The (re-)evolution of vegetative desiccation tolerance in Linderniaceae appears to be linked to the presence of dehydration-responsive cis-elements in the promoters of desiccation-related genes.
Keywords: Desiccation tolerance, dehydration-responsive element, gene regulation, LEA-like protein, resurrection plants, stress protein
Abstract
Reproductive structures of plants (e.g. seeds) and vegetative tissues of resurrection plants can tolerate desiccation. Many genes encoding desiccation-related proteins (DRPs) have been identified in the resurrection plant Craterostigma plantagineum, but the function of these genes remains mainly hypothetical. Here, the importance of the DRP gene pcC13-62 for desiccation tolerance is evaluated by analysing its expression in C. plantagineum and in the closely related desiccation-tolerant species Lindernia brevidens and the desiccation-sensitive species Lindernia subracemosa. Quantitative analysis revealed that pcC13-62 transcripts accumulate at a much lower level in desiccation-sensitive species than in desiccation-tolerant species. The study of pcC13-62 promoters from these species demonstrated a correlation between promoter activity and gene expression levels, suggesting transcriptional regulation of gene expression. Comparison of promoter sequences identified a dehydration-responsive element motif in the promoters of tolerant species that is required for dehydration-induced β-glucuronidase (GUS) accumulation. We hypothesize that variations in the regulatory sequences of the pcC13-62 gene occurred to establish pcC13-62 expression in vegetative tissues, which might be required for desiccation tolerance. The pcC13-62 promoters could also be activated by salt stress in Arabidopsis thaliana plants stably transformed with promoter::GUS constructs.
Introduction
Tolerance to desiccation in plants is found in specialized tissues, such as spores, seeds, or pollen, in most angiosperms, and in vegetative tissues of a small group of resurrection plants. Resurrection plants comprise several distantly related plant lineages that tolerate the loss of most of their cellular water and suspend all metabolic activities, but restart normal physiological processes upon rewatering. The most widely accepted hypothesis locates the evolution of the desiccation tolerance trait at the beginning of land colonization (Cushman and Oliver, 2011). Later during their evolution, plants lost the ability to tolerate vegetative desiccation but retained genes relevant for desiccation tolerance in tissues such as spores, seeds, and pollen. More recently, these genes were apparently reprogrammed to be expressed in the vegetative tissues of angiosperm resurrection plants, restoring their ability to tolerate desiccation. Studies of seeds and resurrection plants support this theory, because it has been shown that similar molecular programs are activated in vegetative tissues and seeds or pollen (Bartels and Salamini, 2001; Giarola et al., 2017). Desiccation tolerance is acquired following a tightly controlled developmental or dehydration-induced process, which involves the accumulation of protective proteins such as the late embryogenesis abundant (LEA) proteins, reactive oxygen species (ROS) scavengers, ultraviolet protective compounds, nitrogen-rich metabolites (e.g. amino acids), polyols, and non-reducing sugars. In conditions of mild dehydration, sugars are required for osmoregulation, whereas during severe dehydration they may participate in hydrogen interactions with other polar macromolecules, leading to the suspension of metabolic activities and the formation of the so-called glassy state (Burke, 1986).
LEA genes are the most abundant group of protective genes activated in desiccation-tolerant tissues. The name LEA was assigned to these genes as they were initially discovered to be abundantly expressed during the final stage of cotton seed development (Dure et al., 1981). However, these genes are also expressed during dehydration in vegetative tissues of both desiccation-tolerant and desiccation-sensitive plants. Conserved amino acid motifs and sequence similarities have been used to divide LEA proteins into different subclasses (Hundertmark and Hincha, 2008; Jaspard et al., 2012). It has been hypothesized that high expression levels of LEAs are required for the acquisition of desiccation tolerance (Bartels and Salamini, 2001). Although LEA genes have similar expression patterns, they encode proteins with variable amino acid compositions and cellular localizations (Tunnacliffe et al., 2010). This suggests that LEA genes may have multiple functions that are required for tissue protection under stress. Protection-related functions such as binding to membranes or macromolecule structures, or scavenging of reactive oxygen species and ions, have been demonstrated for some LEAs (Tunnacliffe et al., 2010). LEAs are predicted to participate in the formation of the glassy state together with sugars (Hoekstra et al., 2001).
The South African resurrection plant Craterostigma plantagineum has been studied to understand the molecular mechanisms underlying desiccation tolerance. Many different mRNAs encoding desiccation-related proteins (DRPs) have been identified (Bartels et al., 1990; Ditzer et al., 2001; Rodrigo et al., 2004). All DRPs are abundantly expressed in desiccated plant leaves. Some DRPs could be assigned to the LEA proteins, as they contain conserved LEA motifs in their amino acid sequences, but others do not contain LEA motifs and thus are generally referred to as LEA-like DRPs. Some C. plantagineum DRPs have been studied in detail. For example, the LEA-like DRP CDeT11-24 was shown to protect enzymes from desiccation-related damage (Petersen et al., 2012). Although the expression of other C. plantagineum DRPs predicts their involvement in desiccation-tolerance-related functions, these functions remain mainly unknown. The LEA-like DRP pcC13-62 was also associated with desiccation tolerance, but no function could be inferred and no sequence similarity was found with other proteins in public repositories (Bartels et al., 1990; Piatkowski et al., 1990).
Since desiccation tolerance genes appear to be ubiquitous in plants, the rewiring of regulatory networks is predicted to play a major role in the (re-)evolution/(re-)activation of vegetative desiccation tolerance. Therefore, the study of regulatory networks is essential to decipher what is required for desiccation tolerance. Several factors may be involved in gene regulation, including cis- and trans-acting elements and short and long non-coding RNAs. The same trans-acting elements appear to be activated upon dehydration in desiccation-tolerant and -sensitive species, and no desiccation-tolerance-specific cis-elements have been identified so far (Giarola et al., 2017). In C. plantagineum, dehydration-specific non-coding RNAs, namely CDT-1 and 28852, have been identified and have been linked to desiccation tolerance, but their function remains mainly unknown (Hilbricht et al., 2008; Giarola and Bartels, 2015). Abscisic acid (ABA) and ABA-related pathways seem to play an important role in regulating gene expression relevant for desiccation tolerance (Bartels and Salamini, 2001). In C. plantagineum, DRP transcripts accumulate and DRP promoters are activated following ABA treatments, suggesting transcriptional regulation of gene expression (Bartels et al., 1990; Michel et al., 1993; Michel et al., 1994; Velasco et al., 1998; Rodrigo et al., 2004; Ditzer and Bartels, 2006). ABA-responsive elements (ABREs) are found in promoters of ABA-inducible genes, including C. plantagineum DRP genes. ABA-independent transcriptional regulation is also found in dehydration-responsive genes and it is mediated by factors binding to the dehydration-responsive element (DRE) promoter motifs (Todaka et al., 2015).
The family of Linderniaceae represents a good source of plants for comparative studies as it groups desiccation-tolerant species such as C. plantagineum and Lindernia brevidens together with species that are mostly sensitive to desiccation, like Lindernia subracemosa. Thus, regulatory desiccation-related signatures can be inferred by comparing the promoter regions of protective genes in these species. Using such an approach, the high expression level observed for the CDeT11-24 LEA-like protective gene in tolerant species was linked to promoter architecture and the presence of cis-elements (van den Dries et al., 2011). This finding supports the role of certain cis-elements in the rewiring of regulatory networks of desiccation-tolerance genes. However, data are still limited to draw general conclusions.
In this study we compared the expression and promoter activity of the DRP gene pcC13-62 in closely related desiccation-tolerant and desiccation-sensitive Linderniaceae. We found that the accumulation of 13-62 transcripts is controlled at the promoter level and involves a DRE motif that occurs only in tolerant species. Our data show the importance of DRE motifs for the regulation of desiccation-tolerant genes in resurrection plants and suggest the contribution of nucleotide variations in regulatory regions for the (re-)establishment of desiccation tolerance in vegetative tissues.
Materials and methods
Plant materials
Craterostigma plantagineum Hochst., Lindernia brevidens Skan, and Lindernia subracemosa De Wild plants were grown as described by Bartels et al. (1990) and Dinakar and Bartels (2012). For the dehydration treatment, fully grown 6- to 8-week-old plants were gradually dehydrated in pots. Relative water content (RWC) measurements were made according to Bernacchia et al. (1996). In vitro cultures of C. plantagineum for transient expression experiments were grown on Murashige and Skoog (MS) medium at day/night temperatures of 22/18 °C with 80 µmol m−2 s−1 light for 16 h per day.
Arabidopsis thaliana (Col-0) wild-type and transgenic seeds were germinated and plants were grown on soil under short-day conditions (120–150 μmol m−2 s−1 light at 22 °C with a light/dark cycle of 8/16 h) for 4 weeks and then moved to long-day conditions (16 h light/8 h dark) to induce flowering.
Transgenic plants were selected on MS medium containing 50 µg ml–1 kanamycin under the same conditions as described for in vitro cultures and transferred to soil.
Molecular biology techniques and DNA sequence analysis
Molecular biology techniques were performed according to Green and Sambrook (2012). DNA sequencing was carried out by GATC Biotech (https://www.gatc-biotech.com/en/index.html) and primer synthesis by Eurofins MWG Operon (http://www.eurofinsgenomics.eu). Signal peptides were predicted using SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) (Nielsen, 2017). Sequence alignment figures were obtained with the Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/index.html) using the alignment file generated with T-Coffee software (Notredame et al., 2000). All primers used are listed in Supplementary Table S1 at JXB online.
Identification of Cp pcC13-62 homologs and phylogenetic analysis
Putative Cp pcC13-62 homologs were identified from L. brevidens and L. subracemosa transcriptome data (data not published) or GenBank using the C. plantagineum pcC13-62 predicted protein sequence (GenBank accession number AAA63616) as the query. The sequence alignment of pcC13-62 homologs was generated with T-Coffee (Notredame et al., 2000) and used for phylogenetic analysis in MEGA6 (Tamura et al., 2013). Phylogenetic analysis was performed using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood is shown in Fig. 1B.
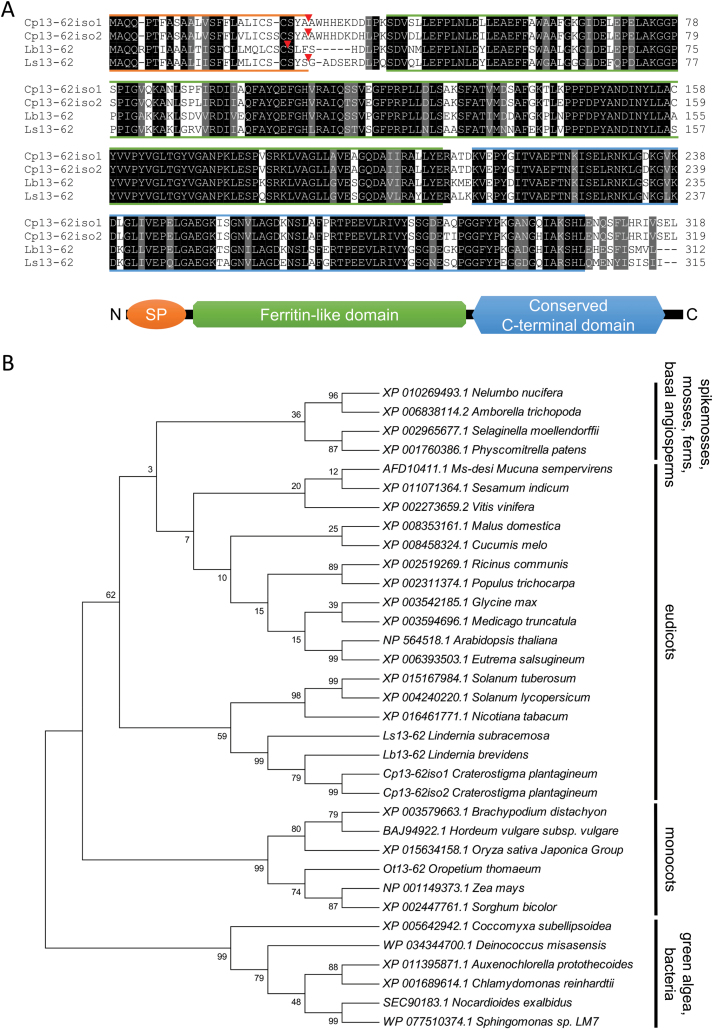
Fig. 1.
Alignment of Craterostigma plantagineum, Lindernia brevidens, and Lindernia subracemosa 13-62 amino acid sequences and phylogenetic analysis of selected 13-62 protein homologs. (A) Alignment of the predicted 13-62 amino acid sequences. Identical (black) and conserved (grey) amino acids are indicated. Coloured lines are used to show the different protein domains. Orange: predicted signal peptide (SP); red triangles indicate the predicted SP cleavage site; green: ferritin-like domain (pfam13668); blue: conserved C-terminal domain. (B) phylogenetic analysis of 13-62 protein homologs. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Phylogenetic analysis was performed using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood is shown. (This figure is available in colour at JXB online.)
RNA isolation and cDNA synthesis
Total RNA was isolated as described by Valenzuela-Avendaño et al. (2005). The concentration and purity of the RNA was determined using a BioSpec-nano spectrophotometer (Shimadzu Biotech, Japan). RNA integrity was verified by 2% (w/v) agarose gel electrophoresis. A 4 µg aliquot of total RNA was treated with DNase I (Thermo Fisher Scientific, St. Leon-Rot, Germany) to remove any DNA; 2 µg of the DNase I-treated RNA was reverse transcribed into cDNA using the RevertAid Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s instructions. The remaining 2 µg of RNA served as a control to monitor genomic DNA contamination in cDNA preparations. The cDNA was diluted 15 times with diethylpyrocarbonate-treated water and 5 µl was used as a template for a 20 µl reverse transcription–quantitative PCR (RT–qPCR).
Primer design and RT–qPCR analysis
RT–qPCR analysis was performed as described by Giarola et al. (2015). The specificity of the primers was verified by melting curve analysis (Supplementary Fig. S1) and sequencing. To prepare template samples for standard curves, the 13-62 amplicons generated with RT–qPCR primers (Supplementary Table S1) were cloned into pJET1.2 vectors (Thermo Fisher Scientific). Plasmids containing the amplicons were linearized using the HindIII restriction enzyme. Linearized DNAs were purified after gel electrophoresis using the NucleoSpin® Gel and PCR Clean-up kit (MACHEREY-NAGEL, Düren, Germany) and quantified with a BioSpec-nano spectrophotometer (Shimadzu Biotech). Standard curves were obtained by the amplification of 1/10 dilutions of linearized vectors, starting from 1010 copies. The amount of linearized plasmid required to obtain 1010 copies was calculated as described in Giarola et al. (2015). The transcript copy number for 5 µl of cDNA template was calculated from Cq values using standard curves.
Isolation of 13-62 5ʹ gene sequences and mutagenesis
Genomic DNA was extracted from leaves according to Rogers and Bendich (1985) and used to prepare genome walking libraries with the GenomeWalker™ universal kit (Clontech, Heidelberg, Germany). Genomic fragments corresponding to the 5ʹ 13-62 gene sequence were amplified from C. plantagineum and L. subracemosa genome walking libraries with gene-specific (Supplementary Table S1) and library-specific (AP1 and AP2; GenomeWalker™ universal kit manual) primers. The amplification from L. brevidens libraries was unsuccessful and thus primers designed to match conserved sequence regions of C. plantagineum and L. subracemosa genomic fragments (Lb13-62g_F and Lb13-62g2_R; Supplementary Table S1) were used to obtain the 5ʹ 13-62 sequence from L. brevidens genomic DNA. The sequence between the β-galactosyltransferase and the 13-62 translational start codons from C. plantagineum (962 bp for isoform1 and 1029 bp for isoform2) and L. subracemosa (723 bp) was used as the 13-62 promoter sequence in the promoter analyses. A transposon is inserted between the β-galactosyltransferase and the 13-62 genes in L. brevidens, and thus the sequence between the transposon terminal repeats and the 13-62 translational start codon was used as the promoter sequence for this species. The promoter sequences were screened for cis-acting regulatory elements using the PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace) (Higo et al., 1999) and TRANSFAC (http://genexplain.com/transfac/) (Wingender et al., 1996) databases.
Preparation of promoter::GUS constructs and mutagenesis
The 13-62 promoter sequences from C. plantagineum (Cp13-62iso1 and Cp13-62iso2), L. brevidens (Lb13-62), and L. subracemosa (Ls13-62) were amplified by PCR from genomic DNA with species-specific GUS primers (listed in Supplementary Table S1) to add restriction sites required for cloning the promoter fragment into pBT10GUS vectors (Sprenger-Haussels and Weisshaar, 2000). The Cp13-62iso1, Cp13-62iso2, and Ls13-62 promoters were cloned using NcoI and XbaI restriction, sites whereas the Lb13-62 promoter was cloned using NcoI and SalI restriction sites. Mutagenesis of predicted cis-elements in the promoter sequences was achieved with primers (Supplementary Table S1) using the Quick-change II Site-Directed Mutagenesis kit (Stratagene, Heidelberg, Germany).
Analysis of promoter activity using transient transformation
Relative promoter activities were determined using the transient transformation method described by van den Dries et al. (2011). Briefly, the Cp13-62, Lb13-62, and Ls13-62 promoter::GUS fusion constructs in pBT10GUS vectors were co-bombarded together with a vector carrying the CaMV35S::GFP construct. The activity of promoter fragments was assayed in both homologous and heterologous genetic backgrounds. The number of green fluorescent protein (GFP)-expressing cells per leaf was determined 16 h after particle bombardment with an inverted confocal laser-scanning microscope (Nikon Eclipse TE2000-U/D-Eclipse C1; Nikon, Düsseldorf, Germany). For dehydration treatments, the bombarded leaves were kept on filter paper for 4 h at room temperature. Leaves were histochemically stained with a solution containing 0.5 mg ml–1 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid (X-Gluc, Gold Biotechnology, St. Louis, USA). The number of β-glucuronidase (GUS) spots per leaf was determined using a stereoscopic microscope with a binocular eyepiece tube (Nikon SMZ 800). Relative promoter activities were calculated by dividing the number of GUS spots by the number of GFP spots. Mean values and standard deviations were calculated from at least four independent replicates.
Analysis of promoter activity in stably transformed A. thaliana plants
Stably transformed A. thaliana plants were generated by the floral dip method (Clough and Bent, 1998) using pBIN19 binary vectors (Bevan, 1984). Promoter::GUS-expressing cassettes were excised from pBT10GUS vectors using the BglII restriction enzyme and inserted within the BamHI site of pBIN19 vectors. The presence of the promoter::GUS cassette in transgenic plants was confirmed by PCR. As 13-62 transcript expression is also induced by salt stress (Supplementary Fig. S2), we analysed the activity of promoter::GUS constructs in transgenic lines subjected to salt stress treatment. This allows testing of a stress response in a more uniform and reproducible way than by dehydration. Transgenic plants at the flowering stage were watered for 2 weeks with 150 mM NaCl or water for salt stress and control experiments, respectively. GUS expression was analysed in rosette leaves, cauline leaves, siliques, roots, open flowers, and floral clusters. Four-leaf-stage seedlings were removed from soil and incubated for 16 h in MS solution (control) or MS solution containing 150 mM NaCl. Plant material was vacuum-infiltrated for 3 min with a 0.5 mg ml–1 X-Gluc solution (Gold Biotechnology) and then incubated for 16 h at 37 °C. After destaining, tissues were observed using a Nikon SMZ 800 stereoscopic microscope (Nikon, Tokyo, Japan) and pictures were taken with a digital camera connected to the microscope (Nikon Digital Sight DS-2Mv).
Results
Genomic organization of 13-62 genes and quantification of 13-62 transcripts
The C. plantagineum DRP pcC13-62 gene has been previously described (Bartels et al., 1990; Piatkowski et al., 1990; Schneider et al., 1993). To gain more insight into the role of this gene in desiccation tolerance, we compared the pcC13-62 gene structures and gene expression in the desiccation-tolerant species C. plantagineum and L. brevidens and in the desiccation-sensitive species L. subracemosa. Previous DNA blot analysis predicted the presence of two to three copies of pcC13-62 in the genome of C. plantagineum (Bartels et al., 1990; Piatkowski et al., 1990). Here, we identified two 13-62 isoforms in the C. plantagineum genome, which were named Cp13-62iso1 and Cp13-62iso2 (GenBank accession numbers MH247237 and MH247238, respectively). The protein sequences predicted for Cp13-62iso1 and Cp13-62iso2 are very similar (98% similarity; Fig. 1A). Cp13-62 homologs were identified in L. brevidens and L. subracemosa and were termed Lb13-62 (GenBank accession number MH247239) and Ls13-62 (GenBank accession number MH247240), respectively. The 13-62 predicted protein sequences from all three species showed more than 74% identity and 83% similarity between each other, with the N-terminal region being the most variable (Fig. 1A). All proteins contain N-terminal signal peptides and ferritin-like domains (pfam13668). The C-terminal regions of these proteins are very conserved. Homologs of 13-62, sometimes referred to as DRPs, are found in the genomes of several groups of bacteria, a few Chlorophyta, and most Embryophyta (Carniel et al., 2016). The overall conservation is quite high in Embryophyta, including the C. plantagineum, L. brevidens, and L. subracemosa genes (Supplementary Fig. S3): all genes contain the conserved domains and form a separate cluster, as shown by phylogenetic analysis (Fig. 1B).
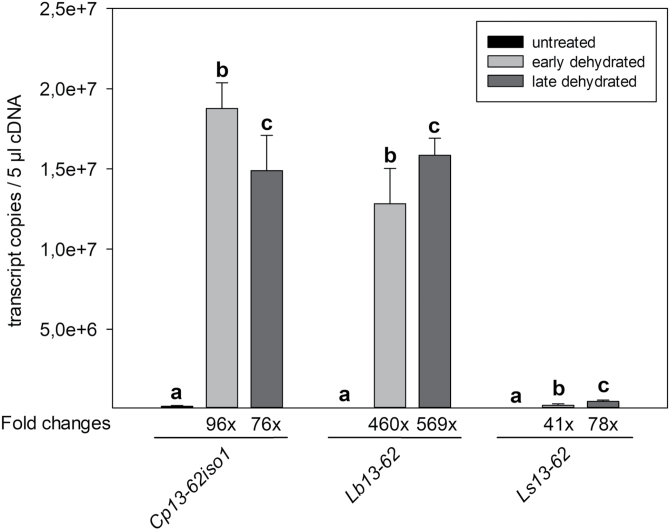
The 13-62 protein coding sequence is conserved across desiccation-tolerant and non-tolerant species. However, abundant expression of the 13-62 transcript is found in desiccation-tolerant tissues. Fig. 2 shows that pcC13-62 transcripts are expressed at a high level in response to dehydration in the desiccation-tolerant species C. plantagineum and L. brevidens, but only weakly in the desiccation-sensitive plant L. subracemosa. pcC13-62 transcripts are also abundant in dehydrated leaves of the monocot resurrection plant Oropetium thomaeum (Supplementary Fig. S4). Correlation of 13-62 expression and desiccation tolerance is supported by transcriptomic studies of selected unrelated monocot and dicot plants, which show high accumulation of pcC13-62 homologs in desiccation-tolerant structures such as seeds and pollen (Table 1). In C. plantagineum, L. brevidens, and L. subracemosa, pcC13-62 transcripts are also expressed in response to mannitol and salt treatments (Supplementary Fig. S2). Some 13-62 homologs also accumulate in response to osmotic, salt, and/or cold stress in various species, but the expression in stressed tissues is much lower than in reproductive organs (data not shown).
Fig. 2.
Absolute quantification of 13-62 transcripts in Craterostigma plantagineum, Lindernia brevidens, and Lindernia subracemosa by RT–qPCR analysis. cDNA was prepared from total RNA isolated from leaves of untreated, partially dehydrated (relative water content 50–60%), and late dehydrated (relative water content 30–40%) plants and amplified using 13-62-specific primers. Transcript copy numbers were calculated from three different biological replicates (mean +SD). Different letters above bars denote statistically significant differences within each group of samples (P<0.05; one-way ANOVA).
Table 1.
Expression patterns of predicted 13-62 homologs in selected dicots and monocots
| Gene name | GenBank Ac. | Species | Expression | References |
|---|---|---|---|---|
| AT1G47980 | NP_564518.1 | Arabidopsis thaliana | Em, Fl, Po, Ro, Se, Sm; | Schmid et al. (2005); Brady et al. (2007) |
| OS, SS leaves | Kilian et al. (2007) | |||
| AT3G62730 | NP_191832.1 | Arabidopsis thaliana | Em, Ro, Se, Si, Sm, St; | Schmid et al. (2005); Brady et al. (2007) |
| OS, SS roots | Kilian et al. (2007) | |||
| Medtr2g033580 | XP_003594696.1 | Medicago truncatula | Se | Benedito et al. (2008) |
| Medtr2g033520 | XP_003594692.1 | Medicago truncatula | Se | Benedito et al. (2008) |
| Glyma15g14720 | XP_003546306.2 | Glycine max | Fl, GP, Se | Libault et al. (2010); Severin et al. (2010) |
| Glyma13g11550 | XP_003542185.1 | Glycine max | Po, Se | Libault et al. (2010); Severin et al. (2010) |
| LOC_Os03g22470 | XP_015632037.1 | Oryza sativa | Em, En, In, Ro, Se, Sg | Jain et al. (2007); Li et al. (2007) |
| LOC_Os04g33150 | XP_015634158.1 | Oryza sativa | Em, En, In, Ro, Se | Jain et al. (2007); Li et al. (2007) |
| GRMZM2g085260 | NP_001149373.1 | Zea mays | En, Ro, Se, Ta | Downs et al. (2013) |
| GRMZM2g327051 | NP_001150304.1 | Zea mays | Ta | Downs et al. (2013) |
CS, Cold stress; Em, Embryo; En, Endosperm; Fl, Flowers; GP, Green pods; In, Inflorescence; OS, Osmotic stress; Pl, Pollen; Po, Pods; Ro, Roots; Se, Seeds; Sg, Stigma; Si, Siliques; Sm, Stamens; SS, Salt stress; St, Stem; Ta, Tassel.
The 13-62 promoter structure
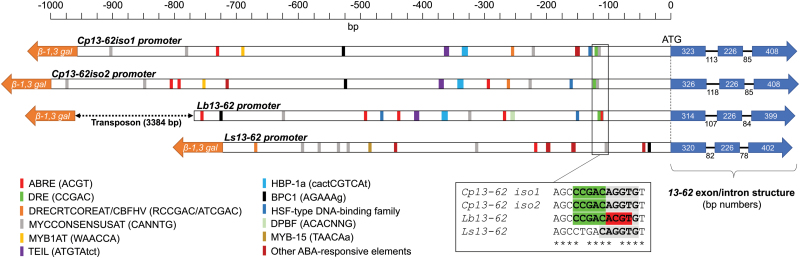
The genomic regions of Cp13-62iso1, Cp13-62iso2, Lb13-62, and Ls13-62 were isolated and analysed to identify regulatory motifs that cause high expression of 13-62 transcripts in vegetative tissues of desiccation-tolerant species (Fig. 3). The genomic regions of the 13-62 locus are conserved and similarly organized: the β-galactosyltransferase protein coding sequence is upstream of the 13-62 gene, and the 13-62 coding sequence has the same exon-intron structure (Fig. 3). The distance between the β-galactosyltransferase coding sequence and the 13-62 coding sequence varies among the different species. The distance is ~700 bp in L. subracemosa and nearly 1000 bp in C. plantagineum, whereas it is more than 4000 bp in L. brevidens (Fig. 3). This large difference is due to the insertion of a transposable element ~800 bp upstream of the Lb13-62 gene translational start codon. The transposable element contains features similar to the maize P instability factor (PIF) such as short terminal inverted repeat sequences and the coding sequence for a transposase (Supplementary Fig. S5) (Zhang et al., 2001).
Fig. 3.
Structure of the 13-62 gene locus in Craterostigma plantagineum, Lindernia brevidens, and Lindernia subracemosa. Putative cis-acting regulatory elements associated with abscisic acid (ABA)- and dehydration-responsive gene expression are indicated by coloured boxes. The box indicates the position of the conserved drought-responsive element (DRE) in the promoter of desiccation-tolerant species.
Elements that are related to ABA and drought promoter responsiveness were predicted upstream of the Cp, Lb, and Ls13-62 gene translational start sites (Fig. 3). Only a few structural elements are conserved in the desiccation-tolerant species. The promoter of the desiccation-sensitive species L. subracemosa showed almost no conservation of cis-elements (Fig. 3). Most of the sequence conservation was observed within the first 170 bp upstream of the translational start site (Supplementary Fig. S6). In this region we identified a putative consensus sequence (CCGAC) for a DRE and binding sites for Myc factors (Fig. 3). The putative DRE motif is conserved in the desiccation-tolerant species C. plantagineum and L. brevidens but not in the desiccation-sensitive species L. subracemosa, in which a single-nucleotide variation was found (Fig. 3).
Functional analysis of Cp, Lb, and Ls13-62 promoters in response to dehydration
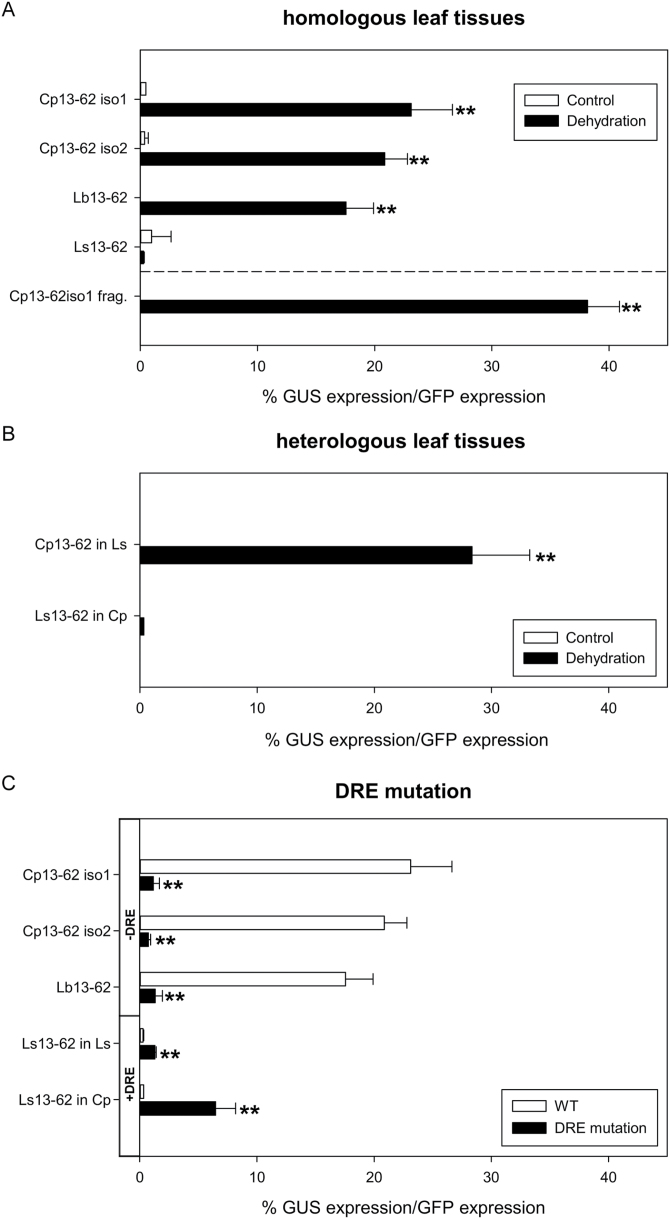
To test which promoter sequences are essential for transcriptional activation of the 13-62 genes, the above-described promoter regions were fused to GUS as a reporter gene and the promoter activities were determined. The response of the Cp13-62iso1, Cp13-62iso2, Lb13-62, and Ls13-62 promoters to dehydration was evaluated by monitoring GUS accumulation in dehydrated leaf tissues transiently transformed with Cp, Lb, and Ls13-62 promoter::GUS constructs. Only promoters from the desiccation-tolerant species (C. plantagineum and L. brevidens) showed activities in response to dehydration when tested in the corresponding species (Fig. 4A). We then analysed the activity of the Ls13-62 promoter in C. plantagineum and the Cp13-62 promoter in L. subracemosa to determine whether the lack of Ls13-62 promoter activity upon dehydration was due to the promoter structure per se or the absence of transcription factors in the desiccation-sensitive species. The Cp13-62 promoter was functional in the L. subracemosa background, which suggests that trans-acting factors are present in L. subracemosa. However, the Ls13-62 promoter did not function in the C. plantagineum background, which indicates a lack of cis-elements essential for promoter activation in response to dehydration (Fig. 4B).
Fig. 4.
Activity of the 13-62 promoter in response to dehydration. Wild-type and mutated Craterostigma plantagineum (Cp), Lindernia brevidens (Lb), and Lindernia subracemosa (Ls) promoter fragments were fused to the GUS reporter gene and tested for their activity upon dehydration in homologous and heterologous leaf tissues using a transient expression assay. (A) Response to dehydration of 13-62 wild-type promoters in homologous tissues. (B) Response to dehydration of Cp13-62iso1 and Ls13-62 promoters in heterologous tissues. (C) Effect of mutation of the DRE motif on activity of the 13-62 promoter in homologous and heterologous tissues. The DRE consensus sequence was either impaired (-DRE; promoters of desiccation-tolerant species) or restored (+DRE; promoter of desiccation-sensitive species) and the response to dehydration was compared using wild-type and mutated promoters. A construct expressing green fluorescence protein (GFP) under the control of the CaMV35S promoter was used to normalize all experiments. Bars indicate the relative promoter activities, expressed as a percentage of 13-62 promoter fragment activity relative to that of the CaMV35S promoter. The values are calculated from at least four independent experiments (mean +SD) for each treatment. Statistically significant differences from the control mean in (A) and (B) or wild-type expression in (C) of each promoter fragment are indicated above the error bars: *P<0.05, **P<0.01 (Student’s t-test).
Next, we tested whether the conserved 170 bp region from the desiccation-tolerant promoters could drive GUS expression in response to dehydration. A promoter::GUS construct containing this region derived from the Cp13-62iso1 promoter was sufficient to obtain dehydration-induced promoter activation in C. plantagineum (Fig. 4A). This fragment contains the DRE motif conserved in the tolerant species (Fig. 3). Thus, the functionality of this element was tested; promoter::GUS constructs with mutated DRE motifs (CCGAC mutated to CTGAC in the tolerant species, or CTGAC into CCGAC in the sensitive species) were created and used in expression experiments. The mutated promoters from the desiccation-tolerant species were unable to drive dehydration-induced GUS expression, suggesting that this motif was essential for the dehydration response (DRE-; Fig. 4C). By contrast, when the DRE element of the desiccation-sensitive species L. subracemosa was restored to CCGAC, a small increase of promoter activity was measured in response to dehydration (DRE+; Fig. 4C). The activity increased when the Ls13-62(+DRE) promoter was tested in the C. plantagineum background, but it did not reach the same level as observed for the wild-type promoters of desiccation-tolerant species (Fig. 4C). Taken together, these results suggest that the DRE element is essential but not sufficient for dehydration-induced activation of the 13-62 promoter.
Cp, Lb, and Ls13-62 promoter activities in A. thaliana
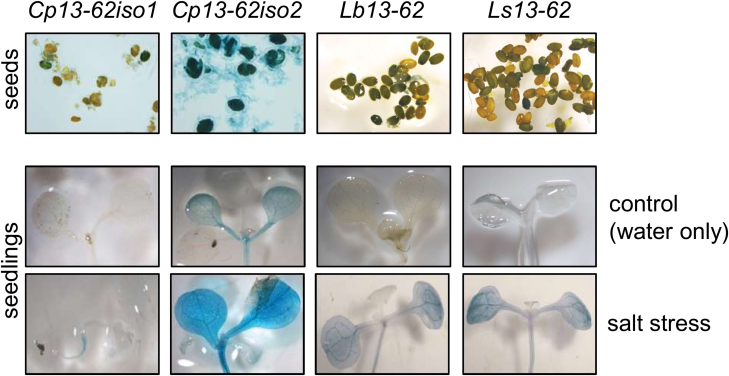
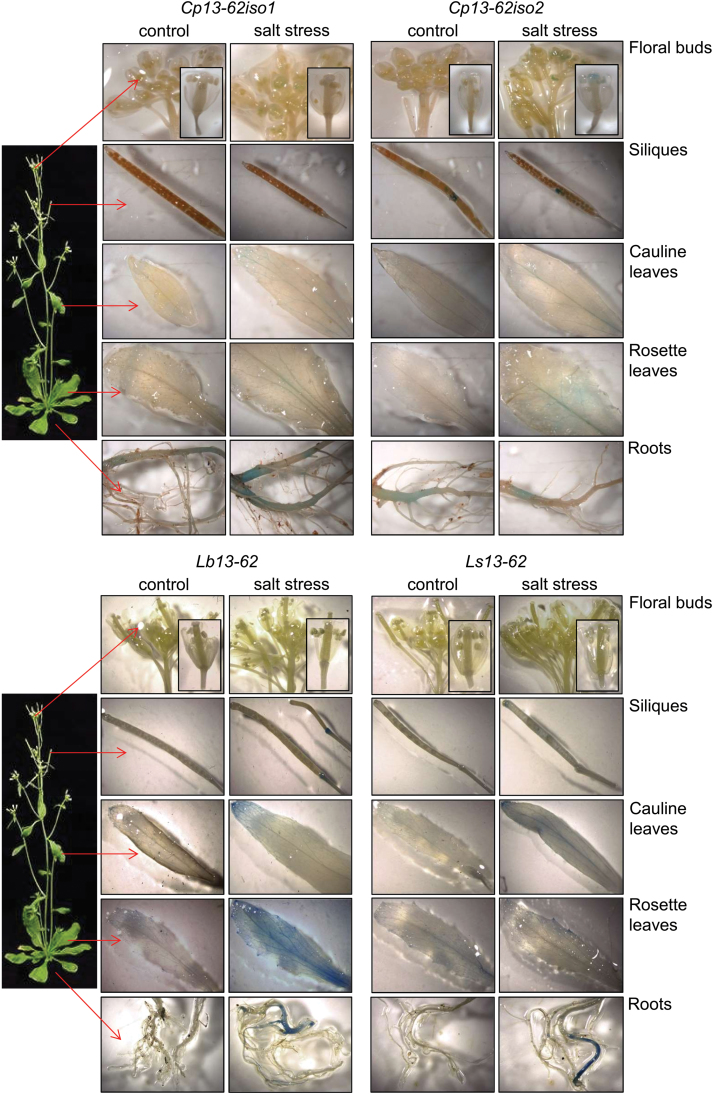
We tested whether the Cp, Lb, and Ls13-62 promoter::GUS constructs were also functional when stably integrated into the A. thaliana genome. Transgenic plants were assayed for GUS activity at different developmental stages and upon salt stress. Salt stress was chosen instead of dehydration because this allowed more robust testing than dehydration and because it was previously shown that pcC13-62 accumulated to high levels following NaCl treatments (Smith-Espinoza et al., 2003). The GUS reporter gene was expressed in transgenic seeds, suggesting promoter activation during seed development (Fig. 5). GUS activity was not detected in seedlings that were germinated under control conditions, but the 13-62 promoters from both desiccation-tolerant and desiccation-sensitive species (except for Cp13-62iso1) could be activated in seedlings subjected to salt stress (Fig. 5). This indicates that trans-acting elements from Arabidopsis that are active upon salt stress can trigger transcription of the GUS reporter gene from 13-62 promoters. Next, transgenic plants were grown to the flowering stage to allow assessment of whether the 13-62 promoters were activated in different plant organs under control and stress conditions. GUS staining was barely detected in plant organs from unstressed control plants (Fig. 6). When plants were subjected to salt stress, mainly leaves but also roots showed GUS accumulation. Most activity was observed in leaves under stress conditions when GUS expression was under the control of the Lb13-62 promoter (Fig. 6). It cannot be concluded from these experiments whether other cis-elements as well as the DRE are involved in the response to salt stress.
Fig. 5.
Activity of the 13-62 promoter in A. thaliana seeds and seedlings. Seeds and seedlings from stably transformed Arabidopsis plants containing the Cp, Lb, and Ls13-62 promoter::GUS constructs were analysed for GUS expression. Seedlings were germinated on soil and incubated with water (control) or 150 mM NaCl (salt stress) for 16 h before staining for GUS activity. Representative pictures from different lines are shown.
Fig. 6.
Activity of the 13-62 promoter in untreated and stressed A. thaliana plants. Stably transformed Arabidopsis plants containing the Cp, Lb, and Ls13-62 promoter::GUS constructs were watered for 2 weeks with water (control) or 150 mM NaCl (salt stress), and tissues were subsequently analysed for GUS expression. Different lines were analysed and representative pictures of plant organs stained for GUS activity are shown.
Discussion
The DRP pcC13-62 gene was originally classified among genes restricted to the resurrection plant C. plantagineum as no significant sequence similarities were identified (Bartels et al., 1990). Advances in genome sequencing and transcriptome analyses revealed that genes similar to pcC13-62 are present in bacteria, green algae, mosses, ferns, and angiosperms (Table 1; Fig. 1B) (Battista et al., 2001; Zha et al., 2013; Carniel et al., 2016; Kitajima et al., 2017). This observation corroborates the hypothesis that protective genes important for desiccation tolerance in resurrection plants are present in the genome of desiccation-sensitive species. All pcC13-62-related proteins contain a ferritin-like domain and a conserved C-terminal domain (Carniel et al., 2016). Enzymatic assays with recombinant pcC13-62-related proteins from sap of the lacquer tree Toxicodendron vernicifluum (TvFe2D) or floral nectar of the bean Mucuna sempervirens (MS-desi) support a biochemical function for this family of proteins (Zha et al., 2013; Kitajima et al., 2017). The C-terminal domain of TvFe2D suppresses laccase and peroxidase colorimetric reactions (Kitajima et al., 2017), whereas the MS-desi protein was able to inhibit citrate synthase activity (Zha et al., 2013).
The expression of pcC13-62 and pcC13-62-related genes is linked to desiccation tolerance. Transcripts of 13-62 accumulated abundantly in desiccated tissues of the desiccation-tolerant plants C. plantagineum and L. brevidens, but only to a very low level in desiccated leaves of the desiccation-sensitive plant L. subracemosa (Fig. 2). This result could be explained by the lack of selective pressure for desiccation tolerance in the habitat of L. subracemosa. The correlation of pcC13-62 and desiccation tolerance is supported by observations from several other species. Deletion of the Deinococcus radiodurans locus DRB0118, which encodes a pcC13-62 homolog, was responsible for the loss of viability of desiccated cultures (Battista et al., 2001). Expression of the pcC13-62 gene increases in dehydrating leaves of the resurrection grass O. thomaeum (Supplementary Fig. S4). The pcC13-62 family underwent large expansions and diversifications in the desiccation-tolerant lichen photobiont Trebouxia gelatinosa. Members of the 13-62 family were among the most up-regulated genes during desiccation of T. gelatinosa (Carniel et al., 2016). One of the two pcC13-62 homologs in A. thaliana (At3g62730) is specifically expressed in immature seeds and progressively accumulates during seed development (Becerra et al., 2006). A pcC13-62–related expressed sequence tag was also found among genes expressed in spores of the aquatic fern Ceratopteris richardii (Salmi et al., 2005). In several non-resurrection species, pcC13-62 genes are mainly expressed in desiccation-tolerant reproductive tissues and organs, including seeds and pollen (Table 1). Taken together, these results suggest that pcC13-62 homologs are highly expressed in bacteria, lichens, spores, seeds, pollen, and vegetative tissues of resurrection plants, and that the expression is linked to desiccation tolerance. The function of the encoded proteins might be relevant for enzymatic reactions, perhaps inhibiting enzymes that are involved in degradation processes.
The Cp, Lb, and Ls13-62 genes showed conserved structural features, supporting their common origin (Fig. 3). Intriguingly, we identified a transposable element similar to members of the PIF/Harbinger superfamily inserted between the Lb13-62 and β-1,3 galactosyltransferase coding sequences. PIF/Harbinger elements are DNA-mediated elements that contain coding sequences for two proteins required for their mobilization, namely a transposase and a Myb-like domain-containing protein (Feschotte and Pritham, 2007). The element in L. brevidens carries mutations in the transposase coding sequence, suggesting a lack of autonomous transposition (data not shown). The Lb13-62 promoter sequence used for the analysis in this study was restricted in its length to the region between the predicted transposon terminal inverted repeat sequence and the ATG start codon of Lb13-62. Our analysis showed that regulatory cis-elements important for dehydration-induced activation appear to be within this sequence (Fig. 4).
Cp, Lb, and Ls13-62 promoter activities correlated with transcript expression levels, suggesting that expression is regulated mostly at the transcriptional level. Cp and Lb13-62 promoters showed comparable dehydration-induced activities, whereas the Ls13-62 promoter could not be activated upon dehydration (Fig. 4A). Promoter activity is the result of the interaction of trans-acting binding elements with their corresponding cis-acting elements. Since the promoter from C. plantagineum was active in dehydrated L. subracemosa leaves but the L. subracemosa promoter was not functional in dehydrated C. plantagineum leaves, the lower activity in L. subracemosa can be explained by the lack of essential cis-acting elements (Fig. 4B). The availability of trans-acting elements required for promoter activation in response to dehydration but a lack of proper cis-acting elements was also demonstrated for the CDeT11-24 gene in L. subracemosa (van den Dries et al., 2011). Transcriptomic studies suggest that most dehydration-induced protective genes of resurrection species are similar to genes whose transcripts accumulate abundantly during the acquisition of desiccation tolerance in seeds (Rodriguez et al., 2010; Costa et al., 2017). pcC13-62 homologs from non-desiccation-tolerant plants are mainly expressed in seeds, and the Cp, Lb, and Ls13-62 promoters showed activity in Arabidopsis seeds. We hypothesize that regulatory elements for seed-specific expression are conserved among species. Hence, pcC13-62 must be linked to desiccation tolerance, and the (re-)activation of pcC13-62 expression in vegetative tissues may have contributed to the (re-)establishment of desiccation tolerance in resurrection plants.
Promoters of dehydration-induced genes contain conserved dehydration-responsive cis-acting DNA elements such as DRE-element/C-repeat (DRE/CRT) and ABA-response elements (ABRE) (Nakashima and Yamaguchi-Shinozaki, 2010). In silico analysis revealed the presence of several ABA- and dehydration-related cis-elements in the Cp, Lb, and Ls13-62 promoters. However, the spatial organizations of these cis-elements mainly differ, indicating limited conservation of the promoter architecture among the three species (Fig. 3). However, some elements, such as TEIL, HBP-1a, HSF, DRE, and MYC, were found in the promoters of the two desiccation-tolerant species (Fig. 3). The first 170 bp upstream of the ATG start codon shows the highest conservation across the three species and it is sufficient for dehydration-responsive gene activation. This sequence contains the only DRE element in the desiccation-tolerant species C. plantagineum and L. brevidens, and the deletion of the DRE element almost completely abolished dehydration-induced promoter activation (Fig. 4C). In L. subracemosa, a single-nucleotide mutation in the corresponding promoter region suppressed part of the dehydration responsiveness. When the DRE element was introduced into the L. subracemosa promoter, dehydration responsiveness was partially restored (Fig. 4C). Therefore, we assume that other cis-acting elements in the first 170 bp are possibly missing in the L. subracemosa promoter and may work as coupling elements for the DRE. The importance of the DRE element in the promoter of desiccation-tolerant species has previously been demonstrated for the CDeT11-24 promoter. The CDeT11-24 gene requires the DRE element for high promoter activity upon stress (van den Dries et al., 2011). It has been hypothesized that desiccation tolerance in vegetative plant tissues (re-)evolved through mutations in the regulatory sequences of desiccation-related genes (van den Dries et al., 2011). Here, we provide support for this hypothesis by showing that the desiccation-related pcC13-62 gene from desiccation-tolerant Linderniaceae was reprogrammed to be expressed at high levels in vegetative tissues. Similar to the 11-24 gene, a DRE motif is involved in dehydration-induced gene expression in the pcC13-62 promoter. This allows us to conclude that such elements, in combination with unknown coupling elements, are important for the regulation of desiccation-tolerance genes in resurrection species.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used in this study.
Fig. S1. Specificity of the primer pairs used to amplify 13-62 in RT–qPCR amplifications.
Fig. S2. RT–PCR analysis of Craterostigma plantagineum, Lindernia brevidens, and Lindernia subracemosa 13-62 expression in response to salt and osmotic stress.
Fig. S3. Alignment of amino acid sequences of 13-62 protein homologs in selected Embryophyta.
Fig. S4. RT–PCR analysis of Oropetium thomaeum 13-62 (Ot13-62) expression during dehydration and rehydration.
Fig. S5. Sequence of the transposable element identified in the Lb13-62 genomic locus.
Fig. S6. Putative cis-acting elements identified in the 13-62 promoter.
Acknowledgements
Part of this work was supported by the DFG project Smartwall BA 712/18-1. The authors thank C. Buchholz for helping to grow the plants.
Glossary
Abbreviations:
- ABA
abscisic acid
- ABRE
ABA-responsive element
- DRE
dehydration-responsive element
- DRP
desiccation-related protein
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- LEA
late embryogenesis abundant
- PIF
P instability factor.
References
- Bartels D, Salamini F. 2001. Desiccation tolerance in the resurrection plant Craterostigma plantagineum. A contribution to the study of drought tolerance at the molecular level. Plant Physiology 127, 1346–1353. [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F. 1990. Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta 181, 27–34. [DOI] [PubMed] [Google Scholar]
- Battista JR, Park MJ, McLemore AE. 2001. Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43, 133–139. [DOI] [PubMed] [Google Scholar]
- Becerra C, Puigdomenech P, Vicient CM. 2006. Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genomics 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, et al. . 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Bernacchia G, Salamini F, Bartels D. 1996. Molecular characterization of the rehydration process in the resurrection plant Craterostigma plantagineum. Plant Physiology 111, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Burke MJ. 1986. The glassy state and survival of anhydrous biological systems. In: Leopold AC, ed. Membranes, metabolism, and dry organisms. Ithaca: Cornell University Press, 358–363. [Google Scholar]
- Carniel FC, Gerdol M, Montagner A, Banchi E, De Moro G, Manfrin C, Muggia L, Pallavicini A, Tretiach M. 2016. New features of desiccation tolerance in the lichen photobiont Trebouxia gelatinosa are revealed by a transcriptomic approach. Plant Molecular Biology 91, 319–339. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Costa MD, Artur MA, Maia J, et al. . 2017. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nature Plants 3, 17038. [DOI] [PubMed] [Google Scholar]
- Cushman JC, Oliver MJ. 2011. Understanding vegetative desiccation tolerance using integrated functional genomics approaches within a comparative evolutionary framework. In: Lüttge U, Beck E, Bartels D, eds. Plant desiccation tolerance, Vol. 215 Heidelberg: Springer-Verlag Berlin Heidelberg, 307–338. [Google Scholar]
- Dinakar C, Bartels D. 2012. Light response, oxidative stress management and nucleic acid stability in closely related Linderniaceae species differing in desiccation tolerance. Planta 236, 541–555. [DOI] [PubMed] [Google Scholar]
- Ditzer A, Bartels D. 2006. Identification of a dehydration and ABA-responsive promoter regulon and isolation of corresponding DNA binding proteins for the group 4 LEA gene CpC2 from C. plantagineum. Plant Molecular Biology 61, 643–663. [DOI] [PubMed] [Google Scholar]
- Ditzer A, Kirch HH, Nair A, Bartels D. 2001. Molecular characterization of two alanine-rich Lea genes abundantly expressed in the resurrection plant C. plantagineum in response to osmotic stress and ABA. Journal of Plant Physiology 158, 623–633. [Google Scholar]
- Downs GS, Bi YM, Colasanti J, Wu W, Chen X, Zhu T, Rothstein SJ, Lukens LN. 2013. A developmental transcriptional network for maize defines coexpression modules. Plant Physiology 161, 1830–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L 3rd, Greenway SC, Galau GA. 1981. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 20, 4162–4168. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics 41, 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giarola V, Bartels D. 2015. What can we learn from the transcriptome of the resurrection plant Craterostigma plantagineum?Planta 242, 427–434. [DOI] [PubMed] [Google Scholar]
- Giarola V, Challabathula D, Bartels D. 2015. Quantification of expression of dehydrin isoforms in the desiccation tolerant plant Craterostigma plantagineum using specifically designed reference genes. Plant Science 236, 103–115. [DOI] [PubMed] [Google Scholar]
- Giarola V, Hou Q, Bartels D. 2017. Angiosperm plant desiccation tolerance: hints from transcriptomics and genome sequencing. Trends in Plant Science 22, 705–717. [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbricht T, Varotto S, Sgaramella V, Bartels D, Salamini F, Furini A. 2008. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytologist 179, 877–887. [DOI] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. 2001. Mechanisms of plant desiccation tolerance. Trends in Plant Science 6, 431–438. [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. 2008. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology 143, 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspard E, Macherel D, Hunault G. 2012. Computational and statistical analyses of amino acid usage and physico-chemical properties of the twelve late embryogenesis abundant protein classes. PLoS One 7, e36968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Imamura T, Iibushi J, Ikenaga M, Tachibana Y, Andoh N, Oyabu H, Hirooka K, Shiina T, Ishizaki Y. 2017. Ferritin 2 domain-containing protein found in lacquer tree (Toxicodendron vernicifluum) sap has negative effects on laccase and peroxidase reactions. Bioscience, Biotechnology, and Biochemistry 81, 1165–1175. [DOI] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y. 2007. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiology 144, 1797–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. 2010. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. The Plant Journal 63, 86–99. [DOI] [PubMed] [Google Scholar]
- Michel D, Furini A, Salamini F, Bartels D. 1994. Structure and regulation of an ABA- and desiccation-responsive gene from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology 24, 549–560. [DOI] [PubMed] [Google Scholar]
- Michel D, Salamini F, Bartels D, Dale P, Baga M, Szalay A. 1993. Analysis of a desiccation and ABA-responsive promoter isolated from the resurrection plant Craterostigma plantagineum. The Plant Journal 4, 29–40. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. 2010. Promoters and transcription factors in abiotic stress-responsive gene expression. In: Pareek A, Sopory SK, Bohnert HJ, eds. Abiotic stress adaptation in plants: physiological, molecular and genomic foundation. Dordrecht: Springer Netherlands, 199–216. [Google Scholar]
- Nielsen H. 2017. Predicting secretory proteins with SignalP. In: Kihara D, ed. Protein function prediction: methods and protocols. New York: Springer New York, 59–73. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Petersen J, Eriksson SK, Harryson P, Pierog S, Colby T, Bartels D, Röhrig H. 2012. The lysine-rich motif of intrinsically disordered stress protein CDeT11-24 from Craterostigma plantagineum is responsible for phosphatidic acid binding and protection of enzymes from damaging effects caused by desiccation. Journal of Experimental Botany 63, 4919–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkowski D, Schneider K, Salamini F, Bartels D. 1990. Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiology 94, 1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Bockel C, Blervacq AS, Bartels D. 2004. The novel gene CpEdi-9 from the resurrection plant C. plantagineum encodes a hydrophilic protein and is expressed in mature seeds as well as in response to dehydration in leaf phloem tissues. Planta 219, 579–589. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Edsgärd D, Hussain SS, Alquezar D, Rasmussen M, Gilbert T, Nielsen BH, Bartels D, Mundy J. 2010. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. The Plant Journal 63, 212–228. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. 1985. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Salmi ML, Bushart TJ, Stout SC, Roux SJ. 2005. Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii. Plant Physiology 138, 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider K, Wells B, Schmelzer E, Salamini F, Bartels D. 1993. Desiccation leads to the rapid accumulation of both cytosolic and chloroplastic proteins in the resurrection plant Craterostigma plantagineum Hochst. Planta 189, 120–131. [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, et al. . 2010. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biology 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Espinoza CJ, Richter A, Salamini F, Bartels D. 2003. Dissecting the response to dehydration and salt (NaCl) in the resurrection plant Craterostigma plantagineum. Plant, Cell & Environment 26, 1307–1315. [Google Scholar]
- Sprenger-Haussels M, Weisshaar B. 2000. Transactivation properties of parsley proline-rich bZIP transcription factors. The Plant Journal 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. 2015. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Frontiers in Plant Science 6, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Hincha DK, Leprince O, Macherel D. 2010. LEA proteins: versatility of form and function. In: Lubzens E, Cerda J, Clark M, eds. Dormancy and resistance in harsh environments, Vol. 21 Heidelberg: Springer-Verlag Berlin Heidelberg, 91–108. [Google Scholar]
- Valenzuela-Avendaño JP, Mota IAE, Uc GL, Perera RS, Valenzuela-Soto EM, Aguilar JJZ. 2005. Use of a simple method to isolate intact RNA from partially hydrated Selaginella lepidophylla plants. Plant Molecular Biology Reporter 23, 199–200. [Google Scholar]
- van den Dries N, Facchinelli F, Giarola V, Phillips JR, Bartels D. 2011. Comparative analysis of LEA-like 11-24 gene expression and regulation in related plant species within the Linderniaceae that differ in desiccation tolerance. New Phytologist 190, 75–88. [DOI] [PubMed] [Google Scholar]
- Velasco R, Salamini F, Bartels D. 1998. Gene structure and expression analysis of the drought- and abscisic acid-responsive CDeT11-24 gene family from the resurrection plant Craterostigma plantagineum Hochst. Planta 204, 459–471. [DOI] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knüppel R. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Research 24, 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha HG, Liu T, Zhou JJ, Sun H. 2013. MS-desi, a desiccation-related protein in the floral nectar of the evergreen velvet bean (Mucuna sempervirens Hemsl): molecular identification and characterization. Planta 238, 77–89. [DOI] [PubMed] [Google Scholar]
- Zhang X, Feschotte C, Zhang Q, Jiang N, Eggleston WB, Wessler SR. 2001. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proceedings of the National Academy of Sciences, USA 98, 12572–12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.