct UPR genes and responses appear decreased in OA chondrocytes (17)
The autophagic process primarily degrades proteins and organelles under hypoxic and low nutrition/energy states, thereby exercising quality control and generating cell energy via recycling (5,7–11). Autophagy is constitutively active in normal articular chondrocytes, promoted by factors including FoxO, AMPK, and regulated in development and DNA damage response 1 (REDD1)(19,20). Autophagy, and AMPK activity, FoxOs, and REDD1, are decreased in aged and OA cartilages (3,4,19,20). Pharmacologic and transgenic inhibition of autophagy potentiates OA in mice (7,8), contrasting with chondroprotective effects of autophagy up-regulation (7,10,11).
The UPS accounts for ~80–90% of cell protein breakdown in proteostasis, via preferential degradation, within proteasomes, of damaged polyubiquinated proteins, and many short-lived, regulatory cytosolic proteins (eg, IκB, cyclins)(12,21). In the UPS, isopeptide bonds are formed between ubiquitin and protein substrates by ubiquitin-activating, -conjugating, and ubiquitin-protein ligases (E1–3, respectively)(21). Other ubiquitin molecules are sequentially conjugated to 1 of 7 lysine residues of attached ubiquitin, most commonly at ubiquitin K48, which targets proteins for proteasomal degradation (21). Alternatively, mono- or polyubiquitination (eg, via K63) affects protein function and trafficking, and reduces pressure on the proteasome by stimulating substrate aggregation and removal by chaperone-mediated autophagy (21–25).
The 26S proteasome, composed of 33 distinct subunits, is a cytosolic, energy-requiring ATP-driven "latent proteolytic machine”, regulated largely via substrate access, subunit expression, assembly, turnover, and function (12,21). Polyubiquitinated proteins bind a multi-unit 19S "regulatory cap". Substrates are partially unfolded, facilitated by deubiquitinating enzymes, to enter the barrel-shaped 20S proteolytic core (12,21). The 20S particle has 4 heptameric rings; the two outer ring α subunits regulate substrate access to the proteolytic core, and two inner ring β subunits have chymotrypsin-, trypsin-, and caspase-like activities (21). The 19S cap ATPases stimulate proteolytic activity by opening the 20S “gate” for substrate access (21). Additional regulation is exerted by proteasome-dedicated chaperones and subunits, including PSMG1 (26) and PSMD11 (27–30). The UPS also networks extensively with autophagy (12,13), FOXOs, and AMPK (31,32).
Differing proteostasis demands, aging patterns, and stressors, are linked with variances in UPS composition and activity between tissues, and with age (33). Robust UPS capacity in youth maintains proteome integrity, and inhibits senescence (34). Aging facilitates UPS failure, via accumulated misfolded proteins, and, in some tissues, UPS collapse under stress predates aging (21). UPS failure promotes multiple hallmarks of aging (12,13,21,22,31), senescence and death in cells, disease in multiple tissues, and decreased organism longevity (21).
Prior studies modulating proteasomal function in OA have been limited to experimental rat and mouse knee OA in wild type young animals (35–38). These works assessed nascent disease triggered by surgical knee instability or monosodium iodoacetate, and principally analyzed effects of potent, broad proteasomal protease pharmacologic inhibition using the peptide aldehyde MG132, started prior to OA onset (35–38). MG132 treatment was chondroprotective, with the integrity of proteasomal function appearing to be permissive for OA development and early disease advancement; decreased proteasomal degradation of the NF-κB inhibitor IκB was implicated in these effects, by lessening inflammation and cartilage matrix catabolism (35–38). Our study characterized proteasomal dysfunction in human chondrocytes from donors with knee OA. Results elucidated novel potential pathophysiologic impact of chondrocyte proteasomal impairment in established human OA, and a molecular mechanism for selective reversibility of some OA-promoting effects in diseased chondrocytes via improved proteasomal function.
MATERIALS AND METHODS
Reagents
Unless otherwise indicated, all chemical reagents were from Sigma-Aldrich (St Louis, MO), Recombinant human IL-1β was from Gemini Bio-Products (Sacramento, CA). High-glucose Dulbecco's modified Eagle's medium (DMEM) with L-glutamine and penicillin/streptomycin solution was from Mediatech (Manassa, VA). The MMP13 ELISA kit was from Thermofisher (Waltham, MA).
Human tissue samples and cell culture
All human subject studies were approved by the San Diego VA and Scripps Research Institute IRBs. Human knee chondrocytes were isolated from autopsy donors that were graded macroscopically, as described (6). Here, we studied Grade 0 normal, and Grade I representing cartilage with an intact surface, and Grade IV representing moderate to severe OA, with overt fibrillation (6). There was no pre-selection of donors or donor samples for any characteristic other than OA or non-OA. Chondrocytes were cultured in Dulbecco's modified Eagle's (DMEM) high glucose medium, with L-glutamine and 100 IU/ml penicillin/100 µg/ml streptomycin (Mediatech), supplemented with 10% fetal calf serum, at 37°C (6). Chondrocytes were studied in first passage, or, where indicated, as primary cells.
Immunohistochemistry
Human knee cartilage paraffin-embedded sections were incubated with primary antibody for 18 h at 4°C. We used rabbit anti-K48-linked proteins (05–1307, EMD Millipore. Temecula, CA), and anti-total ubiquitin (sc-166553, Santa Cruz Biotechnology, Dallas, TX) antibodies. HRP-conjugated secondary antibody employed SuperPicture Polymer Detection Kit (Thermo Scientific), using colorimetric detection with diaminobenzidine (DAB) substrate.
Transfection and expression plasmid construction
Human PSMD11 cDNA was obtained from Addgene.org (pQTEV-PSMD11, ID 31333), amplified by PCR, and subcloned, with and without human influenza hemagglutinin (HA)-tagging at the N-terminal, into pCDNA3.1(+) plasmid vector between Xho1 and BamH1 sites (primers cited in Supplemental Table 1), and verification by sequencing. Human chondrocytes were cultured in 6-well plates for 16–18 h, then transfected with PSMD11 using XtremeGENE HP DNA reagent (Roche) following manufacturer instructions, for 72h. SDS-PAGE in Bis-Tris gels (Thermo Scientific), and Western blotting of cultured chondrocyte lysates, were done as previously detailed (6). Primary antibodies to PSMD11 (#14303), Caspase-3 (#9662), PARP (#9532) and HA-tag (#3724) were from Cell Signal Technologies (Danvers, MA). Secondary antibodies (1:5000) were HRP-conjugated and detected with ECL reagent (Super Pico or Femto, Thermo Scientific), either using X-Ray film or Gel Doc system (BioRad)
Western blotting
SDS-PAGE of cultured chondrocyte lysates in Bis-Tris gels (Thermo Scientific), and Western blotting, were done as previously detailed (6). Primary antibodies to PSMG1 (#13378), PSMD11 (#14303), FOXO1 (#2880), FOXO3a (#12829), FOXO4 (#2499), phosphorylated FOXO1/FOXO3a (p-FOXO1/p-FOXO3A) (#9464), Sox9 (#82630), LC3/A/B (#12741), p62 (#8025), and HA-tag (#3724) were from Cell Signal Technologies (Danvers, MA). Antibodies to p-FOXO4 (#47278) were from Abcam (Cambridge, UK), and to MMP13 (#3533) from BioVision (Milpitas, CA). Secondary antibodies (1:5000) were HRP-conjugated and detected with ECL reagent (Super Pico or Femto, Thermo Scientific). Quantification of band intensity was performed with ImageStudioLite (LiCOR, Lincoln, NE), with intensity of bands calculated relative to the background from the same membrane.
Proteasome Activity Assay
Proteasome activity assays specifically analyzed chymotrypsin-like activity present at the core proteasome particle, utilizing an AMC(7-amino-4-methylcoumarin)-tagged peptide substrate (Succ-LLVY-AMC), that releases free, highly fluorescent AMC in the presence of protease activity. In brief, cells were lysed at 4°C in PBS, containing 0.5% NP-40, followed by centrifugation at 15,000 rpm at 4°C. After protein level determination, supernatant was assayed for chymotrypsin-like activity, using a proteasome activity kit from Abcam (ab107921), per manufacturer instructions. To do so, 10 µg aliquots of protein, including samples and positive control from the manufacturer, were assayed in triplicate, with and without proteasome inhibitor MG132 (200 µM), and incubation for 30 min at 37°C in the dark. Output was measured on a fluorimetric microplate reader at T1 at 350/440 nm (excitation/emission) and T2 25 minutes later. The relative fluorescence units (RFU) were determined as followed: ΔRFU = (RFU2 − iRFU2) − (RFU1 − iRFU1), where iRFU represents wells containing proteasome inhibitor showing non-proteasome activity. Proteasome activity was calculated as: , where: B = Amount of AMC in the sample well (pmol); V = Sample volume added into the reaction well (µL); T1 = Time (min) of the first reading (RFU1 and iRFU1); T2 = Time (min) of the first reading (RFU2 and iRFU2); D = Sample dilution factor. Calibration curves were performed for AMC concentration and purified 20S proteasome activity.
Quantitative reverse transcription (RT)-PCR
RNA was extracted and isolated from cultured human chondrocytes, utilizing RNeasy mini kit (Qiagen, Hilden, GE). RNA levels were quantified using a Nanodrop 1000 spectrophotometer (Thermo Scientific); samples with 260 nm/280 nm absorbance ratios >2.0 were used for RT. Equal amounts of total RNA from each sample underwent RT reaction, using Transcriptor High Fidelity cDNA Synthesis Kit (Roche), which includes anchored oligo(dT)18 and random hexamer primers. Quantitative RT-PCR was done using a Roche480 Light Cycler (Roche) and specific primer sets (Supplementary Table 1). Levels of mRNA for genes of interest were calculated as fold change from control or as ratio correlating copy number of gene of interest versus total housekeeping gene GAPDH copy number. For absolute quantification, pGEM-T Easy vector system (Promega, Madison, WI) was used to clone gene fragments (GAPDH, PSMB5, PSMB3, PSMB8 and PSMD11).
Immunofluorescence
Human chondrocytes were cultured on 6-well plates containing round cover slips (12mm, VWR) for 16–18 h, then transfected (XtremeGENE siRNA transfection reagent, Roche) with PSMD11 siRNA (on-target, L-011367-01-0005, GE Dharmacon) for 72 h. Culture media was removed from plates and cells were fixed-permeabilized with cold methanol (−20°C) for 15min. Serial washes were performed with PBS, followed by blocking with 1% BSA in PBS for 2 h at room temperature. Incubation with primary IgG was conducted overnight at 4°C with rabbit anti-LC3A/B (12741, Cell Signal) or rabbit anti-LAMP1 (ab24170, Abcam) or rabbit anti-p62/SQSTM1 (88588, Cell Signal). Secondary antibody incubation (2 h at 22°C) detected primary antibody with Alexa Fluor 488 goat anti-rabbit IgG (A11034, Thermo Scientific) or with Alexa Flour 555 goat anti-rabbit IgG (21429, Thermo Scientific). Imaging employed a BZ-X700 microscope (Keyence, Itasca, IL).
Chromatin Immunoprecipitation (ChIP) Assay
We tested a defined, specific 48 bp sequence in the Sox9-binding enhancer region of collagen II α1 (COL2A1) for cartilage expression (39). We also assessed a defined, functional SOX9/HIF1a complex binding site in the 5'-UTR of AGC1 (40). To do so, we used primers corresponding to nucleotides 60–82 [forward] and 220–240 [reverse] in the AGC1 −640 to −510 promoter region (GenBank: AF031586). ChIP assay (Millipore) was carried out per manufacturer instructions, using Protein A agarose/Salmon Sperm DNA (50% slurry) reduce nonspecific background, and immunoprecipitation with SOX9 antibody (ab3697, Abcam). Following elution of histone/DNA complexes, and DNA recovery by phenol/chloroform extraction, DNA was resuspended in 10 µL TE buffer. PCR was conducted with 1 µL of eluate, 2 µL of 20 µM primers for Col2a1 and AGCN1 (Supplemental Table 1), 10 µL KAPA2G Fast ReadyMix PCR (Kapa Biosystems reagent from Sigma-Aldrich) and H2O up to 20 µL reaction volume.
Statistical analyses
Statistical analyses were done using GraphPad Prism software. The difference between means of unpaired samples was performed using two-way analysis of variance with Bonferroni's post-test or using unpaired t-test.
RESULTS
Ubiquitinated protein substrates are increased in human OA chondrocytes and OA cartilage in situ
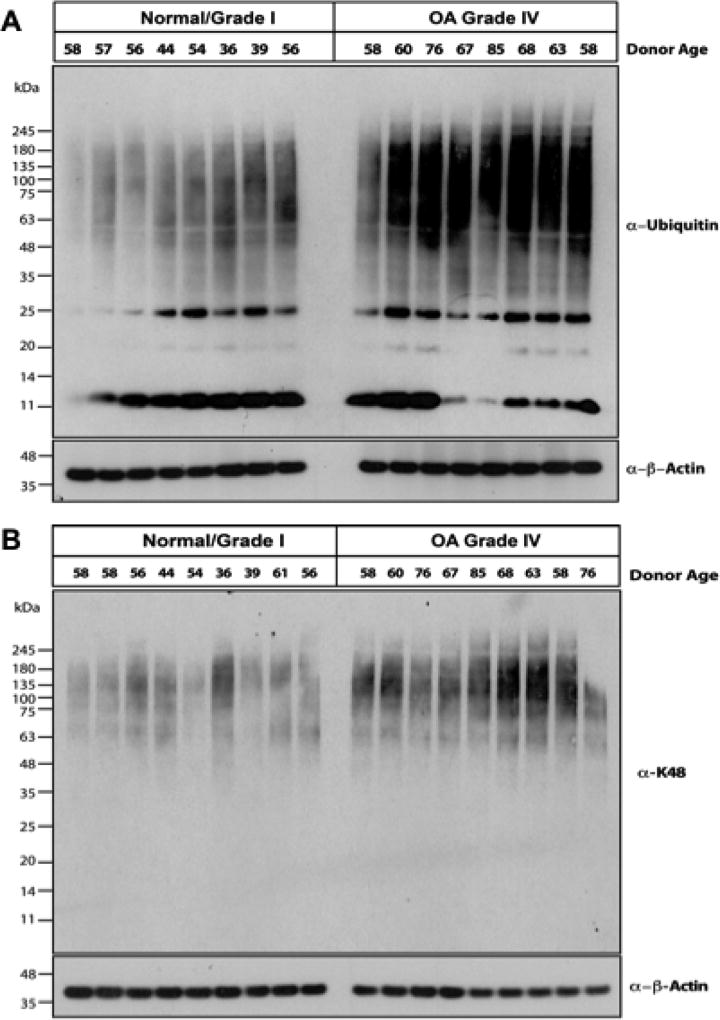
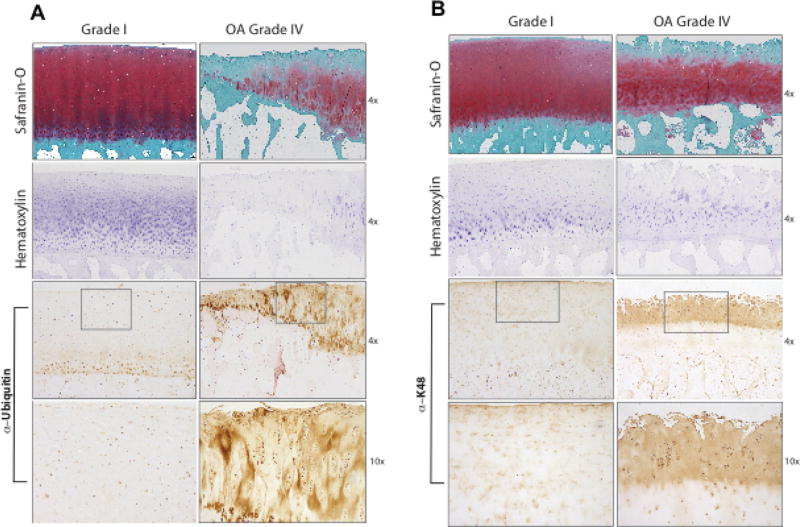
We observed increased levels of ubiquitinated proteins in OA compared to normal chondrocytes, in Western blot of cell lysates analyzed for total ubiquitin (Ub), and K48-linked substrates (Figure 1A, B, respectively). Both ubiquitin (Figure 2A) and K48-linked polyubiquitin levels were increased in human knee OA cartilages, compared to normal cartilages, assessed by immunohistochemistry (Figure 2A, B, respectively). In Grade IV OA cartilages, polyubiquitin staining was particularly intense in chondrocyte clusters (Figure 2B).
Figure 1. Increased ubiquitin and polyubiquitinated substrate levels in human knee OA chondrocytes.
Equal aliquots (10 µg) of lysates from normal (Grade 0/Grade I) and Grade IV OA human articular chondrocytes were used for 4–12% SDS-PAGE/Western blot analysis, employing antibodies directed against ubiquitin (Ub, n=16) (A) and K48-linked ubiquitin (n=18) (B). Equal protein loading was also confirmed by blotting for β-actin.
Figure 2. Increased levels of ubiquitin and polyubiquitinated substrates in human knee OA cartilages in situ.
Immunohistochemistry of paraffin embedded sections from grade IV OA and normal human knee articular cartilages employed antibodies specific for ubiquitin (Ub) (A) and K48-linked ubiquitin (K48) (B). Safranin-O, and hematoxylin cell staining are shown at 4× magnification. For IHC, 4× and 10× magnification results are shown, with the third panels from the top showing a boxed area magnified in the panels immediately below. Representative of 12 separate normal and 12 separate OA donors.
Decreased chondrocyte proteasome activity in OA
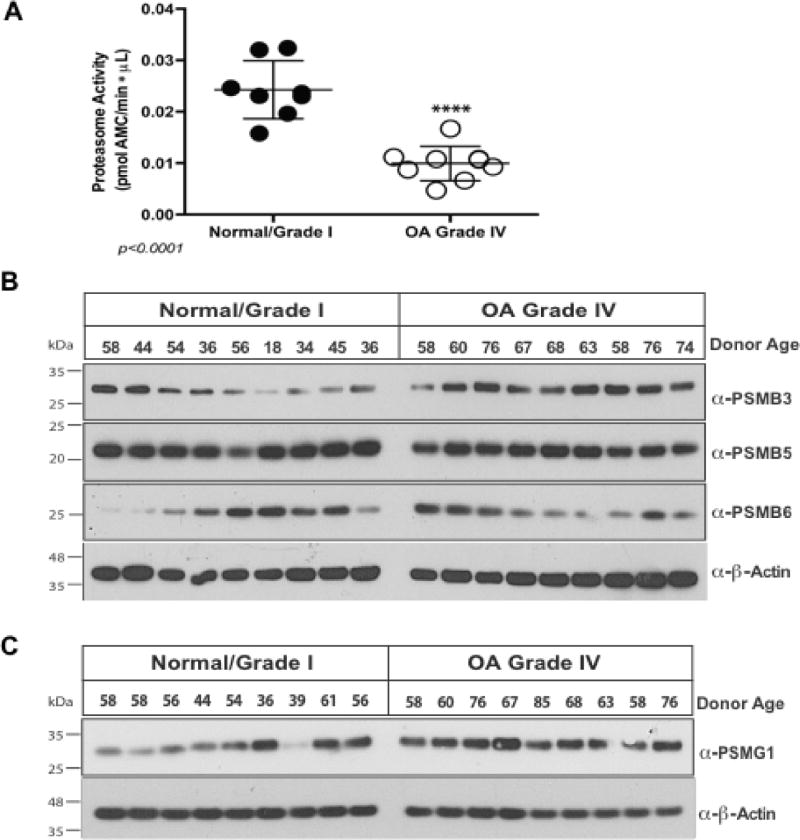
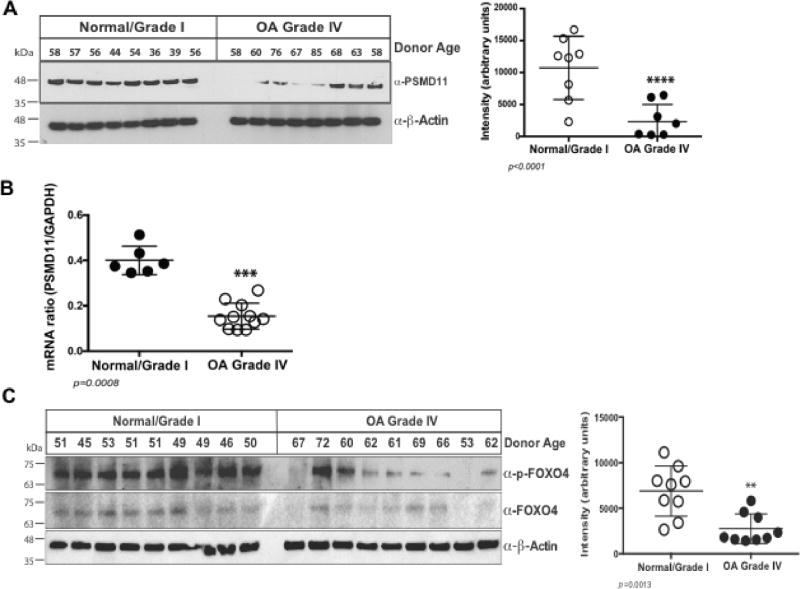
Proteasome activity was significantly decreased in cultured chondrocyte lysates from Grade IV OA compared to normal donors (Figure 3A). Examining three of the seven 20S proteasome core β subunit constituents of the inner ring, where catalytic activity is localized, Western blots revealed no significant decrease in PSMB3, PSMB5 and PSMB6 levels in Grade IV OA chondrocytes (Figure 3B). Hence, we examined the critical proteasome regulatory subunits PSMG1 and PSMD11 (27,28), since PSMG1 facilitates 20S proteasome assembly by promoting grouping of the β subunits into the heteroheptameric α ring (26), and PSMD11 stabilizes the interaction between 19S and 20S subunits of the proteasome (27,28). OA chondrocytes did not have significant decrease in PSMG1 levels (Figure 3C). In contrast, OA chondrocytes demonstrated decreased PSDM11 protein (Figure 4A) and PSMD11 mRNA levels (Figure 4B). Levels of FOXO4, a transcription factor that stimulates PSMD11 mRNA expression (28,31), were grossly comparable in normal and OA chondrocytes. Phosphorylation of FOXOs is required for translocation to the nucleus and subsequent functionality (41). We observed that levels of p-FOXO4 were markedly decreased in OA compared to normal chondrocytes (Figure 4C).
Figure 3. Proteasome activity is diminished in OA chondrocytes, without significant decrease in levels of critical 20S inner ring β subunits and proteasome assembly chaperone PSMG1.
(A) Lysates were collected from normal/grade I (n=8) and grade IV (n=8) chondrocytes, in the absence of protease inhibitors; 10 µg protein aliquots were assayed for 20S proteasome function, using SUC-LLVY-AMC substrate, measuring at 37°C at 351nm/430nm excitation/emission. Replicate controls had proteasome inhibitor MG132 (200 µM). We observed greater than 50% decrease (p<0.0001, t-test) in proteasome protease activity in OA. (B) Aliquots (10 µg) of protein extracted from primary chondrocytes (n=18 donors, 9 normal and 9 grade IV) were analyzed by Western blot to determine levels of critical 20S proteasomal inner ring β-subunits, revealing no significant variation between normal and grade IV. (C) PSMG1 was not appreciably decreased in a panel of OA chondrocytes compared to normal donor cells.
Figure 4. PSMD11 and p-FOXO4 levels are decreased in OA chondrocytes.
(A) Western blots of chondrocyte lysates, with densitometric analysis, revealed decreased PSMD11 in OA grade IV compared to normal donor cells. (B) PSMD11 mRNA levels were significantly decreased in OA compared to normal chondrocytes (p=0.0008, t-test). (C) Western blots, with densitometric analysis, also revealed decreased p-FOXO4 levels, but no consistent changes in total FOXO4, in OA compared to normal donor chondrocytes.
Decreased PSMD11 reduces levels of SOX9 and AGC1 in chondrocytes
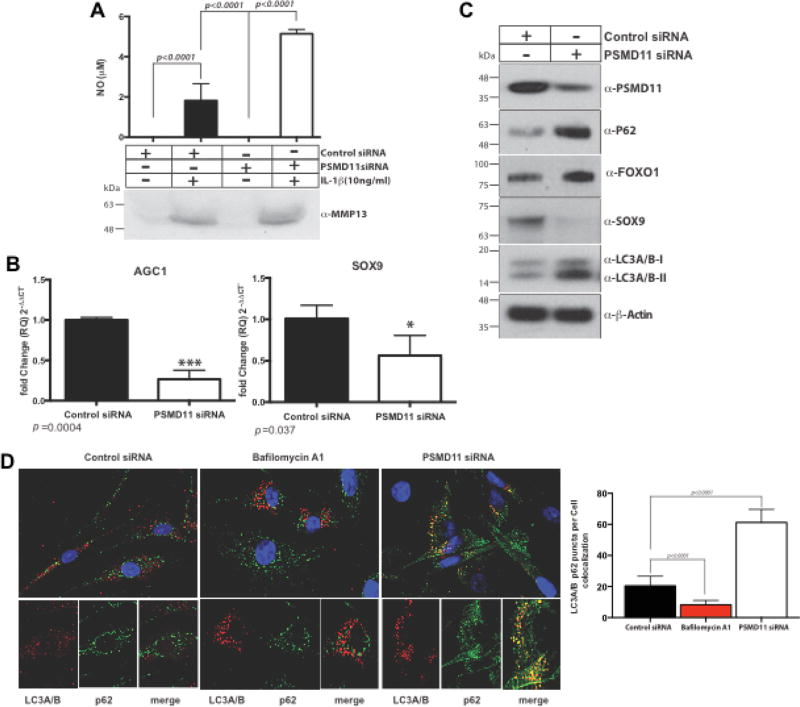
We first assessed, via transfection with PSMD11 siRNA, the functional implications of selective decrease of PSMD11 expression in normal chondrocytes. We validated induction of decrease in level of PSMD11, associated with reduced proteasome activity and increased levels of K48 polyubiquinated peptides (Supplemental Figure 1A–B), without significant changes in levels of total and p-FOXO1, p-FOXO3a, and p-FOXO4 (Supplemental Figure 1C). Under these conditions, in normal chondrocytes, PSMD11 knockdown induced a greater than 70% increase of NO release in response to IL-1β (10 ng/ml), but no significant difference in MMP-13 protein level (Figure 5A). MMP3 and MMP13 mRNA levels were not significantly affected by PSMD11 siRNA (data not shown). In contrast, PSMD11 siRNA knockdown decreased SOX9 and AGC1 mRNA levels (Figure 5B), and attenuated SOX9 protein levels in normal chondrocytes (Figure 5C). In normal chondrocytes, PSMD11 siRNA stimulated multiple elements of the autophagic machinery, specifically evidenced by LC3A/B-I to LC3A/B-II conversion and increased levels of the autophagy-UPS adaptor and LC3-binding protein p62 (13) (Figure 5C), with associated perinuclear co-localization of p62 and LC3A/B in normal chondrocytes (Figure 5D).
Figure 5. PSMD11 siRNA knockdown induces increased NO release, induces decreases in SOX9 and AGC1, and stimulates elements of the autophagy process, in normal chondrocytes.
We studied PSMD11 siRNA knockdown effects on NO and MMP13 release in response to IL-1β (10 ng/ml) in normal human knee chondrocytes (A). We also analyzed PSMD11 siRNA effects on SOX9 and AGC1 (B), and SOX9 protein expression, as well as autophagy-related LC3A/B-I to LC3A/B-II conversion, the autophagy-UPS adaptor and LC3-binding protein p62 (C), and perinuclear co-localization of p62 and LC3A/B-I to LC3A/B-II in normal chondrocytes (D), with 300 cells/treatment counted to quantify structures where LC3A/B and p62 co-localized. Analysis was by 2-way ANOVA (Multiple comparison with Bonferroni post hoc comparison).
Effects of PSMD11 gain of function in human OA chondrocytes
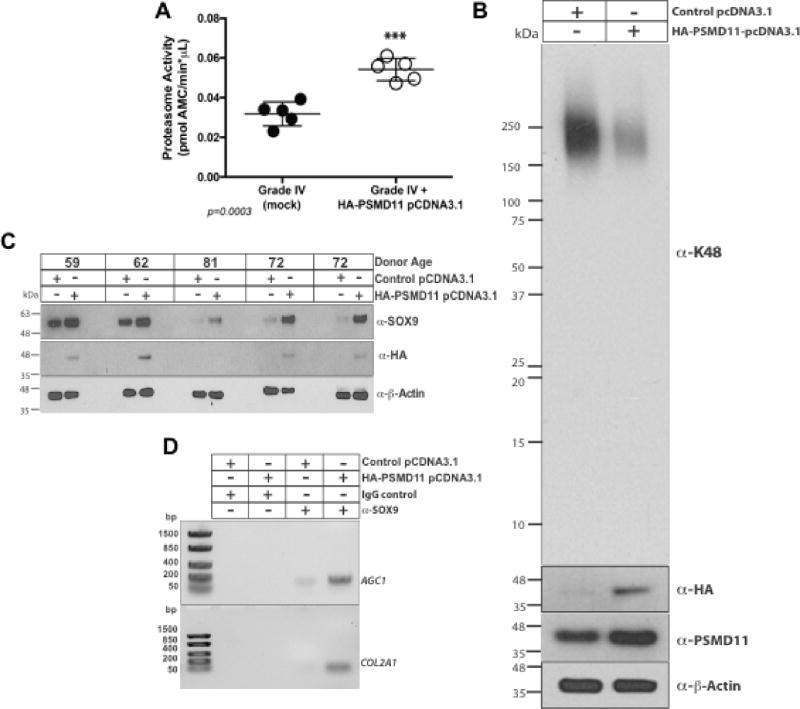
Since PSMD11 was decreased in Grade IV OA chondrocytes, we induced selective PSMD11 gain of function by transfection in OA cells. To do so, we followed an approach previously effective in stem cells (28). Transfecting HA-tagged PSMD11 cDNA, we achieved significant increases in proteasome activity, and observed associated decrease in levels of K48 polyubiquitinated substrates in OA chondrocytes (Figure 6A–B), markedly lower IL-1β-induced NO release (Supplemental Figure 2A), decreased intracellular and released MMP13 (Supplemental Figure 2A–B), and increased SOX9 protein level in 5 of 5 human OA chondrocyte donor samples tested (Figure 6C). Moreover, ChIP assays demonstrated that PSMD11 gain of function increased SOX9 binding to the AGC1 promoter and COL2A1 enhancer (Figure 6D). Under these conditions, PSMD11 gain of function stimulated multiple elements of the autophagic machinery, evidenced by changes in LC3A/B conversion and p62 (Supplemental Figure 3A), and associated increase in perinuclear co-localization of p62 and LC3A/B in 5 out of 5 OA chondrocyte donors tested (data not shown). Last, PSMD11 transfection induced increased Sox9 nuclear localization and levels of SOX9, AGC1, and COL2A1 mRNA in OA chondrocytes (Supplemental Figure 3B–C).
Figure 6. Effects of selective PSMD11 gain of function in cultured human knee OA chondrocytes.
(A) Transfecting Grade IV OA chondrocytes, we assessed effects of HA tag-PSMD11 plasmid, compared to control plasmid transfection, on proteasome activity (n=4 donors), and (B) on accumulation of K48-polyubiquitinated proteins (results representative of 4 donors). (C) PSMD11 gain of function by transfection in OA chondrocytes (n=5 different donors), validated to augment cellular HA, was assessed for effects on SOX9 levels. (D) Using ChIP assay, we separately analyzed the effect of PSMD11 gain of function on nuclear SOX9 binding to the AGC1 promoter and COL2A1 enhancer.
Discussion
In this study, we demonstrated that proteasomal function is impaired in human knee OA chondrocytes, and that integrity of proteasomal function plays a significant role in maintenance of differentiation of cultured chondrocytes. Our studies included analyses of intersections between proteasomal function and autophagy in normal and OA chondrocytes.
Selective induction of proteasomal dysfunction in normal donor chondrocytes, achieved by PSMD11 siRNA, stimulated multiple elements of the autophagic machinery, including p62 and p62 perinuclear co-localization with LC3. This finding may be due to an acute stress response to PSMD11 knockdown in healthy cells able to adaptively increase autophagy to support proteostasis. Gain of proteasomal function in OA donor chondrocytes, achieved selectively by PSMD11 transfection, was able to induce increase in elements of the autophagic machinery. Distinct autophagic pathways (microautophagy, chaperone-mediated autophagy, and macroautophagy) effect degradation of cytosolic proteins, employing multiple enzymes, and differing cargo delivery to lysosomes or vacuoles, and with ordered phases of each process (13). Full examination of cross-talk between the UPS and autophagy processes (13), and evaluation of ultimate autophagic protein degradation, were beyond the scope of this study. However, effectiveness of the autophagic process in OA chondrocytes is known to be decreased once OA is well-established (3,4,19,20). As such, our results suggest that proteasomal function compromise is likely more deleterious in aging and OA chondrocytes (12) by a compound proteostasis defect that includes autophagy impairment (7–11). The impact of proteasomal dysfunction in OA chondrocytes could potentially be amplified by decreased autophagic removal of damaged proteasomes (42).
Our study defined molecular mechanistic factors contributing to deficient proteasomal function in human knee OA chondrocytes, and associated accumulation of K48 polyubiquitinated substrates. Specifically, we linked proteasomal dysfunction in OA chondrocytes to decreases in PSMD11 and of the PSMD11 transcription inducer p-FOXO4, but not total FOXO4. PSMD11, a 19S proteasomal subunit. is required for assembly of a functional 20S proteasomal core (27,28,31). Essentially, 20S particles exist as free complexes, with constitutively latent proteolytic activity primed by 19S particle ATPases that also permit access of proteins to the 20S core (27,28). Two members of the forkhead box O family, FOXO1 and FOXO3, are decreased in aging and OA chondrocytes in vitro, and articular cartilages in situ (3,19). Though FOXO1 and FOXO3 are far more abundant than FOXO4 in human articular chondrocytes, FOXO1 and FOXO3 do not have a defined role in regulating proteasomal activity, unlike FOXO4.
The observed decrease in PSMD11 in OA chondrocytes was at least partially selective, relative to other modulators of proteasome assembly and 20S proteasomal core protease activity. Specifically, 3 inner ring β subunits and the 20S particle assembly chaperone PSMG1 were not decreased in OA compared to normal human knee chondrocytes. Another central finding was that selective PSMD11 gain of function by transfection improved proteasomal function in OA chondrocytes, evidenced by increase in 20S chymotrypsin-like proteolytic activity and decreased K48 polyubiquitinated substrates. PSMD11 is regulated by factors other than FOXO4, including cAMP and PKA signaling, which transcriptionally support PSMD11 expression (30). However, cAMP and PKA signaling have mixed effects on matrix catabolism by articular chondrocytes that would theoretically limit their potential translational utility in increasing proteasomal function in OA chondrocytes.
Selective modulation of PSMD11, beyond effects on proteosomal function, had marked functional impact in normal and OA chondrocytes, with major effects on maintenance of SOX9 and chondrocytic differentiation. We defined affected responses to include levels of SOX9 protein and SOX9 nuclear translocation and binding to the AGC1 and COL2A1 promoters, and levels of AGC1 and COL2A1 mRNA expression. SOX9 is influenced by multiple signals, including those provided by a variety of growth factors, transcription factors, and epigenetic modifications (43). As such, the net contributions of PSMD11 and proteasomal function on SOX9, AGC1, and maintenance of articular chondrocytic differentiation, remain to be determined both in early and late OA.
PSMD11-selective modulation of proteosomal function significantly affected IL-1β-triggered release of NO in both normal and OA chondrocytes, and release of MMP13 by OA chondrocytes. The NO findings carry significance partly due to the capacity of NO to exert both anti-anabolic and pro-catabolic effects on extracellular matrix homeostasis in cartilage, including suppression of proteoglycans and collagen synthesis (44). Moreover, excess NO promotes chondrocyte apoptosis (45,46). NO is generated and degraded by a variety of mechanisms in chondrocytes (46). Moreover, the UPS regulates NO metabolism by complex and paradoxical mechanisms, including regulation of iNOS expression by NF-κB, and of proteasomal degradation of inducible NO synthase (iNOS)(47,48). Hence, the mechanism of the observed capacity of proteasomal dysfunction to modulate NO release in chondrocytes could be multifactorial.
Both UPS dysfunction and OA increase progressively with aging (1,2,21), and this study was not designed powered to separate effects of aging from OA in chondrocytes. That question clearly will require further study. Another limitation is that our studies of protease activity in isolated proteasomes from cytosol, rather than whole cells, could have readily underestimated the extent of decreased proteasomal function in OA chondrocytes. In this context, substrate selection and gate opening are highly regulated processes that are highly dependent on ATP. OA chondrocytes, largely due to mitochondrial dysfunction, have decreased ATP levels (6).
Our results support a model in which impaired chondrocyte proteasomal function in established OA renders cartilage damage more difficult to slow and potentially reverse. However, there could well be substantially less chondrocyte proteasomal impairment in earlier stages of OA, such as those previously studied (35–38). The chondrocyte UPS dysfunction studied here may reflect an advanced stage of disease, given that we focused on chondrocytes from donors with advanced (Grade IV) OA. Further studies on UPS effects in chondrocytes from cartilages early OA would be of interest. In this regard, multiple pharmacologic proteasome protease inhibitors, including MG132, increase autophagy in normal cells (13,49). As such, our results suggest that chondroprotective effects of MG132 administration in nascent experimental knee OA in young rodents (35–38) may have been partly mediated by induction of increased autophagy in the normal chondrocytes of young animals. Other chondroprotective mechanisms in vivo of pharmacologic proteasomal inhibition by MG132 in young animals could include suppression of articular inflammatory processes and cartilage matrix catabolism, and pain (35–38), partially via increased protein stability of the proteasomal degradation-regulated native NF-κB inhibitor IκB (37,38). Proteasomal protease inhibitors such as MG132 are only one means to elucidate proteasome function in OA. Specifically, mice transgenic for K48R-mutated ubiquitin, to study focused interference with polyubiquitination, had only marginal and focal chondroprotection in rodent knee instability-induced OA, albeit in a system with potentially problematic residual effects of endogenous normal ubiquitin (38). In the end, questions concerning the impact of the phenotype of proteasomal dysfunction in OA chondrocytes would benefit most from replication of specific molecular mechanisms causing proteasomal function impairment. This is particularly the case because not only proteasomal core proteases, but also deubiquitinases impart and tune proteasomal functionality (13,21).
In conclusion, we have provided the first evidence, to our knowledge, of impaired proteasomal function in human OA chondrocytes. We mechanistically implicated decreases in chondrocyte PSMD11 and p-FOXO4, with consequent decreases in SOX9 mRNA and SOX9 protein and AGC1 promoter and COL2A1 mRNA levels, along with increased NO release in response to inflammatory stimulation. However, we note that UPS dysfunction promotes disease partly by effects on diverse processes, promoting mitochondrial dysfunction, oxidative stress, and altered IGF receptor, insulin receptor, PI3K, and Akt signaling (12,13,21,22,31). Translationally, our results revealed that proteasomal gain of function, achieved in a highly selective manner by PSMD11 gain of function, stimulated elements of the autophagy process in both normal and advanced knee OA chondrocytes, and ialso increased SOX9, AGC1 promoter and COL2A1 enhancer binding, and AGC1 and COL2A1 mRNA. Pharmacologic means are emerging to up-regulate 20S proteasomal activity, including by use of small molecules such as deubiquitinase inhibitors (50). The chondrocyte proteasome could provide a novel, pharmacologically targetable therapy in OA, potentially more chondroprotective in well-established disease.
Supplementary Material
We transfected normal human chondrocytes with PSMD11 siRNA, and (A) validated induced decreased level of PSMD11, and increased levels of K48 polyuybiquinated peptide substrates, and (B) decreased proteasome protease activity. PSMD11 siRNA knockdown did not induce detectable changes in total and phosphorylated FOXO1, FOXO3a, and FOXO4 (C).
Transfecting Grade IV OA chondrocytes, we assessed effects of HA tag-PSMD11 plasmid, compared to control plasmid transfection, on NO release (n=4 donors) and intracellular MMP13 (A)(data representative of 4 donors studied), and extracellular MMP13 protein released (B)(n=10 donors), in response to IL-1β.
(A) We transfected OA chondrocytes with HA-tagged PSMD11 cDNA, and assessed levels of p62 and LC3A/B conversion (n=4). (B) We assessed levels of SOX9, and of SOX9 translocated to the nucleus. (C) We also assayed SOX9, COL2A1, and AGC1 mRNA.
Acknowledgments
Supported by the NIH (PAGO07996; RT, MKL), and VA Research Service (RT, RLB).
Eric J Bennett, PhD, Biological Sciences, Cell and Experimental Biology, UC San Diego, provided valuable advice in designing proteasomal analyses in this study.
Footnotes
Disclosures : None
References
- 1.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Review. Nat Rev Rheumatol. 2016;12:412–20. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Review. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014;22:162–70. doi: 10.1016/j.joca.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terkeltaub R, Yang B, Lotz M, Liu-Bryan R. Chondrocyte AMP-activated protein kinase activity suppresses matrix degradation responses to proinflammatory cytokines interleukin-1β and tumor necrosis factor α. Arthritis Rheumatol. 2011;63:1928–37. doi: 10.1002/art.30333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López de Figueroa P, Lotz MK, Blanco FJ, Caramés B. Autophagy activation and protection from mitochondrial dysfunction in human chondrocytes. Arthritis Rheumatol. 2015;67:966–76. doi: 10.1002/art.39025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu-Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor γ coactivator 1α. Arthritis Rheumatol. 2015;67:2141–53. doi: 10.1002/art.39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng NT, Guo A, Meng H. The protective role of autophagy in experimental osteoarthritis, and the therapeutic effects of Torin 1 on osteoarthritis by activating autophagy. BMC Musculoskelet Disord. 2016;17:150. doi: 10.1186/s12891-016-0995-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouderlique T, Vuppalapati KK, Newton PT, Li L, Barenius B, Chagin AS. Targeted deletion of Atg5 in chondrocytes promotes age-related osteoarthritis. Ann Rheum Dis. 2016;75:627–31. doi: 10.1136/annrheumdis-2015-207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caramés B, Olmer M, Kiosses WB, Lotz MK. The relationship of autophagy defects to cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015;67:1568–76. doi: 10.1002/art.39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama K, Kawakami Y, Kobayashi M, Greco N, Cummins JH, Matsushita T, et al. Local intra-articular injection of rapamycin delays articular cartilage degeneration in a murine model of osteoarthritis. Arthritis Res Ther. 2014;16:482. doi: 10.1186/s13075-014-0482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2015;74:1432–40. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 12.Vilchez D, Saez I, Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Review. Nat Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 13.Lilienbaum A. Relationship between the proteasomal system and autophagy. Review. Int J Biochem Mol Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 14.Hotamisligil GS, Davis RJ. Cell Signaling and Stress Responses. Review. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husa M, Petursson F, Lotz M, Terkeltaub R, Liu-Bryan R. C/EBP homologous protein drives pro-catabolic responses in chondrocytes. Arthritis Res Ther. 2013;15:R218. doi: 10.1186/ar4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uehara Y, Hirose J, Yamabe S, Okamoto N, Okada T, Oyadomari S, et al. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthritis Cartilage. 2014;22:1007–17. doi: 10.1016/j.joca.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Li YH, Tardif G, Hum D, Kapoor M, Fahmi H, Pelletier JP, et al. The unfolded protein response genes in human osteoarthritic chondrocytes: PERK emerges as a potential therapeutic target. Arthritis Res Ther. 2016;18:172. doi: 10.1186/s13075-016-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo FJ, Xiong Z, Lu X, Ye M, Han X, Jiang R. ATF6 upregulates XBP1S and inhibits ER stress-mediated apoptosis in osteoarthritis cartilage. Cell Signal. 2014;26:332–42. doi: 10.1016/j.cellsig.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Akasaki Y, Alvarez-Garcia O, Saito M, Caramés B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349–58. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Garcia O, Matsuzaki T, Olmer M, Plate L, Kelly JW, Lotz MK. Regulated in Development and DNA Damage Response 1 Deficiency Impairs Autophagy and Mitochondrial Biogenesis in Articular Cartilage and Increases the Severity of Experimental Osteoarthritis. Arthritis Rheumatol. 2017;69:1418–1428. doi: 10.1002/art.40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Review. Annu Rev Biochem. 2015;84:435–64. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löw P. The role of ubiquitin-proteasome system in ageing. Gen Comp Endocrinol. 2011;172:39–43. doi: 10.1016/j.ygcen.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Review. Cells. 2014;3:1027–88. doi: 10.3390/cells3041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira JV, Soares AR, Ramalho JS, Pereira P, Girao H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci Rep. 2015;5:10210. doi: 10.1038/srep10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanduri P, Hao R, Fitzpatrick T, Yao TP. Chaperone-mediated 26S proteasome remodeling facilitates free K63 ubiquitin chain production and aggresome clearance. J Biol Chem. 2015;290:9455–64. doi: 10.1074/jbc.M114.627950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki K, Hamazaki J, Koike M, Hirano Y, Komatsu M, Uchiyama Y, Tanaka K, Murata S. PAC1 gene knockout reveals an essential role of chaperone-mediated 20S proteasome biogenesis and latent 20S proteasomes in cellular homeostasis. Mol Cell Biol. 2010;30:3864–74. doi: 10.1128/MCB.00216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Körner R, Nickell S, Lasker K, Sali A, Tamura T, Nishioka T, Förster F, Baumeister W, Bracher A. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A. 2012;109:149–54. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, Gage FH, Dillin A. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–8. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno D, Viana R, Sanz P. Two-hybrid analysis identifies PSMD11, a non-ATPase subunit of the proteasome, as a novel interaction partner of AMP-activated protein kinase. Int J Biochem Cell Biol. 2009;41:2431–9. doi: 10.1016/j.biocel.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A. 2015;112:E7176–85. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kevei É, Hoppe T. Ubiquitin sets the timer: impacts on aging and longevity. Review. Nat Struct Mol Biol. 2014;21:290–2. doi: 10.1038/nsmb.2806. [DOI] [PubMed] [Google Scholar]
- 32.Ronnebaum SM, Patterson C, Schisler JC. UPS: metabolic and proteolytic homeostasis linked via AMPK and the ubiquitin proteasome system. Review. Mol Endocrinol. 2014;28:1602–15. doi: 10.1210/me.2014-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Review. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schröter F, Adjaye J. The proteasome complex and the maintenance of pluripotency: sustain the fate by mopping up? Stem Cell Res Ther. 2014;5:24. doi: 10.1186/scrt413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed AS, Li J, Erlandsson-Harris H, Stark A, Bakalkin G, Ahmed M. Suppression of pain and joint destruction by inhibition of the proteasome system in experimental osteoarthritis. Pain. 2012;153:18–26. doi: 10.1016/j.pain.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Etienne S, Gaborit N, Henrionnet C, Pinzano A, Galois L, Netter P, et al. Local induction of heat shock protein 70 (Hsp70) by proteasome inhibition confers chondroprotection during surgically induced osteoarthritis in the rat knee. Biomed Mater Eng. 2008;18:253–60. [PubMed] [Google Scholar]
- 37.Ye J, Qing Z, Ma J, Wang Z. Effect of proteasome inhibitor on NF-κB and MMP-9 expression in synovial tissues of osteoarthritis rats. Int J Clin Exp Med. 2017;10:402–09. [Google Scholar]
- 38.Radwan M, Wilkinson DJ, Hui W, Destrument AP, Charlton SH, Barter MJ, et al. Protection against murine osteoarthritis by inhibition of the 26S proteasome and lysine-48 linked ubiquitination. Ann Rheum Dis. 2015;74:1580–7. doi: 10.1136/annrheumdis-2013-204962. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–46. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duval E, Leclercq S, Elissalde JM, Demoor M, Galéra P, Boumédiene K. Hypoxia-inducible factor 1alpha inhibits the fibroblast-like markers type I and type III collagen during hypoxia-induced chondrocyte redifferentiation: hypoxia not only induces type II collagen and aggrecan, but it also inhibits type I and type III collagen in the hypoxia-inducible factor 1alpha-dependent redifferentiation of chondrocytes. Arthritis Rheum. 2009;60:3038–48. doi: 10.1002/art.24851. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Review. Biochem J. 2004;380(Pt 2):297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall RS1, Li F1, Gemperline DC1, Book AJ1, Vierstra RD2. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell. 2015;58:1053–66. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu CF, Samsa WE, Zhou G, Lefebvre V. Transcriptional control of chondrocyte specification and differentiation. Review. Semin Cell Dev Biol. 2017;62:34–49. doi: 10.1016/j.semcdb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao M, Stefanovic-Racic M, Georgescu HI, Miller LA, Evans CH. Generation of nitric oxide by lapine meniscal cells and its effect on matrix metabolism: stimulation of collagen production by arginine. J Orthop Res. 1998;16:104–11. doi: 10.1002/jor.1100160118. [DOI] [PubMed] [Google Scholar]
- 45.Kühn K, Shikhman AR, Lotz M. Role of nitric oxide, reactive oxygen species, and p38 MAP kinase in the regulation of human chondrocyte apoptosis. J Cell Physiol. 2003;197:379–87. doi: 10.1002/jcp.10372. [DOI] [PubMed] [Google Scholar]
- 46.Lotz M. The role of nitric oxide in articular cartilage damage. Review. Rheum Dis Clin North Am. 1999;25:269–82. doi: 10.1016/s0889-857x(05)70067-3. [DOI] [PubMed] [Google Scholar]
- 47.Qureshi AA, Tan X, Reis JC, Badr MZ, Papasian CJ, Morrison DC, Qureshi N. Suppression of nitric oxide induction and pro-inflammatory cytokines by novel proteasome inhibitors in various experimental models. Lipids Health Dis. 2011;10:177. doi: 10.1186/1476-511X-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolodziejski PJ, Musial A, Koo JS, Eissa NT. Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc Natl Acad Sci U S A. 2002;99:12315–20. doi: 10.1073/pnas.192345199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chondrogianni N, Voutetakis K, Kapetanou M, Delitsikou V, Papaevgeniou N, Sakellari M, Lefaki M, et al. Proteasome activation: An innovative promising approach for delaying aging and retarding age-related diseases. Review. Ageing Res Rev. 2015;23(Pt A):37–55. doi: 10.1016/j.arr.2014.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We transfected normal human chondrocytes with PSMD11 siRNA, and (A) validated induced decreased level of PSMD11, and increased levels of K48 polyuybiquinated peptide substrates, and (B) decreased proteasome protease activity. PSMD11 siRNA knockdown did not induce detectable changes in total and phosphorylated FOXO1, FOXO3a, and FOXO4 (C).
Transfecting Grade IV OA chondrocytes, we assessed effects of HA tag-PSMD11 plasmid, compared to control plasmid transfection, on NO release (n=4 donors) and intracellular MMP13 (A)(data representative of 4 donors studied), and extracellular MMP13 protein released (B)(n=10 donors), in response to IL-1β.
(A) We transfected OA chondrocytes with HA-tagged PSMD11 cDNA, and assessed levels of p62 and LC3A/B conversion (n=4). (B) We assessed levels of SOX9, and of SOX9 translocated to the nucleus. (C) We also assayed SOX9, COL2A1, and AGC1 mRNA.