Abstract
c-Fos and c-Jun are members of the AP-1 family of transcriptional activators that regulate the expression of genes during cell proliferation. To facilitate in vitro studies of mechanisms of transcriptional activation by c-Jun and c-Fos we developed a method for obtaining recombinant c-Fos/c-Jun that is highly active in DNA binding and transcriptional activation in vitro. Full-length human c-Fos and c-Jun were expressed in Escherichia coli. The expression of c-Fos was dependent on a helper plasmid that encodes rare ArgtRNAs. Both over-expressed c-Fos and c-Jun were recovered from inclusion bodies. A c-Fos/c-Jun complex was generated by co-renaturation and purified via a His-tag on the full-length human c-Fos. The resulting c-Fos/c-Jun bound DNA with high affinity and specificity, and activated transcription in a reconstituted human RNA polymerase II transcription system. The availability of active recombinant human c-Fos/c-Jun will allow future biochemical studies of these important transcriptional activators.
INTRODUCTION
AP-1 transcription factors activate many genes including those involved in cell growth and proliferation (reviewed in 1). AP-1 proteins of the Jun and Fos families contain extended α helical dimerization/DNA-binding domains of ∼62 amino acids that consist of a basic region found N-terminal of a leucine zipper (2). While there is high sequence homology in the basic leucine zipper domains of AP-1 proteins, there is much less homology in other regions, which contain activation domains (3–5). Members of the Jun family can form homo- and heterodimers. Members of the Fos family cannot form homo- or heterodimers among themselves, but can heterodimerize with Jun proteins. Dimers bind DNA through sequence specific contacts between amino acids in the basic regions and the major groove of DNA (6).
In vitro studies have been useful for characterizing the dimerization, DNA binding and transcriptional activation properties of AP-1 proteins (2,3,5,7–10). Full-length recombinant human c-Fos, however, has not been available for in vitro characterization. We have succeeded in obtaining highly purified full-length recombinant human c-Fos using an E.coli expression system. Heterodimers of recombinant human c-Fos and human c-Jun were highly active in DNA binding and transcriptional activation in vitro. The availability of recombinant human c-Fos will facilitate biochemical studies of mechanisms by which human c-Fos/c-Jun activates transcription.
MATERIALS AND METHODS
Plasmids
pGEX-c-Fos was created by digesting pDKL8 [a kind gift from M. Piechaczyk, Institute de Génétique Moléculaire, France (11)] with EcoRI and HindIII to release a fragment containing the human c-Fos cDNA. The overhanging ends of this fragment were filled in with Klenow and subcloned into the filled in SalI site of pGEX-2TKN (a kind gift from S. Ruppert and R. Tjian, University of California, Berkeley). pET-6His-c-Fos was created by performing a triple ligation that included (i) an NdeI to BamHI fragment containing the human c-Fos cDNA isolated from pGEX-c-Fos, (ii) annealed oligos encoding a His-tag bordered by NcoI and NdeI sites and (iii) pET-19b (Novagen) cut with NcoI and NdeI. The resulting plasmid encodes human c-Fos with 17 additional amino acids fused to its N-terminus (including a six His-tag): MGHHHHHHMSKFRLTAT-c-Fos.
pET-Jun, a plasmid for expressing full-length human c-Jun was a gift from T. Hoey (Tularik Inc.). In creating this plasmid an NcoI site was generated that changed the fourth base pair in the c-Jun coding region from A/T to G/C, resulting in a point mutation in the second amino acid of c-Jun (Thr to Ala).
Plasmid p(AP-1)5-E1b-CAT, used as a template for in vitro transcription, was created by inserting five direct copies of a double stranded oligonucleotide containing the AP-1 element from the human Metallothionein IIA promoter (–105 to –96) into the XbaI site of plasmid E1b-CAT (12). Plasmid p(AP-1)5-E1b-G-less (13), used as template DNA in the in vitro transcription assays, was created by inserting a 377 bp G-less cassette [(excised from p(GAL4)5-E1b-G-less (kind gift from M. G. Peterson and R. Tjian, University of California, Berkeley) with SacI] into the SacI site of p(AP-1)5-E1b-CAT. Plasmid p(AP-1)1-E1b-CAT, used as template for generating DNase I footprinting probes, was created by inserting one copy of the human Metallothionein IIA AP-1 element into the XbaI site of plasmid E1b-CAT.
Expression and purification of recombinant transcriptional activators
His-c-Fos was expressed in Escherichia coli strain BL21:DE3 transformed with both the pET-6His-c-Fos expression plasmid and the plasmid pSBET (14). A culture was grown in Luria–Bertani (LB) medium containing 0.3 mM ampicillin and 10 µg/ml kanamycin at 37°C until the OD600 nm was 0.5, at which point isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM, final) was added to induce protein expression. Cells were harvested 2 h later and resuspended in a solution containing 20 mM Tris (pH 7.9), 20% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.1 M NaCl, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF) and sonicated four times for 15 s. Samples were centrifuged for 30 min in a Sorval SS34 rotor at 18 000 r.p.m. and 4°C. Precipitated material (inclusion bodies) containing his-c-Fos was resuspended in 10 ml of 5 mM DTT and sonicated twice for 30 s. Samples were centrifuged for 10 min in a Sorval SS34 rotor at 15 000 r.p.m. and 4°C. Insoluble material was washed three more times by resuspending in 10 ml of 5 mM DTT, followed by centrifugation. Pellets from the final wash were resuspended in buffer A [20 mM Tris (pH 7.9), 1 mM EDTA, 1 mM DTT and 6 M guanidine–HCl] and protein was extracted by overnight nutation at 4°C. After centrifugation to remove the remaining insoluble material, samples containing His-c-Fos were aliquoted and stored at –80°C. His-c-Fos can be further purified by dialysis and NTA–agarose (Qiagen) affinity chromatography as described below for His-c-Fos/c-Jun heterodimers. Full-length human c-Jun was expressed in E.coli BL21:DE3 transformed with pET-c-Jun using a protocol similar to that described above for his-c-Fos (13).
Recombinant human His-c-Fos/c-Jun was prepared and purified using the following protocol. Equal moles of His-c-Fos and human c-Jun (both in buffer A) were mixed and the solution was diluted with buffer A to give final a concentration of 200 ng/µl total protein. The mixture was then subjected to three sequential dialyses (3 h each) in buffer B [20 mM Tris (pH 7.9), 0.1 mM EDTA, 10% glycerol and 1 mM DTT] containing the following additions: (i) 7 M urea and 1 M NaCl, (ii) 1 M urea and 1 M NaCl and (iii) 1 M NaCl. A fourth dialysis was performed overnight in buffer C [20 mM Tris (pH 7.9), 10% glycerol, 5 mM β-mercaptoethanol, 0.1 M NaCl]. The final dialysate was clarified by centrifugation and applied to an NTA–agarose column (Qiagen) at 0.5 mg protein/ml of packed resin. The resin was washed with 10 column vol of buffer C containing 20 mM imidazole. Protein was eluted first with buffer C containing 100 mM imidazole and then with buffer C containing 500 mM imidazole. Fractions of the 500 mM imidazole elution that contained protein were pooled and dialyzed into TGED(0.1) buffer [20 mM Tris (pH 7.9), 10% glycerol, 1 mM EDTA, 1 mM DTT and 100 mM KCl]. Aliquots of the final solution were stored at –80°C. The preparation of human c-Jun/c-Jun homodimers is described in detail elsewhere (13).
Gel filtration chromatography was performed on Superose 6 PC 3.2/30 column (Amersham/Phamacia) using a Phamacia SMART system. The solvent system consisted of 20 mM Tris (pH 7.9), 1 mM EDTA, 0.2 M NaCl and 1 mM DTT. A Sample (50 µl) of purified c-Fos/c-Jun (1 µg) was loaded on the column. Solvent (2.4 ml) was run over the column and 50 µl fractions were collected. A sample (10 µl) of every odd fraction from the void through the low molecular weight region were resolved by 12% SDS–PAGE. Proteins were visualized by silver staining.
DNase I footprinting
DNase I footprints were performed either with (i) a 214 bp DNA fragment containing a single AP-1 site (region –105 to –96 of the human Metallothionein IIA promoter) and surrounding vector DNA generated by PCR using p(AP-1)1-E1b-CAT as a template, or (ii) a 187 bp DNA fragment containing the –111 to +42 region of the human IL-2 promoter and some surrounding vector DNA. The DNA fragments were generated by PCR and were 32P-labeled on the 5′-end of the non-template strand. Reactions contained poly(dG–dC)·poly(dG–dC) as a non-specific competitor (final concentration of 2 µg/ml). Recombinant c-Fos/c-Jun and c-Jun/c-Jun (amounts indicated in figure legends) were incubated with promoter DNA for 20 min at 30°C under buffer conditions that were identical to those used for transcription in a reaction volume of 20 µl. A solution (2 µl) containing 0.05 U/µl DNase I (Promega) and 11 mM CaCl2 was added to each reaction. After a 30 s incubation at 30°C, reactions were stopped with 83 µl of stop solution containing 25 mM EDTA, 125 mM KCl and 10 µg of carrier yeast RNA. SDS was added to each reaction to a final concentration of 0.5%. Reactions were incubated at 65°C for 15 min and then placed on ice for 10 min. After a 10 min spin at 14 000 r.p.m. in a microcentrifuge, the supernatants were transferred to new tubes. DNA was ethanol precipitated and resuspended in formamide loading buffer. Products were resolved by 8% denaturing PAGE.
In vitro transcription
General transcription factors (TFIIB, -E, -F and -H) and RNA polymerase II were expressed and purified as previously described (15). Human TFIID was immunopurified (16). Recombinant human TFIIA was prepared as previously described (17,18). Transcription reactions using the reconstituted transcription system were performed as previously described (16) with the following modifications. Each reaction contained either p(AP-1)5-E1b-CAT or p(AP-1)5-E1b-G-less. Where indicated, recombinant AP-1 proteins were pre-incubated with promoter DNA for 5 min at 30°C prior to the addition of the remaining general transcription factors (TFIIA, -B, -D, -E, -F and -H) and RNA polymerase II. After the addition of general transcription factors, reactions were incubated at 30°C for 30 min. Nucleoside triphosphates were added and RNA synthesis was allowed to proceed for 20 min at 30°C. Transcription reactions were stopped, RNA transcripts were processed and, where indicated, primer extension was performed as previously described (16). Transcription in a Jurkat nuclear extract was carried out as described above, except that purified general transcription factors were replaced with 4 µl of nuclear extract.
RESULTS
Expression and purification of human c-Fos/c-Jun heterodimers
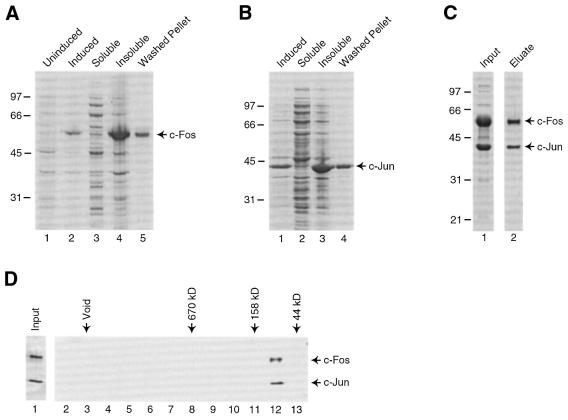
To facilitate studies of mechanisms of transcriptional activation by human c-Fos/c-Jun we developed techniques for the expression of human c-Fos in E.coli and the reconstitution of c-Fos/c-Jun complex that is highly active for DNA binding and transcriptional activation in vitro. We have previously expressed human c-Jun in E.coli using an available expression vector (13). A vector for expressing human c-Fos was not available. We obtained a plasmid containing the human c-Fos cDNA from M. Piechaczyk (11) and subcloned the cDNA downstream of DNA encoding a His-tag in a plasmid useful for expression of proteins in E.coli. We thought that the His-tag would be helpful in purifying c-Fos/c-Jun heterodimers away from c-Jun/c-Jun homodimers. Initial attempts to express c-Fos failed (data not shown). We noticed that the human c-Fos cDNA contained many Arg codons that are used infrequently in E.coli and, therefore, tested the effect of pSBET (14), a plasmid that encodes rare ArgtRNAs, on expression of c-Fos. As shown in Figure 1A, high levels of c-Fos were produced by E.coli transformed with both pET-6His-c-Fos and pSBET (compare the uninduced and induced E.coli extracts shown in lanes 1 and 2, respectively). Nearly all of the over-expressed c-Fos was found in the insoluble fraction of the extract (Fig. 1A, compare lanes 3 and 4) and could be recovered after extensive washing of the inclusion bodies (lane 5). Human c-Jun was expressed separately in E.coli and was also in the insoluble portion of extracts (Fig. 1B).
Figure 1.
Expression and purification of recombinant human c-Fos/c-Jun. (A) Expression of his-tagged human c-Fos in E.coli. His-c-Fos was expressed in E.coli strain BL21:DE3 transformed with both the pET-6His-c-Fos expression plasmid and the plasmid pSBET (14). Cells were lyzed by sonication and inclusion bodies were washed extensively. Samples that were resolved by SDS–PAGE and analyzed by Coomassie staining included: lane 1, E.coli just prior to induction; lane 2, E.coli after induction; lane 3, supernatant after sonication; lane 4, pellet after sonication; lane 5, washed inclusion bodies. (B) Expression of human c-Jun in E.coli. Human c-Jun was expressed in E.coli strain BL21:DE3. Cells were lyzed by sonication and inclusion bodies were washed extensively. Samples that were resolved by SDS–PAGE and analyzed by Coomassie staining included: lane 1, E.coli after induction; lane 2, supernatant after sonication; lane 3, pellet after sonication; lane 4, washed inclusion bodies. (C) Purification of c-Fos/c-Jun. After co-renaturation by stepwise dialysis, recombinant human c-Fos/c-Jun was purified by chromatography on NTA–agarose column (Qiagen). Samples of the material applied to the column (lane 1) and the eluted and dialyzed c-Fos/c-Jun (lane 2) were resolved SDS–PAGE and visualized by staining with Coomassie Brilliant Blue. (D) Gel filtration chromatography on c-Fos/c-Jun. Recombinant c-Fos/c-Jun was subjected to chromatography on a Superose 6 column. Protein present in the fractions collected were resolved by SDS–PAGE and visualized by silver staining. Lane 1 shows 10% of the input. Lanes 2–13 are every odd fraction spanning the void to the low molecular weight range. The positions at which size standards migrated are shown above the gel.
To form a complex of c-Fos and c-Jun, the two proteins were separately solubilized from inclusion bodies using guanidine–HCl, extracts containing the proteins were mixed, and the mixture was subjected to stepwise dialysis to renature the proteins. After renaturation, the mixture was passed over NTA–agarose to bind his-c-Fos/c-Jun. His-c-Fos monomers would also bind the resin; however, c-Jun/c-Jun homodimers that formed during renaturation would not be retained on the NTA–agarose resin because of the lack of a His-tag. Protein was eluted from the column with 500 mM imidazole and dialyzed. Samples of the input and final dialyzed material are shown in Figure 1C. The dialyzed c-Fos/c-Jun appears to contain comparable amounts of the two subunits as assessed by Coomassie staining. Gel filtration chromatography confirmed that the recombinant c-Fos/c-Jun is not aggregated, but instead migrates at an apparent molecular weight expected for the heterodimer (Fig. 1D).
Recombinant human c-Fos/c-Jun is highly active in DNA binding
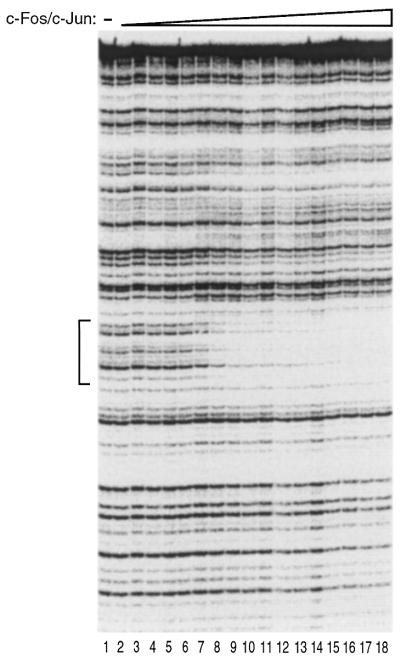
To test the activity and specificity of the recombinant human c-Fos/c-Jun preparation in DNA binding we performed quantitative DNase I footprinting on a DNA fragment containing a single high affinity AP-1 element from the human Metallothionein IIA promoter. Figure 2 shows a titration of c-Fos/c-Jun over a broad concentration range. Protection of the AP-1 element (bracket) was clearly observed. There was no detectable binding of c-Fos/c-Jun to regions of the DNA outside of the known AP-1 element. An appKD of 1.3 nM was determined from quantitation of this data (19). We conclude that the recombinant c-Fos/c-Jun binds specifically to the AP-1 element with high affinity.
Figure 2.
Recombinant human c-Fos/c-Jun binds DNA with high affinity. Increasing amounts of recombinant human c-Fos/c-Jun were added to a DNA fragment containing a single high affinity AP-1 element. The binding of c-Fos/c-Jun to the DNA fragment was analyzed by DNase I footprinting. The final concentrations of c-Fos/c-Jun heterodimer in reactions were: lane 1, 0 nM; lane 2, 0.4 nM; lane 3, 0.6 nM; lane 4, 0.8 nM; lane 5, 1.0 nM; lane 6, 1.3 nM; lane 7, 1.6 nM; lane 8, 2.1 nM; lane 9, 2.8 nM; lane 10, 3.6 nM; lane 11, 4.7 nM; lane 12, 6.1 nM; lane 13, 7.9 nM; lane 14, 10 nM; lane 15, 13 nM; lane 16, 17 nM; lane 17, 22 nM; lane 18, 29 nM. The DNA concentration was 0.5 nM. The protected region is indicated with a bracket.
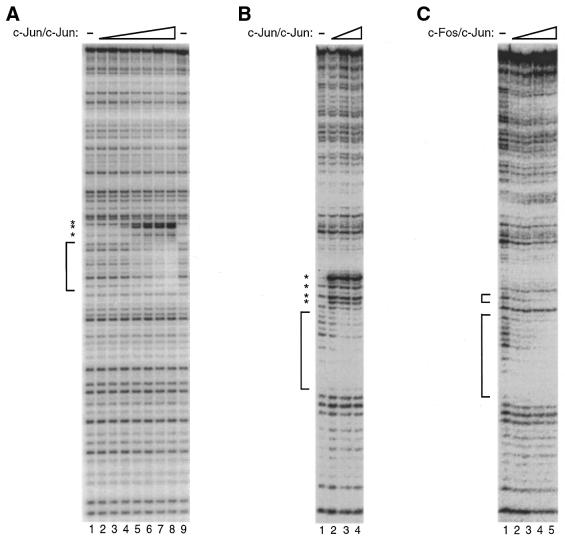
In performing comparative DNase I footprinting studies with c-Fos/c-Jun and c-Jun/c-Jun, we noticed that on binding the AP-1 element from the human Metallothionein IIA promoter c-Jun/c-Jun caused a DNase I hypersensitive site adjacent to the protected region (Fig. 3A). While c-Fos/c-Jun protected the same AP-1 element it did not cause a detectable DNase I hypersensitive site (see Fig. 2). The DNase I hypersensitive site caused by c-Jun/c-Jun was not limited to the high affinity AP-1 element, but was also observed on a weaker AP-1 element found in the human IL-2 promoter (Fig. 3B). Recombinant c-Fos/c-Jun did not cause a DNase I hypersensitive site on the IL-2 promoter (Fig. 3C). Based on these results we conclude that the c-Fos/c-Jun preparation is not contaminated with amounts of c-Jun/c-Jun homodimers capable of competing with c-Fos/c-Jun for DNA binding.
Figure 3.
Recombinant human c-Fos/c-Jun is not contaminated with c-Jun/c-Jun. (A) The binding of c-Jun/c-Jun to a high affinity AP-1 element causes hypersensitivity to DNase I. Increasing amounts of recombinant human c-Jun/c-Jun were added to a DNA fragment containing a single high affinity AP-1 element. The binding of c-Jun/c-Jun to the DNA fragment was analyzed by DNase I footprinting. The final concentrations of c-Jun/c-Jun heterodimer in reactions were: lane 1, 0 nM; lane 2, 1.0 nM; lane 3, 1.8 nM; lane 4, 3.3 nM; lane 5, 5.8 nM; lane 6, 10 nM; lane 7, 19 nM; lane 8, 34 nM; lane 9, 0 nM. The DNA concentration was 0.1 nM. The protected region is indicated with a bracket. Hypersensitive sites are indicated with asterisks. (B) The binding of c-Jun/c-Jun to an AP-1 element in the human IL-2 promoter causes hypersensitivity to DNase I. Increasing amounts of recombinant human c-Jun/c-Jun were added to an IL-2 promoter DNA fragment. The binding of c-Jun/c-Jun to the DNA fragment was analyzed by DNase I footprinting. The final concentrations of c-Jun/c-Jun heterodimer in reactions were: lane 1, 0 nM; lane 2, 20 nM; lane 3, 40 nM; lane 4, 60 nM. The DNA concentration was 1.5 nM. The protected region is indicated with a bracket. Hypersensitive sites are indicated with asterisks. (C) The binding of c-Fos/c-Jun to the human IL-2 promoter does not cause hypersensitivity to DNase I. Increasing amounts of recombinant human c-Fos/c-Jun were added to an IL-2 promoter DNA fragment. The binding of c-Fos/c-Jun to the DNA fragment was analyzed by DNase I footprinting. The final concentrations of c-Fos/c-Jun heterodimer in reactions were: lane 1, 0 nM; lane 2, 3.6 nM; lane 3, 7.2 nM; lane 4, 15 nM; lane 5, 30 nM. The DNA concentration was 1.5 nM. Protected regions are indicated with brackets.
AP-1 dimers are known to bind cooperatively with other transcriptional activators to composite elements in promoters. For example, c-Fos/c-Jun binds cooperatively with NFAT proteins to composite elements in promoters such as the IL-2 promoter (20–24). The cooperativity is mediated by interactions between the basic leucine zipper regions of c-Fos/c-Jun and the DNA binding domain of NFATp. We have previously shown that the recombinant c-Fos/c-Jun is active for cooperative DNA binding with NFATp using DNase I footprinting on the human IL-2 promoter (25).
Recombinant human c-Fos/c-Jun activates transcription in vitro
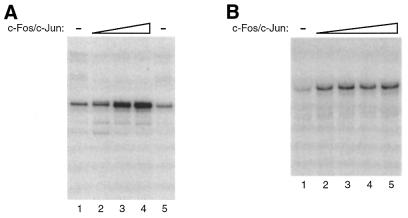
While DNA binding by c-Fos/c-Jun requires only the basic leucine zipper regions of these proteins, transcriptional activation requires the activation domains, which are outside of the dimerization/DNA binding domains (2–5,7,8). To test the ability of the recombinant human c-Fos/c-Jun to activate transcription, we performed in vitro transcription experiments. As shown in Figure 4A, c-Fos/c-Jun activated transcription ∼7-fold in a Jurkat nuclear extract. The promoter used in this experiment contained five high affinity AP-1 elements upstream of the adenovirus E1b TATA box and transcripts were detected by primer extension. One potential problem with this experiment is that the Jurkat nuclear extract contains endogenous AP-1 proteins that could contribute to the observed activation. A more stringent test of the transcriptional activity of c-Fos/c-Jun was performed using a human RNA polymerase II transcription system reconstituted from recombinant and highly purified transcription factors. We have previously shown that this transcription system is devoid of AP-1 proteins (13,25). As shown in Figure 4B, c-Fos/c-Jun activated transcription 4-fold in the reconstituted transcription system. We conclude that c-Fos/c-Jun is not only highly active in DNA binding, but also is a potent activator of transcription in vitro.
Figure 4.
Recombinant c-Fos/c-Jun activates transcription in vitro. (A) Recombinant c-Fos/c-Jun activates transcription in an extract prepared from unstimulated Jurkat cell nuclei. The template used contained five AP-1 elements upstream of the E1b TATA box and the CAT gene. Transcripts were analyzed by primer extension using a CAT specific primer followed by denaturing PAGE. The concentrations of c-Fos/c-Jun in reactions were: lane 1, 0 nM; lane 2, 12 nM; lane 3, 20 nM; lane 4, 40 nM; lane 5, 0 nM. The DNA concentration was 2 nM. (B) Recombinant c-Fos/c-Jun activates transcription in a reconstituted human transcription system. The DNA template [p(AP-1)5-E1b-G-less] contained five AP-1 elements upstream of the E1b TATA box and a 377 bp G-less cassette. Transcripts were analyzed by denaturing PAGE. The concentrations of c-Fos/c-Jun in reactions were: lane 1, 0 nM; lane 2, 2.4 nM; lane 3, 4.8 nM; lane 4, 7.2 nM; lane 5, 9.6 nM. The DNA concentration was 0.8 nM.
DISCUSSION
Here we have described a method for preparing recombinant human c-Fos/c-Jun that is highly active in DNA binding and transcriptional activation in vitro. A plasmid was generated for expressing His-tagged full-length human c-Fos. We devised a method for expressing the His-tagged c-Fos protein that takes advantage of a helper rare ArgtRNA expression plasmid, pSBET. We described the recovery of His-tagged c-Fos from inclusion bodies and a method for forming a c-Fos/c-Jun complex by co-renaturation. c-Fos/c-Jun complexes were purified by nickel-affinity chromatography. Purified recombinant human c-Fos/c-Jun was highly active in DNA binding. Moreover, the c-Fos/c-Jun preparation was not contaminated by significant amounts of c-Jun/c-Jun homodimers, as assessed by the absence of c-Jun/c-Jun dependent DNase I hypersensitivity. Finally, the recombinant c-Fos/c-Jun activated transcription in vitro in both a crude nuclear extract and a highly purified RNA polymerase II transcription system. The methods described here will facilitate future biochemical studies of human c-Fos and c-Jun.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Tricia Lively for reagents and advice and Anita Seto for technical help in generating plasmids. This research was supported by a Public Health Service grant GM-55235 from the National Institutes of Health. J.A.G. is currently a Pew Scholar in the Biomedical Sciences and was a Special Fellow of the Leukemia Society of America during the early stages of this work.
References
- 1.Karin M., Liu,Z. and Zandi,E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol., 9, 240–246. [DOI] [PubMed] [Google Scholar]
- 2.Turner R. and Tjian,R. (1989) Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science, 243, 1689–1694. [DOI] [PubMed] [Google Scholar]
- 3.Abate C., Luk,D., Gagne,E., Roeder,R.G. and Curran,T. (1990) Fos and jun cooperate in transcriptional regulation via heterologous activation domains. Mol. Cell. Biol., 10, 5532–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohmann D. and Tjian,R. (1989) Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell, 59, 709–717. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland J.A., Cook,A., Bannister,A.J. and Kouzarides,T. (1992) Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev., 6, 1810–1819. [DOI] [PubMed] [Google Scholar]
- 6.Glover J.N. and Harrison,S.C. (1995) Crystal structure of the heterodimeric bZIP transcription factor cFos-cJun bound to DNA. Nature, 373, 257–261. [DOI] [PubMed] [Google Scholar]
- 7.Abate C., Luk,D., Gentz,R., Rauscher,F.J.,III and Curran,T. (1990) Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl Acad. Sci. USA, 87, 1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abate C., Luk,D. and Curran,T. (1991) Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol. Cell. Biol., 11, 3624–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentz R., Rauscher,F.J., Abate,C. and Curran,T. (1989) Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science, 243, 1695–1699. [DOI] [PubMed] [Google Scholar]
- 10.Kerppola T.K. and Curran,T. (1991) Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell, 66, 317–326. [DOI] [PubMed] [Google Scholar]
- 11.Roux P., Verrier,B., Klein,B., Niccolino,M., Marty,L., Alexandre,C. and Piechaczyk,M. (1991) Retrovirus-mediated gene transfer of a human c-fos cDNA into mouse bone marrow stromal cells. Oncogene, 6, 2155–2160. [PubMed] [Google Scholar]
- 12.Lillie J.W. and Green,M.R. (1989) Transcription activation by the adenovirus E1a protein. Nature, 338, 39–44. [DOI] [PubMed] [Google Scholar]
- 13.Lively T.N., Ferguson,H.A., Galasinski,S.K., Seto,A.G. and Goodrich,J.A. (2001) c-Jun binds the N terminus of human TAFII250 to derepress RNA polymerase II transcription in vitro. J. Biol. Chem., 276, 25582–25588. [DOI] [PubMed] [Google Scholar]
- 14.Schenk P.M., Baumann,S., Mattes,R. and Steinbiss,H.H. (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques, 19, 196–198. [PubMed] [Google Scholar]
- 15.Kugel J.F. and Goodrich,J.A. (1998) Promoter escape limits the rate of transcription from the adenovirus major late promoter on negatively supercoiled templates. Proc. Natl Acad. Sci. USA, 95, 9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galasinski S.K., Lively,T.N., Grebe de Barron,A. and Goodrich,J.A. (2000) Acetyl-CoA stimulates RNA polymerase II transcription and promoter binding by TFIID in the absence of histones. Mol. Cell. Biol., 20, 1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozer J., Moore,P.A., Bolden,A.H., Lee,A., Rosen,C.A. and Lieberman,P.M. (1994) Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev., 8, 2324–2335. [DOI] [PubMed] [Google Scholar]
- 18.Sun X., Ma,D., Sheldon,M., Yeung,K. and Reinberg,D. (1994) Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev., 8, 2336–2348. [DOI] [PubMed] [Google Scholar]
- 19.Brenowitz M., Senear,D.F., Shea,M.A. and Ackers,G.K. (1986) Quantitative DNase footprint titration: a method for studying protein–DNA interactions. Methods Enzymol., 130, 132–181. [DOI] [PubMed] [Google Scholar]
- 20.Jain J., McCaffrey,P.G., Miner,Z., Kerppola,T.K., Lambert,J.N., Verdine,G.L., Curran,T. and Rao,A. (1993) The T cell transcription factor NFATp is a substrate for calcineurin and interacts with the DNA-binding domains of Fos and Jun. Nature, 365, 352–355. [DOI] [PubMed] [Google Scholar]
- 21.Hoey T., Sun,Y.L., Williamson,K. and Xu,X. (1995) Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity, 2, 461–472. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Rodriguez C., Aramburu,J., Rakeman,A.S. and Rao,A. (1999) NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl Acad. Sci. USA, 96, 7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rooney J.W., Sun,Y.L., Glimcher,L.H. and Hoey,T. (1995) Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol., 15, 6299–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Glover,J.N., Hogan,P.G., Rao,A. and Harrison,S.C. (1998) Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature, 392, 42–48. [DOI] [PubMed] [Google Scholar]
- 25.Kim L.J., Ferguson,H.A., Seto,A.G. and Goodrich,J.A. (2000) Characterization of DNA binding, transcriptional activation and regulated nuclear association of recombinant human NFATp. BMC Immunol., 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]