Abstract
Species identification lies at the heart of biodiversity studies that has in recent years favoured DNA-based approaches. Microbial Biological Resource Centres are a rich source for diverse and high-quality reference materials in microbiology, and yet the strains preserved in these biobanks have been exploited only on a limited scale to generate DNA barcodes. As part of a project funded in the Netherlands to barcode specimens of major national biobanks, sequences of two nuclear ribosomal genetic markers, the Internal Transcribed Spaces and 5.8S gene (ITS) and the D1/D2 domain of the 26S Large Subunit (LSU), were generated as DNA barcode data for ca. 100 000 fungal strains originally assigned to ca. 17 000 species in the CBS fungal biobank maintained at the Westerdijk Fungal Biodiversity Institute, Utrecht. Using more than 24 000 DNA barcode sequences of 12 000 ex-type and manually validated filamentous fungal strains of 7 300 accepted species, the optimal identity thresholds to discriminate filamentous fungal species were predicted as 99.6 % for ITS and 99.8 % for LSU. We showed that 17 % and 18 % of the species could not be discriminated by the ITS and LSU genetic markers, respectively. Among them, ∼8 % were indistinguishable using both genetic markers. ITS has been shown to outperform LSU in filamentous fungal species discrimination with a probability of correct identification of 82 % vs. 77.6 %, and a clustering quality value of 84 % vs. 77.7 %. At higher taxonomic classifications, LSU has been shown to have a better discriminatory power than ITS. With a clustering quality value of 80 %, LSU outperformed ITS in identifying filamentous fungi at the ordinal level. At the generic level, the clustering quality values produced by both genetic markers were low, indicating the necessity for taxonomic revisions at genus level and, likely, for applying more conserved genetic markers or even whole genomes. The taxonomic thresholds predicted for filamentous fungal identification at the genus, family, order and class levels were 94.3 %, 88.5 %, 81.2 % and 80.9 % based on ITS barcodes, and 98.2 %, 96.2 %, 94.7 % and 92.7 % based on LSU barcodes. The DNA barcodes used in this study have been deposited to GenBank and will also be publicly available at the Westerdijk Institute's website as reference sequences for fungal identification, marking an unprecedented data release event in global fungal barcoding efforts to date.
Key words: Automated curation, Biological resource centre, Fungi, ITS, LSU, Taxonomic thresholds
Introduction
Species identification lies at the heart of biodiversity studies, and biodiversity efforts have in recent years favoured DNA-based approaches over morphology-based approaches (Hebert et al., 2003, De Queiroz, 2007, Verkley et al., 2013, Woudenberg et al., 2013, Liu et al., 2016, Wang et al., 2016a, Wang et al., 2016b). The success of DNA barcoding has given rise to characterisation of bacterial biodiversity in every nook and cranny on the planet (Huttenhower et al., 2012, Mau et al., 2014, Afshinnekoo et al., 2015). Knowledge of the microbial composition of such communities aide knowledge of host-microbe interactions and their environmental function, and revealed a complex and sensitive balance that may be easily disturbed (Fuhrman, 2009, Garza et al., 2016, Levy et al., 2017). Such metagenomics studies have been limited in fungi due to the complexity of fungal genomes and the lack of validated databases cataloguing sufficient biodiversity. Fungal genomes are generally larger than prokaryotic genomes with a size ranging from 9 Mb to 178 Mb because they contain large amounts of non-coding and repetitive DNA (Mohanta & Bae 2015). They have been shown to be divergent, even between the members of the same genus (Galagan et al. 2005), and dynamic in nature (Dujon et al., 2004, Kellis et al., 2004, Strope et al., 2015). The studies already conducted into fungal metagenomic communities have often been limited to generic or higher levels (Cui et al., 2013, Geml et al., 2014, Tedersoo et al., 2014, Nguyen et al., 2015, Botschuijver et al., 2017), limiting our understanding of the role of fungal species in our surrounding ecosystems.
The Internal Transcribed Spacer (ITS) nrDNA region has been proposed as a universal barcode for fungi (Schoch et al., 2012, Blaalid et al., 2013, Koljalg et al., 2013). Despite criticism (Kiss 2012), ITS sequences have been shown to be useful in delineating many fungal species (Irinyi et al., 2015, Vu et al., 2016). Together with the D1/D2 domain of the Large Subunit (LSU, 26S) nrDNA sequences, ITS sequence analysis is frequently used as a method to discriminate fungal species (Kurtzman and Robnett, 1998, Fell et al., 2000, Boon et al., 2010, Kooij et al., 2015, Woudenberg et al., 2015). Unfortunately less than 23 000 (0.6 %; Vu et al. 2014) of the estimated 3.8 million species (Hawksworth & Lücking 2017) of fungi have ITS sequences available at GenBank and many of the sequences are often of poor quality (Nilsson et al., 2006, Vu et al., 2016). Furthermore, the optimal similarity range to make informed decisions about species delineation is still highly uncertain compared to bacterial species (Stackebrandt and Ebers, 2006, Edgar, 2018). In most fungal ecology studies, a 97 % threshold is given as default (Geml et al., 2014, Gweon et al., 2015). Although there have been efforts to define a range of taxonomic thresholds from 97 % to 100 % for fungal species identification, such as the UNITE species hypothesis (Koljalg et al. 2013), threshold choice remains subjective.
Mycologists studying fungal strains have deposited reference materials in public culture collections for over a century. Microbial Biological Resource Centres (MBRC) preserve this heritage which constitutes an invaluable source for well-defined and high-quality materials for microbiology. At the Westerdijk Fungal Biodiversity Institute (WI, previously CBS-KNAW), Utrecht, The Netherlands, more than 100 000 living strains of yeasts and filamentous fungi are preserved that were originally assigned to ca. 17 000 species. Information on the publically available strains, are accessible via the website http://www.westerdijkinstitute.nl/Collections/. This includes data on morphology, physiology, molecular markers sequences, growth conditions and safety measures. The CBS filamentous fungal collection currently has approximately 70 000 strains including ca. 8 000 ex-type strains representing more than 12 000 currently recognized species, with the oldest strains isolated around 1895. When accessioned, each identified strain is assigned a taxon name from MycoBank (Robert et al. 2013), an online registration system for fungal species and higher level taxon names. Strain identification is normally achieved with the knowledge and methods available at the time of accession. While names of strains other than ex-type strains are updated following modern taxonomic concepts, it has not been possible to re-evaluate the original identification of each strain in the CBS collection. For most of the older strains, not all characters necessary for identification are expressed in culture or modern molecular studies have not been conducted yet.
In the WI DNA barcoding project, ITS and LSU barcode sequences were generated based on DNA extracted from cultures for the majority of CBS strains. In a previous study, the barcoding project resulted in the release of 8 669 barcode sequences of manually validated CBS strains representing 1 351 yeast species (Vu et al. 2016). In this study, over 24 000 sequences of manually validated strains that belong to 7 300 filamentous fungal species, have been made available by depositing them in GenBank and at the website of the Westerdijk Institute to improve fungal identification in online public databases. Similarly to the results obtained in Vu et al. (2016), we showed that ITS and LSU can be used to classify a large portion of all fungi to species level. Additionally, ITS and LSU sequences were complementary, and could be used together to achieve a better identification performance. Based on this large number of ITS and LSU barcodes, thresholds were predicted for circumscription of species and higher taxa of filamentous fungi. The newly generated ITS barcodes were also compared with the “Top 50 Most Wanted Fungi” (Nilsson et al., 2016, UNITE Community, 2017) dataset to reveal the most frequently sampled environmental sequence types that have been difficult to be assigned to meaningful taxonomic levels.
Material and methods
Generation and management of barcode sequences
The protocols of genomic DNA extraction and generation of the ITS and LSU barcode sequences were given in Eberhardt (2012) and Stielow et al. (2015). The ITS sequences were generated using the forward and backward primers ITS5 and ITS4 for PCR reactions for amplification, containing partial 18S, complete ITS1-5.8S-ITS2, and partial LSU sequences. The LSU sequences were generated in the D1/D2 domain employing the forward and backward primers LROR and LR5 (Stielow et al. 2015). To be able to manage a large amount of sequence data and to keep track of the experimental procedures, a laboratory information management system (LIMS; Vu et al. 2012) was used for the WI DNA barcoding workflow as a module of BioloMICS (Robert et al. 2011), a software package to manage biological databases. The trace files obtained from the sequencer were aligned. The consensus sequences were imported automatically into the WI database, and edited manually by replacing or removing all ambiguous characters. After that, they were checked, compared with the existing sequences from local and public databases such as GenBank, and validated by the curators. For the analyses presented in this paper, only validated sequences that were added to the database until December 2016 were included, and for each strain, only one sequence was selected.
Computing a similarity score between DNA sequences
The similarity score of two DNA sequences was computed using our own implementation (Robert et al. 2011) of the BLAST algorithm (Altschul et al. 1997) as the percentage identity of the most similar region (coverage) between the two sequences.
Selecting representative sequences for species
The representative sequence of a species was taken as the sequence of the ex-type strain if it was available, otherwise as the central representative sequence of all the sequences of the strains associated with the species (Vu et al. 2016). Here, the central representative sequence of a group was the one that had the highest similarity score to the other sequences in the group. The strain associated with the (central) representative sequence was considered to represent the (central) representative strain of the species. The similarity score of two strains was the similarity score of their reference sequences.
Probability of correct identification (PCI)
Given a genetic marker, the identification of a species is correct if, for every strain of the species, there is no other strain from another species for which the similarity score between them is greater or equal to the minimum similarity score between the strains of the species under consideration. The PCI of the given genetic marker is the fraction of species correctly identified. To evaluate the resolving power of multiple genetic markers for species discrimination, the similarity score of two strains using multiple genetic markers was computed as the average similarity score of all similarity scores computed for each genetic marker (CBOL Plant Working Group, 2009, Schoch et al., 2012).
Clustering of strains or DNA sequences
To cluster strains or DNA sequences, we used an algorithm to find connected components implemented in Vu et al. (2018) as it has been shown to be highly accurate in comparison with other clustering algorithms (Vu et al. 2014). Briefly, given a similarity score or threshold, two strains or sequences of a dataset will be connected if there is a path of strains or sequences between them in which the similarity score of a strain or sequence to the next one is equal to or greater than the given threshold. The clustering algorithm places all connected strains or sequences of the dataset in the same group.
Quality of automatic clustering
Different thresholds lead to different groupings of the strains or sequences. The goal is to place the members of the dataset in the correct taxonomic groups and automatically assign them to a taxon name. The F-measure proposed by (Paccanaro et al. 2006) was used to evaluate an automatic clustering compared to the curated taxonomic assignments. Let C = (C1, . . . ,Cl) be the partition of a given set of strains or sequences obtained by taxonomic classification, and K = (K1, . . . ,Km ) be the partitions obtained by clustering the dataset with a given threshold. The quality of clustering is computed by the F-measure function F(K, C), defined as follows.
where |x| is the size of set x. The F-measure ranges from 0 to 1. The higher the value of the F-measure, the more similar the clustering result is to the taxonomic classification. It is equal to 1 when the clustering result matches the taxonomic classification perfectly.
Predicting a taxonomic threshold for species identification
The taxonomic threshold to cluster a dataset of strains or sequences was calculated as the optimal threshold that produces the best quality for clustering in comparison with the taxonomic classification, i.e. the one having the highest F-measure.
Results and discussion
Barcoding dataset captures thousands of strains
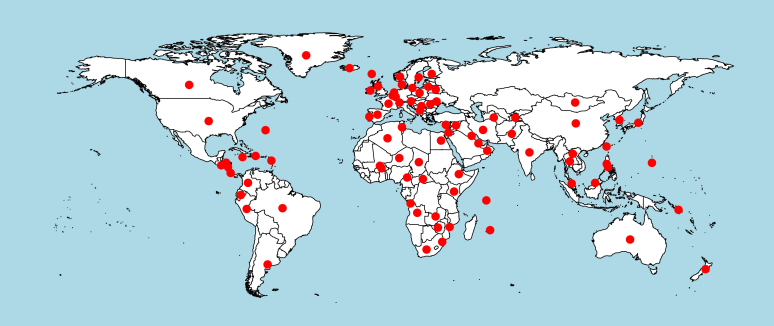
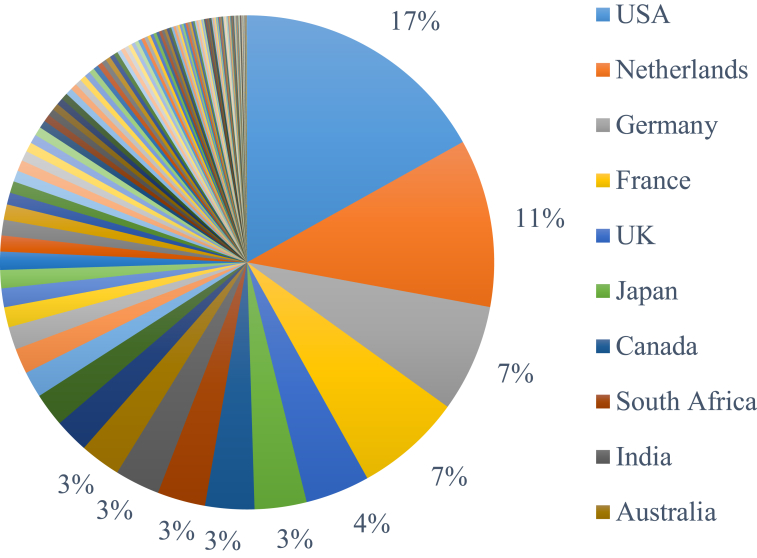
We selected ITS and LSU sequences for 14 859 validated strains of 7 376 accepted species in which 4 490 strains were ex-type. The metadata of these strains such as the type information and the country of collection are given in Supplementary file S1. The strains were collected from all over the world (Fig. 1). The 10 countries having the most strains collected were the USA (17 %), the Netherlands (11 %), Germany (7 %), France (7 %), UK (4 %), Japan (3 %), Canada (3 %), South Africa (3 %), India (3 %), and Australia (3 %) (Fig. 2). Of the 4 490 ex-type strains, 43 % (1 942) were listed as type material (Federhen 2015) in GenBank, of which, 98 % (1 905) had the same taxon name as the name given at GenBank and 37 entries in GenBank had a name of a synonym. The 57 % (2 548) not listed as type material, of which 80 % (2 035) had the same taxon name and 1 % (172) had a synonym listed at GenBank. For 341 CBS ex-type strains, no information was available in GenBank. As the barcode sequences of these ex-type strains have been made available by depositing them in GenBank, this study will improve filamentous fungal barcodes and taxonomy at public databases.
Fig. 1.
Collection locations of 13 173 strains in the current study. Each red dot represents a country of collection. The remaining 1 689 strains had no associated information.
Fig. 2.
The countries of collection together with the percentage of the strains.
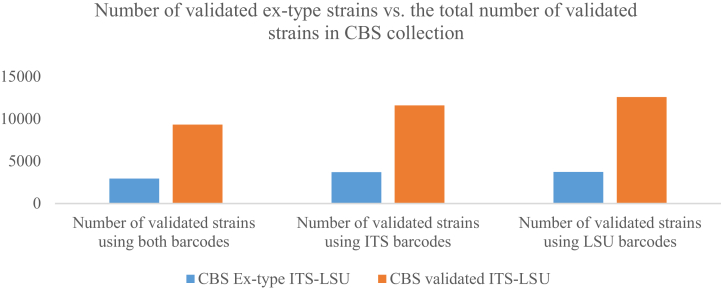
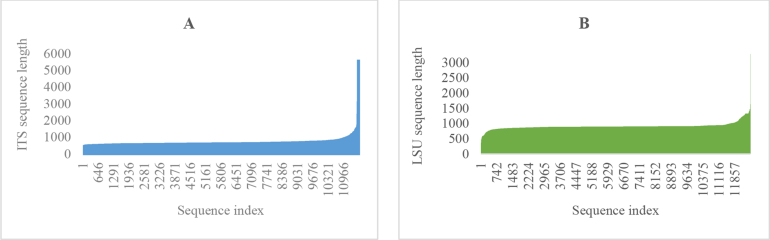
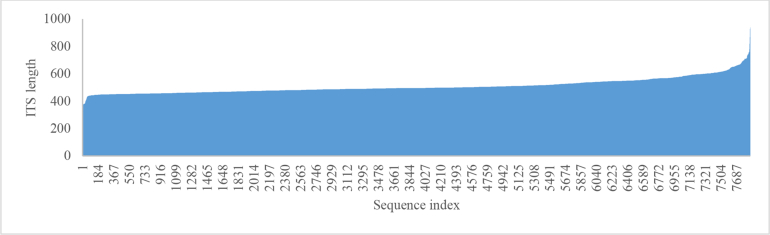
Of the 14 859 validated strains, there were 11 605 (3 718 ex-type) and 12 588 (3 737 ex-type) strains having ITS and LSU barcodes, respectively, in which 9 336 (2 965 ex-type) strains had both barcodes. These values are summarized in Fig. 3. The ITS barcode sequences had a length ranging from 200 bp to 5 530 bp with an average of 622 bp, while the LSU barcode sequences had a length ranging from 242 bp to 3 295 bp with an average of 911 bp (Fig. 4).
Fig. 3.
Number of manually validated ex-type strains using ITS/LSU barcodes versus the total number of manually validated strains in the CBS filamentous fungal collection.
Fig. 4.
The lengths of ITS (A) and LSU (B) barcode sequences.
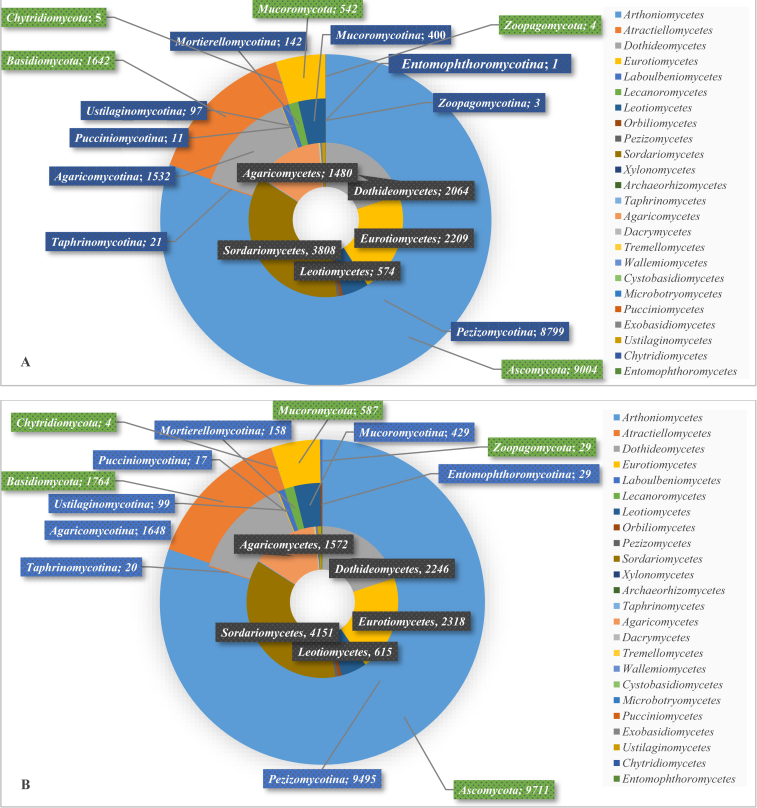
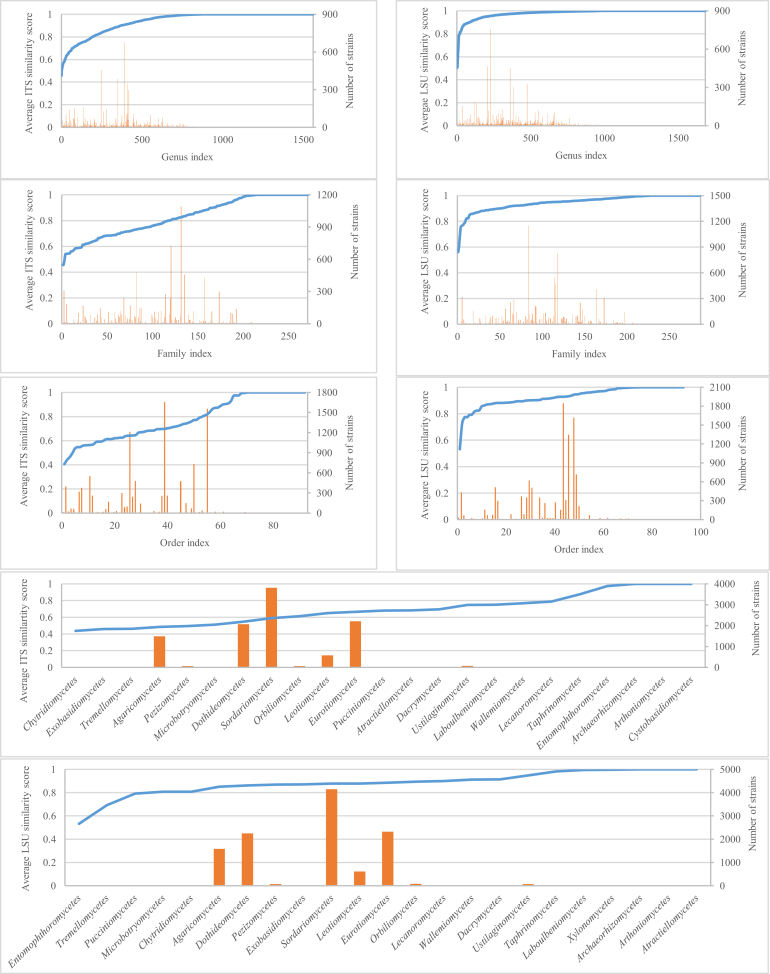
Fig. 5 shows the 5 (5) filamentous fungal phyla, 9 (8) subphyla, 24 (24) classes, 90 (92) orders, 270 (286) families, 1 559 (1 683) genera, and 6 064 (6 469) species of the strains having ITS (LSU) sequences. The average numbers of ITS and LSU sequences per order were 114 and 122, respectively. The average numbers of sequences per family, genus, and species were 38, 7, and 2 for both genetic markers. At the phylum level, the phylum Ascomycota was dominant with 9 004 ITS and 9 711 LSU sequences, followed by Basidiomycota with 1 642 ITS and 1 764 LSU, Mucoromycota with 542 ITS and 587 LSU, Zoopagomycota with 4 ITS and 29 LSU and Chytridiomycota with 5 ITS and 4 LSU sequences. The most dominant subphylum was Pezizomycotina with 8 799 ITS and 9 495 LSU sequences, followed by Agaricomycotina with 1 532 ITS and 1 648 LSU, Mucoromycotina with 400 ITS and 429 LSU, Mortierellomycotina with 142 ITS and 158 LSU, Ustilaginomycotina with 97 ITS and 99 LSU, Entomophthoromycotina with 1 ITS and 29 LSU, Taphrinomycotina with 21 ITS and 20 LSU, Pucciniomycotina with 11 ITS and 17 LSU, and Zoopagomycotina with 3 ITS sequences. There were 24 filamentous fungal classes having both ITS and LSU barcodes. Of these, Sordariomycetes was the most dominant class with 3 810 ITS and 4 151 LSU sequences, followed by the three classes Eurotiomycetes with 2 210 ITS and 2 318 LSU, Dothideomycetes with 2 064 ITS and 2 247 LSU, Agaricomycetes with 1 483 ITS and 1 582 LSU, and Leotiomycetes with 574 ITS and 615 LSU sequences. The remaining classes had a small number (< 100) of barcode sequences. These classes are visualised in Fig. 6. At the order level, Hypocreales was the biggest order with 1 656 ITS and 1 847 LSU sequences, followed by Eurotiales with 1 557 ITS and 1 618 LSU and Pleosporales with 1 213 ITS and 1 349 LSU sequences. At the family level, Trichocomaceae was the dominant family with 1 094 ITS and 1 147 LSU sequences. At the genus level, Penicillium was the largest genus with 680 ITS and 757 LSU sequences. Finally, at the species level, the biggest group was Exophiala dermatitidis with 97 ITS and 77 LSU sequences. Note that there were 411 (495), 602 (695), 1 145 (1 279), 1 212 (1 341), 1 447 (1 675), and 27 (33) ITS (LSU) sequences had no information available at the phylum, subphylum, class, order, family and genus level, respectively. All of these information can also be found in Supplementary file S2. The barcode sequences are organised into different datasets of ex-type and manually validated strains as explained in Table 1.
Fig. 5.
The classes, subphyla and phyla together with the associated number of ITS (A) and LSU (B) sequences.
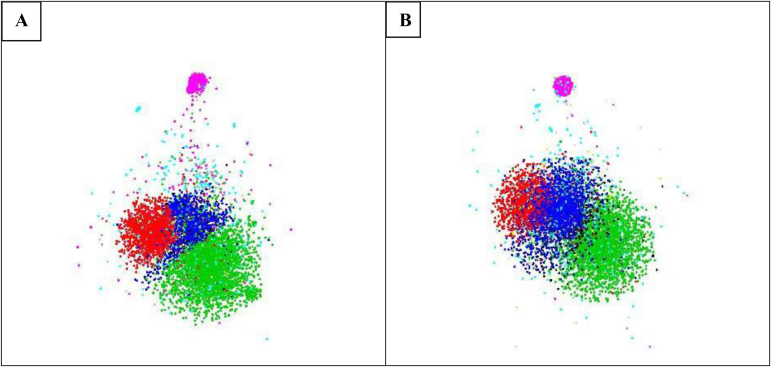
Fig. 6.
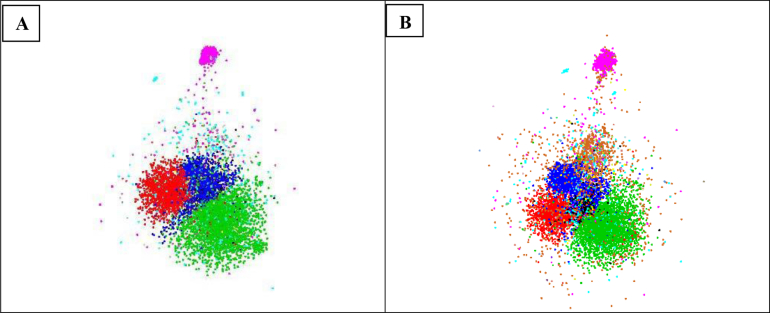
The distributions of the ITS (A) and LSU (B) sequences of the manually validated strains. The sequences of the same colour belong to the same class. The five biggest classes in green, red, blue and pink represent 3 810, 2 210, 2 064, 1 483, 574 ITS and 4 151, 2 318, 2 247, 1 582, 615 LSU sequences of Sordariomycetes, Eurotiomycetes, Dothideomycetes, Agaricomycetes, and Leotiomycetes, respectively. The sequences in turquoise colour are the ones (1 145 for ITS and 1 279 for LSU) without a class name given in the database. The 3D coordinates of the sequences were computed using fMLC (Vu et al. 2018) to compute a complete similarity matrix and LargeVis (Tang et al. 2016) to calculate the coordinates of the sequences. The sequences were visualized using the rgl package in R (https://r-forge.r-project.org/projects/rgl/).
Table 1.
Numbers of filamentous fungal classes, orders, families, genera, species, strains, and sequences of different barcode datasets. The “Validated Ex-type datasets”, abbreviated as ITS-T, LSU-T and ITS/LSU-T, contained all the validated sequences of strains that were designated as ex-type strains for a currently accepted species or of a synonymised species name. The “Validated datasets”, abbreviated as ITS-V, LSU-V and ITS/LSU-V included the ex-type strains as well as all the CBS strains that were checked by the curators to confirm their species assignments using ITS and/or LSU reference sequences.
| Dataset | Dataset abbreviation | Number of phyla | Number of subphyla | Number of classes | Number of orders | Number of families | Number of genera | Number of species | Number of strains |
|---|---|---|---|---|---|---|---|---|---|
| Validated Ex-type ITS | ITS-T | 5 | 9 | 20 | 68 | 188 | 911 | 3 165 | 3 718 |
| Validated ITS | ITS-V | 5 | 9 | 24 | 90 | 271 | 1 559 | 6 064 | 11 605 |
| Validated Ex-type LSU | LSU-T | 5 | 8 | 20 | 69 | 199 | 985 | 3 238 | 3 737 |
| Validated LSU | LSU-V | 5 | 8 | 24 | 92 | 286 | 1 683 | 6 469 | 12 588 |
| Ex-Type having both ITS and LSU | ITS/LSU-T | 5 | 8 | 20 | 63 | 174 | 813 | 2 569 | 2 965 |
| Validated dataset having both ITS and LSU sequences | ITS/LSU-V | 5 | 8 | 24 | 89 | 260 | 1 417 | 5 157 | 9 336 |
ITS and LSU barcodes have high within species similarity scores
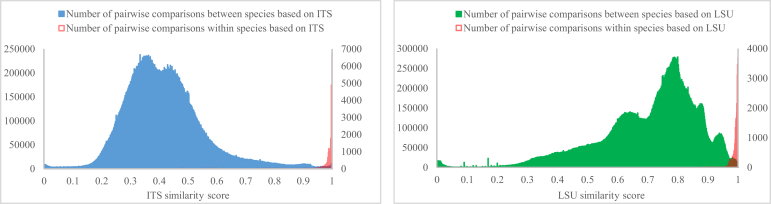
To understand the taxonomic variation captured by our rDNA barcodes, we examined the within and between variability among filamentous fungal species in the manually validated ITS-V and LSU-V datasets (see Table 1). Fig. 7 shows the distribution of DNA similarity scores (Methods) for all pairwise comparisons within and between species of the ITS-V and LSU-V datasets. From the similarity scores of 94 % for ITS-V and 96 % for LSU-V, it is clear that the percentage of pairwise comparisons within species was bigger than the percentage of pairwise comparisons between species. For both datasets, the distributions of within-species similarities were tight, with 99.37 % of all comparisons falling between 94–100 % in ITS-V and 99.21 % falling between 96–100 % in LSU-V. The distributions of between-species DNA similarity scores were much broader than of within-species DNA similarity scores. The distribution of interspecific similarity scores in ITS-V was bimodal, with 98.79 % of all comparisons falling within 15–94 %. In LSU-V, the distribution was polymodal, and 99.3 % of all comparisons had a score between 20–100 %. This indicates that ITS and LSU sequences are highly conserved within species, and that ITS is more divergent between species than LSU. Compared to the result obtained from the analysis of CBS yeast barcodes (Vu et al. 2016), the similarity scores within filamentous fungal species were similar as in yeast species. The similarity scores between filamentous fungal species were more variable than between yeast species using ITS barcodes (15–94 % vs. 15–70 %), and were about the same when using LSU barcodes (20–100 % vs. 30–97 %).
Fig. 7.
The distribution of DNA similarity scores for pairwise comparisons between species and within species, for manually validated strains in the ITS-V (A) and LSU-V (B) datasets.
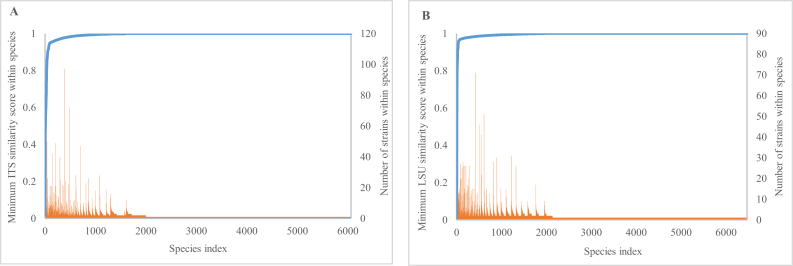
Although the distributions of within-species similarities were tight as seen earlier, there was still a small number of species (0.67 % in the ITS-V and 0.4 % in the LSU-V dataset) that had a minimum similarity score less than 94 % for ITS and 96 % for LSU (see Fig. 8). This was also observed in Vu et al. (2016), and could be caused by: a) some sequences were wrongly declared as ITS or LSU; b) some sequences were wrongly associated with the strain, and/or; c) since ITS and LSU have multiple copies of different lengths and variable sequences, the amplified/stored copy may not be the dominant one or there could be several versions that are very dissimilar from each other (Simon and Weiss, 2008, Kiss, 2012).
Fig. 8.
Minimum ITS (A) and LSU (B) similarity score within species of the ITS-V and LSU-V datasets.
Ex-type strains are well positioned among other strains in the species
As the ex-type strains are the reference points for species naming and identification, it is essential to know if the ex-type strain of a species is also the central representative strain of the species. If the ex-type strain is positioned eccentrically (i.e. the similarity score between the ex-type and representative central strains was not 1) relative to other strains in the species, the identification procedure may produce unstable identifiers. Specifically, based on a sequence comparison with the ex-type strain, the procedure might associate strains that, while highly similar to the ex-type strain, are not highly similar to the species as a whole, and it may exclude others. Using the manually validated CBS barcode datasets, we addressed this issue.
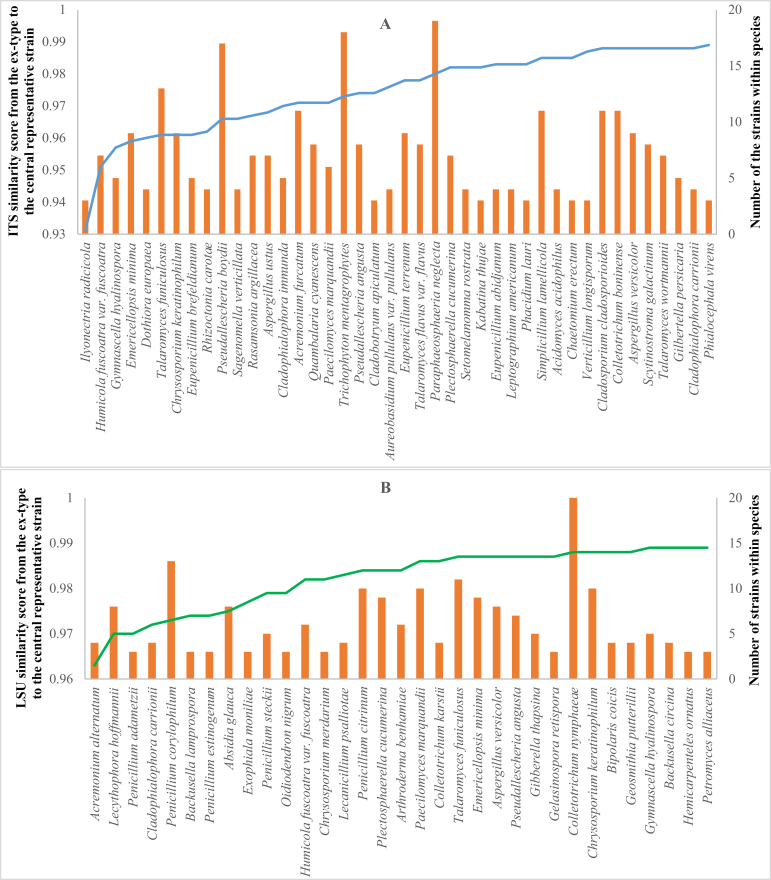
In the ITS-V dataset, there were 2 904 species of 5 358 strains having an ex-type strain, of which 407 species of 2 435 strains had more than two strains. Among them, 139 species of 824 strains had eccentrically positioned ex-type strains. The average ITS similarity score between the ex-type and central representative strains of the 407 species was 99.65 %. In the LSU-V dataset, there were 2 956 species of 5 465 strains having an ex-type strain, of which 410 species of 2 519 strains had more than two strains. Among them, 220 species of 1 473 strains had eccentrically positioned ex-type strains. The average LSU similarity score between the ex-type and central representative strains of the 410 species was 99.69 %. The lists of species that had positioned ex-type strains based on ITS and LSU barcodes are given in Supplementary file S3. Fig. 9 shows the species of more than two strains with a similarity score less than 99 % from the ex-type to the central representative strain (42 and 34 species, for ITS-V and LSU-V, respectively). The low similarity scores indicate higher than usual intraspecific variation of strains in some species, or the concept of the species has shifted away from the traditional species criteria with the accumulation of similar strains over time, without accounting for the similarity to the ex-type strain.
Fig. 9.
Filamentous fungal species with ITS-V (A) and LSU-V (B) similarity score less than 99 % from the ex-type to central representative strain. The number of the strains of the species is displayed in the secondary axis.
ITS and LSU delineate over 80 % of all studied filamentous fungal species
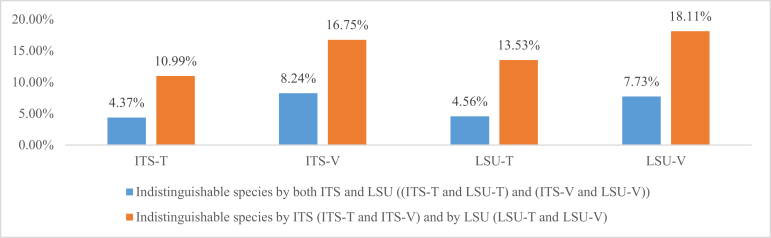
The percentages of pairwise comparisons between species at the similarity scores of 100 % in Fig. 7 indicate that a number of species were either synonyms, or that the ITS and/or LSU regions are not always able to discriminate between some pairs of species. To examine the fraction of species that cannot be distinguished based on ITS or LSU, sequences of ex-type and manually validated barcode datasets (Table 1) were clustered when their sequences had a similarity score of 100 %. The species whose sequences were grouped into the same cluster are indistinguishable by the respective genetic marker. The obtained groups contained species names of nomenclatural synonyms or indistinguishable species (taxonomic synonyms or closely related species). Fig. 10 shows the percentage of indistinguishable species in the different datasets using ITS and LSU barcodes. For the ex-type datasets, 10.99 % and 13.53 % of all species were indistinguishable using ITS and LSU barcodes, respectively. For the manually validated datasets, these numbers were 16.75 % and 18.11 % respectively. These values were much higher than the numbers obtained from the analysis of yeast barcodes (Vu et al. 2016) where only 2 % and 4 % using ITS and LSU ex-type sequences, and 6 % and 8 % using manually validated ITS and LSU sequences were indistinguishable. In addition, the average number of strains per species in this study was smaller 2.01 (14 862/7 377) vs. 3.73 (5 198/1 392), indicating that more redundant species were described in filamentous fungal taxonomy, or ITS and LSU currently works better for species discrimination in yeasts than in filamentous fungi. To resolve which filamentous fungal species need to be reduced to synonymy, further taxonomic studies are required.
Fig. 10.
Percentages of species synonyms and indistinguishable species by using ITS and LSU barcodes using a threshold of 100 %, respectively.
ITS and LSU barcodes are complementary
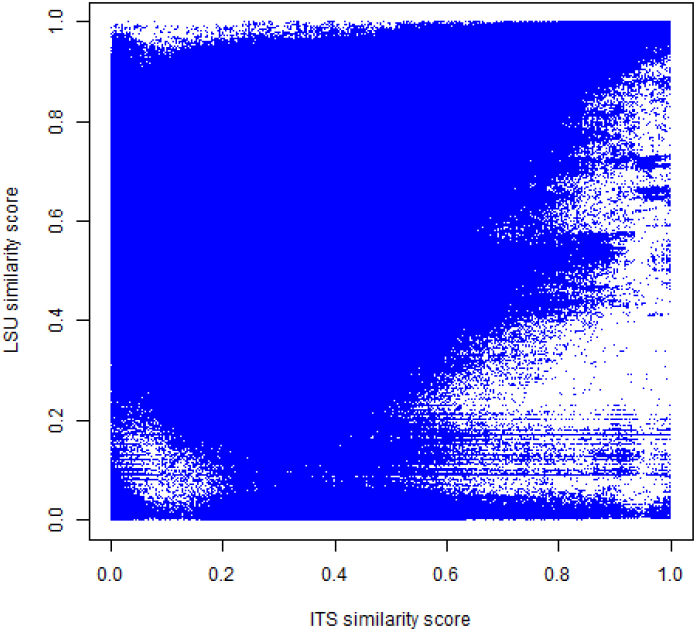
To evaluate the level of correlation between the groupings obtained by both genetic markers, all 9 336 manually validated CBS strains (ITS/LSU-V dataset) belonging to 5 158 species having both ITS and LSU sequences were aligned in a pairwise manner to produce two similarity matrices, one for each genetic marker. Fig. 11 shows the scatter plot of all pairwise comparisons based on ITS and LSU similarity scores. A Mantel test (i.e. a Pearson Correlation moment between two distance matrices calculated on the basis of 999 permutations) between the two matrices was calculated and a correlation of 0.63 between ITS and LSU was observed. Even when the ITS similarity score was greater than 68.5 % and the LSU similarity score greater was than 92.7 % which happened only in 5 % of all cases, the correlation between ITS and LSU was also low at 0.60. These low values are indicative of a partial independence of the two genetic markers, indicating that they can be used together with the advantage of additional discriminative power. These results are slightly different from the correlations obtained for yeasts in (Vu et al. 2016) in which the correlation of the two genetic markers in general was also low at 0.47. However, when the ITS and LSU similarity scores were greater than 60 % and 89 %, a strong correlation of 0.84 between ITS and LSU was observed in the yeast dataset.
Fig. 11.
2D scatter plots of ITS similarity scores versus LSU similarity scores of the ITS/LSU-V dataset.
Combining ITS and LSU barcodes improves species discrimination
We evaluated and compared the resolving powers of ITS and LSU for filamentous fungal species discrimination based on the PCI of ITS and LSU alone, and of the combination of ITS and LSU using the manually validated dataset ITS/LSU-V. The PCIs of ITS and LSU were 82.18 % and 77.57 %, respectively. When combining both genetic markers, the PCI of 83 % is slightly higher than the PCI of ITS alone. These values were higher than the corresponding values computed for fungi given in Schoch et al. (2012) which were 77 % for ITS and 75 % for LSU. However, they were lower than the PCIs of 88.4 % for ITS, 84.6 % for LSU, and 85.83 % for the combination of the two genetic markers that were observed for yeasts (Vu et al. 2016).
ITS and LSU can be used to discriminate filamentous fungal species
To predict taxonomic thresholds to discriminate filamentous fungal species, we clustered the ex-type and manually validated barcode datasets ITS-T, LSU-T, ITS-V, and LSU-V with different thresholds from 0.97 to 1 to see which thresholds produced the best quality (F-measure) for clustering. To optimise the prediction, sequences of species indistinguishable by ITS and LSU were removed. The datasets obtained after removing these sequences were ITS-T-Standard, LSU-T-Standard, ITS-V-Standard, and LSU-V-Standard. To see if the combination of the two genetic markers ITS and LSU works better in species discrimination than a single genetic marker alone, we used the manually validated dataset ITS/LSU-V where the similarity score of two strains was computed as the average of the two ITS and LSU similarity scores.
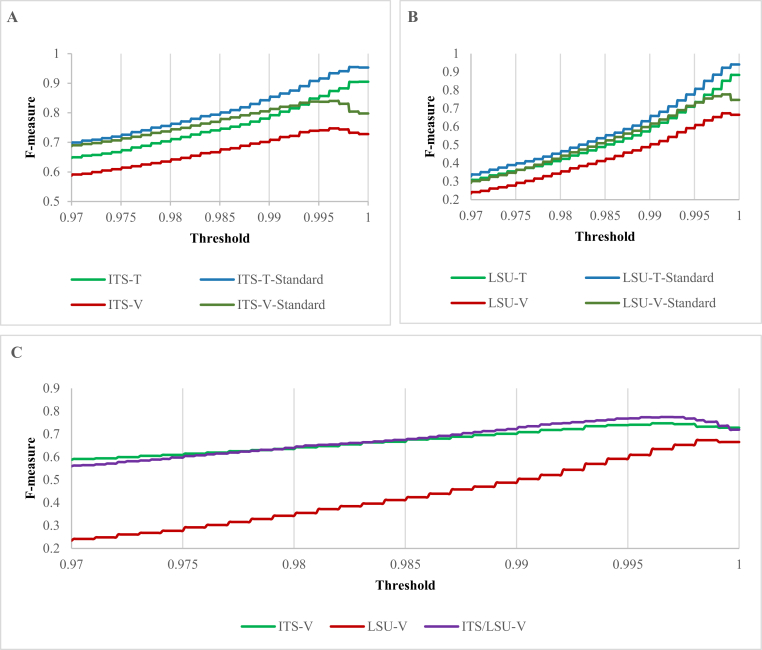
Fig. 12 shows the F-measures obtained by clustering different ITS (A), LSU (B) and combined (C) barcode datasets with thresholds ranging from 0.97 to 1. The line ITS/LSU-V in Fig. 12C shows the F-measures obtained when clustering the strains of the manually validated dataset ITS/LSU-V using the barcodes of both genetic markers. The vertical lines in the figures represent the thresholds proposed by UNITE for the species hypotheses (Koljalg et al. 2013). The optimal thresholds and best F-measures computed for each dataset are displayed in Table 2.
Fig. 12.
Clustering qualities obtained when clustering the different barcode datasets ITS (A), LSU (B) and combined (C) with thresholds ranging from 0.97 to 1 using an incremental step of 0.0001. The vertical lines in the figures represent the thresholds proposed by UNITE for the species hypotheses.
Table 2.
Optimal thresholds and best F-measures obtained by clustering different barcode datasets.
| Dataset | Abbreviation | Optimal threshold | Best F-measure | Number of species | Number of strains |
|---|---|---|---|---|---|
| Ex-Type ITS | ITS-T | 99.91 % | 90.50 % | 3 165 | 3 718 |
| Standard CBS Ex-type ITS | ITS-T-Standard | 99.81 % | 95.46 % | 2 950 | 3 393 |
| Manually validated ITS | ITS-V | 99.61 % | 74.72 % | 6 064 | 11 605 |
| Standard manually validated ITS | ITS-V-Standard | 99.61 % | 84.01 % | 5 375 | 9 338 |
| Ex-Type LSU | LSU-T | 99.91 % | 88.35 % | 3 238 | 3 737 |
| Standard CBS Ex-type LSU | LSU-T-Standard | 99.91 % | 94.06 % | 2 993 | 3 362 |
| Manually validated LSU | LSU-V | 99.81 % | 67.32 % | 6 469 | 12 588 |
| Standard manually validated LSU | LSU-V-Standard | 99.81 % | 77.68 % | 5 588 | 9 671 |
| Manually validated dataset having both ITS and LSU | ITS/LSU-V | 99.65 % | 77.47 % | 5 158 | 9 336 |
The high clustering qualities (best F-measures) obtained by removing indistinguishable species, which were 95.46 % for ITS-T-Standard, 84.01 % for ITS-V-Standard, 94.06 % for LSU-T-Standard and 77.68 % for LSU-V-Standard, showed that ITS can LSU can be used to discriminate filamentous fungal species. In addition, ITS outperformed LSU in species discrimination. The clustering quality of the combination of ITS and LSU genetic markers (ITS/LSU-V) outperformed each individual genetic marker (ITS-V and LSU-V), confirming that the two genetic markers can supplement each other in species identification for filamentous fungal strains.
Although there were a number of species with low similarity scores between their sequences as in Nilsson et al. (2008), the optimal thresholds predicted to discriminate filamentous fungi were rather high. For ITS, the optimal thresholds predicted for the ex-type datasets ITS-T and ITS-T-Standard were 99.91 % and 99.81 %, respectively. When including more data into the analysis (ITS-V and ITS-V-Standard), a slightly lower optimal threshold of 99.61 % was observed. For LSU, the optimal thresholds to discriminate ex-type and manually validated strains were 99.91 % (LSU-T and LSU-T-Standard) and 99.81 % (LSU-V and LSU-V-Standard), respectively. The reason that the optimal thresholds predicted for the ex-type datasets were higher than the optimal thresholds predicted for the validated datasets, is that many species in the validated datasets overlapped each other. The average number of sequences per species in a validated dataset was about 1.5 times more than in the corresponding ex-type dataset.
Compared to our previous investigations in yeasts (Vu et al. 2016), these optimal thresholds were higher than the taxonomic thresholds predicted to discriminate yeast species (99.61 % vs. 98.41 % using ITS and 99.81 % vs. 99.51 % using LSU barcodes). The associated clustering qualities were much lower in filamentous fungi, especially for LSU (84.01 % vs. 90.67 % for ITS and 77.68 % vs. 91.48 % for LSU). However, it must be noted that more fungal species were included in this study than were used in the yeasts study (Vu et al. 2016). The high values of the optimal thresholds imply that sequences can be wrongly assigned to a species name even with a small number of sequencing errors.
Low UNITE thresholds are not sensitive enough for filamentous fungal species identification
The UNITE cut-off values ranging from 97 % to 100 % for species identification have been proposed by Koljalg et al. (2013), and we intended to identify the appropriate UNITE threshold for filamentous fungal species identification. We clustered the ex-type and manually validated ITS datasets with six thresholds defined in UNITE. The obtained F-measures are given in Table 3. It can be seen that these values varied up to 25 % in the type datasets and up to 15 % on the manually validated datasets. The F-measures obtained by the thresholds 97–98.5 % were low when compared with the F-measure obtained by clustering the datasets with the optimal threshold of 99.61 % predicted for species identification in the manually validated dataset, indicating these thresholds may not be the most appropriate for filamentous fungal species identification. The same result was observed in a recent study in Robbertse et al. (2017), where the ITS region was shown to be identical or very close to being identical (99–100 %) for some Trichoderma species.
Table 3.
F-measures computed for UNITE thresholds on the ex-type and manually validated ITS datasets.
| Threshold | F-measure of ITS-T | F-measure of ITS-T-Standard | F-measure of ITS-V | F-measure of ITS-V-Standard |
|---|---|---|---|---|
| 97.00 % | 64.71 % | 69.66 % | 58.76 % | 68.70 % |
| 97.50 % | 66.69 % | 71.95 % | 60.91 % | 70.67 % |
| 98.00 % | 70.33 % | 75.55 % | 63.50 % | 73.68 % |
| 98.50 % | 74.04 % | 79.37 % | 66.66 % | 76.95 % |
| 99.00 % | 78.03 % | 84.25 % | 70.13 % | 80.48 % |
| 99.50 % | 84.81 % | 90.76 % | 73.92 % | 83.79 % |
| 100.00 % | 90.50 % | 95.32 % | 72.28 % | 79.77 % |
| 99.61 % | 87.36 % | 93.34 % | 74.73 % | 84.01 % |
Clustering at higher taxonomic levels reveals the need for a revision of fungal classification
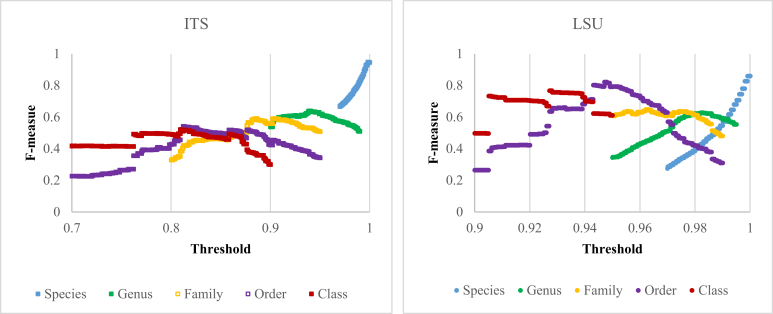
To evaluate the usefulness of ITS and LSU barcodes to recognise higher taxa, such as genera, families, orders and classes, we applied the F-measure to different levels in the taxonomic hierarchy. We clustered the type datasets ITS-T and LSU-T with different thresholds ranging from 70 % (for ITS) and 90 % (for LSU) to 100 % to get the optimal thresholds that produce the best quality for clustering at different taxonomic levels (Fig. 13, Table 4). Although ITS outperformed LSU in species identification, LSU had a better discriminatory power for taxonomic assignment than ITS at family, order and class levels. With a clustering quality (F-measure) of 82 %, LSU outperformed ITS in taxonomic classification at the order level. Nevertheless, the clustering qualities produced by both ITS and LSU at genus and family levels were low (64 % and 59.21 % for ITS, and 62.86 % and 64.91 % for LSU) indicating that there is a necessity to revise the classification of filamentous fungi at these taxonomic levels. As already demonstrated for the yeasts dataset (Vu et al. 2016), taxonomic revisions based on multigene phylogeny, proposed to implement the “one fungus one name” principle, can greatly improve the observed F-measures at genus level (Liu et al., 2016, Wang et al., 2016a, Wang et al., 2016b).
Fig. 13.
Clustering qualities (F-measures) obtained by comparing the clustering results of ITS-T and LSU-T with different taxonomic classifications at higher levels with thresholds ranging from 0.7 (for ITS) and 0.9 (for LSU) to 1 using an incremental step of 0.0001.
Table 4.
The optimal thresholds with the best quality (F-measure) predicted to discriminate filamentous fungal strains at higher taxonomic levels using ITS and LSU barcodes of the type datasets.
| Dataset | Genus Threshold | F-measure | Family Threshold | F-measure | Order Threshold | F-measure | Class Threshold | F-measure |
|---|---|---|---|---|---|---|---|---|
| ITS-T | 94.31 % | 64.00 % | 88.51 % | 59.21 % | 81.21 % | 54.21 % | 80.91 % | 52.16 % |
| LSU-T | 98.21 % | 62.86 % | 96.21 % | 64.91 % | 94.71 % | 82.24 % | 92.71 % | 76.70 % |
To investigate which genera, families, orders and classes are in need of revision, we computed the average similarity score for each genus, family, order and class of the datasets ITS-V and LSU-V. These values together with the associated number of strains are given in the Fig. 14 and Supplementary file S2. It can be seen that 31 % of genera, 62 % of families, 61 % of orders, and 69 % of classes of the dataset ITS-V had an average similarity score less than the associated predicted threshold. For the dataset LSU-V, these values are 30 %, 51 %, 54 % and 63 %. Those genera, families, orders and classes with a low average similarity score compared to the associated predicted threshold and a high number of the strains are the taxa most urgently in need of revision.
Fig. 14.
Average similarity score within genera, families, orders and classes of the ITS-V and LSU-V datasets.
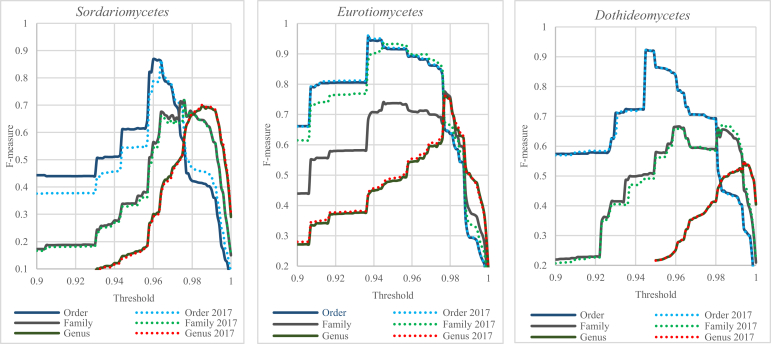
To investigate further the classification of filamentous fungi at higher levels, we computed the best F-measure and predicted an optimal threshold to discriminate strains in the three biggest classes Sordariomycetes represented by 4 151 strains, Eurotiomycetes represented by 2 318 strains and Dothideomycetes represented by 2 247 strains using the LSU-V dataset, as LSU outperformed ITS in classification at higher levels (Fig. 15, Table 5). As the continuous development in taxonomy results in an on-going stream of reclassifications and introduction of new names, we also updated the sequences with the current names present in the MycoBank till May 2017 to see if recent name changes have an impact on the classification of these three classes.
Fig. 15.
Clustering qualities (F-measures) obtained by comparing the clustering results of the LSU sequences of the three classes Sordariomycetes, Eurotiomycetes and Dothideomycetes with the taxonomic classifications at genus, family and order levels before and after updating taxon names.
Table 5.
The optimal thresholds with the best quality (F-measure) predicted to discriminate filamentous fungal strains at higher taxonomic level using LSU barcodes of the LSU-V dataset.
| Dataset | Genus Threshold | F-measure | Family Threshold | F-measure | Order Threshold | F-measure |
|---|---|---|---|---|---|---|
| Sordariomycetes | 98.5 % | 69.48 % | 97.6 % | 71.73 % | 96 % | 86.82 % |
| Sordariomycetes 2017 | 98.5 % | 70.00 % | 97.6 % | 71.76 % | 96.4 % | 86.47 % |
| Eurotiomycetes | 97.7 % | 75.94 % | 97.7 % | 77.47 % | 93.7 % | 95.68 % |
| Eurotiomycetes 2017 | 97.7 % | 76.02 % | 94.5 % | 93.49 % | 93.7 % | 96.17 % |
| Dothideomycetes | 99.4 % | 54.52 % | 96.1 % | 66.58 % | 94.5 % | 92.17 % |
| Dothideomycetes 2017 | 99.4 % | 54.84 % | 98.3 % | 67.10 % | 94.5 % | 92.29 % |
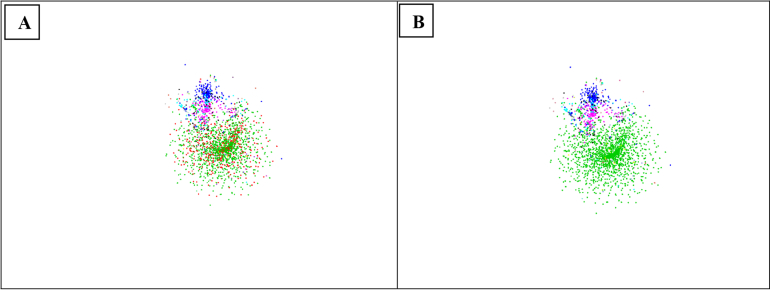
It can be seen from Fig. 15 and Table 5 that recent name changes in these three classes have slightly improved the classifications at genus, family and order levels as the predicted optimal thresholds were the same or slightly different except for the family levels of Eurotiomycetes and Dothideomycetes. Furthermore, the best F-measures obtained in most cases increased up to 0.32 %. In the Sordariomycetes, the best F-measure obtained at the order level was slightly reduced from 86.82 % to 86.47 %. The families Aspergillaceae, Thermoascaceae and Trichocomaceae are accepted families in Eurotiomycetes. A significant improvement in the classification at the family level could be obtained when these three families are merged (Trichocomaceae sensu lato). In that case, the clustering quality increased from 77.47 % to 93.39 %. Fig. 16 shows the distribution of the LSU sequences in Eurotiomycetes before and after the change of the family names. Before the update, the LSU sequences of the three families were intermixed. Based on these data, there is less support to maintain them as separate families than as one. On the other hand, this difference could also be explained by the progressive taxonomy in the Eurotiales, certainly when this is compared with the other orders in this class. This might make this comparison less balanced. It needs to be noted that LSU sequences are not commonly used to study family level relationships in the Eurotiales. These families are delimited by phenotypic characters and using multi-locus sequence datasets, including protein coding genes (Houbraken and Samson, 2011, Quandt et al., 2015).
Fig. 16.
The distribution of the LSU sequences of the class Eurotiomycetes before (A) and after (B) updating sequence names. The sequences of the same colour belong to the same family. The four biggest families represented by the colours green, red, blue and pink in the left picture are Trichocomaceae with 1 147 sequences, Aspergillaceae with 464 sequences, Herpotrichiellaceae with 257 sequences and Arthrodermataceae with 172 sequences, respectively. The family Aspergillaceae has been recently merged into the family Trichocomaceae, as can be seen in the right figure.
At the order level, the best F-measures obtained of the three classes were high, ranging from 86.47 % to 96.17 %, indicating that LSU could separate the strains of the orders in these classes. At the family level, the best F-measure obtained in Eurotiomycetes was 93.39 % as seen earlier, showing that the families of this class could be discriminated using LSU sequences. Dothideomycetes had the lowest best F-measures at the genus and family levels. In addition, the optimal thresholds predicted to separate the LSU sequences in Dothideomycetes at these levels were high (99.4 % and 98.3 %), indicating that LSU might not be the best genetic marker to resolve the classifications or that there is a need for a taxonomic revision at the genus and family levels in Dothideomycetes. Recent studies (Chen et al., 2017, Videira et al., 2017, Yang et al., 2017) have started using the RPB2 gene to resolve genera and families in Dothideomycetes.
ITS and LSU barcodes can contribute to taxonomic reclassifications
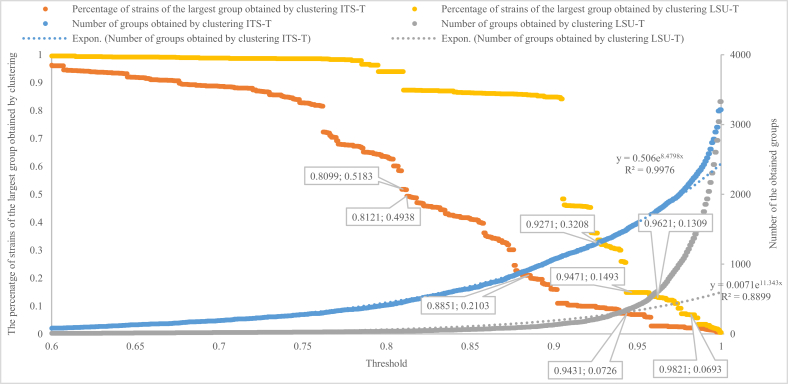
To study the phylogenetic relationships of filamentous strains resulting from ITS and LSU barcodes, we examined the division of the largest group and the total number of the groups obtained over different threshold values (Fig. 17). At low DNA similarity scores, the total number of the groups was small, and many dissimilar strains grouped together. With a similarity score < 80 % for ITS and < 90 % for LSU, the largest group contained > 50 % of the strains. For any threshold, the number of groups obtained by ITS was always higher than by LSU, confirming that ITS is more variable than LSU. While the total numbers of the groups by both genetic markers follow two exponential smoothing curves (with the best-fit exponential trend lines y = 0,506e8,4798x with R2 = 0,9976 for ITS, and y = 0,0071e11,343x with R2 = 0,8899 for LSU) indicating a rather sudden increase in the number of taxa at higher ITS and LSU similarity scores, there are discontinuity points in both lines of the percentage of the strains of the largest group. At these points, the largest groups were split into two or more subgroups. It is interesting to see that the thresholds predicted for species, genus, family, order, and class identifications (Table 4) were either a discontinuity point or close to a discontinuity point (Fig. 17), indicating that ITS and LSU barcodes can contribute to taxonomic reclassifications.
Fig. 17.
The total number of the obtained groups (displayed in the secondary axis) and the percentage of strains of the largest group obtained by clustering ITS-T and LSU-T with thresholds increased from 0.6 to 1 with a step of 0.0001.
ITS barcodes could identify 25 % of the “Top 50 Most Wanted Fungi” to the class level
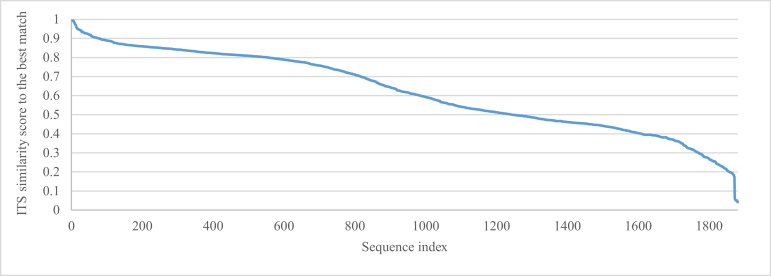
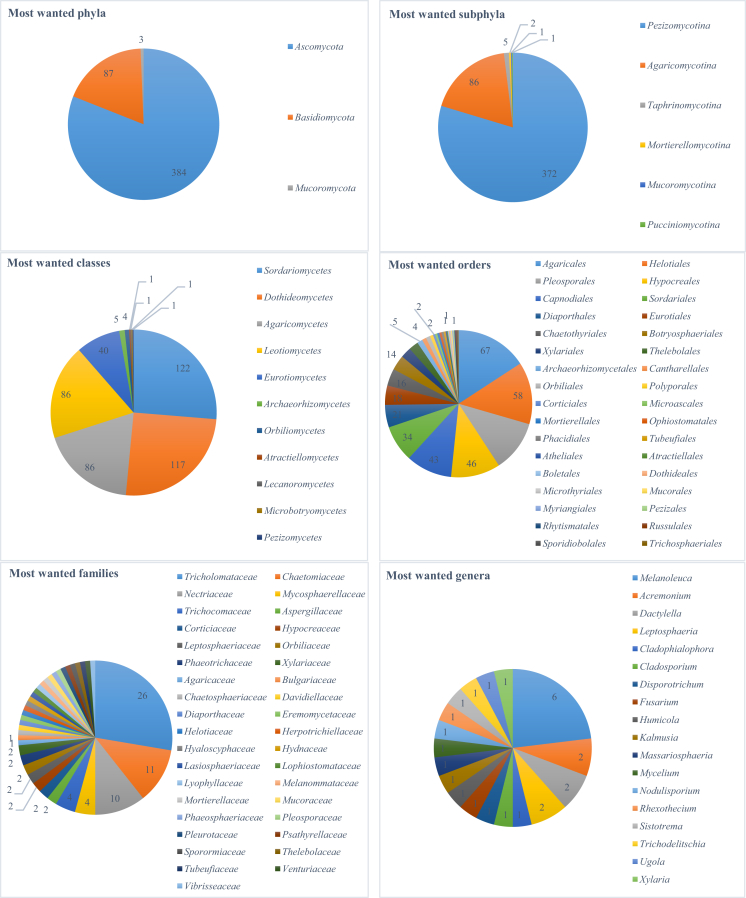
Finally, this section shows how the newly generated barcodes could help to reveal unidentified fungal lineages that comprise the majority of fungi in molecular ecological studies. We took the dataset called “Top 50 Most Wanted Fungi” (UNITE Community 2017) representing 2 024 most frequently sampled environmental sequence types of 1 493 undefined lineages using the fungal genetic marker ITS, created by Nilsson et al. (2016), as an example. These most wanted sequences were visualised in Fig. 18 using fMLC (Vu et al. 2018). It is interesting to see that they formed a big group lying between the two classes Dothideomycetes and Agaricomycetes. The unidentified sequences were then compared with the ITS validated dataset (ITS-V) using BLAST (Altschul et al. 1997). Of the 2 024 sequences, 143 sequences having a coverage with their best match lower than the minimum ITS length (267 bp) obtained by extracting ITS regions of the ITS validated dataset (ITS-V) using the software ITSx (http://microbiology.se/software/itsx/) (Fig. 19), were excluded from the analysis. Fig. 20 shows the ITS similarity scores of the remaining sequences to their best-match barcode sequence, in which there were 500 (24 %), 475 (23 %), 110 (5 %), and 26 (1 %) sequences having a similarity score greater than 80.91 %, 81.21 %, 88.51 % and 94.31 % to their best match, respectively. According to the thresholds predicted to identify ITS sequences at different taxonomic levels given in Table 4, these sequences could be assigned to the same class, family, order and genus of their best match sequence. Fig. 21 shows the taxa together with the associated number of sequences that could be assigned to the “Top 50 Most Wanted Fungi” dataset. Of the 500 sequences of 414 undefined lineages having a best match at the class level, 26 sequences had no associated phylum name available. The remaining 474 sequences were assigned automatically to Ascomycota (384), Basidiomycota (87) and Mucoromycota (3). Furthermore, 33 sequences had no associated subphylum name available. The remaining 467 sequences were assigned to six subphyla: Pezizomycotina (372), Agaricomycotina (86), Taphrinomycotina (5), Mortierellomycotina (2), Mucoromycotina (1), and Pucciniomycotina (1). In addition, 36 sequences had no associated class name available. The remaining 464 sequences were assigned to 11 classes with the five most dominant groups Sordariomycetes (119), Dothideomycetes (114), Leotiomycetes (86), Agaricomycetes (86), and Eurotiomycetes (40). Of the 475 sequences of 393 undefined lineages having a best match at the order level, 51 sequences had no associated order name available. The remaining 424 sequences were assigned to 34 orders with the six dominant groups Agaricales (67), Helotiales (58), Hypocreales (46), Pleosporales (48), Capnodiales (43) and Sordariales (34). Of the 110 sequences of 94 undefined lineages having a best match at the family level, 16 sequences had no associated family name available. The remaining 94 sequences were assigned to 37 families in which Tricholomataceae was the biggest group with 26 sequences, followed by Chaetomiaceae (11) and Nectriaceae (10). The other families had a small number (<5) of sequences. At the genus level, 26 sequences of 25 undefined lineages were assigned to 18 genera. Among them, Melanoleuca, Acremonium, Leptosphaeria and Dactylella had 6, 2, 2 and 2 sequences, respectively. The remaining genera had only one sequence found. Finally, at the species level, there were six undefined sequences EF619883, JX982422, FN397290, KP335575, JX043184, and HQ607985 having a similarity score greater than 99 % to their best matches of the strains CBS 126038 (99.84 %), CBS 284.52 (99.6 %), CBS 953.68 (99.5 %), CBS 955.73 (99.39 %), CBS 612.68 (99.23 %), and CBS 366.90 (99.17 %), respectively. Among them, only EF619883 had the similarity score greater than the optimal threshold 99.61 % predicted to identify ITS sequences at the species level. This sequence was linked to Sistotrema octosporum (CBS 126038). The other five sequences JX982422, FN397290, KP335575, JX043184, and HQ607985 may respectively belong to the same or closely related species, namely Leptosphaeria allorgei, Humicola parvispora, Rhexothecium globosum, Massariosphaeria roumeguerei, and Cladosporium alternicoloratum. The results presented in this section demonstrate the potential of the newly generated barcodes to improve fungal taxonomy in molecular ecology studies. These data can also be found in Supplementary file S4.
Fig. 18.
The distributions of the ITS sequences of the validated dataset ITS-V (left) and its extension with the ITS sequences of the “Top 50 Most Wanted Fungi” dataset (right). The five groups in green, red, blue and pink represent 3 810, 2 210, 2 064, 1 483, and 574 ITS sequences of Sordariomycetes, Eurotiomycetes, Dothideomycetes, Agaricomycetes, and Leotiomycetes, respectively. All 2 024 sequences of the “Top 50 Most Wanted Fungi” dataset are in chocolate colour. The group in turquoise colour contains 1 145 sequences that have no a class name given in the database. The 3D coordinates of the sequences were computed using fMLC (Vu et al. 2018). The sequences were visualized using the rgl package in R (https://r-forge.r-project.org/projects/rgl/).
Fig. 19.
The lengths of the ITS regions extracted from the ITS validated dataset using the software ITSx (http://microbiology.se/software/itsx/). The obtained minimum length was 267.
Fig. 20.
The ITS similarity scores of the most wanted sequences to their best-match ITS barcodes.
Fig. 21.
The phyla, subphyla, classes, orders, families and genera together with the number of the sequences found in the “Top 50 Most Wanted Fungi” dataset.
Conclusions
Microbial Biological Resource Centres can play an important role in providing reference materials for global initiatives such as DNA barcoding and genome sequencing projects. While efforts to collect material for DNA barcoding of higher organisms in the field have been successful, those targeting filamentous fungi and other microorganisms are hampered by high costs and time needed for the process of collecting, isolating and identifying those microorganisms. The microbes that have been deposited in public culture collections over many decades – over 700 public collections are now registered in the CCInfo database of the World Federation of Culture Collections (WFCC) – provide an indispensable and rich source of well-documented microbial material for DNA barcoding which thus far has not been fully explored. The isolation, study and long-term preservation of these materials has only been possible due to major investments by society. Barcoding strains preserved in these collections disclose primary information on their genetic constitution and enables the scientific community to exploit the diversity and quality of these permanently available resources for the benefit of health, food security and sustainable development.
In a previous study, the barcoding project at the Westerdijk Institute resulted in the release of 8 669 barcode sequences of manually validated CBS strains representing 1 351 yeast species (Vu et al. 2016). In the current study, 24 193 manually validated ITS and LSU sequences of CBS ex-type, and other reference strains of filamentous fungi representing 7 376 species have been released to GenBank. Additional information on type materials of these strains are also shared with GenBank. At the same time, the data have been incorporated into the identification tools available on the webpages of the Westerdijk Institute (www.westerdijkinstitute.nl). The release of such a number of sequences from a MBRC is unprecedented, and will lead to downstream improvements in fungal identification in associated public databases. All strains involved in this study are publicly available as reference materials for research from the CBS Collection of the Westerdijk Institute, an ISO 9001:2015 certified public collection that maintains the highest standards of the OECD Guidelines for MBRC's. More sequences that were and are still being produced in the barcoding project are in the process of annotation and validation and will be released to publicly available sequence databases in the future. Establishing the authenticity of the sequences is challenging, especially for asexual taxa. Often these strains in the CBS Collection are unique and no previous sequence data exist to be used as a point of reference.
Computational approaches to estimating rational taxonomic boundaries and the quality of higher classifications based on large datasets of ribosomal RNA gene sequences have been broadly applied to bacteria and archaea based on 16S sequences (Yarza et al., 2014, Kim et al., 2014). However, they have rarely been applied to filamentous fungi. The set of validated data built up in our study allowed us to replicate this approach. In our study, which covers a broad sampling of filamentous fungal species, it was confirmed that ITS sequences are better in discriminating between species than LSU which was similarly observed for yeasts (Vu et al. 2016). Except for the species that cannot be discriminated (17 % by ITS and 18 % by LSU), ITS and LSU could be used to separate filamentous fungal strains at species level with a threshold of 99.61 % for ITS and 99.81 % for LSU. The clustering quality value of ITS at species level was 84 % while for LSU this was 78 %. It was also shown that the low ITS thresholds (less than 99 %) was not sensitive enough for filamentous fungal species identification. At the genus level, the clustering quality values obtained by both ITS and LSU were low, indicating a necessity to revise the generic taxonomy of many filamentous fungi. At family and higher taxonomic levels, LSU had a better discriminatory power of taxonomic assignment than ITS. With a clustering quality value of 80 %, LSU was shown to be suitable for identifying filamentous fungi at the order level.
Together with the previous study on yeasts (Vu et al. 2016), the released barcodes and the appropriately predicted similarity thresholds can be employed to flag potentially new species of the estimated ∼3.8 million unknown fungal species (Hawksworth & Lücking 2017). In addition, they aid in the documentation of fungal diversity observed using metagenomics approaches (Handelsman, 2004, Hibbett, 2016). Using the newly generated ITS barcodes, six sequences the “Top 50 Most Wanted Fungi” dataset could be assigned to the same or closely related species of their best match barcode sequence, 24 %, 23 %, 5 %, and 1 % of which could be identified to the class, order, family and genus levels, respectively, demonstrating the potential to advance fungal taxonomy in molecular ecology studies.
Despite their ability to delineate a large number of fungal species, ITS and LSU are not sufficient as a barcode for all fungi. In addition, they may be inaccurate in validating genome sequences as genome assemblies often do not include sequence data from ribosomal cistron, and if included, the regions are not always correctly assembled (Robbertse et al. 2017). To overcome this limitation, secondary DNA barcodes for the fungal kingdom have been suggested (Stielow et al. 2015). More attention should be given to single copy protein coding genes, or even whole genome sequences (Coissac et al., 2016, Robbertse et al., 2017). Until such a major effort is realised, ITS and LSU sequences will continue to play an important role in fungal identification and classification. The effort to select specimens for generating whole genome sequences can be guided by phylogenetic inferences based on ITS and LSU sequences. A prioritisation of fungal species for whole genome sequencing can be guided by the presence of fungi in metagenomics communities, such as soil, human and water samples. Fungi commonly present in clinical, environmental, or economical relevant communities can often be identified to species level by their ITS and LSU barcodes, and can be used to prioritize the next phase of sequence-based fungal identification methodologies.
Data availability
The barcode sequences present in this study have been submitted to GenBank (www.ncbi.nlm.nih.gov) under the RefSeq Targeted Loci BioProject: http://www.ncbi.nlm.nih.gov/bioproject/PRJNA422523. They will also be available at the Westerdijk institute's website www.westerdijkinstitute.nl/Collections/ as reference sequences for fungal identification.
Acknowledgements
This study was financially supported by the “Fonds Economische Structuurversterking (FES)”, Dutch Ministery of Education, Culture and Science grant BEK/BPR-2009/137964-U, “Making the Tree of Life Work”, and financial support from the Royal Academy of Arts and Sciences in the Netherlands (KNAW). We are grateful to the barcoding team including Geert van Haalem, Sarina de Roos, Marloes Wouters, Desiree Smits, Romana Renfurm, Maryam Saradeghi Keisari, Janneke Bloem, and the researchers at the Westerdijk Fungal Biodiversity Institute who contributed to this study by generating, analysing and validating a selection of the sequences included.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.simyco.2018.05.001.
Contributor Information
D. Vu, Email: d.vu@westerdijkinstitute.nl.
G.J.M. Verkley, Email: g.verkleij@westerdijkinstitute.nl.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Afshinnekoo E., Meydan C., Chowdhury S. Geospatial Resolution of Human and Bacterial Diversity with City-Scale Metagenomics. Cell Systems. 2015;1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaalid R., Kumar S., Nilsson R.H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Molecular Ecology Resources. 2013;13:218–224. doi: 10.1111/1755-0998.12065. [DOI] [PubMed] [Google Scholar]
- Boon E., Zimmerman E., Lang B.F. Intra-isolate genome variation in arbuscular mycorrhizal fungi persists in the transcriptome. Journal of Evolutionary Biology. 2010;23:1519–1527. doi: 10.1111/j.1420-9101.2010.02019.x. [DOI] [PubMed] [Google Scholar]
- Botschuijver S., Roeselers G., Levin E. Intestinal Fungal Dysbiosis Associates With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology. 2017;153:1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group A DNA barcode for land plants. PNAS. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Hou L.W., Duan W.J. Didymellaceae revisited. Studies in Mycology. 2017;87:105–159. doi: 10.1016/j.simyco.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coissac E., Hollingsworth P.M., Lavergne S. From barcodes to genomes: Extending the concept of DNA barcoding. Molecular Ecology. 2016;25:1423–1428. doi: 10.1111/mec.13549. [DOI] [PubMed] [Google Scholar]
- Cui L., Morris A., Ghedin E. The human mycobiome in health and disease. Genome Medicine. 2013;5:1–12. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Queiroz K. Species concepts and species delimitation. Systematic Botany. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Eberhardt U. Methods for DNA barcoding of fungi. DNA Barcodes: Methods and Protocols. 2012;858:183–205. doi: 10.1007/978-1-61779-591-6_9. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. Updating the 97 % identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- Federhen S. Type material in the NCBI Taxonomy Database. Nucleic Acids Research. 2015;43:D1086–D1098. doi: 10.1093/nar/gku1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1 / D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Henn M.R., Ma L.-J. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Research. 2005;15:1620–1631. doi: 10.1101/gr.3767105. [DOI] [PubMed] [Google Scholar]
- Garza D.R., Van Verk M.C., Huynen M.A. Bottom-up ecology of the human microbiome: from metagenomes to metabolomes. BioRxiv. 2016 [Google Scholar]
- Geml J., Gravendeel B., Van Der Gaag KJ. The contribution of DNA metabarcoding to fungal conservation: diversity assessment, habitat partitioning and mapping red-listed fungi in protected coastal Salix repens communities in the Netherlands. PLoS One. 2014;9:e99852. doi: 10.1371/journal.pone.0099852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon H.S., Oliver A., Taylor J. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods in Ecology and Evolution. 2015;6:973–980. doi: 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiology and Molecular Biology Reviews. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D.L., Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology Spectrum. 2017;5:1–2. doi: 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PubMed] [Google Scholar]
- Hebert P.D., Cywinska A., Ball S.L. Biological identifications through DNA barcodes. Proceedings of the Royal Society B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett D. The invisible dimension of fungal diversity. Science. 2016;351:1150–1151. doi: 10.1126/science.aae0380. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C., Fah Sathirapongsasuti J., Segata N. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irinyi L., Serena C., Garcia-Hermoso D. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database – The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycology. 2015;53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kim M., Oh H.S., Park S.C. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- Kiss L. Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. PNAS. 2012;109 doi: 10.1073/pnas.1207143109. E1811–E1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg U., Nilsson R.H., Abarenkov K. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kooij P.W., Aanen D.K., Schiøtt M., Boomsma J.J. Evolutionarily advanced ant farmers rear polyploid fungal crops. Journal of Evolutionary Biology. 2015;28:1911–1924. doi: 10.1111/jeb.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C.P., Robnett C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- Levy M., Blacher E., Elinav E. Microbiome, metabolites and host immunity. Current Opinion in Microbiology. 2017;35:8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang Q., Theelen B. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology. 2016;81:1–26. doi: 10.1016/j.simyco.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau R.L., Liu C.M., Aziz M. Linking soil bacterial biodiversity and soil carbon stability. The ISME Journal. 2014;9:1477–1480. doi: 10.1038/ismej.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T.K., Bae H. The diversity of fungal genome. Biological Procedures Online. 2015;17:8. doi: 10.1186/s12575-015-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.D.N., Viscogliosi E., Delhaes L. The lung mycobiome: An emerging field of the human respiratory microbiome. Frontiers in Microbiology. 2015;6:1–9. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R.H., Kristiansson E., Ryberg M. Intraspecific ITS variability in the Kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolutionary Bioinformatics. 2008;4:193–201. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R.H., Ryberg M., Kristiansson E. Taxonomic reliability of DNA sequences in public sequences databases: A fungal perspective. PLoS One. 2006;1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R.H., Wurzbacher C., Bahram M. Top 50 most wanted fungi. MycoKeys. 2016;12:29. [Google Scholar]
- Paccanaro A., Casbon J.A., Saqi M.A.S. Spectral clustering of protein sequences. Nucleic Acids Research. 2006;34:1571–1580. doi: 10.1093/nar/gkj515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt C.A., Kohler A., Hesse C.N. Metagenome sequence of Elaphomyces granulatus from sporocarp tissue reveals Ascomycota ectomycorrhizal fingerprints of genome expansion and a Proteobacteria-rich microbiome. Environmental Microbiology. 2015;17:2952–2968. doi: 10.1111/1462-2920.12840. [DOI] [PubMed] [Google Scholar]
- Robbertse B., Strope P.K., Chaverri P. Improving taxonomic accuracy for fungi in public sequence databases: applying 'one name one species' in well-defined genera with Trichoderma/Hypocrea as a test case. Database. 2017 doi: 10.1093/database/bax072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V., Szoke S., Jabas B. BioloMICS Software: Biological data management, identification, classification and statistics. The Open Applied Informatics Journal. 2011;5:87–98. [Google Scholar]
- Robert V., Vu D., Amor A.B. MycoBank gearing up for new horizons. IMA Fungus. 2013;4:371–379. doi: 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS. 2012;109:1–6. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon U.K., Weiss M. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Molecular Biology and Evolution. 2008;25:2251–2254. doi: 10.1093/molbev/msn188. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today. 2006;33:152–155. [Google Scholar]
- Stielow J.B., Lévesque C.A., Seifert K.A. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015;35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope P.K., Skelly D.A., Kozmin S.G. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Research. 2015;125:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Liu J., Zhang M., Mei Q. Proceedings of the 25th International Conference on WWW, Montreal, Canada. 2016. Visualizing Large-scale and High-dimensional Data; pp. 287–297. [Google Scholar]
- Tedersoo L., Bahram M., Põlme S. Global diversity and geography of soil fungi. Science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- UNITE Community . UNITE Community; 2017. UNITE top50 release. Version 01.12.2017. [Google Scholar]
- Verkley G.J.M., Quaedvlieg W., Shin H. A new approach to species delimitation in Septoria. Studies in Mycology. 2013;75:213–305. doi: 10.3114/sim0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira S.I.R., Groenewald J.Z., Nakashima C. Mycosphaerellaceae – chaos or clarity? Studies in Mycology. 2017;87:257–421. doi: 10.1016/j.simyco.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Georgievska S., Szoke S. fMLC: Fast multi-level clustering and visualization of large molecular datasets. Bioinformatics. 2018;34:1577–1579. doi: 10.1093/bioinformatics/btx810. [DOI] [PubMed] [Google Scholar]
- Vu D., Groenewald M., Szöke S. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Studies in Mycology. 2016;85:91–105. doi: 10.1016/j.simyco.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Szöke S., Wiwie C. Massive fungal biodiversity data re-annotation with multi-level clustering. Scientific Reports. 2014;4:6837. doi: 10.1038/srep06837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Eberhardt U., Szöke S. A laboratory information management system for DNA barcoding workflows. Integrative Biology. 2012;4:744–755. doi: 10.1039/c2ib00146b. [DOI] [PubMed] [Google Scholar]
- Wang Q., Begerow D., Groenewald M. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology. 2016;81:55–83. doi: 10.1016/j.simyco.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Groenewald M., Takashima M. Multigene phylogeny and reclassification of yeasts and related filamentous taxa in Basidiomycota. Studies in Mycology. 2016;81:27–53. doi: 10.1016/j.simyco.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg J.H.C., Groenewald J.Z., Binder M. Alternaria redefined. Studies in Mycology. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg J.H.C., Seidl M.F., Groenewald J.Z. Alternaria section Alternaria: Species, formae speciales or pathotypes? Studies in Mycology. 2015;82:1–21. doi: 10.1016/j.simyco.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Groenewald J.Z., Cheewangkoon R. Families, genera, and species of Botryosphaeriales. Fungal Biology. 2017;121:322–346. doi: 10.1016/j.funbio.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The barcode sequences present in this study have been submitted to GenBank (www.ncbi.nlm.nih.gov) under the RefSeq Targeted Loci BioProject: http://www.ncbi.nlm.nih.gov/bioproject/PRJNA422523. They will also be available at the Westerdijk institute's website www.westerdijkinstitute.nl/Collections/ as reference sequences for fungal identification.