Abstract
Despite substantial declines in mortality following myocardial infarction (MI), subsequent left ventricular remodeling (LVRm) remains a significant long-term complication. Extracellular small non-coding RNAs (exRNAs) have been associated with cardiac inflammation and fibrosis and we hypothesized that they are associated with post-MI LVRm phenotypes. RNA sequencing of exRNAs was performed on plasma samples from patients with “beneficial” (decrease LVESVI ≥ 20%, n = 11) and “adverse” (increase LVESVI ≥ 15%, n = 11) LVRm. Selected differentially expressed exRNAs were validated by RT-qPCR (n = 331) and analyzed for their association with LVRm determined by cardiac MRI. Principal components of exRNAs were associated with LVRm phenotypes post-MI; specifically, LV mass, LV ejection fraction, LV end systolic volume index, and fibrosis. We then investigated the temporal regulation and cellular origin of exRNAs in murine and cell models and found that: 1) plasma and tissue miRNA expression was temporally regulated; 2) the majority of the miRNAs were increased acutely in tissue and at sub-acute or chronic time-points in plasma; 3) miRNA expression was cell-specific; and 4) cardiomyocytes release a subset of the identified miRNAs packaged in exosomes into culture media in response to hypoxia/reoxygenation. In conclusion, we find that plasma exRNAs are temporally regulated and are associated with measures of post-MI LVRm.
Keywords: Left ventricular remodeling, Myocardial infarction, microRNA, Extracellular RNA, Cardiac magnetic resonance imaging, RNA sequencing, And inflammation
Highlights
-
•
Plasma exRNA signatures were associated with post-MI remodeling as assessed by cardiac MRI.
-
•
Candidate exRNAs originated from different cell types, and were dynamically regulated in experimental models of ischemia.
-
•
Candidate exRNAs were predicted to regulate pathways of inflammation and fibrosis by bioinformatics analysis.
Plasma RNA signatures are associated with heart remodeling following a myocardial infarction, and are predicted to regulate inflammation pathways. These extracellular RNAs are derived from different types of cells in the heart and are altered in the heart tissue as well as plasma in a mouse model of myocardial infarction, reflecting the complex and dynamic process of heart remodeling after myocardial infarction.
1. Introduction
There have been substantial improvements in acute myocardial infarction (MI) care in recent decades, resulting in reduced mortality rates during the acute post-MI phase. Despite this, post-MI cardiac remodeling and the subsequent development of heart failure (HF) remains a key long-term source of morbidity for this patient group (Ezekowitz et al., 2009). Left ventricular remodeling (LVRm) post-MI is a multifaceted and dynamic process that is the product of the complex interplay between infarct size, genetic and epigenetic influences on cell biology, and effects of neurohormonal antagonism (Heusch et al., 2014). Recently, there has also been growing interest in the role of inflammation in post-MI LVRm (Abbate et al., 2015). Early identification of pathological LVRm is a significant clinical challenge in the treatment of post-MI patients. Natriuretic peptides are the current gold standard biomarker for HF; however, a limitation of their use is that levels are typically only elevated once there is a change in LV function and significant remodeling has already occurred (Talwar et al., 2000). Although there is an inverse relationship between LV ejection fraction (LVEF) and likelihood of adverse remodeling, many patients with a preserved LVEF post-MI will still go on to develop adverse remodeling (Abbate et al., 2015). In fact, the majority of post-MI HF cases in the modern era occur with preserved LV function and may not be captured by alterations in natriuretic peptides (Wijk et al., 2015).
Post-MI LVRm consists of distinct phases involving multiple cell types in the heart including cardiomyocytes, fibroblasts, endothelial cells, and leukocytes (Burchfield et al., 2013). The acute post-MI phase is hallmarked by cardiomyocyte death and subsequent recruitment of inflammatory cells to remove dead cells and begin the repair process. Distinct from this period is a sub-acute phase where the inflammatory response is resolved and fibroblast proliferation and secretion of extracellular matrix proteins leads to the formation of scar. Chronically, continuation of these processes and the global impact of molecular changes on cardiac function leads to what is termed “beneficial” or “adverse” LVRm (Burchfield et al., 2013). Differences in cellular response to events, timing of the resolution of inflammatory response, and the degree of fibrosis all contribute to driving LVRm down either beneficial or adverse paths (Anzai, 2013). The molecular mechanisms and markers of these stages and processes have not been fully characterized. An increased understanding of the molecular architecture of post-MI remodeling offers the opportunity to identify potential early markers of adverse LVRm and therapeutic targets to prevent the development of post-MI HF.
The past decade has seen the phenomenon of the non-coding RNA revolution, where previously disregarded RNA species have been rediscovered as critical regulators of physiological and pathological processes (Cech and Steitz, 2014). Small non-coding RNAs are short regulatory transcripts (<200 nt) including the extensively studied microRNAs (miRNA) (Rajan et al., 2014), and more recently re-discovered piwi-interacting RNA (piRNA) (Rajan et al., 2014), small nucleolar RNA (snoRNA) (Stepanov et al., 2015), tRNA fragments (Cozen et al., 2015), and yRNA fragments (Cambier et al., 2017). These RNA species can affect the expression of genes and pathways important in cardiovascular diseases (CVD) including myocardial fibrosis (Thum, 2014), coronary atherosclerosis (Aryal et al., 2014), and cardiac arrhythmias (Kim, 2013). Importantly, they are stably expressed in the circulation (extracellular RNA; exRNA) as well as in tissue, leading to potential utility as diagnostic and prognostic biomarkers of disease. Significant work has been directed to the study of plasma exRNAs in acute coronary syndrome and in coronary artery disease. However, the majority of studies have been focused on candidate plasma miRNAs (Jancovicova et al., 2017, Zile et al., 2011), rather than using an unbiased discovery approach. Hence few studies have investigated the association of other circulating non-coding RNAs with MI or LVRm in humans and the only other reported non-coding RNA species to date are long non-coding RNA (Li et al., 2017, Gao et al., 2017b, Piccoli et al., 2017).
In this study, we examine the landscape of exRNAs in human circulation by RNA sequencing (RNAseq) to determine whether changes in plasma exRNAs post-MI are associated with LVRm. Furthermore, the cellular origin and temporal regulation of candidate exRNAs is examined in animal and cell models of cardiac ischemia. This is intended as an exploratory study to identify exRNA signatures associated with LVRm, and increase our understanding of the molecular architecture and biology of LVRm. Future studies in expanded cohorts for validation are important next steps for the development of these exRNA signatures as clinical biomarkers for post-MI LVRm.
2. Materials and Methods
2.1. Patient Population (OMEGA-REMODEL Trial)
The Omega-3 Acid Ethyl Esters on LV Remodeling After Acute MI (OMEGA-REMODEL; clinicaltrials.gov identifier NCT00729430) trial was a prospective, multicenter, double-blind, placebo-controlled trial testing the effect of omega-3 fatty acid supplementation (4 g/day) for 6 months after acute MI on adverse LVRm by cardiac magnetic resonance imaging (CMR, full study report in Heydari et al., 2016). The Institutional Review Boards of the respective institutions approved all protocols involving human patients and all participants provided written informed consent. This study conformed to standards indicated by the Declaration of Helsinki. Venous blood from OMEGA-REMODEL was collected in EDTA vacutainers at the time of baseline CMR imaging (2–4 weeks post-MI), centrifuged at 2000 ×g for 10 min for plasma separation, and immediately stored at −80 °C. CMR phenotyping (detailed in previous work by our group; (Heydari et al., 2016)) included LV end-systolic volume index (LVESVI), LVEF, myocardial mass, infarct size, and extracellular volume fraction (ECV; a validated surrogate of myocardial interstitial expansion (Haaf et al., 2016)). Of the 331 subjects in the overall study, 238 underwent post-treatment follow-up CMR for serial comparison for LV remodeling. Baseline characteristics were compared via chi-squared (categorical) or Wilcoxon tests (continuous).
2.2. Identification of Patients with Favorable and Adverse LVRm and their Respective Propensity-Matched Controls
Reduction of LVESVI during the convalescent phase after MI has remained the most robust risk marker across early (White et al., 1987, Migrino et al., 1997) and recent clinical trials (St John Sutton et al., 2017, Doughty et al., 2004). Consistent with other acute MI clinical trials, there was a high proportion of favorable structural LVRm in patients of the randomized OMEGA-REMODEL study. We therefore defined beneficial LVRm as >20% reduction of indexed LVESV (LVRmfav = 1) given its high specificity towards favorable post-MI HF prognosis to be consistent with the published literature (St John Sutton et al., 2017). Patients with an expansion of indexed LVESV of >15% were defined as adverse remodelers (LVRmfav = 0). A logistic regression was then performed to identify the strongest propensity-matched patient pairs based on patient age, gender, baseline LVEF, baseline infarct size by late gadolinium enhancement, and a history of diabetes. A validated propensity score-based 5 to 1 greedy matching algorithm was used to minimize matching bias (http://www2.sas.com/proceedings/sugi26/p214-26.pdf). The 11 best-matched patient pairs were then chosen.
In addition to patients with favorable and adverse LVRm, patients with adverse electrical remodeling (n = 10) were identified and matched to patients without adverse electrical remodeling (n = 10) as described above. Adverse electrical remodeling was defined as sudden cardiac arrest or the detection of ambient ventricular arrhythmia on clinical follow-up.
2.3. RNASeq of Plasma Samples
RNAseq was performed on the 11 best-matched patient pairs with beneficial and adverse LVRm and on the 10 best-matched patients with and without adverse electrical remodeling. RNA was extracted from 1 mL plasma and libraries constructed according to our previously published methods (Danielson et al., 2017). Briefly, plasma RNA was isolated using the miRCURY RNA Isolation kit for Biofluids (Exiqon) with modified protocol and libraries were constructed and amplified from approximately 10 ng RNA using the NEBNext small RNA library prep set for Illumina (NEB). Size selection of libraries was performed by gel electrophoresis on a 10% Novex TBE gel with excision of the 140 to 160 nucleotide bands (corresponding to 21–40 nucleotide RNA fragments). Libraries were diluted to a final concentration of 2 nM, pooled, and sequenced on an Illumina HiSeq 2000 for single read 50 cycles at the Center for Cancer Computational Biology at Dana-Farber Cancer Institute.
Healthy controls were used from a different study in our laboratory and were prepared using an alternative method (Shah et al., 2017). Briefly, plasma exRNA from 26 subjects was isolated using a modified mirVana PARIS protocol (AM1556; Life Technologies) with sequential phenol–chloroform extractions (Burgos et al., 2013). RNA was concentrated using the Zymo RNA Clean & Concentrator kit (Zymo Research) and libraries were prepared using the NEXTflex Small RNA Sequencing Kit v2 by Bioo-Scientific. Pools of 15 samples were created, denatured and clustered on either a single read Illumina V3 flowcell (GD-401-3001; Illumina) or a single read rapid Illumina V2 flowcell (GD-402-4002; Illumina). The flowcells were run on the Illumina HiSeq 2500 (Illumina) for 50 cycles with a 7 cycle indexing read.
2.4. Next Generation Sequence Analysis
The BCL files were de-multiplexed using CASAVA v1.82, and the adaptor sequences within the read sequences were trimmed by FastX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). The processed sequences were filtered for small RNAs >16 nucleotides in length. The sequences were then aligned, quantified, and annotated using sRNABench 1.0 pipeline (Barturen et al., 2014). Briefly, the pipeline implemented hierarchical sequence mapping strategy that first mapped and removed spike-in library, contaminants, and rRNA. Next, RNA species were sequentially mapped to known mature miRNA, tRNA, snoRNA, piRNA, and yRNA on the human genome sequence (hg19) using Bowtie2 (Langmead et al., 2009) with parameters that allow for 1 mismatch in seed alignment (-N 1), try two set of seeds (-R 2), and set the length of seed substrings to be 16 (-L 16). Mapped small RNA species were quantified to read counts and normalized to reads per kilobase per million reads mapped (RPKM). Differential expression analysis was performed using edgeR (Robinson et al., 2010) from Bioconductor (Gentleman et al., 2004) for all contrasts. The p-values were adjusted for multiple-test correction using False Discovery Rate (FDR) (Benjamini and Hochberg, 1995). All fastq files have been deposited into the exRNA Atlas (http://exrna.org/resources/data/), accession number EXR-SADAS1UJ0CzW-AN as part of Extracellular RNA Communication Consortium data sharing, and is widely available. For all reads mapped to the human miRNAs, isomers of miRNAs (isomiRs) were identified with respect to the canonical miRNA sequence as found in miRBase v21. All isomiRs depicted in this Fig. S4C are length variations – extensions (titled “ext”) or shortening (titled “trimmed”) and sequence variations (“Unmatched”). The 5′ or 3′ refers to the end of the miRNA where the sequence length alteration is observed. These isomiRs are isoforms (both sequence and length variations) based upon the canonical miRNA defined by miRBase v21.
2.5. High Throughput RT-qPCR
RNA was extracted from plasma (331 patients, OMEGA-REMODEL trial) using miRCURY RNA Isolation Kit – Biofluids (Exiqon) and reverse transcribed using miScript II RT Kit (Qiagen, Fredrick, MD, USA). cDNA samples were pre-amplified with miScript Microfluidics PreAMP Kit (Qiagen, Fredrick, MD, USA) and RT-qPCR miRNA profiling analyses performed on the BioMark System (Fluidigm Corp. South San Francisco, CA) using miScript miRNA Assays (Qiagen, Fredrick, MD, USA) and Dynamic Array 96.96 (Fluidigm Corp. South San Francisco, CA). A Ct value of 23 was set as the cut-off point for detectable miRNA species. Measurement of RNAs on the Fluidigm platform has previously been extensively validated against conventional RT-qPCR (Jang et al., 2011). The qPCR raw data has been deposited in GEO (accession number: GSE112690).
2.6. Principal Components Analysis (PCA)
We utilized PCA, an unsupervised learning technique that statistically groups correlated miRNAs together into components, for analysis of both RNAseq data and RT-qPCR validation data. For analysis of differences in miRNA profiles between post-MI patients and healthy controls (RNAseq data), miRNA reads per million (RPM) for all subjects were computed and entered into PCA after they were mean-centered, standardized (mean 0, variance 1), and log-transformed; the first two principal components were plotted for all subjects. For the validation data (RT-qPCR), miRNA values from high-throughput RT-qPCR were mean-centered and standardized, and entered into the PCA. Values that were below-detection limit were treated as “23” in the analysis (the upper limit the assay, reflecting very low plasma RNA expression level). Varimax rotation was used to determine final PC loadings. We included 4 principal components (PCs) based on (1) examination of an eigenvalue scree plot, and (2) inclusion of PCs that explain up to 60% of population variance (61.3% for 4 PCs). PC scores based on loadings and miRNA expression values were included in linear regression models for association with CMR measures (see below).
2.7. Ingenuity Pathway Analysis (IPA)
mRNA targets of the miRNAs from PC1, PC2, and PC3 (validation data) that had a loading >60% were obtained through IPA microRNA Target Filter. Only those mRNAs that were experimentally observed targets of the miRNAs from each PC were chosen to be included in the core pathway analysis form IPA.
2.8. Association between exRNAs and LV Remodeling by CMR
We performed serial quantitation of LV volumes (end-diastole and end-systole), ECV in the non-infarct myocardium as a surrogate measure of interstitial fibrosis, and infarct size by late gadolinium enhancement (Heydari et al., 2016). A 16-segmental map was used in quantitation of ECV and infarct size and segments with late gadolinium enhancement were not included in the measurement of non-infarct ECV. We estimated linear regression models for several cross-sectional (at the time of the blood draw, baseline at 2–4 weeks post-MI) and longitudinal LV phenotypes (changes in LV phenotypes from baseline to 6 months post-MI): (1) LVESVI; (2) LVEF; (3) LV mass index. In addition, we examined the association of baseline non-infarct myocardial fibrosis by ECV with each PC. Each regression model was specified with each of the 3 CMR parameters as the outcome (dependent variable), included all 4 PC scores, as well as adjustment for age, sex, race, presence of diabetes, a binary variable denoting randomization (to omega-3 fatty acid therapy vs. placebo), and a binary variable representing use of any LVRm medication (beta-blockade, angiotensin converting enzyme inhibition, or angiotensin-II receptor blockade). For the exRNAs other than miRNAs (yRNAs, snoRNAs and piRNAs), linear regression was carried out for mean centered and standardized Ct values as a function of changes in LV phenotypes from baseline to 6 months post-MI (LVESVI, LV Mass, LVEF) after adjustment for all the variables used in the PC regression analysis.
2.9. Animal and Cell Models of Ischemia/Reperfusion and Hypoxia
2.9.1. Murine Model of Ischemia/Reperfusion
Adult wild-type C57BL/6 mice were subjected to ischemia reperfusion (I/R) consisting of 30 min left anterior coronary artery (LAD) ligation followed by reperfusion, as previously described (n = 5; Matsui et al., 2002). “Sham” surgeries without ligation were performed as a control (n = 4; Fig. S1). Animals were survived for 24 h, 1 week, or 4 weeks prior to sacrifice and blood and heart tissue was collected.
2.9.2. Adult Murine Cell Isolation and Culture
Cell specific miRNA expression was investigated in cells isolated from healthy adult wildy type mice. Hearts were collected from mice to isolate cardiomyocytes and non-cardiomyocytes (Liu et al., 2015), and primary endothelial cells and fibroblasts were cultured from mouse hearts of as previously described (Ashida et al., 2011). Isolated cardiomyocytes, non-cardiomyocytes, and cultured primary endothelial cells were confirmed by RT-qPCR analysis cell-specific markers including cTnI, CD31, and Col1a1 (Fig. S2).
2.9.3. Cell Model of Hypoxia/Reoxygenation in Neonatal Cells
Neonatal rat ventricular myocytes (NRVMs) were isolated from 1 day postnatal Sprague-Dawley pups (Melman et al., 2015) and subjected to 24 h hypoxia (2% oxygen) followed by 12 h re-oxygenation. Cells were harvested in lysis buffer (miRVANA PARIS kit) and media was collected and sequentially centrifuged at 2000 ×g for 10 min and 3000 ×g for 10 min. All cell experiments were run in duplicate across at least 3 separate NRVM isolations.
2.9.4. RNA Extraction and RT-qPCR for Animal and Cell Models
RNA from cultured or isolated murine cells and mouse tissue samples were isolated using TRIzol following the manufactures' protocol. Plasma samples were “spiked” with 5 pmol/mL exogenous cel-miR-39-3p and RNA was extracted using the miRCURY RNA Isolation kit for Biofluids (Exiqon) according to manufacturer's protocol. Extracellular vesicle (EV) associated RNA was extracted from NRVM culture media using the exoRNEASY kit (Qiagen) and cellular RNA extracted with the miRVANA PARIS kit according to manufacturer's protocol (Ambion). All RNA samples were reverse transcribed by Universal cDNA Synthesis kit (Exiqon) and RT-qPCR performed for specific miRNA species using the LNA-based Exiqon platform.
2.9.5. Statistical Analysis
U6 (mouse tissue and NRVM cells), cel-miR-39-3p (plasma), or hsa-miR-191-5p (isolated murine cells) was used for RT-qPCR normalization; due to a lack of reliable housekeeper genes, culture media EV miRNA was normalized to cell number and media volume input. Statistical analysis was performed on ΔCt values using Graphpad Prism software. Data from the mouse model was analyzed by one-way ANOVA with Tukey post-hoc analysis, and cell data by Student's t-test. Additionally, the RT-qPCR ΔCt values for mouse tissue, plasma, and cell data were exponentiated as 2–1/Ct, scaled, and centered before unsupervised agglomerative hierarchical clustering with the Manhattan distance measure and the Ward method of clustering was performed. The clustering results were plotted as a heatmap using a modified version of heatmap.2 in R.
3. Results
3.1. exRNAs in Post-MI Patient Population
3.1.1. Baseline Characteristics of the Study Population
Baseline characteristics of the study population are presented in Table 1. Blood was collected from patients at time of initial CMR (2–4 weeks post-MI) and 238 underwent post-treatment follow-up CMR 6 months later (Table 2). The vast majority of patients in our analytic cohort had beneficial LVRm and preserved LVEF (mean 56.6 ± 7.9%). Histograms of the percent change in LVESV and LVM (Fig. S3) demonstrate that the majority had little change in these variables and more importantly, very few had adverse LVRm as seen in the right-tail of the distribution in these histograms. From this cohort we selected n = 11 patients with beneficial and n = 11 patients with adverse LVRm for RNAseq analysis, which were well matched for baseline demographics and disease severity (Table S1 for description of study subpopulations for sequencing). In addition, n = 10 patients with adverse electrical remodeling were matched to n = 10 patients without adverse electrical remodeling for RNAseq analysis. However, follow up clinical data revealed that only 2 of the 331 patients in this cohort experienced sudden cardiac death and so differentially expressed RNAs between these patient groups were not analyzed further. However, sequence data is available for the adverse electrical remodeling group (exRNA Atlas accession number: EXR-SADAS1UJ0CzW-AN).
Table 1.
Baseline clinical and biochemical characteristics of individuals in the RT-qPCR cohort (N ≤ 331). Values for continuous covariates expressed as mean ± standard deviation; categorical covariates are expressed as a fraction of the overall cohort.
| Variable | N | Value |
|---|---|---|
| Age (years) | 331 | 58.9 ± 10.1 |
| Female, % | 331 | 64 (19%) |
| White race, % | 331 | 268 (81%) |
| Diabetes, % | 331 | 86 (26%) |
| Current or prior smoking, % | 331 | 166 (50%) |
| Hypertension, % | 331 | 213 (64%) |
| Hyperlipidemia, % | 331 | 234 (71%) |
| Prior myocardial infarction, % | 331 | 196 (59%) |
| Body mass index (kg/m2) | 331 | 29.0 ± 5.4 |
| Systolic blood pressure (mmHg) | 329 | 121 ± 16 |
| Diastolic blood pressure (mmHg) | 329 | 70 ± 10 |
| ST-elevation myocardial infarction, % | 331 | 196 (60%) |
| Aspirin use | 331 | 325 (98%) |
| Beta blocker use | 331 | 303 (92%) |
| Angiotensin converting enzyme inhibitor use | 331 | 244 (74%) |
| Statin use | 331 | 320 (97%) |
| Randomized to omega-3 fatty acid, % | 331 | 162 (49%) |
| Serum total cholesterol, mg/dl | 305 | 138 ± 67 |
| Serum high density lipoprotein, mg/dl | 305 | 37 ± 27 |
| Serum low density lipoprotein, mg/dl | 304 | 80 ± 47 |
| Serum triglycerides, mg/dl | 305 | 121 ± 95 |
Table 2.
Baseline (2–4 week) and final (6 month) CMR imaging characteristics. All morphologic parameters are indexed to body surface area, calculated at the time of CMR. The “Δ” (change) indices are calculated as final (6-month) value minus baseline (4-week) value divided by initial value. Late gadolinium enhancement was measured by the full-width half-maximum technique.
| CMR index | Baseline (N ≤ 331) |
Follow-up (N ≤ 238. 72%) |
||
|---|---|---|---|---|
| N | Mean (or N) | N | Mean (or N) | |
| LVESVI (ml/m2) | 331 | 39.4 ± 16.5 | 238 | 36.8 ± 13.1 |
| LV mass index (g/m2) | 319 | 60.0 ± 14.5 | 226 | 57.5 ± 13.0 |
| LV ejection fraction, % | 331 | 53.9 ± 9.4 | 238 | 56.6 ± 7.9 |
| RV ejection fraction, % | 331 | 53.3 ± 7.1 | 239 | 52.6 ± 7.7 |
| Late gadolinium enhancement, g | 327 | 17.4 ± 17.2 | 244 | 14.0 ± 13.9 |
| Average extracellular volume fraction | 255 | 0.34 ± 0.05 | 192 | 0.35 ± 0.05 |
| ΔLVESVI | – | 238 | −2 ± 19% | |
| ΔLV mass index | – | 219 | −2 ± 17% | |
| ΔLV ejection fraction | – | 238 | 3 ± 12% | |
3.1.2. RNAseq and exRNA Diversity
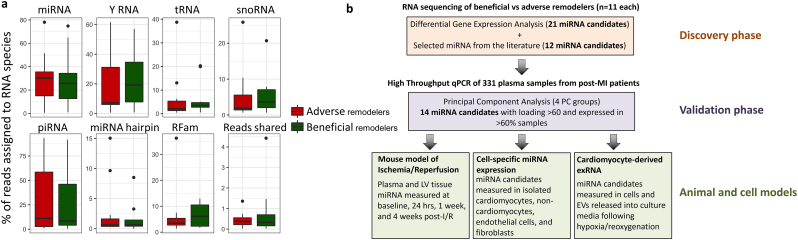
RNAseq was performed on plasma from these 22 patients (n = 11 beneficial and n = 11 adverse LVRm as above). Sequencing quality statistics shown in Table S2. Top RNA species detected by sequencing included miRNA, piRNA, yRNA fragments, tRNA fragments, and snoRNA (Fig. 1a), and the percentage of reads assigned to each species was not significantly different between the two groups. In this study we primarily focused on miRNA, of which 188 different species were expressed in at least 50% of all samples, with 42 different species expressed across all samples. To identify miRNA candidates differentially expressed between beneficial and adverse LVRm, we performed differential expression analysis between the two subgroups. To focus on miRNAs ubiquitously abundant in plasma and suitable as biomarker candidates, we selected miRNA candidates with a fold change in expression of ≥2 (or ≤ 0.5), a nominal p value of ≤0.1, and expression (RPKM > 0) across ≥50% of samples. This yielded 21 miRNA candidates from the discovery cohort for validation (miR-30a-5p, miR-100-5p, miR-146a-5p, miR-146b-5p, miR-744-5p, miR-98-5p, miR-194-5p, miR-29b-3p, miR-29c-3p, miR-378c, miR-378e, miR-381-3p, miR-658, miR-15a-5p, miR-193b-5p, miR-200a-3p, miR-215-5p, miR-3168, miR-4306, miR-4326, and miR-4443). We included 12 additional miRNAs candidates selected from the literature that had previously reported aberrant expression or relevance in cardiovascular disease (miR-21-5p (Li et al., 2016), miR-30d-5p (Melman et al., 2015), miR-1 (Melman et al., 2014), miR-423-5p (Thum, 2014), miR-150-5p (Devaux et al., 2013), miR-223-3p, (Zampetaki et al., 2012), miR-29a-3p (Roncarati et al., 2014), miR-29a-5p (Roncarati et al., 2014), miR-155-5p (Seok et al., 2014), miR-208b-3p (Lv et al., 2014), miR-208a (Melman et al., 2014), miR-133a-5p (Sayed et al., 2009), miR-499a-5p (Zaseeva et al., 2016)). A diagram outlining the analysis approach utilized in this study is shown in Fig. 1b.
Fig. 1.
(a) Box plot representing percentage of reads assigned to different RNA species from sequencing. (b) Flow chart describing experimental outline and analysis approach utilized in this study.
To determine how miRNA expression differed between post-MI patients and healthy controls, this data was compared to RNAseq data from a cohort of 26 healthy controls (age in years: 44 ± 11.8; female: 38%) (Shah et al., 2017). PC analysis on miRNA expression for these three groups shows robust separation between healthy controls and post-MI patients, while the beneficial and adverse LVRm groups shows a more subtle separation (Fig. S4a). It should be noted that the post-MI and healthy control cohorts were sequenced using different preparation kits (NEBNext small RNA library prep set for Illumina and NEXTflex Small RNA Sequencing Kit v2 by Bioo-Scientific, respectively) which could inflate differences between the two groups due to kit biases. However, a recent study published by our group has shown that while RNA expression differences exist between different kits, this is low compared to biological differences observed (Yeri et al., 2018).
Identification and quantification of isomiRs was conducted using an in-house built analysis pipeline. Several isomiR categories were identified, including extensions and shortenings at both the 5′ and 3′ ends of the miRNA and sequence variations (Fig. S4C). However, there were no significant differences in isomiR frequencies between our beneficial and adverse remodeling groups. Due to this fact, and a lack of understanding of the relevance of plasma exRNAs and the absence of universally agreed upon reliable RT-qPCR methodologies for isomiR analysis (Magee et al., 2017), further analysis of specific isomiRs was not conducted in our larger patient cohort.
In addition to miRNA, statistically significant differential expression was detected for 4 annotated piRNAs, 1 snoRNA, and differential expression trending towards significance for 1 yRNA fragment (corresponding to the 5′ fragment of yRNA 1; Fig. S4b). We considered non-miRNA exRNAs as exploratory given their limited and variable annotation and unclear function, and thus focused on miRNA candidates for further analysis. Nonetheless, we did assess the association of these exRNAs with LVRm phenotypes in the validation cohort.
3.1.3. Association of Candidate exRNAs with Baseline and Serial LV Phenotypes by CMR
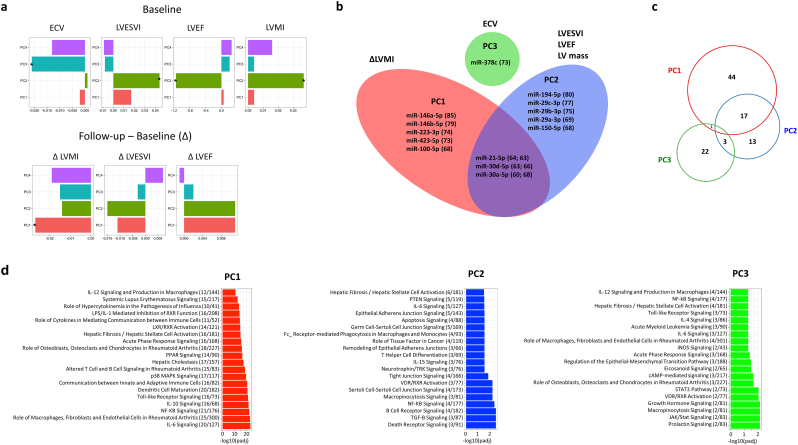
The miRNA candidates selected above were measured in 331 participants of the OMEGA-REMODEL study by high-throughput RT-qPCR (Table S3). Due to the vast majority of these patients exhibiting relatively favorable LVRm, which would result in reduced effect sizes between the miRNAs and serial LV phenotypes, and to eliminate collinearity of the miRNAs, we chose to analyze the RT-qPCR data by PC regression. This allowed us to examine the correlation of PC factors consisting of distinct sets of miRNA correlated with distinct CMR measures and biological processes. PCA demonstrated 4 PCs accounting for 61.3% of total variance. We used PC loading, which indicates the amount of variation explained by a variable, as a measurement of influence by a specific miRNA in a given PC. miRNAs with ≥60% loading in a given PC and with >60% expression across samples (14 total miRNAs) are shown in Fig. 2b and Table S4. In linear models for association of PCs with baseline and follow-up CMR parameters (Table S5), we found statistically significant associations between several PCs and myocardial structure/function (Fig. 2a–b): PC1 vs. change in LV Mass; PC2 vs. LVESVI, LVEF, and LV mass; PC3 vs. ECV.
Fig. 2.
(a) Linear regression models for CMR parameters as a function of RNA PC score, *p < 0.05. (b) miRNA with loading >60 and detectable expression in >60% of samples for PC1-3 (numbers in brackets represent PC loading). (c) Venn diagram showing the overlap in the number of mRNA targets for miRNAs in each PC group. (d) Top 20 significant pathways from Ingenuity Pathway analysis for mRNA targets of miRNAs from PC1 (red), PC2 (blue), and PC3 (green).
Using IPA, we generated predicted functional pathways associated with validated mRNA targets of the miRNAs in each of these PC groups (Fig. 2c–d). Top pathway hits included interleukin signaling, PTEN signaling, and toll-like receptor signaling, with a strong overall theme of inflammatory and fibrotic pathways represented. Furthermore, apoptosis and death receptor signaling were amongst the significant target pathways for PC2, the PC group associated with LVEF. While there was some overlap in mRNA targets between groups, a greater proportion of the mRNA targets identified were unique to each PC group, suggesting that these represent clusters of miRNA with distinct functionality that may contribute to the observed CMR parameter correlations.
Finally, of the other exRNAs assessed in the 331 patients, the yRNA 1 fragment had a significant correlation with change in LV Mass, with a β coefficient of −0.043 and a Benjamini-Hochberg adjusted P value of 0.007. Here, a negative β coefficient means greater increase in LVM over time is associated with higher circulating yRNA 1 fragment concentration.
3.2. Candidate miRNAs in Animal and Cell Models of MI
3.2.1. Temporal Regulation of Candidate miRNA in Animal Models
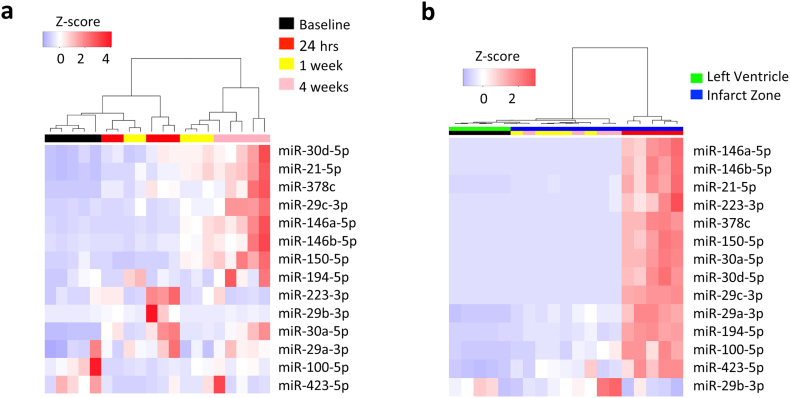
To determine the temporal regulation of exRNAs, candidate miRNAs that were highly loaded on each PC and highly expressed were selected for analysis in the murine I/R model. Time-points chosen were pre-ischemic (baseline), acute to sub-acute phase (24 h and 1 week) and chronic post-infarct phase (4 weeks). Both plasma and tissue had temporally distinct expression of miRNA (Fig. 3; Table 3). At 24 h, miR-223-3p and miR-30a-5p alone increased in plasma (Fig. 3a), while 13 of the 14 miRNA measured were upregulated in tissue (Fig. 3b; miR-100-5p, miR-146a-5p, miR-150-5p, miR-146b-5p, miR-194-5p, miR-223-3p, miR-29a-3p, miR-29c-3p, miR-30a-5p, miR-30d-5p, miR-378c, miR-423-5p, miR-744-5p). Conversely, the majority of plasma changes were seen at the chronic time-point (upregulation of miR-146a-5p, miR-150-5p, miR-146b-5p, miR-21-5p, miR-29c-3p, miR-30a-5p, miR-30d-5p, and miR-378c) with no significant alterations in tissue miRNA expression at this time. Fewer changes were observed at the sub-acute (1 week) time-point: miR-21-5p increased in plasma and miR-194-5p, miR-21-5p, and miR-423-5p were up-regulated in tissue. Notably, miRNAs were not significantly changed in plasma or tissue in sham-treated animals (Fig. S1).
Fig. 3.
Heatmap of selected miRNAs in mouse plasma (a) and tissue (b) at baseline and following ischemia with 24 h, 1 week, and 4 weeks reperfusion (n = 5).
Table 3.
Summary of human, animal, and cell findings from this study for miRNA associated with PC groups.
| RNAseq of beneficial |
RT-qPCR validation |
Mouse model I/R |
In vitro |
Cell type |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vs adverse remodelers (n = 11 patients per group) |
(331 patients) |
LV tissue |
Plasma |
hypox/reox (NRVMs) |
Expression (Adult wild-type mice) |
||||||||
| Fold change | P value | PC1 (ΔLV) | PC2 (LVEF) | PC3 (ECV) | 24 h | 1 week | 4 weeks | 24 h | 1 week | 4 weeks | Media EVs | Predominant cell type | |
| miR-21-5p | 1.96 | 0.13 | X | X | 7.2**** | 8.5* | 16.3**** | 10.3* | FB/EC | ||||
| miR-29a-3p | 0.49 | 0.09 | X | 10.4*** | FB/EC | ||||||||
| miR-29b-3p | 0.23 | 0.09 | X | 4.7* | CM/EC | ||||||||
| miR-29c-3p | 0.24 | 0.05 | X | 374.1**** | 52.8**** | CM | |||||||
| miR-30a-5p | 0.4 | 0.05 | X | X | 162.0**** | 20.2** | 15.8* | CM | |||||
| miR-30d-5p | 0.94 | 0.86 | X | X | 92.3**** | 10.8** | 8.7* | CM | |||||
| miR-100-5p | 0.4 | 0.09 | X | 9.0*** | 6.7* | CM/FB | |||||||
| miR-146a-5p | 2.36 | 0.04 | X | 133.0**** | 18.3*** | Non-CM | |||||||
| miR-146b-5p | 2.36 | 0.08 | X | 377.3**** | 15.2* | Non-CM | |||||||
| miR-150-5p | 0.08 | 0.04 | X | 165.2**** | 8.3 | Non-CM | |||||||
| miR-194-5p | 0.18 | 0.04 | X | 14.8**** | 2.6* | CM/non-CM | |||||||
| miR-223-3p | 0.6 | 0.58 | X | 325.4*** | 3.9** | Non-CM | |||||||
| miR-378c | 0.26 | 0.09 | X | 78.4**** | 31.3** | CM | |||||||
| miR-423-5p | 0.91 | 0.85 | X | 9.7**** | 4.3* | FB/EC | |||||||
| miR-744-5p | 2.11 | 0.06 | X | 169.2*** | EC | ||||||||
In relation to poor remodellers (i.e. positive number = increased in poor remodeller).
Fold change vs baseline, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, Student's t-test.
3.2.2. Cell Type Expression of miRNA and Hypoxia/Reoxygenation Model
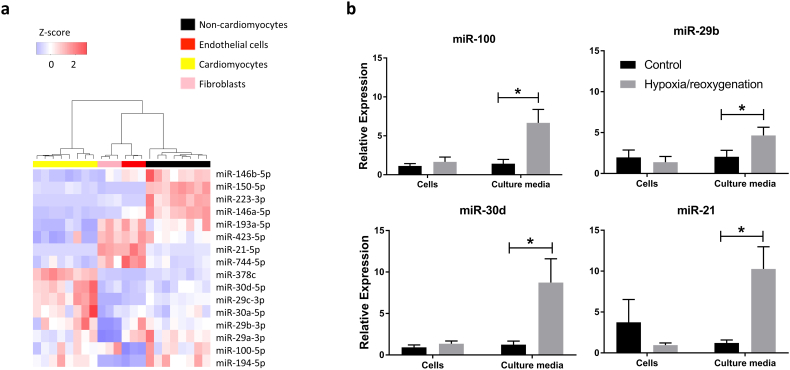
We next assessed cell type sources of these miRNAs in dissociated cells from adult wild type mice and found distinct clustering of cardiomyocytes vs non-cardiomyocytes (Fig. 4a). Specifically, miR-378c, and members of the miR-30 and miR-29 family were highly expressed in cardiomyocytes compared to non-cardiomyocytess; miR-423-5p, miR-21-5p, and miR-744-5p were highly expressed in fibroblasts and endothelial cells; and miR-146b-5p, miR-150-5p, miR-223-3p, and miR146a-5p had the highest expression in the non-cardiomyocyte group (representing all non-cardiomyocyte cardiac cells, including immune cells). Stimulation of expression and release of miRNA by hypoxia/reoxygenation in cardiomyocytes was investigated in NRVMs. miR-100-5p, miR-30d-5p, miR-21-5p, and miR-29b-3p were up-regulated in EVs released in the media (Fig. 4b).
Fig. 4.
(a) Heatmap of candidate miRNAs in isolated murine cells. (b) Candidate EV-miRNAs released by neonatal rat ventricular myocytes into the media in response to hypoxia/reoxygenation (2% oxygen for 24 h hypoxia followed by 12 h reoxygenation). Data represents mean ± SEM. *p < 0.05, Student's t-test.
4. Discussion
In this study, we found that plasma circulating exRNA profiles that include miRNAs and a YRNA fragment, are associated with post-MI LVRm phenotypes. Interestingly, a murine model of MI showed that the majority of our candidate miRNAs were not increased in plasma at acute time-points and that temporal expression of miRNA differed in tissue and plasma (results summarized in Table 3). Furthermore, given the variety of cell types in the myocardium affected during acute ischemic injury, we investigated whether expression of miRNAs was cell-specific, uncovering that cells responsible for “myocardial” miRNA expression are not exclusively cardiomyocytes. Nevertheless, in studies of cardiomyocytes exposed to hypoxia/reoxygenation (a cellular model of reperfused MI), we found several of our candidate miRNAs released into the media and associated with extracellular vesicles, suggesting that cardiomyocytes remain an important source of non-coding RNA expression in response to injury and stress. Collectively, these data add to a burgeoning literature on the post-MI molecular architecture and provide additional information on the complex interplay between different cell types in orchestrating LVRm.
Due to their role in communication across cell types and organs, miRNAs have been a rich source of investigation for novel biomarker and mechanistic pathway discovery in CVD. Specifically, given their dual role as a measurable, circulating biomarker and function in gene silencing in target cells (Creemers et al., 2012, Tijsen et al., 2010), exRNAs have been proposed as mediators of early myocardial remodeling (e.g., fibrosis and hypertrophy) (Gao et al., 2017a), complementary to natriuretic and other peptide-based biomarkers (Lee et al., 2006). In this exploratory study, we show an association between different groups of miRNAs with LVRm phenotypes by CMR (including measures related to myocardial extracellular matrix expansion and hypertrophy) in the sub-acute post-MI phase.
It is important to note that the purpose of this study was not to identify a definitive marker for predicting the development of LVRm in post-MI patients, but to better understand the associations of exRNA signatures with structural phenotypes (as assessed by CMR) in a well-treated patient cohort, and garner insight into the biology driving differential LVRm. The patient group examined here received rapid revascularization and optimal medical therapy resulting in a lack of extreme LVRm phenotypes. Given the lack of sufficient patients with adverse remodeling in this cohort (see Fig. S3), exRNA signatures perform similarly to clinical models (that include age, gender, diabetes, NT-proBNP and LV mass by CMR) and neither have sufficient discrimination (AUC range 0.54–0.59 range). Interestingly, when we measure exRNAs identified in this study in a population with chronic heart failure and a wide range of LVRm by echocardiography, we can improve discrimination for remodeling over clinical models (Shah et al., 2018). Future studies in larger cohorts of post-MI patients with a wider range of LVRm would be needed to further develop these exRNA signatures as clinically relevant biomarkers for prognostic use. Nonetheless, this study, which includes the largest cohort of post-MI patients with detailed CMR phenotyping, demonstrated the association of exRNAs with specific features of LVRm suggesting a possible role for exRNAs in molecular processes implicated in LVRm.
We suggest that these groups of miRNAs may be functionally implicated in cellular pathways relevant to LVRm, including apoptosis signaling, immune cell function and signaling, and fibrotic pathways (demonstrated by IPA). Of interest is the relationship between each of the principal components and the inflammatory signaling pathways. Each PCA is related to NF-kB signaling which ultimately leads to increased IL-1β activity. This pathway is of particular interest because of the recent use of IL-1β pathway antagonist in clinical trials of post-MI patients. The IL-1β receptor antagonist, anakinra, has been shown to reduce adverse post-MI LVRm, and decrease the risk of HF hospitalization in a post-MI population (Abbate et al., 2013). Similarly, there are associations between each of the principal components and IL-6 signaling, which is downstream of IL-1β activity. These findings underscore the potential role of inflammation in post-MI remodeling, and are hypothesis generating in regard to whether specific miRNAs exert their effect on post-MI remodeling through the IL-1β pathway, and whether dynamic changes in these miRNAs in response to anti-inflammatory therapies may serve as novel markers of response.
In this work we used an unbiased approach (RNAseq) on archived post-MI plasma samples to demonstrate a wide array of RNAs in circulation. In addition to miRNA, we identified several other exRNA species (4 piRNAs, and 1 snoRNA) differentially expressed in individuals with adverse vs beneficial LVRm post-MI. Interestingly, a fragment of yRNA 1 was also differentially expressed in the RNAseq cohort (trending towards statistical significance, p = 0.15) and significantly correlated with change in LV mass in the RT-qPCR validation cohort, with higher levels associated with adverse LVRm. A recent paper has described a functional role for yRNA fragments that appears largely related to inflammation; specifically, secretion of a 5′ fragment of yRNA 4 in EVs from cardiosphere-derived cells and its cardioprotective role in a rat model of I/R (Cambier et al., 2017). The present study is the first to describe perturbation of yRNA fragment expression in human CVD. The discovery of such non-coding RNAs highlights the importance of unbiased approaches (e.g., deep RNAseq) for non-coding RNA and biomarker discovery in patient samples.
To further characterize exRNA candidates that were associated with LVRm phenotypes, we utilized a mouse model of I/R to investigate the temporal regulation of miRNA in plasma and tissue. Previous mouse model studies have described a generalized increase in extracellular RNA (not specific to microRNA) at 30 min and 2 h post permanent LAD ligation (Stieger et al., 2017) and an increase in circulating myocardium-associated miRNAs (miR-208a, miR-499, miR-1, and miR-133) following 45 min ischemia and 3 h reperfusion (Chen et al., 2014). While interesting, this likely reflects RNA released in response to myocardial damage similar to the release of troponins in the acute period following MI. The current study aimed to evaluate miRNA across a broader time period that assessed exRNA release in response to cellular changes occurring in the acute, sub-acute, and chronic phases following I/R. We observed distinct clustering of time-points by miRNA expression; baseline and acute (24 h) samples clustered distinctly in tissue, whereas baseline and chronic (4 week) samples showed particularly strong separation in plasma. Eleven of the 15 miRNA measured were significantly up-regulated in tissue at the 24 h time-point with only 3 up-regulated in plasma. During the sub-acute period (1 week) 3 miRNA were up-regulated in tissue and 2 in plasma, with one species (miR-21-5p) up-regulated in both tissue and plasma. Finally, no miRNAs were up-regulated in tissue at the chronic time point but 8 of the miRNA candidates were up-regulated in plasma. Together, this demonstrates a highly complex temporal regulation of post-MI miRNA expression and suggests that when comparing data from different studies, the time-point at which samples were collected should be taken into consideration.
The patient samples used in this study most closely align with the sub-acute time-point (collected 2–4 weeks post-MI), however, it is difficult to draw precise comparisons between the human and mouse studies presented here. We have recently shown that plasma exRNAs altered in human heart failure are concordantly regulated in myocardial tissue and plasma in a murine model of pressure-overload hypertrophy and heart failure (Shah et al., 2018), suggesting the value of animal models in derivation of mechanistic insights about exRNAs. Due to the nature of murine I/R models, there is not significant enough distinction between animals to classify them as “beneficial” or “adverse” remodelers and thus it is difficult to directly compare changes in expression in the mouse model vs the patient cohort (where differential expression was measured in beneficial vs adverse remodelers). Instead, we have compared post-MI miRNA expression levels to baseline expression levels in order to describe the temporal patterning of post-MI miRNA expression.
The lack of correlation between tissue and plasma expression suggests that either a significant proportion of the miRNAs dysregulated post-MI do not originate from cardiac tissue, or that there is a delay in their release. Expression of our candidate miRNAs was cell-type specific and miRNAs from each PC group were expressed across different cell types, highlighting the importance of multiple cell types in the cardiac remodeling processes. Interestingly, miRNA with high expression in the “non-cardiomyocyte” cells included those previously associated with inflammation and inflammatory cells (miR-146a-5p (Croci et al., 2016, Feng et al., 2017), miR-146b-5p (Croci et al., 2016, Hulsmans et al., 2012), and miR-150-5p (Croci et al., 2016)). While there are no good cell culture models for LVRm, we did demonstrate that cardiomyocytes release extracellular vesicle-associated miRNA in response to hypoxia/reoxygenation in vitro, suggesting that stressors can trigger the release of remodeling-associated exRNAs. Overall this data suggests that multiple cell types contribute to the post-MI exRNA milieu and that further studies will be required to determine the exact reasons for the temporally distinct tissue and plasma expression patterns.
The strengths of our study include the use of plasma small non-coding RNAseq in a well-defined population, unbiased initial measurements, the availability of CMR measurements of LVRm, and the use of animal and cellular models to interrogate temporal and cellular expression of RNA. Nevertheless, there are several limitations to our study that merit comment. First, the size of the available discovery cohort limits our ability to comprehensively identify more miRNA candidates. Second, the “archival” nature of the samples utilized for RNAseq may make them susceptible to RNA degradation, limiting RNA extraction yield and our ability to comprehensively map and quantify known RNAs. Indeed, several miRNA species detected from RNAseq data were not expressed across all samples. Nonetheless, while we did identify a panel of miRNA candidates from the discovery cohort that were differentially expressed, we supplemented the panel with literature-curated miRNAs to improve generalizability in the validation. Third, since the start of the study, methodologies in biofluids RNA isolation (Filant et al., 2018) and RNA sequencing platforms (Giraldez et al., 2017) have advanced rapidly in the field to decrease bias and improve the scope of exRNA discovery. Finally, while the PCA-based approach allowed us to take advantage of groups of similar RNAs to limit type 1 error in regression, we recognize that the effect size of the association between each PC and each CMR parameter was modest. However, the general degree of myocardial impairment post-MI (and its change over 6 months) was itself modest in the individuals in this study (Fig. S3), limiting the ability of regression techniques to “bring out” associations between miRNAs and phenotypes. This nonetheless represents the current state of clinical care in our center, and the population in which improved disease risk stratification is necessary.
Future studies utilizing fresh samples with careful phenotyping and an unbiased screening method (next-generation sequencing) will be necessary to validate the findings presented here. This is particularly important for the consideration of any of these identified exRNAs as biomarkers for the development of LVRm in post-MI patients, which will require a much larger patient group. Furthermore, mechanistic studies are required to determine the extent to which these candidates play a functional role in the development of LVRm and to confirm their involvement in inflammatory pathways suggested by IPA.
In conclusion, this study has demonstrated that: 1) exRNA signatures, are associated with CMR-defined LVRm phenotypes; 2) plasma and tissue levels of the candidate remodeling-associated miRNAs are dynamically regulated in a murine model of I/R; and 3) distinct cell types in the heart contribute to these miRNAs, pointing to the complex intercellular interactions in the pathogenesis of LVRm. Overall, this data adds to a growing literature that implicates plasma miRNAs as functional mediators of cardiac remodeling processes and phenotypes.
Funding Sources
RK is funded by the NIH (R01-HL091157). A.R. is funded by the NIH (HL122987, HL135886, TR000901) and the AHA (14CSA20500002, 16SFRN31720000). SD is funded by NIH (UH3-TR000901, HL122547) and AHA (16SFRN31720000). This publication is part of the NIH Extracellular RNA Communication Consortium paper package and was supported by the NIH Common Fund's exRNA Communication Program.
Conflicts of Interest
RVS is a consultant for MyoKardia, Inc., Amgen, Inc., Best Doctors, Inc., and KOLGroups, Inc., none of which participated in this study. SD is a founding member of Dyrnamix which had no role in this study. There are no conflicts for any of the authors related to design, execution or analysis of the experiments presented in this manuscript.
Author Contributions
Conceptualization, KMD, RS, SD, AR; Methodology, KMD, CX, XL, MJH, BH, SA, YEW, KT; Formal Analysis, KMD, YEW, AY; Investigation, KMD, XL, FCG, KT, AD, CX; Resources, RYK, JEF, AR, SD; Data Curation, MJH, BH, SA, YEW, RS; Writing – Original Draft, KMD, RS, AY; Writing – Review & Editing, XL, FCG, MS, KT, AD, CX, MJH, BH, SA, KJ, JEF, YEW, AR, RYK, SD; Visualization, KMD, AY; Supervision, SD, TR, RYK, YEW, JEF; Funding Acquisition, SD, TR, RYK.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.05.013.
Appendix A. Supplementary data
Supplementary tables and figures.
References
- Abbate A., Kontos M.C., Abouzaki N.A., Melchior R.D., Thomas C., Van Tassell B.W., Oddi C., Carbone S., Trankle C.R., Roberts C.S., Mueller G.H., Gambill M.L., Christopher S., Markley R., Vetrovec G.W., Dinarello C.A., Biondi-Zoccai G. Comparative safety of interleukin-1 blockade with anakinra in patients with ST-segment elevation acute myocardial infarction (from the VCU-ART and VCU-ART2 pilot studies) Am. J. Cardiol. 2015;115:288–292. doi: 10.1016/j.amjcard.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Abbate A., Van Tassell B.W., Biondi-Zoccai G., Kontos M.C., Grizzard J.D., Spillman D.W., Oddi C., Roberts C.S., Melchior R.D., Mueller G.H., Abouzaki N.A., Rengel L.R., Varma A., Gambill M.L., Falcao R.A., Voelkel N.F., Dinarello C.A., Vetrovec G.W. Effects of interleukin-1 blockade with anakinra on adverse cardiac remodeling and heart failure after acute myocardial infarction [from the Virginia Commonwealth University-Anakinra remodeling trial (2) (VCU-ART2) pilot study] Am. J. Cardiol. 2013;111:1394–1400. doi: 10.1016/j.amjcard.2013.01.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai T. Post-infarction inflammation and left ventricular remodeling: a double-edged sword. Circ. J. 2013;77:580–587. doi: 10.1253/circj.cj-13-0013. [DOI] [PubMed] [Google Scholar]
- Aryal B., Rotllan N., Fernández-Hernando C. Noncoding RNAs and atherosclerosis. Curr Atheroscler Rep. 2014;16:407. doi: 10.1007/s11883-014-0407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida N., Senbanerjee S., Kodama S., Foo S.Y., Coggins M., Spencer J.A., Zamiri P., Shen D.X., Li L., Sciuto T., Dvorak A., Gerszten R.E., Lin C.P., Karin M., Rosenzweig A. IKK beta regulates essential functions of the vascular endothelium through kinase-dependent and -independent pathways. Nat. Commun. 2011:2. doi: 10.1038/ncomms1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barturen G., Rueda A., Hamberg M., Alganza A., Lebron R., Kotsyfakis M., Shi B.-J., Koppers-Lalic D., Hackenberg M. 2014. sRNAbench: Profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. [Methods in Next Generation Sequencing] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Burchfield J.S., Xie M., Hill J.A. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation. 2013;128:388–400. doi: 10.1161/CIRCULATIONAHA.113.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos K.L., Javaherian A., Bomprezzi R., Ghaffari L., Rhodes S., Courtright A., Tembe W., Kim S., Metpally R., Van Keuren-Jensen K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA. 2013;19:712–722. doi: 10.1261/rna.036863.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier L., De Couto G., Ibrahim A., Echavez A.K., Valle J., Liu W., Kreke M., Smith R.R., Marbán L., Marbán E. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol. Med. 2017;9:337–352. doi: 10.15252/emmm.201606924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chen C., Feng Y., Zou L., Wang L., Chen H.H., Cai J.Y., Xu J.M., Sosnovik D.E., Chao W. Role of extracellular RNA and TLR3-Trif signaling in myocardial ischemia-reperfusion injury. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- Croci S., Zerbini A., Boiardi L., Muratore F., Bisagni A., Nicoli D., Farnetti E., Pazzola G., Cimino L., Moramarco A., Cavazza A., Casali B., Parmeggiani M., Salvarani C. MicroRNA markers of inflammation and remodelling in temporal arteries from patients with giant cell arteritis. Ann. Rheum. Dis. 2016;75:1527–1533. doi: 10.1136/annrheumdis-2015-207846. [DOI] [PubMed] [Google Scholar]
- Danielson K.M., Rubio R., Abderazzaq F., Das S., Wang Y.Y.E. High throughput sequencing of extracellular RNA from human plasma. PLoS One. 2017;12 doi: 10.1371/journal.pone.0164644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y., Vausort M., Mccann G.P., Zangrando J., Kelly D., Razvi N., Zhang L., Ng L.L., Wagner D.R., Squire I.B. MicroRNA-150 a novel marker of left ventricular remodeling after acute myocardial infarction. Circ. Cardiovasc. Genet. 2013;6:290–298. doi: 10.1161/CIRCGENETICS.113.000077. [DOI] [PubMed] [Google Scholar]
- Doughty R.N., Whalley G.A., Walsh H.A., Gamble G.D., Lopez-Sendon J., Sharpe N. Effects of carvedilol on left ventricular remodeling after acute myocardial infarction: the CAPRICORN Echo Substudy. Circulation. 2004;109:201–206. doi: 10.1161/01.CIR.0000108928.25690.94. [DOI] [PubMed] [Google Scholar]
- Ezekowitz J.A., Kaul P., Bakal J.A., Armstrong P.W., Welsh R.C., Mcalister F.A. Declining in-hospital mortality and increasing heart failure incidence in elderly patients with first myocardial infarction. J. Am. Coll. Cardiol. 2009;53:13–20. doi: 10.1016/j.jacc.2008.08.067. [DOI] [PubMed] [Google Scholar]
- Feng B., Chen S.L., Gordon A.D., Chakrabarti S. miR-146a mediates inflammatory changes and fibrosis in the heart in diabetes. J. Mol. Cell. Cardiol. 2017;105:70–76. doi: 10.1016/j.yjmcc.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Filant J., Nejad P., Paul A., Simonson B., Srinivasan S., Zhang X., Balaj L., Das S., Gandhi R., Laurent L.C., Sood A.K. Isolation of extracellular RNA from serum/plasma. Methods Mol. Biol. 2018;1740:43–57. doi: 10.1007/978-1-4939-7652-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Xu W., Wang J., Wang K., Li P. The role and molecular mechanism of non-coding RNAs in pathological cardiac remodeling. Int. J. Mol. Sci. 2017:18. doi: 10.3390/ijms18030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Liu Y., Guo S., Yao R., Wu L., Xiao L., Wang Z., Liu Y., Zhang Y. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell. Physiol. Biochem. 2017;44:1497–1508. doi: 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y.C., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y.H., Zhang J.H. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004:5. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez, M., Spengler, R., Etheridge, A., Godoy, P., Barczak, A., Srinivasan, S., De Hoff, P., Tanriverdi, K., Courtright, A., Lu, S., Khoory, J., Rubio, R., Baxter, D., Driedonks, T., Buermans, H., Nolte-T Hoen, E., Jiang, H., Wang, K., Ghiran, I., Wang, Y., Van Keuren-Jensen, K., Freedman, J., Woodruff, P., Laurent, L., Erle, D., Galas, D. & Tewari, M. 2017. Systematic assessment of next generation sequencing for quantitative small RNA profiling: a multiple protocol study across multiple laboratories bioRxiv preprint, May 17 2017.
- Haaf P., Garg P., Messroghli D.R., Broadbent D.A., Greenwood J.P., Plein S. Cardiac T1 mapping and extracellular volume (ECV) in clinical practice: a comprehensive review. J. Cardiovasc. Magn. Reson. 2016;18:89. doi: 10.1186/s12968-016-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusch G., Libby P., Gersh B., Yellon D., Böhm M., Lopaschuk G., Opie L. Cardiovascular remodelling in coronary artery disease and heart failure. Lancet. 2014;383:1933–1943. doi: 10.1016/S0140-6736(14)60107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari B., Abdullah S., Pottala J.V., Shah R., Abbasi S., Mandry D., Francis S.A., Lumish H., Ghoshhajra B.B., Hoffmann U., Appelbaum E., Feng J.Z.H., Blankstein R., Steigner M., Mcconnell J.P., Harris W., Antman E.M., Jerosch-Herold M., Kwong R.Y. Effect of omega-3 acid ethyl esters on left ventricular remodeling after acute myocardial infarction the OMEGA-REMODEL randomized clinical trial. Circulation. 2016;134:378. doi: 10.1161/CIRCULATIONAHA.115.019949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans M., Van Dooren E., Mathieu C., Holvoet P. Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. Plos One. 2012;7 doi: 10.1371/journal.pone.0032794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovicova J., Hlozankova M., Dvorakova J., Buresova C., Hamanova M., Chalupova L., Cuchnova K., Kappel A., Keller A., Ruzicka V. Microrna-based diagnostics in cardiovascular disease. Atherosclerosis. 2017;263 (e35-e36) [Google Scholar]
- Jang J.S., Simon V.A., Feddersen R.M., Rakhsham F., Schultz D.A., Zschnke M.A., Lingle W.L., Kolbert C.P., Jen J. Quantitative miRNA expression analysis using Fluidigm microfluidics dynamic arrays. BMC Genom. 2011;12:144. doi: 10.1186/1471-2164-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.H. MicroRNA regulation of cardiac conduction and arrhythmias. Transl. Res. 2013;161:381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.S., Wang T.J., Vasan R.S. Screening for ventricular remodeling. Curr. Heart Fail. Rep. 2006;3:5–13. doi: 10.1007/s11897-006-0025-7. [DOI] [PubMed] [Google Scholar]
- Li H.P., Zhang X.R., Wang F., Zhou L., Yin Z.W., Fan J.H., Nie X., Wang P.H., Fu X.D., Chen C., Wang D.W. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by Upregulating mitochondrial translation. Circulation. 2016;134:734–751. doi: 10.1161/CIRCULATIONAHA.116.023926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Cong Y., Gao X., Wang Y., Lin P. Differential expression profiles of long non-coding RNAs as potential biomarkers for the early diagnosis of acute myocardial infarction. Oncotarget. 2017;8:88613–88621. doi: 10.18632/oncotarget.20101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.J., Xiao J.J., Zhu H., Wei X., Platt C., Damilano F., Xiao C.Y., Bezzerides V., Bostrom P., Che L., Zhang C.X., Spiegelman B.M., Rosenzweig A. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv P., Zhou M.X., He J., Meng W.W., Ma X.H., Dong S.L., Meng X.C., Zhao X., Wang X., He F.C. Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int. J. Mol. Sci. 2014;15:5774–5788. doi: 10.3390/ijms15045774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee R., Telonis A.G., Cherlin T., Rigoutsos I., Londin E. Assessment of isomiR discrimination using commercial qPCR methods. Noncoding RNA. 2017;3 doi: 10.3390/ncrna3020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Li L., Wu J.C., cook S.A., Nagoshi T., Picard M.H., Liao R.L., Rosenzweig A. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J. Biol. Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- Melman Y.F., Shah R., Danielson K., Xiao J.J., Simonson B., Barth A., Chakir K., Lewis G.D., Lavender Z., Truong Q.A., Kleber A., Das R., rosenzweig A., Wang Y.Y., Kass D.A., SINGH J.P., Das S. Circulating MicroRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis a translational pilot study. Circulation. 2015;131:2202–2216. doi: 10.1161/CIRCULATIONAHA.114.013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman Y.F., Shah R., Das S. MicroRNAs in heart failure is the picture becoming less miRky? Circ. Heart Fail. 2014;7:203–214. doi: 10.1161/CIRCHEARTFAILURE.113.000266. [DOI] [PubMed] [Google Scholar]
- Migrino R.Q., Young J.B., Ellis S.G., White H.D., Lundergan C.F., Miller D.P., Granger C.B., Ross A.M., Califf R.M., Topol E.J. End-systolic volume index at 90 to 180 minutes into reperfusion therapy for acute myocardial infarction is a strong predictor of early and late mortality. The Global Utilization of Streptokinase and t-PA for Occluded Coronary Arteries (GUSTO)-I Angiographic Investigators. Circulation. 1997;96:116–121. doi: 10.1161/01.cir.96.1.116. [DOI] [PubMed] [Google Scholar]
- Piccoli M.T., Gupta S.K., Viereck J., Foinquinos A., Samolovac S., Kramer F.L., Garg A., Remke J., Zimmer K., Batkai S., Thum T. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- Rajan K.S., Velmurugan G., Pandi G., Ramasamy S. miRNA and piRNA mediated Akt pathway in heart: Antisense expands to survive. Int. J. Biochem. Cell Biol. 2014;55:153–156. doi: 10.1016/j.biocel.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., Mccarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati R., Anselmi C.V., Losi M.A., Papa L., Cavarretta E., Martins P.D., Contaldi C., Jotti G.S., Franzone A., Galastri L., Latronico M.V.G., Imbriaco M., Esposito G., DE Windt L., Betocchi S., Condorelli G. Circulating miR-29a, among other up-regulated MicroRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2014;63:920–927. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- Sayed D., Rane S., Abdellatif M. MicroRNAs challenge the status quo of therapeutic targeting. J. Cardiovasc. Transl. Res. 2009;2:100–107. doi: 10.1007/s12265-008-9052-y. [DOI] [PubMed] [Google Scholar]
- Seok H.Y., Chen J.H., Kataoka M., Huang Z.P., Ding J., Yan J.L., Hu X.Y., Wang D.Z. Loss of MicroRNA-155 protects the heart from pathological cardiac hypertrophy. Circ. Res. 2014;114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Yeri A., Das A., Courtright-Lim A., Ziegler O., Gervino E., Ocel J., Quintero-Pinzon P., Wooster L., Bailey C.S., Tanriverdi K., Beaulieu L.M., Freedman J.E., Ghiran I., Lewis G.D., Van Keuren-Jensen K., Das S. Small RNA-seq during acute maximal exercise reveal RNAs involved in vascular inflammation and cardiometabolic health: brief report. Am. J. Physiol. Heart Circ. Physiol. 2017;313:H1162–h1167. doi: 10.1152/ajpheart.00500.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Ziegler O., Yeri A., Liu X., Murthy V., Rabideau D., Xiao C.Y., Hanspers K., Belcher A., Tackett M., Rosenzweig A., Pico A.R., Januzzi J.L., Das S. MicroRNAs associated with reverse left ventricular remodeling in humans identify pathways of heart failure progression. Circ. Heart Fail. 2018;11 doi: 10.1161/CIRCHEARTFAILURE.117.004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John Sutton M., Linde C., Gold M.R., Abraham W.T., Ghio S., Cerkvenik J., Daubert J.C. Left ventricular architecture, long-term reverse remodeling, and clinical outcome in mild heart failure with cardiac resynchronization: results from the REVERSE trial. JACC Heart Fail. 2017;5:169–178. doi: 10.1016/j.jchf.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Stieger P., Daniel J.M., Tholen C., Dutzmann J., Knopp K., Gunduz D., Aslam M., Kampschulte M., Langheinrich A., Fischer S., Cabrera-Fuentes H., Wang Y., Wollert K.C., Bauersachs J., Braun-Dullaeus R., Preissner K.T., Sedding D.G. Targeting of extracellular RNA reduces edema formation and infarct size and improves survival after myocardial infarction in mice. J. Am. Heart Assoc. 2017:6. doi: 10.1161/JAHA.116.004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov G.A., Filippova J.A., Komissarov A.B., Kuligina E.V., Richter V.A., Semenov D.V. Regulatory role of small nucleolar RNAs in human diseases. Biomed. Res. Int. 2015;2015:10. doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwar S., Squire I.B., Downie P.F., Mccullough A.M., Campton M.C., Davies J.E., Barnett D.B., Ng L.L. Profile of plasma N-terminal proBNP following acute myocardial infarction; correlation with left ventricular systolic dysfunction. Eur. Heart J. 2000;21:1514–1521. doi: 10.1053/euhj.1999.2045. [DOI] [PubMed] [Google Scholar]
- Thum T. Noncoding RNAs and myocardial fibrosis. Nat. Rev. Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- Tijsen A.J., Creemers E.E., Moerland P.D., Pinto Y.M. MicroRNAs as circulating biomarkers for heart failure: questions about MiR-423-5p response. Circ. Res. 2010;106 doi: 10.1161/CIRCRESAHA.110.220616. [DOI] [PubMed] [Google Scholar]
- White H.D., Norris R.M., Brown M.A., Brandt P.W., Whitlock R.M., Wild C.J. Left ventricular end-systolic volume as the major determinant of survival after recovery from myocardial infarction. Circulation. 1987;76:44–51. doi: 10.1161/01.cir.76.1.44. [DOI] [PubMed] [Google Scholar]
- Wijk S.S.V., Van Empel V., Davarzani N., Maeder M.T., Handschin R., Pfisterer M.E., Hans-Peter Brunner-La Rocca H.P., Investigators, T. C Circulating biomarkers of distinct pathophysiological pathways in heart failure with preserved vs. reduced left ventricular ejection fraction. Eur. J. Heart Fail. 2015;17:1006–1014. doi: 10.1002/ejhf.414. [DOI] [PubMed] [Google Scholar]
- Yeri A., Courtright A., Danielson K.M., Hutchins E., Alsop E., Carlson E., Hsieh M., Ziegler O., Das A., Shah R.V., Rozowsky J., Das S., Van Keuren-Jesen K. Evaluation of commercially available small RNA Seq library preparation kits using low input RNA. BMC Genomics. 2018;19:331. doi: 10.1186/s12864-018-4726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A., Willeit P., Tilling L., Drozdov I., Prokopi M., Renard J.M., Mayr A., Weger S., Schett G., Shah A., Boulanger C.M., Willeit J., Chowienczyk P.J., Kiechl S., Mayr M. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J. Am. Coll. Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- Zaseeva A.V., Zhirov I.V., Tereschenko S.N., Scvortcov A.A., Masenko V.P., Kochetov A.G., Gimadiev R.R., Abramov A.A., Lyang O.V. The role of circulating miR-21, miR-34a, miR-423, miR-208a and miR-499a in ischemic and dilated cardiomyopathy. Eur. J. Heart Fail. 2016;18:492. [Google Scholar]
- Zile M.R., Mehurg S.M., Arroyo J.E., Stroud R.E., Desantis S.M., Spinale F.G. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ. Cardiovasc. Genet. 2011;4:614–619. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables and figures.