Abstract
Purpose
Atopic dermatitis (AD) is a common and chronic inflammatory skin disease affecting up to 20% of children and 3% of adults worldwide. Although previous reports including genome-wide association study (GWAS) approaches have identified several risk factors that appear to be associated with AD development, replication studies are lacking. In our current study, we replicated the associations between candidate susceptibility loci and AD.
Methods
A total of 885 Korean subjects (425 AD patients and 460 unaffected controls) were genotyped for 17 single nucleotide polymorphisms (SNPs) from previous GWASs and meta-analyses of AD and from immune-related genes.
Results
Several SNPs showed significant associations with AD in the case-control analysis (minimum P=0.005 at rs17389644), suggesting that these polymorphisms may be related to this disease. In addition, several SNPs showed significant signals (minimum P=0.004 at rs6473227) in severe AD compared to unaffected controls. In additional linear regression analysis, a few genotypes appeared to have potential effects on the SCORing AD (SCORAD) values (minimum P=0.003 at rs13361382 on TMEM232) and immunoglobulin E (IgE) levels (minimum P<0.0001 at rs4713555 near HLA-DRB1 and HLA-DQA1) in AD patients.
Conclusions
Our replication and extended study provide additional supporting information on the genetic associations (especially, variants in TMEM232 and nearby to IL21 and HLA-DRB1/HLA-DQA1) related to AD, its clinical severity and IgE involvement.
Keywords: Atopic dermatitis; single nucleotide polymorphism, severity
INTRODUCTION
Atopic dermatitis (AD) is a common, chronic and relapsing inflammatory skin disease characterized by itchiness, pruritis, erythema, scaling and papulovesicles.1 AD is the most common skin disease worldwide, affecting up to 30% of children and 3% of adults.2 A number of familial studies and twin studies have also demonstrated that AD is a highly heritable disease.3
AD belongs to a group of atopic diseases with common characteristics including allergen sensitization, epithelial barrier abnormalities and Type 2 immune responses.4 It is widely assumed that atopy is related to AD in children because immunoglobulin E (IgE) plays a pivotal role in its pathogenesis. Hence, measurements of the total IgE levels have been used to evaluate allergic subjects in clinical practice and to determine the risk factors for severe AD.5
It is now known that AD is a complex disease affected by interactions among multiple genetic factors and environmental components. Several genome-wide association studies (GWASs) on AD in European, Chinese and Japanese populations have identified numerous potential susceptibility loci.6,7,8,9 The first meta-analysis of AD in subjects of European descent identified 3 additional risk loci (rs479844 at 11q13.1, rs2164983 at 19p13.2, rs2897442 on KIF3A at 5q31).10 More recently, a multi-ancestry meta-analysis in the largest number of samples studied to date from European, African, Japanese and Latino populations identified 31 susceptibility loci, including 10 new risk loci, for AD.11 Studies of immune-related candidate genes (such as IL23R, IL5RA, and IL2) have also reported the associations between genetic variants and immune-mediated diseases including atopic eczema.12,13,14 For instance, a promoter single nucleotide polymorphism (SNP; rs2069762) of IL2 was found to be significantly associated with allergic disease through the mediation of the type 1 T helper (Th1)/type 2 T helper (Th2) cells balance.14 However, although they are important for substantiating previously identified GWAS and candidate genes, replication studies in different AD cohorts are lacking, and therefore the need for a comprehensive etiology of AD still remains.
The aim of our present study was to investigate whether the genetic variants previously identified by genome-wide and candidate genes studies would be replicated in our own AD cohort and to perform an extended analysis of the association of these variants with the clinical severity and IgE in AD.
MATERIALS AND METHODS
Subjects
AD subjects (n=425) were recruited solely from our tertiary referral hospital, having been examined at the Childhood Asthma Atopy Center of Asan Medical Center in Seoul, Korea and diagnosed according to the criteria of Hanifin and Rajka.15 The severity of AD was assessed in these cases using the SCOring AD (SCORAD) classification as follows: mild <15, 15≤ moderate <40, severe ≥40.16 Patients with moderate to severe AD were included in the AD group. A control group (n=460) was also recruited from 9 primary schools and 16 kindergartens in the Seoul area. Subjects in the control group had no history of AD, food allergy, allergic rhinitis, asthma or any parental AD history, and all gave a negative skin prick test result.
The total serum IgE levels in the peripheral blood were measured in all subjects using a fluorescent enzyme immunoassay (ImmunoCAP system; Phadia AB, Uppsala, Sweden). Subjects were also tested for their sensitivity to the following 16 allergens: Dermatophagoides pteronyssinus, Dermatophagoides farinae, dog epithelium, cat epithelium, cockroach, grass, mixed tree pollen 1 and 2, Alternaria, Aspergillus, ragweed, mugwort, milk, egg white, peanut and soybean. A positive skin reaction was defined as a wheal size ≥3 mm after subtraction of a negative control. The Korean version of the International Study of Asthma and Allergies in Childhood (ISAAC) questionnaire was used.17 Approval to conduct the study was obtained from the human ethics committees of Asan Medical Center and from the principals of the schools attended by the children (Institutional Review Board No. 2015-1031). Written informed consent was obtained from the parents of all children who participated.
SNP genotyping
Seventeen SNPs with a previously reported AD association were selected from a panel of immune-related genes, and from the results of previous GWASs and meta-analysis. More detailed information on each SNP was shown in Supplementary Table 1. DNA was isolated from the peripheral blood of our 885 study participants (425 AD patients and 460 unaffected controls) using the WizPrepTM gDNA Mini Kit (Wizbiosolutions, Seongnam, Korea). Genotyping of these blood samples was performed using the high-throughput Fluidigm EP1 system (Fluidigm, South San Francisco, CA, USA) with a Fluidigm SNP Type™ assay platform. According to the manufacturer's instructions, a specific target amplification reaction was used to increase the copy number of targeted genomic regions using Qiagen 2X Multiplex PCR Master Mix (Qiagen, Hilden, Germany). Following the amplification reactions on the dynamic array Integrated Fluidic Circuits (IFCs; Fluidigm), fluorescence intensities were measured with the EP1 reader. Genotypes were determined using the Fluidigm SNP Genotyping Analysis program. Visual inspections were performed for all SNP determinations, and call rate over 95% was applied to further SNP analysis.
Statistical analysis
Logistic and linear regression analyses using Statistical Analysis System software (SAS v9.4; SAS Inc., Cary, NC, USA) were performed to determine associations. For the multivariate analysis, age and sex were adjusted as covariates. A P value of <0.05 was considered statistically significant. In silico analyses including the Signal Scan program (http://www-bimas.cit.nih.gov/molbio/signal/) and the SNP functional predictions program (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process =home) were performed to investigate the potential functions of any SNPs found to be significantly associated with AD. The SNPSpD program (http://gump.qimr.edu.au/general/daleN/SNPSpD) was used to correct for multiple testing errors.
RESULTS
Characteristics of the study subjects
A cohort comprising 425 AD patients and 460 unaffected controls was recruited. The mean age was lower in the AD patients than in the control subjects (6.95±3.58 vs 11.84±2.09, P<0.01). The total IgE levels were significantly higher in the AD group (732.3±1,483.0 IU/mL vs 119.8±224.0 IU/mL, P<0.01). The detailed characteristics of these subjects are summarized in Table 1.
Table 1. Clinical profiles of the study subjects.
| Variables | AD | Unaffected control | P value |
|---|---|---|---|
| No. of subjects | 425 | 460 | - |
| Age (yr) | 6.95 ± 3.58 | 11.84 ± 2.09 | < 0.01 |
| Sex (male, %) | 54.8 | 41.7 | - |
| SCORAD | 69.66 ± 18.28 | NA | - |
| Total IgE (IU/mL) | 732.3 ± 1,483.0 | 119.8 ± 224.0 | < 0.01 |
| Severity of AD | |||
| Severe (≥ 40) | 161 | NA | - |
| Moderate (≥ 15, < 40) | 264 | NA | - |
AD, atopic dermatitis; SCORAD, SCORing AD; IgE, immunoglobulin E; NA, not applicable.
Association analysis with AD development and severity
To conduct a replication study of previously described susceptible loci in AD, we selected a total of 17 SNPs (10 located at gene regions and 7 at nearby genes) that had been identified by previous GWASs and meta-analyses of AD and also from a panel of immune-related genes (Supplementary Table 2). These SNPs were then successfully genotyped in our AD and control populations and were found to be common variants in these subjects with a minor allele frequency (MAF) above 0.05.
A case-control analysis adjusted for age and sex as covariates was performed. The results replicated significant associations between 8 SNPs (rs6682925, rs7622183, rs17454584, rs17389644, rs11741861, rs6473227, rs2212434 and rs2143950) and AD development (minimum P=0.005 at rs17389644 under a dominant model, Table 2). In addition, most of these significant SNPs found in the case-control analysis and an additional polymorphism, rs11741861, that is co-located at IRGM and ZNF300 showed significant signals in the association analysis with AD severity (minimum P=0.004 at rs6473227 under a dominant model, Table 3).
Table 2. Association analysis between genetic polymorphisms and AD.
| SNP | (Nearby) gene, full name | Minor allele | MAF | Genetic model | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additive | Dominant | Recessive | |||||||||||
| AD cases (n=425) | Controls (n=460) | OR (95% CI) | P | Pcor. | OR (95% CI) | P | Pcor. | OR (95% CI) | P | Pcor. | |||
| rs6682925 | IL23R, Interleukin 23 receptor | C | 0.424 | 0.403 | 1.15 (0.89–1.48) | 0.29 | - | 0.96 (0.66–1.41) | 0.84 | - | 1.69 (1.06–2.70) | 0.03 | NS |
| rs7622183 | IL5RA, Interleukin 5 receptor subunit alpha | T | 0.464 | 0.517 | 0.74 (0.57–0.96) | 0.02 | NS | 0.66 (0.44–0.99) | 0.05 | - | 0.69 (0.45–1.06) | 0.09 | - |
| rs17454584 | (IL2, Interleukin 2) | G | 0.127 | 0.101 | 1.69 (1.14–2.50) | 0.009 | NS | 1.78 (1.16–2.72) | 0.008 | NS | 1.76 (0.34–9.06) | 0.50 | - |
| rs17389644 | (IL21, Interleukin 21) | A | 0.127 | 0.103 | 1.75 (1.17–2.61) | 0.006 | NS | 1.86 (1.20–2.87) | 0.005 | NS | 1.79 (0.34–9.35) | 0.49 | - |
| rs10214237 | (IL7R, Interleukin 7 receptor) | C | 0.173 | 0.179 | 1.00 (0.71–1.41) | 0.99 | - | 1.00 (0.76–1.31) | 0.99 | - | 1.00 (1.00–1.01) | 0.97 | - |
| rs13361382 | TMEM232, Transmembrane protein 232 | A | 0.125 | 0.106 | 1.02 (0.67–1.55) | 0.92 | - | 1.05 (0.67–1.64) | 0.85 | - | 0.59 (0.11–3.21) | 0.54 | - |
| rs6871536 | RAD50, RAD50 double strand break repair protein/TH2LCRR, T helper type 2 locus control region associated RNA | C | 0.211 | 0.185 | 1.23 (0.89–1.71) | 0.21 | - | 1.35 (0.92–1.98) | 0.12 | - | 0.91 (0.34–2.40) | 0.84 | - |
| rs2569190 | CD14, CD14 molecule/TMCO6, Transmembrane and coiled-coil domains 6 | G | 0.364 | 0.384 | 0.90 (0.68–1.18) | 0.45 | - | 0.83 (0.57–1.19) | 0.31 | - | 1.00 (0.57–1.74) | 0.99 | - |
| rs11741861 | IRGM, Immunity related GTPase M/ZNF300, Zinc finger protein 300 | G | 0.376 | 0.365 | 1.26 (0.96–1.64) | 0.09 | - | 1.19 (0.82–1.72) | 0.36 | - | 1.72 (1.02–2.90) | 0.04 | NS |
| rs4713555 | (HLA-DRB1, Major histocompatibility complex class II DR beta 1/HLA-DQA1, Major histocompatibility complex class II DQ alpha 1) | T | 0.326 | 0.281 | 1.32 (1.00–1.76) | 0.05 | - | 1.34 (0.93–1.92) | 0.12 | - | 1.73 (0.91–3.28) | 0.09 | - |
| rs2275913 | IL17A, Interleukin 17A | A | 0.443 | 0.431 | 0.97 (0.75–1.27) | 0.83 | - | 0.84 (0.57–1.25) | 0.39 | - | 1.17 (0.73–1.87) | 0.51 | - |
| rs9357733 | EFHC1, EF-hand domain containing 1 | G | 0.309 | 0.338 | 0.95 (0.73–1.25) | 0.72 | - | 0.91 (0.63–1.32) | 0.62 | - | 1.00 (0.57–1.75) | 0.99 | - |
| rs4271002 | NAT2, N-acethyltransferase 2 | C | 0.221 | 0.223 | 0.98 (0.72–1.34) | 0.91 | - | 1.03 (0.71–1.48) | 0.89 | - | 0.75 (0.31–1.82) | 0.53 | - |
| rs6473227 | (ZBTB10, Zinc finger and BTB domain containing 10) | A | 0.358 | 0.427 | 0.72 (0.56–0.93) | 0.01 | NS | 0.74 (0.51–1.07) | 0.10 | - | 0.50 (0.30–0.83) | 0.008 | |
| rs4246905 | TNFSF15, TNF superfamily member 15 | T | 0.329 | 0.330 | 0.94 (0.72–1.23) | 0.66 | - | 0.96 (0.67–1.38) | 0.82 | - | 0.84 (0.47–1.50) | 0.56 | - |
| rs2212434 | (EMSY, EMSY BRCA2 interacting transcriptional repressor) | C | 0.474 | 0.516 | 0.79 (0.61–1.01) | 0.06 | - | 0.81 (0.53–1.22) | 0.32 | - | 0.64 (0.42–0.98) | 0.04 | NS |
| rs2143950 | (PPP2R3C, Protein phosphatase 2 regulatory | T | 0.403 | 0.353 | 1.30 (1.00–1.71) | 0.05 | - | 1.51 (1.04–2.19) | 0.03 | NS | 1.21 (0.71–2.06) | 0.49 | - |
P value of logistic regression analysis by adjusting age and sex as covariates. Bold values indicate the statistical significance of P<0.05. Pcor. value corrected for multiple testing (effective number of correction=15.98) using the SNPSpD program.
AD, atopic dermatitis; SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; NS, not significant.
Table 3. Associations between genetic polymorphisms and AD severity.
| SNP | (Nearby) gene | Minor allele | MAF | AD severity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (n = 460) | AD cases | Control vs Moderate | Control vs Severe | Moderate vs Severe | |||||||
| Moderate (n = 264) | Severe (n = 161) | P | Pcor. | P | Pcor. | P | Pcor. | ||||
| rs6682925 | IL23R | C | 0.403 | 0.407 | 0.453 | 0.33 | - | 0.19 | - | 0.21 | - |
| rs7622183 | IL5RA | T | 0.517 | 0.471 | 0.451 | 0.05 | - | 0.15 | - | 0.66 | - |
| rs17454584 | (IL2) | G | 0.101 | 0.128 | 0.124 | 0.02 | NS | 0.07 | - | 0.58 | - |
| rs17389644 | (IL21) | A | 0.103 | 0.126 | 0.129 | 0.02 | NS | 0.03 | NS | 0.81 | - |
| rs10214237 | (IL7R) | C | 0.179 | 0.183 | 0.156 | 0.98 | - | 0.99 | - | 0.26 | - |
| rs13361382 | TMEM232 | A | 0.106 | 0.118 | 0.137 | 0.97 | - | 0.48 | - | 0.23 | - |
| rs6871536 | RAD50/TH2LCRR | C | 0.185 | 0.198 | 0.232 | 0.39 | - | 0.26 | - | 0.34 | - |
| rs2569190 | CD14/TMCO6 | G | 0.384 | 0.374 | 0.346 | 0.90 | - | 0.38 | - | 0.52 | - |
| rs11741861 | IRGM/ZNF300 | G | 0.365 | 0.362 | 0.399 | 0.60 | - | 0.03 | 0.45 | - | |
| rs4713555 | (HLA-DRB1/HLA-DQA1) | T | 0.281 | 0.322 | 0.331 | 0.12 | - | 0.18 | - | 0.90 | - |
| rs2275913 | IL17A | A | 0.431 | 0.447 | 0.436 | 0.98 | - | 0.83 | - | 0.87 | - |
| rs9357733 | EFHC1 | G | 0.338 | 0.322 | 0.287 | 0.50 | - | 0.52 | - | 0.32 | - |
| rs4271002 | NAT2 | C | 0.223 | 0.232 | 0.203 | 0.68 | - | 0.41 | - | 0.28 | - |
| rs6473227 | (ZBTB10) | A | 0.427 | 0.367 | 0.343 | 0.14 | - | 0.004 | NS | 0.58 | - |
| rs4246905 | TNFSF15 | T | 0.330 | 0.324 | 0.338 | 0.60 | - | 0.77 | - | 0.54 | - |
| rs2212434 | (EMSY) | C | 0.516 | 0.500 | 0.430 | 0.29 | - | 0.03 | NS | 0.06 | - |
| rs2143950 | (PPP2R3C) | T | 0.353 | 0.405 | 0.399 | 0.01 | NS | 0.47 | - | 0.86 | - |
P value of logistic analysis under additive model by adjusting age and sex as covariates. Bold values indicate the statistical significance of P<0.05. Pcor. value corrected for multiple testing (effective number of correction=15.98) using the SNPSpD program.
AD, atopic dermatitis; SNP, single nucleotide polymorphism; MAF, minor allele frequency; NS, not significant.
Association analysis with SCORAD and total IgE
Additional linear regression analysis using SCORAD and total IgE, as important indices of AD assessment, was performed. In SCORAD association analysis in AD patients (n=189), 4 SNPs showed significant signals (rs13361382 on TMEM232, rs6871536 co-located at RAD50 and TH2LCRR, rs11741861 co-located at IRGM and ZNF300, rs2275913 on IL17A; minimum P=0.003 at rs13361382 under additive model, Table 4) even after correction for multiple testing (Pcor=0.04). Analysis using the total IgE level revealed that rs17454584 near IL2, rs17389644 near IL21 and rs4713555 near HLA-DRB1 and HLA-DQA1 had a potential association in AD patients (minimum P<0.0001 at rs4713555 under a recessive model, Pcor<0.001, Table 4).
Table 4. Associations of genetic polymorphisms with SCORAD and total IgE.
| SNP | SCORAD | Total IgE | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Score | Genetic model | Score | Genetic model | |||||||||||||||
| Additive | Dominant | Recessive | Additive | Dominant | Recessive | |||||||||||||
| M/M (No.) | M/m (No.) | m/m (No.) | P | Pcor. | P | Pcor. | P | Pcor. | M/M (No.) | M/m (No.) | m/m (No.) | P | Pcor. | P | Pcor. | P | Pcor. | |
| rs6682925 | 71.03 ± 9.02 (62) | 69.77 ± 7.66 (83) | 67.00 ± 10.34 (42) | 0.21 | - | 0.34 | - | 0.26 | - | 715.1 ± 60.7 (139) | 612.9 ± 45.1 (185) | 624.7 ± 69.8 (80) | 0.21 | - | 0.16 | - | 0.55 | - |
| rs7622183 | 67.78 ± 9.14 (55) | 68.55 ± 7.39 (86) | 73.79 ± 10.88 (46) | 0.14 | - | 0.42 | - | 0.10 | - | 707.9 ± 64.6 (120) | 620.8 ± 44.3 (196) | 665.4 ± 70.5 (89) | 0.52 | - | 0.87 | - | 0.19 | - |
| rs17454584 | 68.77 ± 5.88 (137) | 71.99 ± 9.98 (52) | - (0) | 0.21 | - | 0.21 | - | - | - | 610.6 ± 34.6 (312) | 702.5 ± 71.7 (96) | 2,439.5 ± 1,219.8 (4) | 0.28 | - | 0.55 | - | 0.01 | NS |
| rs17389644 | 68.77 ± 5.88 (137) | 71.67 ± 10.04 (51) | - (0) | 0.26 | - | 0.26 | - | - | - | 608.1 ± 34.7 (308) | 695.0 ± 70.9 (96) | 2,439.5 ± 1,219.8 (4) | 0.30 | - | 0.58 | - | 0.01 | NS |
| rs10214237 | 70.36 ± 6.17 (130) | 67.66 ± 9.29 (53) | 87.00 ± 50.23 (3) | 0.91 | - | 0.56 | - | 0.09 | - | 610.9 ± 37.0 (273) | 759.7 ± 69.4 (120) | 332.6 ± 105.2 (10) | 0.94 | - | 0.69 | - | 0.34 | - |
| rs13361382 | 67.81 ± 5.71 (141) | 76.52 ± 12.10 (40) | 90.40 (1) | 0.003 | 0.04 | 0.004 | NS | 0.22 | - | 565.3 ± 32.9 (295) | 769.8 ± 81.1 (90) | 821.5 ± 410.8 (4) | 0.13 | - | 0.13 | - | 0.64 | - |
| rs6871536 | 69.20 ± 6.45 (115) | 68.77 ± 8.88 (60) | 82.81 ± 26.19 (10) | 0.21 | - | 0.62 | - | 0.03 | NS | 601.2 ± 38.3 (247) | 718.6 ± 63.0 (130) | 532.0 ± 125.4 (18) | 0.39 | - | 0.22 | - | 0.63 | - |
| rs2569190 | 69.85 ± 7.96 (77) | 66.86 ± 7.43 (81) | 77.01 ± 15.10 (26) | 0.35 | - | 0.79 | - | 0.02 | - | 568.0 ± 44.5 (163) | 792.4 ± 58.1 (186) | 427.6 ± 58.2 (54) | 0.75 | - | 0.57 | - | 0.14 | - |
| rs11741861 | 73.50 ± 8.07 (83) | 65.63 ± 7.29 (81) | 69.97 ± 13.99 (25) | 0.06 | - | 0.009 | NS | 0.96 | - | 552.7 ± 43.2 (164) | 736.8 ± 54.3 (184) | 654.9 ± 82.5 (63) | 0.98 | - | 0.68 | - | 0.53 | - |
| rs4713555 | 70.49 ± 8.03 (77) | 69.66 ± 7.51 (86) | 68.51 ± 14.29 (23) | 0.61 | - | 0.66 | - | 0.70 | - | 679.7 ± 50.4 (182) | 532.6 ± 39.6 (181) | 1,081.6 ± 168.9 (41) | 0.08 | - | 0.95 | - | <0.0001 | <0.001 |
| rs2275913 | 64.62 ± 8.79 (54) | 72.71 ± 7.71 (89) | 71.47 ± 10.90 (43) | 0.06 | - | 0.01 | NS | 0.66 | - | 569.9 ± 51.0 (125) | 700.6 ± 50.4 (193) | 600.9 ± 67.2 (80) | 0.81 | - | 0.91 | - | 0.59 | - |
| rs9357733 | 69.87 ± 6.99 (100) | 71.53 ± 8.67 (68) | 63.57 ± 14.58 (19) | 0.47 | - | 0.99 | - | 0.11 | - | 615.4 ± 43.6 (199) | 742.2 ± 59.4 (156) | 447.1 ± 65.9 (46) | 0.50 | - | 0.91 | - | 0.20 | - |
| rs4271002 | 68.84 ± 6.62 (108) | 70.78 ± 8.28 (73) | 70.70 ± 26.72 (7) | 0.43 | - | 0.40 | - | 0.87 | - | 683.7 ± 44.1 (240) | 604.1 ± 48.8 (153) | 482.8 ± 133.9 (13) | 0.22 | - | 0.23 | - | 0.59 | - |
| rs6473227 | 70.60 ± 8.38 (71) | 68.74 ± 7.02 (96) | 70.59 ± 15.40 (21) | 0.74 | - | 0.59 | - | 0.89 | - | 643.2 ± 49.2 (171) | 701.5 ± 51.3 (187) | 503.5 ± 69.8 (52) | 0.98 | - | 0.49 | - | 0.33 | - |
| rs4246905 | 69.24 ± 7.51 (85) | 69.53 ± 7.59 (84) | 74.53 ± 17.57 (18) | 0.35 | - | 0.59 | - | 0.24 | - | 704.1 ± 52.2 (182) | 620.3 ± 46.4 (179) | 420.9 ± 64.2 (43) | 0.40 | - | 0.78 | - | 0.18 | - |
| rs2212434 | 74.00 ± 11.03 (45) | 67.88 ± 6.93 (96) | 69.18 ± 10.55 (43) | 0.19 | - | 0.07 | - | 0.77 | - | 742.2 ± 72.8 (104) | 617.4 ± 42.8 (208) | 590.8 ± 63.7 (86) | 0.47 | - | 0.64 | - | 0.48 | - |
| rs2143950 | 70.24 ± 8.71 (65) | 68.87 ± 7.22 (91) | 70.21 ± 12.41 (30) | 0.87 | - | 0.73 | - | 0.89 | - | 599.2 ± 50.1 (143) | 647.1 ± 45.9 (199) | 812.5 ± 102.4 (63) | 0.22 | - | 0.38 | - | 0.24 | - |
P value of linear regression analysis under additive model by adjusting age and sex as covariates. Pcor. value corrected for multiple testing (effective number of correction=15.98) using the SNPSpD program. M/M, M/m, and m/m indicate the homozygote of the major allele, heterozygote, and homozygote of the minor allele, respectively. Bold values indicate the statistical significance of P<0.05.
AD, atopic dermatitis; SCORAD, SCORing AD; IgE, immunoglobulin E; NS, not significant; n, number of subjects.
Meta-analysis and in silico analyses of significant SNPs
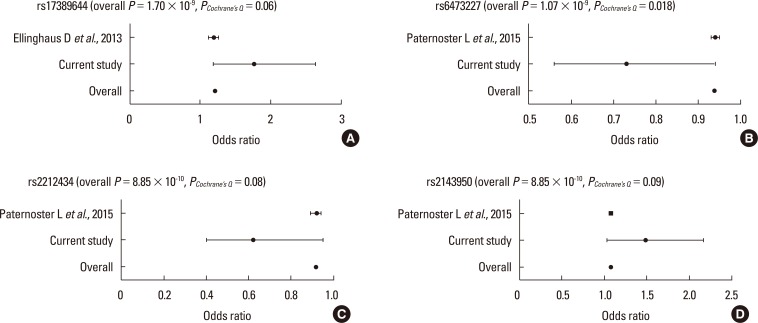
Based on the SNPs identified from previous studies of AD, a meta-analysis was performed. Among the variants that were replicated with significance in our analysis, 4 SNPs (rs17389644, rs6473227, rs2212434 and rs2143950) appeared to have positive associations with AD (minimum overall P=8.85×10−10 for rs2212434 and rs2143950, Figure). However, among the positive SNPs, only rs6473227 still retained positive significance in the Cochran's Q test established by the presence of heterogeneity (PCochrane's Q=0.018).
Figure. Result of meta-analysis. Plots of the replicated SNPs of (A) rs17389644, (B) rs6473227, (C) rs2212434 and (D) rs2143950 with significance in this study in relation to the published studies on AD are calculated using the software package PLINK. The association of each SNP with AD is evaluated through the fixed-effect meta-analysis P value. Additional significance of statistical heterogeneity measured using the χ2-based Cochran's Q test is considered (PCochrane's Q<0.05). Bold values indicate the statistical significance of P<0.05. AD, atopic dermatitis; SNP, single nucleotide polymorphism.
We then investigated the potential functions of the significant SNPs using in silico analyses. Intriguingly, the minor C allele of rs6682925T>C in the regulatory upstream region was predicted to be a putative binding site for the GATA-1 and NF-E transcription factors, but not the sequence including the major T allele of this SNP (Supplementary Table 3). In the analysis to predict the functions of these SNPs, rs6682925, rs7622183 and rs6473227 were estimated to be potential exonic splicing enhancer (ESE) sites that were dependent on the major and minor alleles of each SNP (Supplementary Table 3).
DISCUSSION
AD is a complex disease caused by a combination of multiple genetic and interacting environmental factors. The identification of the genetic factors that contribute to AD is therefore important developing of new therapeutic and prevention strategies for this condition. Although GWASs and meta-analyses have been performed in a number of studies and identified many susceptibility loci for AD,6,7,8,9,10,11 replication studies for these findings are still lacking and reliable markers for AD thus remain to be identified. In our current replication and additional analysis, several previously described genetic variants again showed significant associations with AD, with the top signals found at rs13361382 in TMEM232, rs17389644 near IL21, and rs4713555 near HLA-DRB1/HLA-DQA1 for SCORAD, AD development and the total IgE level, respectively. These results suggest that these genetic variants may contribute to a predisposition for AD.
A recent multi-ancestry meta-analysis of the largest number of samples yet analyzed (21,399 AD cases and 95,464 controls) identified 31 susceptibility loci including 10 novel loci related to innate immune signaling and T cell function.11 Variants of immune system genes (such as IL23R, IL5RA, and IL2) have also been reported as candidates for immune-mediated disorders.12,13,14 However, many previous studies have not fully replicated the associations of SNPs with AD described by previous reports, including variants identified by GWASs or meta-analyses for reasons such as small sample sizes, differences in study designs and an inappropriate reliance on standard significance thresholds.18,19 Hence, replication studies are needed to provide more accurate estimates of the association of genetic variations with the development of diseases and related phenotypes. In our current study, the rs6473227 and rs2143950 SNPs, which were previously highlighted in a meta-analysis,11 were found to have a significant association with AD and its severity, respectively, suggesting that they could serve as genetic markers for AD.
In our present replication study in an AD and control population, 8 previously identified SNPs again showed significant associations with AD in our case-control analysis (minimum P=0.005 at rs17389644 under dominant model, Table 2). However, several known SNPs (rs6682925, rs7622183, rs11741861, rs2212434, and rs2143950) showed only nominal association with AD. On the other hand, despite higher MAFs of a few SNPs (rs17454584, rs17389644, and rs2143950) in both moderate and severe AD cases than those of controls, the significance of these SNPs was not increased depending on AD severity. This may be due to the insufficient sample size (in particular, the low number of severe AD cases) and/or involvement of other regulators; therefore, further replication and studies are needed.
The rs17389644 variant near the IL21 locus showing the most significant signal among our AD subjects and notably was previously identified as a novel susceptibility locus for AD using genome-wide immune chip analysis and meta-analysis.20 Several previous studies have reported that IL21 contributes to the pathogenesis of allergic diseases. The serum IL21 levels were found to increase during the acute exacerbation of asthma and to fall again after treatment.21 In other recent studies, it was found that both IL21 and IL21R expression was higher in acute skin lesions of AD patients,22 and that IL21 levels were are higher in adult AD patients than in unaffected controls.23 Although little is known about the biological mechanism of IL21 on IgE production in allergic diseases, additional recent studies have suggested that IL21 may suppress serum IgE production, with possible involvement of Th2 cells that produce IL4 and/or IL13.24,25,26 Our current results have indicated that the minor A allele of rs17389644 near IL21 is associated with higher levels of IgE in AD patients (P=0.01 under recessive model, Table 4), again suggesting that this variant plays a role in altering the serum IgE level in AD with hope of further functional studies elsewhere. Additional functional studies that investigate the effects of the minor A allele of rs17389644 on IL21 expression or on its trans-acting activity in relation to AD are needed to elucidate this mechanism.
There has been some conflicting evidence as to whether previously reported susceptibility markers on candidate genes are in fact associated with AD. For instance, some previous studies have reported that SNPs (rs2040704 on RAD50, rs3091307 co-located within TH2LCRR and near RAD50) at chromosome 5q31.1 are significantly associated with AD.8,27 However, another study reported only a nominal association between this rs3091307 locus and an AD patient group and no association at all with the allergic type of AD.28 In our result, the rs6871536 SNP co-located at the TH2LCRR and RAD50 was found to have no association signal, with the exception of a nominal signal only in the association with SCORAD These conflicting results may be due to insufficient sample sizes (in particular, the low number of AD cases with SCORAD) and/or different genetic backgrounds among populations that have been used thus far. Further replication studies of large cohorts comprising different populations are warranted.
SCORAD is an important AD assessment index, and the IgE level is one of the central players in allergic diseases including AD. The serum total IgE level, as a useful endophenotype, is generally increased in patients with AD.29 Different associations of genetic variants with the SCORAD and IgE parameters have been reported in AD cohorts: for instance, no associations have previously been reported between the FLG R501X mutation and the SCORAD or IgE levels in AD, whereas significant associations have been described between the -1112 C/T SNP of IL13 with both SCORAD and IgE.30 Our current findings also identified significant association signals for several SNPs with SCORAD (at rs13361382 on TMEM232) and IgE (at rs4713555 near HLA-DRB1 and HLA-DQA1). Another recent study has also reported a potential association between genetic variations of TMEM232 and AD in a Chinese population.31 In addition, considering the association between SNPs located near to the HLA-DRB1 locus and total IgE in asthma and the involvement between human leucocyte HLA class II and IgE responses,32,33 rs4713555 may also play a role in regulating the IgE levels in AD. Hence, although further validation is needed, it is possible that SCORAD or IgE-associated SNPs could be predictive markers for AD.
We performed a meta-analysis of 4 SNPs (rs17389644, rs6473227, rs2212434, and rs2143950) that have been identified as risk loci for AD in previous genome-wide studies.11,20 As shown in Figure, rs2212434 near to EMSA and rs2143950 near to PPP2R3C showed the most significant signal. However, when Cochran's Q test was used to assess heterogeneity of the estimated effect-sizes from the individual studies,34 only rs6473227 near to ZBTB10 still retained positive significance (PCochrane's Q=0.018). There are few clues in the current literature that might explain the direct relationship between the potential SNP and allergic/immune responses. Therefore, further evaluation will be needed to identify whether these 2 SNPs could indeed be useful markers for AD.
To estimate the potential functions of the SNPs we evaluated in AD, we employed in silico analysis of the variants found to be significantly associated with this disorder. Using the Signal Scan program to identify the putative transcription factor's binding sites, the CTATCA and CTATC sequences including the ‘C’ allele of rs6682925 in the promoter region of IL23R were estimated to be putative binding elements for the GATA-1 and NF-E regulators. However, the major ‘T’ allele of this SNP did not show any results in this regard, suggesting that that the minor ‘C’ allele of rs6682925 may affect gene expression (Supplementary Table 3). In our additional search for potential ESE sites for splicing machinery using the SNP Function Prediction program, 3 SNPs (rs6682925, rs7622183, and rs6473227) were observed to have different binding scores for splicing factors depending on the major and minor alleles of each (Supplementary Table 3). In additional search of the expression quantitative trait loci (eQTL) based on the conditional eQTL analysis (https://eqtl.onderzoek.io/index.php?page=info), rs7622183 was also found to act as cis-acting eQTL, suggesting that this variant might act as a potential cis-regulator for the gene. In the case of functional relevance for rs2275913, this variant is positioned in the promoter region of IL17A and within a binding motif for the critical regulator of nuclear factor-activated T cells (NFAT), leading to a higher promoter activity and production of IL17A in the minor A allele than the major G allele of rs2275913.35,36 In addition, IL17A rs2275913 has been reported to be associated with the risk of several diseases including inflammatory diseases (for instance, rheumatoid arthritis), allergic asthma and rhinitis.37,38 Therefore, further functional studies are required.
In conclusion, we have replicated the associations between previously identified SNPs and AD in our current study and performed extended analyses of these variants to better understand the clinical phenotypes of this condition. Although this study has limitations of small number of samples and lack of functional evaluation, our results suggest that several genetic variants may indeed be associated with AD and its clinical phenotypes (SCORAD and total IgE). However, further studies that include functional evaluations of the significantly associated SNPs identified herein will be needed.
ACKNOWLEDGMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (NRF-2014R1A2A1A10050687).
Footnotes
There are no financial or other issues that might lead to conflict of interest.
SUPPLEMENTARY MATERIALS
Detailed information of each SNP
References for our selection of 17 SNPs
Functional predictions of significant SNPs using in silico analyses
References
- 1.Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003;361:151–160. doi: 10.1016/S0140-6736(03)12193-9. [DOI] [PubMed] [Google Scholar]
- 2.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387:1109–1122. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- 3.van Beijsterveldt CE, Boomsma DI. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur Respir J. 2007;29:516–521. doi: 10.1183/09031936.00065706. [DOI] [PubMed] [Google Scholar]
- 4.Spergel JM. From atopic dermatitis to asthma: the atopic march. Ann Allergy Asthma Immunol. 2010;105:99–106. doi: 10.1016/j.anai.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Stone SP, Muller SA, Gleich GJ. IgE levels in atopic dermatitis. Arch Dermatol. 1973;108:806–811. [PubMed] [Google Scholar]
- 6.Esparza-Gordillo J, Weidinger S, Fölster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 7.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 8.Schaarschmidt H, Ellinghaus D, Rodríguez E, Kretschmer A, Baurecht H, Lipinski S, et al. A genome-wide association study reveals 2 new susceptibility loci for atopic dermatitis. J Allergy Clin Immunol. 2015;136:802–806. doi: 10.1016/j.jaci.2015.01.047. [DOI] [PubMed] [Google Scholar]
- 9.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–694. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 10.Paternoster L, Standl M, Chen CM, Ramasamy A, Bønnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2011;44:187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capon F, Di Meglio P, Szaub J, Prescott NJ, Dunster C, Baumber L, et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum Genet. 2007;122:201–206. doi: 10.1007/s00439-007-0397-0. [DOI] [PubMed] [Google Scholar]
- 13.Cheong HS, Kim LH, Park BL, Choi YH, Park HS, Hong SJ, et al. Association analysis of interleukin 5 receptor alpha subunit (IL5RA) polymorphisms and asthma. J Hum Genet. 2005;50:628–634. doi: 10.1007/s10038-005-0304-2. [DOI] [PubMed] [Google Scholar]
- 14.Christensen U, Haagerup A, Binderup HG, Vestbo J, Kruse TA, Børglum AD. Family based association analysis of the IL2 and IL15 genes in allergic disorders. Eur J Hum Genet. 2006;14:227–235. doi: 10.1038/sj.ejhg.5201541. [DOI] [PubMed] [Google Scholar]
- 15.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol Suppl (Stockh) 1980;92:44–47. [Google Scholar]
- 16.Kunz B, Oranje AP, Labrèze L, Stalder JF, Ring J, Taïeb A. Clinical validation and guidelines for the SCORAD index: consensus report of the European Task Force on Atopic Dermatitis. Dermatology. 1997;195:10–19. doi: 10.1159/000245677. [DOI] [PubMed] [Google Scholar]
- 17.Asher MI, Keil U, Anderson HR, Beasley R, Crane J, Martinez F, et al. International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur Respir J. 1995;8:483–491. doi: 10.1183/09031936.95.08030483. [DOI] [PubMed] [Google Scholar]
- 18.Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLOS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodríguez E, Matanovic A, Marenholz I, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet. 2013;45:808–812. doi: 10.1038/ng.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng S, Gao S, Hu H, Hu Y, Yu S, Zhao S. The differentiation and clinical significance of follicular helper T cells during acute exacerbation in asthma patients. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2015;31:1383–1386. [PubMed] [Google Scholar]
- 22.Jin H, Oyoshi MK, Le Y, Bianchi T, Koduru S, Mathias CB, et al. IL-21R is essential for epicutaneous sensitization and allergic skin inflammation in humans and mice. J Clin Invest. 2009;119:47–60. doi: 10.1172/JCI32310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizutani H, Tamagawa-Mineoka R, Nakamura N, Masuda K, Katoh N. Serum IL-21 levels are elevated in atopic dermatitis patients with acute skin lesions. Allergol Int. 2017;66:440–444. doi: 10.1016/j.alit.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Lin PY, Jen HY, Chiang BL, Sheu F, Chuang YH. Interleukin-21 suppresses the differentiation and functions of T helper 2 cells. Immunology. 2015;144:668–676. doi: 10.1111/imm.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamagawa-Mineoka R, Kishida T, Mazda O, Katoh N. IL-21 reduces immediate hypersensitivity reactions in mouse skin by suppressing mast cell activation or IgE production. J Invest Dermatol. 2011;131:1513–1520. doi: 10.1038/jid.2011.73. [DOI] [PubMed] [Google Scholar]
- 26.Wood N, Bourque K, Donaldson DD, Collins M, Vercelli D, Goldman SJ, et al. IL-21 effects on human IgE production in response to IL-4 or IL-13. Cell Immunol. 2004;231:133–145. doi: 10.1016/j.cellimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Esparza-Gordillo J, Schaarschmidt H, Liang L, Cookson W, Bauerfeind A, Lee-Kirsch MA, et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J Allergy Clin Immunol. 2013;132:371–377. doi: 10.1016/j.jaci.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 28.Namkung JH, Lee JE, Kim E, Kim HJ, Seo EY, Jang HY, et al. Association of polymorphisms in genes encoding IL-4, IL-13 and their receptors with atopic dermatitis in a Korean population. Exp Dermatol. 2011;20:915–919. doi: 10.1111/j.1600-0625.2011.01357.x. [DOI] [PubMed] [Google Scholar]
- 29.Furue M. Atopic dermatitis--immunological abnormality and its background. J Dermatol Sci. 1994;7:159–168. doi: 10.1016/0923-1811(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 30.Trzeciak M, Gleń J, Rębała K, Bandurski T, Sikorska M, Nowicki R. Coexistence of 2282del4 FLG gene mutation and IL-18 -137G/C gene polymorphism enhances the risk of atopic dermatitis. Postepy Dermatol Alergol. 2016;33:57–62. doi: 10.5114/pdia.2015.48050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu YY, Tang JP, Liu Q, Zheng XD, Fang L, Yin XY, et al. Scanning indels in the 5q22.1 region and identification of the TMEM232 susceptibility gene that is associated with atopic dermatitis in the Chinese Han population. Gene. 2017;617:17–23. doi: 10.1016/j.gene.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 32.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 33.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoaglin DC. Misunderstandings about Q and ‘Cochran's Q test’ in meta-analysis. Stat Med. 2016;35:485–495. doi: 10.1002/sim.6632. [DOI] [PubMed] [Google Scholar]
- 35.Espinoza JL, Takami A, Nakata K, Onizuka M, Kawase T, Akiyama H, et al. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS One. 2011;6:e26229. doi: 10.1371/journal.pone.0026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279:52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- 37.Eskandari-Nasab E, Moghadampour M, Tahmasebi A. Meta-analysis of risk association between interleukin-17A and F gene polymorphisms and inflammatory diseases. J Interferon Cytokine Res. 2017;37:165–174. doi: 10.1089/jir.2016.0088. [DOI] [PubMed] [Google Scholar]
- 38.Resende EP, Todo-Bom A, Loureiro C, Mota Pinto A, Oliveiros B, Mesquita L, et al. Asthma and rhinitis have different genetic profiles for IL13, IL17A and GSTP1 polymorphisms. Rev Port Pneumol (2006) 2017;23:10–16. doi: 10.1016/j.rppnen.2016.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed information of each SNP
References for our selection of 17 SNPs
Functional predictions of significant SNPs using in silico analyses